Abstract
We have inserted two expression cassettes at tagged reference chromosomal sites by using recombinase-mediated cassette exchange in mammalian cells. The three sites of integration displayed either stable or silencing position effects that were dominant over the different enhancers present in the cassettes. These position effects were strongly dependent on the orientation of the construct within the locus, with one orientation being permissive for expression and the other being nonpermissive. Orientation-specific silencing, which was observed at two of the three site tested, was associated with hypermethylation but not with changes in chromatin structure, as judged by DNase I hypersensitivity assays. Using CRE recombinase, we were able to switch in vivo the orientation of the transgenes from the permissive to the nonpermissive orientation and vice versa. Switching from the permissive to the nonpermissive orientation led to silencing, but switching from the nonpermissive to the permissive orientation did not lead to reactivation of the transgene. Instead, transgene expression occurred dynamically by transcriptional oscillations, with 10 to 20% of the cells expressing at any given time. This result suggested that the cassette had been imprinted (epigenetically tagged) while it was in the nonpermissive orientation. Methylation analysis revealed that the methylation state of the inverted cassettes resembled that of silenced cassettes except that the enhancer had selectively lost some of its methylation. Sorting of the expressing and nonexpressing cell populations provided evidence that the transcriptional oscillations of the epigenetically tagged cassette are associated with changes in the methylation status of regulatory elements in the transgene. This suggests that transgene methylation is more dynamic than was previously assumed.
Stably integrated transgenes are often poorly expressed because of position effects that are caused by the influence of the site of chromosomal integration (4, 12). In cultured cells, two categories of position effects have been recognized: stable and silencing (29, 39). Stable position effects are characterized by pancellular expression of the transgene (expression in every cell of a given tissue or cell population) at levels that are dictated by the site of integration and are different from the expression level of the endogenous gene or of similar transgenes integrated at other sites. Silencing position effects are characterized by progressive silencing of the transgene at a rate characteristic of the site of integration. During the process of silencing, expression occurs in only a fraction of the cell population and can therefore be described as heterocellular. Silencing position effects in cultured cells have some similarity to position effect variegation (PEV) in Drosophila and mammals (14, 36, 42), which is characterized by clonally inherited silencing of expression in a fraction of the cells of a given tissue. PEV thereby results in heterocellular expression of the transgenes. PEV can be temporally stable, or the proportion of expressing cells can decrease with the age of the animal (32).
Attempts to overcome position effects have generally focused on including strong native regulators such as locus control regions (LCRs) (16) in the transgenic construct or on making the transgene so large that it is likely to include all the sequences required to establish its native epigenetic organization (28, 30). However, neither strategy is fully effective, highlighting the fact that even following years of experimental analysis of transgenes, the fundamental mechanisms of position effects remain largely unknown (2). DNA elements called chromatin insulators have been reported to decrease silencing and variegating position effects in mammalian cells (7).
The major obstacle to understanding position effects has been the technical inability to integrate reporter constructs into a defined genomic locus. To overcome this problem, we have recently developed a targeting approach that we call recombinase-mediated cassette exchange (RMCE) (5, 11). RMCE uses site-specific recombinases to integrate single-copy transgenes without selectable markers into previously tagged sites in mammalian cells, allowing the sensitive and accurate dissection of the elements within the transgene that influence expression and epigenetic organization. In addition, since the RMCE system used here causes the integration of the transgene in each of the two possible orientations, we are able to test the effect of this variable for the first time.
We have previously used RMCE in mouse erythroleukemia (MEL) cells to study the LCR of the human β-globin gene locus, a group of five DNase I-hypersensitive sites (HS) that controls the expression of all the β-like globin genes (6, 15). In these experiments, constructs consisting of a β-globin promoter linked to a lacZ reporter and to various HSs of the LCR were targeted to a randomly selected integration site, named RL1. We were able to show that components of the β-globin LCR not only helped to overcome position effects by increasing the proportion of expressing cells, a previously described property of enhancers (12), but also could increase the level of transcription in each expressing cell (5). A striking observation was that with some of the DNase I HSs of the LCR, expression was heterocellular, and that apparently inactive cells could, with time, start to transcribe while, conversely, active cells could become silent. We referred to these temporally dynamic expression patterns as transcriptional oscillations (10). The rate of transcriptional oscillation was dependent on the strength of the enhancer included in the transgene.
We have now extended our previous analysis by comparing at three loci the expression of two cassettes containing different cis-regulatory sequences driving an enhanced green fluorescence protein (EGFP) reporter gene, integrated in both possible orientations. We show that each integration site causes distinct EGFP expression patterns that can be categorized as either stable or silencing position effects, confirming the critical importance of the site of integration on the level of transgene expression. Surprisingly, we found that the orientation of the cassette within each site is a critical determinant of the type of position effect observed. We also report that silencing of the construct in the nonpermissive orientation can epigenetically imprint the transgene and that an escape from repression, leading to transcriptional oscillations, is associated with changes in methylation at specific sites within the regulatory elements of the transgenes, suggesting that methylation can be a more dynamic DNA modification than has previously been assumed.
MATERIALS AND METHODS
Plasmids.
Plasmid construction was performed by standard methods. The sequences of all plasmids are available on request. Plasmid pL1HYTK1L was described previously (11). Plasmid L1CMVEGFP1L contains a NheI-NotI fragment from pEGFP-N1 (Clontech, Palo Alto, Calif.) cloned between the L1 and 1L LoxP511 sites of pL1HYTK1L. Plasmid L1234EGFP1L contains HS4 (HUMHBB positions 951 to 2199), HS3 (HUMHBB positions 4273 to 5122), HS2 (HUMHBB positions 7764 to 9172), and the β-globin promoter (positions −374 to +44 relative to cap site) linked to the EGFP coding sequence (the AgeI-NotI fragment of pEGFP-N1).
Cell culture and transfections.
Cell culture and transfections were performed as described previously (11). Induction of differentiation with 2% dimethyl sulfoxide (DMSO) for 5 days resulted in 70 to 95% benzidine positivity depending on the clones.
Creation of the reference loci.
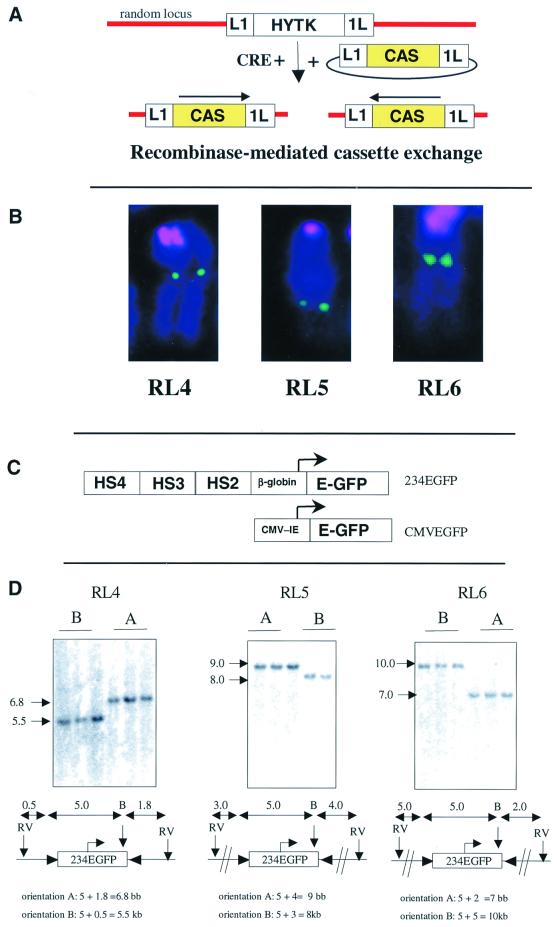
Loci RL4, RL5, and RL6 were created by transfecting 10 μg of a BspHI fragment of plasmid pL1HYTK1L containing L1 and 1L sites flanking the PGK-HYTK gene plus about 500 bp of vector sequences on both sides (to protect the lox sites from exonuclease digestion before integration). After selection with 1.1 mg of hygromycin per ml for 12 days, clones were isolated and single-copy integration events were identified by Southern blotting after digestion with EcoRV, an enzyme that does not cut into pL1HYTK1L. Twelve single-copy clones were then tested for ganciclovir sensitivity. Three clones that were strongly sensitive to ganciclovir (>99% cell death in 48 h in 10 μM ganciclovir) were termed RL4, RL5, and RL6 and were used throughout this study. The orientation of the integration was determined relative to chromosomal EcoRV sites located close to the integration sites by Southern blotting. The orientation of the cassettes was also determined relative to the residual plasmid sequences at the three loci in reference to AlwNI or NdeI sites. The letters A and B were assigned according to the orientation relative to the residual plasmid sequences.
Flow cytometry and cell sorting.
Flow cytometry and cell sorting was performed on Beckton Dickinson instruments (FACSCAN, FACS-STAR plus, and FACS SCALIBUR). EGFP was quantitated under standardized conditions using untransfected cells and GFP or fluorescein isothiocyanate (FITC)-labeled beads as standards. To ensure that all measurements were performed during log phase, the cells were fed for two consecutive days before being subjected to fluorescence-activated cell sorter (FACS) analysis. Dead cells were gated out on the basis on propidium iodide exclusion. Comparisons between different clones were always performed on the same day. The percentage of expressing cells was determined relative to a cutoff value set at the level of autofluorescence of the 99.5th percentile of untransfected control cells analyzed on the same day. The level of EGFP was defined as the linearized mean fluorescence of 10,000 cells in the FL1 (green) channel.
The FACS-gal procedure and the enrichment for transfected cells with the CMV-LacZ plasmid was performed as described previously (5).
Mapping of HSs with DNase I.
HS analysis was performed as described previously (10). Genomic DNA samples were digested with PvuII and probed with the EGFP coding sequence.
Methylation analysis.
Southern blots were quantified with a Molecular Dynamics PhosphorImager. All Southern blot analyses were performed at least twice. The genomic DNA was digested with PvuII (a methylation-insensitive enzyme that yields a 5.5-kb fragment regardless of the site of integration) plus one methylation-sensitive enzyme. Genomic samples exhibiting signs of partial digestion (a band larger than 5.5 kb) were discarded.
Bisulfite conversion was conducted as described previously (20). Nested PCR of converted genomic DNA was carried out with primer pairs +bisHS2-1 (GTTATATTTTTGTGTGTTTTTATTAGTGAT) and −bisHS2-1 (ATTTTCATACCTTCCTCTTCCATATCCTTA) in the first round (30 cycles with an annealing temperature of 50°C) and +bisHS2-2 (TATAGTTTAAGTATGAGTAGTTTTGGTTAG) and −bisHS2-2 (TACACATATATTAATAAAACCTAATTCTAC) in the second round (29 cycles with an annealing temperature of 50°C). The converted PCR products were cloned using the TA cloning kit (Invitrogen, Carlsbad, Calif.), and individuals clones were sequenced on an automatic sequencer.
FISH analysis.
Fluorescent in situ hybridization (FISH) was performed as described previously (2) using the entire plasmid pL1234EGFP1L as a probe. MEL cells were hypotonically swollen in 75 mM KCl for 9 min, at which time 1/40 volume of fixative (fresh methanol-acetic acid at 3:1) was added to the suspension, the tube was inverted, and the cells were centrifuged gently for 7 min. The cells were washed a further four times in fixative, and slides were prepared by pipetting 30 μl of cell suspension onto a slide and stored in 100% ethanol at −20°C. The major satellite probe was prepared by amplifying mouse genomic DNA with PCR primers from the conserved part of the repeat (43). The primers used were 5′-TAGAAATGTCCACTGTAGG-3′ and 5′-CAGTTTTCTTGCCATATTC-3′, with annealing at 39.8°C. This PCR product and the pL1234EGFP1L plasmid were separately nick translated to incorporate CY3.5 and biotin, respectively. By using standard hybridization conditions, the biotin signal was detected with avidin-fluorescein isothiocyanate (FITC). Images were captured and processed as previously described (2).
RESULTS
Creation of the tagged RL4, RL5, and RL6 loci.
The sites into which the transgenes were targeted were created using pL1HYTK1L, a plasmid that contains two inverted Lox sites (referred to as L1 and 1L) flanking the hygromycin-thymidine kinase (HYTK) gene, a fusion gene that can be selected for positively with hygromycin and against negatively with ganciclovir (Fig. 1A). Linearized pL1HYTK1L was transfected into MEL cells, hygromycin-resistant clones were isolated, and Southern blotting was performed to identify clones with single-copy integration (data not shown). Three single-copy clones that proved particularly sensitive to ganciclovir were designated RL4, RL5, and RL6 (for “random locus 4, 5, and 6). To determine the approximate chromosomal locations of RL4, RL5, and RL6, three-color FISH was performed using the pL1HYTK1L plasmid and mouse major satellite repeat DNA as probes. This revealed that all three integration sites were far from the potentially repressive environment of the centromere (Fig. 1B).
FIG. 1.
Site-specific integration at RL4, RL5, and RL6. (A) RMCE. Reference loci are created by insertion of plasmid L1HYTK1L at random integration sites in MEL cells. Expression cassettes are then integrated using RMCE by cotransfection of a Cre expression plasmid and a plasmid containing the expression cassette of interest. Cells that undergo an exchange can be selected for resistance to ganciclovir. Site-specific integration occurs in both possible orientations. (B) Three reference loci (random loci RL4, RL5, and RL6) were created and localized on the chromosomes by FISH. The chromosomes were stained with DAPI (blue pseudocolor), and the centromere were visualized with a CY3.5-labeled probe (red pseudocolor), and the reference loci were visualized indirectly with FITC-labeled avidin (green pseudocolor). None of the loci is near the centromere or telomere. (C) Cassette 234EGFP contains the coding sequence of the EGFP genes linked to a human β-globin promoter fragment and to HS2, HS3, and HS4 of the human β-globin LCR. Cassette CMVEGFP contains the same reporter linked to the CMV IE promoter-enhancer. (D) Southern blots demonstrating site-specific integration of cassette 234EGFP at RL4, RL5, and RL6 in both orientations. Three subclones with integration in each orientation at all three loci are shown (except RL5 orientation B, for which only two subclones are shown). Orientation was determined using chromosomal EcoRV sites for reference (see Materials and Methods).
Recombinase-mediated cassette exchange.
Two plasmids containing L1 and 1L Lox sites flanking the expression cassettes 234EGFP and CMVEGFP (Fig. 1C) were created. The first cassette, 234EGFP, contains HS2, HS3, and HS4 of the β-globin LCR 5′ to the β-globin promoter driving the EGFP reporter gene. The second cassette, CMVEGFP, contains the cytomegalovirus (CMV) immediate-early (IE) promoter-enhancer driving the EGFP reporter (25, 35).
Cassettes 234EGFP and CMVEGFP were then integrated in both possible orientations at the three reference loci RL4, RL5, and RL6 by a cassette exchange mediated by recombination between the inverted Lox sites (11) (Fig. 1D). At all three integration sites, the frequency of exchange was very high: between 80 and 100% of the ganciclovir-resistant clones had a transfected cassette integrated in one of the two possible orientations.
Comparison of cassettes 234EGFP and CMVEGFP.
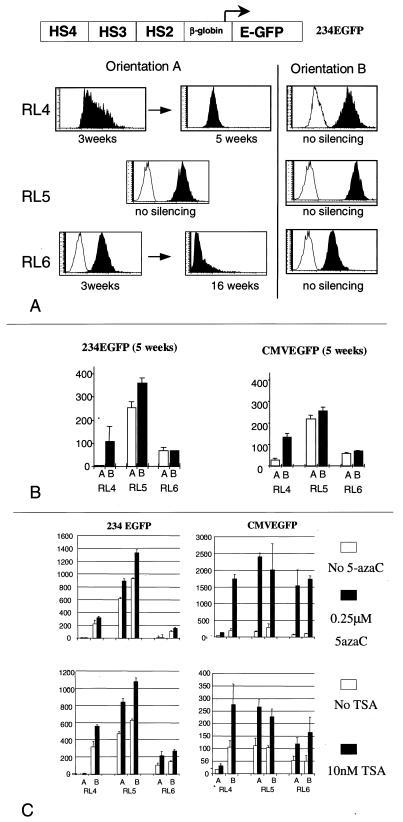
For each locus, the level of expression (defined as the mean green fluorescence of the cell population) and the proportion of expressing cells (from which the rate of silencing can be evaluated) of at least six subclones (three of each orientation) were monitored regularly for several months by flow cytometry (Fig. 2A and B). As indicated by the small error bars, the levels of expression of different subclones with identical cassettes in the same orientation at the same chromosomal site were remarkably similar, demonstrating that the use of RMCE successfully eliminates the variables due to the site of integration. By contrast, comparisons of the level of EGFP expression with the cassettes in different orientations and at different loci revealed striking position effects.
FIG. 2.
Expression at RL4, RL5, and RL6. (A) Representative histograms obtained by flow cytometry of cells containing cassettes 234EGFP at RL4, RL5, and RL6 at various times postintegration. The white histogram overlays were obtained from control untransfected cells. The x axis shows the number of cells; the y axis shows the mean green fluorescence on a relative scale. Orientation-dependent stable and silencing position effects can be observed (see the text). (B) Bar graphs summarizing the expression levels (defined as the mean level of green fluorescence in 10,000 cells) of cassettes 234EGFP and CMVEGFP 5 to 6 weeks after integration at loci RL4, RL5, and RL6. The white bars show expression in orientation A, and the black bars show expression in orientation B. Each bar represents the average of two independent determinations of the expression level of three clones, and error bars indicate standard deviation. Silencing was not a factor in comparing expression levels between the various cell lines, since at 5 weeks postintegration the expression in all the samples tested was pancellular or absent (silencing at RL4 in orientation A was already complete but was minimal at RL6 in orientation A). (C) Cells with 234EGFP or CMVEGFP at RL4, RL5, and RL6 were incubated with 0.25 μM 5-azaC for 24 h (top panels) or with TSA for 48 h (bottom panels). Each bar represents the average (and standard deviation) of two independent experiments in which the expression level of three subclones containing one of the cassettes in each orientation was determined. The locus of integration and the orientation of the cassette are indicated below the bars.
At RL4, 3 weeks after the transfection, expression of cassette 234EGFP in orientation A was very low and heterocellular. At 1 month later, the cassette was completely silent in all clones examined, demonstrating a high rate of silencing at this locus in this orientation. Expression in orientation B was much higher than in orientation A and was pancellular. No signs of silencing were detected over a 6-month monitoring period. At the same RL4 locus, expression of the CMVEGFP cassette was similar to that of the 234EGFP cassette. In orientation A, heterocellular, low-level expression was detected 3 weeks postintegration. Rapid silencing was then observed. In orientation B, expression occurred at a higher level and did not silence over time.
At RL5, expression of cassette 234EGFP inserted in orientations A and B occurred at high levels and was pancellular. No silencing could be detected over at least 6 months of culture. Nevertheless, the level of expression in orientation B was about 1.5-fold higher than in orientation A. The Student t test revealed that this difference was statistically significant (P < 0.0001). Expression of cassette CMVEGFP in orientations A and B was also pancellular and occurred at high level, with expression in orientation B being slightly higher than in orientation A. This difference did not achieve statistical significance.
At RL6, pancellular expression of cassette 234EGFP was initially observed at similar levels in both orientations, but expression in orientation A slowly silenced over a 4-month period while expression in orientation B remained stable. Expression of cassette CMVEGFP followed the same pattern, with silencing of expression in orientation A being observed after 3 to 5 months in culture in eight of nine clones.
Comparison of expression levels of the same cassette at the three loci revealed 10-fold differences in the average GFP fluorescence for either the 234EGFP or the CMVEGFP cassette, presumably a consequence of different transcription rates. For both cassettes, expression was weakest at RL6 and strongest at RL5. Furthermore, for both cassettes, silencing occurred rapidly at RL4 and more slowly at RL6. This illustrates the strong influence of the site of integration on both the level of transgene expression and the rate of silencing and indicates that the determinants of expression present at the sites of integration influence both the CMV enhancer and the β-globin LCR fragments similarly.
MEL cells can be induced to mature into late-stage erythrocytes by a variety of treatments, including exposure to DMSO (22). This differentiation generally strongly activates constructs with β-globin regulatory elements. Induction of differentiation with DMSO led to a two- to threefold increase in the level of expression of cassette 234EGFP in the cells that were expressing but did not reactivate the silenced loci (data not shown).
TSA and 5-azaC stimulate expression at active loci.
To investigate the role of cytosine methylation in the control of gene expression in these cells, clones with cassettes 234EGFP or CMVEGFP in each orientation at loci RL4, RL5, and RL6 were treated with 5-azacytidine (5-azaC), a demethylating agent (Fig. 2C) (31). After incubation of the cells containing cassette 234EGFP with 5-azaC, the levels of expression increased 1.5- to 2-fold in all the clones with pancellular expression. Importantly, silenced loci could not be reactivated. With the CMVEGFP cassette, the results were more striking, with a 5- to 10-fold-greater response than for the 234EGFP cassette and minimal effects on the silenced RL4 locus in orientation A.
To determine if histone acetylation plays a role in transgene regulation, the same panel of clones were treated with trichostatin A (TSA), a histone deacetylase inhibitor (Fig. 2C) (44). Incubation of the 234EGFP cells with 10 nM TSA for 48 h was associated with a doubling of the level of expression at all loci in which expression was already occurring but had little effect at the silenced RL4 locus in the nonpermissive A orientation. Treatment of the CMVEGFP cassette with TSA resulted in a stronger activation than did treatment of the 234EGFP cassette. In both cases, this increase in expression could either be due to a direct action of TSA on the chromatin at the integration sites or be indirect. Treatment with a combination of 10 nM TSA and 0.25 μM 5-azaC also failed to reactivate the silenced RL4 and RL6 loci (data not shown).
Levels of expression correlate with levels of methylation.
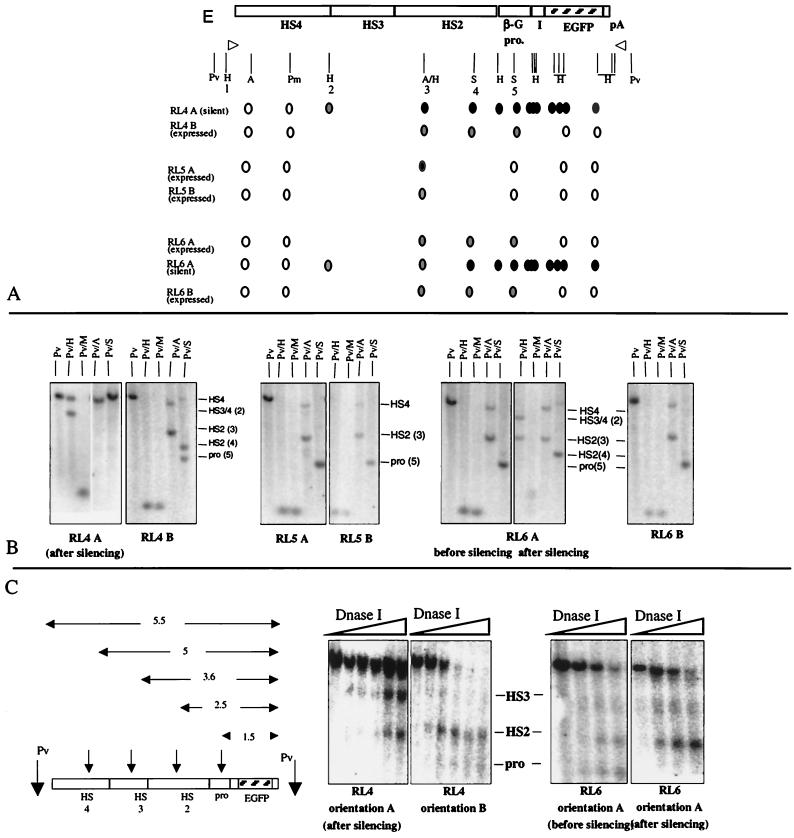
To test the relationship between levels of methylation and expression patterns, we digested genomic DNA with methylation-sensitive restriction endonucleases (and their methylation-insensitive isoschizomers when available) and performed Southern blot analyses (Fig. 3A and B). At RL5, the locus with the highest expression levels, cassette 234EGFP in both orientations was almost completely unmethylated except for a small amount of methylation in HS2, detected using AvaI. At RL4 and RL6 in orientation B (the orientation in which expression is stable), the results were similar to those for RL5, with an overall paucity of methylation except at HS2 and the β-globin promoter, which were partially methylated (Fig. 3A and B). Overall, the level of expression correlated inversely with the degree of methylation of the cis-regulatory sequences.
FIG. 3.
Methylation and DNase I studies. (A) The first line represents the functional map of the cassette (the hatched bar in the EGFP coding sequence is the probe used for hybridization). The vertical lines are restriction sites (A, AvaI; H, HpaII; Pm, PmlI; Pv, PvuII; S, SnaBI). The open circles represent unmethylated CpGs (sites at which digestion was between 95 and 100% complete), the solid circles represent methylated CpGs (sites at which digestion was between 0 and 5% complete), and the shaded circles represent partially methylated CpG (sites at which digestion was between 5 and 95% complete). Expressed cassettes were largely unmethylated, while silenced cassettes were almost fully methylated at all the testable CpG in HS2, the promoter, and the EGFP coding sequence but unmethylated at HS4. The HpaII sites are numbered for reference in the text and in Fig. 3, 4, and 5. (B) Representative Southern blots. Genomic DNA was double digested with PvuII plus one of the methylation-sensitive enzymes and probed with the coding sequence of EGFP (Pv, PvuII, H, HpaII; M, MspI; S, SnaBI; A, AvaI). Number in parentheses refers to the restriction sites numbered in panel A. (C) DNase I HS mapping. Nuclei were digested for increasing times with DNase I, and genomic DNA was digested with PvuII (Pv) and probed with the EGFP coding sequence (hatched bar). The HSs are formed at least as well in the silenced constructs as in the active constructs. In contrast, an HS that maps at the promoter can be detected only in actively expressing cells.
In the silenced cell lines (RL4, orientation A, and RL6, orientation A, after several months in culture), hypermethylation was found at all the CpG dinucleotides tested in the EGFP coding sequences, the β-globin promoter, and HS2. Methylation of the two CpG dinucleotides in HS3 could not be assessed due to the lack of suitable restriction endonucleases, but an HpaII site introduced as part of the cloning of HS2 and HS3 was partially methylated. Interestingly, HS4 was not methylated. We concluded that at both RL4 and RL6, silencing was associated with complete methylation of HS2, the promoter, and the coding sequence.
DNase I hypersensitivity studies.
To determine whether the differences in expression levels and rate of silencing between loci were associated with different chromatin structures, we performed DNase I hypersensitivity mapping on clones with cassette 234EGFP at loci RL4, RL5, and RL6. These experiments revealed that strong DNase I HSs that map to HS2 and HS3 formed at all three loci and in both orientations, including after silencing of expression at RL4 and RL6 (Fig. 3C). Silencing at loci RL4 and RL6 is therefore not due to the failure of HS formation. By contrast, the presence of an HS that maps to the β-globin promoter correlated with expression, showing that silencing in these cells correlates with the failure of transcriptional initiation.
Stable epigenetic imprinting of nonexpressing cassettes.
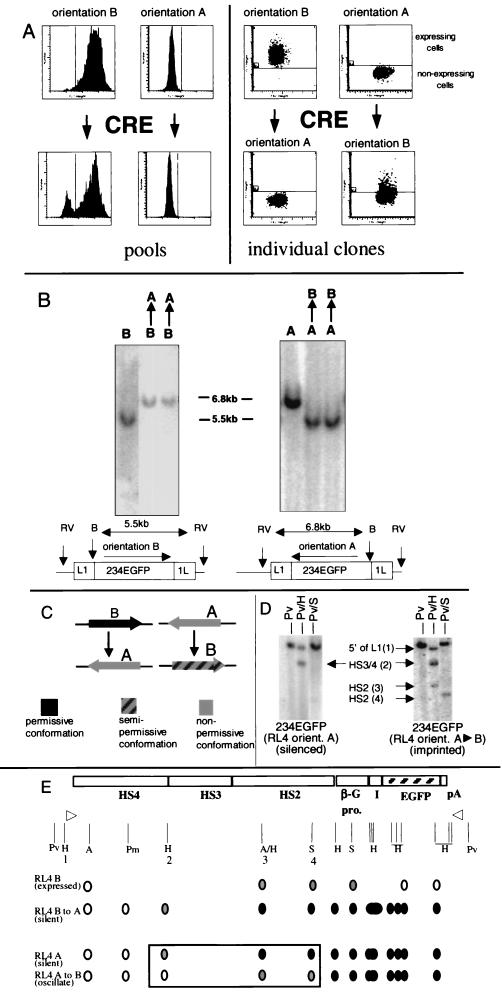
To gain further insight into the cause of silencing and of the orientation dependence of expression, we inverted in vivo the 234EGFP cassette integrated at RL4, the locus with the most pronounced effect of orientation. This was possible because the transgene remains flanked by L1 Lox sites in opposite orientations following RMCE, allowing inversion of the transgene if Cre recombinase is reexpressed in the cell (1). Cells containing 234EGFP inserted at RL4 in the permissive orientation (orientation B) were transiently transfected with a Cre expression plasmid and were examined as pools 4 weeks later. FACS analysis revealed that about 10% of the cells had lost expression of EGFP (Fig. 4A). To verify that in vivo inversion of the cassette had indeed taken place after reexpression of Cre, cells that had lost EGFP expression following transfection were purified by flow cytometry, individual clones were isolated, and Southern blot analyses were performed. This revealed that all 20 silenced clones tested had undergone an in vivo inversion (Fig. 4B). A total of 35 clones that were still expressing EGFP after the transient transfection of Cre were also tested by Southern blot analyses. None of these clones had undergone an in vivo cassette inversion. Therefore, a perfect correlation was found between in vivo inversion and the loss of transgene expression.
FIG. 4.
In vivo inversion of the cassette does not fully reactivate the silenced cassette. (A) In vivo inversion of cassette 234EGFP at RL4 by transient transfection of a Cre expression plasmid. The four histograms on the left show flow cytometry analyses of pools of cells before and 4 weeks after transient transfection with a Cre expression plasmid. The appearance of the GFP-negative cell subpopulation after transient Cre expression in cells containing 234EGFP in orientation B (the permissive orientation) suggests that B-to-A inversions lead to silencing of the cassette. The absence of a GFP-positive cell subpopulation after transient CRE expression in cells with a cassette in orientation A suggests that A-to-B inversions do not fully reactivate the cassette. The four dot plots on the right show flow cytometry analyses of individual clones of cells having undergone B-to-A or A-to-B inversions as determined by Southern blot analyses (see Fig. 5B). Analysis of individual clones with a B-to-A inversion confirms that active cassettes are silenced by inversion. Analysis of individual clones with an A-to-B inversion reveals that silenced cassettes are not fully reactivated by inversion; instead, a low level of heterocellular expression is observed. (B) Southern blots demonstrating the A-to-B and B-to-A inversions. (C) Summary of the in vivo inversion experiments. Inversion from the permissive orientation (orientation B) to the nonpermissive orientation (orientation A) leads to transcriptional silencing. Inversion from the nonpermissive orientation to the permissive orientation leads to a heterocellular pattern expression instead of complete reactivation, indicating that the cassette was epigenetically tagged while it was silenced. (D) Southern blots illustrating the methylation analysis of the RL4 cassettes. After in vivo inversion, the cassette remains mostly methylated (compare with Fig. 3B, RL4 orientation B results). However, HS2 is subject to specific demethylation compared with the RL4 orientation A transgene. (E) Summary of methylation analysis after in vivo inversion of cassette 234EGFP at RL4. The vertical lines are restriction sites (H, HpaII-MspI; A, AvaI; P, PmlI; S, SnaBI). The open circles represent unmethylated CpGs, and the solid circles represent methylated CpGs; the shaded areas represent partially methylated CpGs. Epigenetic tagging of the A-to-B inverted cassette is associated with retention of methylation of the promoter and the EGFP coding sequence and with selective partial demethylation of the HS2 region.
To determine if inversion from the nonpermissive to the permissive orientation would reactivate expression, cells with cassette 234EGFP in orientation A were transiently transfected with the Cre expression plasmid and examined by FACS analysis 2 to 4 weeks later. No reactivation of EGFP could be detected (Fig. 4A). To determine if in vivo inversions were occurring, individual clones were generated and Southern blot analyses were performed, identifying one clone that had undergone an inversion (Fig. 4B). Expression analysis revealed that although this clone was now in the permissive orientation, it had not acquired the phenotype of cells that had integrated the transgene in the permissive orientation in the first place. Instead, expression was heterocellular, with only 10 to 20% of the cells expressing EGFP at levels in excess of those found in a negative control (Fig. 4A). To generate more clones with an inversion from the nonpermissive to the permissive orientation, cells with cassette 234EGFP in orientation A were cotransfected with a Cre expression plasmid and a CMV-LacZ expression plasmid. At 48 h following transfection, successfully transfected cells were isolated by flow cytometry after staining for LacZ expression by the FACS-gal procedure (27). Two clones with an inversion were recovered (Fig. 4B). These clones had expression patterns similar to that of the previously isolated A-to-B inversion clone, with only a minority of expressing cells. We conclude that inversion from the nonpermissive to the permissive orientation does not fully reactivate the transgene, indicating that an epigenetic modification is present on cassette 234EGFP in the nonpermissive orientation which is not lost upon inversion (Fig. 4C). Epigenetic tagging is known to occur in the germ line at many loci and is referred to as genomic imprinting (26). Since the experimentally induced phenomenon that we report here in cell culture is a superficially similar process of epigenetic tagging, we termed it inversion-mediated imprinting.
Inversion-mediated imprinting of cassette 234EGFP is associated with persistence of methylation in the EGFP coding sequence and the β-globin promoter and with partial demethylation of HS2.
To test the methylation status of the cassettes after in vivo inversion, we performed experiments similar to that described above (Fig. 4D and E). After inversion from the permissive to the nonpermissive orientation, the methylation pattern observed was indistinguishable from that of clones directly recovered in the nonpermissive orientation (Fig. 3), suggesting that in the nonpermissive chromatin context, cassettes are efficiently de novo methylated and that no irreversible epigenetic tags were attached to the cassette while it was in the permissive orientation.
It was of particular interest to test the methylation pattern of the imprinted cassette (A-to-B inverted clones) because these clones allowed us to analyze whether the failure to reactivate expression in the majority of cells was due to the methylation of the construct. Restriction enzyme methylation assays indicated that the hypermethylation of the EGFP coding sequence and the β-globin promoter was unaltered by the in vivo inversion, suggesting that maintenance of the silent state in the otherwise permissive orientation may be due to the conservation of the preexisting methylation patterns. However, the degree of methylation of HS2 in the cassette imprinted by inversion was markedly reduced compared with that in the silenced, noninverted clones (Fig. 4D), suggesting that the small amount of expression in the imprinted cells might be due to this partial demethylation. This prompted us to study these cells in greater detail.
Escape from inactivation is associated with transcriptional oscillations and methylation changes at HS2.
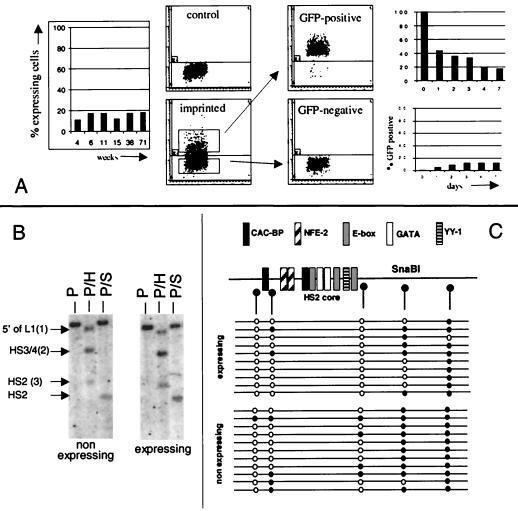
To determine whether the inversion-mediated imprinting was stable or whether it would be eliminated by serial passages in culture, we monitored these cells over a 70-week period. The percentage of expressing cells in the imprinted population was analyzed by measuring the proportion of cells with fluorescence greater than a threshold defined as the 99.5th percentile of the background fluorescence of untransfected control cells analyzed simultaneously. This revealed that the inversion-mediated imprinting was stable, since the proportion of expressing cells in RL4 A-to-B inversion clones remained between 10 and 20% throughout that time in culture (Fig. 5A). These expression characteristics giving rise to the heterocellular pattern of expression of this cell population were strikingly similar to the transcriptional oscillations that we had previously observed at locus RL1 (10).
FIG. 5.
Transcriptional oscillation of the 234EGFP cassette imprinted by inversion is associated with selective methylation changes at HS2. (A) In the left panel, cells with the imprinted cassette at RL4 were grown in culture for 71 weeks and the percentage of GFP-positive cells was tested periodically. The epigenetic tag was stable over this period since the GFP-positive fraction of cells remains between 10 and 20%. In the four middle panels, GFP-positive and GFP-negative cells were purified by flow cytometry (x axis, forward side scatter; y axis, green fluorescence). In the two right panels, bar graphs are shown summarizing the percent GFP-positive cells as a function of time when the GFP-positive and GFP-negative cell fractions were returned to culture and monitored daily for GFP expression. Both populations rapidly returned to the parental phenotype, demonstrating that expression in these cells occurs by transcriptional oscillations. (B) Methylation analyses of the two sorted fractions with methylation-sensitive restriction nucleases: H, HpaII; S, SnaBI; Pv, PvuII. Both the SnaBI site in HS2 and the HpaII site 3 (see Fig. 3) are methylated twice as much in the GFP-negative cells as in the GFP-positive cells. HpaII sites 2 and 3 (see Fig. 3) are comethylated in 45% of the GFP-negative cells but in only 30% of the GFP-positive cells. (C) Summary of bisulfite sequencing analysis of the plus strand of the core of HS2. Each line represents the methylation status of the five CpG dinucleotides of an allele (a single molecule) of HS2. The open circles represent unmethylated CpGs, and the solid circles represent methylated CpGs. The GFP-negative cells are generally less methylated than the GFP-positive cells. The three CpGs near the core of HS2 are less methylated than the two CpGs on the 3′ side. Each CpG dinucleotide therefore appears to have a specific probability of methylation.
To test whether transcriptional oscillations were occurring, 16 individual subclones of each of these inverted clones were generated and analyzed by flow cytometry. All the subclones displayed heterocellular expression patterns similar to those of the parental clones (data not shown), supporting the possibility that expression in these cells is oscillating (10). To generate definitive data, GFP-positive and GFP-negative cells from one of the inverted clones were sorted and placed back in culture for 1 week and expression of EGFP was monitored by FACS daily (Fig. 5A). Within 48 h of the start of culture, about 10% of the cells from the GFP-negative fraction had become positive for GFP expression. Similarly, within 7 days of the start of culture, 82% of the cells from the GFP-positive fraction had lost expression and were fluorescing below the 99.5th percentile of the control cells. Expression had therefore been silenced in the majority of the expressing cells, and expression had been activated in the corresponding minority of the nonexpressing cells. Both populations had reverted to heterocellular expression patterns similar to the parent population. Together, these data demonstrate that the heterocellular expression patterns of the cassette imprinted by inversion are caused by transcriptional oscillations.
To assess the role of methylation and acetylation in determining transcriptional oscillation of the cassette imprinted by inversion, we treated the inverted clones with 5-azaC or with TSA for 2 to 8 days. These treatments increased the proportion of expressing cells by up to fourfold (from about 10–20 to 40% of the cells) but did not lead to complete reactivation of expression (data not shown). The response to these drugs was therefore similar to what we previously reported for RL1 (10), suggesting that the oscillations at RL1 and RL4 are caused by similar molecular mechanisms.
Methylation analysis with methylation-sensitive restriction nucleases was performed at three time points during the 70-week growth period of the imprinted cells (Fig. 4D and data not shown). The results of all three analyses concurred, showing that HS2 was partly methylated in these cells and demonstrating that the partial methylation was not merely a transient phenomenon due to the strand breakage and religation of the DNA sequence that occurred during the inversion but was a temporally stable characteristic of these oscillating cells. Again, this suggests that oscillations at RL1 and at RL4 are caused by similar molecular mechanisms, since we had previously observed that the transgenes that oscillate at RL1 are partially methylated in their regulatory regions (E.E.B., unpublished data).
We then performed methylation analysis on sorted GFP-positive and GFP-negative cells. Because of the long half-life of EGFP in these cells (>24 h), we could only enrich rather than purify for cells with the state of transcription of interest at the transgene locus. Despite this caveat, the methylation analysis was informative.
Sorted GFP-positive and GFP-negative cells from the population of cells with the imprinted 234EGFP cassette at RL4 were analyzed using methylation-sensitive enzymes. Quantitative analysis of the restriction patterns observed with these enzymes revealed that the coding sequence of the EGFP reporter and the promoter were completely methylated in both expressing and nonexpressing cells (Fig. 5B). By contrast, three CpG sites located within HS2 and between HS2 and HS3 were partially methylated in both populations of cells but were about 50% less methylated in the GFP-positive than in the GFP-negative cells. Incomplete digestion could be ruled out because complete digestion by HpaII (site 1) could be detected on the same blots (Fig. 5B) and because a repeat of these experiments with genomic DNA extracted from cells sorted on a different day and with fresh enzymes yielded the same results.
To extend these results, the methylation status of five CpG dinucleotides within a 500-bp fragment spanning the core of HS2 was determined by bisulfite sequencing (8), a method that allows the determination of the methylation status of all the CpG nucleotides within the segment tested. The sequencing of 10 clones (each representing the positive strand of one “allele” of HS2 that was originally present in a single cell) from each population revealed that 32% of the CpGs in the HS2 fragment from the GFP-positive cells were methylated whereas 56% of the CpGs in the HS2 fragment from the GFP-negative cells were methylated (Fig. 5C). Hypomethylation in GFP-expressing cells was detectable at each of the five CpGs, but the three CpGs located closer to the core of HS2 were on average distinctly less methylated than the two downstream CpGs closer to the hypermethylated promoter and EGFP coding sequences. Therefore, two independent methods clearly indicate that the GFP-positive cells were hypomethylated with respect to the GFP-negative fraction. Since expression in these cells is oscillating, the simplest interpretation of these results is that methylation of the inverted cassette also oscillates.
DISCUSSION
We have tagged three genomic sites with pairs of inverted L1 Lox sites and inserted two expression cassettes at each of these reference loci by using RMCE. Pancellular variation in expression levels from the same cassette integrated at different loci (stable position effects) was observed in the presence of two unrelated enhancers (HS2, HS3, and HS4 of the β-globin LCR and the CMV IE). Strikingly, the levels of expression of both cassettes were affected in the same manner by the locus of integration, with the RL5 locus allowing the strongest expression and RL6 allowing the weakest. These results demonstrate the presence at most integration sites of unknown determinants of transcription rate that are dominant over two completely distinct enhancers that both promote high levels of transcription in erythroid cells. Silencing of expression was observed at two of the three loci tested and occurred regardless of the enhancers present, again demonstrating the presence at these integration sites of determinants of transcriptional competence that are dominant over the LCR and the CMV IE.
Orientation dependence of expression.
Surprisingly, expression of the two cassettes was dependent on their orientations within the reference locus. At two of the three loci (RL4 and RL6), expression was temporally stable in one orientation but gradually silenced in the other. At the third locus (RL5), expression was temporally stable in both orientations but was nevertheless higher in one of the two orientations. The nature of the sequences in the flanking chromatin responsible for the orientation dependence is unknown. Such sequences are probably not large blocks of heterochromatin from which chromatin condensation spreads over large distances, as has been proposed for PEV in Drosophila (9), since inversion would not significantly change the distance between the regulatory sequences within the transgene and distant heterochromatic blocks that are believed to induce silencing at distances greater than 1 Mb (41). The simplest explanation is that the unknown determinants of transgene expression are sequences located within a few hundred bases of the sites of integration, combining with the regulatory sequences of the expression cassette to cause either stable or silencing position effects. Orientation-dependent position effect variegation has been previously reported for the brown transgene in Drosophila by Sabl and Henikoff (33). These authors proposed that the orientation of the transgene affected somatic pairing with repetitive elements that led to inactivation. Although the transgenes inserted at RL1 are present at a single copy in the cell, a similar phenomenon could conceivably be occurring in MEL cells. The direction of DNA replication or unidirectional transcription into the transgene from endogenous promoters (24) could also be the source of the polarity of the position effect that we observed.
Silencing in MEL cells is associated with methylation but not with changes in DNase I HS formation.
The observation that methylation at RL4 and RL6 correlates strongly with the level of GFP expression and with silencing is in accordance with previous reports on silencing of globin constructs and retroviruses in cell culture and in transgenic mice (13, 19–21, 29). However, the formation of strong DNase I HSs even after silencing has not been reported before, and contrasts with previous reports by us and others that position effect variegation on globin transgenes in mice is associated with loss of DNase I sensitivity at the LCR (2, 13). This difference suggests that silencing in MEL cells occurs by a mechanism that differs from silencing of globin transgenes that go through the mouse germ line. Interestingly, HS4 was not methylated, even in completely silenced cells, suggesting that HS4 might be more resistant to methylation than HS2 is. Whether this has any functional consequences is not clear.
Inversion-mediated imprinting.
In vivo inversion toward the permissive orientation of a silenced cassette did not lead to complete reactivation of the cassette but instead led to a temporally stable heterocellular pattern of expression caused by transcriptional oscillations associated with dynamic activation and inactivation of the transgenes. Two independent methylation assays, Southern blot and bisulfite sequencing analyses, revealed that the GFP-positive and GFP-negative cells have different levels of methylation of HS2 but not of the promoter or the GFP coding sequence. Since GFP expression is dynamic in these cells, it probably follows that the methylation of HS2 is itself dynamic. To our knowledge, dynamic methylation of an enhancer has not been previously documented but several reports have demonstrated selective demethylation of viral enhancers in transgenes and of regulatory sequences (18). It has been proposed that demethylation of regulatory elements is associated with binding of protein factors such as Sp1 in CpG islands or NF-κB in B cells (3, 34). Studies with Xenopus and mammalian cells suggest that factor-mediated selective demethylation is replication dependent (17, 23). Changes in the methylation of HS2 in the oscillating cells might therefore be mediated by temporally unstable binding of factors capable of inducing demethylation at HS2.
Although the causes of the instability of both expression and methylation of the imprinted cells are not known, our results suggest that inversion-mediated imprinting is probably related to the methylation of the promoter and EGFP coding sequence. We previously reported that expression of a nonimprinted transgene at a different integration site (RL1) could oscillate at different rates depending on the enhancer present. Transgenes therefore oscillate when they are partly methylated because of inversion-mediated imprinting or when the enhancer is too weak for a given integration site. We propose a model in which competition between de novo methylation and demethylation is a critical determinant of transgene expression. In this model, stable position effects are due to variable and reversible levels of de novo methylation associated with various integration sites. Silencing position effects, on the other hand, are due to stochastic variations in the recruitment of either the demethylation or the de novo methylation activity, causing an individual cell to reach a threshold of methyl-CpG density that cannot be reversed, leading in turn to permanent silencing of the gene. In this model, the known abilities of enhancers to prevent silencing (38), to set expression levels (5), and to determine the rate of transcriptional oscillation (10) would be related, at least in part, to the capacity of these regulatory elements to bind protein factors that promote demethylation.
The overall role of methylation in vertebrates is still debated (37), but it is clear that methylation is involved in X-inactivation, genomic imprinting, silencing of parasitic DNA, and maybe silencing of tissue-specific genes. Whether the imprinting of the cassette that we artificially created by inversion and the resulting transcriptional oscillations that we observed mimic physiological processes, such as those that might occur when regulatory regions are demethylated as part of normal development, after a chromosomal rearrangement, or after activation of an oncogene, will have to be tested experimentally. Such a “somatic” imprinting could represent an additional means of keeping a gene repressed even if its regulatory sequences are demethylated, for instance by ectopic activation of a transcription factor or by the insertion of a retrovirus. The oscillations that appear to result from such imprinting could potentially explain the erratic behavior of many tumor cells as well as the extreme plasticity of stem cells in response to different environments (40).
ACKNOWLEDGMENTS
Y.Q.F. and E.E.B. are supported by NIH grants HL55435, HL38655, and DK56845; M.L. is supported by NIH grant GM19767; and J.M.G. is supported by grants DK02467 and DK56786.
We thank Judith Dunai, Yale University, for technical assistance with the FISH assay.
REFERENCES
- 1.Abremski K, Frommer B, Hoess R H. Linking-number changes in the DNA substrate during Cre-mediated loxP site-specific recombination. J Mol Biol. 1986;192:17–26. doi: 10.1016/0022-2836(86)90460-2. [DOI] [PubMed] [Google Scholar]
- 2.Alami R, Greally J M, Tanimoto K, Hwang S, Feng Y Q, Engel J D, Fiering S, Bouhassira E E. Beta-globin YAC transgenes exhibit uniform expression levels but position effect variegation in mice. Hum Mol Genet. 2000;9:631–636. doi: 10.1093/hmg/9.4.631. [DOI] [PubMed] [Google Scholar]
- 3.Bergman Y, Mostoslavsky R. DNA demethylation: turning genes on. Biol Chem. 1998;379:401–407. doi: 10.1515/bchm.1998.379.4-5.401. [DOI] [PubMed] [Google Scholar]
- 4.Bestor T H. Gene silencing as a threat to the success of gene therapy. J Clin Investig. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouhassira E E, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 6.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 7.Chung J H, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effects in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 8.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobie K, Mehtali M, McClenaghan M, Lathe R. Variegated gene expression in mice. Trends Genet. 1997;13:127–130. doi: 10.1016/s0168-9525(97)01097-4. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y Q, Alami R, Bouhassira E E. Enhancer-dependent transcription oscillations in mouse erythroleukemia cells. Mol Cell Biol. 1999;19:4907–4917. doi: 10.1128/mcb.19.7.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y Q, Seibler J, Alami R, Eisen A, Fiering S N, Bouhassira E E. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J Mol Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 12.Fiering S, Whitelaw E, Martin D I. To be or not to be active: the stochastic nature of enhancer action. Bioessays. 2000;22:381–387. doi: 10.1002/(SICI)1521-1878(200004)22:4<381::AID-BIES8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Garrick D, Sutherland H, Robertson G, Whitelaw E. Variegated expression of a globin transgene correlates with chromatin accessibility but not methylation status. Nucleic Acids Res. 1996;24:4902–4909. doi: 10.1093/nar/24.24.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graubert T A, Hug B A, Wesselschmidt R, Hsieh C L, Ryan T M, Townes T M, Ley T J. Stochastic, stage-specific mechanisms account for the variegation of a human globin transgene. Nucleic Acids Res. 1998;26:2849–2858. doi: 10.1093/nar/26.12.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosveld F. Activation by locus control regions? Curr Opin Genet Dev. 1999;9:152–157. doi: 10.1016/S0959-437X(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 16.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh C L. Evidence that protein binding specifies sites of DNA demethylation. Mol Cell Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh C L. Dynamics of DNA methylation pattern. Curr Opin Genet Dev. 2000;10:224–228. doi: 10.1016/s0959-437x(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 19.Jahner D, Stuhlmann H, Stewart C L, Harbers K, Lohler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 20.Lorincz M C, Schubeler D, Goeke S C, Walters M, Groudine M, Martin D I. Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol Cell Biol. 2000;20:842–850. doi: 10.1128/mcb.20.3.842-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo H Y, Liang X L, Frye C, Wonio M, Hankins G V, Chui D K, Alter B P. Embryonic hemoglobins are expressed in definitive cells. Blood. 1999;94:359–361. [PubMed] [Google Scholar]
- 22.Marks P A, Rifkind R A. Erythroleukemic differentiation. Annu Rev Biochem. 1978;47:419–448. doi: 10.1146/annurev.bi.47.070178.002223. [DOI] [PubMed] [Google Scholar]
- 23.Matsuo K, Silke J, Georgiev O, Marti P, Giovannini N, Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moser T, Harbers K, Kratochwil K. Tissue- and stage-specific activation of an endogenous provirus after transcription through its integration site in the opposite orientation. Mol Cell Biol. 1996;16:384–389. doi: 10.1128/mcb.16.1.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson J A, Gnann J W J, Ghazal P. Regulation and tissue-specific expression of human cytomegalovirus. Curr Top Microbiol Immunol. 1990;154:75–100. doi: 10.1007/978-3-642-74980-3_4. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls R D, Saitoh S, Horsthemke B. Imprinting in Prader-Willi and Angelman syndromes. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 27.Nolan G P, Fiering S, Nicolas J F, Herzenberg L A. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-d-galactosidase activity after transduction of Escherichia coli lacZ. Proc Natl Acad Sci USA. 1988;85:2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson K R, Clegg C H, Huxley C, Josephson B M, Haugen H S, Furukawa T. Transgenic mice containing a 248-kb yeast artificial chromosome carrying the human beta-globin locus display proper developmental control of human globin genes. Proc Natl Acad Sci USA. 1993;90:7593–7597. doi: 10.1073/pnas.90.16.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pikaart M J, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porcu S, Kitamura M, Witkowska E, Zhang Z, Mutero A, Lin C, Chang J, Gaensler K M L. The human beta globin locus introduced by YAC transfer exhibits a specific and reproducible pattern of developmental regulation in transgenic mice. Blood. 1997;90:4602–4609. [PubMed] [Google Scholar]
- 31.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson G, Garrick D, Wilson M, Martin D I K, Whitelaw E. Age-dependent silencing of globin transgenes in the mouse. Nucleic Acids Res. 1996;24:1465–1471. doi: 10.1093/nar/24.8.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabl J F, Henikoff S. Copy number and orientation determine the susceptibility of a gene to silencing by nearby heterochromatin in Drosophila. Genetics. 1996;142:447–458. doi: 10.1093/genetics/142.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi B Z, Cedar H. DNA methylation represses transcription in vivo. Nat Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- 35.Stamminger T, Fleckenstein B. Immediate-early transcription regulation of human cytomegalovirus. Curr Top Microbiol Immunol. 1990;154:3–19. doi: 10.1007/978-3-642-74980-3_1. [DOI] [PubMed] [Google Scholar]
- 36.Wakimoto B T. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 37.Walsh C P, Bestor T H. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walters M C, Fiering S, Eidemiller J, Magis W, Groudine M, Martin D I K. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I K. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 40.Watt F M, Hogan B L. Out of Eden: stem cells and their niches. Science. 2000;287:1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 41.Weiler K S, Wakimoto B T. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 42.Widlund H R, Cao H, Simonsson S, Magnusson E, Simonsson T, Nielsen P E, Kahn J D, Crothers D M, Kubista M. Identification and characterization of genomic nucleosome-positioning sequences. J Mol Biol. 1997;267:807–817. doi: 10.1006/jmbi.1997.0916. [DOI] [PubMed] [Google Scholar]
- 43.Wong A K, Rattner J B. Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res. 1988;16:11645–11661. doi: 10.1093/nar/16.24.11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]