Abstract
Proteolysis targeting chimera (PROTAC) small-molecule degraders have emerged as a promising new type of therapeutic agents, but the design of PROTAC degraders with excellent oral pharmacokinetics is a major challenge. In this study, we present our strategies toward the discovery of highly potent PROTAC degraders of androgen receptor (AR) with excellent oral pharmacokinetics. Employing thalidomide to recruit cereblon/cullin 4A E3 ligase and through the rigidification of the linker, we discovered highly potent AR degraders with good oral pharmacokinetic properties in mice with ARD-2128 being the best compound. ARD-2128 achieves 67% oral bioavailability in mice, effectively reduces AR protein and suppresses AR-regulated genes in tumor tissues with oral administration, leading to the effective inhibition of tumor growth in mice without signs of toxicity. This study supports the development of an orally active PROTAC AR degrader for the treatment of prostate cancer and provides insights and guidance into the design of orally active PROTAC degraders.
Graphical Abstract

INTRODUCTION
Prostate cancer is the second most common malignancy among men in the US and the second most common cause of cancer-related death worldwide. The development of effective therapeutics for the treatment of prostate cancer has been a major focus of scientific research.1–5 The androgen receptor (AR) and its downstream signaling play a critical role in the development and progression of both localized and metastatic prostate cancer.6–10 A number of AR-targeted agents (Figure 1) have been developed to treat advanced prostate cancer, including abiraterone (1), which blocks androgen synthesis and enzalutamide (2) and apalutamide (3, ARN-509), which bind to AR and function as AR antagonists,11–14 but resistance to these AR targeted agents typically develops within 18 months of treatment. In the majority of prostate cancers resistant to these AR targeted agents, AR signaling remains functionally active and continues to play a key role in tumor progression. AR gene amplification, point mutations, and alternate splicing15–18 have been identified as some of the major mechanisms of resistance to these agents targeting AR. There is an urgent need to develop new therapeutic strategies targeting AR for the treatment of prostate cancer, particularly metastatic castration-resistant prostate cancer (mCRPC).10,19,20
Figure 1.
Representative AR antagonists and PROTAC AR degraders.
In recent years, protein degradation based on the proteolysis targeting chimera (PROTAC) strategy has emerged as a promising approach to drug discovery.21–36 PROTAC small-molecule degraders can achieve a more complete target inhibition than traditional small-molecule inhibitors through reducing target protein levels in disease tissues and are predicted to be therapeutically more efficacious.37–43 To date, a number of classes of PROTAC AR degraders have been reported (Figure 1).44–59 In 2008, Crews et al. reported a class of AR degraders designed using a ligand to recruit the MDM2 E3 ligase.44 In 2017, Naito et al. reported specific and non-genetic IAP-dependent protein eraser (SNIPER) compounds that degrade AR using a ligand to recruit the cellular inhibitor of apoptosis protein 1 (cIAP1) E3 ligase.49 Salami et al. reported ARCC-4 as a potent AR degrader that operates by recruiting the VHL-1/cullin 2 E3 ligase complex.52 Our laboratory has reported the discovery of potent AR degraders by recruiting the VHL-1/cullin 2 E3 ligase complex.54–57 Takwale et al. reported the discovery of TD-802 as a potent AR degrader with good liver microsomal stability and pharmacokinetic properties.60 Scientists from Arvinas has disclosed a number of classes of PROTAC AR degraders in patents and patent applications61–63 and has advanced ARV-110 (9) into Phase I clinical development,58,59 whose chemical structure was recently disclosed.64
A PROTAC degrader molecule consists of a ligand for the target protein tethered through a linker to a second ligand, which binds to and recruits an E3 ligase, and consequently, it has a much higher molecular weight than a typical oral drug. Despite the potential promise of PROTAC degraders as a completely new class of therapeutics, the design of PROTAC degraders to achieve excellent oral pharmacokinetics has been a major challenge for the entire PROTAC field. In the present study, we present our strategies toward discovering potent PROTAC degraders of AR with excellent oral pharmacokinetics in mice. Our efforts yielded a number of highly potent PROTAC AR degraders with excellent oral pharmacokinetics.
RESULTS AND DISCUSSION
Design and Synthesis of New AR Degraders with a Cereblon Ligand.
We previously reported a series of highly potent AR degraders, as exemplified by ARD-61, which employ VHL E3 ligands.54–57 ARD-61 has a molecular weight of 1095.8, a large polar surface area (calculated tPSA = 189.2 Å2), and high lipophilicity (calculated CLogP = 8.2) (Figure 2), which are properties all detrimental to oral bioavailability. While ARD-61 potently and effectively induces AR degradation in AR-positive (AR+) prostate cancer cells in vitro and in prostate cancer xenograft tumor tissues in vivo by intraperitoneal injection, it has no oral bioavailability in mice (data not shown).
Figure 2.
Design of new AR degraders employing a cereblon ligand.
In the design of ARD-61, we employed a VHL ligand, which has a molecular weight of nearly 500 and is a peptidomimetic. To obtain potential orally bioavailable AR degraders, we decided to employ thalidomide, which is a cereblon ligand with a low molecular weight (258) and excellent druglike properties.
We first designed and synthesized compounds 10–15 (Table 1) using the AR antagonist employed in ARD-61 (shown in red in Figure 2), tethered to thalidomide through a linear, flexible linker. The aim was to identify the linker length optimal for AR degradation. We evaluated these compounds by Western blotting to determine their ability to reduce the level of AR protein in the VCaP prostate cancer cell line, which has a very high expression level of wild-type AR protein due to AR gene amplification. The AR protein levels were quantified by densitometry, providing the data summarized in Table 1. Compound 10, with a linker of three methylene groups, effectively reduces the AR protein level at 0.1 and 1 μM by 63 and 70%, respectively, but has a minimal effect on the AR protein at 1 and 10 nM. Compound 11, containing a linker with four methylene groups, effectively reduces the AR protein by 42% at 1 nM and by 76% at 10 nM. Interestingly, compound 11 at 0.1 and 1 μM reduces the AR protein level by 51 and 21%, respectively, which are less than the reduction at 10 nM. This reveals the classical “hook” effect that is often observed with PROTAC degraders.60 Compounds 12–15 containing a linker with 5–8 methylene groups all effectively reduce AR protein at all the four concentrations tested (1, 10, 100, and 1000 nM).
Table 1.
Determination of Optimal Linker Lengths Using AR Degraders with Flexible and Semi-Rigid Linkersa

| |||||
|---|---|---|---|---|---|
| Compound | Linker | % AR protein degradation in VCaP Cells (μM) |
|||
| 0.001 | 0.01 | 0.1 | 1 | ||
|
| |||||
| DMSO | -- | 0 | 0 | 0 | 0 |
| 10 |
|
0 | 25 | 63 | 70 |
| 11 |

|
42 | 76 | 55 | 21 |
| 12 |
|
49 | 84 | 89 | 97 |
| 13 |
|
69 | 85 | 77 | 79 |
| 14 |
|
28 | 58 | 79 | 62 |
| 15 |
|
48 | 76 | 82 | 73 |
| 16 |
|
58 | 83 | 79 | 76 |
| 17 |
|
22 | 37 | 54 | 53 |
| 18 |
|
42 | 63 | 84 | 82 |
| 19 |
|
12 | 59 | 85 | 60 |
All the data were the average of three independent experiments.
Next, we synthesized compounds 16–19, which employ linkers with more conformational constraints and improved solubility. Western blotting showed that compounds 16–19 are all very potent and effective in reducing the AR protein level in the VCaP cells, indicating that soluble and conformationally constrained linkers can be employed for the design of potent AR degraders.
We synthesized compounds 20–24 by changing the linking position from the meta- to the ortho-position in the phenyl ring of the cereblon ligand in compounds 12, 13, 16, 17, and 18 (Table 2). Compounds 20–24 are much less potent and effective than compounds 12, 13, 16, 17, and 18 in reducing AR protein in the VCaP cell line. These data indicate that the tethering position in the cereblon ligand for our designed AR degraders is critical for the potent and effective reduction of AR protein.
Table 2.
Investigation of Different Linking Positions of the Cereblon liganda

| |||||
|---|---|---|---|---|---|
| Compound | Linker | % AR protein degradation in VCaP Cells (μM) |
|||
| 0.001 | 0.01 | 0.1 | 1 | ||
|
| |||||
| DMSO | -- | 0 | 0 | 0 | 0 |
| 20 |
|
<5 | 9 | 36 | 38 |
| 21 |
|
10 | 22 | 43 | 48 |
| 22 |
|
<5 | 23 | 41 | 32 |
| 23 |
|
<5 | <5 | <5 | 9 |
| 24 |
|
7 | 40 | 47 | 32 |
All the data are the average of three independent experiments.
Compounds 12, 13, 16, and 17 potently and effectively reduce AR protein in the VCaP cell line. We next assessed the plasma exposure for compounds 12, 13, 16, and 17 in mice with a single oral administration at 10 mg/kg with the data summarized in Table 3. The data show that while compounds 12 and 13 with a very flexible linker have low oral exposures, compounds 16 and 17 with a semi-rigid linker display an improved oral plasma exposure over that of compounds 12 and 13. In particularly, compound 16 has an excellent oral exposure with plasma concentrations of 1242.1, 919.3, and 603.5 ng/mL with 10 mg/kg PO dosing at 1, 3, and 6 h time points or with plasma concentrations of 1.6, 1.2, and 0.8 μM, respectively.
Table 3.
Assessment of Oral Exposure of Compounds 12, 13, 16, and 17 in Micea
| plasma drug concentration (mean ± SD, ng/ ml) |
|||
|---|---|---|---|
| compound | 1 h | 3 h | 6 h |
| 12 | 111.2 ± 41.2 | 55.9 ± 42.0 | 2.9 ± 1.3 |
| 13 | 16.9 ± 6.9 | 8.0 ± 7.7 | 1.6 ± 1.4 |
| 16 | 1242.1 ± 467.2 | 919.3 ± 256.7 | 603.5 ± 231.9 |
| 17 | 267.0 ± 104.5 | 254.5 ± 39.2 | 96.9 ± 33.9 |
Each compound was administered with a single dose at 10 mg/kg via oral gavage using 100% PEG 200 as the formulation. Plasma samples were collected at 1, 3, and 6 h time points with three mice for each time point and analyzed by LC-MS/MS. Plasma concentrations are presented as mean ± standard deviation (SD).
Encouraged by the oral exposure data for compound 16, we synthesized compounds 25–42 by employing even more conformational constrained linkers with lengths similar to that in compound 16 (Table 4). All the linkers in compounds 25–40 also retain a positively charged amine group to maintain good physiochemical properties in the degrader molecules. These compounds were evaluated for AR degradation in VCaP cells by Western blotting at four different concentrations (1, 10, 100, and 1000 nM) with a 24 h treatment time. The data are summarized in Table 4.
Table 4.
Investigation of the Effect of Different Rigid Linkers on the AR Degradersa

| |||||
|---|---|---|---|---|---|
| Compound | Linker | % AR protein degradation in VCaP Cells (μM) |
|||
| 0.001 | 0.01 | 0.1 | 1 | ||
|
| |||||
| DMSO | -- | 0 | 0 | 0 | 0 |
| 25 |
|
54 | 77 | 91 | 73 |
| 26 |

|
64 | 96 | 98 | 90 |
| 27 |
|
79 | 93 | 85 | 55 |
| 28 |
|
62 | 94 | 98 | 96 |
| 29 |
|
71 | 83 | 95 | 83 |
| 30 |

|
65 | 92 | 96 | 79 |
| 31 |
|
85 | 97 | 96 | 83 |
| 32 |
|
76 | 97 | 97 | 96 |
| 33 |
|
57 | 93 | 96 | 85 |
| 34 |
|
40 | 96 | 99 | 99 |
| 35 |
|
37 | 78 | 93 | 88 |
| 36 |
|
30 | 82 | 97 | 95 |
| 37 |
|
38 | 80 | 87 | 84 |
| 38 |
|
16 | 56 | 71 | 51 |
| 39 |
|
14 | 38 | 67 | 80 |
| 40 |
|
15 | 48 | 73 | 82 |
| 41 |
|
0 | 16 | 82 | 88 |
| 42 |
|
13 | 30 | 38 | 57 |
All the data are the average of three independent experiments.
Compounds 25–33 effectively reduce the AR protein level by >50% at 1 nM and achieve a maximum AR degradation of >90%. Compounds 34–37 are less potent than compounds 25–33 but are still capable of reducing the AR protein level by >50% at 10 nM. Compounds 38–41 are less potent than compounds 34–37 and reduce the AR protein level by 50% at 100 nM. Compound 42 is the least potent degrader among this series of compounds.
Modification of the AR Antagonist Portion in AR Degraders.
Compound 28 is a highly potent AR degrader. We next performed modifications of the AR antagonist portion in compound 28 and obtained compounds 43–47 (Table 5). These compounds were also evaluated for AR degradation in VCaP cells by Western blotting at four different concentrations (1, 10, 100, and 1000 nM) with a 24 h treatment time, delivering the data summarized in Table 5.
Table 5.
Investigation of the Effect of Different AR Antagonists on the AR Degradersa

| |||||
|---|---|---|---|---|---|
| Compound | AR antagonist portion | % AR protein degradation in VCaP Cells (μM) |
|||
| 0.001 | 0.01 | 0.1 | 1 | ||
|
| |||||
| DMSO | -- | 0 | 0 | 0 | 0 |
| 28 |
|
62 | 94 | 98 | 96 |
| 43 |
|
<5 | 19 | 75 | 90 |
| 44 |
|
<5 | 30 | 25 | 39 |
| 45 |

|
<5 | 15 | 75 | 80 |
| 46 |
|
22 | 45 | 87 | 90 |
| 47 |
|
6 | 12 | 33 | 41 |
All the data are the average of three independent experiments..
Removal of dimethyl or tetramethyl groups from the tetramethylcyclobutanyl moiety in compound 28 resulted in compounds 43 or 44, respectively. While compound 43 effectively reduces the AR protein at 100 nM and 1 μM, it is approximately 100 times less potent than compound 28. Compound 44 has no effect on the level of AR protein at 1 nM and reduces AR protein level by 25–39% at 10–1000 nM. We replaced 2-C1 substituent on the phenyl with 2-F or 2-CF3 in compound 28, yielding compounds 45 or 46, respectively. Compound 45 effectively reduces AR protein at 100 nM and 1 μM, but it is ineffective at 1 and 10 nM. Compound 46 is less potent than compound 45. Both compounds 45 and 46 are therefore much less potent than compound 28 in reducing the level of AR protein. We generated compound 47 by replacing the substituted phenyl group in compound 46 with a substituted pyridine, which had been used in apalutamide (3, ARN-509, Figure 1). Compound 47 is much less potent and effective than compound 46 in reducing AR protein and is thus a weak AR degrader. These data establish that the AR antagonist portion plays a critical role for the highly potent and effective AR protein degradation by compound 28.
Further Assessment of the Degradation of AR Protein in AR+ Prostate Cancer Cell Lines.
Based on our initial screening data in the VCaP cell line, compounds 26, 27, 28, 33, and 34 are highly potent and effective AR degraders. The potency of these compounds in the VCaP prostate cancer cell line was further evaluated.
Western blotting data showed that these compounds are highly potent and effective in reducing AR protein in a dose-dependent manner in the VCaP cell line (Figure 3). Compounds 26, 27, 28, 33, and 34 achieve DC50 values of 0.2, 0.8, 0.3, 0.1, and 0.4 nM, respectively, in the VCaP cell line. Compounds 26, 27, 28, 33, and 34 are capable of reducing AR protein by >90% with DC90 values of 3, 2, 3, 2, and 2 nM in the VCaP cell line, respectively. The hook effect was observed at high concentrations for compounds 26 and 27.
Figure 3.
Western blotting analysis of AR protein in the AR+ VCaP cell line.
We next tested these compounds for their potencies in inducing AR degradation in the LNCaP cell line carrying an AR T878A mutation (Figure 4). Compounds 26, 27, 28, 33, and 34 achieve DC50 values of 5.1, 1.4, 8.3, 1.4, and 3.4 nM, respectively, in the LNCaP cell line. Furthermore, compounds 26, 27, 28, 33, and 34 are capable of reducing AR protein by >90% with DC90 values of 41, 42, 25, 10, and 10 nM, respectively, in the LNCaP cell line. No hook effect was observed for all these compounds at concentrations up to 1 μM.
Figure 4.
Western blotting analysis of AR protein in the AR+ LNCaP cell line.
We evaluated their degradation kinetics in both VCaP and LNCaP cell lines (Figure 5). Compounds 26, 27, 28, 33, and 34 effectively reduce the AR protein level within 3 h and achieve near-complete AR depletion after a 6 h treatment. The kinetic data show that induced AR degradation by these PROTAC degraders in both LNCaP and VCaP cells is fairly rapid.
Figure 5.
Western blotting analysis of AR protein in the AR+ VCaP and LNCaP cell lines.
Evaluation of Potent AR Degraders for Cell Growth Inhibition in VCaP and LNCaP Cell Lines.
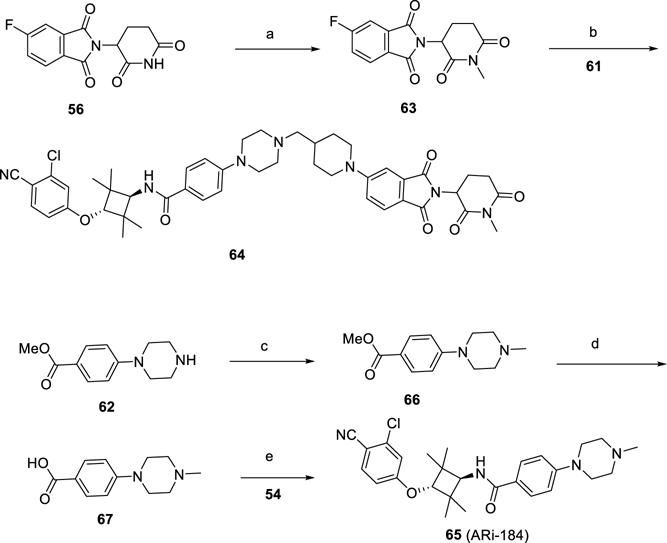
We evaluated a number of highly potent AR degraders for their cell growth inhibition in the VCaP and LNCaP cell lines. We synthesized 64 (Scheme 3), in which a methyl group was installed on the amino group of the piperidine-2,6-dione moiety in the cereblon ligand portion in 28 and included 64 as a control compound in the cell growth assay. We synthesized compound 65 (ARi-184, Scheme 3), which is the AR antagonist used for the design of these potent AR degraders and included it as an additional control compound in the cell growth assay. We also included enzalutamide as another control compound in the cell growth assay. The results are shown in Figure 6.
Scheme 3. Synthesis of Compounds 64 and 65a.
a(a) Mel, K2CO3, DMF, 60 °C; (b) DIPEA, DMSO, 100 °C; (c) Mel, K2CO3, DMF, 60 °C; (d) NaOH, MeOH/H2O, rt; (e) HATU, DIPEA, DMF, rt.
Figure 6.
Cell growth inhibition in LNCaP and VCaP cells treated with AR degraders. LNCaP and VCaP cells were treated with different compounds in a charcoal-stripped medium in the presence of 0.1 nM of AR agonist R1881 for 4 days. Cell viability was determined with a WST-8 assay.
In the VCaP cell line, enzalutamide has an IC50 value of 394 nM, while 64 and 65 have IC50 values of 783 and 49 nM, respectively. Compounds 26, 27, 28, 33, and 34 are all highly potent and effective in the inhibition of cell growth in the VCaP cell line and achieve IC50 values of 7, 8, 4, 4, and 5 nM, respectively.
In the LNCaP cell line, enzalutamide has an IC50 value of 133 nM, whereas 64 and 65 have IC50 values of 237 and 81 nM, respectively. Compounds 26, 27, 28, 33, and 34 are also highly potent and effective in the inhibition of cell growth in the LNCaP cell line and achieve IC50 values of 10, 11, 5, 14, and 13 nM, respectively.
We investigated the mechanism of AR degradation induced by 28 in LNCaP and VCaP cells. Our data showed that AR degradation induced by 28 can be effectively blocked by pretreatment with the AR antagonist (65), cereblon ligand (thalidomide), proteasome inhibitor (MG132), or NEDD8 activating El enzyme inhibitor (MLN4924) in both VCaP and LNCaP cell lines (Figure 7). These mechanistic data demonstrate that 28 is a bona fide PROTAC AR degrader.
Figure 7.
Mechanistic investigation of AR degradation induced by 28 (ARD-2128) in VCaP and LNCaP cells. Cells were pretreated with AR antagonist 65 (ARi-184), thalidomide, MG132, and MLN4924 followed by 3 h treatment with ARD-2128 at 100 nM.
We next investigated the ability of 28 to suppress AR-regulated gene expression in the VCaP and LNCaP cell lines, with a potent AR antagonist (65) included as the control (Figure 8). Our data showed that compound 28 effectively suppresses the expression of PSA and TMPRSS2 genes in both VCaP and LNCaP cell lines in a dose-dependent manner. It is effective in reducing the mRNA levels of PSA and TMPRSS2 genes at concentrations as low as 10 nM. In direct comparison, compound 28 at 10 nM is more effective in reducing the mRNA levels of PSA and TMPRSS2 genes than the corresponding AR antagonist (65) at 1 μM in both VCaP and LNCaP cell lines.
Figure 8.
Suppression of AR-regulated gene expression in the VCaP and LNCaP cell lines by the AR degrader 28 (ARD-2128) and the AR antagonist (65) (ARi-184). VCaP and LNCaP cells were treated for 24 h, and a quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed to determine the mRNA levels for AR and AR-regulated genes.
Pharmacokinetic Studies of AR Degraders in Mice.
We evaluated the pharmacokinetics (PK) of five highly potent AR degraders (compounds 26, 27, 28, 33, and 34) in mice with both intravenous and oral administration, obtaining the data summarized in Table 6.
Table 6.
Summary of PK Data for Compounds 26, 27, 28, 33, and 34 in Male ICR Mice
| compound | route | dose (mg/kg) | T1/2 (h) | AUC0–t (h·ng/ml) | Cl (ml/min/kg) | Vss (L/kg) | route | dose (mg/kg) | T1/2 (h) | Tmax (h) | Cmax (ng/ml) | AUC0–t (h·ng/ml) | F (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 26 | IV | 2 | 17.8 | 11,035 | 1.9 | 2.7 | PO | 5 | 12.0 | 4.0 | 1389 | 20,600 | 75 |
| 27 | IV | 2 | 11.5 | 15,759 | 1.7 | 1.5 | PO | 5 | 11.2 | 4.0 | 980 | 14,588 | 37 |
| 28 (ARD-2128) | IV | 2 | 27.6 | 13,299 | 1.2 | 2.7 | PO | 5 | 18.8 | 4.7 | 1304 | 22,361 | 67 |
| 33 | IV | 1 | 21.0 | 4334 | 2.2 | 3.8 | PO | 3 | 12.4 | 6.0 | 207 | 3127 | 24 |
| 34 | IV | 1 | 25.5 | 2565 | 3.2 | 6.8 | PO | 3 | 67.8 | 4.7 | 134 | 2550 | 33 |
Cmax, maximum drug concentration; AUC0–24 h, area-under-the-curve between 0 and 24 h; Cl = plasma clearance rate; Vss = steady state volume of distribution; T1/2 = terminal half-life, Tmax = the time take to reach Cmax; F = oral bioavailability; IV, intravenous administration; PO, oral administration.
The PK data show that 26, 27, 28, 33, and 34 achieve good to excellent overall PK profiles. They all have low clearance (1.2–3.2 mL/min/kg) and a moderate to high steady-state volume of distribution (Vss) of between 1.5 and 6.8 L/kg. Each compound has a long T1/2 following intravenous administration, ranging between 11.5 and 27.6 h, and oral administration, ranging between 11.2 and 67.8 h. To compare their Cmax and AUC in oral administration, we converted their Cmax and AUC values to per mg/kg dosing. For each mg/kg oral administration, 26, 27, 28, 33, and 34 achieve Cmax values of 278, 196, 261, 69, and 45 ng/mL, respectively, and AUC0–24 h of 4120, 2918, 4472, 1042, 850 h·ng/mL, respectively. Compounds 26, 27, 28, 33, and 34 achieve overall oral bioavailabilities of 75, 37, 67, 24, and 33%, respectively.
Pharmacodynamic Evaluation of Several AR Degraders in VCaP Xenograft Tumors.
Based on their excellent PK profiles, we evaluated 26, 27, and 28 for their pharmacodynamics (PD) effect in reducing AR protein in the VCaP Xenograft tumor tissue in mice with a single oral administration. The PD results are shown in Figure 9.
Figure 9.
Western blot analysis of AR protein in VCaP xenograft tumors in SCID mice. SCID mice bearing VCaP tumors were treated with a single oral administration with each compound (26, 27, and ARD-2128) at 20 mg/kg. Mice were euthanized at indicated time points, and tumor tissues were collected for Western blot analysis of AR protein. GAPDH was used as the loading control.
Our Western blotting analysis of the VCaP tumor tissues showed that a single oral administration of 26 and 28 at 20 mg/kg is effective in reducing the levels of AR protein in mice after 24 h but has a modest effect at the 6 h time point. However, compound 27 reduces the level of AR protein only modestly at both time points.
To gain further insights into the PD data, we determined the drug concentrations in both plasma and tumor tissue for compounds 26, 27, and 28. The data obtained are summarized in Table 7. Our data showed that with a single dose PO administration, all three compounds achieve good drug exposures in both plasma and tumor tissues. However, compounds 28 and 26 have two times higher drug concentrations than 27 at both 6 and 24 h time points in both plasma and tumor tissue. Furthermore, both compounds 28 and 26 have higher drug concentrations at the 24 h time point than at the 6 h time point. These data suggest that with daily administration, these AR degraders could be expected to accumulate in both plasma and tumor tissues.
Table 7.
Analysis of Drug Concentrations in Plasma and VCaP Tumor Tissue in SCID Mice for 26, 27, and 28 (ARD-2128)a
| compound | time point (h) | plasma concentration (ng/ml) | tumor concentration (ng/kg) |
|---|---|---|---|
| mean ± SD | mean ± SD | ||
| 26 | 6 | 1365.0 ± 1102.9 | 718.3 ± 110.6 |
| 24 | 1666.0 ± 691.6 | 1743.3 ± 448.6 | |
| 27 | 6 | 803.3 ± 203.0 | 314.8 ± 93.7 |
| 24 | 775.7 ± 346.6 | 685.0 ± 232.5 | |
| 28 (ARD-2128) | 6 | 1513.3 ± 41.6 | 635.0 ± 83.2 |
| 24 | 1659.3 ± 846.0 | 1506.7 ± 705.0 |
Each compound was administered with a single dose at 20 mg/kg with 100% PEG200 as the formulation in mice bearing VCaP tumors with one tumor per mouse. Plasma and tumor tissue were collected at 6 and 24 h time points for each compound with three mice for each time point.
We next evaluated 28 (ARD-2128) for its pharmacodynamics (PD) effect in reducing AR protein in the VCaP xenograft tumor tissue in mice with daily oral administration for 3 consecutive days. The PD results are shown in Figure 10.
Figure 10.
Pharmacodynamic analysis for 28 (ARD-2128) in VCaP xenograft tumors. SCID mice bearing VCaP tumors were treated with daily, PO dose of ARD-2128 at 10 mg/kg for 3 consecutive days. Mice were euthanized at indicated time points after the last dose, and tumor tissues were collected for Western blot and qRT-PCR analysis. GAPDH was used as the loading control in the Western blot analysis.
Western blotting analysis of the VCaP tumor tissues showed that daily oral administration of 28 at 10 mg/kg for 3 days effectively reduces the levels of AR protein in mice at 3 and 6 h but has only a modest effect at the 24 h time point. We also investigated the ability of 28 to suppress AR regulated gene expression in vivo. Our data showed that 28 effectively suppresses the expression of PSA, TMPRSS2, and FKBP5 genes in vivo at 3 and 6 h and is capable of reducing the mRNA level in all three genes by >50% at 3 h with daily oral administration for 3 days at 10 mg/kg (Figure 10).
Antitumor Activity of ARD-2128 in the VCaP Xenograft Model in Mice.
Based on the PD data and also the drug exposure data in the VCaP tumors, we evaluated 28 (ARD-2128) for its antitumor efficacy in the VCaP xenograft tumor model in mice. We included enzalutamide, a FDA approved AR antagonist as a control compound in this efficacy experiment. The efficacy data are summarized in Figure 11.
Figure 11.
Antitumor activity of ARD-2128 in the VCaP xenograft tumor model in SCID mice. Enzalutamide was included as a control. Each compound was dosed via oral gavage daily for a total of 21 days.
Enzalutamide at 40 mg/kg oral administration inhibits tumor growth by 39% over the vehicle control at the end of the 21 day treatment (p = 0.0075). In comparison, ARD-2128 at 10, 20, and 40 mg/kg inhibits tumor growth by 46, 69, and 63%, respectively, at the end of the 21 day treatment over the vehicle control (p < 0.0001 for all three doses vs control). While ARD-2128 at 10 mg/kg is slightly more effective than enzalutamide at 40 mg/kg, it is significantly more effective at both 20 and 40 mg/kg, than enzalutamide at 40 mg/kg in the inhibition of tumor growth at the end of treatment (p < 0.05 for ARD-2128 at both doses vs enzalutamide).
At all the three dose levels, ARD-2128 and enzalutamide are well tolerated in mice, causing no more than 5% of maximum weight loss during the entire experiment.
Evaluation of ARD-2128 for Its Plasma Stability and Microsomal Stability.
We evaluated ARD-2128 for its plasma and microsomal stability in mouse, rat, dog, monkey, and humans. The data showed that ARD-2128 has excellent plasma and microsomal stability in all the five species (Table 8).
Table 8.
Liver Microsomal and Plasma Stability of ARD-2128 in Five Species (Human, Mouse, Rat, Dog, and Monkey)
| species | liver microsomal stability (T1/2, min) | plasma stability (T1/2, min) |
|---|---|---|
| mouse | >120 | >120 |
| rat | >120 | >120 |
| dog | >120 | >120 |
| monkey | >120 | >120 |
| human | >120 | >120 |
CHEMISTRY
The synthesis of compounds 10–15, 20, and 21 is shown in Scheme 1. Compound 51 was synthesized from the reaction of compounds 48 and 49 with subsequent hydrolysis by NaOH. Compound 54 was produced by the substitution reaction of 52 with 53 followed by deprotection by TFA of the NHBoc group. The key intermediates 55 were synthesized by the amidation of a series of benzoic acids (51) by compound 54. Finally, the target compounds were obtained through the substitution reaction of intermediate 55 with 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione and compound 56 with 2-(2,6-dioxopiperidin-3-yl)-4-fluoroiso-indoline-1,3-dione.
Scheme 1. Synthesis of Compounds 10–15, 20, and 21a.
a(a) DIPEA, DMSO, 100 °C; (b) NaOH, MeOH/H2O, rt; (c) NaH, DMF, 0 °C to rt; TFA, DCM, rt; (d) HATU, DIPEA, DMF, rt; TFA, DCM, rt; (e) DIPEA, DMSO, 100 °C; (f) DIPEA, DMSO, 100 °C.
Compounds 16–19, 22–28, and 35–40 were synthesized according to the method shown in Scheme 2. Amidation of compound 54 with 58 gave the key intermediate 59 after subsequent deprotection by TFA. The intermediate 61 was made by the reductive animation of compound 59 with different aldehydes and ketones (60) and subsequent deprotection by TFA. The target compounds were obtained by the substitution reaction of compound 61 with either 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione (56) or 2-(2,6-dioxopiperidin-3-yl)-4-fluoro-isoindoline-1,3-dione (57).
Scheme 2. Synthesis of Compounds 16–19, 22–28, and 35–40a.
a(a) HATU, DIPEA, DMF, rt; TFA, DCM, rt; (b) NaBH(OAc)3, AcOH, DCE, rt; TFA, DCM, rt; (c) DIPEA, DMSO, 100 °C; (d) DIPEA, DMSO, 100 °C.
The synthesis of negative control compounds (64 and 65) is shown in Scheme 3. Compound 63 was produced by methylation of the intermediate 56. Compound 64 was made by the reaction of compound 61 with 63. As shown in Scheme 3, intermediate 67 was synthesized from the methylation of intermediate 62 and Mel with subsequent hydrolysis by NaOH. Compound 65 was made by the amidation of compounds 67 and 54.
The synthesis of compounds 29–32 is shown in Scheme 4. Compound 68 was produced by the amidation reaction between 54 and 66 and subsequent hydrolysis by NaOH. The key intermediate 70 was synthesized from the amidation of compound 68 by 69. The intermediates 71 were made by the reductive amination of compound 70 with different aldehydes or ketones. Finally, the target compounds were obtained by a substitution reaction between 71 and 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione (56).
Scheme 4. Synthesis of Compounds 29–32a.
a(a) HATU, DIPEA, DMF, rt; (b) NaOH, MeOH/H2O, rt; (c) HATU, DIPEA, DMF, rt; TFA, DCM, rt; (d) NaBH(OAc)3, AcOH, DCE, rt; TFA, DCM, rt; (e) DIPEA, DMSO, 100 °C.
Compounds 33 and 34 were synthesized according to the method in Scheme 5. The key intermediate (72) was synthesized with the substitution reaction of compound 56 with 69 and subsequent deprotection by TFA. Intermediate 76 was synthesized from the substitution reaction of compound 73 with the alcohol (74) and hydrolysis by NaOH. Intermediate 77 was synthesized by the amidation of compound 76 by compound 54. The oxidation of compound 77 gave intermediate 78, the reductive amination of which produced the target compounds 33 and 34.
Scheme 5. Synthesis of Compounds 33 and 34a.
a(a) DIPEA, DMSO, 100 °C; TFA, DCM, rt; (b) DIPEA, DMSO, 100 °C; (c) NaOH, MeOH/H2O, rt; (d) HATU, DIPEA, DMF, rt; (e) DMP, DCM, rt; (f) NaBH(OAc)3, AcOH, DCE, rt.
The synthesis of compounds 41 and 42 is shown in Scheme 6. Compound 81 was produced by the substitution reaction of compounds 73 and 79 followed by hydrolysis by NaOH. Intermediate 82 was synthesized by the amidation of compounds 54 with 81 followed by deprotection with TFA. Finally, the target compounds were obtained by the substitution reaction of 82 with 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione (56).
Scheme 6. Synthesis of Compounds 41 and 42a.
a(a) DIPEA, DMSO, 100 °C; (b) NaOH, MeOH/H2O, rt; (c) HATU, DIPEA, DMF, rt; TFA, DCM, rt; (d) DIPEA, DMSO, 100 °C.
The synthesis of compounds 43 and 44 is shown in Scheme 7. Compound 84 was produced by the substitution reaction of compounds 52 and 83 with subsequent deprotection by TFA. The key intermediate 85 was synthesized by the amidation of compound 84 with 58 followed by deprotection by TFA. Compound 87 was obtained through the reductive amination of intermediates 86 and 85 and subsequent deprotection by TFA. Finally, the target compounds were obtained by a substitution of 87 with 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione 56.
Scheme 7. Synthesis of Compounds 43 and 44a.
a(a) NaH, DMF, 0 °C to rt; TFA, DCM, rt; (b) HATU, DIPEA, DMF, rt; TFA, DCM, rt; (c) NaBH(OAc)3, AcOH, DCE, rt; TFA, DCM, rt; (d) DIPEA, DMSO, 100 °C.
Compounds 45–47 were synthesized according to the method in Scheme 8. Compound 89 was synthesized from the substitution reaction of compounds 88 and 53 followed by deprotection by TFA. Amidation of compound 89 with 58 with subsequent deprotection by TFA gave intermediate 90. Intermediate 91 was synthesized from the reductive amination of compound 90 with 86. Finally, the target compounds were obtained from the substitution reaction of 91 with 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione (56).
Scheme 8. Synthesis of Compounds 45–47a.
a(a) NaH, DMF, 0 °C to rt; TFA, DCM, rt; (b) HATU, DIPEA, DMF, rt; TFA, DCM, rt; (c) NaBH(OAc)3, AcOH, DCE, rt; TFA, DCM, rt; (d) DIPEA, DMSO, 100 °C.
DISCUSSION AND CONCLUSIONS
In this study, we report the design, synthesis, and evaluation of a series of PROTAC AR degraders using a potent AR antagonist and thalidomide as the cereblon ligand with the objective of discovering potent and orally bioavailable AR degraders. Following determination of the optimal linker lengths using flexible linkers, a series of AR degraders were designed and synthesized by employing conformational constrained linkers containing a positively charged amine. This led to identification of a number of highly potent AR degraders, which display excellent pharmacokinetics and oral bioavailability with ARD-2128 being the best compound. Mechanistic studies showed that ARD-2128 is a bona fide PROTAC AR degrader and strongly suppresses AR-regulated genes in a dose- and time-dependent manner in AR+ prostate cancer cell lines. ARD-2128 also potently inhibits cell growth in the VCaP prostate cancer cell line with AR gene amplification and the LNCaP cell line with an AR mutation. As compared to our previously reported AR degrader ARD-61, which has a molecular weight of 1095.8, a calculated tPSA of 189.2 Å2, and a calculated CLogP of 8.2, ARD-2128 has a much reduced molecular weight (820.4), a reduced calculated tPSA (155.4 Å2), and a lower calculated CLogP (6.9). ARD-2128 achieves an overall excellent pharmacokinetic profile and has excellent plasma and tissue exposures in both native prostate tissue and xenograft tumor tissue after a single oral administration in mice. A single oral administration of ARD-2128 effectively reduces AR protein in the VCaP tumor tissue. Importantly, oral administration of ARD-2128 is effective in the inhibition of tumor growth in the VCaP xenograft tumor model without any signs of toxicity. In a direct comparison, ARD-2128 at 20 or 40 mg/kg is more efficacious than enzalutamide at 40 mg/kg. ARD-2128 is therefore a potent and orally efficacious PROTAC AR degrader.
While ARD-2128 achieves a DC90 of 3.5 nM in the VCaP cell line in vitro, much higher concentrations of ARD-2128 are needed to achieve effective AR degradation in the VCaP tumor tissue in mice. ARD-2128 was found to have a plasma protein binding of 99.8% in mouse plasma (data not shown), suggesting that significant improvement in mouse plasma protein binding for ARD-2128 may further improve its in vivo potency in AR degradation and tumor growth inhibition.
It is important to note that Arvinas scientists have employed the same class of AR antagonist and thalidomide as the cereblon ligand for the synthesis of a large number of PROTAC AR degraders.61–64 In fact, the chemical structure for ARD-2128 and associated chemical data were disclosed in one of these patents (example 316).61 However, although AR degradation activity in the VCaP cell line for ARD-2128 was reported in the patent,61 detailed biological characterization and pharmacological data were not reported.
In summary, we presented our strategies toward discovering highly potent AR degraders with excellent oral pharmacokinetics in mice. These general strategies employed in our study provide insights and guidance for the design of potent and orally bioavailable PROTAC degraders for other therapeutically important protein targets.
EXPERIMENTAL SECTION
Chemistry.
General Experiment and Information.
Unless otherwise noted, all purchased reagents were used as received without further purification. 1H NMR and 13C NMR spectra were recorded on a Bruker Advance 400 MHz spectrometer. 1H NMR spectra are reported in parts per million (ppm) downfield from tetramethylsilane (TMS). All 13C NMR spectral peaks are reported in ppm and measured with 1H decoupling. In reported spectral data, the format (δ) chemical shift (multiplicity, J values in Hz, integration) was used with the following abbreviations: s = singlet, d = doublet, t = triplet, q = quartet, and m = multiplet. Mass spectrometric (MS) analysis was carried out with a Waters UPLC mass spectrometer. The final compounds were all purified by a C18 reverse phase preparative HPLC column with solvent A (0.1% TFA in H2O) and solvent B (0.1% TFA in CH3CN) as eluents. The purity of all the final compounds was shown to be >95% by UPLC–MS or UPLC.
General Procedure for the Synthesis of Compounds 10–15, 20, and 21.
DIPEA (3 equiv) was added to a solution of compounds 48 (1 mmol) and 49 (1.1 equiv each) in DMSO. After stirring at 100 °C for 2 h, water was added and the reaction mixture was extracted with EtOAc; it was then washed with water, and the organic phase was dried over Na2SO4. Compound 50 was obtained by removing the solvent under vacuum and purified by flash column with 84% yield. Then, the desired intermediate (51) was obtained by hydrolysis with NaOH solid (2.0 equiv) in MeOH/H2O at r.t. in 88% yield.
NaH (1.2 equiv) was added to a solution of 53 (10 mmol) in dry DMF at 0 °C. After stirring the mixture at 0 °C for 20 min, 52 (1.0 equiv) was added and the mixture was stirred at rt for 4 h. After UPLC-MS demonstrated full conversion of the starting materials, H2O was added and the mixture was extracted three times with EtOAc. The combined organic layers were washed with brine and then dried over anhydrous Na2SO4. The solvent was removed on a rotary evaporator. The Boc-protected intermediate was obtained by flash column chromatography. The desired intermediate (54) was obtained by deprotection with TFA in DCM in 86% yield.
DIPEA (5 equiv) and HATU (1.2 equiv) were added to a solution of compounds 54 (1 mmol) and 51 (1.1 equiv) in DMF (2 mL). After stirring at rt for 1 h, water was added and the reaction mixture was extracted by EtOAc; it was then washed by water, and the organic phase was dried by Na2SO4. Compound 55 was obtained by removing the solvent under vacuum and purified by flash column with subsequent deprotection with TFA with 80% yield.
DIPEA (5 equiv) was added to a solution of 55 (0.8 mmol) and 2-(2,6-dioxopiperidin-3-yl)-5-fluoroiso-indoline-1,3-dione 56 (1.1 equiv) in DMSO (2 mL). After 4 h at 100 °C, the mixture was subject to HPLC purification to afford compounds 10–15 with 70–85% yields.
DIPEA (5 equiv) was added to a solution of the compounds 55 (0.5 mmol) and 2-(2,6-dioxopiperidin-3-yl)-4-fluoroisoindoline-1,3-dione (10) (1.1 equiv) in DMSO (2 mL). After 4 h at 100 °C, the mixture was subject to HPLC purification to afford compounds 20–21 in yields of 75–86%.
4-((1r,3r)-3-Amino-2,2,4,4-tetramethylcyclobutoxy)-2-chlorobenzonitrile (INT-54).
1H NMR (400 MHz, DMSO-d6) δ 7.87 (d, J = 8.7 Hz, 1H), 7.19 (d, J = 2.3 Hz, 1H), 6.99 (dd, J = 8.8, 2.3 Hz, 1H), 4.37 (s, 1H), 3.76 (s, 1H), 3.11 (s, 1H), 1.29 (s, 6H), 1.08 (s, 6H). UPLC–MS calculated for C15H20ClN2O [M + H]+: 279.13, found: 279.07. UPLC-retention time: 2.0 min, purity >95%.
N-((1r,3r)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(piperazin-1-yl)benzamide (INT-59).
1H NMR (400 MHz, DMSO-d6) δ 9.08 (s, 1H), 7.89 (d, J = 8.7 Hz, 1H), 7.81 (s, 1H), 7.60 (d, J = 9.2 Hz, 1H), 7.18 (d, J = 2.3 Hz, 1H), 7.05 (d, J = 8.9 Hz, 2H), 7.01–6.98 (m, 1H), 4.33 (s, 1H), 4.07 (d, J = 9.1 Hz, 1H), 3.54–3.44 (m, 4H), 3.26 (s, 4H), 1.23 (s, 6H), 1.13 (s, 6H). UPLC–MS calculated for C26H32ClN4O2 [M + H]+: 467.22, found: 467.24. UPLC-retention time: 4.1 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-((3-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)propyl)amino)benzamide (10).
1H NMR (400 MHz, MeOH-d4) δ 7.74 (d, J = 8.7 Hz, 1H), 7.67 (d, J = 8.8 Hz, 2H), 7.57 (d, J = 8.4 Hz, 1H), 7.14 (d, J = 2.4 Hz, 1H), 7.03–6.98 (m, 2H), 6.87 (dd, J = 8.4, 2.2 Hz, 1H), 6.68 (d, J = 8.8 Hz, 2H), 5.05 (dd, J = 12.4, 5.4 Hz, 1H), 4.30 (s, 1H), 4.14 (s, 1H), 3.37 (t, J = 6.9 Hz, 3H), 2.90–2.66 (m, 4H), 2.14–2.08 (m, 1H), 2.04–1.95 (m, 2H), 1.29 (s, 6H), 1.23 (s, 6H). UPLC–MS calculated for C38H40ClN6O6 [M + H]+: 711.27, found: 711.30. UPLC-retention time: 5.9 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-((4-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)butyl)amino)benzamide (11).
1H NMR (400 MHz, MeOH-d4) δ 7.73–7.67 (m, 3H), 7.54 (d, J = 8.4 Hz, 1H), 7.12 (d, J = 2.4 Hz, 1H), 6.97 (td, J = 4.6, 2.5 Hz, 2H), 6.82 (dd, J = 8.4, 2.2 Hz, 1H), 6.79–6.74 (m, 2H), 5.03 (dd, J = 12.4, 5.5 Hz, 1H), 4.27 (s, 1H), 4.12 (s, 1H), 3.28–3.21 (m, 4H), 2.87–2.68 (m, 3H), 2.65 (s, 1H), 2.12–2.05 (m, 1H), 1.77 (d, J = 2.8 Hz, 4H), 1.27 (s, 6H), 1.21 (s, 6H). UPLC–MS calculated for C39H42C1N6O6 [M + H]+: 725.29, found: 725.23. UPLC-retention time: 5.9 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-((5-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)pentyl)amino)benzamide (12).
1H NMR (400 MHz, MeOH-d4) δ 7.71 (dd, J = 8.7, 4.9 Hz, 3H), 7.53 (dd, J = 8.3, 3.7 Hz, 1H), 7.12 (d, J = 2.4 Hz, 1H), 6.97 (dd, J = 9.1, 2.3 Hz, 2H), 6.81 (ddd, J = 19.2, 11.4, 6.1 Hz, 3H), 5.03 (dd, J = 12.4, 5.4 Hz, 1H), 4.28 (s, 1H), 4.13 (s, 1H), 3.25–3.17 (m, 4H), 2.88–2.65 (m, 3H), 2.09 (ddd, J = 12.7, 6.7, 3.8 Hz, 1H), 1.69 (dt, J = 13.9, 7.0 Hz, 4H), 1.56 (dd, J = 15.0, 8.3 Hz, 2H), 1.41 (dd, J = 11.0, 4.7 Hz, 1H), 1.28 (s, 6H), 1.22 (s, 6H). UPLC–MS calculated for C40H44ClN6O6 [M + H]+: 739.30, found: 739.28. UPLC-retention time: 6.3 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-((6-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)hexyl)amino)benzamide (13).
1H NMR (400 MHz, MeOH-d4) δ 7.75–7.70 (m, 2H), 7.64 (d, J = 8.7 Hz, 1H), 7.47 (d, J = 8.4 Hz, 1H), 7.05 (d, J = 2.4 Hz, 1H), 6.98–6.93 (m, 2H), 6.90 (dd, J = 9.3, 2.2 Hz, 2H), 6.75 (dd, J = 8.4, 2.2 Hz, 1H), 5.00–4.94 (m, 1H), 4.21 (s, 1H), 4.07 (s, 1H), 3.17 (dt, J = 22.5, 7.1 Hz, 4H), 2.81–2.60 (m, 3H), 2.05–1.99 (m, 1H), 1.62 (dd, J = 11.8, 5.4 Hz, 4H), 1.43 (d, J = 6.9 Hz, 4H), 1.21 (s, 6H), 1.15 (s, 6H). UPLC–MS calculated for C41H46ClN6O6 [M + H]+: 753.32, found: 753.30. UPLC-retention time: 5.4 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-((7-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)heptyl)amino)benzamide (14).
1H NMR (400 MHz, MeOH-d4) δ 7.76–7.69 (m, 3H), 7.53 (d, J = 8.4 Hz, 1H), 7.11 (d, J = 2.4 Hz, 1H), 7.00–6.95 (m, 2H), 6.89–6.85 (m, 2H), 6.81 (dd, J = 8.4, 2.2 Hz, 1H), 5.03 (dd, J = 12.4, 5.5 Hz, 1H), 4.27 (s, 1H), 4.13 (s, 1H), 3.20 (td, J = 7.0, 5.3 Hz, 4H), 2.77 (qdd, J = 14.4, 8.6, 6.3 Hz, 3H), 2.12–2.03 (m, 1H), 1.65 (d, J = 5.3 Hz, 4H), 1.44 (d, J = 2.2 Hz, 6H), 1.27 (s, 6H), 1.21 (s, 6H). UPLC–MS calculated for C42H48ClN6O6 [M + H]+: 767.33, found: 767.30. UPLC-retention time: 6.8 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-((8-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)octyl)amino)benzamide (15).
1H NMR (400 MHz, MeOH-d4) δ 7.73 (dd, J = 8.7, 3.9 Hz, 3H), 7.55 (d, J = 8.4 Hz, 1H), 7.13 (d, J = 2.4 Hz, 1H), 6.98 (dd, J = 8.6, 2.3 Hz, 2H), 6.88–6.81 (m, 3H), 5.04 (dd, J = 12.4, 5.5 Hz, 1H), 4.29 (s, 1H), 4.14 (s, 1H), 3.21 (t, J = 7.2 Hz, 4H), 2.88–2.67 (m, 3H), 2.09 (ddd, J = 10.1, 5.3, 2.4 Hz, 1H), 1.66 (dd, J = 14.1, 7.1 Hz, 4H), 1.43 (d, J = 8.5 Hz, 8H), 1.29 (s, 6H), 1.23 (s, 6H). UPLC-MS calculated for C43H50ClN6O6 [M + H]+: 781.35, found: 781.22. UPLC-retention time: 6.9 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-((5-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)pentyl)amino)benzamide (20).
1H NMR (400 MHz, MeOH-d4) δ 7.73–7.68 (m, 3H), 7.51 (dd, J = 8.5, 7.2 Hz, 1H), 7.10 (d, J = 2.4 Hz, 1H), 7.04–6.99 (m, 2H), 6.96 (dd, J = 8.8, 2.4 Hz, 1H), 6.83 (d, J = 8.7 Hz, 2H), 5.07–5.01 (m, 1H), 4.26 (s, 1H), 4.11 (s, 1H), 3.21 (t, J = 7.1 Hz, 2H), 2.88–2.65 (m, 3H), 2.13–2.04 (m, 1H), 1.75–1.65 (m, 4H), 1.58–1.49 (m, 2H), 1.26 (s, 6H), 1.20 (s, 6H). UPLC–MS calculated for C40H44ClN6O6 [M + H]+: 739.30, found: 739.31. UPLC-retention time: 6.6 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-((6-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)hexyl)amino)benzamide (21).
1H NMR (400 MHz, MeOH-d4) δ 7.71 (dt, J = 9.5, 2.4 Hz, 3H), 7.54 (dd, J = 8.6, 7.1 Hz, 1H), 7.13 (d, J = 2.4 Hz, 1H), 7.06–7.01 (m, 2H), 6.98 (dd, J = 8.8, 2.4 Hz, 1H), 6.84–6.80 (m, 2H), 5.05 (dd, J = 12.5, 5.5 Hz, 1H), 4.28 (s, 1H), 4.14 (s, 1H), 3.21 (t, J = 7.2 Hz, 2H), 2.77 (dddd, J = 17.6, 13.2, 4.8, 3.4 Hz, 3H), 2.14–2.07 (m, 1H), 1.75–1.65 (m, 4H), 1.50 (dd, J = 8.7, 5.3 Hz, 4H), 1.28 (s, 6H), 1.22 (s, 6H). UPLC–MS calculated for C41H46ClN6O6 [M + H]+: 753.32, found: 753.31. UPLC-retention time: 6.4 min, purity >95%.
General Procedure for the Synthesis of Compounds 16–19, 22–28, and 35–40.
DIPEA (5 equiv) and HATU (1.2 equiv) were added to a solution of compounds 54 (10 mmol) and 58 (1.1 equiv) in DMF (2 mL). After stirring at rt for 1 h, water was added and the reaction mixture was extracted with EtOAc; it was then washed with water, and the organic phase was dried over Na2SO4. Compound 59 was obtained by removing the solvent under vacuum and purified by flash column chromatography with subsequent deprotection by TFA with 84% yield.
AcOH (10%) in DCE was added to a solution of compounds 59 (1 mmol) and various aldehydes or ketones. After the mixture was stirred at rt for 10 min, NaBH(OAc)3 (1.2 equiv) was added and the mixture was stirred at rt for a further 2 h. The Boc-protected intermediates 61 were obtained by flash column chromatography. The desired intermediates (61) were obtained by deprotection with TFA in DCM in 70–80% yields.
DIPEA (5 equiv) was added to a solution of the compounds 61 (0.5 mmol) and 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione 56 (1.1 equiv) or 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoIine-1,3-dione 57 (1.1 equiv) in DMSO (2 mL). After stirring at 100 °C for 4 h, the mixture was subject to HPLC purification to afford compounds 16–19, 22–28, and 35–40 with 70–90% yields.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)ethyl)piperazin-1-yl)benzamide (16).
1H NMR (400 MHz, DMSO-d6) δ 11.07 (s, 1H), 7.91 (d, J = 8.7 Hz, 1H), 7.82 (d, J = 8.4 Hz, 2H), 7.64 (d, J = 8.3 Hz, 1H), 7.59 (d, J = 9.1 Hz, 1H), 7.21 (s, 1H), 7.10 (s, 2H), 7.03–6.96 (m, 2H), 5.08 (d, J = 5.2 Hz, 1H), 4.34 (s, 1H), 4.07 (d, J = 9.1 Hz, 2H), 3.67 (d, J = 17.3 Hz, 4H), 3.43–3.38 (m, 2H), 3.17 (dd, J = 28.7, 21.7 Hz, 4H), 2.88 (dd, J = 21.8, 9.5 Hz, 1H), 2.60 (d, J = 16.7 Hz, 1H), 2.51 (d, J = 1.4 Hz, 2H), 2.08–1.98 (m, 1H), 1.23 (s, 6H), 1.14 (s, 6H). UPLC–MS calculated for C41H45ClN7O6 [M + H]+: 766.31, found: 766.18. UPLC-retention time: 4.6 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(3-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)propyl)piperazin-1-yl)benzamide (17).
1H NMR (400 MHz, DMSO-d6) δ 11.07 (s, 1H), 9.71 (d, J = 104.2 Hz, 2H), 7.91 (d, J = 8.7 Hz, 1H), 7.82 (d, J = 8.4 Hz, 2H), 7.59 (d, J = 9.3 Hz, 1H), 7.21 (s, 1H), 7.07 (d, J = 8.5 Hz, 2H), 7.02 (d, J = 8.5 Hz, 1H), 4.34 (s, 1H), 4.11–3.98 (m, 3H), 3.59 (s, 4H), 3.32–3.27 (m, 3H), 3.18 (s, 3H), 3.07 (s, 1H), 2.09 (s, 4H), 1.95 (d, J = 7.9 Hz, 2H), 1.23 (s, 6H), 1.14 (s, 6H). UPLC–MS calculated for C42H47ClN7O6 [M + H]+: 780.33, found: 780.20. UPLC-retention time: 4.6 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(4-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)butyl)piperazin-1-yl)benzamide (18).
1H NMR (400 MHz, DMSO-d6) δ 11.06 (s, 1H), 7.90 (d, J = 8.7 Hz, 1H), 7.82 (d, J = 8.7 Hz, 2H), 7.64–7.57 (m, 2H), 7.21 (d, J = 2.3 Hz, 1H), 7.07 (d, J = 8.8 Hz, 2H), 7.03–6.97 (m, 2H), 5.04 (dd, J = 12.9, 5.3 Hz, 1H), 4.34 (s, 1H), 4.12–3.99 (m, 3H), 3.39 (s, 1H), 3.18 (ddd, J = 15.7, 13.7, 6.9 Hz, 8H), 2.89 (ddd, J = 17.5, 14.3, 5.2 Hz, 1H), 2.62–2.54 (m, 1H), 2.02 (dd, J = 19.0, 13.7 Hz, 1H), 1.80 (d, J = 6.8 Hz, 1H), 1.71–1.61 (m, 2H), 1.54 (dd, J = 14.5, 7.4 Hz, 1H), 1.22 (s, 6H), 1.14 (s, 6H). UPLC–MS calculated for C43H49ClN7O6 [M + H]+: 794.34, found: 794.18. UPLC-retention time: 4.9 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(5-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)pentyl)piperazin-1-yl)benzamide (19).
1H NMR (400 MHz, DMSO-d6) δ 11.06 (s, 1H), 7.90 (d, J = 8.7 Hz, 1H), 7.82 (d, J = 8.3 Hz, 2H), 7.59 (dd, J = 8.2, 6.3 Hz, 2H), 7.20 (d, J = 1.4 Hz, 1H), 7.07 (d, J = 8.4 Hz, 2H), 7.00 (dd, J = 14.2, 6.8 Hz, 2H), 5.04 (dd, J = 12.8, 5.2 Hz, 1H), 4.34 (s, 1H), 4.07 (d, J = 9.1 Hz, 2H), 3.99 (s, 2H), 3.91 (s, 3H), 3.60 (s, 2H), 3.17 (d, J = 12.1 Hz, 5H), 2.87 (d, J = 13.0 Hz, 1H), 2.59 (d, J = 17.0 Hz, 1H), 2.04–1.97 (m, 1H), 1.74 (d, J = 7.0 Hz, 2H), 1.63 (d, J = 7.1 Hz, 2H), 1.57–1.52 (m, 1H), 1.44 (d, J = 6.8 Hz, 1H), 1.23 (s, 6H), 1.14 (s, 6H). UPLC–MS calculated for C44H51ClN7O6 [M + H]+: 808.36, found: 808.19. UPLC-retention time: 4.8 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethyl)piperazin-1-yl)benzamide (22).
1H NMR (400 MHz, DMSO-d6) δ 7.82 (d, J = 8.8 Hz, 2H), 7.73 (d, J = 8.7 Hz, 1H), 7.64 (dd, J = 8.3, 7.3 Hz, 1H), 7.14 (ddd, J = 16.4, 9.7, 5.1 Hz, 5H), 7.01–6.97 (m, 1H), 5.09 (dd, J = 12.5, 5.4 Hz, 1H), 4.30 (s, 1H), 4.16 (s, 1H), 3.89 (t, J = 6.2 Hz, 2H), 3.81–3.76 (m, 1H), 3.51 (ddd, J = 25.6, 16.0, 9.9 Hz, 9H), 2.80 (tdd, J = 9.6, 8.0, 4.3 Hz, 3H), 2.17–2.10 (m, 1H), 1.30 (s, 6H), 1.24 (s, 6H). UPLC–MS calculated for C41H45ClN7O6 [M + H]+: 766.31, found: 766.18. UPLC-retention time: 4.7 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(3-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)propyl)piperazin-1-yl)benzamide (23).
1H NMR (400 MHz, MeOH-d4) δ 7.81 (d, J = 2.3 Hz, 2H), 7.72 (s, 1H), 7.61 (dd, J = 8.5, 7.1 Hz, 1H), 7.12 (ddd, J = 5.8, 5.4, 2.3 Hz, 5H), 7.00 (d, J = 6.4 Hz, 1H), 5.08 (dd, J = 12.5, 5.5 Hz, 1H), 4.31 (s, 1H), 4.16 (s, 1H), 3.93 (dd, J = 31.9, 25.4 Hz, 2H), 3.69 (s, 2H), 3.52 (dt, J = 6.8, 4.2 Hz, 3H), 3.45 (d, J = 6.8 Hz, 1H), 3.29 (d, J = 2.9 Hz, 1H), 2.90–2.68 (m, 3H), 2.15 (dddd, J = 11.2, 9.6, 9.0, 4.4 Hz, 4H), 1.30 (s, 6H), 1.24 (s, 6H). UPLC–MS calculated for C42H47ClN706 [M + H]+: 780.33, found: UPLC-retention time: 4.8 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(4-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)butyl)piperazin-1-yl)benzamide (24).
1H NMR (400 MHz, MeOH-d4) δ 7.84–7.79 (m, 2H), 7.73 (d, J = 8.7 Hz, 1H), 7.58 (dd, J = 8.6, 7.1 Hz, 1H), 7.15–7.07 (m, 5H), 6.99 (dd, J = 8.8, 2.4 Hz, 1H), 5.07 (dd, J = 12.5, 5.5 Hz, 1H), 4.30 (s, 1H), 4.16 (s, 1H), 4.06 (d, J = 34.1 Hz, 2H), 3.69 (s, 2H), 3.46 (t, J = 6.7 Hz, 2H), 3.22 (ddd, J = 59.9, 21.4, 19.4 Hz, 6H), 2.92–2.67 (m, 3H), 2.16–2.10 (m, 1H), 1.93 (ddd, J = 11.5, 10.2, 6.8 Hz, 2H), 1.83–1.74 (m, 2H). UPLC–MS calculated for C43H49ClN7O6 [M + H]+: 794.34, found: 794.18. UPLC-retention time: 4.9 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)azetidin-3-yl)piperazin-1-yl)benzamide (25).
1H NMR (400 MHz, MeOH-d4) δ 10.74 (s, 1H), 7.77 (d, J = 8.9 Hz, 2H), 7.65 (dd, J = 8.4, 7.3 Hz, 2H), 7.06 (t, J = 5.8 Hz, 3H), 6.93 (dd, J = 8.8, 2.4 Hz, 1H), 6.88 (d, J = 2.0 Hz, 1H), 6.72 (dd, J = 8.3, 2.1 Hz, 1H), 5.07–5.01 (m, 1H), 4.40–4.30 (m, 5H), 4.24 (s, 1H), 4.11 (s, 1H), 3.55 (d, J = 57.2 Hz, 8H), 2.82–2.62 (m, 3H), 2.08 (ddd, J = 10.2, 8.9, 5.4 Hz, 1H), 1.25 (s, 6H), 1.18 (s, 6H). 13C NMR (100 MHz, MeOH-d4) δ 173.24, 170.32, 169.15, 167.68, 167.68, 163.00, 154.40, 151.98, 137.56, 135.35, 134.12, 128.81, 125.78, 124.66, 119.54, 116.61, 115.83, 115.02, 114.95, 114.32, 105.05, 104.31, 84.42, 58.90, 54.84, 53.12, 49.26, 49.09, 45.18, 40.34, 30.82, 23.07, 22.37, 22.31. UPLC–MS calculated for C42H46ClN7O6 [M + H]+: 777.30, found: 777.25. UPLC-retention time: 4.8 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-((1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)azetidin-3-yl)methyl)piperazin-1-yl)benzamide (26).
1H NMR (400 MHz, DMSO-d6) δ 11.11 (s, 1H), 7.92 (d, J = 8.7 Hz, 1H), 7.86 (dd, J = 8.7, 3.8 Hz, 2H), 7.63 (d, J = 8.5 Hz, 2H), 7.22 (d, J = 2.4 Hz, 1H), 7.11 (dd, J = 9.6, 4.4 Hz, 3H), 7.05–7.02 (m, 1H), 5.14–5.03 (m, 1H), 4.40–4.24 (m, 2H), 4.20–3.85 (m, 5H), 3.63 (ddd, J = 44.7, 25.1, 5.8 Hz, 5H), 3.23–3.14 (m, 2H), 2.97–2.86 (m, 1H), 2.66–2.49 (m, 5H), 2.43–2.32 (m, 1H), 2.11–2.00 (m, 1H), 1.26 (s, 6H), 1.17 (s, 6H). UPLC–MS calculated for C43H46ClN7O6 [M + H]+: 792.33, found: 792.23. UPLC-retention time: 4.7 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperidin-4-yl)piperazin-1-yl)benzamide (27).
1H NMR (400 MHz, MeOH-d4) δ 10.82 (s, 1H), 7.83 (d, J = 8.9 Hz, 2H), 7.74 (dd, J = 8.6, 1.0 Hz, 2H), 7.44 (d, J = 2.2 Hz, 1H), 7.32 (dd, J = 8.6, 2.3 Hz, 1H), 7.13 (dd, J = 11.8, 5.7 Hz, 3H), 7.00 (dd, J = 8.8, 2.4 Hz, 1H), 5.10 (dd, J = 12.5, 5.5 Hz, 1H), 4.33–4.22 (m, 3H), 4.16 (s, 1H), 4.07 (d, J = 34.3 Hz, 1H), 3.63 (ddd, J = 11.9, 8.3, 3.8 Hz, 5H), 3.31–3.04 (m, 4H), 2.96–2.62 (m, 4H), 2.33 (d, J = 10.8 Hz, 2H), 2.18–2.08 (m, 1H), 1.89 (tt, J = 12.2, 6.0 Hz, 2H), 1.30 (s, 6H), 1.24 (s, 6H). 13C NMR (100 MHz, MeOH-d4) δ 173.23 (s), 170.24 (s), 169.14 (s), 167.89 (s), 167.43 (s), 163.00 (s), 154.72 (s), 152.00 (s), 137.57 (s), 135.33 (s), 134.35 (s), 128.78 (s), 125.80 (s), 124.81 (s), 119.83 (s), 118.49 (s), 116.57 (s), 115.74 (s), 114.85 (s), 114.33 (s), 108.48 (s), 104.35 (s), 84.42 (s), 63.49 (s), 58.88 (s), 49.09 (s), 48.64 (s), 46.17 (s), 45.47 (s), 40.32 (s), 30.79 (s), 25.74 (s), 23.02 (s), 22.26 (s). UPLC–MS calculated for C44H48ClN7O6 [M + H]+: 806.35, found: 806.32. UPLC-retention time: 4.9 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-((1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperidin-4-yl)methyl)piperazin-1-yl)benzamide (28).
1H NMR (400 MHz, MeOH-d4) δ 10.81 (s, 1H), 7.83 (d, J = 8.9 Hz, 2H), 7.72 (dd, J = 14.6, 8.6 Hz, 2H), 7.38 (d, J = 2.2 Hz, 1H), 7.26 (dd, J = 8.6, 2.3 Hz, 1H), 7.13 (dd, J = 10.5, 5.7 Hz, 3H), 7.00 (dd, J = 8.8, 2.4 Hz, 1H), 5.09 (dd, J = 12.5, 5.5 Hz, 1H), 4.31 (s, 1H), 4.17 (s, 1H), 4.09 (t, J = 20.0 Hz, 4H), 3.70 (d, J = 32.5 Hz, 3H), 3.24 (dd, J = 20.6, 12.6 Hz, 4H), 3.08 (t, J = 11.7 Hz, 2H), 2.91–2.82 (m, 1H), 2.80–2.65 (m, 2H), 2.27 (ddd, J = 11.1, 7.4, 4.0 Hz, 1H), 2.18–2.08 (m, 1H), 2.03–1.93 (m, 2H), 1.53–1.43 (m, 3H), 1.31 (s, 6H), 1.25 (s, 6H). 13C NMR (100 MHz, MeOH-d4) δ 173.24 (s), 170.29 (s), 169.15 (s), 168.08 (s), 167.55 (s), 163.00 (s), 155.39 (s), 152.05 (s), 137.57 (s), 135.34 (s), 134.33 (s), 128.79 (s), 125.72 (s), 124.77 (s), 118.92 (s), 118.05 (s), 116.58 (s), 115.74 (s), 114.84 (s), 114.33 (s), 108.10 (s), 104.35 (s), 100.00 (s), 84.42 (s), 61.64 (s), 58.88 (s), 51.98 (s), 49.05 (s), 44.99 (s), 40.33 (s), 30.84 (d, J = 8.2 Hz), 28.84 (s), 23.02 (s), 22.33 (d, J = 13.2 Hz). UPLC–MS calculated for C45H50ClN7O6 [M + H]+: 820.36, found: 820.24. UPLC-retention time: 4.7 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(2-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)-2-azaspiro[3.3]heptan-6-yl)piperazin-1-yl)benzamide (35).
1H NMR (400 MHz, DMSO-d6) δ 11.33 (s, 1H), 8.16 (d, J = 8.7 Hz, 1H), 8.08 (d, J = 8.7 Hz, 2H), 7.92 (d, J = 8.3 Hz, 1H), 7.86 (d, J = 9.2 Hz, 1H), 7.46 (d, J = 2.3 Hz, 1H), 7.34 (d, J = 8.8 Hz, 2H), 7.27 (dd, J = 8.8, 2.3 Hz, 1H), 7.06 (d, J = 1.6 Hz, 1H), 6.93 (dd, J = 8.4, 1.8 Hz, 1H), 5.32 (dd, J = 12.8, 5.3 Hz, 1H), 4.68–4.57 (m, 3H), 4.32 (s, 3H), 4.07 (dd, J = 16.1, 8.1 Hz, 3H), 3.77 (t, J = 32.8 Hz, 3H), 3.24–3.07 (m, 2H), 2.96–2.78 (m, 7H), 2.48–2.24 (m, 2H), 1.48 (s, 6H), 1.40 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 173.28 (s), 170.56 (s), 167.94 (s), 167.64 (s), 167.09 (s), 163.10 (s), 158.67 (s), 155.42 (s), 152.01 (s), 137.34 (s), 136.53 (s), 134.23 (s), 129.47 (s), 125.74 (s), 125.28 (s), 117.68 (s), 117.27 (s), 116.75 (s), 115.04 (d, J = 24.4 Hz), 114.74–114.42 (m), 105.27 (s), 104.09 (s), 84.37 (s), 63.61 (s), 62.65 (s), 58.58 (s), 53.99 (s), 49.21 (s), 48.33 (s), 45.15 (s), 35.56 (s), 31.64 (s), 24.44 (s), 23.58 (s). UPLC–MS calculated for C45H49ClN7O6 [M + H]+: 818.34, found: 818.16. UPLC-retention time: 4.8 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-((2-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)-2-azaspiro[3.3]heptan-6-yl)methyl)piperazin-1-yl)benzamide (36).
1H NMR (400 MHz, DMSO-d6) δ 11.26 (s, 1H), 8.09 (d, J = 8.7 Hz, 1H), 8.01 (d, J = 8.8 Hz, 2H), 7.83 (s, 1H), 7.78 (d, J = 9.1 Hz, 1H), 7.39 (d, J = 2.4 Hz, 1H), 7.25 (d, J = 8.9 Hz, 2H), 7.21–7.19 (m, 1H), 6.97 (d, J = 1.9 Hz, 1H), 6.84 (dd, J = 8.4, 2.0 Hz, 1H), 5.25 (dd, J = 12.8, 5.4 Hz, 1H), 4.52 (s, 1H), 4.35–4.17 (m, 9H), 3.46 (d, J = 6.6 Hz, 3H), 3.12–3.03 (m, 1H), 2.92–2.71 (m, 5H), 2.37–2.14 (m, 4H), 1.41 (s, 6H), 1.32 (s, 6H). UPLC–MS calculated for C46H51ClN7O6 [M + H]+: 832.36, found: 832.19. UPLC-retention time: 4.9 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(6-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)-6-azaspiro[3.4]octan-2-yl)piperazin-1-yl)benzamide (37).
1H NMR (400 MHz, DMSO-d6) δ 11.08 (s, 1H), 7.93–7.88 (m, 1H), 7.83 (d, J = 8.6 Hz, 2H), 7.66 (d, J = 8.4 Hz, 1H), 7.60 (d, J = 9.2 Hz, 1H), 7.20 (s, 1H), 7.08 (d, J = 8.6 Hz, 2H), 7.01 (d, J = 8.8 Hz, 1H), 6.90 (d, J = 9.9 Hz, 1H), 6.80 (t, J = 7.7 Hz, 1H), 5.09–5.04 (m, 1H), 4.34 (s, 2H), 4.07 (d, J = 9.1 Hz, 2H), 3.95–3.89 (m, 1H), 3.55–3.40 (m, 6H), 3.08 (s, 3H), 2.90 (dd, J = 15.3, 10.5 Hz, 1H), 2.61 (d, J = 8.5 Hz, 1H), 2.51 (s, 1H), 2.40 (d, J = 7.5 Hz, 3H), 2.07 (ddd, J = 18.8, 12.9, 5.9 Hz, 3H), 1.23 (s, 6H), 1.14 (s, 6H). UPLC–MS calculated for C46H51ClN7O6 [M + H]+: 832.36, found: 832.21. UPLC-retention time: 5.0 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-((6-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)-6-azaspiro[3.4]octan-2-yl)methyl)piperazin-1-yl)benzamide (38).
1H NMR (400 MHz, DMSO-d6) δ 11.05 (s, 1H), 7.87 (d, J = 8.7 Hz, 1H), 7.79 (d, J = 8.7 Hz, 2H), 7.62 (dd, J = 8.4, 4.3 Hz, 1H), 7.57 (d, J = 9.2 Hz, 1H), 7.17 (d, J = 2.3 Hz, 1H), 7.04 (d, J = 8.7 Hz, 2H), 6.98 (dd, J = 8.8, 2.4 Hz, 1H), 6.87 (dd, J = 12.7, 1.7 Hz, 1H), 6.81–6.72 (m, 1H), 5.03 (dd, J = 13.0, 4.9 Hz, 1H), 4.31 (s, 1H), 4.04 (d, J = 9.1 Hz, 1H), 3.97 (d, J = 8.5 Hz, 2H), 3.58–3.24 (m, 9H), 3.07 (d, J = 25.8 Hz, 4H), 2.83 (ddd, J = 22.7, 13.2, 8.3 Hz, 2H), 2.58 (d, J = 7.3 Hz, 1H), 2.48 (dd, J = 3.5, 1.7 Hz, 1H), 2.22–2.07 (m, 3H), 2.01–1.87 (m, 4H), 1.20 (s, 6H), 1.11 (s, 6H). UPLC–MS calculated for C47H53ClN7O6 [M + H]+: 846.37, found: 846.22. UPLC-retention time: 4.9 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(3-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)-3-azaspiro[5.5]undecan-9-yl)piperazin-1-yljbenzamide (39).
1H NMR (400 MHz, DMSO-d6) δ 11.18 (s, 1H), 8.00 (d, J = 8.7 Hz, 1H), 7.92 (d, J = 8.7 Hz, 2H), 7.77 (d, J = 8.5 Hz, 1H), 7.70 (d, J = 9.2 Hz, 1H), 7.42 (s, 1H), 7.36–7.29 (m, 2H), 7.18 (d, J = 8.9 Hz, 2H), 7.11 (dd, J = 8.8, 2.4 Hz, 1H), 5.17 (dd, J = 12.8, 5.4 Hz, 1H), 4.15 (dd, J = 17.4, 10.7 Hz, 3H), 3.75 (d, J = 10.3 Hz, 2H), 3.58 (s, 4H), 3.35 (d, J = 12.6 Hz, 3H), 3.19 (t, J = 11.8 Hz, 2H), 3.05–2.94 (m, 1H), 2.74–2.65 (m, 2H), 2.61 (dd, J = 5.5, 3.7 Hz, 2H), 2.18–1.89 (m, 6H), 1.80–1.69 (m, 4H), 1.54 (s, 2H), 1.33 (s, 6H), 1.24 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 173.29 (s), 170.59 (s), 168.14 (s), 167.46 (s), 167.10 (s), 163.10 (s), 159.00 (s), 158.65 (s), 155.41 (s), 152.02 (s), 137.34 (s), 136.53 (s), 134.47 (s), 129.47 (s), 125.74 (s), 125.44 (s), 117.83 (s), 117.27 (s), 116.75 (s), 115.16 (s), 114.83 (s), 104.09 (s), 84.37 (s), 58.58 (s), 49.22 (s), 48.32 (s), 45.46 (s), 40.78 (s), 33.94 (s), 30.48 (s), 24.44 (s), 23.58 (s), 21.86 (s). UPLC–MS calculated for C49H57CIN7O6 [M + H]+: 874.41, found: 874.25. UPLC-retention time: 5.0 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(2-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)-2-azaspiro[3.5]nonan-7-yl)piperazin-1-yl)benzamide (40).
1H NMR (400 MHz, MeOH-d4) δ 10.80 (s, 1H), 7.85–7.82 (m, 2H), 7.74 (d, J = 8.7 Hz, 1H), 7.66 (d, J = 8.3 Hz, 1H), 7.15–7.10 (m, 3H), 7.01–6.98 (m, 1H), 6.84 (s, 1H), 6.68 (dd, J = 8.3, 1.7 Hz, 1H), 5.07 (dd, J = 12.4, 5.4 Hz, 1H), 4.31 (s, 1H), 4.17 (s, 1H), 4.10 (d, J = 11.4 Hz, 1H), 3.93–3.86 (m, 2H), 3.78 (s, 2H), 3.68 (d, J = 12.7 Hz, 2H), 3.38 (dd, J = 8.2, 3.2 Hz, 2H), 3.27–3.16 (m, 2H), 2.90–2.82 (m, 1H), 2.75 (dt, J = 14.2, 4.0 Hz, 2H), 2.23 (d, J = 9.4 Hz, 3H), 2.12 (ddd, J = 10.0, 5.3, 2.4 Hz, 1H), 1.85–1.61 (m, 5H), 1.31 (s, 6H), 1.25 (s, 6H). UPLC–MS calculated for C47H53ClN7O6 [M + H]+: 846.37, found: 846.19. UPLC-retention time: 5.0 min, purity >95%.
General Procedure for the Synthesis of Compounds 64 and 65.
Compounds 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione (56) (0.5 mmol) and methyl iodide (1.1 equiv) were dissolved in DMF. K2CO3 (1.2 equiv) was added to the solution, and the reaction mixture was stirred at 60 °C for 2 h. Water was added; the reaction mixture was extracted by EtOAc and washed with water; and the organic phase was dried over Na2SO4. The compound Me-protected (63) (5-fluoro-2-(1-methyl-2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione) can be obtained by removing the solvent under vacuum and purified by flash column in 85% yield.
DIPEA (5 equiv) was added to a solution of the compound 61 (0.3 mmol) and 5-fluoro-2-(1-methyl-2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione 63 (1.1 equiv) in DMSO (2 mL). After 4 h at 100 °C, the mixture was subject to HPLC purification to afford compounds 64 with an 81% yield.
Methyl 4-(piperazin-1-yl)benzoate (62) (1 mmol) and methyl iodide (1.1 equiv) were dissolved in DMF. K2CO3 (1.2 equiv) was added to the solution, and the reaction mixture was stirred at 60 °C for 2 h. Water was added; the reaction mixture was extracted by EtOAc and washed by water; and the organic phase was dried by Na2SO4. The product, methyl 4-(4-methylpiperazin-1-yl)benzoate (66) can be obtained by removing the solvent under vacuum and purified by flash column in 83% yield.
NaOH (2 equiv) was added to a solution of methyl 4-(4-methylpiperazin-1-yl)benzoate (66) (0.8 mmol) in MeOH/H2O, and the mixture was stirred at rt for 2 h. Then, the MeOH was removed under reduced pressure; the pH was adjusted to acidity with 2 M HCl; and the mixture was extracted with EtOAc. The solvent was removed to afford the product 4-(4-methylpiperazin-1-yl)benzoic acid (67), which was used without further purification.
DIPEA (5 equiv) and HATU (1.2 equiv) were added to a solution of compounds 4-((1R,3R)-3-amino-2,2,4,4-tetramethylcyclobutoxy)-2-chlorobenzonitrile (54) (0.5 mmol) and 4-(4-methylpiperazin-1-yl)benzoic acid (67) (1.1 equiv) in DMF (2 mL). After stirring at rt for 1 h, water was added and the reaction mixture was extracted by EtOAc; it was then washed by water, and the organic phase was dried by Na2SO4. The product (65) can be obtained by removing the solvent under vacuum and purified by flash column chromatography with 88% yield.
5-Fluoro-2-(1-methyl-2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione (INT-63).
1H NMR (400 MHz, DMSO-d6) δ 8.01 (dd, J = 8.3, 4.5 Hz, 1H), 7.84 (dd, J = 7.4, 2.2 Hz, 1H), 7.73 (ddd, J = 9.5, 8.3, 2.3 Hz, 1H), 5.24 (dd, J = 13.1, 5.4 Hz, 1H), 3.03 (s, 3H), 2.99–2.91 (m, 1H), 2.78 (ddd, J = 17.2, 4.3, 2.5 Hz, 1H), 2.62–2.53 (m, 1H), 2.10 (dtd, J = 12.9, 5.3, 2.5 Hz, 1H). UPLC–MS calculated for C14H11FN2O4 [M + H]+: 291.08, found: 290.94. UPLC-retention time: 3.1 min, purity >95%.
Methyl 4-(4-methylpiperazin-1-yl)benzoate (INT-66).
1H NMR (400 MHz, DMSO) δ 7.82–7.75 (m, 2H), 6.97 (d, J = 9.0 Hz, 2H), 3.78 (s, 3H), 2.25 (s, 3H), 1.92 (s, 1H). UPLC–MS calculated for C13H14N2O2 [M + H]+: 235.15, found: 235.08. UPLC-retention time: 0.6 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-((1-(2-(1-methyl-2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperidin-4-yl)methyl)piperazin-1-yl)benzamide (64).
1H NMR (400 MHz, MeOH-d4) δ 7.83 (d, J = 8.9 Hz, 2H), 7.72 (dd, J = 12.7, 8.6 Hz, 2H), 7.39 (d, J = 2.2 Hz, 1H), 7.27 (dd, J = 8.6, 2.3 Hz, 1H), 7.13 (dd, J = 10.6, 5.7 Hz, 3H), 7.00 (dd, J = 8.8, 2.4 Hz, 1H), 5.12 (dd, J = 12.9, 5.4 Hz, 1H), 4.31 (s, 1H), 4.19–4.07 (m, 4H), 3.21 (d, J = 6.9 Hz, 2H), 3.16 (s, 3H), 3.09 (t, J = 11.7 Hz, 2H), 2.90 (dd, J = 7.0, 3.8 Hz, 2H), 2.77–2.70 (m, 1H), 2.68 (s, 1H), 2.32–2.23 (m, 1H), 2.14–2.08 (m, 1H), 1.98 (d, J = 12.6 Hz, 2H), 1.54–1.36 (m, 3H), 1.30 (s, 6H), 1.24 (s, 6H). UPLC–MS calculated for C46H53ClN7O6 [M + H]+: 834.37, found: 834.36. UPLC-retention time: 4.8 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-methylpiperazin-1-yl)benzamide (65).
1H NMR (400 MHz, MeOH-d4) δ 7.75–7.69 (m, 2H), 7.61 (d, J = 8.7 Hz, 1H), 7.00 (dd, J = 9.5, 5.7 Hz, 3H), 6.88 (dd, J = 8.8, 2.4 Hz, 1H), 4.20 (s, 1H), 4.06 (s, 1H), 3.94 (d, J = 12.7 Hz, 2H), 3.57 (dd, J = 13.3, 8.8 Hz, 2H), 3.22–3.06 (m, 4H), 2.89 (s, 3H), 1.20 (s, 6H), 1.13 (s, 6H). UPLC–MS calculated for C27H34ClN7O7 [M + H]+: 481.24, found: 481.26. UPLC-retention time: 3.5 min, purity >95%.
General Procedure for the Synthesis of Compounds 29–32.
DIPEA (5 equiv) and HATU (1.2 equiv) were added to a solution of compounds 54 (2 mmol) and 66 (1.1 equiv) in DMF (2 mL). After stirring at rt for 1 h, water was added and the reaction mixture was extracted by EtOAc; it was then washed by water, and the organic phase was dried by Na2SO4. Compound 67 was obtained by removing the solvent under vacuum and purified by flash column with 84% yield.
NaOH (2 equiv) was added to a solution of 67 (1 mmol) in MeOH/H2O and stirred at rt for 2 h. Then, the MeOH was removed under reduced pressure; the pH was adjusted to <7 with 2 M HCl; and the mixture was extracted with EtOAc. The solvent was removed to afford the product 68, which was used without further purification.
Compounds 68 (0.8 mmol) and 69 (1.2 equiv) were dissolved in DMF. DIPEA (5 equiv) and HATU (1.2 equiv) was added to the solution, and the reaction mixture was stirred at rt for 1 h. Water was then added,; the reaction mixture was extracted by EtOAc and washed with water; and the organic phase was dried by Na2SO4. The compound, Boc-protected-70 was obtained by removing the solvent under vacuum and purified by flash column chromatography. This desired intermediate 70 was obtained by deprotection with TFA in DCM in 89% yield.
A solution of compounds 70 (0.5 mmol) and a series of aldehydes or ketones was added AcOH (10%) in DCE. After the mixture was stirred at rt for 10 min, NaBH(OAc)3 (1.2 equiv) was added and the mixture was stirred at rt for a further 2 h. The compound Boc-protected-71 was obtained by removing the solvent under vacuum and purified by flash column chromatography. Intermediate 71 was obtained by deprotection with TFA in DCM in 70–85% yields in total.
DIPEA (5 equiv) was added to a solution of compounds 71 (0.3 mmol) and 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione 56 (1.1 equiv) in DMSO (2 mL). After 4 h at 100 °C, the mixture was subject to HPLC purification to afford compounds 29–32 with 70–90% yields.
4-(((1r,3r)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)carbamoyl)benzoic acid (INT-68).
1H NMR (400 MHz, DMSO-d6) δ 13.22 (s, 1H), 8.03 (t, J = 5.8 Hz, 3H), 7.95–7.90 (m, 2H), 7.21 (d, J = 2.3 Hz, 1H), 7.01 (dd, J = 8.8, 2.3 Hz, 1H), 4.33 (s, 1H), 4.09 (d, J = 9.1 Hz, 1H), 1.25 (s, 6H), 1.15 (s, 6H). UPLC–MS calculated for C23H24ClN2O4 [M + H]+: 427.14, found: 427.18. UPLC-retention time: 5.4 min, purity >95%.
N-((1r,3r)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(piperazine-1-carbonyl)benzamide (INT-70).
1H NMR (400 MHz, DMSO-d6) δ 9.18 (s, 1H), 7.99–7.91 (m, 3H), 7.57 (d, J = 8.3 Hz, 2H), 7.20 (d, J =2.4 Hz, 1H), 7.01 (dd, J = 8.8, 2.4 Hz, 1H), 4.34 (s, 1H), 4.10 (d, J = 9.1 Hz, 1H), 3.69 (d, J = 106.1 Hz, 4H), 3.20 (s, 4H), 1.25 (s, 6H), 1.15 (s, 6H). UPLC–MS calculated for C27H32ClN4O3 [M + H]+: 495.22, found: 495.24. UPLC-retention time: 3.8 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcy-ciobutyi)-4-(4-(1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)azetidin-3-yl)piperazine-1-carbonyl)-benzamide (29).
1H NMR (400 MHz, DMSO-d6) δ 11.17 (s, 1H), 8.03 (dd, J = 14.4, 6.2 Hz, 3H), 7.81 (d, J = 8.3 Hz, 1H), 7.67 (d, J = 8.2 Hz, 2H), 7.30 (d, J = 2.3 Hz, 1H), 7.10 (dd, J = 8.8, 2.4 Hz, 1H), 7.00 (d, J = 1.7 Hz, 1H), 6.85 (dd, J = 8.4, 1.9 Hz, 1H), 5.16 (dd, J = 12.8, 5.3 Hz, 1H), 4.53–4.25 (m, 6H), 4.18 (d, J = 9.0 Hz, 1H), 4.06–3.89 (m, 1H), 3.71 (s, 2H), 3.30 (d, J = 30.5 Hz, 3H), 3.06–2.90 (m, 1H), 2.79–2.51 (m, 3H), 2.18–2.06 (m, 1H), 1.45–1.35 (m, 1H), 1.33 (s, 6H), 1.24 (s, 6H). UPLC–MS calculated for C43H45ClN7O7 [M + H]+: 806.31, found: 806.23. UPLC-retention time: 5.0 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcy-ciobutyi)-4-(4-(1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperidin-4-yl)piperazine-1-carbonyl)enzamide (30).
1H NMR (400 MHz, MeOH-d4) δ 10.79 (s, 1H), 7.96–7.92 (m, 2H), 7.72 (dd, J = 11.5, 6.1 Hz, 2H), 7.64–7.60 (m, 2H), 7.40 (d, J = 2.2 Hz, 1H), 7.28 (dd, J = 8.6, 2.3 Hz, 1H), 7.13 (d, J = 2.4 Hz, 1H), 6.99 (dd, J = 8.8, 2.4 Hz, 1H), 5.11–5.06 (m, 1H), 4.31–4.18 (m, 4H), 3.77 (dd, J = 36.2, 29.7 Hz, 2H), 3.65–3.36 (m, 5H), 3.21 (d, J = 7.3 Hz, 1H), 3.06 (t, J = 12.2 Hz, 2H), 2.93–2.82 (m, 1H), 2.79–2.67 (m, 2H), 2.27 (d, J = 10.5 Hz, 2H), 2.17–2.06 (m, 1H), 1.85 (td, J = 12.1, 8.4 Hz, 2H), 1.32 (d, J = 7.3 Hz, 2H), 1.30 (s, 6H), 1.24 (s, 6H). UPLC–MS calculated for C45H49ClN7O7 [M + H]+: 834.34, found: 834.18. UPLC-retention time: 4.3 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-((1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)azetidin-3-yl)methyl)piperazine-1-carbonyl)benzamide (31).
1H NMR (400 MHz, MeOH-d4) δ 7.77 (d, J = 8.9 Hz, 2H), 7.65 (dd, J = 8.4, 7.3 Hz, 2H), 7.06 (t, J = 5.8 Hz, 3H), 6.93 (dd, J = 8.8, 2.4 Hz, 1H), 6.88 (d, J = 2.0 Hz, 1H), 6.72 (dd, J = 8.3, 2.1 Hz, 1H), 5.07–5.01 (m, 1H), 4.40–4.30 (m, 5H), 4.24 (s, 1H), 4.11 (s, 1H), 3.55 (d, J = 57.2 Hz, 8H), 2.82–2.62 (m, 3H), 2.08 (ddd, J = 10.2, 8.9, 5.4 Hz, 1H), 1.25 (s, 6H), 1.18 (s, 6H). 13C NMR (100 MHz, MeOH-d4) δ 173.23 (s), 170.25 (s), 170.02 (s), 168.85 (s), 167.89 (s), 167.45 (s), 162.96 (s), 154.70 (s), 137.57 (s), 136.91 (s), 136.53 (s), 135.35 (s), 134.32 (s), 127.74 (s), 127.13 (s), 124.80 (s), 119.80 (s), 118.48 (s), 116.60 (s), 115.74 (s), 114.31 (s), 108.47 (s), 104.39 (s), 84.37 (s), 63.82 (s), 59.14 (s), 49.08 (s), 48.41 (s), 46.16 (s), 40.33 (s), 30.79 (s), 25.67 (s), 23.04 (s), 22.33 (d, J = 7.3 Hz), 7.80 (s). UPLC–MS calculated for C44H47ClN7O7 [M + H]+: 820.32, found: 820.19. UPLC-retention time: 4.4 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-((1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperidin-4-yl)methyl)piperazine-1-carbonyljbenzamide (32).
1H NMR (400 MHz, MeOH-d4) δ 10.80 (s, 1H), 7.96 (d, J = 8.0 Hz, 2H), 7.75 (d, J = 8.7 Hz, 1H), 7.69 (d, J = 8.5 Hz, 1H), 7.64 (d, J = 8.1 Hz, 2H), 7.36 (d, J = 1.9 Hz, 1H), 7.25 (dd, J = 8.6, 2.0 Hz, 1H), 7.15 (d, J = 2.3 Hz, 1H), 7.01 (dd, J = 8.7, 2.3 Hz, 1H), 5.09 (dd, J = 12.4, 5.4 Hz, 1H), 4.32 (s, 1H), 4.21 (s, 1H), 4.09 (d, J = 13.3 Hz, 2H), 3.86 (d, J = 30.7 Hz, 3H), 3.42 (d, J = 26.0 Hz, 3H), 3.27–3.22 (m, 1H), 3.18 (d, J = 6.8 Hz, 2H), 3.05 (dd, J = 14.6, 9.3 Hz, 2H), 2.92–2.83 (m, 1H), 2.80–2.68 (m, 2H), 2.29–2.20 (m, 1H), 2.18–2.10 (m, 1H), 2.01 1.93 (m, 2H), 1.45 (dq, J = 20.7, 9.1 Hz, 3H), 1.33 (s, 6H), 1.26 (s, 6H). 13C NMR (100 MHz, MeOH-d4) δ 173.26 (s), 170.21 (d, J = 19.5 Hz), 168.85 (s), 168.07 (s), 167.55 (s), 162.96 (s), 155.37 (s), 137.57 (s), 136.96 (s), 136.53 (s), 135.36 (s), 134.31 (s), 127.77 (s), 127.07 (s), 124.77 (s), 118.86 (s), 118.03 (s), 116.61 (s), 114.31 (s), 108.09 (s), 104.38 (s), 84.37 (s), 61.81 (s), 59.14 (s), 51.75 (s), 49.05 (s), 40.34 (s), 30.82 (s), 28.81 (s), 23.05 (s), 22.35 (d, J = 8.4 Hz), 7.80 (s). UPLC–MS calculated for C46H51ClN7O7 [M + H]+: 848.35, found: 848.19 UPLC-retention time: 4.6 min, purity >95%.
General Procedure for the Synthesis of Compounds 33 and 34.
DIPEA (5 equiv) was added to a solution of compounds 69 (2 mmol) and 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione (56) (1.1 equiv) in DMSO. After stirring at 80 °C for 4 h, water was added; the reaction mixture was extracted by EtOAc and washed by water; and the organic phase was dried by Na2SO4. The product (72) can be obtained by removing the solvent under vacuum and purified by flash column chromatography with following deprotection by TFA with 82% yield.
DIPEA (5 equiv) was added to a solution of compounds 73 (1.6 mmol) and 74 (1.1 equiv) in DMSO. After stirring at 80 °C for 4 h, water was added and the reaction mixture was extracted by EtOAc, washed by water and the organic phase was dried by Na2SO4. Compound 75 can be obtained by removing the solvent under vacuum and purified by flash column chromatography with 85–90% yields.
NaOH (2 equiv) was added to a solution of 75 (1 mmol) in MeOH/H2O and stirred at rt for 2 h. Then, the MeOH was removed under reduced pressure; the pH was adjusted to <7 with 2 M HCl; and the mixture was extracted with EtOAc. The solvent was removed to afford the product (76), which was used without further purification.
DIPEA (5 equiv) and HATU (1.2 equiv) were added to a solution of compounds 54 (0.8 mmol) and 76 (1.1 equiv) in DMF. After stirring at rt for 1 h, water was added; the reaction mixture was extracted with EtOAc and washed by water; and the organic phase was dried by Na2SO4. Compound 77 can be obtained by removing the solvent under vacuum and purified by flash column with 88% yield.
DMP (2 equiv) was added to a solution of compound 77 (0.5 mmol) in DCM. After the reaction was stirred at rt for 2 h, compound 78 was obtained by removing the solvent under vacuum and purified by flash column with 80–90% yields.
A solution of compounds 78 (0.5 mmol) and 72 (1.2 equiv) was added to AcOH (10%) in DCE. After the mixture was stirred at rt for 10 min, NaBH(OAc)3 (1.2 equiv) was added and the mixture was stirred at rt for another 2 h. Then, the desired compounds 33 and 34 can be obtained by removing the solvent under vacuum and purified by flash column chromatography with 85–90% yields.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-(4-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperazin-1-yl)piperidin-1-yl)benzamide (33).
1H NMR (400 MHz, MeOH-d4) δ 10.76 (s, 1H), 7.75–7.64 (m, 4H), 7.40 (d, J = 2.2 Hz, 1H), 7.28 (dd, J = 8.5, 2.3 Hz, 1H), 7.07 (d, J = 2.4 Hz, 1H), 7.01 (d, J = 9.0 Hz, 2H), 6.93 (dd, J = 8.8, 2.4 Hz, 1H), 5.05 (dd, J = 12.5, 5.5 Hz, 1H), 4.24 (s, 1H), 4.09 (d, J = 0.5 Hz, 1H), 4.06 (d, J = 13.3 Hz, 3H), 3.78–3.26 (m, 8H), 2.95–2.85 (m, 2H), 2.84–2.77 (m, 1H), 2.74–2.61 (m, 2H), 2.25 (d, J = 11.5 Hz, 2H), 2.07 (ddd, J = 10.0, 5.3, 2.5 Hz, 1H), 1.84 (qd, J = 12.1, 3.7 Hz, 2H), 1.24 (s, 6H), 1.17 (s, 6H). 13C NMR (100 MHz, MeOH-d4) δ 173.21 (s), 170.23 (s), 169.25 (s), 167.59 (s), 167.30 (s), 163.01 (s), 154.32 (s), 152.50 (s), 137.56 (s), 135.33 (s), 134.14 (s), 128.76 (s), 124.74 (s), 124.28 (s), 121.55 (s), 119.15 (s), 116.59 (s), 115.79 (s), 114.56 (s), 114.32 (s), 109.08 (s), 104.32 (s), 84.43 (s), 63.76 (s), 58.83 (s), 49.16 (s), 48.33 (s), 46.72 (s), 44.93 (s), 40.33 (s), 30.79 (s), 25.80 (s), 23.03 (s), 22.30 (d, J = 6.1 Hz). UPLC–MS calculated for C44H49ClN7O6 [M + H]+: 806.34, found: 806.18. UPLC-retention time: 4.5 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-((4-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperazin-1-yl)methyl)piperidin-1-yl)benzamide (34).
1H NMR (400 MHz, MeOH-d4) δ 10.84 (s, 1H), 7.84–7.72 (m, 4H), 7.51 (d, J = 2.2 Hz, 1H), 7.38 (dd, J = 8.5, 2.3 Hz, 1H), 7.15 (d, J =2.4 Hz, 1H), 7.06 (d, J = 9.0 Hz, 2H), 7.01 (dd, J = 8.8, 2.4 Hz, 1H), 5.14–5.10 (m, 1H), 4.32 (s, 1H), 4.16 (s, 1H), 3.98 (d, J = 13.0 Hz, 3H), 3.87–3.35 (m, 7H), 3.22 (d, J = 7.0 Hz, 2H), 3.01–2.85 (m, 3H), 2.83–2.70 (m, 2H), 2.26–2.11 (m, 2H), 2.06–1.94 (m, 2H), 1.59–1.44 (m, 2H), 1.31 (s, 6H), 1.25 (s, 6H). UPLC–MS calculated for C45H51ClN7O6 [M + H]+: 820.36, found: 820.22. UPLC-retention time: 4.7 min, purity >95%.
General Procedure for the Synthesis of Compounds 41 and 42.
DIPEA (5 equiv) was added to a solution of compounds 73 (2 mmol) and 79 (1.1 equiv) in DMSO. After stirring at 80 °C for 4 h, water was added; the reaction mixture was extracted by EtOAc and washed by water; and the organic phase was dried by Na2SO4. Compound 80 was obtained by removing the solvent under vacuum and purified by flash column chromatography with 88% yield.
NaOH (2 equiv) was added to a solution of 80 (1 mmol) in MeOH/H2O and stirred at rt for 2 h. Then, the MeOH was removed under reduced pressure; the pH was adjusted to acidity with 2 M HCl; and the mixture was extracted with EtOAc. The solvent was removed to afford product 81, which was used without further purification.
DIPEA (5 equiv) and HATU (1.2 equiv) were added to a solution of compounds 54 (0.8 mmol) and 81 (1.1 equiv) in DMF. After stirring at rt for 1 h, water was added; the reaction mixture was extracted by EtOAc and washed by water; and the organic phase was dried by Na2SO4. Compound 82 can be obtained by removing the solvent under vacuum and purified by flash column with following deprotection with 80–90% yield.
DIPEA (5 equiv) was added to a solution of compounds 82 (0.5 mmol) and 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione (56) (1.1 equiv) in DMSO (2 mL). After 4 h at 100 °C, the mixture was subject to HPLC purification to afford compounds 41 and 42 with 70–80% yields.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)-7-azaspiro[3.5]nonan-7-yl)benzamide (41).
1H NMR (400 MHz, MeOH-d4) δ 10.80 (s, 1H), 7.83 (d, J = 8.9 Hz, 2H), 7.74 (d, J = 8.7 Hz, 1H), 7.59 (d, J = 8.4 Hz, 1H), 7.23 (d, J = 8.9 Hz, 2H), 8.09–6.16 (m, 14H), 7.15 (d, J = 2.4 Hz, 1H), 7.01 (dd, J = 8.8, 2.4 Hz, 1H), 6.96 (d, J = 2.0 Hz, 1H), 6.83 (dd, J = 8.4, 2.1 Hz, 1H), 5.09–5.05 (m, 1H), 4.31 (s, 1H), 4.17 (s, 1H), 4.13–4.07 (m, 1H), 3.50–3.45 (m, 2H), 3.40 (d, J = 5.4 Hz, 2H), 2.94–2.67 (m, 4H), 2.59–2.51 (m, 2H), 2.16–2.08 (m, 1H), 1.96 (dd, J = 6.3, 4.4 Hz, 2H), 1.89–1.82 (m, 4H), 1.31 (s, 6H), 1.25 (s, 6H). UPLC–MS calculated for C43H46ClN6O6 [M + H]+: 777.32, found: 777.16. UPLC-retention time: 6.6 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(9-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)amino)-3-azaspiro[5.5]undecan-3-yl)benzamide (42).
1H NMR (400 MHz, DMSO-d6) δ 11.08 (s, 1H), 7.91 (dd, J = 11.9, 8.9 Hz, 1H), 7.79 (s, 1H), 7.56 (d, J = 5.5 Hz, 2H), 7.21 (d, J = 11.5 Hz, 1H), 7.11 6.93 (m, 4H), 6.89 (d, J = 11.8 Hz, 1H), 5.04 (d, J = 5.2 Hz, 1H), 4.33 (d, J = 12.0 Hz, 2H), 4.11–4.04 (m, 1H), 3.46 (s, 1H), 3.33 (s, 3H), 3.03 (d, J= 12.1 Hz, 1H), 2.90 (d, J = 12.0 Hz, 1H), 2.66–2.49 (m, 3H), 2.02–1.91 (m, 1H), 1.75 (dd, J = 37.5, 23.0 Hz, 5H), 1.52 (s, 1H), 1.37 (dd, J = 24.9, 12.3 Hz, 3H), 1.22 (s, 6H), 1.15 (s, 6H), 1.03 (s, 1H). UPLC–MS calculated for C45H50ClN6O6 [M + H]+: 805.35, found: 805.20. UPLC-retention time: 7.0 min, purity >95%.
General Procedure for the Synthesis of Compounds 43–47.
NaH (1.2 equiv) was added to a solution of 83 (10 mmol) in dry DMF at 0 °C. After stirring the mixture at 0 °C for 20 min, 52 was added and the mixture was stirred at rt for 4 h. After UPLC-MS demonstrated the full conversion of starting materials, H2O was added; the mixture was extracted three times with EtOAc; and the combined organic layers were washed with brine and then dried over anhydrous Na2SO4. The solvent was removed on a rotary evaporator. The Boc-protected intermediate was obtained by flash column chromatography. The desired intermediate (84) was obtained by deprotection with TFA in DCM in 85% yield.
DIPEA (5 equiv) and HATU (1.2 equiv) were added to a solution of compounds 84 (2 mmol) and 58 (1.1 equiv) in DMF (2 mL). After stirring at rt for 1 h, water was added; the reaction mixture was extracted by EtOAc and washed by water; and the organic phase was dried by Na2SO4. The Boc protected compound (85) can be obtained by removing the solvent under vacuum and purified by flash column chromatography with 80% yield. Then, the desired intermediate (85) was obtained by deprotection with TFA in DCM in 70–80% yields in total.
AcOH (10%) was added to a solution of compounds 85 (1 mmol) and aldehyde 86 in DCE. After the mixture was stirred at rt for 10 min, NaBH(OAc)3 (1.2 equiv) was added and the mixture was stirred at rt for another 2 h. The Boc-protected intermediates 87 were obtained by flash column chromatography. Then, the desired intermediate 87 were obtained by deprotection with TFA in DCM in 73–84% yields in total.
DIPEA (5 equiv) was added to a solution of the compounds 87 (0.5 mmol) and 2-(2,6-dioxopiperidin-3-yl)-5-fluoroisoindoline-1,3-dione (56) (1.1 equiv) in DMSO (2 mL). After stirring at 100 °C for 4 h, the mixture was subject to HPLC purification to afford compounds 43 and 44 with 70–80% yields. Following the procedures used to prepare compounds 43 and 44, compounds 45–47 were obtained using the same methods.
N-((1S,3S)-3-(3-Chloro-4-cyanophenoxy)-2,2-dimethylcyclobutyl)-4-(4-((1-(2-(2,6-dioxopiperidin-3-yi)-1,3-dioxoisoindolin-5-yl)-piperidin-4-yl)methyl)piperazin-1-yl)benzamide (43).
1H NMR (400 MHz, MeOH-d4) δ 7.77 (d, J = 8.9 Hz, 2H), 7.65 (dd, J = 8.4, 7.3 Hz, 2H), 7.06 (t, J = 5.8 Hz, 3H), 6.93 (dd, J = 8.8, 2.4 Hz, 1H), 6.88 (d, J = 2.0 Hz, 1H), 6.72 (dd, J = 8.3, 2.1 Hz, 1H), 5.07–5.01 (m, 1H), 4.40–4.30 (m, 5H), 4.24 (s, 1H), 4.11 (s, 1H), 3.55 (d, J = 57.2 Hz, 8H), 2.82–2.62 (m, 3H), 2.08 (ddd, J = 10.2, 8.9, 5.4 Hz, 1H), 1.25 (s, 6H), 1.18 (s, 6H). UPLC–MS calculated for C43H47ClN7O6 [M + H]+: 792.33, found: 792.22. UPLC-retention time: 4.5 min, purity >95%.
N-((1R,3R)-3-(3-Chloro-4-cyanophenoxy)cyclobu1yl)-4-(4-((4-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperazin-1-yl)-methyl)piperazin-1-yl)benzamide (44).
1H NMR (400 MHz, DMSO-d6) δ 11.08 (s, 1H), 8.14 (d, J = 7.2 Hz, 1H), 7.90 (dd, J = 8.7, 4.4 Hz, 1H), 7.83 (d, J = 8.7 Hz, 2H), 7.68 (t, J = 7.0 Hz, 1H), 7.37 (s, 1H), 7.27 (dd, J = 11.0, 6.4 Hz, 2H), 7.09–7.02 (m, 3H), 4.52 (t, J = 7.2 Hz, 1H), 4.10 (d, J = 12.7 Hz, 2H), 3.95 (dd, J = 16.9, 8.0 Hz, 3H), 3.69–3.57 (m, 2H), 3.12 (d, J = 5.5 Hz, 3H), 3.02 (t, J = 11.8 Hz, 2H), 2.90 (t, J = 13.0 Hz, 1H), 2.75 (dt, J = 11.5, 7.7 Hz, 1H), 2.65–2.46 (m, 6H), 2.33–2.17 (m, 2H), 2.05–1.99 (m, 1H), 1.88 (d, J = 11.6 Hz, 2H), 1.29–1.25 (m, 2H). UPLC–MS calculated for C41H42ClN7O6 [M + H]+: 764.30, found: 764.24. UPLC-retention time: 3.8 min, purity >95%.
N-((1R,3R)-3-(4-Cyano-3-fluorophenoxy)-2,2,4,4-tetramethylcyclobutyl)-4-(4-((1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperidin-4-yl)methyl)piperazin-1-yl)benzamide (45).
1H NMR (400 MHz, MeOH-d4) δ 7.77 (d, J = 8.9 Hz, 2H), 7.65 (dd, J = 8.4, 7.3 Hz, 2H), 7.06 (t, J = 5.8 Hz, 3H), 6.93 (dd, J = 8.8, 2.4 Hz, 1H), 6.88 (d, J = 2.0 Hz, 1H), 6.72 (dd, J = 8.3, 2.1 Hz, 1H), 5.07–5.01 (m, 1H), 4.40–4.30 (m, 5H), 4.24 (s, 1H), 4.11 (s, 1H), 3.55 (d, J = 57.2 Hz, 8H), 2.82–2.62 (m, 3H), 2.08 (ddd, J = 10.2, 8.9, 5.4 Hz, 1H), 1.25 (s, 6H), 1.18 (s, 6H). UPLC–MS calculated for C45H51FN7O6 [M + H]+: 804.39, found: 804.30. UPLC-retention time: 5.6 min, purity >95%.
N-((1R,3R)-3-(4-Cyano-3-(trifluoromethyl)phenoxy)-2,2,4,4-tetra-methylcyclobutyl)-4-(4-((1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperidin-4-yl)methyl)piperazin-1-yl)benzamide (46).
1H NMR (400 MHz, MeOH-d4) δ 7.77 (d, J = 8.9 Hz, 2H), 7.65 (dd, J = 8.4, 7.3 Hz, 2H), 7.06 (t, J = 5.8 Hz, 3H), 6.93 (dd, J = 8.8, 2.4 Hz, 1H), 6.88 (d, J = 2.0 Hz, 1H), 6.72 (dd, J = 8.3, 2.1 Hz, 1H), 5.07–5.01 (m, 1H), 4.40–4.30 (m, 5H), 4.24 (s, 1H), 4.11 (s, 1H), 3.55 (d, J = 57.2 Hz, 8H), 2.82–2.62 (m, 3H), 2.08 (ddd, J = 10.2, 8.9, 5.4 Hz, 1H), 1.25 (s, 6H), 1.18 (s, 6H). UPLC–MS calculated for C46H51F3N7O6 [M + H]+: 854.39, found: 854.28. UPLC-retention time: 6.2 min, purity >95%.
N-((1R,3R)-3-((6-Cyano-5-(trifluoromethyl)pyridin-3-yl)oxy)-2,2,4,4-tetramethyl-cyclobutyl)-4-(4-((1-(2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-5-yl)piperidin-4-yl)methyl)piperazin-1-yl)-benzamide (47).
1H NMR (400 MHz, MeOH-d4) δ 7.77 (d, J = 8.9 Hz, 2H), 7.65 (dd, J = 8.4, 7.3 Hz, 2H), 7.06 (t, J = 5.8 Hz, 3H), 6.93 (dd, J = 8.8, 2.4 Hz, 1H), 6.88 (d, J = 2.0 Hz, 1H), 6.72 (dd, J = 8.3, 2.1 Hz, 1H), 5.07–5.01 (m, 1H), 4.40–4.30 (m, 5H), 4.24 (s, 1H), 4.11 (s, 1H), 3.55 (d, J = 57.2 Hz, 8H), 2.82–2.62 (m, 3H), 2.08 (ddd, J = 10.2, 8.9, 5.4 Hz, 1H), 1.25 (s, 6H), 1.18 (s, 6H). UPLC–MS calculated for C45H50F3N8O6 [M + H]+: 855.38, found: 855.30. UPLC-retention time: 5.1 min, purity >95%.
Cell Lines and Cell Culture.
All the LNCaP and VCaP cells used were purchased from American Type Culture Collection (ATCC). LNCaP and VCaP cells were grown in RPMI 1640 (Invitrogen), and VCaP cells were grown in DMEM with Glutamax (Invitrogen). All of the cells were supplemented with 10% fetal bovine serum (Invitrogen) at 37 °C in a humidified 5% CO2 incubator. Cell viability was evaluated by a WST-8 assay (Dojindo) following the manufacturer’s instructions. Western blot analysis was performed as previously described.55,57
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR).
Real-time PCR was performed using a QuantStudio 7 Flex Real-Time PCR system as described previously.55,57 RNA was purified using the Qiagen RNase-Free DNase set, and then after quantification, the extracted RNA was converted to cDNA using a High Capacity RNA-to-cDNA Kit from Applied Biosystems (Thermo Fisher Scientific). The levels of AR, TMPRSS2, FKBP5, PSA (KLK3), and GAPDH were quantified using a TaqMan Fast Advanced Master Mix from Applied Biosystems. The level of gene expression was evaluated using a comparative CT method, which compares the CT value to GAPDH (ΔCT) and then to vehicle control (ΔΔCT).
Western Blotting.
Treated cells were lysed by RIPA buffer supplemented with protease and phosphatase inhibitors. The cell lysates were separated by 4–12% SDS-PAGE gels and blotted into PVDF membranes. Software ImageJ was used to quantify the percentage of AR degradation. The net protein bands and loading controls are calculated by deducting the background from the inverted band value. The final relative quantification values are the ratio of net band to net loading control.65
PK, PK/PD, and Efficacy Studies in Mice.
All in vivo studies were performed under animal protocol (PRO00009463) approved by the Institutional Animal Care & Use Committee (IACUC) of the University of Michigan, in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
To grow VCaP xenograft tumors, male CB17 SCID mice (Charles River Laboratories) were injected subcutaneously with 5 × 106 VCaP cells (ATCC) in 5 mg/mL Matrigel (Coming).
For the determination of oral exposures for AR degraders, each compound was administered in non-tumor-bearing male mice via oral gavage using 100% PEG200 as the dosing vehicle. Animals were sacrificed at indicated time points with three mice for each time point for each compound, and 300 μL of blood was collected from each animal and were stored at −80 °C until analysis.
For PK/PD studies in tumor-bearing male SCID mice, each compound was administered in animals via oral gavage using 100% PEG200 as the dosing vehicle when the VCaP tumors reached approximately 200 mm3. Animals were sacrificed at indicated time points with three mice for each compound at each time point, and blood (300 μL) and tumor were collected from each animal for analysis. Isolated tumor samples were placed in a Precellys tube (CK28-R) and immediately frozen in liquid nitrogen. All plasma and tumor samples were stored at −80 °C until analysis. For the analysis of AR protein levels in tumor samples, resected VCaP xenograft tumor tissues were ground into powder in liquid nitrogen and lysed in CST lysis buffer with halt proteinase inhibitors. Twenty micrograms of whole tumor clarified lysates were separated on 4–20% or 4–12% Novex gels. Western blots were performed as detailed in the previous section.
All animal experiments in this study were approved by the University of Michigan Committee on Use and Care of Animals and Unit for Laboratory Animal Medicine (ULAM). The pharmacokinetics of ARD-2128 and analogs was determined in normal male SCID mice or with VCaP tumor following oral gavage (PO) single dosing at 10 or 20 mg/kg. The solid compounds were dissolved in a vehicle containing 100% PEG200. The animals (total 9 mice/compound or 6 mice/compound) were sacrificed at 1, 3, and 6 h, or 6 and 24 h after the final administration of the chemicals and then followed by the collection of blood samples (300 μL) and tumor samples. The blood samples were centrifuged at 15000 rpm for 10 min, and then the supernatant plasma was saved for analysis. Isolated tumor samples were immediately frozen and ground with a mortar and pestle in liquid nitrogen. All plasma and tumor samples were stored at −80 °C until analysis. To prepare tumor samples for LCMS analysis, mixed ultrapure water and acetonitrile solution (4:1) were added to the defrosted tumor tissue samples 5:1, v/w, in order to facilitate homogenization with a Precellys evolution homogenizer under 4 °C. The homogenized tissues solution was denatured using cold acetonitrile (1:3, v/v) with vortex and centrifuged at 13000 rpm and 4 °C for 10 min. Following protein precipitation, the final supernatants were collected for LC-MS analysis.
To determine drug concentrations in plasma and tumor samples, an LC–MS/MS method was developed and validated. The LC–MS/MS method consisted of a Shimadzu HPLC system, and chromatographic separation of a test compound was achieved using a Waters XBridge-C18 column (5 cm × 2.1 mm, 3.5 μm). An AB Sciex QTrap 5500 mass spectrometer equipped with an electrospray ionization source (Applied biosystems, Toronto, Canada) in the positive-ion multiple reaction monitoring (MRM) mode was used for detection. For example, the precursor/product ion transitions were monitored at m/z 820.3–542.2 and 455.2–425.2 for ARD-2128 and internal standard, respectively, in the positive electrospray ionization mode. The mobile phases used on HPLC were 0.1% formic acid in purified water (A) and 0.1% formic acid in acetonitrile (B). The gradient (B) was held at 10% (0–0.3 min), increased to 95% at 0.7 min, then stayed at isocratic 95% B for 2.3 min, and then immediately stepped back down to 10% for 2 min re-equilibration. The flow rate was set at 0.4 mL/min. All pharmacokinetic parameters were calculated by noncompartmental methods using WinNonlin, version 3.2 (Pharsight Corporation, Mountain View, CA, USA).
For the in vivo efficacy experiments, when VCaP tumors reached an average volume of 150 mm3, mice were tumor size matched and randomly assigned to different experimental groups with seven mice for each group. Drugs or vehicle control were given at the dose schedule as indicated using 100% PEG200 as the dosing vehicle. Tumor sizes and animal weights were measured 2–3 times per week. Tumor volume (mm3) = (length × width2)/2. Tumor growth inhibition was calculated as TGI (%) = (Vc − Vt)/(Vc − Vo) × 100, where Vc and Vt are the medians of the control and treated groups at the end of the treatment, respectively, and Vo is that at the start. The tumor volumes at the end of treatment were statistically analyzed using a two-tailed, unpaired t-test (GraphPad Prism 8.0).
PK Studies in Mice.
Pharmacokinetic (PK) studies were performed in Shanghai Medicilon Inc. Shanghai, 201200, China. Male ICR mice, weighing 18–20 g, were purchased from Sino-British SIPPR/BK Lab Animal Ltd., Shanghai, China. One group of three mice was dosed intravenously (IV) via a bolus in the tail vein with a dose level of 2 mg/kg, and a second group of three mice was dosed orally as a single esophageal gavage with a dose level of 5 mg/kg. The drug solution was freshly prepared before administration. For the IV route, each compound was formulated in 5% DMSO, 10% solutol, 85% saline as a clear solution, a dosage volume of 5 mL/kg, and a theoretical concentration of 0.4 mg/mL. For the oral route, each was formulated in 5% DMSO, 10% solutol, 85% saline as a clear solution, a dosage volume of 10 mL/kg, and a theoretical concentration of 0.5 mg/mL. Blood samples were collected at the following time points: IV, 0.083, 0.25, 0.5, 1, 2, 4, 6, 8, and 24 h after dosing (n = 3 mice for ach per sampling time); PO, 0.083, 0.25, 0.5, 1, 2, 4, 6, 8, and 24 h after dosing (n = 3 mice for each per sampling time). Each mouse was sampled once and then euthanized. Blood was kept on ice. Within 1 h after sampling, blood was centrifuged at 3900 rpm for 15 min, and the supernatant was diluted three times with water. A total of 5 μL of diluted supernatant was injected into the LC/MS/MS system for quantitative analysis.
Microsomal Metabolic Stability Studies.
In vitro metabolism study of a test compound was performed in human, mouse, rat, dog, and monkey liver microsomes to evaluate its cytochrome P450-mediated metabolism. The metabolic stability was assessed using pooled mouse, rat, dog, monkey, and human liver microsomes, which were purchased from XenoTech (Lenexa, Kansas).
A total of 1 μM of the test compound was incubated with 0.5 mg/mL of the respective Ever microsome and 1.7 mM cofactor-NADPH in 0.1 M K-phosphate buffer (pH = 7.4) containing 5 mM MgCl2 at 37 °C, with the acetonitrile concentration less than 0.1% in the final incubation solution. After 0, 5, 10, 15, 30, and 45 min of incubation, the reaction was stopped immediately by adding 150 μL cold acetonitrile containing IS to each 45 μL incubation solution in the wells of the corresponding plates. The incubation without the addition of NADPH was used as the negative control. Ketanserin was incubated similarly as the positive control. After quenching, the plate was shaken far 10 min (600 rpm/min) and centrifuged at 6000 rpm for 15 min. A total of 80 μL of the supernatant was then transferred from each well into a 96-well plate containing 140 μL of water for LC–MS/MS analysis, from which the remaining amount of the test compound was determined. The natural log of the remaining amount of the test compound was plotted against time to determine the disappearance rate and the half-life of the test compound.
Plasma Stability Studies.
The in vitro stability of a test compound was studied in human, mouse, rat, dog, and monkey plasmas in Medicilon Inc. (Shanghai, China). Human plasma was purchased from ZenBio (Durham, NC, USA), and other plasmas were prepared inhouse. A test compound was dissolved in DMSO to a final concentration of 10 mM and then diluted to 10 μM in 0.1 M K/Mg buffer. A total of 90 μL of pre-warmed plasma at 37 °C was added to the wells of a 96-well plate before spiking them with 10 μL of 10 μM test compound to make the final concentration of the test compound at 1 μM. The spiked plasma samples were incubated at 37 °C for 2 h. Reactions were terminated at 0, 5, 15, 30, 60, and 120 min by adding 400 μL of acetonitrile containing IS. After quenching, the plates were shaken for 5 min at 600 rpm and stored at −20 °C if necessary before analysis by LC/MS. Before LC/MS analysis, the samples were thawed at room temperature and centrifuged at 6000 rpm for 20 min. A total of 100 μL of the supernatant from each well was transferred into a 96-well sample plate containing 100 μL of water for LC/MS analysis. Procaine was used as the reference control compound for human, mouse, dog, and monkey plasma stability studies, and Benfluorex was used as the reference control compound for rat plasma stability studies. The in vitro plasma half-life (t1/2) was calculated using the expression t1/2 = 0.693/b, where b is the slope found in the linear fit of the natural logarithm of the fraction remaining of the test compound vs incubation time.
Supplementary Material
ACKNOWLEDGMENTS
This study is supported in part by funding from Oncopia Therapeutics, Inc. (now part of Roivant Sciences), the National Cancer Institute, NIH (P50 CA186786), and the University of Michigan Comprehensive Cancer Center Core Grant from the National Cancer Institute, NIH (P30CA046592).
ABBREVIATIONS USED
- AR
androgen receptor
- mCRPC
metastatic castration-resistant prostate cancer
- PROTAC
proteolysis targeting chimera
- cIAP1
cellular inhibitor of apoptosis protein 1
- V ss
steady-state volume of distribution
- C max
maximum drug concentration
- AUC0–24 h
area-under-the-curve between 0 and 24 hr.
- Cl
plasma clearance rate
- T 1/2
terminal half-life
- F
oral bioavailability
- IV
intravenous administration
- PO
oral administration
- PD
pharmacodynamics
- CYP
cytochrome P450
- ATCC
American-type culture collection
- qRT-PCR
quantitative real-time polymerase chain reaction
- SCID
severe combined immunodeficient
Footnotes
The authors declare the following competing financial interest(s): The University of Michigan has filed patent applications on these AR degraders, which have been licensed to Oncopia Therapeutics, Inc. X.H., L.Z., W.X., C.Q., B.M., and B.W. are co-inventors on these patent applications and receive royalties from the University of Michigan. S.W. was a co-founder and served as a paid consultant to Oncopia. S.W. and the University of Michigan also owned equity in Oncopia, which was acquired by Roivant Sciences. S.W. is a paid consultant to Roivant Sciences. The University of Michigan has received a research contract from Oncopia and Roivant for which S.W. serves as the principal investigator.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.1c00882.
Western blotting of AR protein for compounds 10–47 in VCaP cells; cell growth inhibition curves of selected AR degraders in LNCaP and VCaP cell lines; concentrations of compounds 12, 13, 16, and 17 in plasma in SCID mice with a single oral administration; concentrations of compounds 26, 27, and 28 (ARD-2128) in plasma and tumor tissues in SCID mice; 1H and 13C NMR spectra for ARD-2128; HPLC purity spectra for representative AR degraders; and microsomal metabolic and plasma stability data of ARD-2128 (PDF)
Molecular string file for all the final target compounds (CSV)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jmedchem.1c00882
Contributor Information
Xin Han, The Rogel Cancer Center, and Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan 48109, United States.
Lijie Zhao, The Rogel Cancer Center, and Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan 48109, United States.
Weiguo Xiang, The Rogel Cancer Center, and Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan 48109, United States.
Chong Qin, The Rogel Cancer Center, and Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan 48109, United States.
Bukeyan Miao, The Rogel Cancer Center, and Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan 48109, United States.
Donna McEachern, The Rogel Cancer Center, and Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan 48109, United States.
Yu Wang, The Rogel Cancer Center, and Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan 48109, United States.
Hoda Metwally, The Rogel Cancer Center, and Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan 48109, United States.
Lu Wang, Department of Pharmaceutical Sciences, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States.
Aleksas Matvekas, Department of Pharmaceutical Sciences, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States.
Bo Wen, Department of Pharmaceutical Sciences, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States.
Duxin Sun, Department of Pharmaceutical Sciences, College of Pharmacy, University of Michigan, Ann Arbor, Michigan 48109, United States.
Shaomeng Wang, The Rogel Cancer Center,, Department of Internal Medicine, Department of Pharmacology, and Department of Medicinal Chemistry, University of Michigan, Ann Arbor, Michigan 48109, United States.
REFERENCES
- (1).Sternberg CN; Fizazi K; Saad F; Shore ND; De Giorgi U; Penson DF; Ferreira U; Efstathiou E; Madziarska K; Kolinsky MP; Cubero DIG; Noerby B; Zohren F; Lin X; Modelska K; Sugg J; Steinberg J; Hussain M Enzalutamide and survival in nonmetastatic, castration-resistant prostate cancer. N Engl J Med. 2020, 382, 2197–2206. [DOI] [PubMed] [Google Scholar]
- (2).Harris WP; Mostaghel EA; Nelson PS; Montgomery B Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Prod Urol. 2009, 6, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wang C; Peng G; Huang H; Liu F; Kong DP; Dong KQ; Dai LH; Zhou Z; Wang KJ; Yang J; Cheng YQ; Gao X; Qu M; Wang HR; Zhu F; Tian QQ; Liu D; Cao L; Cui XG; Xu CL; Xu DF; Sun YH Blocking the feedback loop between neuroendocrine differentiation and macrophages improves the therapeutic effects of enzalutamide (MDV3100) on prostate cancer. Clin Cancer Res. 2018, 24, 708–723. [DOI] [PubMed] [Google Scholar]
- (4).Watson PA; Arora VK; Sawyers CL Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015, 15, 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Myung JK; Banuelos CA; Fernandez JG; Mawji NR; Wang J; Tien AH; Yang YC; Tavakoli I; Haile S; Watt K; McEwan IJ; Plymate S; Andersen RJ; Sadar MD An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest. 2013, 123, 2948–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Guo C; Linton A; Kephart S; Ornelas M; Pairish M; Gonzalez J; Greasley S; Nagata A; Burke BJ; Edwards M; Hosea N; Kang P; Hu W; Engebretsen J; Briere D; Shi M; Gukasyan H; Richardson P; Dack K; Underwood T; Johnson P; Morell A; Felstead R; Kuruma H; Matsimoto H; Zoubeidi A; Gleave M; Los G; Fanjul AN Discovery of aryloxy tetramethylcyclobutanes as novel androgen receptor antagonists. J. Med. Chem. 2011, 54, 7693–7704. [DOI] [PubMed] [Google Scholar]
- (7).Guerrini A; Tesei A; Ferroni C; Paganelli G; Zamagni A; Carloni SD; Donato M; Castoria G; Leonetti C; Porru MD; Cesare M; Zaffaroni N; Beretta GLD; Rio A; Varchi G A new avenue toward androgen receptor pan-antagonists: C2 sterically hindered substitution of hydroxy-propanamides. J. Med. Chem. 2014, 57, 7263–7279. [DOI] [PubMed] [Google Scholar]
- (8).Tran C; Ouk S; Clegg NJ; Chen Y; Watson PA; Arora V; Wongvipat J; Smith-Jones PM; Yoo D; Kwon A; Wasielewska T; Welshie D; Chen CD; Higano CS; Beer TM; Hung DT; Scher HI; Jung ME; Sawyers CL Development of a second generation antiandrogen for treatment of advanced prostate cancer. Science 2009, 324, 787–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Pal SK; Patel J; He M; Foulk B; Kraft K; Smirnov DA; Twardowski P; Kortylewski M; Bhargava V; Jones JO Identification of mechanisms of resistance to treatment with abiraterone acetate or enzalutamide in patients with castration-resistant prostate cancer (CRPC). Cancer. 2018, 124, 1216–1224. [DOI] [PubMed] [Google Scholar]
- (10).Karantanos T; Com PG; Thompson TC Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene 2013, 32, 5501–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yu J; Zhang L; Yan G; Zhou P; Cao C; Zhou F; Li X; Chen Y Discovery and biological evaluation of novel androgen receptor antagonist for castration-resistant prostate cancer. Eur. J. Med. Chem. 2019, 171, 265–281. [DOI] [PubMed] [Google Scholar]
- (12).Clegg NJ; Wongvipat J; Joseph JD; Tran C; Ouk S; Dilhas A; Chen Y; Grillot K; Bischoff ED; Cal L; Aparicio A; Dorow S; Arora V; Shao G; Qian J; Zhao H; Yang GB; Cao CY; Sensintaffar J; Wasielewska T; Herbert MR; Bonnefous C; Darimont B; Scher HI; Smith-Jones P; Klang M; Smith ND; De Stanchina E; Wu N; Ouerfelli O; Rix PJ; Heyman RA; Jung ME; Sawyers CL; Hager JH ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer Res. 2012, 72, 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Munuganti RS; Hassona MD; Leblanc E; Frewin K; Singh K; Ma D; Ban F; Hsing M; Adomat H; Lallous N; Andre C; Jonadass JP; Zoubeidi A; Young RN; Guns ET; Rennie PS; Cherkasov A Identification of a potent antiandrogen that targets the BF3 site of the androgen receptor and inhibits enzalutamide-resistant prostate cancer. Chem. Biol. 2014, 21, 1476–1485. [DOI] [PubMed] [Google Scholar]
- (14).Rodriguez-vida A; Galazi M; Rudman S; Chowdhury S; Sternberg CN enzalutamide for the treatment of metastatic castration-resistant prostate cancer. Drug Des. Dev. Ther. 2015, 9, 3325–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Esther J; Dorff TB; Maughan BL Recent developments in the treatment of non-metastatic castration resistant prostate cancer. Cancer Treat Res Commun. 2020, 24, 100181. [DOI] [PubMed] [Google Scholar]
- (16).Xu H; Sun Y; Huang C-P; You B; Ye D; Chang C Preclinical study using ABT263 to increase enzalutamide sensitivity to suppress prostate cancer progression via targeting BCL2/ROS/USP26 axis through altering ARv7 protein degradation. Cancers 2020, 12, 831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hamdy FC; Donovan JL; Lane JA; Mason M; Metcalfe C; Holding P; Davis M; Peters TJ; Turner EL; Martin RM; Oxley J; Robinson M; Staffurth J; Walsh E; Bollina P; Catto J; Doble A; Doherty A; Gillatt D; Kockelbergh R; Kynaston H; Paul A; Powell P; Prescott S; Rosario DJ; Rowe E; Neal DE 10-year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med. 2016, 375, 1415–1424. [DOI] [PubMed] [Google Scholar]
- (18).Moilanen AM; Riikonen R; Oksala R; Ravanti L; Aho E; Wohlfahrt G; Nykänen PS; Törmäkangas OP; Palvimo JJ; Kallio PJ Discovery of ODM-201, a new generation androgen receptor inhibitor targeting resistance mechanisms to androgen signaling-directed prostate cancer therapies. Sci. Rep. 2015, 5, 12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Balbas MD; Evans MJ; Hosfield DJ; Wongvipat J; Arora VK; Watson PA; Chen Y; Greene GL; Shen Y; Sawyers CL Overcoming mutation-based resistance to antiandrogens with rational drug design. Elife. 2013, 2, No. e00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Heidegger I; Massoner P; Eder IE; Pircher A; Pichler R; Agner F; Bektic J; Hominger W; Klocker H Novel therapeutic approaches for the treatment of castration-resistant prostate cancer. J Steroid Biochem 2013, 138, 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hansen JD; Condroski K; Correa M; Muller G; Man HW; Ruchelman A; Zhang W; Vocanson F; Crea T; Liu W; Lu G; Baculi F; LeBrun L; Mahmoudi A; Carmel G; Hickman M; Lu CC Protein degradation via CRL4crbn ubiquitin ligase: discovery and structure-activity relationships of novel glutarimide analogs that promote degradation of aiolos and/or GSPT1. J. Med. Chem. 2018, 61, 492–503. [DOI] [PubMed] [Google Scholar]
- (22).Toure M; Crews CM Small-molecule PROTACS: new approaches to protein degradation. Angew. Chem., Int. Ed. 2016, 55, 1966–1973. [DOI] [PubMed] [Google Scholar]
- (23).Ishida T; Chilli A E3 ligase ligands for PROTACs: how they were found and how to discover new ones. SLAS Discov. 2021, 26, 484–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gu S; Cui D; Chen X; Xiong X; Zhao Y PROTACs: An Emerging Targeting Technique for Protein Degradation in Drug Discovery. BioEssays 2018, 40, 1700247. [DOI] [PubMed] [Google Scholar]
- (25).Bondeson DP; Mares A; Smith IE; Ko E; Campos S; Miah AH; Mulholland KE; Routly N; Buckley DL; Gustafson JL; Zinn N; Grandi P; Shimamura S; Bergamini G; Faelth-Savitski M; Bantscheff M; Cox C; Gordon DA; Willard RR; Flanagan JJ; Casillas LN; Votta BJ; den Besten W; Famm K; Kruidenier L; Carter PS; Harling JD; Churcher I; Crews CM Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Churcher I PROTAC-induced protein degradation in drug discovery: breaking the rules or Just making new ones? J. Med. Chem. 2018, 61, 444–452. [DOI] [PubMed] [Google Scholar]
- (27).Burslem GM; Crews CM Small-molecule modulation of protein homeostasis. Chem. Rev. 2017, 117, 11269–11301. [DOI] [PubMed] [Google Scholar]
- (28).Zou Y; Ma D; Wang Y The PROTAC technology in drug development. Cell Biochem. Fund. 2019, 37, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lu J; Qian Y; Altieri M; Dong H; Wang J; Raina K; Hines J; Winkler JD; Crew AP; Coleman K; Crews CM Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 2015, 22, 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ohoka N; Shibata N; Hattori T; Naito M Protein knockdown technology: application of ubiquitin ligase to cancer therapy. Curr. Cancer Drug Targets 2016, 16, 136–146. [DOI] [PubMed] [Google Scholar]
- (31).An S; Fu L Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. EBioMedicine. 2018, 36, 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zhou B; Hu J; Xu F; Chen Z; Bai L; Fernandez-Salas E; Lin M; Liu L; Yang CY; Zhao Y; McEachern D; Przybranowski S; Wen B; Sun D; Wang S Discovery of a small-molecule degrader of bromodomain and extra-terminal (BET) proteins with picomolar cellular potencies and capable of achieving tumor regression. J. Med. Chem. 2018, 61, 462–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Qin C; Hu Y; Zhou B; Fernandez-Salas E; Yang CY; Liu L; McEachern D; Przybranowski S; Wang M; Stuckey J; Meagher J; Bai L; Chen Z; Lin M; Yang J; Ziazadeh DN; Xu F; Hu J; Xang W; Huang L; Li S; Wen B; Sun D; Wang S Discovery of QCA570 as an exceptionally potent and efficacious proteolysis targeting chimera (PROTAC) degrader of the bromodomain and extra-terminal (BET) proteins capable of inducing complete and durable tumor regression. J. Med. Chem. 2018, 61, 6685–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bai L; Zhou B; Yang CY; Ji J; McEachern D; Przybranowski S; Jiang H; Hu J; Xu F; Zhao Y; Liu L; Fernandez-Salas E; Xu J; Dou Y; Wen B; Sun D; Meagher J; Stuckey J; Hayes DF; Li S; Ellis MJ; Wang S Targeted degradation of BET proteins in triple-negative breast cancer. Cancer Res. 2017, 77, 2476–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Li Y; Yang J; Aguilar A; McEachern D; Przybranowski S; Liu L; Yang CY; Wang M; Han X; Wang S Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J. Med. Chem. 2019, 62, 448–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hu JT; Hu B; Wang ML; Xu FM; Miao B. K. Ys.; Yang CY; Wang M; Liu ZM; Hayes DF; Chinnaswamy K; Delproposto J; Stuckey J; Wang SM Discovery of ERD-308 as a highly potent proteolysis targeting chimera (PROTAC) degrader of estrogen receptor (ER). J. Med. Chem. 2019, 62, 1420–1442. [DOI] [PubMed] [Google Scholar]
- (37).Winter GE; Buckley DL; Paulk J; Roberts JM; Souza A; Dhe-Paganon S; Bradner JE Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015, 348, 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Cromm PM; Samarasinghe KTG; Hines J; Crews CM Addressing kinase-independent functions of fak via PROTAC-mediated degradation. J. Am. Chem. Soc. 2018, 140, 17019–17026. [DOI] [PubMed] [Google Scholar]
- (39).Burslem GM; Song J; Chen X; Hines J; Crews CM Enhancing antiproliferative activity and selectivity of a FLT-3 inhibitor by proteolysis targeting chimera conversion. J. Am. Chem. Soc. 2018, 140, 16428–16432. [DOI] [PubMed] [Google Scholar]
- (40).Raina K; Lu J; Qian Y; Altieri M; Gordon D; Rossi AM; Wang J; Chen X; Dong H; Siu K; Winkler JD; Crew AP; Crews CM; Coleman KG PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci. 2016,113, 7124–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lai AC; Crews CM Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2017, 16, 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bai L; Zhou H; Xu R; Zhao Y; Chinnaswamy K; McEachern D; Chen JY; Yang CY; Liu ZM; Wang M; Liu L; Jiang H; Wen B; Kumar P; Meagher JL; Sun DX; Stuckey JA; Wang S A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell 2019, 36, 498–511.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Khan S; Zhang X; Lv D; Zhang Q; He Y; Zhang P; Liu X; Thummuri D; Yuan Y; Wiegand JS; Pei J; Zhang WZ; Sharma A; McCurdy CR; Kuruvilla VM; Baran N; Ferrando AA; Kim YM; Rogojina A; Houghton PJ; Huang G; Hromas R; Konopleva M; Zheng G; Zhou D A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 2019, 25, 1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Schneekloth AR; Pucheault M; Tae HS; Crews CM Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg. Med. Chem. Lett. 2008, 18, 5904–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Sakamoto KM; Kim KB; Verma R; Ransick A; Stein B; Crews CM; Deshaies RJ Development of PROTACs to target cancer-promoting proteins for ubiquitination and degradation. Mol. Cell Proteomics 2003, 2, 1350–1358. [DOI] [PubMed] [Google Scholar]
- (46).Schneekloth JS; Fonseca FN; Koldobskiy M; Mandal A; Deshaies R; Sakamoto K; Crews CM Chemical genetic control of protein levels: selective in vivo targeted degradation. J. Am. Chem. Soc. 2004, 126, 3748–3754. [DOI] [PubMed] [Google Scholar]
- (47).Rodriguez-Gonzalez A; Cyrus K; Salcius M; Kim K; Crews CM; Deshaies RJ; Sakamoto KM Targeting steroid hormone receptors for ubiquitination and degradation in breast and prostate cancer. Oncogene 2008, 27, 7201–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Crew AP; Homberger KR; Snyder LB; Zimmermann K; Wang J; Berlin M; Crews CM; Dong H Compounds and methods for the targeted degradation of androgen receptor. US 2018/0099940 A1, Apr. 12, 2018.
- (49).Shibata N; Nagai K; Morita Y; Ujikawa O; Ohoka N; Hattori T; Koyama R; Sano O; Imaeda Y; Nara H; Cho N; Naito M Development of protein degradation inducers of androgen receptor by conjugation of androgen receptor ligands and inhibitor of apoptosis protein ligands. J. Med. Chem. 2018, 61, 543–575. [DOI] [PubMed] [Google Scholar]
- (50).Duncan ES; Rooney TPC; Bayle ED; Mirza T; Willems HMG; Clarke JH; Stephen P Andrews, and John Skidmore. Systematic Investigation of the Permeability of Androgen Receptor PROTACs. ACS Med. Chem. Lett. 2020, 11, 1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Itoh Y; Kitaguchi R; Ishikawa M; Naito M; Hashimoto Y Design, synthesis and biological evaluation of nuclear receptor-degradation inducers. Bioorg. Med. Chem. 2011, 19, 6768–6778. [DOI] [PubMed] [Google Scholar]
- (52).Salami J; Alabi S; Willard RR; Vitale NJ; Wang J; Dong H; Jin M; McDonnell DP; Crew AP; Neklesa TK; Crews CM Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun Biol. 2018, 1, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Kargbo RB Treatment of prostate cancers and kennedy’s disease by PROTAC-androgen receptor degradation. ACS Med. Chem. Lett. 2019, 10, 701–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kregel S; Wang C; Han X; Xiao L; Fernandez-Salas E; Bawa P; McCollum BL; Wilder-Romans K; Apel IJ; Cao X; Speers C; Wang S; Chinnaiyan AM Androgen receptor degraders overcome common resistance mechanisms developed during prostate cancer treatment. Neoplasia 2020, 22, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Han X; Wang C; Qin C; Xiang W; Fernandez-Salas E; Yang CY; Wang M; Zhao LJ; Xu TF; Chinnaswamy K; Delproposto J; Stuckey J; Wang SM Discovery of ARD-69 as a highly potent proteolysis targeting chimera (PROTAC) degrader of androgen receptor (AR) for die treatment of prostate cancer. J. Med. Chem. 2019, 62, 941–964. [DOI] [PubMed] [Google Scholar]
- (56).Zhao L; Han X; Lu J; McEachern D; Wang S A highly potent PROTAC androgen receptor (AR) degrader ARD-61 effectively inhibits AR-positive breast cancer cell growth in vitro and tumor growth in vivo. Neoplasia 2020, 22, 522–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Han X; Zhao LJ; Xiang WG; Qin C; Miao B; Xu T; Wang M; Yang CY; Chinnaswamy K; Stuckey J; Wang S Discovery of highly potent and efficient PROTAC degraders of androgen receptor (AR) by employing weak binding affinity VHL E3 ligase ligands. J. Med. Chem. 2019, 62, 11218–11231. [DOI] [PubMed] [Google Scholar]
- (58).Mullard A First targeted protein degrader hits the clinic. Nat. Rev. Drug. Discov. 2019,18, 237–239. [DOI] [PubMed] [Google Scholar]
- (59).Mullard A Arvinas’s PROTACs pass first safety and PK analysis. Nat. Rev. Drug. Discov. 2019, 18, 895–895. [DOI] [PubMed] [Google Scholar]
- (60).Takwale AD; Jo SH; Jeon YU; Kim HS; Shin CH; Lee HK; Ahn S; Lee CO; Ha JD; Kim JH; Hwang JY Design and characterization of cereblon-mediated androgen receptor proteolysis-targeting chimeras. Eur. J. Med. Chem. 2020, 208, 112769. [DOI] [PubMed] [Google Scholar]
- (61).Crew AP; Snyder LB; Wang J Compounds and methods for the targeted degradation of androgen receptor. US patent 10584101B2.
- (62).Crew AP; Snyder LB; Wang J; Haskell RJ; Moore MD Methods of treating prostate cancer. US patent application, 20210113556A1. [Google Scholar]
- (63).Crew AP; Berlin M; Chen X; Crews CM; Dong HQ; Qian YM; Snyder L; Wang J; Zimmer-mann K Compounds and methods for the targeted degradation of androgen receptor. WO 2019023553A1.
- (64).Snyder LB Discovery of ARV-110, a first in class androgen receptor degrading PROTAC® for the treatment of men with metastatic castration resistant prostate cancer. American Association for Cancer Research (AACR), 2021, April 10–15. [Google Scholar]
- (65).Gadd MS; Testa A; Lucas X; Chan KH; Chen W; Lamont DJ; Zengerle M; Ciulli A Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol 2017, 13, 514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.