Abstract

Free radicals are crucial indicators for stress and appear in all kinds of pathogenic conditions, including cancer, cardiovascular diseases, and infection. However, they are difficult to detect due to their reactivity and low abundance. We use relaxometry for the detection of radicals with subcellular resolution. This method is based on a fluorescent defect in a diamond, which changes its optical properties on the basis of the magnetic surroundings. This technique allows nanoscale MRI with unprecedented sensitivity and spatial resolution. Recently, this technique was used inside living cells from a cell line. Cell lines differ in terms of endocytic capability and radical production from primary cells derived from patients. Here we provide the first measurements of phagocytic radical production by the NADPH oxidase (NOX2) in primary dendritic cells from healthy donors. The radical production of these cells differs greatly between donors. We investigated the cell response to stimulation or inhibition.
Keywords: magnetometry, relaxometry measurements (T1), NV centers, nanodiamonds
Excessive production of free radicals leads to oxidative stress. Such oxidative stress is related to inflammatory diseases (arthritis, vasculitis), ischemic diseases (heart diseases, intestinal ischemia), neurological disorders (Alzheimer’s disease, Parkinson’s disease), and many others.1 On the other hand, free radicals also fulfill important functions in cell signaling and are vital for the immune system.2 There they play vital roles in immune cell maturation and function. In dendritic cells (DC; cells of the immune system that are specialized in antigen presentation to naive T cells3) they can for example affect the secretion of cytokines and the antigen-presenting capacity and thereby alter their ability to combat pathogens.4
Within DCs there are several processes that lead to radical formation. In the presence of pathogens the NADPH oxidase NOX2 is activated, which leads to the generation of superoxide radicals in the lumen of endosomes, phagosomes, and the extracellular space. Superoxide radicals can convert rapidly to a toxic peroxide. This compound can interact with nitric oxide to generate peroxynitrate anions. Hydrogen peroxide can in turn be converted to hydroxyl radicals.5 These radicals are highly reactive and have a short lifetime. They can damage genomic DNA, proteins, and lipids or interfere in electron transport. What we know so far about free radical generation in DCs mostly comes from ensemble experiments from large cell populations. The results have been obtained either indirectly or directly. Indirect methods include the detection of damage to DNA or lipids by free radicals. Direct methods are based on fluorescent or spin label probes that react with the radicals to form a visible compound.6,7 However, these methods are usually not specific for radicals but detect reactive oxygen species (ROS) in general, real-time measurements are problematic, and the probes themselves often are at least somewhat toxic and react with the radicals and thus interfere in the process.8,9 Additionally, the process is irreversible and thus these probes measure the accumulated ROS production rather than the current status.
Herein, we use diamonds containing nitrogen-vacancy (NV) centers for quantum sensing to detect radicals. The technique is based on defects (NV centers), which “feel” their magnetic environment and convert magnetic resonance signals into optical signals. Thus, nanoscale resolution magnetic resonance measurements are possible. This technique has already achieved impressive results in physics, including measurements of magnetic particles,10 domain walls,11 and even the spin of a single electron12 or nuclear spins.13−15
Unlike many earlier works, which were conducted with bulk diamonds, we use nanodiamonds. These are excellently biocompatible and can be ingested into cells.16−19 In the biomedical field the application of nanoscale resolution magnetic resonance is rather new.20 Ermakova et al., for instance, measured ferritin proteins on a nanodiamond surface.21 This was further used by Wang et al. to measure ferritin in cells.22 There have also been some reports on successful diamond magnetometry measurements in cells. Steinert et al. measured gadolinium-labeled slices of embedded cells.23 Le Sage et al. were able to detect magnetic particles, the so-called magnetosomes, in living bacteria.24 Also, tracking particle orientation and temperature sensing have been achieved with diamond magnetometry.25,26
More specifically, we make use of a specific mode of diamond magnetometry called T1 relaxometry. This sequence, which is sensitive to spin noise (in our case from unpaired electrons in free radicals), has also been used successfully for several applications, including the detection of different chemicals.20,27−30 Morita et al. recently performed the first free radical measurements within living yeast cells.31 While such demonstrations in cell cultures are essential, measurements in primary cells are a necessary step toward clinical applications for two reasons. First, this will enable measurements of free radical production in cells isolated from patients suffering from ROS-related diseases.32−35 Second, the variation in radical production differs a great deal among different types of immune cells (e.g., DC, macrophage, neutrophil) and this is important for their immune functions.36,37 This heterogeneity among cell types cannot be assessed in uniform cell lines.
In this paper, we investigate free radical generation following activation and inhibition of NOX2 during phagocytosis in primary human monocyte derived dendritic cells.
In order to perform relaxometry in cells (in this case specifically in phagosomes), diamond particles should be first internalized by cells. We first investigated the uptake ability of dendritic cells on bare FNDs. Human primary dendritic cells are able to uptake diamond particles, as shown in Figure 1a. Additionally, we used a liposome coating to facilitate the uptake and stabilize the particle size. Liposome coating was performed as described previously by Morita et al.38 To coat diamonds with a cationic liposome containing zwitterionic phosphatidylcholines, 1 mg/mL of FNDs from the stock were added to 1 mL of a liposomal formulation (9 μmol of cholesterol, 63 μmol of l-α-phosphatidylcholine (egg yolk), and 18 μmol of stearylamine dissolved in 1 mL of distilled water) to make a 2 μg/mL FND-liposome solution, followed by 30 s of vortexing. Liposome-coated diamond particles were characterized by dynamic light scattering (DLS) and cryo transmission electron microscopy (TEM). DLS data showed no differences in diamond sizes before and after liposome coating, TEM revealed a liposome layer thickness of 4.8 ± 1.2 nm on the surface of diamond particles. By using a liposome coating, we were able to track the movement of diamonds in cells. Confocal Z-stack images showed that, after 1 h of incubation, there are many diamond particles inside cells (Figure 1a). We found that coating FNDs with liposomes had several consequences. We found a slight aggregation of the FND, as shown in Figure 1b, which is actually beneficial for the spin measurements due to a slowdown in the movement speed of diamond particles inside cells. The effect of a liposome coating on diamond uptake is shown in Figure 1b. More liposome-coated FNDs are found inside cells in comparison to bare particles. These FNDs are mostly located close to the cell membrane after 1 h of incubation, as confirmed by Z-stack confocal images and phalloidin-FITC staining of the cell membrane. Figure 1c shows a comparison between the particle numbers.
Figure 1.
Diamond uptake by primary human DCs: (a) DCs incubated with 2 μg/mL bare FNDs or (b) 2 μg/mL liposome coated FNDs. (c) Quantitative analysis of FND uptake per cell. The experiment was repeated three times on cells from each donor, and three to four donors were tested; 50 cells were counted. Color code: green, Phalloidin-FITC, staining actin filaments (also known as F-actin); blue: DAPI (staining DNA); red: FNDs. Error bars stand for mean ± SD.
To measure radical production inside a cell, diamond particles should be in the region of interest. Thus, we evaluated whether the FNDs colocalize with endosomes/phagosomes. Bare FNDs or liposome-coated FNDs were internalized into human primary DCs (see Figure 1). Next, we investigated the intracellular location of diamond particles. To this end, pHrodo Green conjugated E. coli particles were used to label endophagosomes. These pHrodo Green conjugated E. coli particles were designed to observe the phagocytosis processes. The particles emit green fluorescence when they are in an acidic environment, as for example in phagosomes (pH 4.5).
Particles lack fluorescence outside the cell. Figure 2a shows that FNDs colocalize with pHrodo Green E. coli particles very well inside cells. Using Z-stack confocal images, diamonds were located within the cells and in endophagosomes after 1 h of incubation. These images were further deconvoluted, and statistical analysis was performed by the Huygens software. Manders coefficients (MCs) are well-established colocalization measures that calculate the percentage of the total signal from one channel which overlaps with the signal from the other channel.39,40 We found that M1 is 1.00 ± 0.01, indicating that FNDs are completely overlapping with pHrodo Green conjugates (M2 was 0.54 ± 0.26, since there are fewer diamonds than endosomes). Thus, we can conclude that FNDs were located inside endophagosomes.
Figure 2.

Subcellular location of FNDs revealed by confocal microscopy. (a) FNDs colocalize with pHrodo Green conjugate E. coli particles inside human primary DCs. DCs were incubated with pHrodo Green conjugate E. coli particles to stain endosomes and lysosomes, and FNDs were added separately. Color code: green, pHrodo Green conjugate E. coli particles; red, FNDs. The scale bar is 20 μm.
Cell viability was tested by an MTT assay (Figure 3). Cells were incubated with different concentrations of bare FNDs (1, 10, and 20 μg/mL), liposome-coated FNDs (FND final concentration was 2 μg/mL; this amount of liposome-coated FND solution is identical with what we used for T1 measurements), or 5% DMSO for 24 h. DMSO was used as a positive control, as it induces cell death. We found no differences between the control and the cells exposed to bare FNDs. The cell viability test indicated that both FNDs and liposome-coated FNDs show a very good cytocompatibility in human primary DCs.
Figure 3.
Cell viability test by an MTT assay. 100% represents a control with no exposure to FNDs. The experiment was repeated for cells from three donors, and error bars represent the standard deviations. The data were analyzed by using one-way ANOVA in comparison to the control group. ***p ≤ 0.001 is defined as significant.
NOX2 is the major enzyme that generates superoxide in immune cells. Zymosan A is often used to activate NOX2 at phagosomes and induce radical production in neutrophils and phagocytes.41,42 To investigate free radical generation inside dendritic cells, in this work, zymosan A was used to activate the NOX2 complex in the phagosome.42 NV centers are only sensitive to spins within a few nanometers. Thus, when FNDs are used to measure radical generation, they should be located within a few nanometers from the region of interest within a cell.
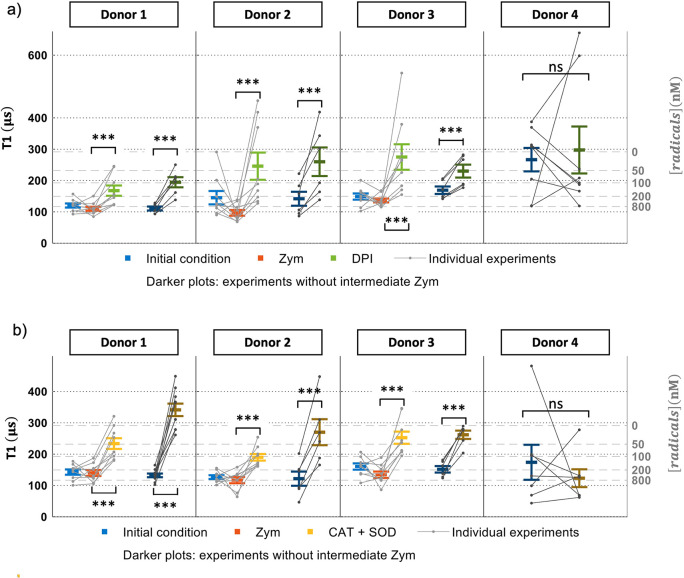
In our T1 experiment, cells were first incubated with liposome-coated FNDs for 1 h. After that, we collected initial T1 values, followed by stimulation with zymosan A. The resulting T1 values are shown in Figure 4. There is no difference in T1 values between the initial measurements and those after addition of zymosan A, which implies that our FNDs (or more accurately bacterial contaminations from nonsterile production; see Figure S8 for a proof of endotoxin contaminants) might already induce free radical generation by activating phagosomes. As a result, phagosomes already contain free radicals. To further investigate this, we produced endotoxin-free particles by harsh oxidation of all organic material on the surface. The results are shown in Figure S7. In this case we indeed observe the increase in free radical production after addition of zymosan A that results from the activation of cells.
Figure 4.
Free radical generation in single human DCs from each donor determined by T1. Prior to T1 measurements FNDs were ingested by cells. The gray lines connect data points from an experiment. Every gray sequence was measured in a single particle within a single cell and repeated six to nine independent times for each donor. The different lines were recorded from a different cell and a different particle from the same donor. The averages from multiple particles for each donor are shown in color. (a) Initial T1 values were collected from cells, and then zymosan (zym) was added and T1 was recorded. Finally, DPI was added. In (b) the same sequence is performed as in (a), with the difference being that SOD/CAT was added instead of DPI. For donors 1–3, T1 measurements were performed with or without zymosan A stimulation. For donor 4, T1 measurements were performed without zymosan A. Each T1 measurement takes 10 min. Error bars represent mean ± standard deviation, and data between each group were analyzed by a paired t test: ns,no significant difference. ***p < 0.0001 represents a significant difference.
DPI, a NOX inhibitor,42 was used to inhibit NOX2 in this work. In presence of DPI, T1 increased by 166 ± 82% in comparison to the initial T1 values in Figure 4a. This indicates that fewer radicals are generated. As another control, we added the antioxidant enzymes SOD and CAT. When NOX2 is active, the phagosomes start to produce O2•– when there is oxygen; SOD and CAT can convert this radical to H2O2. Thus, we expect an increase in T1 after addition of SOD and CAT.43,44
In Figure 4b, after SOD and CAT were added to the sample, radical production dropped by 157 ± 37% in comparison to the initial T1 values. From T1 values we estimated free radical concentrations on the basis of a calibration conducted with known concentrations of radicals under controlled conditions.45 We found concentrations in the nanomolar range. It was reported that human monocyte derived DCs can produce 0.6 mM/s of ROS in phagosomes during phagocytosis.41 It is known that only a small fraction of ROS are radicals, which agrees well with what we found.44
To check if our diamonds are activating NOX2, T1 measurements were performed without using zymosan A. Here we added SOD, CAT, and DPI to inhibit NOX2 directly after the initial measurement (see darker graphs of Figure 4). In Figure 5, cells show the same response as was found after the stimulation of zymosan A. Of note, Figure 4 shows T1 measurements collected from cells of four different donors, and data of T1 measurement from these four donors showed the same response to zymosan A, DPI, and SOD/CAT. In Figure S1a–c, we tested the effect of zymosan A and SOD/CAT alone (without cells) on diamond (See Figure S2). There are no differences between the initial T1 and T1 after the chemicals were added. Another control we performed was to measure radical formation outside the cells. In this case we did not see any significant changes (see Figure S6).
Figure 5.
Cellular ROS measurement by a DCFDA assay. (a) Cellular ROS measurements after exposure to stimuli or inhibitors. (b) Cellular ROS measurements after cells are stimulated by FNDs or zymosan A for 2 h. Legend: control, cells without DCFDA staining; initial, cells without any treatment but stained with DCFDA; DPI, diphenyleneiodonium chloride (final concentration 50 μM/mL); SOD, superoxide dismutase (final concentration 1000 U/mL); CAT, catalase (final concentration 600 U/mL). The experiment was repeated three to four times on cells from three donors. Error bars represent mean ± standard deviation. The data were analyzed by a paired t test between each group, and *p < 0.05, **p < 0.01, and ***p < 0.001 represent significant differences.
While cells of three of the donors were essentially the same in their response to stimuli or inhibitors, one was clearly different. As Figure 4 shows, cells from this donor do not respond to DPI. In the presence of SOD/CAT (Figure 4), we did not see a significant difference as in the other donors. Similarly, large variability between donors was found for ROS production in bulk experiments.36 We used DCFDA, a fluorescent dye, to measure the total amount of cellular reactive oxygen species (ROS) production. The result of the DCFDA assay shows a similar response to different chemicals. After incubation with FNDs, the total amount of ROS increased in comparison to the initial measurement. When cells are incubated with bare FNDs, there is a slight difference (p < 0.001) from the initial groups, which indicates that FNDs (or more accurately contaminants within them) can trigger an immune response and thus produce ROS. There is also no difference between the FND group and the group after adding zymosan A. The cells respond similarly to DPI and SOD/CAT as in our T1 measurements. After cells are treated with DPI and SOD/CAT, ROS generation is significantly decreased. In comparison to our T1 measurements the DCFDA assay is a traditional method which detects ROS from an ensemble of cells. Additionally, this assay is not designed for long-term measurement, as the dye bleaches and reveals the history of a sample rather than the current state. Finally, the DCFDA assay is not specific for radicals but rather detects the sum of all ROS (including radicals and nonradicals). To address whether adding FNDs resulted in the activation of DCs, we tested for cytokine production (see Figure S3 in the Supporting Information). On the basis of the ELISA results, FND does not show to any proinflammatory phenotype in DCs (based on IL6 and TNF-α) and it does not activate the inflammasomal pathway (based on IL1beta (IL1β)). More than any other cytokine family, the interleukin-1 family members are closely linked to damaging inflammation; however, the same members also function to increase nonspecific resistance to infection and development of the immune response to foreign antigens.46,47 It is likely to see rapid release of large quantities of active IL-1β directly across a disintegrating plasma membrane.48 Whether cell membranes were still intact after incubation with FNDs for 24 or 48 h was checked by a LIVE/DEAD Viability/Cytotoxicity Kit (see Figure S4). We did not observe any signs that the membrane integrity was compromised in the experiments.
Finally, we also eliminated the concern of a potential temperature increase during the measurement. To exclude this, we measured the temperature in cells before and after a T1 measurement with a thermal camera (see Figure S5). We did not observe any temperature changes within the accuracy of the instrument.
We have demonstrated the first free radical measurements with relaxometry in primary human DCs. In contrast to previous work on cell lines, primary cells more closely resemble the in vivo situation. Additionally, it is only possible in primary cells to determine heterogeneity between donors as well as different cells from the same donor, which we observed here. While traditional fluorescent probes for ROS measurements reveal the overall concentration of ROS (dominated by more abundant nonradicals), our method is specific to radicals, because they contain an unpaired electron. Given that free radicals are the most reactive molecules, these have the greatest potential to damage biomolecules and thus might be a good diagnostic indicator. Our technique is able to measure the current radical load, while ROS probes reveal the history of the sample. With our system, we can follow an increase or decrease in radical concentrations in a phagosome in single cells with subcellular resolution in real time. This is, for instance, relevant for identifying sources of radical production. This can be a specific cell within a population or even a specific organelle within a cell. Since we provide single-particle measurements, it is also possible to identify heterogeneities between different cells from one sample. While these findings are interesting from a fundamental standpoint, they might also aid drug development if their working mechanism or mechanisms of disease can be understood better. The fact that it is possible to follow individual cells over the course of an experiment is a powerful tool to differentiate between the biological variability of the original cells (and particles) and the effect of the intervention. In sum, we have provided unique data by our technique, which has distinct advantages over the state of the art conventional techniques.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.1c03021.
Additional details of experimental materials and methods, control experiment of T1 measurements on bare FND particles, and cytokine production assay results (PDF)
Author Contributions
L.N. designed the study under the supervision of R.S. L.N. and A.C.N. conceived the study and acquired and analyzed the data. L.N., A.C.N., and R.S. wrote the manuscript. V.G.D. and T.H. assisted with some T1 measurements. M.B. prepared human primary DCs, performed all the ELISA measurement, and analyzed data in this work under the supervision of G.v.d.B. M.C. processed T1 data. C.R,-S,-M, performed the endotoxin test. M.G. and C.P.E. prepared endotoxin-free FNDs. P.C. supervised this preparation.
Author Contributions
⊥ L.N. and A.C,N. contributed equally.
This work was financially supported by an ERC starting grant (ERC-2016-STG Stress Imaging 714289). We also thank the China Scholarship Council for supporting us with a scholarship (No.201706170089) as well as the Stichting De Cock-Hadders (projectnumber 2021–06) for L.N. A.C.N. acknowledges the Kolff Institute for her Ph.D. scholarship. Confocal images shown in this paper were acquired from the UMCG Imaging and Microscopy Center (UMIC) under NWO grant 175-010-2009-023 for imaging work in the paper. The work was further supported by the Czech Science Foundation Project No. 18-17071S (to P.C.), MSM Project No. 8C18004 (NanoSpin) (to P.C.), and European Regional Development Fund, OP RDE, Projects: ChemBioDrug (No. CZ.02.1.01/0.0/0.0/16_019/0000729) (to P.C.) and CARAT (No. CZ.02.1.01/0.0/0.0/16_026/0008382) (to M.G. and P.C.). M.C. thanks the Swiss National Science Foundation for supporting this work with grant PZ00P2 185824. The authors are grateful to Dr. Jan Stursa for irradiation of the FNDs.
The authors declare no competing financial interest.
Supplementary Material
References
- Lobo V.; Patil A.; Phatak A.; Chandra N. N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4 (8), 118. 10.4103/0973-7847.70902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight J. A. Review: Free radicals, antioxidants, and the immune system. Annals of Clinical & Laboratory Science. 2000, 30 (2), 145–158. [PubMed] [Google Scholar]
- Cechim G.; Chies J. A. In vitro generation of human monocyte-derived dendritic cells methodological aspects in a comprehensive review. An. Acad. Bras. Cienc. 2019, 91 (4), 1. 10.1590/0001-3765201920190310. [DOI] [Google Scholar]
- Karlsson A.; Nygren E.; Karlsson J.; Nordström I.; Dahlgren C.; Eriksson K. Ability of Monocyte-Derived Dendritic Cells To Secrete Oxygen Radicals in Response to Formyl Peptide Receptor Family Agonists Compared to That of Myeloid and Plasmacytoid Dendritic Cells. Clin. Vaccine Immunol. 2007, 14 (3), 328–330. 10.1128/CVI.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton M. B.; Kettle A. J.; Winterbourn C. C. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 1998, 92 (9), 3007–3017. 10.1182/blood.V92.9.3007.421k47_3007_3017. [DOI] [PubMed] [Google Scholar]
- Hardy M.; Zielonka J.; Karoui H.; Sikora A.; Michalski R.; Podsiadły R.; Lopez M.; Vasquez-Vivar J.; Kalyanaraman B.; Ouari O. Detection and Characterization of Reactive Oxygen and Nitrogen Species in Biological Systems by Monitoring Species-Specific Products. Antioxid. Redox Signaling 2018, 28 (15), 1416–1432. 10.1089/ars.2017.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz H. M.; Khan N.; Khramtsov V. V. Use of Electron Paramagnetic Resonance Spectroscopy to Evaluate the Redox State In Vivo. Antioxid. Redox Signaling 2007, 9 (10), 1757–1772. 10.1089/ars.2007.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardman P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: Progress, pitfalls, and prospects. Free Radical Biol. Med. 2007, 43 (7), 995–1022. 10.1016/j.freeradbiomed.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Yi K.; Lin Q.; Yang L.; Chen X.; Chen H.; Liu Y.; Wei D. Free radical sensors based on inner-cutting graphene field-effect transistors. Nat. Commun. 2019, 10 (1), 1–10. 10.1038/s41467-019-09573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel L.; Wang Z.; Tschudin M. A.; Rohner D.; Gutiérrez-Lezama I.; Ubrig N.; Gibertini M.; Giannini E.; Morpurgo A. F.; Maletinsky P. Probing magnetism in 2D materials at the nanoscale with single-spin microscopy. Science 2019, 364 (6444), 973–976. 10.1126/science.aav6926. [DOI] [PubMed] [Google Scholar]
- Juraschek D. M.; Meier Q. N.; Trassin M.; Trolier-McKinstry S. E.; Degen C. L.; Spaldin N. A. Dynamical Magnetic Field Accompanying the Motion of Ferroelectric Domain Walls. Phys. Rev. Lett. 2019, 123 (12), 127601. 10.1103/PhysRevLett.123.127601. [DOI] [PubMed] [Google Scholar]
- Grinolds M. S.; Hong S.; Maletinsky P.; Luan L.; Lukin M. D.; Walsworth R. L.; Yacoby A. Nanoscale magnetic imaging of a single electron spin under ambient conditions. Nat. Phys. 2013, 9 (4), 215–219. 10.1038/nphys2543. [DOI] [Google Scholar]
- Cujia K. S.; Boss J. M.; Herb K.; Zopes J.; Degen C. L. Tracking the precession of single nuclear spins by weak measurements. Nature 2019, 571 (7764), 230–233. 10.1038/s41586-019-1334-9. [DOI] [PubMed] [Google Scholar]
- Mamin H. J.; Kim M.; Sherwood M. H.; Rettner C. T.; Ohno K.; Awschalom D. D.; Rugar D. Nanoscale Nuclear Magnetic Resonance with a Nitrogen-Vacancy Spin Sensor. Science 2013, 339 (6119), 557–560. 10.1126/science.1231540. [DOI] [PubMed] [Google Scholar]
- Müller C.; Kong X.; Cai J. M.; Melentijević K.; Stacey A.; Markham M.; Twitchen D.; Isoya J.; Pezzagna S.; Meijer J.; Du J. F.; et al. Nuclear magnetic resonance spectroscopy with single spin sensitivity. Nat. Commun. 2014, 5 (1), 1–6. 10.1038/ncomms5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan K.; Hasani M.; Zheng T.; Schirhagl R. Nanodiamonds for In Vivo Applications. Small 2018, 14 (19), 1703838. 10.1002/smll.201703838. [DOI] [PubMed] [Google Scholar]
- Chipaux M.; van der Laan K. J.; Hemelaar S. R.; Hasani M.; Zheng T.; Schirhagl R. Nanodiamonds and Their Applications in Cells. Small 2018, 14 (24), 1704263. 10.1002/smll.201704263. [DOI] [PubMed] [Google Scholar]
- Faklaris O.; Joshi V.; Irinopoulou T.; Tauc P.; Sennour M.; Girard H.; Gesset C.; Arnault J. C.; Thorel A.; Boudou J. P.; Curmi P. A.; et al. Photoluminescent diamond nanoparticles for cell labeling: study of the uptake mechanism in mammalian cells. ACS Nano 2009, 3 (12), 3955–3962. 10.1021/nn901014j. [DOI] [PubMed] [Google Scholar]
- Faklaris O.; Garrot D.; Joshi V.; Druon F.; Boudou J. P.; Sauvage T.; Georges P.; Curmi P. A.; Treussart F. Detection of single photoluminescent diamond nanoparticles in cells and study of the internalization pathway. Small 2008, 4 (12), 2236–2239. 10.1002/smll.200800655. [DOI] [PubMed] [Google Scholar]
- Zhang T.; Pramanik G.; Zhang K.; Gulka M.; Wang L.; Jing J.; Chu Z.; et al. Toward Quantitative Bio-sensing with Nitrogen-Vacancy Center in Diamond. ACS sensors. 2021, 6 (6), 2077–2107. 10.1021/acssensors.1c00415. [DOI] [PubMed] [Google Scholar]
- Ermakova A.; Pramanik G.; Cai J. M.; Algara-Siller G.; Kaiser U.; Weil T.; Tzeng Y. K.; Chang H. C.; McGuinness L. P.; Plenio M. B.; Naydenov B.; et al. Detection of a few metallo-protein molecules using color centers in nanodiamonds. Nano Lett. 2013, 13 (7), 3305–3309. 10.1021/nl4015233. [DOI] [PubMed] [Google Scholar]
- Wang P.; Chen S.; Guo M.; Peng S.; Wang M.; Chen M.; Ma W.; Zhang R.; Su J.; Rong X.; Shi F.; et al. Nanoscale magnetic imaging of ferritins in a single cell. Science advances. 2019, 5 (4), eaau8038 10.1126/sciadv.aau8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert S.; Ziem F.; Hall L. T.; Zappe A.; Schweikert M.; Götz N.; Aird A.; Balasubramanian G.; Hollenberg L.; Wrachtrup J. Magnetic spin imaging under ambient conditions with sub-cellular resolution. Nat. Commun. 2013, 4 (1), 1–6. 10.1038/ncomms2588. [DOI] [PubMed] [Google Scholar]
- Le Sage D.; Arai K.; Glenn D. R.; DeVience S. J.; Pham L. M.; Rahn-Lee L.; Lukin M. D.; Yacoby A.; Komeili A.; Walsworth R. L. Optical magnetic imaging of living cells. Nature 2013, 496 (7446), 486–489. 10.1038/nature12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness L. P.; Yan Y.; Stacey A.; Simpson D. A.; Hall L. T.; Maclaurin D.; Prawer S.; Mulvaney P.; Wrachtrup J.; Caruso F.; Scholten R. E.; et al. Quantum measurement and orientation tracking of fluorescent nanodiamonds inside living cells. Nat. Nanotechnol. 2011, 6 (6), 358–363. 10.1038/nnano.2011.64. [DOI] [PubMed] [Google Scholar]
- Kucsko G.; Maurer P. C.; Yao N. Y.; Kubo M.; Noh H. J.; Lo P. K.; Park H.; Lukin M. D. Nanometre-scale thermometry in a living cell. Nature 2013, 500 (7460), 54–58. 10.1038/nature12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetienne J. P.; Hingant T.; Rondin L.; Cavaillès A.; Mayer L.; Dantelle G.; Gacoin T.; Wrachtrup J.; Roch J. F.; Jacques V. Spin relaxometry of single nitrogen-vacancy defects in diamond nanocrystals for magnetic noise sensing. Phys. Rev. B: Condens. Matter Mater. Phys. 2013, 87 (23), 235436. 10.1103/PhysRevB.87.235436. [DOI] [Google Scholar]
- Hopper D. A.; Grote R. R.; Parks S. M.; Bassett L. C. Amplified Sensitivity of Nitrogen-Vacancy Spins in Nanodiamonds Using All-Optical Charge Readout. ACS Nano 2018, 12 (5), 4678–4686. 10.1021/acsnano.8b01265. [DOI] [PubMed] [Google Scholar]
- Pelliccione M.; Myers B. A.; Pascal L. M. A.; Das A.; Jayich A. B. Two-Dimensional Nanoscale Imaging of Gadolinium Spins via Scanning Probe Relaxometry with a Single Spin in Diamond. Physical Review Applied 2014, 2 (5), 054014. 10.1103/PhysRevApplied.2.054014. [DOI] [Google Scholar]
- Barton J.; Gulka M.; Tarabek J.; Mindarava Y.; Wang Z.; Schimer J.; Raabova H.; Bednar J.; Plenio M. B.; Jelezko F.; Nesladek M.; et al. Nanoscale Dynamic Readout of a Chemical Redox Process Using Radicals Coupled with Nitrogen-Vacancy Centers in Nanodiamonds. ACS Nano 2020, 14 (10), 12938–12950. 10.1021/acsnano.0c04010. [DOI] [PubMed] [Google Scholar]
- Morita A.; Nusantara A. C.; Martinez F. P.; Hamoh T.; Damle V. G.; van der Laan K. J.; Sigaeva A.; Vedelaar T.; Chang M.; Chipaux M.; Schirhagl R.. Quantum monitoring the metabolism of individual yeast mutant strain cells when aged, stressed or treated with antioxidant. arXiv preprint arXiv:2007.16130. 2020. Jul 31.
- Deffert C.; Carnesecchi S.; Yuan H.; Rougemont A. L.; Kelkka T.; Holmdahl R.; Krause K. H.; Schäppi M. G. Hyperinflammation of chronic granulomatous disease is abolished by NOX2 reconstitution in macrophages and dendritic cells†. Journal of pathology 2012, 228 (3), 341–350. 10.1002/path.4061. [DOI] [PubMed] [Google Scholar]
- Olagnier D.; Peri S.; Steel C.; van Montfoort N.; Chiang C.; Beljanski V.; Slifker M.; He Z.; Nichols C. N.; Lin R.; Balachandran S.; et al. Cellular Oxidative Stress Response Controls the Antiviral and Apoptotic Programs in Dengue Virus-Infected Dendritic Cells. PLoS Pathog. 2014, 10 (12), e1004566 10.1371/journal.ppat.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Shi M.; Qiu Q.; Huang M.; Zeng S.; Zou Y.; Zhan Z.; Liang L.; Yang X.; Xu H. Piperlongumine Suppresses Dendritic Cell Maturation by Reducing Production of Reactive Oxygen Species and Has Therapeutic Potential for Rheumatoid Arthritis. J. Immunol. 2016, 196 (12), 4925–4934. 10.4049/jimmunol.1501281. [DOI] [PubMed] [Google Scholar]
- Grassi F.; Tell G.; Robbie-Ryan M.; Gao Y.; Terauchi M.; Yang X.; Romanello M.; Jones D. P.; Weitzmann M. N.; Pacifici R. Oxidative stress causes bone loss in estrogen-deficient mice through enhanced bone marrow dendritic cell activation. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (38), 15087–15092. 10.1073/pnas.0703610104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantegazza A. R.; Savina A.; Vermeulen M.; Pérez L.; Geffner J.; Hermine O.; Rosenzweig S. D.; Faure F.; Amigorena S. NADPH oxidase controls phagosomal pH and antigen cross-presentation in human dendritic cells. Blood 2008, 112 (12), 4712–4722. 10.1182/blood-2008-01-134791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paardekooper L. M.; Dingjan I.; Linders P. T.; Staal A. H.; Cristescu S. M.; Verberk W. C.; van den Bogaart G. Human Mono-cyte-Derived Dendritic Cells Produce Millimolar Concentra-tions of ROS in Phagosomes Per Second. Front. Immunol. 2019, 10, 1216. 10.3389/fimmu.2019.01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita A.; Martinez F. P. P.; Chipaux M.; Jamot N.; Hemelaar S. R.; van der Laan K. J.; Schirhagl R. Cell Uptake of Lipid-Coated Diamond. Part. Part. Syst. Charact. 2019, 36 (8), 1900116. 10.1002/ppsc.201900116. [DOI] [Google Scholar]
- Pike J. A.; Styles I. B.; Rappoport J. Z.; Heath J. K. Quantifying receptor trafficking and colocalization with confocal microscopy. Methods 2017, 115, 42–54. 10.1016/j.ymeth.2017.01.005. [DOI] [PubMed] [Google Scholar]
- McDonald J. H.; Dunn K. W. Statistical tests for measures of colocalization in biological microscopy. J. Microsc. 2013, 252 (3), 295–302. 10.1111/jmi.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paardekooper L. M.; Dingjan I.; Linders P. T.; Staal A. H.; Cristescu S. M.; Verberk W. C.; van den Bogaart G. Human Monocyte-Derived Dendritic Cells Produce Millimolar Concentrations of ROS in Phagosomes Per Second. Front. Immunol. 2019, 10, 1216. 10.3389/fimmu.2019.01216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y.; Park J.; Lee J. E.; Yenari M. A. NOX inhibitors - A promising avenue for ischemic stroke. Experimental neurobiology 2017, 26 (4), 195–205. 10.5607/en.2017.26.4.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singel K. L.; Segal B. H. NOX2-dependent regulation of inflammation. Clin. Sci. 2016, 130 (7), 479–490. 10.1042/CS20150660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herb M.; Schramm M. Functions of ROS in macrophages and antimicrobial immunity.. Antioxidants 2021, 10 (2), 313. 10.3390/antiox10020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona Martínez F.; Nusantara A. C.; Chipaux M.; Padamati S. K.; Schirhagl R. Nanodiamond relaxometry-based detection of free-radical species when produced in chemical reactions in biologically relevant conditions. ACS sensors 2020, 5 (12), 3862–3869. 10.1021/acssensors.0c01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H.; Jones D. P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21 (7), 363–383. 10.1038/s41580-020-0230-3. [DOI] [PubMed] [Google Scholar]
- Dinarello C. A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunological reviews 2018, 281 (1), 8–27. 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Castejon G.; Brough D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22 (4), 189–195. 10.1016/j.cytogfr.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.