Abstract
Surface-enhanced Raman scattering (SERS) is a powerful technique for sensitive label-free analysis of chemical and biological samples. While much recent work has established sophisticated automation routines using machine learning and related artificial intelligence methods, these efforts have largely focused on downstream processing (e.g., classification tasks) of previously collected data. While fully automated analysis pipelines are desirable, current progress is limited by cumbersome and manually intensive sample preparation and data collection steps. Specifically, a typical lab-scale SERS experiment requires the user to evaluate the quality and reliability of the measurement (i.e., the spectra) as the data are being collected. This need for expert user-intuition is a major bottleneck that limits applicability of SERS-based diagnostics for point-of-care clinical applications, where trained spectroscopists are likely unavailable. While application-agnostic numerical approaches (e.g., signal-to-noise thresholding) are useful, there is an urgent need to develop algorithms that leverage expert user intuition and domain knowledge to simplify and accelerate data collection steps. To address this challenge, in this work, we introduce a machine learning-assisted method at the acquisition stage. We tested six common algorithms to measure best performance in the context of spectral quality judgment. For adoption into future automation platforms, we developed an open-source python package tailored for rapid expert user annotation to train machine learning algorithms. We expect that this new approach to use machine learning to assist in data acquisition can serve as a useful building block for point-of-care SERS diagnostic platforms.
Keywords: Diagnostics, automation, plasmonics, surface-enhanced Raman scattering, SERS, artificial intelligence, XGBoost
Introduction
Surface-enhanced Raman scattering (SERS) is a powerful label-free detection and analysis technique that exploits the near-field enhancement of inelastically scattered Raman signal via nanostructured plasmonic surfaces.1 SERS is highly sensitive, capable of even single molecule detection, with broad applicability in detection and monitoring of disease, particularly for cancer. While many proof-of-concept SERS studies emerge annually, and technologies to enable point-of-use and even wearable devices are now a reality, widespread adoption of the technique to replace or supplement existing sensing platforms has not come to fruition. A major bottleneck of this goal is that application of SERS currently requires expert users to collect and interpret data.
In a typical SERS data acquisition process, whether it is a clinical diagnostic platform or a characterization of an unknown chemical entity, hundreds to thousands of spectra are typically collected, preprocessed, and subjected to downstream analyses, e.g., principal component analysis (PCA), hierarchical clustering, or other types of classification routines. Much literature has been devoted to the pre-processing considerations,2–4 including de-noising, smoothing, baseline correction algorithms, background subtraction methods, and cosmic ray removal.
Machine learning (ML) and artificial intelligence (AI) methods (e.g., convolutional neural networks, or CNNs, deep neural networks, random forest classifiers, etc.) have been widely applied to various classification tasks following preprocessing. For instance, such methods have enabled classification of small molecules5 and their mixtures,6 various minerals,7 bacteria,8,9 and viruses.10 Discrimination of esophageal cancer,11 non-small-cell lung cancer,12 and nasopharyngeal and liver cancer,13 has also been demonstrated. CNNs have been applied to Raman/SERS spectra of circulating biomarkers as well, such as extracellular vesicles (EVs) in prostate,14 lung,15 and pancreatic cancer,16 as well as general cancer biomarker identification.17 Diabetes mellitus detection,18 applications in cytopathology,19 AI-based discrimination of tumor suppressor genes,20 nitroxoline quantification,21 and caffeine and associated metabolites detection22 have also been proposed. Overall, many ML algorithms have emerged to complement or replace traditional methods (e.g., multivariate classification) for data analysis in vibrational spectroscopy.
While it is apparent that ML greatly improves prediction accuracy and automated spectral processing improves the efficiency of SERS platforms in general, we posit that progress in developing SERS-based diagnostics is not limited by the lack of state-of-the-art ML algorithms, but instead by the absence of automated data collection and sampling protocols. For example, following spectral preprocessing steps, the user has to decide which spectra are adequate for further downstream analyses (e.g., biological sample classification for diagnostic purposes). A question remains at this stage, whether the analyte of interest and the SERS substrate have been sampled exhaustively enough to produce meaningful and statistically representative data. This step arguably creates the largest barriers to automation of SERS platforms as it (1) requires significant user expertise and domain knowledge, (2) assumes minimal user bias, and (3) relies on several related, but not identical measurements. Recognizing the ability of ML algorithms to translate user intuition to diverse classification problems,23,24 it is clear that ML methods will provide high value to aid in such expert-driven sampling decisions, even during measurement. To the best of our knowledge, such approaches for SERS data collections have not yet been reported in the field.
Surface-enhanced Raman spectroscopy data are highly dynamic in nature,1,25–27 manifesting in the heterogeneous fluctuation of spectra, even for a single analyte measured on a high-quality, geometrically ordered substrate.28,29 For typical measurements, multiple spots need to be sampled many times to account for heterogeneity, arising from pre-measurement parameters (sample exposure time, data collection frequency, laser power, etc.), spatial differences in analyte concentration and orientation, ionic composition of the solution, osmotic and elastic potentials and material-related heterogeneities of the SERS substrate itself, impurities present on the surface,30–32 etc. These issues, unfortunately, have led to doubt in the ability to perform truly quantitative SERS.1,33–38
In light of the above discussion, the main objective of this work is to develop a robust and automated ML-SERS approach to ‘‘sufficiently’’ sample the substrate, i.e., to automatically collect a statistically representative quantity of high-quality spectra for a given substrate and analyte(s). Such an approach offers minimal operator intervention for SERS spectra acquisition, increasing the efficiency of measurement and reproducibility of the downstream analyses.
The hierarchical data sampling scheme currently used in SERS experiments (Fig. S1, Supplemental Material) is designed to collect representative spectra with high signal-to-noise ratios. For a given sample, spectra are collected at separate spots (e.g., x, y coordinates) to capture the spectral diversity of the sample. To increase the signal-to-noise ratio and reduce variance, multiple spectra at each spot are typically collected and averaged. By excluding negative spectra from the averaging, the signal-to-noise ratio can be increased. Manual exclusion of negative spectra can be accomplished by an expert but is cumbersome due to the thousands of spectra generated in a typical SERS experiment. Automatic ‘‘bad’’ (i.e., negative) spectra identification is thus a significant objective to improving SERS experimental data. As a valuable step towards this goal, in this study, we develop a suite of ML algorithms to classify spectra as either ‘‘good’’ or ‘‘bad’’ (i.e., negative) and critically assess their performance. The highest performing XGBoost model was identified and utilized to characterize both in-sample and out-of-sample datasets. This model was then utilized to characterize an out-of-sample dataset and offer the potential for automated data collection, removing the need to monitor the collection procedure completely.
Methods
Sample Preparation
Two commercial plasmonic substrates were chosen for this study, from Moxtek (Moxtek Inc., USA) and Plasmore (Plasmore S.R.L, Italy). A well-characterized SERS standard reporter, Rhodamine 6G (R6G), was selected as a model compound for surface scanning. Two different concentrations were prepared in ultrapure water to demonstrate a high (3 mM) and low (10 nM) R6G concentration. The plasmonic substrates were characterized by scanning electron microscope (SEM), using a ThermoFisher Quattro S (ThermoFisher Scientific, USA). For SEM measurement, substrates were mounted on metal studs using two-sided black carbon tape, and the following imaging parameters were applied: working distance 11.4−12.0 mm, spot size 2.5, accelerating voltage 10.0 kV, and chamber pressure 100 Pa.
SERS Spectra Acquisition
The SERS spectra were acquired using a custom-built inverted Raman scanning confocal microscope with an excitation wavelength of 785 nm and a 60×, 1.2 numerical aperture (NA) water immersion objective on an inverted IX73 Olympus microscope. Raman spectra were captured via an Andor Kymera3281-C spectrophotometer and Newton DU920P-BR-DD charge-coupled device (CCD) camera. Initial in situ data processing and cosmic ray removal were carried out using Solis v.4.31.30005.0 software. All SERS measurements were acquired using exposure time 1 s per scan with a laser power of ~10−20 mW. Moxtek or Plasmore substrates were scanned on a 20 × 20 pixels area thus yielding total 400 spectra per one scanned area. The step size was adjusted to 400 nm, resulting in the total scanned area of 8 µm × 8 µm. To simulate a real scanning procedure performed by a non-trained operator, the scanned areas were selected randomly without any pre-search for ‘‘good’’ signals. Unless elsewhere otherwise described SERS spectra preprocessing was performed using custom scripts written in Matlab v.2020a (The Mathworks, Inc.). Spectral preprocessing included penalized least-squares (PLS) background correction, smoothing, and normalization. Where stated throughout the study, these preprocessed spectral sets were further subjected to principal component analysis (PCA) based on the corresponding Matlab built-in functions.
Results
Sample Collection
High and low concentrations of a common SERS-active reporter molecule, R6G, were dried out on to two high-quality, lithographically formed commercial SERS substrates (Moxtek and Plasmore), as schematized in Fig. 1. SEM micrographs displaying the plasmonic nanostructures on either surface are shown in Fig. 1b. In total, five samples were prepared. Two different concentrations of high 3 mM and low 10 nM R6G concentrations were prepared on either Moxtek or Plasmore substrates. Substrates were either scanned using 10 mW laser power for high concentration or 20 mW laser power for low concentration. A fifth sample was created to investigate the effect of laser power on the recorded SERS signals, therefore a ‘‘low’’ concentration Plasmore was also scanned using lower 10mW power. Prepared substrates were subjected to SERS measurements using a custom confocal scanning Raman microscope to yield several random 20 × 20-pixel areas (total scanned area of 8 µm × 8 µm). Figure 1c shows a representative spectra average and standard for high concentration of R6G deposited on a Moxtek substrate.
Figure 1.
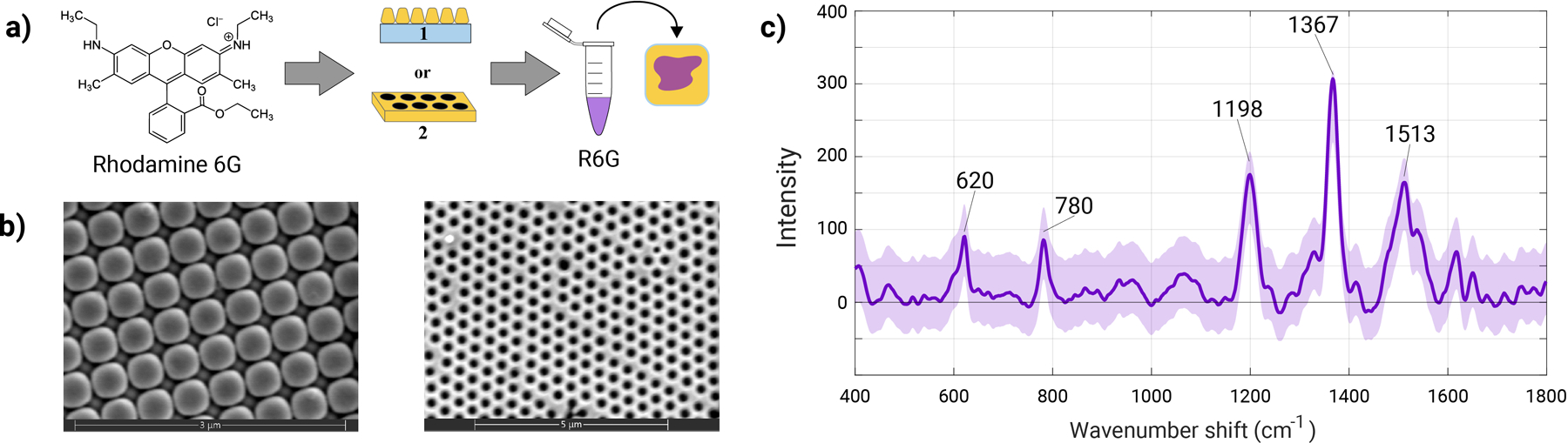
Experimental workflow of the ML-SERS platform. (a) Rhodamine 6G was used as a SERS reported molecule on plasmonic Moxtek (1) or Plasmore (2) substrates. Varying solutions of R6G were pipetted (~20 µL total) onto the surface and 20 × 20 pixels surface areas were scanned. (b) SEM micrographs illustrate the structure of the Moxtek and Plasmore substrates. (c) Representative SERS spectrum of R6G; the highlighted peaks at 620, 780, 1198, 1367, and 1513 cm−1 are characteristic spectral features of R6G.
A conventional approach to classify spectral data is to carry out PCA following manual selection of quality spectra (and/or through iterative use of PCA to screen out low quality or outlier data). An example of this process is illustrated in Fig. S2, Supplemental Material. Use of thresholds or intuitive interpretation using PC score plot and principal component loading spectra are relatively systematic methods to guide the spectra selection procedure. However, for more complex datasets featuring mixtures of chemicals, where the PCs do not cleanly correspond to single entities, it is tedious and time-consuming to apply such manual selection routines for hundreds or thousands of spectra, and further adds a notable source of inter-operator bias. Therefore, we endeavored to explore application of ML algorithms to recognize quality spectra following expert user training.
Data Organization and Parsing for ML Input
To utilize the spectra data for ML, we established a python-based data pipeline that converts plaintext Raman spectrum files as input, preprocesses the spectra, and converts them into a binary format that can be utilized for visualization, data labeling, and model training. The three stages of the data pipeline are shown in Fig. 2. All code utilized for this data pipeline is available under the open-source MIT license on GitHub (see Data Accessibility Statement).
Figure 2.
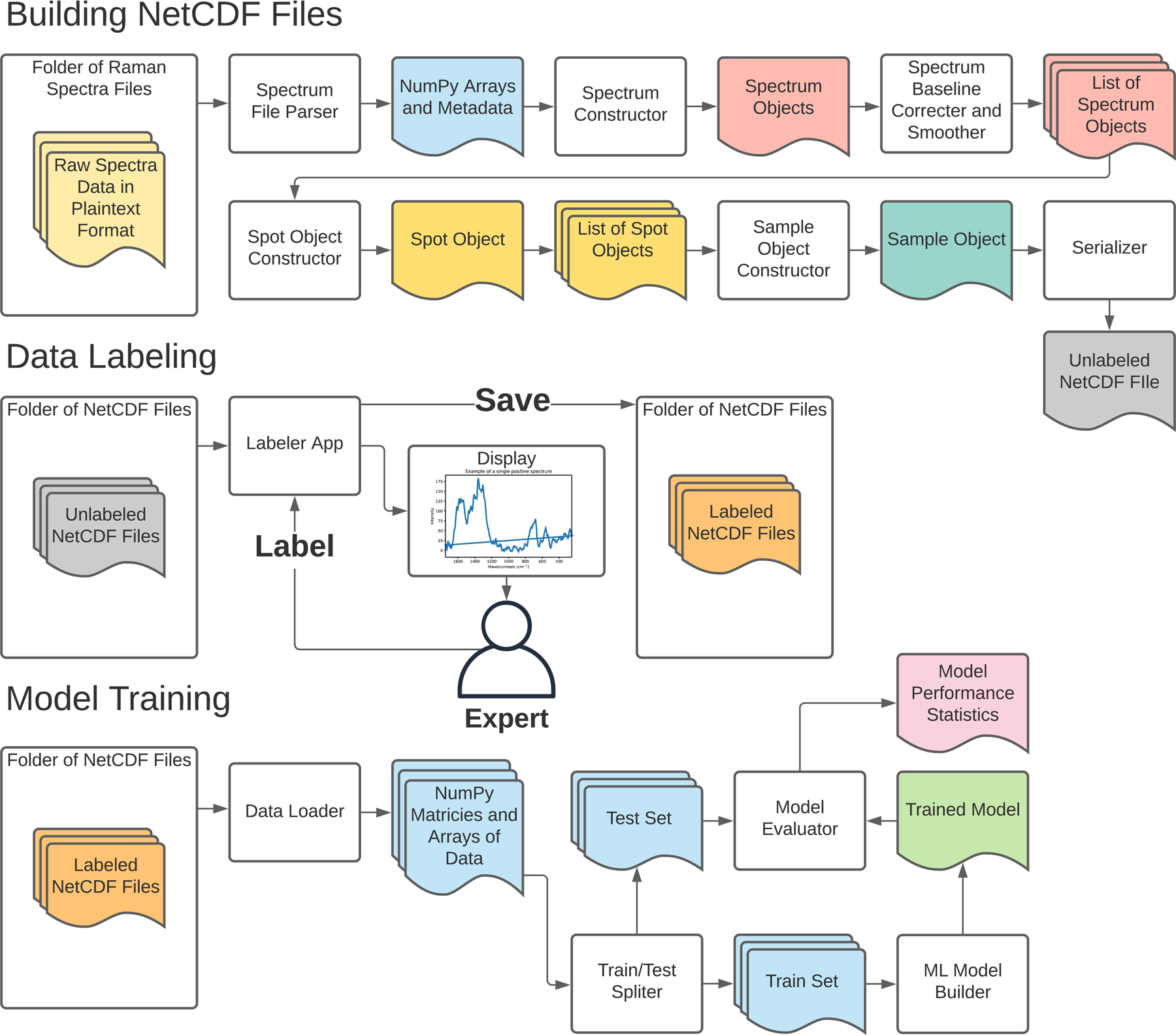
Open-source data pipeline developed for this study. The first stage of the pipeline converts and processes raw spectra files into a binary NetCDF file format. The second data labeling stage employs a custom Python “Labeler” app, allowing an expert Raman user to quickly assign labels (e.g., “good”, “bad”, or “maybe”) to the spectra serialized in the netCDF files. After labels have been assigned, the last stage of the pipeline is model training, where the binary files are loaded into NumPy arrays to train and test various ML models.
The first stage converts plaintext Raman spectrum files into a binary NetCDF file. The NetCDF format (short for network common data form) is a machine independent data storage scheme designed for efficiently saving multi-dimensional scientific data, and well suited for storing spectral datasets.39 An essential part of this stage is the baseline correction and smoothing, which was performed with the airPLS baseline correction algorithm and Whittaker smoothing function, respectively, using code ported to Python 3.40,41 This modified code is available on GitHub in compliance with the LGPL license.
The second stage of the pipeline utilizes expert data labeling to train the ML models in the third stage. To implement a supervised learning algorithm for ‘‘good’’ and ‘‘bad’’ spectra classification, labels need to be associated with each spectrum. Given the need to quickly and easily label thousands of spectra for training (2000 different individual spectra needed to be labeled for this study), a python-based labeling program was created. This program takes a series of netCDF files as input, displays each spectrum to the user and allows for rapid labeling, and then saves the dataset with the applied labels. A screenshot of the Labeler program interface is shown in Fig. S3 (Supplemental Material). The premise is to establish three different bins for the classification purposes: (a) ‘‘good’’ and chemically representative R6G spectrum (also termed as ‘‘positive’’ in this context), (b) ‘‘maybe’’ adequate R6G spectrum where a clear decision could not be made by an expert user, and (c) ‘‘bad’’ (also termed as ‘‘negative’’ in this context), unrepresentative R6G spectrum (e.g., very low signal-to-noise ratios, or S/N). Labeling was based on expert user intuition and experience, focusing on feature-rich spectra with clear sharp peaks and minimal noise. For this study, the Labeler program was used to tag a total of 1995 spectra (940 good, 936 bad, rest 119).
Model Selection and Performance Analysis
Following the labeling task, the acquired spectra were evaluated using an assortment of popular ML-assisted classification routines. The labeled datasets were shuffled and split into train, validation, and test sets (72.25%, 12.75%, and 15%, respectively). The percent positive and negative in each subset was calculated and found to be within ± 5% of 50% for both classes. Building on the hypothesis that existing ML classifiers are well-suited to distinguish between good and bad spectra, we evaluated six distinct methods: logistic regression stochastic gradient descent (LR-SGD), support vector machines stochastic gradient descent (SVM-SGD), decision trees (DT), linear discriminate analysis (LDA), random forest (RF), and XGBoost (XGB). The first five models are implemented within the Scikit-learn package,42 whereas a custom package for XGBoost is used.43 For each model, performance is assessed by calculating resulting receiver operator characteristics (ROC) curve and associated area of the curve (AUC). The ROC curve quantifies the diagnostic ability of a classifier for different discrimination thresholds, while the AUC is independent of the classification cutoff, and thus can give a better overall picture of a model’s performance.44
After hyperparameter tuning using the AUC score (walkthrough can be found in our Jupyter notebooks on GitHub), all six models were assessed by training on the training dataset and their performance validated using the validation set. The ROC plots and associated classification metrics for these six models are shown in Fig. 3. A full description of the calculation for these classification metrics can be found in section 1.4, Supplemental Material. Associated calibration plots are shown in Fig. S4 (Supplemental Material).
Figure 3.
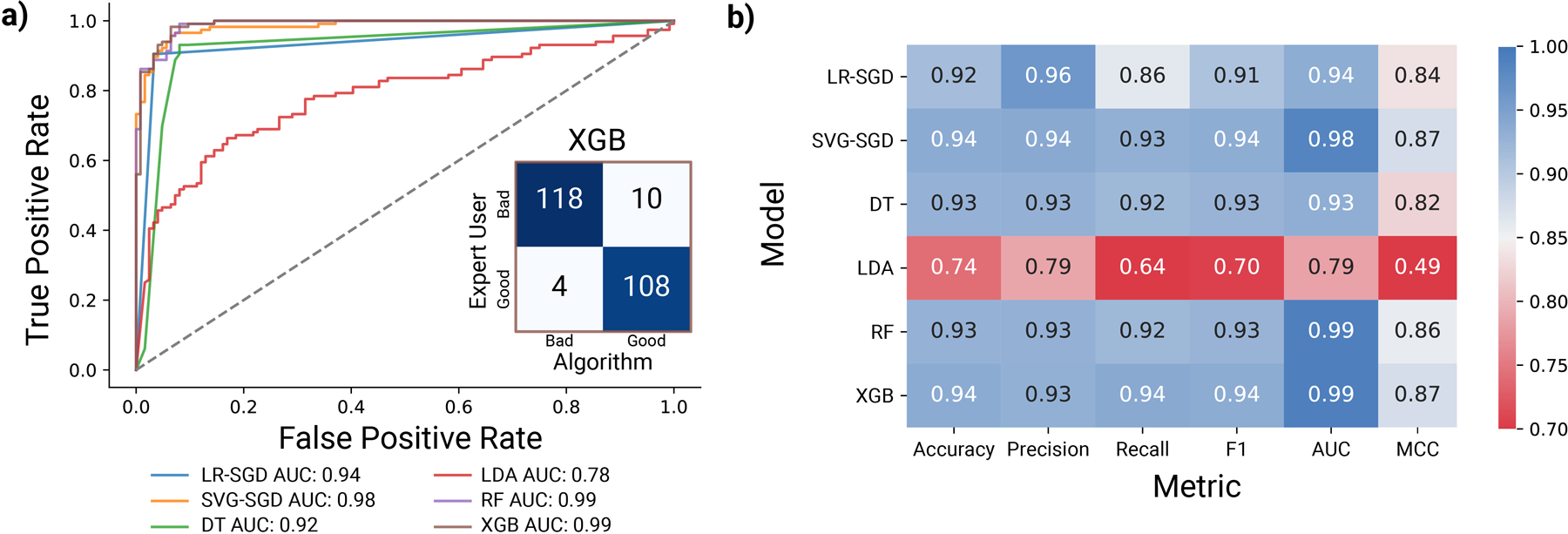
(a) ROC curves for six tested models, logistic regression (LR-SGD), support vector machines (SVG-SGD), decision trees (DT), linear discriminate analysis (LDA), random forest (RF), and XGBoost (XGB). Random guessing is represented by the y=x line, whereas higher performing models lie closer to the left corners of the plot. Inset: Confusion matrix for XGBoost algorithm trained on train and validation set and tested on the test set. The trace of the matrix indicates correct predictions, while the offset values indicate incorrect predictions. (b) Comparison of hyperparameter-tuned model performance on validation data set.
The LDA model was the worst performing in all categories, likely due to the large number of features and lack of regularization. The next worst performing model (by AUC) was the DT model, which consisted of a series of Boolean decisions arranged into a tree structure. This was followed by an LR-SGD and SVM-SGD. Unlike the default logistic regression and SVM solvers, the SGD classifier was not affected by the high correlation between adjacent features (inherent to spectral data, adjacent wavenumber shifts are correlated) and was able to provide stable solutions. Despite their simplicity, the SGD-based models performed well, with the SVM-SGD providing the highest precision of all tested models. The final two tree-based models tested were RF and XGB. The RF model outperformed XGB by 0.0006 AUC units, yet the XGB model was better correlated and scored higher in the accuracy, recall, Matthews correlation coefficient (MCC), and F1 categories (Fig. S4, Supplemental Material). The confusion matrix for the XGB model is shown as an inset in Fig. 3a. These results are consistent with other studies45 which identify XGBoost as a top performing algorithm for binary classification tasks. Therefore, we focus on the XGBoost algorithm in the remainder of this work.
Testing Model Performance
Recognizing the favorable performance of XGB on the validation set, we proceed to investigating the efficacy for insample and out-of-sample tests sets. The in-sample test sets, which involve intermixing the spectra from all samples into the test, train, and validation sets, give a better overall picture of the model performance by capturing the spot-to-spot heterogeneity of the spectra. The true in-sample performance of XGB is estimated by training it on the combined train and validation set and assessing its ability to predict the labels of the 282 spectra in the test set. The metrics derived for the performance of the XGB model are shown in Table 1. Representative classifications for the XGB model on specific spectra are shown in Fig. S5 (Supplemental Material). Overall, we observe good performance of the model with 95% accuracy.
Table 1.
Model performance parameters for the XGBoost algorithm trained on the training set and validation set and tested on the test set.
| Accuracy | Precision | Recall | F1 | MCC | AUC |
|---|---|---|---|---|---|
| 0.95 | 0.93 | 0.97 | 0.95 | 0.87 | 0.99 |
Variable importance
In this experiment, no feature engineering or variable selection was attempted, such that the entire 1024 features of the spectra (i.e., 1024 data points, arising from the CCD dimensions collecting the photons following dispersion) were used to train the XGB algorithm. We note that this feature makes the ML approaches for analytical spectroscopy methods like SERS notably powerful as there is no need to perform a priori dimensionality reduction (e.g., PCA) but rather the full feature space can be used in the training phase. To determine what features were most important in predicting the label of the spectra the importance score for different regions were aggregated together and plotted together. These bar charts give the overall importance of a certain region to the prediction (Fig. 4). Unsurprisingly, the region 1510–1350 cm−1 has the highest importance, as the peaks present in positive spectra tend to cluster around that region. Interestingly, the region 200–400 cm−1 also has some high importance. It is likely that the intensities of these variables give an indication of the overall noise of the spectra, and act as a proxy for estimating the signal-to-noise ratio.
Figure 4.
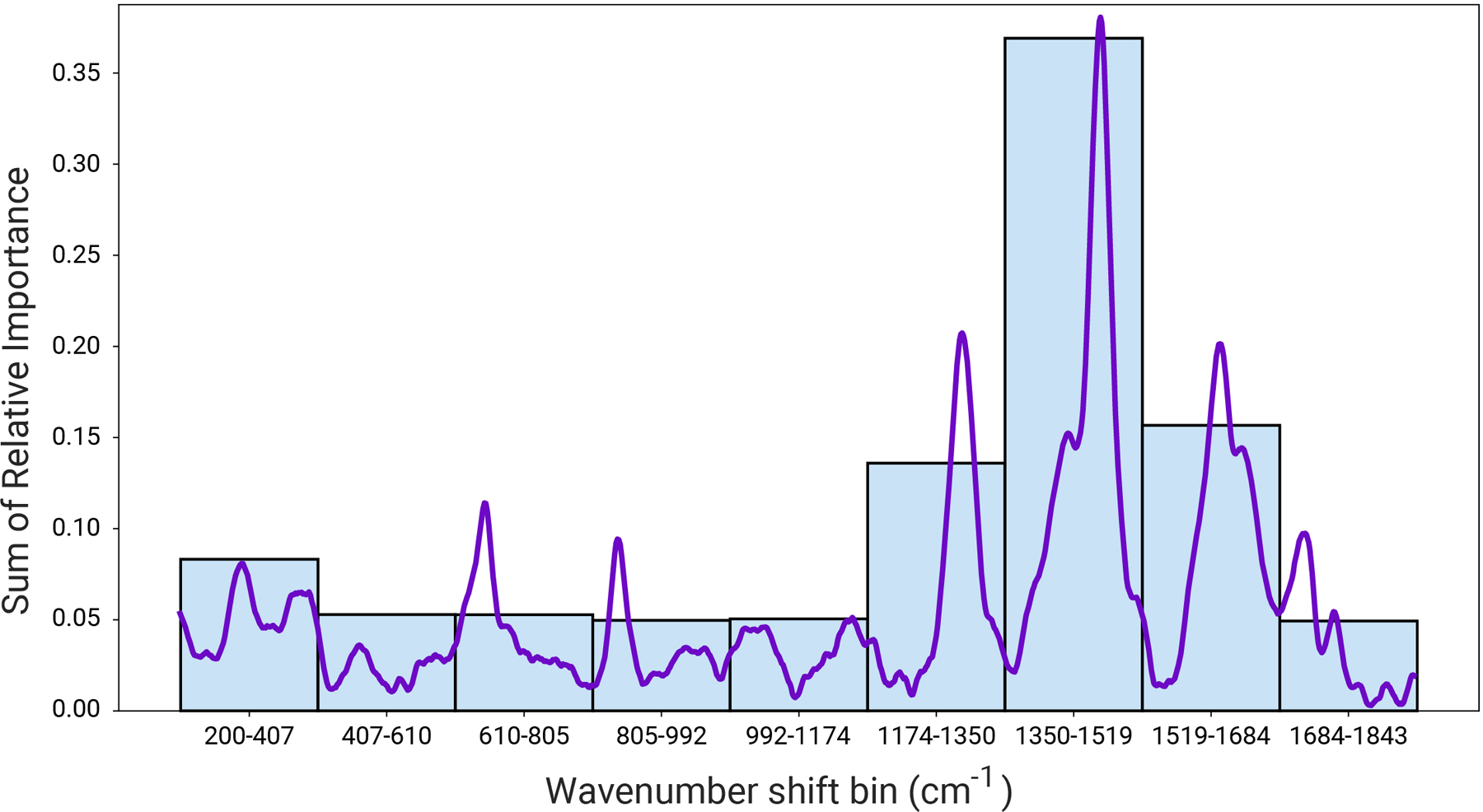
Relative importance of different wavenumber regions for the XGBoost model trained on the in-sample train and validation data. Each bin contains the sum of the importance scores of the wavenumbers in the range (start wavenumber, end wavenumber). An R6G spectrum is overlaid for reference, where it is apparent that relative importance correlates with spectral features of R6G.
Out-of-Sample Testing
To test the out-of-sample performance of XGB, the model was trained on four out of the five substrates and then used to predict the labels of the left-out test substrate (Table 2). The hyperparameters were previously tuned using data from the test substrates, but the data were otherwise unseen by the model. This process was repeated for all five samples. The predictions were assessed with standard metrics (Table 2).
Table 2.
Out-of-sample performance for XGBoost model on five substrates.
| Test sample | Substrate | Dye | Concentration (M) | Power (mW) | Accuracy | Precision | Recall | F1 | AUC | MCC | Fraction positive |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | Moxtek | R6G | 1.00E-08 | 20 | 0.90 | NA | NA | NA | 0.96 | NA | 0.10 |
| 2 | Moxtek | R6G | 3.00 E-03 | 10 | 0.77 | 0.78 | 0.98 | 0.87 | 0.83 | 0.27 | 0.75 |
| 3 | Plasmore | R6G | 1.00E-08 | 20 | 0.95 | 0.91 | 0.64 | 0.75 | 0.99 | 0.74 | 0.12 |
| 4 | Plasmore | R6G | 3.00 E-03 | 10 | 0.72 | 0.78 | 0.73 | 0.75 | 0.80 | 0.42 | 0.60 |
| 5 | Plasmore | R6G | 1.00E-08 | 10 | 0.98 | 0.98 | 1.00 | 0.99 | 0.69 | 0.00 | 0.98 |
For sample 1, no false positives or true negatives were generated making the precision, recall, F1, and MCC formula inapplicable.
To further evaluate out-of-sample performance, we overlay the labeled data onto their spatial coordinates in Fig. 5a, given that these datasets were collected by scanning over a 20 by 20-pixel grid (8 µm × 8 µm) in even increments (400 nm pixel width, approximately diffraction limited). Green represents a ‘‘good’’ spectrum label, while red represents a ‘‘bad’’ spectrum label and purple represents an ambiguous ‘‘maybe’’ spectrum where a label could not be accurately assigned by the expert user. For each of these labeled samples, the model was used to predict the corresponding labels, with colors again plotted in Fig. 5b. Ambiguous spectra were excluded from training the model and were not predicted by the model but are still shown in purple. Incorrect predictions of false negatives and false positives are coded by warm colors, light red, and pink, respectively. Similarly, the true negative was represented by a cool color, i.e., light blue and the true positive by a dark blue. More generally, a perfect prediction would correspond to all true positives and/or true negatives, which corresponds to blue shades.
Figure 5.
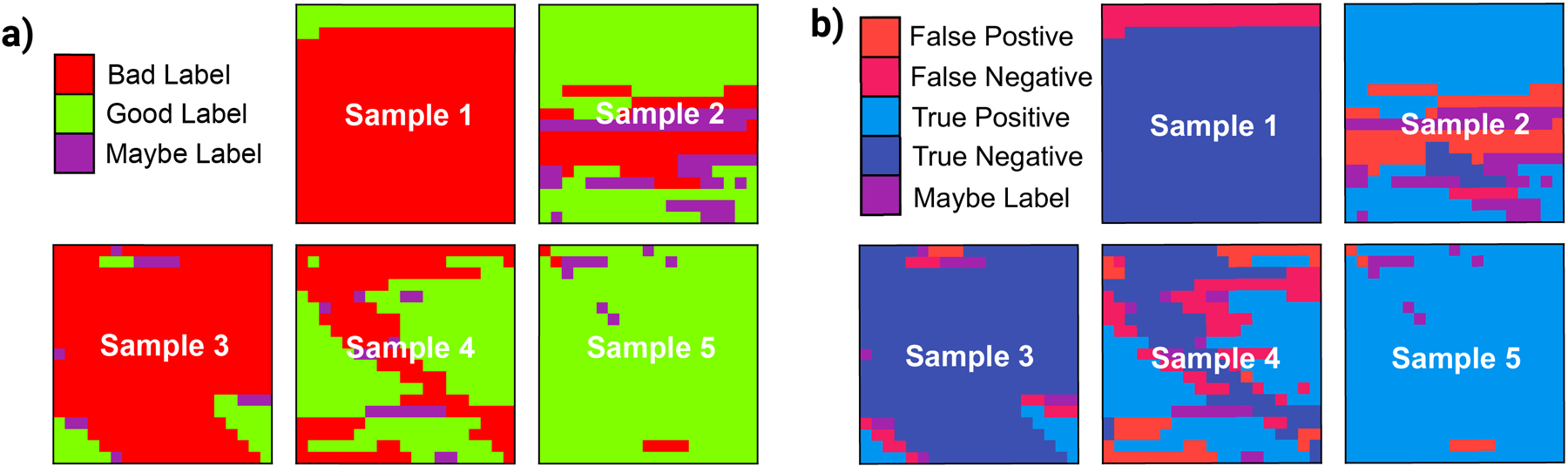
(a) Visualization of the sampled region from the five substrates used in experiment along with the corresponding labels. Red squares represent a negative spectrum, green squares represent a positive spectrum, and purple squares represent spectra with ambiguous labels, which were excluded from fitting and prediction. (b) Comparison of actual and predicted labels for five test samples. The comparison is represented by the color. False positive (red), false negative (pink), true positive (light blue), true negative (dark blue), and ambiguous, maybe labeled, spectra (purple) pixels are shown.
The performance of the XGB model on the in-sample and out-of-sample test set demonstrates that the categorization of spectra can be automated. With the in-sample test set, the XGBoost model achieves an AUC score of 0.99 similar to its performance on the in-sample validation set, indicating minimal overfitting. The 95% accuracy and similarly high precision and recall score prove that the majority of the predictions are correct. The out-of-sample performance is lower and more variable, yet still high. The average out-of-sample AUC score is 0.85 ± 0.12 and an average accuracy is 0.87 ± 0.12 demonstrating that the model is still highly successful at classifying spectra in unseen substrates in different conditions. After learning the labeled spectra from four R6G samples collected at various concentrations, substrates, and power, the model is able to correctly predict the label for spectra 87% of the time. This feat unambiguously shows the potential of ML-assisted sampling to automate spectra categorization.
Discussion
Here we introduced ML-assisted SERS spectra classification methodology to streamline acquisition and efficiently classify recorded spectra. The exclusion of negative signals from the SERS analysis already takes place during normal experimentation. When scanning a substrate, the majority of signals are negative (e.g., noisy, not representative of the typical sampled areas, out of focus, capture cosmic rays). Typically, a trained experimentalist makes the determination of when a ‘‘good’’ signal is collected. Although subjective, this strategy utilizes our impressive pattern matching ability, which is challenging to replicate with structured algorithms. This technique excludes the majority of negative spectra by avoiding their initial collection, but it is not perfect and negative spectra occasionally creep into the recorded dataset. Although experimentalists can easily distinguish between good and bad spectra, the large datasets collected using typical SERS experiments make manual excluding the negative spectra post-collection onerous. In addition to presenting a barrier to large data set collection, these expert user-driven decisions also limit the application of SERS in a clinical setting. For SERS technology to transfer from the research laboratory to the clinic, these subjective labor intensive steps must be eliminated.
The scholarly literature encompassing automatization endeavors of Raman and SERS measurements predominantly demonstrates approaches to automate either (i) the collection or (ii) the data preprocessing phase, e.g., baseline correction, cosmic ray-induced spike removal, noise reduction, scaling and normalization, background subtraction, including various thresholding techniques to harness signal-to-noise ratio for spectra selection.2–4 The main limitations of the current preprocessing techniques are that they either rely on tuning the processing parameters (e.g., fitting parameters) or require calculating and thresholding the S/N, which is not possible if the underlying analyte signal is not known or highly fluctuating. Work by Dallaire et al. discusses the importance of spectral quality for further downstream analyses.46 In particular, they also note the caveat in current literature reports; the spectral quality assessment is largely made offline based on qualitative visual inspection instead of using unbiased and systematic quantitative criteria. The authors elaborately demonstrate the effectiveness of excluding ‘‘bad’’ spectra in cancer detection application.46 Therefore, there exists a clear niche to design robust workflows to select spectra for the downstream analyses. In essence, our strategy is independent of the spectral preprocessing approaches and S/N thresholding, rendering it a promising means to be applied in a wide variety of different platforms.
The underlying reasons why a SERS spectrum may be classified as ‘‘good’’ or ‘‘bad’’ are likely arising from either (1) variations in local analyte concentration or (2) the SERS hotspot phenomena, i.e., localized regions of extreme electromagnetic fields that are highly dependent on underlying substrate geometry and analyte orientation.47–49 Even at the single junction scale, hotspots are highly dynamic in spatial dimension and in time,28 and thus majorly contribute to the dynamic nature of the observed SERS signals. A pertinent yet rarely addressed phenomenon is the spot-to-spot SERS reproducibility. Variation of EF across hotspots is conventionally determined by the substrate uniformity, i.e., the controlled sizes and spacings of the plasmonic features, and also by the total number of hotspots in the detection area. It is possible that this ML approach can help in elucidating the characteristics of SERS hotspots as well as the spectra variability. For example, ML methods can be used for efficient SERS substrate development, since the role of various physical and chemical parameters can be systematically evaluated at the substrate engineering phase. Essentially, our approach can be adapted to characterize the signal-to-noise across a given sample and over time. Until now this development and optimization has traditionally been carried out by finite element modeling (FEM), but the complementary ML approaches can greatly contribute to these processes by allowing for rapid and concise scrutiny of many spectra.
ML algorithms are able to codify human intuition by learning from labeled training data and are well-suited to identify noisy, feature-poor spectra. The use of a ML algorithm has several advantages over a traditional structured algorithm. A trained ML model requires no parameter tuning once trained and can learn from the extensive experience of trained experimentalists. With the availability of open-source ML packages,42 training and integration are straightforward. A plethora of different classes of ML algorithms exist and new ones are frequently being invented. Of the existing classes, they can roughly be divided into two domains, classical ML algorithms and deep learning algorithms. Classical ML algorithms include tree-based algorithms such as random forest and XGBoost, as well as more established classifiers like support vector machines. Deep learning algorithms encompass the tremendous diversity of multilayered neural network models, such as CNNs. Both classical and deep learning models can achieve similarly high performance, but classic ML algorithms can perform well on smaller datasets, whereas deep learning architectures typically require tens of thousands of data points to converge. In this current work, the complete dataset consisted of only 2000 different spectra, thus the tested models were confined to classical models, yet the methods presented here are easily extendable to deep learning models when working with larger datasets.
Amongst all the models tested here, the XGBoost model performed best across both the in-sample and Wout-of-sample datasets. Its performance in this dataset matches our expectation that it is performing akin to a user expert making an intuitive decision. To detail this, consider the major inter-sample variation in the fraction of expert assigned positive labels, likely due to the inhomogeneous covering of dried R6G on the SERS substrates. In sample one, 90% of the spectra are negatively labeled and the classifier predicts a negative label for all of them. In sample 5, the reverse situation occurs; 98% of the spectra are positively labeled and the XGB model assigns a positive label to all the spectra. In these extreme cases, XGB is essentially learning from the out-of-sample labels and not taking into consideration the unique characteristics of the substrate. In this case, an expert experimentalist would adjust their own threshold of classification based on the observed signal-to-noise in a specified sample. For example, if many weak signals were observed, the threshold for collecting a spectrum would be lower than in the case where the majority of spectra had an apparent high signal-to-noise ratio. In the intermediate case of sample 4 with a 60% positive rate, the algorithm performs well (AUC = 0.80), although lower than in the test case (AUC = 0.99). Nevertheless, the algorithm is still successful in categorizing samples with a range of positivity rates.
For the purpose of this study to develop versatile and efficient ML-assisted tools for SERS spectra classification, we chose a known chemical standard molecule R6G. This model compound provided a combination of adequate spectral complexity and variability (e.g., uneven distribution of solution on the substrates resulting in varying degrees of noise) to simulate a typical SERS experiment and subsequent spectra processing. Regarding broader generalizability to more complex analytes such as biological matrices, the best performing ML models (RF and XGBoost) in this work are nonparametric models that do not make any assumptions of the functional form of the classifier. This flexibility ensures that the shape of the dividing lines between the classes, i.e., hyperplanes, deployed by these models can take arbitrary forms, contrasted to logistic regression models where hyperplanes would be linear. Thus, even complex spectra can be efficiently separated from each other since the success of classification does not depend on the complexity or level of noise in the spectral data but instead on the experimentalist’s capability to consistently label spectra as ‘‘good’’ or ‘‘bad’’, based on their own interpretation of data quality, e.g., S/N or presence of trace element peaks.
Additionally, our analyses demonstrated that the ML algorithm is robust at classifying out-of-sample spectra, even across different substrates. However, this experiment does not provide proof that the model is generalizable to all situations, and users wishing to adopt this methodology would need to train new models on a given substrate/analyte pair of interest using the Labeler app. In general, the main limitation of the current approach will be the need to re-train the ML models for varying instrument- and measurement-related parameters. For example, it is likely that the model is dependent on a given laser power, magnification, and acquisition time. However, if experimental parameters are standardized, our out-of-sample performance experiments suggest that inter-substrate and -sample performance is stable. While here we tested highly ordered SERS substrates, we expect that our ML classification approach would perform equally well on SERS platforms comprising nanoparticles in colloidal solutions, even though they typically exhibit more geometrical and topological variation.1,50 Such nanoparticle-based SERS experiments are typically carried out either directly in colloidal solutions or after nanoparticles precipitate or self-assemble on supports. This ultimately leads to a ‘‘metastable’’ environment, especially for colloidal solutions that are highly dynamic. Yet the optical near-field signal amplification is dependent on the local plasmonic field at any given point in the sample, thus we expect individual spectra to still be produced that can be classified as ‘‘bad’’ or ‘‘good’’. As long as the experimentalist can carry out concise pre-classification and model training with the Labeler app, the classification performance is conserved despite the geometry or constituent properties of the underlying substrate.
Future work using this approach will involve automatically tuning the classification threshold based on the number of positively classified spectra in a sample. We also will explore the feedback of this trained algorithm to control stage movement and automate measurement of full datasets.
Conclusion
This work describes application of an ML algorithm to address a central challenge for adapting SERS to automated platforms: the current dependency of expert user-driven endpoints for sampling. The elimination of bad spectra from a collected dataset can increase the signal-to-noise ratio by reducing the variance. Especially in SERS applications, it is desired to collect and analyze as homogeneous sets of spectra as possible, which is accomplished by the ML-assisted spectra selection. By applying this algorithm to the acquisition stage, the labor required to collect many spectra can be reduced making collecting larger and more comprehensive datasets feasible. Furthermore, by automating the acquisition stage of the SERS experiment, another barrier to the clinical application of this technology can be broken down. Given the exponential growth of acquired data (e.g., spectra, images, or videos), there is an immense demand for integrating reliable, automated, and fast analysis methods to the experimental procedures for SERS instrumentation. We envision that the workflow described here will allow for more robust automated SERS analyses. We foresee that the introduced platform can be further expanded to quantitative analyses of chemicals as well as complex biological and clinical samples such as patient-derived EVs or crude serum for modern diagnostic purposes.
Supplementary Material
Funding
This work was supported by the UC Davis Center for Data Science and Artificial Intelligence Research (CeDAR) Innovative Data Science Seed Funding Program; the Ovarian Cancer Education and Research Network, Inc. (OCERN); the NIH/NCI [R01CA241666]. The ThermoFisher Quattro ESEM was funded through the US National Science Foundation under award DMR-1725618.
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplemental Material
All supplemental material mentioned in the text is available in the online version of the journal.
Data Accessibility Statement
All data collected for this study, including SERS datasets and the Labeler program files, can be downloaded from the following open repository: https://doi.org/10.5281/zenodo.3994784. All open-source Python code will be maintained at: https://www.github.com/kul-group/ramanbox.
References
- 1.Langer J, de Aberasturi DJ, Aizpurua J, Alvarez-Puebla RA, et al. “Present and Future of Surface-Enhanced Raman Scattering”. ACS Nano 2020. 14(1): 28–117. doi: 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esmonde-White FWL, Schulmerich MV, Esmonde-White KA, Morris MD “Automated Raman Spectral Preprocessing of Bone and Other Musculoskeletal Tissues”. Proc. SPIE. Int. Soc. Opt. Eng 2009. 7166: 716605. doi: 10.1117/12.809436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Reyes G, Pérez FR “A Method for the Automated Raman Spectra Acquisition”. J. Raman Spectrosc 2017. 48(11): 1654–1664. doi: 10.1002/jrs.5185. [DOI] [Google Scholar]
- 4.Schulze HG, Rangan S, Piret JM, Blades MW, et al. “Developing Fully Automated Quality Control Methods for Preprocessing Raman Spectra of Biomedical and Biological Samples”. Appl. Spectrosc 2018. 72(9): 1322–1340. doi: 10.1177/0003702818778031. [DOI] [PubMed] [Google Scholar]
- 5.Hu W, Ye S, Zhang Y, Li T, et al. “Machine Learning Protocol for Surface-Enhanced Raman Spectroscopy”. J. Phys. Chem. Lett 2019. 10(20): 6026–6031. doi: 10.1021/acs.jpclett.9b02517. [DOI] [PubMed] [Google Scholar]
- 6.Fan X, Ming W, Zeng H, Zhang Z, et al. “Deep Learning-Based Component Identification for the Raman Spectra of Mixtures”. Analyst 2019. 144(5): 1789–1798. doi: 10.1039/C8AN02212G. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Osadchy M, Ashton L, Foster M, et al. “Deep Convolutional Neural Networks for Raman Spectrum Recognition: A Unified Solution”. Analyst 2017. 142(21): 4067–4074. doi: 10.1039/C7AN01371J. [DOI] [PubMed] [Google Scholar]
- 8.Moawad AA, Silge A, Bocklitz T, Fischer K, et al. “Machine Learning-Based Raman Spectroscopic Assay for the Identification of Burkholderia Mallei and Related Species”. Molecules 2019. 24(24): 4516. doi: 10.3390/molecules24244516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarvis RM, Goodacre R “Discrimination of Bacteria Using Surface-Enhanced Raman Spectroscopy”. Anal. Chem 2004. 76(1): 40–47. doi: 10.1021/ac034689c. [DOI] [PubMed] [Google Scholar]
- 10.Deng B, Luo X, Zhang M, Ye L, et al. “Quantitative Detection of Acyclovir by Surface Enhanced Raman Spectroscopy Using a Portable Raman Spectrometer Coupled with Multivariate Data Analysis”. Colloids Surf., B 2019. 173: 286–294. doi: 10.1016/j.colsurfb.2018.09.058. [DOI] [PubMed] [Google Scholar]
- 11.Li S-X, Zeng Q-Y, Li L-F, Zhang Y-J, et al. “Study of Support Vector Machine and Serum Surface-Enhanced Raman Spectroscopy for Noninvasive Esophageal Cancer Detection”. J. Biomed. Opt 2013. 18(2): 27008. doi: 10.1117/1.JBO.18.2.027008. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Ye X, Xu G, Jin X, et al. “Identification and Distinction of Non-Small-Cell Lung Cancer Cells by Intracellular SERS Nanoprobes”. RSC Adv 2016. 6(7): 5401–5407. doi: 10.1039/C5RA21758J. [DOI] [Google Scholar]
- 13.Yu Y, Lin Y, Xu C, Lin K, et al. “Label-free detection of nasopharyngeal and liver cancer using surface-enhanced Raman spectroscopy and partial lease squares combined with support vector machine”. Biomed. Opt. Express 2018. 9(12): 6053–6066. doi: 10.1364/BOE.9.006053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee W, Lenferink ATM, Otto C, Offerhaus HL “Classifying Raman Spectra of Extracellular Vesicles Based on Convolutional Neural Networks for Prostate Cancer Detection”. J. Raman Spectrosc 2020. 51(2): 293–300. doi: 10.1002/jrs.5770. [DOI] [Google Scholar]
- 15.Park J, Hwang M, Choi B, Jeong H, et al. “Exosome Classification by Pattern Analysis of Surface-Enhanced Raman Spectroscopy Data for Lung Cancer Diagnosis”. Anal. Chem 2017. 89(12): 6695–6701. doi: 10.1021/acs.analchem.7b00911. [DOI] [PubMed] [Google Scholar]
- 16.Carmicheal J, Hayashi C, Huang X, Liu L, et al. “Label-Free Characterization of Exosome Via Surface Enhanced Raman Spectroscopy for the Early Detection of Pancreatic Cancer”. Nanomedicine 2019. 16: 88–96. doi: 10.1016/j.nano.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banaei N, Moshfegh J, Mohseni-Kabir A, Houghton JM, et al. “Machine Learning Algorithms Enhance the Specificity of Cancer Biomarker Detection Using SERS-Based Immunoassays in Microfluidic Chips”. RSC Adv 2019. 9(4): 1859–1868. doi: 10.1039/C8RA08930B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guevara E, Torres-Galván JC, Ramírez-Elías MG, Luevano-Contreras C, et al. “Use of Raman Spectroscopy to Screen Diabetes Mellitus with Machine Learning Tools”. Biomed. Opt. Express 2018. 9(10): 4998–5010. doi: 10.1364/BOE.10.004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krauß SD, Roy R, Yosef HK, Lechtonen T, et al. “Hierarchical Deep Convolutional Neural Networks Combine Spectral and Spatial Information for Highly Accurate Raman-Microscopy-Based Cytopathology”. J. Biophotonics 2018. 11(10): e201800022. doi: 10.1002/jbio.201800022. [DOI] [PubMed] [Google Scholar]
- 20.Shi H, Wang H, Meng X, Chen R, et al. “Setting Up a Surface-Enhanced Raman Scattering Database for Artificial-Intelligence-Based Label-Free Discrimination of Tumor Suppressor Genes”. Anal. Chem 2018. 90(24): 14216–14221. doi: 10.1021/acs.analchem.8b03080. [DOI] [PubMed] [Google Scholar]
- 21.Hidi IJ, Jahn M, Weber K, Bocklitz T, et al. “Lab-on-a-Chip-Surface Enhanced Raman Scattering Combined with the Standard Addition Method: Toward the Quantification of Nitroxoline in Spiked Human Urine Samples”. Anal. Chem 2016. 88(18): 9173–9180. doi: 10.1021/acs.analchem.6b02316. [DOI] [PubMed] [Google Scholar]
- 22.Alharbi O, Xu Y, Goodacre R “Simultaneous Multiplexed Quantification of Caffeine and its Major Metabolites Theobromine and Paraxanthine Using Surface-Enhanced Raman Scattering”. Anal. Bioanal. Chem 2015. 407(27): 8253–8261. doi: 10.1007/s00216-015-9004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moosavi SM, Chidambaram A, Talirz L, Haranczyk M, et al. “Capturing Chemical Intuition in Synthesis of Metal-Organic Frameworks”. Nat. Commun 2019. 10(1): 539. doi: 10.1038/s41467-019-08483-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen D, Wu G, Suk H-I “Deep Learning in Medical Image Analysis”. Annu. Rev. Biomed. Eng 2017. 19(1): 221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeanmaire DL, Van Duyne RP “Surface Raman Spectroelectrochemistry: Part I. Heterocyclic, Aromatic, and Aliphatic Amines Adsorbed on the Anodized Silver Electrode”. J. Electroanal. Chem. Interfacial Electrochem 1977. 84(1): 1–20. doi: 10.1016/S0022-0728(77)80224-6. [DOI] [Google Scholar]
- 26.Fleischmann M, Hendra PJ, McQuillan AJ “Raman Spectra of Pyridine Adsorbed at a Silver Electrode”. Chem. Phys. Lett 1974. 26(2): 163–166. doi: 10.1016/0009-2614(74)85388-1. [DOI] [Google Scholar]
- 27.Albrecht MG, Creighton JA “Anomalously Intense Raman Spectra of Pyridine at a Silver Electrode”. J. Am. Chem. Soc 1977. 99(15): 5215–5217. doi: 10.1021/ja00457a071. [DOI] [Google Scholar]
- 28.Michaels AM, Jiang J, Brus L “Ag Nanocrystal Junctions as the Site for Surface-Enhanced Raman Scattering of Single Rhodamine 6G Molecules”. J. Phys. Chem. B 2000. 104(50): 11965–11971. doi: 10.1021/jp0025476. [DOI] [Google Scholar]
- 29.Xu X, Kim K, Li H, Fan DL “Ordered Arrays of Raman Nanosensors for Ultrasensitive and Location Predictable Biochemical Detection”. Adv. Mater 2012. 24: 5457–5463. doi: 10.1002/adma.201201820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vincent B, Edwards J, Emmett S, Jones A “Depletion Flocculation in Dispersions of Sterically-Stabilised Particles (“Soft Spheres”)”. Colloids Surf 1986. 18(2): 261–281. doi: 10.1016/0166-6622(86)80317-1. [DOI] [Google Scholar]
- 31.Wijenayaka LA, Ivanov MR, Cheatum CM, Haes AJ “Improved Parametrization for Extended Derjaguin, Landau, Verwey, and Overbeek Predictions of Functionalized Gold Nanosphere Stability”. J. Phys. Chem. C 2015. 119(18): 10064–10075. doi: 10.1021/acs.jpcc.5b00483. [DOI] [Google Scholar]
- 32.Saunders SR, Eden MR, Roberts CB “Modeling the Precipitation of Polydisperse Nanoparticles Using a Total Interaction Energy Model”. J. Phys. Chem. C 2011. 115(11): 4603–4610. doi: 10.1021/jp200116a. [DOI] [Google Scholar]
- 33.Almehmadi LM, Curley SM, Tokranova NA, Tenenbaum SA, et al. “Surface Enhanced Raman Spectroscopy for Single Molecule Protein Detection. Sci. Rep 2019. 9(1): 12356. doi: 10.1038/s41598-019-48650-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosnick KA, Jiang, Brus LE “Fluctuations and Local Symmetry in Single-Molecule Rhodamine 6G Raman Scattering on Silver Nanocrystal Aggregates”. J. Phys. Chem. B 2002. 106(33): 8096–8099. doi: 10.1021/jp0256241. [DOI] [Google Scholar]
- 35.Zrimsek AB, Chiang N, Mattei M, Zaleski S, et al. “Single-Molecule Chemistry with Surface- and Tip-Enhanced Raman Spectroscopy”. Chem. Rev 2017. 117(11): 7583–7613. doi: 10.1021/acs.chemrev.6b00552. [DOI] [PubMed] [Google Scholar]
- 36.Ru ECL, Etchegoin PG “Single-Molecule Surface-Enhanced Raman Spectroscopy”. Annu. Rev. Phys. Chem 2012. 63(1): 65–87. doi: 10.1146/annurev-physchem-032511-143757. [DOI] [PubMed] [Google Scholar]
- 37.Szeghalmi A, Kaminskyj S, Rösch P, Popp J, et al. “Time Fluctuations and Imaging in the SERS Spectra of Fungal Hypha Grown on Nanostructured Substrates”. J. Phys. Chem. B 2007. 111(44): 12916–24. doi: 10.1021/jp075422a. [DOI] [PubMed] [Google Scholar]
- 38.Taylor J, Huefner A, Li L, Wingfield J, et al. “Nanoparticles and Intracellular Applications of Surface-Enhanced Raman Spectroscopy”. Analyst 2016. 141(17): 5037–5055. doi: 10.1039/C6AN01003B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unidata. netCDF4 Version 1.5.6 Boulder, CO: UCAR/Unidata, 2021. doi: 10.5065/D6H70CW6. [DOI] [Google Scholar]
- 40.Zhang Z-M, Chen S, Liang Y-Z “Baseline Correction Using Adaptive Iteratively Reweighted Penalized Least Squares”. Analyst 2010. 135(5): 1138–1146. doi: 10.1039/b922045c. [DOI] [PubMed] [Google Scholar]
- 41.Eilers PHC “A Perfect Smoother”. Anal. Chem 2003. 75(14): 3631–3636. doi: 10.1021/ac034173t. [DOI] [PubMed] [Google Scholar]
- 42.Pedregosa F, Varoquaux G, Gramfort A, Michel V, et al. “Scikit-Learn: Machine Learning in Python”. J. Mach. Learn. Res 2011. 12: 2825–2830. [Google Scholar]
- 43.Chen T, Guestrin C “XGBoost: A Scalable Tree Boosting System”. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Association for Computing Machinery, San Francisco, CA: August 13, 2016, 2016. Pp. 785–794. doi: 10.1145/2939672.2939785. [DOI] [Google Scholar]
- 44.Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L “The Use of Receiver Operating Characteristic Curves in Biomedical Informatics”. J. Biomed. Inf 2005. 38(5): 404–415. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Borstelmann SM “Machine Learning Principles for Radiology Investigators”. Acad. Radiol 2020. 27(1): 13–25. doi: 10.1016/j.acra.2019.07.030. [DOI] [PubMed] [Google Scholar]
- 46.Dallaire F, Picot F, Tremblay J-P, Sheehy G, et al. “Quantitative Spectral Quality Assessment Technique Validated Using Intraoperative In Vivo Raman Spectroscopy Measurements. J. Biomed. Opt 2020. 25(4): 040501. doi: 10.1117/1.JBO.25.4.040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Creighton JA, Blatchford CG, Albrecht MG “Plasma Resonance Enhancement of Raman Scattering by Pyridine Adsorbed on Silver or Gold Sol Particles of Size Comparable to the Excitation Wavelength”. J. Chem. Soc., Faraday Trans. 2 1979. 75: 790–798. doi: 10.1039/F29797500790. [DOI] [Google Scholar]
- 48.Cang H, Labno A, Lu C, Yin X, et al. “Probing the Electromagnetic Field of a 15-Nanometre Hotspot by Single Molecule Imaging”. Nature 2011. 469: 385–388. doi: 10.1038/nature09698. [DOI] [PubMed] [Google Scholar]
- 49.Radziuk D, Moehwald H “Prospects for Plasmonic Hot Spots in Single Molecule SERS Towards the Chemical Imaging of Live Cells”. Phys. Chem. Chem. Phys 2015. 17: 21072–21093. doi: 10.1039/C4CP04946B. [DOI] [PubMed] [Google Scholar]
- 50.Solís DM, Taboada JM, Obelleiro F, Liz-Marzán LM, et al. “Optimization of Nanoparticle-Based SERS Substrates Through Large-Scale Realistic Simulations”. ACS Photonics 2017. 4(2): 329–337. doi: 10.1021/acsphotonics.6b00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data collected for this study, including SERS datasets and the Labeler program files, can be downloaded from the following open repository: https://doi.org/10.5281/zenodo.3994784. All open-source Python code will be maintained at: https://www.github.com/kul-group/ramanbox.


