Abstract
Gut microbiota succession overlaps with intensive growth in infancy and early childhood. The multitude of functions performed by intestinal microbes, including participation in metabolic, hormonal, and immune pathways, makes the gut bacterial community an important player in cross-talk between intestinal processes and growth. Long-term disturbances in the colonization pattern may affect the growth trajectory, resulting in stunting or wasting. In this review, we summarize the evidence on the mediating role of gut microbiota in the mechanisms controlling the growth of children.
Keywords: gut microbiota, growth, infants, children
1. Introduction
The gut microbiota is a group of microorganisms, mainly bacteria, yeast, fungi, bacteriophages, and other viruses [1], as well as protozoa and archaea [2,3], which form a complex ecosystem in the human gastrointestinal (GI) tract. The intestinal microbiota performs several functions, including participating in the activation and maturation of the immune system, regulating the immune response, maintaining intestinal epithelial homeostasis, providing protection against the invasion of opportunistic pathogens, synthesizing enzymes necessary for the breakdown of complex plant polysaccharides, and taking part in hormonal regulation [4,5,6,7]. Dysbiosis, reduced diversity in the gut microbial community, or microbial immaturity are involved in several intestinal and metabolic diseases that affect growth [8]. It is essential that an intense increases in body size, especially in infancy, occur in parallel with the dynamic succession of gut microbiota [9]. In recent years, more attention has been paid to the study of the mediating role of gut microbiota in shaping the process of physical growth in children. In this paper, we review the mechanisms through which the gut microbiota affects the process of growth in children.
2. Physical Growth of Children
The physical growth of children is defined as an irreversible and constant increase in body size (length or height and weight) and in the size of organs [10]. It is a dynamic, complicated, and long process that continues throughout infancy, childhood, and adolescence [11]. Tracking the growth trajectories of children is important, as they provide essential indicators of infant and childhood development and can predict potential adult health outcomes [12]. A vast body of evidence suggests that physical growth in the earliest stages of ontogenesis not only sets the pattern for adult size but also establishes a biological scaffold for adult health through the profound effects that nutrition, illness, ecology, and social environment have on early development [13]. Undoubtedly, the group of factors affecting children’s development in the first years of life also includes the set of microorganisms that colonize the intestines at that time. Based on the standards developed by the World Health Organization [14], there are three types of growth trajectory in children: standard or normal growth, delayed growth, and rapid growth. The assessment of growth is performed based on anthropometric measurements of the child and a comparison of the obtained values to growth standards. The standards depict normal human growth under optimal environmental conditions and can be used to assess children regardless of ethnicity, socioeconomic status, and type of feeding [14]. Research suggests that children with normal growth trajectories are more likely to experience better health outcomes than those with abnormal growth [15,16]. Abnormalities in child growth refer to deviations in height, weight, or head circumference. Delayed growth is recognized when these deviations are below the standards [17]. Stunting refers to a child with a height-for-age Z-score (HAZ) two standard deviations (−2 SD) below the standard. Wasting, on the other hand, refers to a child with a weight-for-height Z-score (WHZ) < −2 SD below the standard. More specifically, moderate acute malnutrition (MAM) is recognized when the WHZ of the child is between −2 and −3 SD below the standard, whereas severe acute malnutrition (SAM) is recognized when the child’s WAZ ranks below −3 SD from the median of the WHO reference growth standards [18]. Both SAM and MAM usually develop between 3 and 24 months of life [19], which is the period in which the pattern of the gut microbiota is established. Rapid growth is defined when the deviations are above the standards [14,17]. Other very useful tools for assessing the nutritional status of children are the cut-off points developed by Cole et al. [20,21], who defined the ranges of underweight, norm, overweight, and obesity.
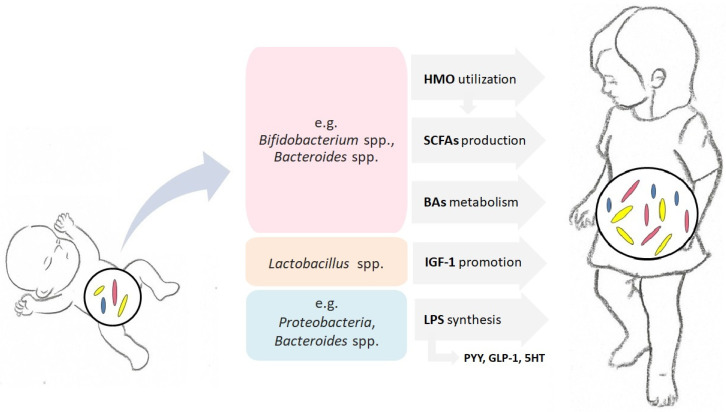
The consequences of abnormal growth rates are varied. Evidence suggests that rapid weight gain in early life is associated with several determinants of metabolic syndrome (e.g., cardiovascular disease and type 2 diabetes) in early adulthood [22,23]. The subsequent consequences of delayed growth include poor cognitive and psychosocial outcomes [24]. The best documented causes of impaired linear growth in children include lowered or increased body mass index, unfavorable prenatal conditions, and infections [25]. All the aforementioned factors affecting the course of growth in children have been well explored. In recent years, it has become clear that a complex of intestinal microbes is also involved in the process of shaping the growth trajectories of children. The intestinal microbiota takes part in a wide range of mechanisms regulating the process of growth, which is reflected in the rate and magnitude of changes in child body size over time (Figure 1).
Figure 1.
Selected mechanisms through which gut microbes affect the process of growth in children. The mediating role of intestinal microbiota in child growth consists of participation in metabolic processes through the production of biologically active metabolites, mainly short-chain fatty acids (SCFAs), in the metabolism of human milk oligosaccharides (HMO) and bile acids (Bas), participation in the hormonal activity of the host through indirect impact on the hormone levels, including insulin-like growth factor-1 (IGF-1), peptide YY (PYY), glucagon-like peptide-1 (GLP-1), or serotonin (5HT), and stimulation of the innate immune system, as well as the initiation of the inflammatory process (through the synthesis of lipopolysaccharides, LPS).
3. Gut Microbiota in the Early Stages of Human Growth
There are presumptions that the process of microbiota colonization begins in utero, most likely through the translocation of bacteria or their antigens to the fetus from the maternal gastrointestinal tract, then from the mother and the environment at delivery, and that it continues throughout the first three years of life [26,27,28]. The development and maturation of the intestinal microbiota are influenced by many factors, such as placental inflammation, maternal infection during pregnancy [29], course of pregnancy, length of pregnancy, type of delivery [30], perinatal condition, hospital environment and length of hospitalization [31], feeding method, use of antibiotics, [32], lifestyle, and geographical factors [33]. From birth to the age of three years, the gut microbiota gradually develops into the adult microbiota [34]. After the third year of life, the intestinal microbiota reaches maturity. Therefore, the fetal period and the first three years of life are considered the critical window for the formation of the microbial colonization pattern. Any disturbances in the process of colonization of the gut microbiota may have a long-term effect on host growth, development, and health later in life [35,36,37]. This is also a key period for infant growth, with significant gains in body length and weight as well as head and chest circumference [37,38]. Both prenatal development and development in early childhood are characterized by the rapid maturation of metabolic, endocrine, nervous, and immune pathways that strongly influence and support the growth and development of the child. These pathways run in parallel and are highly interdependent, and their disturbance by adverse environmental factors, such as infection, may distort the trajectory of a child’s growth and development [37]. As demonstrated by Stewart et al. [39], there are three distinct phases of microbiome succession: a developmental phase (months 3–14), a transitional phase (months 15–30), and a stable phase (months 31–46). Recent studies have shown that the predominant early colonizers of the healthy infant’s gut are maternal fecal bacteria, mainly Bifidobacterium and Bacteroides, and Clostridium during the following six months [26,40,41,42,43]. Studies have also demonstrated that Bacteroides are associated with increased gut diversity and faster intestine maturation [39]. Natural childbirth has been shown to be significantly related to microorganisms reflecting the mother’s vaginal flora, such as Bacteroides, Lactobacillus, and Prevotella [30,39]. Vaginal microbiota with low diversity and rich in Lactobacillus has been connected to birth at term and appropriate birth weight [37]. In turn, in children born by cesarean section, bacteria inhabiting mainly the skin surface, Staphylococcus, Corynebacterium, and Propionibacterium, predominate [30]. Cesarean section is one of the most significant disrupting factors of the proper colonization process [44]. Another important factor associated with infant gut microbiota colonization is breast milk [42]. Approximately 25–30% of the infant’s gut microbiota originates from the mother’s milk [45]. Breastfeeding is related to the predominance of Bifidobacterium and Lactobacillus [46], and the cessation of breastfeeding results in faster gut microbiome maturation, marked by Firmicutes [39]. When modified milk is used, the dominance of Bacteroides and Clostridium is noticeable [46].
4. Microbiota Succession and Growth of Children
The results of the studies carried out thus far allow us to conclude that the course of colonization of the intestinal microbiota in the early stages of ontogenesis has far-reaching consequences for the linear and ponderal growth of children. The most extensive research exploring the aforementioned relationship was performed in the context of malnutrition. Smith et al. [47] conducted an elegant long-term study on the role of the gut microbiota in Malawian twin pairs discordant for kwashiorkor, a form of SAM. Both children in these pairs of twins were treated with a ready-to-use therapeutic food (RUTF) based on peanuts, which resulted in a transient maturation of metabolic function in the kwashiorkor microbiome. In the next stage of the research, fecal microbiota were transplanted from several discordant pairs into gnotobiotic mice. It was noticed that mice that received transplants from children with kwashiorkor and were fed the Malawian diet experienced significant weight loss and disturbances in carbohydrate and amino acid metabolism. The administration of RUTF only transiently ameliorated the observed effects. This finding led to the conclusion that the intestinal microbiome is a casual factor in this type of severe malnutrition, which was also confirmed by Pham et al. [48].
In the study by Subramanian et al. [49], the process of intestinal microbial succession in a cohort of children living in the urban slum of Dhaka, Bangladesh, who exhibited consistently healthy growth during the first two years of life, was analyzed. With the use of a machine-learning-based approach to 16S rRNA datasets, the authors assessed the most age-discriminatory taxa, including Bifidobacterium longum and Streptococcus thermophilus. In the next step, they developed the index of gut microbiota maturity, the so-called microbiota-for-age Z-score (MAZ). In the same study, the MAZ among children with severe acute malnutrition (SAM) (WHZ < −3) was also analyzed. The results revealed that compared to the healthy group, children with SAM had a significantly lower MAZ. This indicates that the gut microbiota of the children with SAM was significantly less mature than that of the healthy children [49].
The results of a longitudinal birth cohort study conducted by Dinh et al. [50] on a small group of children in the urban slum community in Vellore, India showed that a reduced relative abundance of Bifidobacterium longum and Lactobacillus mucosae, in addition to an elevated relative abundance of Desulfovibrio ssp., was associated with stunting. It has been shown that stunted children possess a different gut microbiota composition to non-stunted children. The intestinal microbial ecosystem of stunted children was enriched in inflammogenic taxa, whereas that of the non-stunted group was enriched in probiotic bacterial species. The results of other studies conducted in the suburbs of Chandigarh, India also indicated that the pattern of gut microbiota profile in malnourished children differs from that of healthy subjects [51]. It has been documented that the intestinal microbiota of healthy children is characterized by a significantly higher abundance of Bifidobacterium in comparison to children suffering from SAM. Interesting results were also obtained by Gough et al. [52], who conducted research on a group of children from Malawi and Bangladesh. It has been shown that reduced microbiota diversity is associated with stunting severity and that overgrowth of Acidaminococcus, as well as elevated glutamate-fermenting microbes, may contribute to future growth deficits in already malnourished children. In contrast to the aforementioned studies, Méndez-Salazar et al. [53] investigated differences in the composition of the gut microbiota in both undernourished and obese children from Mexico. The undernourished children demonstrated a significantly higher abundance of Firmicutes and Lachnospiraceae than the obese children, while the Proteobacteria was overrepresented in the obese subjects. Therefore, it can be concluded that the course of intestinal microbiota succession plays an important role in the regulation of early-life growth.
5. Microbiota and Host Metabolism Regulation
Several studies have shown that the early-life microbiota plays a mediating role in host metabolic processes. Gut microbes are essential intermediaries in a wide range of mechanisms, including energy harvest, fat storage, regulation of lipid and glucose metabolism, induction of low-grade inflammation, gut barrier function, control of satiety through gut hormones, and interactions with host genetics [54,55]. To put it simply, the contribution of microbiota to the aforementioned mechanisms is based both on its metabolites, mainly (but not only) short-chain fatty acids (SCFAs), and on the activity of lipopolysaccharides (LPS) and bile acids [55].
5.1. Microbiota and Short-Chain Fatty Acids
Some taxa of enteric microbiota possess the ability to ferment nondigestible carbohydrates. The major products of this fermentation are short-chain fatty acids, primarily acetate, propionate, and butyrate, which play various roles not only in the gastrointestinal tract but also distantly [56,57]. It has been documented that SCFAs can regulate host cell activity via two mechanisms: by binding to G-protein-coupled receptors (GPCRs) or by acting as inhibitors of histone deacetylase [58]. Receptors and transporters for SCFAs are expressed in a vast range of cells from the gastrointestinal tract to the immune and nervous systems [59,60]. The effects of GPCR activation differ greatly depending on the cell in which they are expressed [61]. Butyrate serves as a major energy source for enterocytes [62] and acts as a histone deacetylase inhibitor [63], contributing to the maintenance of barrier integrity and reducing the levels of intestinal inflammation markers [64,65,66]. Butyrate, acetate, and propionate activate GPCRs and thus trigger a wide range of intracellular transduction cascades, including those that modulate hormonal activity [67]. It has been documented that concentrations of SCFAs in the cecum are higher in conventionally raised mice than in germ-free (GF) animals [64], whereas the colonization of GF mice results in increases in SCFA concentrations [68]. At the same time, the use of broad-spectrum antibiotics results in a decrease in SCFA concentrations in conventionally raised mice [68]. This proves that SCFA concentrations are associated with the presence of gut microbes.
5.2. Microbiota and Lipopolysaccharides
The most significant events in the development of host immunity occur in the earliest stages of development [69]. The intense changes in the composition of the microbial community that take place in the infant gut make this period a “window of opportunity” [42]. For this reason, any abnormalities in intestinal succession may have far-reaching consequences for the functioning of the immune system, manifested by increased sensitivity to infections or inflammatory diseases [70,71], which may impair growth. The maturation of innate immunity is characterized by the colonization of gram-negative bacteria, mainly Proteobacteria and Bacteroides spp. The common feature of these two taxa is the presence of lipopolysaccharides (LPS) in the outer cell membrane [72], a pathogen-associated molecular pattern (PAMP) that can stimulate both the innate immune system and nonimmune cells and initiate the inflammatory process [73,74,75]. LPS are potent ligands for Toll-like receptors (TLRs), mainly Toll-like receptor 4 (TLR4), which are expressed in many cells of the body [76], including epithelial enteroendocrine (EE) cells. LPS can activate TLRs in EEs and thus stimulate the secretion of metabolically active hormones, such as peptide YY (PYY) [77], glucagon-like peptide-1 (GLP-1) [78], or serotonin (5HT) [79]. PYY inhibits intestinal motility and increases the absorption rate of nutrients through the intestinal epithelium [80]. GLP-1 potentiates glucose-stimulated insulin secretion in β-cells [81], whereas 5HT participates in the regulation of intestinal motility and fluid secretion [79]. Elevated concentrations of LPS in the blood trigger a condition termed “metabolic endotoxemia” [82,83]. Numerous metabolic disorders, including insulin resistance, type II diabetes, and obesity, are possibly related to this state [84]. Elevated LPS concentrations may result from increased intestinal permeability associated with compositional disturbances in gut microbiota [83]. Endotoxemia can, therefore, contribute to low-grade inflammation, disturbed mucosal barrier integrity, and impaired glycemic control through perturbations in gut hormone secretion, which characterize metabolic syndrome [76,84]. It has been documented that obese children present higher levels of Proteobacteria and that there is a positive correlation between Proteobacteria, Bacteroides, and fat intake [53]. Therefore, it can be concluded that the excessive ponderal growth that characterizes obese children may be associated with an increased abundance of bacteria producing LPS. However, it is not entirely clear what role different LPS-producing bacteria play in the pathophysiology of excessive weight gain or early education of immunity in children [55].
5.3. Microbiota and Bile Acids
Bile acids (BAs) constitute the primary components of bile and are secreted into the small intestine in conjugated forms. The main function of BAs is to aid the solubilization, and thus absorption, of dietary lipids [76]. Most bile acids are reabsorbed in the ileum and returned to the liver, whereas unabsorbed BAs enter the large intestine and are deconjugated and metabolized into secondary BAs by gut microbes [85] via bile salt hydrolase (BSH), which is widely present among intestinal microbiota [86]. Moreover, BAs act as ligands for a nuclear farnesoid X receptor (FXR) as well as for G-protein-coupled bile acid receptor-1 (TGR5). The activation of these receptors initiates a variety of signaling cascades relevant to the regulation of lipid, sugar, and energy metabolism [87]. Tanaka et al. [88] conducted a longitudinal study on a group of Japanese children in which they monitored the succession of the gut bacterial community and its association with the fecal BA profile in the first three years of life. Evidence from this research provided insights that neonates undergo a massive transition in their BA profiles during lactation, which overlaps with the transition of the gut microbiota profiles. Moreover, it has been documented that perturbations of BAs or microbiota profiles (or both) during the first three years of life may result in negative metabolic consequences. However, the associations between bile acids and microbiota succession in infants need to be further explored.
6. Microbiota and Human Milk Oligosaccharides
In early infancy, the diversity of microbiota is low, especially in breastfed babies. During this period, the composition of the gut microbial ecosystem is dominated by species whose role is to participate in the metabolism of human milk oligosaccharide (HMO) [45]. HMOs are a unique-to-humans, structurally complex, and diverse group of glycans, which are present in human milk in high quantities [89]. They are key bioactive components of mothers’ milk [90] and provide numerous health-promoting effects by inhibiting pathogen attachment to epithelial cells (ECs) [91], promoting the growth of specific microbes, modulating intestinal immune responses and patterns of gene expression in intestinal epithelial cells [92]. HMOs can indirectly increase SCFA production, and these elevated levels are mediated by bifidobacterial species [93]. HMOs are composed of five monosaccharides: glucose (Glc), galactose (Gal), N-acetylglucosamine (GlcNAc), fucose (Fuc), and sialic acid N-acetyl-neuraminic acid (Neu5Ac). As HMO compositions can vary, breastfed infants are exposed to structurally different HMOs [94]. The degradation of HMOs to low-molecular-weight oligosaccharides depends on the enzymes produced by bacteria. Next, low-molecular-weight oligosaccharides can be reduced to monosaccharides via carbohydrate-degrading enzymes and converted to SCFAs [95].
The presence of bacteria capable of HMO enzymatic degradation, which the host organism cannot perform on its own, is, therefore, crucial in the production of SCFAs or other organic compounds that may be beneficial to the host during infancy. Moreover, the HMOs present in mothers’ milk stimulate the growth of bacteria that possess the ability to break down these compounds (the so-called bifidus factor). Breastfed infants exhibit significantly higher levels of Bifidobacterium and Lactobacillus and lower levels of pathogenic bacteria than formula-fed infants [96]. Additionally, breastfed infants and formula-fed infants differ in their SCFA concentrations. Specifically, breastfed infants are characterized by higher levels of acetate and lactate and lower levels of propionate and low or absent levels of butyrate [94]. Until recently, HMOs were not present in the formulas administered to children who, for some reason, could not be breastfed, which resulted in a decrease in the percentage of Bifidobacterium in the developing community of gut microbes (due to the lack of a factor stimulating their development).
An excellent study by Charbonneau et al. [97] shed light on the mechanisms through which HMOs interact with gut microbes to regulate growth. The content of breast milk HMOs from mothers in two Malawian birth cohorts was characterized. One cohort included infants who exhibited healthy growth, whereas the other cohort included infants who were severely stunted. The mothers of the stunted infants exhibited a significantly lower abundance of HMOs in breast milk at six months. The reduced HMO content mainly concerned sialylated HMOs, including sialyllacto-N-tetraose b, which was revealed to be the most growth-discriminatory [97]. Next, GF mice and piglets were colonized with a consortium of cultured bacterial strains from the stool of a severely stunted infant. The animals were fed a representative Malawian diet. One group of animals received supplementation of sialylated bovine milk oligosaccharides (S-BMO) that was structurally similar to sialylated HMOs, whereas the other group did not. Supplementation of the animals with S-BMO stimulated weight gain and bone volume. This effect was not evident in the GF animals. The results established that sialylated HMOs trigger a microbiota-dependent promotion of growth.
HMO composition depends on maternal genetics. Mothers who are carriers of an active fucosyltransferase 2 (FUT2) gene, categorized as secretors, produce more HMOs [94]. The composition of the infant gut community depends on the maternal secretor status; in the children of secretors, Bifidobacterium is more abundant [98]. However, the fact that the mother is a carrier of FUT2 per se is not associated with infant growth [99]. The relationship between HMO composition and the growth of children has already been demonstrated in many studies [89,100,101]
7. Microbiota and Growth Hormone/Insulin-like Growth Factor-1 Axis
The major controller of growth in children is the activity of the somatotropic axis. When a child experiences a nutritional deficiency over a long period of time, growth hormone (GH) resistance develops, and the child becomes stunted. The liver and peripheral tissues, including muscles, produce insulin-like growth factor-1 (IGF-1), which promotes systemic and organ growth. IGF-1 is a key mediator of skeletal growth, acting in an endocrine, paracrine, and autocrine manner [102]. The production of IGF-1 is controlled by GH, secreted by the anterior pituitary gland. One of the most interesting mechanisms that could explain the relationship between gut microbiota composition and linear and ponderal growth of children involves the role of intestinal microbes in the promotion of growth hormones. The evidence for this concept comes from the research conducted by Shin et al. [103] and Storelli et al. [104] on Drosophila melanogaster. In the first study, the colonization of germ-free (GF) larvae with Acetobacter pomorum was shown to restore the developmental rate and body size of the host [103]. The results of the second study showed that the strain of Lactobacillus plantarum was sufficient to stimulate the growth of the host under nutrient-poor conditions [104]. Schwarzer et al. [105] documented the impact of gut microbiota on the growth of mice in the neonatal period. The studies compared the growth parameters of wild-type (WT) and germ-free (GF) infant male mice fed a standard breeding diet through early adulthood. The GF mice exhibited significantly lower rates of growth and weight gain than the WT animals. Bone mass and femur length were also lower in the GF mice. Furthermore, the GF mice presented reduced IGF-1 and insulin-like growth factor binding protein-3 (IGFBP-3) levels and reduced expression of Igf1 and Igfbp3 in the liver compared to the WT animals. When the mice were fed a nutrient-depleted diet, the GF animals exhibited greater weight loss and a significant decrease in the expression of Igf1 and Ghr genes in the liver and muscles compared to the WT mice. This evidence indicates that the GF mice developed GH resistance. Additionally, as a result of injections of recombinant IGF-1 (rIGH-1) in the GF mice, there was an increase in body weight and body length, whereas in the WT mice, this effect was not observed. This led to the conclusion that the gut microbiota stimulates growth by facilitating IGF-1 production and activity. In addition, the colonization of GF Drosophila [104,106,107] and GF mice [68,105] with conventional microbiota, including a specific strain of L. plantarum, restored normal growth and production, as well as IGF-1 activity. Moreover, in GF animals, in which GH resistance was observed, it also restored the sensitivity of peripheral tissues to this hormone. A similar effect of growth stimulation in the presence of Lactobacillus rhamnosus was also observed in zebrafish [108].
It is not fully understood how the microbiota stimulates IGF-1 production. It seems, however, that the link between the gut microbiota and the regulation of IGF-1 of the host are byproducts produced by these microbes, whose effects on host IGF-1 levels have been observed [102]. This link has become apparent since Lactobacillus emerged as one of the key microbes affecting IGF-1 levels, and the members of this genus are well-known SCFA producers [109]. In the study of Yan et al. [68], supplementation of SCFAs from antibiotic-treated mice increased the IGF-1 level. Therefore, SCFAs, which are metabolized by gut bacteria from otherwise indigestible-fiber-rich diets, seem to be the most important link between intestinal microbes and the host IGF-1 level, but other mechanisms through which gut microbes affect bone growth probably exist [68,103].
Microbiota and Other Hormones
In addition to its mediating role in regulating the activity of the somatotropic axis, the gut microbiota indirectly influences the levels of other hormones in the host, such as PYY, GLP-1, leptin, ghrelin, and serotonin, and it also plays a mediating role in the activity of the adrenal axis [109,110]. Therefore, it is not an exaggeration to conclude that the gut microbiota can be considered an endocrine organ.
8. Conclusions
The intestinal microbiota is involved in a wide range of processes that regulate the growth of children. The mediating role of gut microbes in growth consists of participation in metabolic processes through the secretion of enzymes and the production of biologically active metabolites (e.g., SCFAs) as well as in the metabolism of HMO and BAs, participation in the hormonal activity of the host through indirect impact on the hormone levels (including IGF-1, PYY or GLP-1), and participation in immune responses and processes leading to inflammation (LPSs). All the aforementioned mechanisms do not exhaust the complexity of the relationship between the course of intestinal succession and the course of children’s growth. There is no doubt, however, that due to the wide range of interactions between the gut microbiota and growth processes, disturbances in gut microbial succession may negatively affect patterns of child growth.
Author Contributions
M.D.-M. conceptualized the paper and drafted the manuscript. J.C.-B. and I.M. reviewed and summarized available research. A.S. drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
All the authors declare that this research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors, and thus there is no conflict of interest regarding this paper.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Scarpellini E., Ianiro G., Attili F., Bassanelli C., De Santis A., Gasbarrini A. The human gut microbiota and virome: Potential therapeutic implications. Dig. Liver Dis. 2015;47:1007–1012. doi: 10.1016/j.dld.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koskinen K., Pausan M.-R., Perras A.K., Beck M., Bang C., Mora M., Schilhabel A., Schmitz R., Moissl-Eichinger C. First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio. 2017;8:e00824-17. doi: 10.1128/mBio.00824-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laforest-Lapointe I., Arrieta M.-C. Microbial Eukaryotes: A Missing Link in Gut Microbiome Studies. mSystems. 2018;3:e00201-17. doi: 10.1128/mSystems.00201-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajzer M., Seeley R.J. Obesity and gut flora. Nature. 2006;444:1009–1010. doi: 10.1038/4441009a. [DOI] [PubMed] [Google Scholar]
- 5.Smith K., McCoy K.D., Macpherson A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Flint H.J., Bayer E.A., Rincon M.T., Lamed R., White B.A. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 7.Belkaid Y., Hand T.W. Role of the Microbiota in Immunity and Inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanton L.V., Charbonneau M.R., Salih T., Barratt M.J., Venkatesh S., Ilkaveya O., Subramanian S., Manary M.J., Trehan I., Jorgensen J.M., et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351:aad3311. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodríguez J.M., Murphy K., Stanton C., Ross R.P., Kober O.I., Juge N., Avershina E., Rudi K., Narbad A., Jenmalm M.C., et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015;26:26050. doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balasundaram P., Avulakunta I.D. StatPearls. StatPearls Publishing; Treasure Island, FL, USA: 2021. Human Growth and Development. [PubMed] [Google Scholar]
- 11.Beunen G., Malina R.M. Growth and Biologic Maturation: Relevance to Athletic Performance. In: Hebestreit H., Bar-Or O., editors. The Young Athlete. Blackwell Publishing Ltd.; Oxford, UK: 2007. pp. 3–17. [Google Scholar]
- 12.Faye C.M., Fonn S., Levin J., Kimani-Murage E. Analysing child linear growth trajectories among under-5 children in two Nairobi informal settlements. Public Health Nutr. 2019;22:2001–2011. doi: 10.1017/S1368980019000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haith M.M., Benson J.B. Encyclopedia of Infant and Early Childhood Development. Elsevier; Amsterdam, The Netherlands: 2008. [Google Scholar]
- 14.WHO Team . Nutrition and Food Safety WHO Child Growth Standards: Length/Height-for-Age, Weight-for-Age, Weight-for-Length, Weight-for-Height and Body Mass Index-for-Age: Methods and Development 2006. WHO; Geneve, Switzerland: 2006. [Google Scholar]
- 15.Haymond M., Kappelgaard A.-M., Czernichow P., Biller B.M.K., Takano K., Kiess W. The participants in the global advisory panel meeting on the effects of growth hormone Early recognition of growth abnormalities permitting early intervention. Acta Paediatr. 2013;102:787–796. doi: 10.1111/apa.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Victora C.G., Adair L., Fall C., Hallal P.C., Martorell R., Richter L., Sachdev H.S., Maternal and Child Undernutrition Study Group Maternal and child undernutrition: Consequences for adult health and human capital. Lancet. 2008;371:340–357. doi: 10.1016/S0140-6736(07)61692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legler J.D., Rose L.C. Assessment of abnormal growth curves. Am. Fam. Physician. 1998;58:153. [PubMed] [Google Scholar]
- 18.WHO Team . Nutrition and Food Safety WHO Child Growth Standards: Growth Velocity Based on Weight, Length and Head Circumference: Methods and Development 2009. WHO; Geneve, Switzerland: 2009. [Google Scholar]
- 19.Victora C.G., de Onis M., Hallal P.C., Blössner M., Shrimpton R. Worldwide Timing of Growth Faltering: Revisiting Implications for Interventions. Pediatrics. 2010;125:e473–e480. doi: 10.1542/peds.2009-1519. [DOI] [PubMed] [Google Scholar]
- 20.Cole T.J., Bellizzi M.C., Flegal K.M., Dietz W.H. Establishing a standard definition for child overweight and obesity worldwide: International survey. BMJ. 2000;320:1240. doi: 10.1136/bmj.320.7244.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole T.J., Flegal K.M., Nicholls D., Jackson A.A. Body mass index cut offs to define thinness in children and adolescents: International survey. BMJ. 2007;335:194–197. doi: 10.1136/bmj.39238.399444.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adair L.S., Cole T.J. Rapid Child Growth Raises Blood Pressure in Adolescent Boys Who Were Thin at Birth. Hypertension. 2003;41:451–456. doi: 10.1161/01.HYP.0000054212.23528.B2. [DOI] [PubMed] [Google Scholar]
- 23.Ong K.K.L., Ahmed M.L., Emmett P.M., Preece M.A., Dunger P.D.B. Association between postnatal catch-up growth and obesity in childhood: Prospective cohort study. BMJ. 2000;320:967–971. doi: 10.1136/bmj.320.7240.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black R.e., Victora C.g., Walker S.P., Bhutta Z.A., Christian P., de Onis M., Ezzati M., Grantham-McGregor S., Katz J., Martorell R., et al. Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet. 2013;382:427–451. doi: 10.1016/S0140-6736(13)60937-X. [DOI] [PubMed] [Google Scholar]
- 25.Graber E., Rapaport R. Growth and Growth Disorders in Children and Adolescents. Pediatr. Ann. 2012;41:e65–e72. doi: 10.3928/00904481-20120307-07. [DOI] [PubMed] [Google Scholar]
- 26.Ferretti P., Pasolli E., Tett A., Asnicar F., Gorfer V., Fedi S., Armanini F., Truong D.T., Manara S., Zolfo M., et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe. 2018;24:133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiménez E., Marín M.L., Martín R., Odriozola J.M., Olivares M., Xaus J., Fernández L., Rodríguez J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Ardissone A.N., De La Cruz D.M., Davis-Richardson A.G., Rechcigl K.T., Li N., Drew J.C., Murgas-Torrazza R., Sharma R., Hudak M.L., Triplett E.W., et al. Meconium Microbiome Analysis Identifies Bacteria Correlated with Premature Birth. PLoS ONE. 2014;9:e90784. doi: 10.1371/journal.pone.0090784. Erratum in PLoS ONE 2014, 9, e101399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prince A.L., Ma J., Kannan P.S., Alvarez M., Gisslen T., Harris R.A., Sweeney E.L., Knox C.L., Lambers D.S., Jobe A.H., et al. The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am. J. Obstet. Gynecol. 2016;214:627.e1–627.e16. doi: 10.1016/j.ajog.2016.01.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominguez-Bello M.G., Costello E.K., Contreras M., Magris M., Hidalgo G., Fierer N., Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks B., Olm M.R., Firek B.A., Baker R., Geller-McGrath D., Reimer S.R., Soenjoyo K.R., Yip J.S., Dahan D., Thomas B.C., et al. The developing premature infant gut microbiome is a major factor shaping the microbiome of neonatal intensive care unit rooms. Microbiome. 2018;6:1–12. doi: 10.1186/s40168-018-0493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pabst O., Cerovic V., Hornef M. Secretory IgA in the Coordination of Establishment and Maintenance of the Microbiota. Trends Immunol. 2016;37:287–296. doi: 10.1016/j.it.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Korpela K., de Vos W.M. Early life colonization of the human gut: Microbes matter everywhere. Curr. Opin. Microbiol. 2018;44:70–78. doi: 10.1016/j.mib.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Palmer C., Bik E.M., DiGiulio D.B., Relman D.A., Brown P.O. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maynard C.L., Elson C.O., Hatton R.D., Weaver C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gensollen T., Iyer S.S., Kasper D.L., Blumberg R.S. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson R.C., Manges A.R., Finlay B.B., Prendergast A.J. The Human Microbiome and Child Growth – First 1000 Days and Beyond. Trends Microbiol. 2018;27:131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Naik D.B., Kulkarni A.P., Aswar N.R. Birth weight and anthropometry of newborns. Indian J. Pediatr. 2003;70:145–146. doi: 10.1007/BF02723742. [DOI] [PubMed] [Google Scholar]
- 39.Stewart C.J., Ajami N.J., O’Brien J.L., Hutchinson D.S., Smith D.P., Wong M.C., Ross M.C., Lloyd R.E., Doddapaneni H., Metcalf G.A., et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asnicar F., Manara S., Zolfo M., Truong D.T., Scholz M., Armanini F., Ferretti P., Gorfer V., Pedrotti A., Tett A., et al. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. mSystems. 2017;2:e00164-16. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korpela K., Costea P.I., Coelho L.P., Kandels-Lewis S., Willemsen G., Boomsma D.I., Segata N., Bork P. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018;28:561–568. doi: 10.1101/gr.233940.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H., et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Koenig J.E., Spor A., Scalfone N., Fricker A.D., Stombaugh J., Knight R., Angenent L.T., Ley R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA. 2011;108((Suppl. 1)):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korpela K., Helve O., Kolho K.-L., Saisto T., Skogberg K., Dikareva E., Stefanovic V., Salonen A., Andersson S., de Vos W.M. Maternal Fecal Microbiota Transplantation in Cesarean-Born Infants Rapidly Restores Normal Gut Microbial Development: A Proof-of-Concept Study. Cell. 2020;183:324–334.e5. doi: 10.1016/j.cell.2020.08.047. [DOI] [PubMed] [Google Scholar]
- 45.Pannaraj P.S., Li F., Cerini C., Bender J.M., Yang S., Rollie A., Adisetiyo H., Zabih S., Lincez P.J., Bittinger K., et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. 2017;171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Festi D., Schiumerini R., Birtolo C., Marzi L., Montrone L., Scaioli E., Di Biase A.R., Colecchia A. Gut Microbiota and Its Pathophysiology in Disease Paradigms. Dig. Dis. 2011;29:518–524. doi: 10.1159/000332975. [DOI] [PubMed] [Google Scholar]
- 47.Smith M.I., Yatsunenko T., Manary M.J., Trehan I., Mkakosya R., Cheng J., Kau A.L., Rich S.S., Concannon P., Mychaleckyj J.C., et al. Gut Microbiomes of Malawian Twin Pairs Discordant for Kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pham T.-P.-T., Alou M.T., Bachar D., Levasseur A., Brah S., Alhousseini D., Sokhna C., Diallo A., Wieringa F., Million M., et al. Gut Microbiota Alteration is Characterized by a Proteobacteria and Fusobacteria Bloom in Kwashiorkor and a Bacteroidetes Paucity in Marasmus. Sci. Rep. 2019;9:9084. doi: 10.1038/s41598-019-45611-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Subramanian S., Huq S., Yatsunenko T., Haque R., Mahfuz M., Alam M.A., Benezra A., DeStefano J., Meier M.F., Muegge B., et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dinh D.M., Ramadass B., Kattula D., Sarkar R., Braunstein P., Tai A., Wanke C.A., Hassoun S., Kane A.V., Naumova E.N., et al. Longitudinal Analysis of the Intestinal Microbiota in Persistently Stunted Young Children in South India. PLoS ONE. 2016;11:e0155405. doi: 10.1371/journal.pone.0155405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balasubramaniam C., Mallappa R.H., Singh D.K., Chaudhary P., Bharti B., Muniyappa S.K., Grover S. Gut bacterial profile in Indian children of varying nutritional status: A comparative pilot study. Eur. J. Nutr. 2021;60:3971–3985. doi: 10.1007/s00394-021-02571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gough E.K., Stephens D.A., Moodie E.E.M., Prendergast A.J., Stoltzfus R.J., Humphrey J.H., Manges A.R. Linear growth faltering in infants is associated with Acidaminococcus sp. and community-level changes in the gut microbiota. Microbiome. 2015;3:24. doi: 10.1186/s40168-015-0089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Méndez-Salazar E.O., Ortiz-López M.G., Granados-Silvestre M.D.L., Palacios-González B., Menjivar M. Altered Gut Microbiota and Compositional Changes in Firmicutes and Proteobacteria in Mexican Undernourished and Obese Children. Front. Microbiol. 2018;9:2494. doi: 10.3389/fmicb.2018.02494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun L., Ma L., Ma Y., Zhang F., Zhao C., Nie Y. Insights into the role of gut microbiota in obesity: Pathogenesis, mechanisms, and therapeutic perspectives. Protein Cell. 2018;9:397–403. doi: 10.1007/s13238-018-0546-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jian C., Carpén N., Helve O., de Vos W.M., Korpela K., Salonen A. Early-life gut microbiota and its connection to metabolic health in children: Perspective on ecological drivers and need for quantitative approach. EBioMedicine. 2021;69:103475. doi: 10.1016/j.ebiom.2021.103475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kovatcheva-Datchary P., Arora T. Nutrition, the gut microbiome and the metabolic syndrome. Best Pract. Res. Clin. Gastroenterol. 2013;27:59–72. doi: 10.1016/j.bpg.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 57.Holmes Z.C., Silverman J.D., Dressman H.K., Wei Z., Dallow E.P., Armstrong S.C., Seed P.C., Rawls J.F., David L.A. Short-Chain Fatty Acid Production by Gut Microbiota from Children with Obesity Differs According to Prebiotic Choice and Bacterial Community Composition. mBio. 2020;11:e00914-20. doi: 10.1128/mBio.00914-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 59.Mohajeri M.H., Brummer R.J.M., Rastall R.A., Weersma R.K., Harmsen H.J.M., Faas M., Eggersdorfer M. The role of the microbiome for human health: From basic science to clinical applications. Eur. J. Nutr. 2018;57:1–14. doi: 10.1007/s00394-018-1703-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolognini D., Tobin A.B., Milligan G., Moss C.E. The Pharmacology and Function of Receptors for Short-Chain Fatty Acids. Mol. Pharmacol. 2015;89:388–398. doi: 10.1124/mol.115.102301. [DOI] [PubMed] [Google Scholar]
- 61.Silva Y.P., Bernardi A., Frozza R.L. The Role of Short-Chain Fatty Acids from Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020;11:25. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roediger W.E. Utilization of Nutrients by Isolated Epithelial Cells of the Rat Colon. Gastroenterology. 1982;83:424–429. doi: 10.1016/S0016-5085(82)80339-9. [DOI] [PubMed] [Google Scholar]
- 63.Davie J.R. Inhibition of Histone Deacetylase Activity by Butyrate. J. Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 64.Smith P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly-Y M., Glickman J.N., Garrett W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee C., Kim B.G., Kim J.H., Chun J., Im J.P., Kim J.S. Sodium butyrate inhibits the NF-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int. Immunopharmacol. 2017;51:47–56. doi: 10.1016/j.intimp.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 66.Yan H., Ajuwon K.M. Butyrate modifies intestinal barrier function in IPEC-J2 cells through a selective upregulation of tight junction proteins and activation of the Akt signaling pathway. PLoS ONE. 2017;12:e0179586. doi: 10.1371/journal.pone.0179586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alfonso-Prieto M., Navarini L., Carloni P. Understanding Ligand Binding to G-Protein Coupled Receptors Using Multiscale Simulations. Front. Mol. Biosci. 2019;6:29. doi: 10.3389/fmolb.2019.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan J., Herzog J.W., Tsang K., Brennan C.A., Bower M.A., Garrett W.S., Sartor B.R., Aliprantis A.O., Charles J.F. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl. Acad. Sci. USA. 2016;113:E7554–E7563. doi: 10.1073/pnas.1607235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Russell S.L., Gold M.J., Hartmann M., Willing B.P., Thorson L., Wlodarska M., Gill N., Blanchet M.-R., Mohn W.W., McNagny K.M., et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X., Zhivaki D., Lo-Man D.Z.R. Unique aspects of the perinatal immune system. Nat. Rev. Immunol. 2017;17:495–507. doi: 10.1038/nri.2017.54. [DOI] [PubMed] [Google Scholar]
- 72.Gordon S. Pattern recognition receptors: Doubling up for the innate immune response. Cell. 2002;111:927–930. doi: 10.1016/S0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- 73.Yücel G., Zhao Z., El-Battrawy I., Lan H., Lang S., Li X., Buljubasic F., Zimmermann W.-H., Cyganek L., Utikal J., et al. Lipopolysaccharides induced inflammatory responses and electrophysiological dysfunctions in human-induced pluripotent stem cell derived cardiomyocytes. Sci. Rep. 2017;7:2935. doi: 10.1038/s41598-017-03147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lancaster G.I., Langley K.G., Berglund N.A., Kammoun H.L., Reibe S., Estevez E., Weir J., Mellett N.A., Pernes G., Conway J.R.W., et al. Evidence that TLR4 Is Not a Receptor for Saturated Fatty Acids but Mediates Lipid-Induced Inflammation by Reprogramming Macrophage Metabolism. Cell Metab. 2018;27:1096–1110.e5. doi: 10.1016/j.cmet.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 75.Takeuchi O., Akira S. Genetic approaches to the study of Toll-like receptor function. Microbes Infect. 2002;4:887–895. doi: 10.1016/S1286-4579(02)01615-5. [DOI] [PubMed] [Google Scholar]
- 76.Martin A.M., Sun E.W., Rogers G.B., Keating D.J. The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front. Physiol. 2019;10:428. doi: 10.3389/fphys.2019.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Larraufie P., Martin-Gallausiaux C., Lapaque N., Dore J., Gribble F.M., Reimann F., Blottiere H.M. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci. Rep. 2018;8:74. doi: 10.1038/s41598-017-18259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lebrun L.J., Lenaerts K., Kiers D., de Barros J.-P.P., Le Guern N., Plesnik J., Thomas C., Bourgeois T., Dejong C.H., Kox M., et al. Enteroendocrine L Cells Sense LPS after Gut Barrier Injury to Enhance GLP-1 Secretion. Cell Rep. 2017;21:1160–1168. doi: 10.1016/j.celrep.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 79.Kidd M., Gustafsson B.I., Drozdov I., Modlin I.M. IL1β- and LPS-induced serotonin secretion is increased in EC cells derived from Crohn’s disease. Neurogastroenterol. Motil. 2009;21:439–450. doi: 10.1111/j.1365-2982.2008.01210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roager H.M., Licht T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.De León D.D., Crutchlow M.F., Ham J.-Y.N., Stoffers D.A. Role of glucagon-like peptide-1 in the pathogenesis and treatment of diabetes mellitus. Int. J. Biochem. Cell Biol. 2006;38:845–859. doi: 10.1016/j.biocel.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 82.Cani P.D., Amar J., Iglesias M.A., Poggi M., Knauf C., Bastelica D., Neyrinck A.M., Fava F., Tuohy K.M., Chabo C.W., et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 83.Brun P., Castagliuolo I., Di Leo V., Buda A., Pinzani M., Palù G., Martines D. Increased intestinal permeability in obese mice: New evidence in the pathogenesis of nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G518–G525. doi: 10.1152/ajpgi.00024.2006. [DOI] [PubMed] [Google Scholar]
- 84.Krajmalnik-Brown R., Ilhan Z.-E., Kang D.-W., DiBaise J.K. Effects of Gut Microbes on Nutrient Absorption and Energy Regulation. Nutr. Clin. Pract. 2012;27:201–214. doi: 10.1177/0884533611436116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wahlström A., Sayin S.I., Marschall H.-U., Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 86.Jones B.V., Begley M., Hill C., Gahan C.G.M., Marchesi J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hylemon P.B., Zhou H., Pandak W.M., Ren S., Gil G., Dent P. Bile acids as regulatory molecules. J. Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanaka M., Sanefuji M., Morokuma S., Yoden M., Momoda R., Sonomoto K., Ogawa M., Kato K., Nakayama J. The Association between Gut Microbiota Development and Maturation of Intestinal Bile Acid Metabolism in the First 3 y of Healthy Japanese Infants. [(accessed on 18 November 2021)];Gut Microbes. 2020 11:205–216. doi: 10.1080/19490976.2019.1650997. Available online: https://pubmed.ncbi.nlm.nih.gov/31550982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lagström H., Rautava S., Ollila H., Kaljonen A., Turta O., Mäkelä J., Yonemitsu C., Gupta J., Bode L. Associations between human milk oligosaccharides and growth in infancy and early childhood. Am. J. Clin. Nutr. 2020;111:769–778. doi: 10.1093/ajcn/nqaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng L., Kiewiet M.B.G., Logtenberg M.J., Groeneveld A., Nauta A., Schols H.A., Walvoort M.T.C., Harmsen H.J.M., De Vos P. Effects of Different Human Milk Oligosaccharides on Growth of Bifidobacteria in Monoculture and Co-culture With Faecalibacterium prausnitzii. Front. Microbiol. 2020;11:569700. doi: 10.3389/fmicb.2020.569700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morrow A.L., Ruiz-Palacios G.M., Altaye M., Jiang X., Guerrero M.L., Meinzen-Derr J.K., Farkas T., Chaturvedi P., Pickering L.K., Newburg D.S. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J. Pediatr. 2004;145:297–303. doi: 10.1016/j.jpeds.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 92.Bode L. The functional biology of human milk oligosaccharides. Early Hum. Dev. 2015;91:619–622. doi: 10.1016/j.earlhumdev.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 93.Wiciński M., Sawicka E., Gębalski J., Kubiak K., Malinowski B. Human Milk Oligosaccharides: Health Benefits, Potential Applications in Infant Formulas, and Pharmacology. Nutrients. 2020;12:266. doi: 10.3390/nu12010266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J., Belzer C., Delgado Palacio S., Arboleya Montes S., Mancabelli L., et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017;81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pokusaeva K., Fitzgerald G.F., Van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harmsen H.J.M., Wildeboer–Veloo A.C.M., Raangs G.C., Wagendorp A.A., Klijn N., Bindels J.G., Welling G.W. Analysis of Intestinal Flora Development in Breast-Fed and Formula-Fed Infants by Using Molecular Identification and Detection Methods. J. Pediatr. Gastroenterol. Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 97.Charbonneau M.R., O’Donnell D., Blanton L.V., Totten S.M., Davis J.C.C., Barratt M.J., Cheng J., Guruge J., Talcott M., Bain J.R., et al. Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell. 2016;164:859–871. doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith-Brown P., Morrison M., Krause L., Davies P.S.W. Mothers Secretor Status Affects Development of Childrens Microbiota Composition and Function: A Pilot Study. PLoS ONE. 2016;11:e0161211. doi: 10.1371/journal.pone.0161211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sprenger N., Lee L.Y., De Castro C.A., Steenhout P., Thakkar S.K. Longitudinal change of selected human milk oligosaccharides and association to infants’ growth, an observatory, single center, longitudinal cohort study. PLoS ONE. 2017;12:e0171814. doi: 10.1371/journal.pone.0171814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Larsson M.W., Lind M.V., Laursen R.P., Yonemitsu C., Larnkjær A., Mølgaard C., Michaelsen K.F., Bode L. Human Milk Oligosaccharide Composition Is Associated with Excessive Weight Gain During Exclusive Breastfeeding—An Explorative Study. Front. Pediatr. 2019;7:297. doi: 10.3389/fped.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alderete T.L., Autran C., Brekke B.E., Knight R., Bode L., Goran M.I., Fields D.A. Associations between human milk oligosaccharides and infant body composition in the first 6 mo of life. Am. J. Clin. Nutr. 2015;102:1381–1388. doi: 10.3945/ajcn.115.115451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yan J., Charles J.F. Gut Microbiota and IGF-1. Calcif. Tissue Int. 2018;102:406–414. doi: 10.1007/s00223-018-0395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shin S.C., Kim S.-H., You H., Kim B., Kim A.C., Lee K.-A., Yoon J.-H., Ryu J.-H., Lee W.-J. Drosophila Microbiome Modulates Host Developmental and Metabolic Homeostasis via Insulin Signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 104.Storelli G., Defaye A., Erkosar B., Hols P., Royet J., Leulier F. Lactobacillus plantarum Promotes Drosophila Systemic Growth by Modulating Hormonal Signals through TOR-Dependent Nutrient Sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 105.Schwarzer M., Makki K., Storelli G., Machuca-Gayet I., Srutkova D., Hermanova P., Martino M.E., Balmand S., Hudcovic T., Heddi A., et al. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016;351:854–857. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- 106.Hyun S. Body size regulation and insulin-like growth factor signaling. Cell. Mol. Life Sci. 2013;70:2351–2365. doi: 10.1007/s00018-013-1313-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Erkosar B., Storelli G., Defaye A., Leulier F. Host-Intestinal Microbiota Mutualism: “Learning on the Fly”. Cell Host Microbe. 2013;13:8–14. doi: 10.1016/j.chom.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 108.Avella M.A., Place A., Du S.-J., Williams E., Silvi S., Zohar Y., Carnevali O. Lactobacillus rhamnosus Accelerates Zebrafish Backbone Calcification and Gonadal Differentiation through Effects on the GnRH and IGF Systems. PLoS ONE. 2012;7:e45572. doi: 10.1371/journal.pone.0045572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jensen E.A., Young J.A., Mathes S.C., List E.O., Carroll R.K., Kuhn J., Onusko M., Kopchick J.J., Murphy E.R., Berryman D.E. Crosstalk between the growth hormone/insulin-like growth factor-1 axis and the gut microbiome: A new frontier for microbial endocrinology. Growth Horm. IGF Res. 2020;53:101333. doi: 10.1016/j.ghir.2020.101333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clarke G., Stilling R.M., Kennedy P.J., Stanton C., Cryan J.F., Dinan T.G. Minireview: Gut Microbiota: The Neglected Endocrine Organ. Mol. Endocrinol. 2014;28:1221–1238. doi: 10.1210/me.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.