Abstract
An outbreak of SARS-CoV-2 coronavirus (COVID-19) first detected in Wuhan, China, has created a public health emergency all over the world. The pandemic has caused more than 340 million confirmed cases and 5.57 million deaths as of 23 January 2022. Although carbohydrates have been found to play a role in coronavirus binding and infection, the role of cell surface glycans in SARS-CoV-2 infection and pathogenesis is still not understood. Herein, we report that the SARS-CoV-2 spike protein S1 subunit binds specifically to blood group A and B antigens, and that the spike protein S2 subunit has a binding preference for Lea antigens. Further examination of the binding preference for different types of red blood cells (RBCs) indicated that the spike protein S1 subunit preferentially binds with blood group A RBCs, whereas the spike protein S2 subunit prefers to interact with blood group Lea RBCs. Angiotensin converting enzyme 2 (ACE2), a known target of SARS-CoV-2 spike proteins, was identified to be a blood group A antigen-containing glycoprotein. Additionally, 6-sulfo N-acetyllactosamine was found to inhibit the binding of the spike protein S1 subunit with blood group A RBCs and reduce the interaction between the spike protein S1 subunit and ACE2.
Keywords: blood group, carbohydrate ligand, COVID-19, spike protein
1. Introduction
Since emerging from Wuhan, China in January 2020, the SARS-CoV-2 coronavirus has widely spread around the globe [1]. As of 23 January 2022, more than 210 countries have been affected by the pandemic, 340 million people have been diagnosed with the virus, and more than 5.57 million people have died. Although the number of new cases has been gradually decreasing, daily activities in most parts of the world have remained restricted. Therefore, deciphering the viral infection mechanism is of great urgency to facilitate the control and treatment of COVID-19.
The densely glycosylated spike (S) protein of SARS-CoV-2, a trimeric class I fusion protein with a metastable prefusion conformation [2,3], docks to enter host cells. When binding to a host-cell receptor, the S1 subunit triggers a dramatic structural rearrangement to fuse the viral membrane with the host-cell membrane, leading to receptor-dependent endocytosis [4,5]. These interactions destabilize the prefusion trimer and result in shedding of the S1 subunit and the transition of the S2 subunit into a stable postfusion conformation [6]. Several studies have shown the possible interactions between SARS-CoV-2 or the spike protein with the host receptors (angiotensin-converting enzyme 2 (ACE2) [7,8,9,10]; dipeptidyl peptidase 4 (DPP4) [11,12]; glucose-regulated protein 7 (GRP78) [13,14]); however, the discussion of cell surface glycan receptors used for SARS-CoV-2 viral binding and entry remained vague [15].
Cell surface glycan receptors are known to play a key role in mediating viral binding and infection. Jackson et al. indicated that the entry of foot-and-mouth disease virus (FMDV) into cells is initiated by the contact with heparin sulfate on the cell surface [16]. Sulfated polysaccharides extracted from sea algae have shown potential to prevent the infection of viruses, including herpes simplex virus (HSV), cytomegalovirus (CMV), human immunodeficiency virus (HIV), and enterovirus (EV) [17,18,19,20,21]. Lactoferrin, an 80 kDa iron-binding glycoprotein existing in several mucosal secretions [22,23], has been reported to inhibit the interaction of the capsid protein VP1 of EV71 with rhabdomyosarcoma cells [24,25]. In addition, sialic acids were reported as cell-surface ligands for many viral proteins of influenza virus, parainfluenza virus, reovirus type 3, adenovirus type 37, human rhinovirus 87, human enterovirus type 70 [26], EV-A71 [27], coxsackievirus A24 [28], and hepatitis A virus [29]. Human coronaviruses OC43, HKU1, and MERS were also shown to interact with sialic acids [30,31,32,33]. Clausen et al. indicated that the SARS-CoV-2 spike protein interacts with both cellular heparan sulfate and ACE2 through its receptor-binding domain [34]. Jayaprakash et al. also showed that the S1A domain of the SARS-CoV-2 spike protein may interact with sialosides by molecular modeling [35]. Furthermore, the N-glycans of DC-SIGN and L-SIGN that were identified to be the receptors for SARS-CoV-2 could influence the entry of coronavirus [36]. Therefore, it is important to study the role and function of cell surface glycans in the infection of SARS-CoV-2, which appears to be indispensable for understanding viral attachment, infection, and pathogenesis.
We employed a solution carbohydrate microarray to analyze the glycan recognition preference of SARS-CoV-2 spike proteins. The results showed that SARS-CoV-2 spike proteins interacted with several glycans, including blood group antigens, which was coherent with the binding with groups A and B human red blood cells (RBCs). In addition, the effects of carbohydrate analogs in the binding of spike proteins with RBCs were also evaluated.
2. Materials and Methods
2.1. Glycan Microarray Analysis
Glycan array screening was carried out using a rapid, nonwashing solution carbohydrate array as previously described [37]. Briefly, donor beads (500 ng/well) and biotin-polyacrylamide (PAA)-sugars (20 ng/well) (GlycoTech, Gaithersburg, MD, USA) mixed with SARS-CoV-2 spike protein S1 or S2 subunits (10–20 ng/well, REC31806-100 and REC31807-100 from The Native Antigen Company, Kidlington, Oxford, UK) were incubated at room temperature for 1 h (a total of 15 μL of reaction solution). A mixture of acceptor beads (500 ng/well) and rabbit antisheep IgG Fc antibodies (10 ng/well, ab102297 from Abcam, Cambridge, UK) was added to the reaction to reach a final volume of 25 μL. All reactions were performed in the dark. After incubation for 2 h, the binding signals were measured from a PerkinElmer EnVision instrument and analyzed using the AlphaScreenTM detection program. The results are expressed as relative fluorescence intensities and based on the average from three independent assays.
2.2. Binding of Spike Proteins to RBCs Evaluated by Fluorescence-Activated Cell Sorting
The prewashed A, B, and O blood type RBCs were obtained from the Blood Bank of Department of Pathology, National Cheng Kung University Hospital. Two micrograms of spike protein S1 or S2 subunits was incubated with different blood groups of RBCs at 4 °C for 24 h. After incubation, RBCs were isolated from the reaction mixture by centrifugation to remove the nonbinding spike proteins. Rabbit antisheep IgG Fc antibodies (5 μg, purchased from Abcam, ab102297) were added to RBCs and the mixture was incubated at 4 °C for 24 h. RBC samples were then washed three times, followed by the addition of Alexa fluor-488-conjugated goat antirabbit IgG F(ab′)2 (2 μg, 23901 from Leadgene, Tainan, Taiwan) and incubated for 1 h at 37 °C. The fluorescence-activated cell sorting (FACS) analysis of spike protein bound RBCs was performed on a FACSCalibur flow cytometer (BD Biosciences). The percentages of blood group A, B, and O RBCs with and without bound spike proteins are indicated in histograms.
2.3. Inhibition Assays by Flow Cytometry
Lactose and three sulfated carbohydrate derivatives (20 μg/200 μL) were incubated with spike protein S1 subunits (2 µg) for 12 h and were then mixed with blood group A RBCs at 4 °C for an additional 24 h. After incubation, RBCs were isolated from the reaction mixture by centrifugation then mixed with rabbit anti-sheep Fc antibody (5 μg) at 4 °C for 24 h. Alexa fluor-488-conjugated goat antirabbit IgG F(ab′)2 (2 μg) was added after three washes and incubated at 37 °C for 1 h. The percentage of blood group A RBCs with bound spike protein S1 subunits was determined from the FL1 channel in flow cytometry.
2.4. Preparation of Lung Tissue Lysate
This study was approved by the National Cheng Kung University Hospital Institutional Review Board (IRB No: A-ER-107-085). The lung cancer tissues were obtained from the Human Biobank of National Cheng Kung University Hospital. The cancer tissues were cut into small pieces and then homogenized thoroughly with the addition of RIPA buffer (Leadgene) and protease inhibitor (Roche). The homogenized tissue samples were centrifuged at 13,000 rpm at 4 °C for 20 min. The supernatant was collected as the whole-cell extract of lung tissue lysate. The protein concentration of the tissue lysate was determined using the Lowry method.
2.5. Co-Immunoprecipitation Assays
The lung tissue lysate was precleaned by adding 10 μL of Protein A/G Plus Agarose (Sigma, Merck KGaA, Darmstadt, Germany) and incubated at 4 °C for 1 h. Rabbit anti-ACE2 monoclonal antibody (Proteintech, Chicago, USA), rabbit antiblood group A antibody (ARP), or antirabbit IgG (Genetex, CA, USA) was mixed with Protein A/G Plus Agarose and incubated at 4 °C for 4 h. The antibody–agarose mixtures were individually mixed with precleaned tissue lysate and incubated at 4 °C overnight. After incubation, agaroses were washed 5 times with lysis buffer and the bound proteins were eluted with SDS-PAGE sample buffer.
2.6. Western Blotting
The co-immunoprecipitated proteins were mixed with SDS-PAGE sample buffer 6 times, denatured for 5 min at 95 °C, and separated by 6% SDS-PAGE using electrophoresis. Proteins were transferred to PVDF membranes (Millipore, Merck KGaA, Darmstadt, Germany) for 90 min at 300 A and the membranes were blocked for 1 h at 25 °C in 5% milk. Rabbit anti-ACE2 antibody (Proteintech) or mouse antiblood group A antibody (ARP) was incubated with membranes at 4 °C overnight. After being washed 3 times, membranes were incubated with goat antirabbit IgG or goat anti-mouse IgG for 1 h at 25 °C. The detection of the signal was performed with an enhanced chemiluminescence detection kit (Millipore). The gels were digitally photographed and scanned using a gel documentation system (ImageQuant™ LAS 4000).
2.7. ELISA
Spike protein S1 subunits (1 μg/mL) were coated on 96-well ELISA plate at 4 °C for 16 h. A blocking procedure was performed by the addition of 0.5% BSA in TBST at 25 °C for 1 h. For inhibition assays, lactose or sulfated carbohydrate derivatives (20 μg/mL) were added into wells and incubated at 25 °C for 1 h followed by two washes. Lung tissue lysates (10 and 1 μg/mL) were added to the well (with and without inhibiter incubation) and incubated at 25 °C for 2 h. After incubation, the reaction mixture in each well was removed and then washed 3 times with PBS. In order to detect the bound ACE2, human ACE2-specific rabbit antibodies (1:1000, Proteintech, Chicago, USA) were added for incubation at 25 °C for 2 h. After washing, HRP-conjugated anti-rabbit IgG antibodies (1:5000, Leadgene) were added for incubation at 37 °C for 1 h. After incubation, the unbound HRP-conjugated antirabbit IgG antibody was washed away and the substrate 3,3′,5,5′-tetramethylbenzidine (TMB, Sigma, St. Louis, MO, USA) was added for incubation at 25 °C for 30 min. Reactions were quenched by adding H2SO4 (1 N) and the absorbance at 450 nm (OD450) was measured by an ELISA reader (Epoch BioTek) in order to determine the quantity of ACE2 in each well.
3. Results
3.1. Sugar-Binding Profiling Analysis of SARS-CoV-2 Spike Proteins
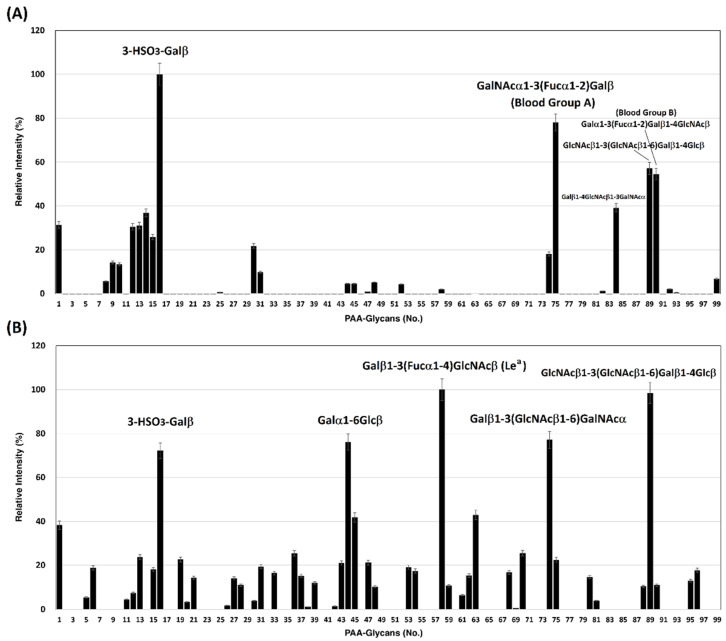
We previously developed a homogeneous solution carbohydrate microarray in which polyacrylamide-based glycans are used to offer a multivalent environment to screen for specific carbohydrates. There are two advantages to this microarray. This platform can be carried out in a high-throughput manner because the washing step is not required during the screening [37] and is suitable to measure weak binding events that are typical in carbohydrate–protein interactions. So far, this platform has successfully demonstrated the carbohydrate-binding specificities of lectins, antibodies, influenza virus hemagglutinins, and influenza viral particles [38,39]. Using this solution carbohydrate microarray that contains 97 different glycans (Table 1), SARS-CoV-2 spike protein S1 subunits were bound specifically to 3-HSO3-Galβ (#16), GalNAcα1-3(Fucα1-2)Galβ (Blood Group A trisaccharide, #75), GlcNAcβ1-3(GlcNAcβ1-6)Galβ1-4Glcβ (#89), and Galα1-3(Fucα1-2)Galβ1-4GlcNAcβ (Blood Group B type 2 tetrasaccharide, #90) (Figure 1A). SARS-CoV-2 spike protein S2 subunits displayed preferential interactions with 3-HSO3-Galβ (#16), Galβ1-6Glcβ (melibiose, #44), Galβ1-3(Fucα1-4)GlcNAcβ (Lea, #58), Galβ1-3(GlcNAcβ1-6)GalNAcα (#74), and GlcNAcβ1-3(GlcNAcβ1-6)Galβ1-4Glcβ (#89) (Figure 1B). The carbohydrate binding preferences of spike protein S1 and S2 subunits with a relative intensity cutoff of 50% are listed in Table 2.
Table 1.
List of PAA-glycans.
| No. | Glycans | No. | Glycans | No. | Glycans |
|---|---|---|---|---|---|
| 1 | PAA-biotin (backbone) | 34 | GlcNAcβ1-3Galβ | 67 | GlcNAcβ1-2Galβ1-3GalNAcα |
| 2 | β-GlcNAc | 35 | Galα1-4GlcNAcβ (αLacNAc) | 68 | NeuAcα2-3Galβ1-4GlcNAcβ |
| 3 | α-Glucose | 36 | Glcα1-4Glcβ | 69 | NeuAcα2-3Galβ1-3Glcβn (3’Sialyl Lec) |
| 4 | β-Glucose | 37 | Galβ1-3GalNAcα, sp = -p-OC6H4- | 70 | Galα1-3Galβ1-4GlcNAcβ, sp = -NHCOCH2NH- |
| 5 | α-Galactose | 38 | Galα1-2Galβ | 71 | GlcNAcα1-3Galβ1-3GalNAcα |
| 6 | β-Galactose | 39 | GlcNAcβ1-4GlcNAcβ | 72 | NeuAcα2-8NeuAcα2-8NeuAc, (NeuAcα2-8)3 |
| 7 | α-Man-6-phosphate | 40 | GlcNAcβ1-4GlcNAcβ, sp = -NHCOCH2NH- | 73 | GlcNAcβ1-3Galβ1-3GalNAcα |
| 8 | α-L-Rhamnose | 41 | NeuAcα2-6GalNAcα | 74 | Galβ1-3(GlcNAcβ1-6)GalNAcα |
| 9 | β-GlcNAc | 42 | 3-HSO3-Galβ1-4GlcNAcβ | 75 | GalNAcα1-3(Fucα1-2)Galβ (Blood Group A), sp = (CH2)3NHCO(CH2)5NH- |
| 10 | α-GalNAc | 43 | 3-HSO3-Galβ1-3GlcNAcβ | 76 | GlcNAcβ1-3Galβ1-4GlcNAcβ |
| 11 | β-GalNAc | 44 | Galα1-6Glcβ (melibiose) | 77 | NeuAcα2-3Galβ1-3GalNAcα |
| 12 | α-Fuc | 45 | NeuAcα2-8NeuAcα, (NeuAcα2-8)2 | 78 | GlcNAcβ1-3(GlcNAcβ1-6)GalNAcα |
| 13 | α-NeuAc | 46 | Galβ1-2Galβ | 79 | Galα1-4Galβ1-4GlcNAcβ |
| 14 | α-NeuAc-OCH2C6H4-p-NHCOOCH2 | 47 | 6-HSO3-Galβ1-4GlcNAcβ | 80 | GlcNAcβ1-4GlcNAcβ1-4GlcNAcβ, sp = -NHCOCH2NH- |
| 15 | MurNAc-lactic acid-L-Ala-D-isoGln | 48 | NeuAcα2-3Gal | 81 | Galβ1-3(NeuAcα2-6)GalNAcα |
| 16 | 3-HSO3-Galβ | 49 | 3-HSO3-Galβ1-3GalNAcβ (sulfate-TF) | 82 | Galβ1-3(NeuAcβ2-6)GalNAcα |
| 17 | β-Mannose-PAA-biotin | 50 | GlcNAcβ1-3GalNAcα | 83 | NeuAcα2-3(NeuAcα2-6)GalNAcα |
| 18 | α-NeuGc | 51 | GlcNAcβ1-6GalNAcα | 84 | Galβ1-4GlcNAcβ1-3GalNAcα |
| 19 | 6-HSO3-GlcNAcβ | 52 | NeuGcα2-6GalNAcα | 85 | Fucα1-2Galβ1-3(Fucα1-4)GlcNAcβ (Leb) |
| 20 | GalNAcα1-3Galβ | 53 | NeuAcβ2-6GalNAcα | 86 | Fucα1-2Galβ1-4(Fucα1-3)GlcNAcβ (Ley) |
| 21 | Galα1-3Galβ | 54 | NeuAcα2-3GalNAcα | 87 | NeuAcα2-3Galβ1-3(Fucα1-4) GlcNAcβ (sialyl Lea) |
| 22 | Fucα1-2Galβ | 55 | GalNAcα1-3(Fucα1-2)Galβ (Blood Group A) | 88 | NeuAcα2-3Galβ1-4(Fucα1-3) GlcNAcβ (sialyl Lex) |
| 23 | Galβ1-3GlcNAc (Lec) | 56 | Galα1-3(Fucα1-2)Galβ (Blood Group B) | 89 | GlcNAcβ1-3(GlcNAcβ1-6)Galβ1-4Glcβ |
| 24 | Galβ1-4Glcβ (Lactose) | 57 | Fucα1-2Galβ1-4GlcNAcβ (H type2) | 90 | Galα1-3(Fucα1-2)Galβ1-4GlcNAcβ |
| 25 | Galβ1-4GlcNAcβ (LacNAc) |
58 | Galβ1-3(Fucα1-4)GlcNAcβ (Lea) | 91 | Galβ1-3GlcNAcβ1-3Galβ1-4Glcβ |
| 26 | Galβ1-3GalNAcα | 59 | Galβ1-4(Fucα1-3)GlcNAcβ (Lex) | 92 | (NeuAcα2-8)5-6 |
| 27 | Fucα1-3GlcNAcβ | 60 | Fucα1-2Galβ1-3GlcNAcβ, Led (H type1) | 93 | Galβ1-4GlcNAcβ1-3(Galβ1- 4GlcNAcβ1-6)GalNAcα |
| 28 | Fucα1-4GlcNAcβ | 61 | NeuAcα2-3Galβ1-4Glcβ (3’Sialyl Lactose) | 94 | (NeuAcα2-6Galβ1-4GlcNAcβ1-2Man)2α1-3,6Manβ1-4GlcNAcβ1-4GlcNAcβ |
| 29 | GalNAcα1-3GalNAcβ | 62 | NeuAcα2-6Galβ1-4Glcβ (3’Sialyl Lactose) | 95 | GalNAcα-Ser |
| 30 | GalNAcα1-3GalNAcα | 63 | 3-HSO3-Galβ1-4(Fucα1-3)GlcNAcβ (3’sulfate Lex) | 96 | GalNAcα1-3(Fucα1-2)Galβ1- 4GlcNAc |
| 31 | Galα1-3GalNAcα | 64 | 3-HSO3-Galβ1-3(Fucα1-4)GlcNAcβ (3’sulfate Lea) | 97 | Neu5Acα2-3(6-HSO3)Galβ1-4(Fucα1-3)GlcNAcβ (6Gal-HSO3-SiaLex) |
| 32 | Galα1-3GalNAcβ | 65 | Galα1-4Galβ1-4Glcβ | 98 | Neu5Acα2-3Galβ1-4(Fucα1-3)(6-HSO3)GlcNAcβ (6GlcNAc-HSO3- SiaLex) |
| 33 | Galβ1-3Galβ | 66 | Galα1-3Galβ1-4Glcβ | 99 | H2O |
Note: sp: spacer.
Figure 1.
Binding profile of SARS-CoV-2 spike proteins with 97 biotin-PAA-sugars. (A) spike protein S1 subunit and (B) spike protein S2 subunit.
Table 2.
The carbohydrate binding preferences of spike protein S1 and S2 subunits (cut off: relative intensity >50%).
| SARS-CoV-2 Spike Protein S1 Subunit | SARS-CoV-2 Spike Protein S2 Subunit | ||
|---|---|---|---|
| No. | Glycan | No. | Glycan |
| 16 | 3-HSO3-Galβ | 16 | 3-HSO3-Galβ |
| 75 | GalNAcα1-3(Fucα1-2)Galβ (Blood Group A trisaccharide) |
44 | Galα1-6Glcβ (melibiose) |
| 89 | GlcNAcβ1-3(GlcNAcβ1-6)Galβ1-4Glcβ | 58 | Galβ1-3(Fucα1-4)GlcNAcβ (Lea) |
| 90 | Galα1-3(Fucα1-2)Galβ1-4GlcNAcβ (Blood Group B trisaccharide) |
74 | Galβ1-3(GlcNAcβ1-6)GalNAcα |
| 89 | GlcNAcβ1-3(GlcNAcβ1-6)Galβ1-4Glcβ | ||
3.2. SARS-CoV-2 Spike Proteins Interact with RBCs
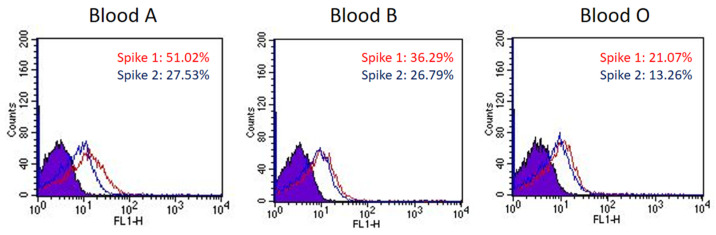
The sugar-binding profiling analysis indicated that SARS-CoV-2 spike proteins displayed binding preference for blood-type antigens, including Group A (#75 in Figure 1A), blood Group B (#90 in Figure 1A), and Lea (#58 in Figure 1B). To investigate if the binding preference of SARS-CoV-2 spike proteins correlated with the viral infection or pathogenesis, we examined RBCs that are known to express different blood groups, including group A (Lea+/Leb−), B (Lea+/Leb−), and O (Lea−/Leb+). The binding assay was conducted using fluorescence-activated cell sorting (FACS). The results indicated that the SARS-CoV-2 spike protein S1 subunit binds strongly to group A RBCs, moderately to group B RBCs, and relatively weakly to group O RBCs (Figure 2). The SARS-CoV-2 spike protein S2 subunit displayed higher binding signals with Lea+ RBCs than with Lea− RBCs (Figure 2). This observation was consistent with the analysis of carbohydrate microarray, which showed that the spike protein S1 subunit shows a higher preference for blood group A and B RBCs. The binding preference is related to the glycan structures existing on the surface of RBCs.
Figure 2.
The binding preference of SARS-CoV-2 spike proteins to blood group A, B, and O RBCs. The histograms of spike protein S1 subunits binding with blood group A, B, and O RBCs show a 51.02%, 36.29%, and 21.07% shift in mean fluorescence intensity (MFI), respectively (red line). The histograms of spike protein S2 subunits binding with blood group A, B, and O RBCs show a 27.53%, 26.79%, and 13.26% shift in MFI, respectively (blue line).
3.3. Carbohydrate Derivatives Interfere with Interaction of SARS-CoV-2 Spike Protein S1 Subunit and Blood Group A RBCs
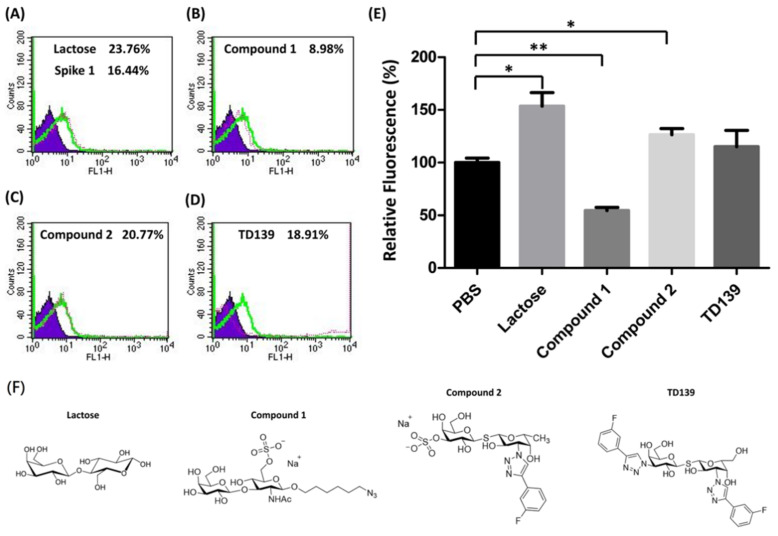
Carbohydrate analogs are able to interrupt the interaction between microorganisms and host cells by associating with glycoproteins on the surface of either host cells or microorganisms. For example, heparin sulfate mimetics exhibit antiviral activity against dengue virus by inhibiting the virus adsorption on host cells to prevent virus entry [40]. Neuraminidase inhibitors are used for anti-influenza therapy by inhibiting the neuraminidase activity to modify the cell surface glycans, which results in prevention of virions spreading to neighboring cells [41]. Since SARS-CoV-2 spike proteins show a binding preference to blood groups A and B, it is important to examine whether carbohydrate analogs interfere with the interaction between spike proteins and RBCs, especially galactin-3 inhibitors [42]. Lactose and three other carbohydrate derivatives were examined, including compounds 1 and 2 and TD-139. Each of them was preincubated with SARS-CoV-2 spike protein S1 subunit, followed by the addition of blood group A RBCs to the assay mixture (Figure 3A–D). The FACS analysis indicated that compound 1 significantly prevented the binding of spike protein S1 subunit with RBCs up to 45% (Figure 3E, p < 0.01). However, lactose and compound 2 enhanced the interaction of spike protein S1 subunit to RBCs by 53% and 26%, respectively (Figure 3E). Interestingly, TD139, a potent inhibitor of galactin-3, exhibited no effect on the spike protein S1 subunit–RBC interaction. The binding inhibition results indicated that carbohydrate analogs containing both sulfate and LacNAc groups reduce the binding affinity between the SARS-CoV-2 spike protein S1 subunit and host cells.
Figure 3.
The binding efficiency of the SARS-CoV-2 spike protein S1 subunit to blood group A RBCs affected by carbohydrate derivatives. (A) The histogram of spike protein S1 subunit binding with blood group A RBC cells shows a 16.44% shift in MFI without carbohydrate inhibitors. Lactose and three sulfated carbohydrate derivatives were preincubated with spike protein S1 subunit then subjected to the binding assay. The histograms of spike protein S1 subunit binding with blood group A RBC cells are shown (green line). The MFI shifts of blood group A RBC cells with carbohydrate derivatives tested are (A) lactose, 23.76%; (B) compound 1, 8.98%; (C) compound 2, 20.77%; and (D) TD139, 18.91%. (E) The relative fluorescence shows the binding efficiency of the spike protein S1 subunit to blood group A RBCs influenced by carbohydrate derivatives. (F) Structures of lactose and three sulfated carbohydrate derivatives. * indicated p < 0.05; ** indicated p < 0.01.
3.4. Blood Group A Antigen on ACE2
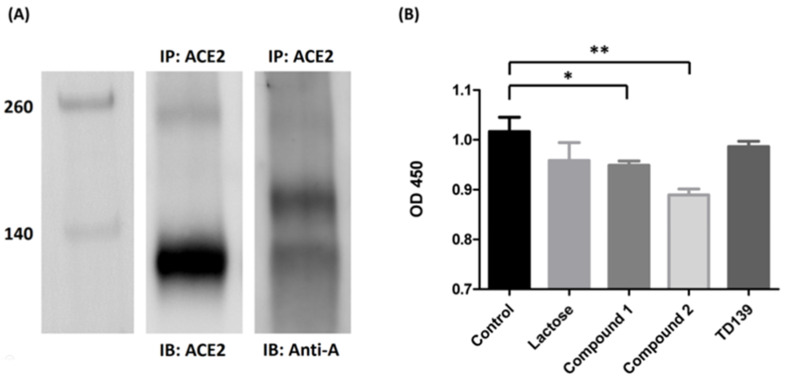
Since ACE2 is widely recognized as the major binding target for SARS-CoV-2 spike proteins, it is worth investigating whether the host receptor ACE2 contains the blood group A antigen. ACE2 protein was obtained from the extraction of lung tissues of a blood group A person. The ACE2 protein was immunoprecipitated with anti-ACE2 antibody. Western blotting analysis indicated that the glycoprotein ACE2 in the lung tissue of the blood group A person contained the carbohydrate chains of the blood group A antigen (Figure 4A).
Figure 4.
ACE2 contains blood group A antigen. (A) Western blotting analysis of ACE2 from the lung tissue of a blood group A patient. Lane 1: markers. Lane 2: the Western blotting of the lung tissue from blood group A patient immunoprecipitated with anti-ACE2 antibody and immunoblotted with anti-ACE2 antibody. Lane 3: the Western blotting of the lung tissue from blood group A patient immunoprecipitated with anti-ACE2 antibody and immunoblotted with anti-A antibody. (B) The binding inhibition assay of lactose or sulfated carbohydrate derivatives in blocking of the SARS-CoV-2 spike protein S1 subunit to ACE2. Compounds 1 and 2 showed a reduction in binding efficiency of ACE2 with the spike protein S1 subunit by 6.7% and 12.5%, respectively. TD139 exhibited no effects in spike protein S1 subunit–ACE2 interaction. * indicated p < 0.05; ** indicated p < 0.01.
3.5. Carbohydrate Derivatives Interfere with Interaction of SARS-CoV-2 Spike Protein S1 Subunit and ACE2
To study if it is possible to disrupt the interaction between SARS-CoV-2 spike protein and ACE2, an ELISA assay was performed to determine the binding inhibition efficiency of carbohydrate derivatives. SARS-CoV-2 spike protein S1 subunits were initially coated on 96-well microplates and incubated with carbohydrate derivatives. After washing away the nonbinding carbohydrate derivatives, human ACE2 proteins prepared from lung tissue lysate by immunoprecipitation were added. The quantities of ACE2 bound on spike protein S1 subunits were determined by ELISA assay. Both compounds 1 and 2 showed a significant decrease in the binding efficiency of ACE2 with the spike protein S1 subunit by 6.7% and 12.5%, respectively (Figure 4B). However, TD139 exhibited no effect on the interaction between spike protein S1 subunit and ACE2. Our results suggested that the specific carbohydrate modifications on ACE2 might be responsible for its binding to SARS-CoV-2 spike protein S1 subunit. Further studies are in progress to decipher the inhibitive effects of these carbohydrate derivatives on the interaction of spike protein S1 subunit and ACE2.
4. Discussion
Coronaviruses represent a large family of single-stranded enveloped RNA viruses and can be divided into four major genera [43,44]. Both SARS-CoV and SARS-CoV-2 belong to the β-genus. An envelope-anchored spike protein mediates the entry of the coronavirus into host cells by first binding to a host receptor and then fusing viral and host membranes [4]. A defined receptor-binding domain of the SARS-CoV-2 spike protein was reported to specifically recognize its host receptor ACE2 [7,8,9,10]. The spike protein of SARS-CoV-2 is a glycosylated, trimeric class I fusion protein with a metastable prefusion conformation [45,46]. According to Clausen et al., the SARS-CoV-2 spike protein interacts with both cellular heparan sulfate and ACE2 through its receptor-binding domain [34]. Li et al. showed that the N-terminal domain of β-coronaviridae spike protein S1 subunits (including SARS-CoV2) may potentially interact with unknown glycans [47]. Jayaprakash et al. also indicated that the N-terminal domain of SARS-CoV-2 spike protein binds with sialosides by molecular modeling [35].
ABO blood group antigens have been reported to be associated with diagnosis, prognosis, and survival of various diseases [48,49,50,51,52]. The relationships between ABO blood group antigens and COVID-19 have also been investigated by many researchers. Jawdat et al. found that blood group B is a risk factor for COVID-19 and blood group O is protective factor for COVID-19 infection [53]. Zhao et al. analyzed the ABO blood group distribution among 2173 COVID-19 patients. They found that an increased risk of infection is associated with blood group A and a decreased risk of infection is associated with blood group O [54]. The same findings were also reported in [55,56,57,58,59,60,61]. However, researchers also observed opposite results in other investigations. Zietz et al. showed that non-O blood group types represented slightly increased infection prevalence compared to the O blood group [62]. The results of Ishaq et al. indicated that blood groups have no significant association with severity of COVID-19 disease or COVID-19-associated mortality [63]. Kim et al. also reported that no relationships between blood type and COVID-19-related mortality or severity of illness were observed [64].
Using the solution carbohydrate microarray, we first demonstrated that the SARS-CoV-2 spike protein S1 subunit binds specifically to blood group A and B antigens, and that the SARS-CoV-2 spike protein S2 subunit exhibits a binding preference for Lea antigen (Figure 1). The glycan-binding feature was further investigated by examining how the spike proteins recognize RBCs (Figure 2). Like many C-type lectin domains, the spike protein of SARS-CoV-2 exhibited trimeric fusion protein structure which can enhance protein–glycan interaction by multivalency [65]. Since blood group antigens are available on the surface of RBCs and all human organs/tissues [66] (including saliva [67]), SARS-CoV-2 could easily attach to cell surfaces or droplets and could then be spread and transmitted through the air over time and distance.
Interestingly, COVID-19-induced coagulopathy and a high incidence of thromboembolic events was found in COVID-19-positive deaths [68]. An unknown blood-clotting complication was also reported, even in the patients that were treated with anticoagulants [69]. Unlike Ebola, Dengue, Lassa, and other hemorrhagic fevers that lead to uncontrolled bleeding, SARS-CoV-2 infection was reported to exhibit red, liquid, gel-like blood clots, as well as tiny clots throughout the lungs of dead patients [70]. Our findings may explain why SARS-CoV-2 viral particles potentially agglutinate RBCs by spike–glycan interaction. The proposed unusual agglutination in blood vessels may lead to blood clot formation. Thus, further studies are necessary to demonstrate the correlation and unravel the mechanistic details.
Carbohydrate derivatives such as swainsonine [71,72], 1,4-dideoxy-1,4-imino-D-mannitol [73], deoxymannojirimycin [74,75], and castanospermine [76,77] have shown anticancer activity in different cancer types. Nucleoside analogs are well-developed antiviral drugs for many viral infections including HIV, HBV, dengue virus, yellow fever virus, JEV, and Zika virus [78,79,80]. According to our findings from the solution carbohydrate microarray, the SARS-CoV-2 spike protein S1 subunit preferentially binds to blood group A/B antigens and specific terminal sugar moieties including galactose, N-acetylgalactosamine, and sulfated galactose. We evaluated the effects of lactose and sulfated glycan analogs (compounds 1 and 2, and TD139) on the interaction between the spike protein S1 subunit and RBCs. Compound 1, a galectin-3 inhibitor, was shown to blockade the interaction of the spike protein S1 subunit with RBCs. However, the inhibition activity was not found in lactose, the other sulfated glycan compound 2, or TD139. This result suggested that the position of a sulfate group is important, and that sulfated glycans play an important role in the spike protein–glycan interaction.
Since we demonstrated that the SARS-CoV-2 spike protein binds to host cells through interaction with the blood group A antigen and this interaction could be interfered with by glycan analogs, it is worth noting whether the well-known host receptor ACE2 contains the blood type A antigen. ACE2 extracted from the lung tissue of a blood group A patient has shown the expression of the blood type A antigen in Western blotting, indicating that the blood group A antigen is present on ACE2 in the lung tissue of the blood group A person. Though compound 1 showed a significant binding inhibition of spike proteins and RBCs, it poorly inhibited the interaction of the spike protein S1 subunit with ACE2. Compound 2, however, showed no binding inhibition for spike proteins and RBCs, but displayed better blocking efficiency than compound 1 in the interaction of the spike protein S1 subunit and ACE2. The binding inhibition experiment implied that the interaction of spike protein and RBCs is not only through group A antigen but also involves other groups of blood antigens. The interaction mechanism between these glycan analogs to blood group A antigen or ACE2 needs to be further investigated in the future.
5. Conclusions
In conclusion, we identified the carbohydrate ligands for SARS-CoV-2 spike proteins and demonstrated the interactions between SARS-CoV-2 spike proteins with blood group antigens on RBCs. Blood type A antigen serves as one of the possible binding targets of SARS-CoV-2 spike protein S1 subunit, suggesting that blood group A patients may be associated with a higher risk of contracting COVID-19 compared to non-A blood groups. Since we found carbohydrate derivatives to prevent the binding of the SARS-CoV-2 spike protein S1 subunit with RBCs, our results are expected to shed light on SARS-CoV-2 drug discovery.
Acknowledgments
We are grateful for the support from the Human Biobank, the Research Center of Clinical Medicine, and National Cheng Kung University Hospital.
Author Contributions
Conceptualization, P.-H.H. and C.-F.C.; data curation, Y.-K.L., W.-C.C., Y.-J.P. and Z.-J.T.; formal analysis, Y.-K.L. and Z.T.; funding acquisition, C.-F.C.; resources, E.P.; writing—original draft, C.-H.L., P.-H.H. and C.-F.C.; writing—review and editing, C.-H.L., P.-H.H. and C.-F.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the Ministry of Science and Technology, Taipei, Taiwan (MOST 110-2327-B-006-004 and MOST 108-2320-B-006-042).
Institutional Review Board Statement
This study was approved by the National Cheng Kung University Hospital Institutional Review Board (IRB No: A-ER-107-085).
Informed Consent Statement
Informed consent was waived because of the analysis used clinical samples from human biobank.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birtles D., Lee J. Identifying distinct structural features of the SARS-CoV-2 spike protein fusion domain essential for membrane interaction. Biochemistry. 2021;60:2978–2986. doi: 10.1021/acs.biochem.1c00543. [DOI] [PubMed] [Google Scholar]
- 4.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;525:135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han Y., Kral P. Computational Design of ACE2-Based Peptide Inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saavedra J.M. Angiotensin receptor blockers and COVID-19. Pharmacol. Res. 2020;156:104832. doi: 10.1016/j.phrs.2020.104832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alhenc-Gelas F., Drueke T.B. Blockade of SARS-CoV-2 infection by recombinant soluble ACE2. Kidney Int. 2020;97:1091–1093. doi: 10.1016/j.kint.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barchetta I., Gisella Cavallo M., Giorgio Baroni M. COVID-19 and diabetes: Is this association driven by the DPP4 receptor? Potential clinical and therapeutic implications. Diabetes Res. Clin. Pract. 2020;163:108165. doi: 10.1016/j.diabres.2020.108165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strollo R., Pozzilli P. DPP4 inhibition: Preventing SARS-CoV-2 infection and/or progression of COVID-19? Diabetes Metab. Res. Rev. 2020;36:e3330. doi: 10.1002/dmrr.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elfiky A.A. Natural products may interfere with SARS-CoV-2 attachment to the host cell. J. Biomol. Struct. Dyn. 2020;39:3194–3203. doi: 10.1080/07391102.2020.1761881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang T.J., Chang Y.C., Ko T.P., Draczkowski P., Chien Y.C., Chang Y.C., Wu K.P., Khoo K.H., Chang H.W., Hsu S.D. Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans. Proc. Natl. Acad. Sci. USA. 2020;117:1438–1446. doi: 10.1073/pnas.1908898117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson T., Ellard F.M., Ghazaleh R.A., Brookes S.M., Blakemore W.E., Corteyn A.H., Stuart D.I., Newman J.W., King A.M. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J. Virol. 1996;70:5282–5287. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu A., Kanda T., Beyene A., Saito K., Meyer K., Ray R. Sulfated homologues of heparin inhibit hepatitis C virus entry into mammalian cells. J. Virol. 2007;81:3933–3941. doi: 10.1128/JVI.02622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee E., Pavy M., Young N., Freeman C., Lobigs M. Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antivir. Res. 2006;69:31–38. doi: 10.1016/j.antiviral.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Escribano-Romero E., Jimenez-Clavero M.A., Gomes P., Garcia-Ranea J.A., Ley V. Heparan sulphate mediates swine vesicular disease virus attachment to the host cell. J. Gen. Virol. 2004;85:653–663. doi: 10.1099/vir.0.19603-0. [DOI] [PubMed] [Google Scholar]
- 20.Witvrouw M., De Clercq E. Sulfated polysaccharides extracted from sea algae as potential antiviral drugs. Gen. Pharmacol. 1997;29:497–511. doi: 10.1016/S0306-3623(96)00563-0. [DOI] [PubMed] [Google Scholar]
- 21.Kuipers M.E., Huisman J.G., Swart P.J., de Bethune M.P., Pauwels R., Schuitemaker H., De Clercq E., Meijer D.K. Mechanism of anti-HIV activity of negatively charged albumins: Biomolecular interaction with the HIV-1 envelope protein gp120. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1996;11:419–429. doi: 10.1097/00042560-199604150-00001. [DOI] [PubMed] [Google Scholar]
- 22.Rey M.W., Woloshuk S.L., deBoer H.A., Pieper F.R. Complete nucleotide sequence of human mammary gland lactoferrin. Nucleic Acids Res. 1990;18:5288. doi: 10.1093/nar/18.17.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell M.J., Ogden J.E. Nucleotide sequence of human lactoferrin cDNA. Nucleic Acids Res. 1990;18:4013. doi: 10.1093/nar/18.13.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin T.Y., Chu C., Chiu C.H. Lactoferrin inhibits enterovirus 71 infection of human embryonal rhabdomyosarcoma cells in vitro. J. Infect. Dis. 2002;186:1161–1164. doi: 10.1086/343809. [DOI] [PubMed] [Google Scholar]
- 25.Weng T.Y., Chen L.C., Shyu H.W., Chen S.H., Wang J.R., Yu C.K., Lei H.Y., Yeh T.M. Lactoferrin inhibits enterovirus 71 infection by binding to VP1 protein and host cells. Antivir. Res. 2005;67:31–37. doi: 10.1016/j.antiviral.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Alexander D.A., Dimock K. Sialic acid functions in enterovirus 70 binding and infection. J. Virol. 2002;76:11265–11272. doi: 10.1128/JVI.76.22.11265-11272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su P.Y., Liu Y.T., Chang H.Y., Huang S.W., Wang Y.F., Yu C.K., Wang J.R., Chang C.F. Cell surface sialylation affects binding of enterovirus 71 to rhabdomyosarcoma and neuroblastoma cells. BMC Microbiol. 2012;12:162. doi: 10.1186/1471-2180-12-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson E.C., Jamshidi F., Johansson S.M., Oberste M.S., Arnberg N. Sialic acid is a cellular receptor for coxsackievirus A24 variant, an emerging virus with pandemic potential. J. Virol. 2008;82:3061–3068. doi: 10.1128/JVI.02470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehmann F., Tiralongo E., Tiralongo J. Sialic acid-specific lectins: Occurrence, specificity and function. Cell. Mol. Life Sci. 2006;63:1331–1354. doi: 10.1007/s00018-005-5589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hulswit R.J.G., Lang Y., Bakkers M.J.G., Li W., Li Z., Schouten A., Ophorst B., van Kuppeveld F.J.M., Boons G.J., Bosch B.J., et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA. 2019;116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.J., Bosch B.J., Rey F.A., de Groot R.J., et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W., Widagdo W., Tortorici M.A., van Dieren B., Lang Y., et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwegmann-Wessels C., Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 2006;23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jayaprakash N.G., Surolia A. Spike Protein and the Various Cell-Surface Carbohydrates: An Interaction Study. ACS Chem. Biol. 2022;17:103–117. doi: 10.1021/acschembio.1c00691. [DOI] [PubMed] [Google Scholar]
- 36.Han D.P., Lohani M., Cho M.W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007;81:12029–12039. doi: 10.1128/JVI.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang C.F., Pan J.F., Lin C.N., Wu I.L., Wong C.H., Lin C.H. Rapid characterization of sugar-binding specificity by in-solution proximity binding with photosensitizers. Glycobiology. 2011;21:895–902. doi: 10.1093/glycob/cwr021. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y.F., Chang C.F., Chi C.Y., Wang H.C., Wang J.R., Su I.J. Characterization of glycan binding specificities of influenza B viruses with correlation with hemagglutinin genotypes and clinical features. J. Med. Virol. 2012;84:679–685. doi: 10.1002/jmv.23219. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y.F., Chang C.F., Tsai H.P., Chi C.Y., Su I.J., Wang J.R. Glycan-binding preferences and genetic evolution of human seasonal influenza A(H3N2) viruses during 1999-2007 in Taiwan. PLoS ONE. 2018;13:e0196727. doi: 10.1371/journal.pone.0196727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hidari K.I., Abe T., Suzuki T. Carbohydrate-related inhibitors of dengue virus entry. Viruses. 2013;5:605–618. doi: 10.3390/v5020605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bassetti M., Castaldo N., Carnelutti A. Neuraminidase inhibitors as a strategy for influenza treatment: Pros, cons and future perspectives. Expert Opin. Pharm. 2019;20:1711–1718. doi: 10.1080/14656566.2019.1626824. [DOI] [PubMed] [Google Scholar]
- 42.Caniglia J.L., Guda M.R., Asuthkar S., Tsung A.J., Velpula K.K. A potential role for Galectin-3 inhibitors in the treatment of COVID-19. PeerJ. 2020;8:e9392. doi: 10.7717/peerj.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perlman S., Netland J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 47.Li F. Receptor recognition mechanisms of coronaviruses: A decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang G., Wang H., Shen Y., Dong J., Wang X., Wang X., Zheng Y., Guo S. Association between ABO blood group and venous thrombosis related to the peripherally inserted central catheters in cancer patients. J. Vasc. Access. 2021;22:590–596. doi: 10.1177/1129729820954721. [DOI] [PubMed] [Google Scholar]
- 49.Gilmiyarova F.N., Kolotyeva N.A., Kuzmicheva V.I., Gusyakova O.A., Borodina I.A., Baisheva G.M., Selezneva I.A. Blood group and human diseases (review of literature) Klin. Lab. Diagn. 2020;65:216–221. doi: 10.18821/0869-2084-2020-65-4-216-221. [DOI] [PubMed] [Google Scholar]
- 50.Wei J., Zhang Y., Zheng J., Feng X., Wang X., Du K., Wang W., Wu G., Zhao Q., Fan D., et al. Prognostic value of ABO blood group in a Chinese population in Northwest China region with curatively resected rectal cancer. J. Cancer. 2019;10:6584–6593. doi: 10.7150/jca.32407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandato V.D., Torricelli F., Mastrofilippo V., Ciarlini G., Pirillo D., Annunziata G., Casali B., Abrate M., Sala G.B., Aguzzoli L. AB0 Blood Group and Ovarian Cancer Survival. J. Cancer. 2019;10:1949–1957. doi: 10.7150/jca.29272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulger A.F., Keklik T., Kumbasar O.O., Arbak P., Demirkazyk A., Gungor A., Erekul S., Alper D. Prognostic significance of blood group antigen expression of tumor tissue in lung cancer patients. Tumori J. 2002;88:395–399. doi: 10.1177/030089160208800509. [DOI] [PubMed] [Google Scholar]
- 53.Jawdat D., Hajeer A., Massadeh S., Aljawini N., Abedalthagafi M.S., Alaamery M. Correlation between ABO Blood Group Phenotype and the Risk of COVID-19 Infection and Severity of Disease in a Saudi Arabian Cohort. J. Epidemiol. Glob. Health. 2022 doi: 10.1007/s44197-021-00023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J., Yang Y., Huang H., Li D., Gu D., Lu X., Zhang Z., Liu L., Liu T., Liu Y., et al. Relationship Between the ABO Blood Group and the Coronavirus Disease 2019 (COVID-19) Susceptibility. Clin. Infect. Dis. 2021;73:328–331. doi: 10.1093/cid/ciaa1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muniz-Diaz E., Llopis J., Parra R., Roig I., Ferrer G., Grifols J., Millan A., Ene G., Ramiro L., Maglio L., et al. Relationship between the ABO blood group and COVID-19 susceptibility, severity and mortality in two cohorts of patients. Blood Transfus. 2021;19:54–63. doi: 10.2450/2020.0256-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nauffal V., Achanta A., Goldhaber S.Z., Piazza G. Association of ABO blood group type with cardiovascular events in COVID-19. J. Thromb. Thrombolysis. 2021;51:584–586. doi: 10.1007/s11239-020-02364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu N., Zhang T., Ma L., Zhang H., Wang H., Wei W., Pei H., Li H. The impact of ABO blood group on COVID-19 infection risk and mortality: A systematic review and meta-analysis. Blood Rev. 2021;48:100785. doi: 10.1016/j.blre.2020.100785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goel R., Bloch E.M., Pirenne F., Al-Riyami A.Z., Crowe E., Dau L., Land K., Townsend M., Jecko T., Rahimi-Levene N., et al. ABO blood group and COVID-19: A review on behalf of the ISBT COVID-19 Working Group. Vox Sang. 2021;116:849–861. doi: 10.1111/vox.13076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernandez J., Prati D., Baselli G., Asselta R., et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Latz C.A., DeCarlo C., Boitano L., Png C.Y.M., Patell R., Conrad M.F., Eagleton M., Dua A. Blood type and outcomes in patients with COVID-19. Ann. Hematol. 2020;99:2113–2118. doi: 10.1007/s00277-020-04169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garibaldi P.M.M., Oliveira L.C., da Fonseca B.A., Auxiliadora-Martins M., Miranda C.H., Almado C.E.L., Langhi D.M., Gilio R.N., Palma L.C., Gomes B.B.M., et al. Histo-blood group A is a risk factor for severe COVID-19. Transfus. Med. 2021 doi: 10.1111/tme.12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zietz M., Zucker J., Tatonetti N.P. Associations between blood type and COVID-19 infection, intubation, and death. Nat. Commun. 2020;11:5761. doi: 10.1038/s41467-020-19623-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishaq U., Malik A., Malik J., Mehmood A., Qureshi A., Laique T., Zaidi S.M.J., Javaid M., Rana A.S. Association of ABO blood group with COVID-19 severity, acute phase reactants and mortality. PLoS ONE. 2021;16:e0261432. doi: 10.1371/journal.pone.0261432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim Y., Latz C., DeCarlo C., Lee S., Png C., Kibrik P., Sung E., Alabi O., Dua A. Relationship between blood type and outcomes following COVID-19 infection. Semin. Vasc. Surg. 2021;34:125–131. doi: 10.1053/j.semvascsurg.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mammen M., Choi S.K., Whitesides G.M. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew. Chem. Int. Ed. Engl. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 66.Sanchez-Mora N., Cebollero Presmanes M., Monroy V., Herranz Aladro M., Alvarez-Fernandez E. Expression of histo-blood group antigens in bronchial squamous metaplasia. Eur. Respir. J. 2007;29:268–272. doi: 10.1183/09031936.00009806. [DOI] [PubMed] [Google Scholar]
- 67.Slomiany B.L., Slomiany A. ABH-blood-group antigens and glycolipids of human saliva. Eur. J. Biochem. 1978;85:249–254. doi: 10.1111/j.1432-1033.1978.tb12233.x. [DOI] [PubMed] [Google Scholar]
- 68.Wichmann D., Sperhake J.P., Lutgehetmann M., Steurer S., Edler C., Heinemann A., Heinrich F., Mushumba H., Kniep I., Schroder A.S., et al. Autopsy findings and venous Thromboembolism in patients with COVID-19. Ann. Intern. Med. 2020;173:268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cha A.E. A mysterious blood-clotting complication is killing coronavirus patients. The Washington Post. Apr 22, 2020. [(accessed on 22 April 2020)]. Available online: https://www.washingtonpost.com/health/2020/04/22/coronavirus-blood-clots/
- 70.Willyard C. Coronavirus blood-clot mystery intensifies. Nature. 2020;581:250. doi: 10.1038/d41586-020-01403-8. [DOI] [PubMed] [Google Scholar]
- 71.Silveira C.R.F., Cipelli M., Manzine C., Rabelo-Santos S.H., Zeferino L.C., Rodriguez Rodriguez G., de Assis J.B., Hebster S., Bernadinelli I., Laginha F., et al. Swainsonine, an alpha-mannosidase inhibitor, may worsen cervical cancer progression through the increase in myeloid derived suppressor cells population. PLoS ONE. 2019;14:e0213184. doi: 10.1371/journal.pone.0213184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu B., Lin Y., Yin H. Inhibition effect of Swainsonine on the growth and metastasis of gastric cancer in vivo. Zhonghua Zhong Liu Za Zhi. 1998;20:168–170. [PubMed] [Google Scholar]
- 73.Yang L.F., Shimadate Y., Kato A., Li Y.X., Jia Y.M., Fleet G.W.J., Yu C.Y. Synthesis and glycosidase inhibition of N-substituted derivatives of 1,4-dideoxy-1,4-imino-d-mannitol (DIM) Org. Biomol. Chem. 2020;18:999–1011. doi: 10.1039/C9OB02029B. [DOI] [PubMed] [Google Scholar]
- 74.Balzarini J. The alpha(1,2)-mannosidase I inhibitor 1-deoxymannojirimycin potentiates the antiviral activity of carbohydrate-binding agents against wild-type and mutant HIV-1 strains containing glycan deletions in gp120. FEBS Lett. 2007;581:2060–2064. doi: 10.1016/j.febslet.2007.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ogier-Denis E., Trugnan G., Sapin C., Aubery M., Codogno P. Dual effect of 1-deoxymannojirimycin on the mannose uptake and on the N-glycan processing of the human colon cancer cell line HT-29. J. Biol. Chem. 1990;265:5366–5369. doi: 10.1016/S0021-9258(19)39367-6. [DOI] [PubMed] [Google Scholar]
- 76.Allan G., Ouadid-Ahidouch H., Sanchez-Fernandez E.M., Risquez-Cuadro R., Fernandez J.M., Ortiz-Mellet C., Ahidouch A. New castanospermine glycoside analogues inhibit breast cancer cell proliferation and induce apoptosis without affecting normal cells. PLoS ONE. 2013;8:e76411. doi: 10.1371/journal.pone.0076411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yee C.S., Schwab E.D., Lehr J.E., Quigley M., Pienta K.J. The effect of castanospermine on the metastatic properties of prostate cancer cells. Anticancer Res. 1997;17:3659–3663. [PubMed] [Google Scholar]
- 78.Kasthuri M., Li C., Verma K., Russell O.O., Dickson L., McCormick L., Bassit L., Amblard F., Schinazi R.F. Synthesis of 4′-Substituted-2′-Deoxy-2′-alpha-Fluoro Nucleoside Analogs as Potential Antiviral Agents. Molecules. 2020;25:1258. doi: 10.3390/molecules25061258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zandi K., Bassit L., Amblard F., Cox B.D., Hassandarvish P., Moghaddam E., Yueh A., Libanio Rodrigues G.O., Passos I., Costa V.V., et al. Nucleoside Analogs with Selective Antiviral Activity against Dengue Fever and Japanese Encephalitis Viruses. Antimicrob. Agents Chemother. 2019;63:e00397-19. doi: 10.1128/AAC.00397-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sari O., Bassit L., Gavegnano C., McBrayer T.R., McCormick L., Cox B., Coats S.J., Amblard F., Schinazi R.F. Synthesis and antiviral evaluation of 2′,2′,3′,3′-tetrafluoro nucleoside analogs. Tetrahedron Lett. 2017;58:642–644. doi: 10.1016/j.tetlet.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]