Abstract
A diversity of mRNAs containing only short open reading frames (sORF-RNAs; encoding less than 30 amino acids) have been shown to be induced in growth and differentiation processes. The early nodulin gene enod40, coding for a 0.7-kb sORF-RNA, is expressed in the nodule primordium developing in the root cortex of leguminous plants after infection by symbiotic bacteria. Ballistic microtargeting of this gene into Medicago roots induced division of cortical cells. Translation of two sORFs (I and II, 13 and 27 amino acids, respectively) present in the conserved 5′ and 3′ regions of enod40 was required for this biological activity. These sORFs may be translated in roots via a reinitiation mechanism. In vitro translation products starting from the ATG of sORF I were detectable by mutating enod40 to yield peptides larger than 38 amino acids. Deletion of a Medicago truncatula enod40 region between the sORFs, spanning a predicted RNA structure, did not affect their translation but resulted in significantly decreased biological activity. Our data reveal a complex regulation of enod40 action, pointing to a role of sORF-encoded peptides and structured RNA signals in developmental processes involving sORF-RNAs.
RNAs encoding only short open reading frames (sORF-RNAs) have received considerable attention in recent years because they show a striking diversity in many cell types from various organisms. Several RNAs exhibit a function without being translated into proteins, for example, tRNAs, rRNAs, RNAs in ribozymes, and small nuclear RNAs from spliceosomes (reviewed in reference 8). However, a pentapeptide-encoding sORF present in the 23S rRNA from Escherichia coli was recently shown to render this bacterium resistant to a specific antibiotic (40). In eukaryotes, several sORF-RNAs are induced at specific stages of development, suggesting their participation in various differentiation processes (7, 14, 18, 31, 38, 40, 44, 46). Eukaryotic cells may use sORF-RNAs for the regulation of several cellular processes, as suggested by a thorough analysis of the yeast genome leading to identification of several new noncoding and sORF-containing RNAs (31). In eukaryotes, sORFs present in mRNAs are likely to be translated, since translation of mRNAs is achieved by a scanning mechanism in a 5′-to-3′ direction. Indeed, there are several examples where translation of upstream sORFs regulates expression of the 3′ ORF corresponding to the gene product (15, 43). At the same time, very little is known about the fate of the encoded oligopeptides in the cell and whether translation of sORFs present in sORF-RNAs is relevant for gene activity. For example, even though a putative protein product of the H19 RNA was detected using immunological approaches (23), the main function of H19 seems to lie in the mRNA molecule rather than in the encoded protein (24).
Leguminous plants have the ability to enter into symbiosis with N2-fixing bacteria (collectively called rhizobia) to form the root nodule. Development of this symbiotic organ depends on the coordinate expression of plant and bacterial genes and starts with the induction of root cortical cell divisions (38). The early nodulin gene enod40 is rapidly induced by rhizobia in the root pericycle and then in the dividing cortical cells of the nodule primordium (1, 12, 21, 45). The enod40 genes are highly conserved in various leguminous species and have also been found in tobacco and rice (22, 42). They lack a common long ORF, and computer predictions suggest that they code for structured RNAs (12). In the enod40 genes, two highly conserved regions were distinguished: box I in the 5′ end, spanning a conserved sORF (sORF I), and box II in the central part of the gene (42) (Fig. 1A). We demonstrated that under nitrogen-limiting conditions overexpression of Medicago truncatula (a model leguminous plant) enod40 (Mtenod40) induces cortical cell division in Medicago roots (9). These experiments also showed that transient expression of either region 1 carrying box I or a 3′ sequence (region 2) spanning box II evoked a response similar to that evoked by the complete gene in alfalfa (Medicago sativa) roots.
FIG. 1.
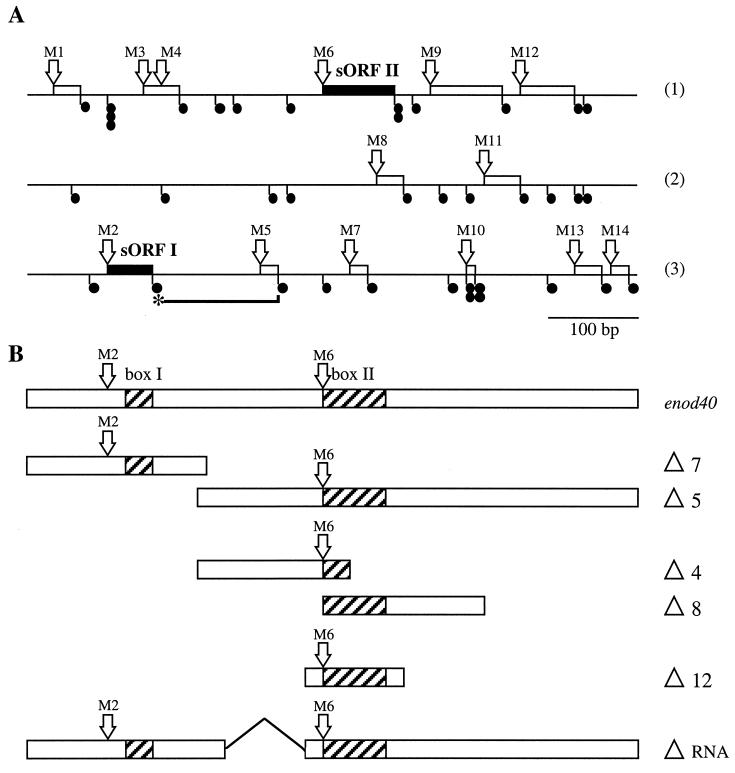
Schematic representation of the Mtenod40 cDNA sequence. (A) Each of the three possible reading frames is depicted separately (1 to 3). Open arrows marked M1 to M14 correspond to the different ATG codons (methionines) of the Mtenod40 gene. Solid circles are stop codons (TAG, TAA, or TGA). ORFs are depicted as boxes, and the conserved sORF I and sORF II are marked. The complete Mtenod40 transcript harboring the two conserved boxes and the two ATG codons of sORF I and II is shown. (B) Deletion series of Mtenod40. Boxes I and II as well as the ATG codons M2 and M6 are indicated for each deletion. For the ΔRNA construct, the region marked by a line was deleted. Note that M6 is absent from Δ8.
In order to gain insight into the molecular mechanism of enod40 action, we used microtargeting to introduce different enod40 constructs into alfalfa roots. This technique uses gold microprojectiles to deliver soluble substances into cells within a very localized area (down to 150 μm) (34) and has previously been applied to study the effects of chitin oligosaccharides and Nod factors on nodule initiation (26, 36). We demonstrate here that two sORFs (I and II) as well as an inter-ORF region spanning a predicted RNA structure are involved in the regulation of enod40 activity in Medicago roots. Using reporter gene fusions, translation of the two sORFs as well as the influence of the 5′ sORF I on translation of the 3′ sORF II was demonstrated, suggesting that this unusual nodulin gene may have a polycistronic nature. In vitro translation of purified Mtenod40 transcripts containing point mutations allowed detection of a specific product starting from the initiation codon of the 5′ sORF I. These data indicate that sORF translation is required for enod40 function in leguminous roots and suggest that sORF-encoded peptides may be important elements in regulatory mechanisms involving sORF-RNAs.
MATERIALS AND METHODS
Plant material and bacterial strains.
Seedlings of M. sativa cultivar Sitel were germinated in water and grown aseptically. Inoculation with Sinorhizobium meliloti strain Rm41 for large-scale preparation of young nodule extracts in aeroponic tanks was performed as described earlier (12).
Escherichia coli CC118 [Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1] (20) and E. coli DH5α (supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1) (17) were used for subcloning. E. coli XL1-Blue [recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac {F′ proAB lacIqZΔM15 Tn10 (Tetr)}] (6) was used as the recipient of all plasmids with point mutations.
Construction of translational fusions of enod40 sORFs and the uidA gene.
A series of translational fusions to the uidA gene driven by the constitutive cauliflower mosaic virus 35S promoter were constructed. A vector (pDH51 5), containing the cauliflower mosaic virus (CaMV) 35S promoter fused to the reporter gene without the initiating methionine and instead having a polylinker, was used to clone different portions of the Mtenod40 and M. sativa enod40 (Msenod40) transcripts.
The constructs diagrammed in Fig. 2 were obtained by insertion of PCR products, using different enod40 oligonucleotides, pMtenod40/pMsenod40 DNAs, and their derivatives as the template. In the case of plasmid M1GUS, an oligonucleotide (AATTCCAACTTCCCCACTACCTTTCTATGT) corresponding to the Mtenod40 cDNA sequence up to and including part of the first sORF, was inserted in the vector. In order to find out whether the 3′ region might inhibit translation of sORF I, we introduced the 3′ region (Δ5; nucleotides 204 to 614 of the Mtenod40 gene) immediately behind the uidA sequence in the in-frame translational fusion (pTra40M2, Fig. 2C). pTra40M2-Δ5 and pTra40M2-inv.Δ5 were prepared by subcloning Δ5 into pTra40M2 in the sense and antisense orientations, respectively (Fig. 2C). The sequences of all constructions were confirmed as mentioned below. Further details on plasmid construction are available upon request.
FIG. 2.
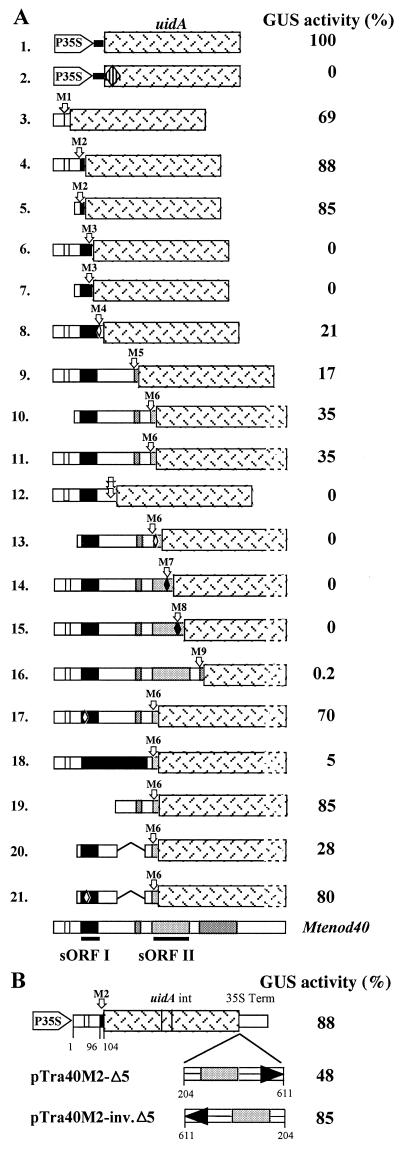
Reporter gene activity induced by Mtenod40 derivatives, with different ATG codons (methionines) fused in frame to the uidA coding sequence. Translational activity was measured after particle gun bombardment of the DNA constructs into seedlings. The activity values are the means of three to five independent experiments and were calculated relative to the expression value for the positive control (P35S-uidA; with its own ATG, 100%). The relative expression level of a given construct varied within a maximum of 20% of the indicated value between different experiments.(e.g., for AUG6, 35% ± 7%; for AUG5, 17% ± 4%). (A) The striped diamond in the p35SuidA construct indicates a polylinker replacing the initiating ATG codon (methionine) of the uidA coding sequence. This vector was used to clone the different Mtenod40 regions up to the indicated ATG codon (M1 to M9). Arrows marked M1 to M9 correspond to the different ATG codons (methionines) of the Mtenod40 gene that were fused to uidA. The triple arrow (construct 12) indicates fusions in the three reading frames after sORF I to test whether its stop codon arrests translation. Solid boxes indicate selected sORFs present in the Mtenod40 gene. The open diamonds indicate a point mutation in the ATG start codon of sORF I or sORF II (M2 or M6; changing ATG to ACG) or the internal ATG codon (methionine) of M3 sORF (M4), and the solid diamonds show ATG codons of sORFs in other frames (M7 and M8). Deletions in the Mtenod40 gene are indicated as lines. Note that M4 is internal to the M3 sORF but lies 8 bp after the stop codon of sORF I. The M4 sORF codes for a 7-amino-acid peptide. (B) Constructs used to test the influence of the 3′ region (Δ5) on translation of sORF I. This sequence was inserted in the 5′-to-3′ (pTra40M2-Δ5) and 3′-to-5′ (pTra40M2-inv.Δ5) direction, downstream of the uidA sequence. int, intron; 35S Term, 35S terminator [poly(A) signal].
Construction of enod40 mutants.
All point mutations were introduced by the Quickchange site-directed mutagenesis kit (Stratagene), using synthetic oligonucleotide primers containing the desired mutation. For pMtenod40-ACG sORF I, the primers were 5′-GTAATAAGGACGAAGCTTCTTTGTTGGG-3′ and 5′-CCCAACAAAGAAGCTTCGTCCTTATTAC-3′. For pMtenod40-TCA sORF I (5.8 kDa), the primers were 5′-ATCCATGGTTCTTCAAACAAACATGGAG-3′ and 5′-CTCCATGTTTGTTTGAAGAACCATGGAT-3′. For pMtenod40-mod.sORF I, the primers were 5′-GGGAAAAATCAATCCACGGCTCGTAAAACAAACATGG-3′ and 5′-CCATGTTTGTTTTACGAGCCGTGGATTGATTTTTCCC-3′. For pMtenod40-ACG sORF II, the primers were 5′-GCTTTTGTTATAGCACGGCAAACCGGCAAGTC-3′ and 5′-GACTTGCCGGTTTGCCGTGCTATAACAAAAGC-3′. For pMtenod40-TCA sORF I/TAG (3.8 kDa), the primers were 5′-CCTAAACAGTTAGCTTTGTGCTTTAGC-3′ and 5′-GCTAAAGCACAAAGCTAACTGTTTAGG-3′. (Underlined nucleotides indicate point mutations.)
Replacements were done in pBluescript pSK(+), and the sequences of the mutants were confirmed as described below.
pMtenod40/pMsenod40-ACG sORF I and pMtenod40-ACG sORF II are similar to pMtenod40 and pMsenod40 but with a mutation in the ATG of sORF I and sORF II, respectively. pMtenod40/pMsenod40-TCA sORF I and pMtenod40 mod.RNA are also similar to pMtenod40 and pMsenod40, but with a mutation in the stop codon of sORF I or modifications in the nucleotide sequence of sORF I (without changes to the encoded amino acid sequence), respectively. Plasmids p35S-Mtenod40/Msenod40, p35S-Mtenod40/Msenod40-ACG sORF I, p35S-Mtenod40-ACG sORF II, p35S-Mtenod40/Msenod40-TCA sORF I, and p35S-Mtenod40 mod.RNA are similar to the plasmids described above but under control of the CaMV rRNA promoter.
Replacement of the Mtenod40 sORF I sequence with the soybean Glycine max Gmenod40 sORF I was done using primers 5′-GATCCTTGTTTGTAATAAGGATGGAGCTTTGTTGGCTCACAACCATC-3′ and 5′-CATGGATGGTTGTGAGCCAACAAAGCTCCATCCTTATTACAAACAAG-3′.
Primers were hybridized in vitro, and the double-stranded oligonucleotide was cloned in the BamHI and NcoI sites of pMtenod40 to yield pSoyMtenod40 carrying the 12-amino-acid-encoding sORF I of the soybean Gmenod40 gene (31) followed by the complete 3′ region of Mtenod40. p35S-SoyMtenod40 was constructed for expression of this RNA in plant cells (see above).
Construction of enod40 deletions.
For construction of the deletion series (Fig. 1B), we used plasmid pDH51 as the vector. These constructs were made by insertion of PCR products, using different enod40 oligonucleotides and the pMtenod40 and pMsenod40 DNAs and derivatives as the template. We obtained the following deletions: p35S-Mtenod40-Δ4 (nucleotides 204 to 350), p35S-Mtenod40-Δ6 (nucleotides 333 to 614), p35S-Mtenod40-Δ8 (nucleotides 333 to 460), and p35S-Mtenod40-Δ12 (nucleotides 310 to 460). p35S-Mtenod40-Δ5 (nucleotides 204 to 614) and p35S-Mtenod40-Δ7 (nucleotides 31 to 200) have been described previously (9). pMtenod40-Δ5 and pMtenod40-Δ7 are derived from pSK(+) and are similar to p35S-Mtenod40-Δ5 and p35S-Mtenod40-Δ7, respectively, but under control of the T3 promoter. The p35S-Mtenod40-Δ5-ACG sORF II and p35S-Mtenod40-Δ7-ACG sORF I are similar to p35S-Mtenod40-Δ5 and p35S-Mtenod40-Δ7, respectively, but with a mutation in the ATG of sORF II and sORF I, respectively. A series of constructions were carried out in which nucleotides 221 to 311 (corresponding to the RNA structure located between sORF I and sORF II) of the Mtenod40 transcript were deleted. These constructions, pMtenod40-ΔRNA and pMsenod40-ACG sORF I-ΔRNA, were introduced into pDH51, pSK(+), and the vector with the uidA gene. We used pMsenod40 and derivatives as templates. The sequences of the deletions were confirmed as described below.
Nucleic acid techniques and in vitro translation assays.
All steps in the cloning procedures were performed as recommended by the suppliers of the enzymes or as described before (33). Plasmids were analyzed by sequencing using a 373A automatic sequencer (Applied Biosystems) and the Pharmacia sequencing kit.
In vitro transcription of the pMtenod40 insert cloned in pSK(+) was performed with T3 polymerase under conditions for the generation of capped RNA using the Trans probe kit (Pharmacia). In vitro translation of purified transcripts was done using rabbit reticulocyte lysates and wheat germ extracts (Combination System; Promega) and either methionine or cysteine as the labeled amino acid. High-resolution sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the in vitro translation products was performed as described below.
The constructs used for these experiments were described above with the exception of pMsenod40-TCA sORF I Δ3′ region, pMsenod40-TCA sORF I ΔpolyA, and pMsenod40-TCA sORF I/TGA. pMsenod40-TCA sORF I Δ3′ and pMsenod40-TCA sORF I ΔpolyA correspond to the pMsenod40 gene but have a mutation in the stop codon of sORF I and lack either the 3′ region or the poly(A) tail of the gene, respectively. pMsenod40-TCA sORF I/TGA is similar to pMsenod40 but contains a frameshift mutation in sORF I (NcoI digested and Klenow repaired), resulting in a translation product of 2.2 kDa.
The amount of enod40 mRNA in mature nodules of M. sativa cv. Sitel was estimated on Northern blots using known amounts of in vitro-transcribed Mtenod40 as the standard. Northern blotting using Mtenod40 and Msc27 probes was performed as described before (9).
Conventional particle gun bombardment.
Particle bombardment using a Biolistic PDS-100/He particle gun (Bio-Rad) and β-glucuronidase (GUS) activity histochemical staining were performed as described before (9). M. sativa A2 suspension cells (5 days after subculture) were diluted to a cell density corresponding to a packed cell volume of 15% (vol/vol) and plated on Murashige-Skoog-based medium (Sigma M 5519; 3% [wt/vol] sucrose and 1% [wt/vol] Bacto-agar). Cells were grown overnight before bombardment.
Seedlings (5 days after germination) or plated cells from an alfalfa suspension culture were bombarded and stained for GUS activity 2 days later. At least 15 seedlings and three cell plates were bombarded per construct, and experiments were performed at least twice. GUS staining was done for at least 5 h at 37°C, and the number of blue spots was counted under a stereomicroscope. Vibratome (Micro-cut H1200; Bio-Rad) 80-μm sections were cut after embedding in 6% agarose.
Microtargeting.
Microtargeting was performed using the setup described by Sautter et al. (34). Seedlings were plasmolysed by addition of 10% (wt/vol) sucrose and 3% (wt/vol) agar for 1 h prior to bombardment and transferred to Gibson medium 1 day after bombardment. Roots were bombarded in the area of emerging root hairs. A suspension of 1.4-μm gold particles was used at ca 0.5 × 106 particles/μl. DNA was used at a concentration of 1 μg/μl. Pressure was 100 bar, vacuum was −900 mb, and the size of restriction was 140 μm. Two days later, roots were cut and cleared by treatment with commercial bleach as described (9). Whole roots were analyzed under the light microscope (Polyvar; Reichert) for foci of dividing cortical cells by changing the focus to scan through the entire depth of the root. At least two separate experiments (testing a minimum of 15 plants) were performed for each construct. Embedding in paraffin and sectioning were performed as described (9).
Extraction and fractionation of peptides.
Plant tissue was ground in liquid nitrogen and homogenized in extraction buffer (100 mM Tris-HCl [pH 8.0], 100 mM KCl, 10% [vol/vol] glycerol, 10 mM EDTA, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). Homogenates were fractionated by centrifugation at 1,000 × g for 10 min. The pellet was resuspended in Laemmli sample buffer for Western blots (Protocols and Applications Guide; Promega), and the supernatant was recentrifuged at 10,000 × g for 10 min after the addition of 2% (vol/vol) Triton X-100. The resulting pellet was resuspended as above for Western blots, while the supernatant was used for both Western blots and enzyme-linked immunosorbent assay (ELISA).
High-resolution SDS-PAGE for the separation of peptides was performed as described (Protocols and Applications Guide; Promega), and blots were transferred onto 0.45-μm nitrocellulose membranes (Schleicher & Schuell) for Western analysis. High-pressure liquid chromatography (HPLC) for peptide detection was carried out using a reverse-phase column (Beckman Ultrasphere C18; 2 by 150 mm; particle size, 5 μm) with a Beckman System Gold. Samples were eluted at 0.3 ml/min for 60 min with a linear 0 to 70% gradient (A, H2O–0.05% trifluoroacetic acid [TFA]; B, 80% CH3CN–0.05% trifluoroacetic acid). Eluted compounds were detected by their absorption at 214 nm.
Immunodetection of the MtENOD40 peptide.
Polyclonal antibodies were produced in a rabbit by injection of synthetic sORF I peptide coupled to ovalbumin and affinity purified using the synthetic peptide and Affi-gel 10 (Bio-Rad) as described by the manufacturer. Secondary anti-rabbit immunoglobulin G antibodies coupled to alkaline phosphatase (Sigma) were used at a 1:2,000 dilution.
Western blot membranes were incubated for 1 h in blocking solution (5% dry milk in phosphate-buffered saline [pH 7.4]–0.1% Tween 20) before incubation overnight at 4°C in the primary anti-MtENOD40 antibody (diluted 1:200 in blocking solution). Incubation in the secondary antibody solution was for 2 h at 4°C. Signals were detected using 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (Sigma). For the supernatant fraction, 70 μg of total protein was loaded per lane.
For ELISA, the synthetic MtENOD40 peptide, nodule protein supernatants, or a combination of the two were fixed onto ELISA plates and incubated with the primary antibody at a dilution of 1:1,000 for 30 min at room temperature. Incubation with the secondary antibody was performed for 2 h at room temperature. Fluorometric detection of the signal was performed using 1 mg of para-nitrophenyl phosphate (Sigma) per ml. Up to 600 μg of total protein was loaded per microtiter well. Immunocompetition assays were done by fixing 10 ng of the MtENOD40 synthetic peptide on an ELISA plate and incubating with anti-sORF I antibodies overnight in the presence of purified peptide or soluble extracts of roots or nodules. After washing and incubation with the secondary antibody, detection of the signal was done as above.
Sequence analysis.
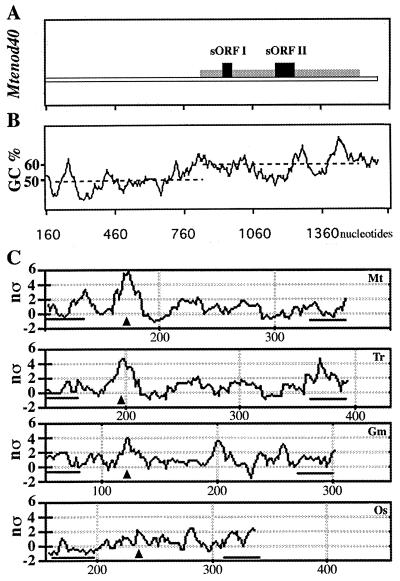
The genomic enod40 sequence was analyzed using GCG programs (University of Wisconsin, Madison) to estimate the percentage of GC. For prediction of the RNA secondary structure, we used the method described before (12), but analyzing window sizes of 30 to 301 nucleotides and scanning with a increment path of 2 bp for each calculation of the number of standard deviations (nς) at different gene positions.
RESULTS
Analysis of Mtenod40 sORF translation in alfalfa roots.
The Mtenod40 transcript is approximately 700 bp long and contains several sORFs in all three reading frames (Fig. 1A). One of these, sORF I, is highly conserved and corresponds to peptides of 10 to 13 amino acids in different species (42). The Mtenod40 transcript contains the longest 5′ region upstream of nucleotide box I published so far (12), as demonstrated by reverse transcription-PCR (data not shown). To investigate the translational capacity of Mtenod40, a series of translational fusions to the uidA gene driven by the constitutive cauliflower mosaic virus 35S promoter were constructed using a vector without an initiating methionine and having instead a polylinker. In this way, the different AUG codons present in the Mtenod40 sequence could be assayed for their capacity to initiate translation. After conventional particle gun bombardment, the constructs were transiently expressed in alfalfa seedlings. Two days later, GUS activity, converting X-d-glucuronic acid (X-d-GlcA) into a blue insoluble product, was estimated as the number of blue spots on roots compared to a 35S-uidA control (Fig. 2). The relative values given in Fig. 2 reflect the observed number of blue cells (in which GUS activity reached the threshold level required for detecting a blue spot on the root), related to the number of cells obtained when the same amount of 35S-uidA control DNA was bombarded (35S-uidA, 100%; Fig. 2A, construct 1). The vector used had no activity itself (Fig. 2A, construct 2). In addition, bombarding a translational fusion of the Mtenod40 promoter and uidA also yielded GUS activity in alfalfa cells, although at reduced levels compared to the 35S CaMV promoter fusion (data not shown).
The fusion with the start codon of the 13-amino-acid oligopeptide (sORF I) sequence of Mtenod40 in frame with the uidA gene (nucleotide 103 of Mtenod40; pTra40-M2) was efficiently translated (Fig. 2A, construct 4) despite the presence of a preceding 8-amino-acid sORF. Translation was observed in epidermal and outer cortical cells of intact roots (Fig. 3A and B) as well as in cultured cells (data not shown). In contrast, translation of uidA was abolished when the reporter gene was fused to a second out-of-frame ATG located within the 13-amino-acid sequence (nucleotide 130 of Mtenod40; pTra40-M3; Fig. 2A, constructs 6 and 7). Thus, the AUG of sORF I is recognized as an efficient translation start site, in contrast to the internal AUG codon, which is present in another frame. Furthermore, translation of the preceding 8-amino-acid sORF was also very efficient, suggesting that sORFs do not prevent reinitiation at the following ORF (Fig. 2A, construct 3). Deletion of the first sORF did not affect translation of sORF I.
FIG. 3.
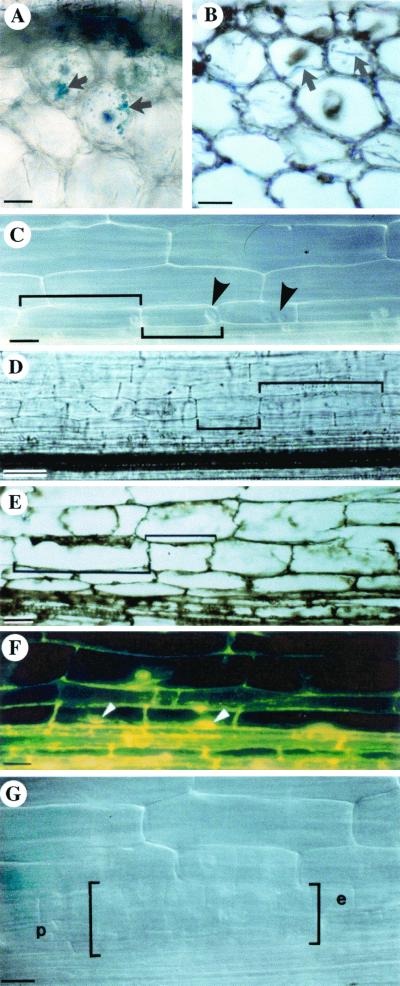
Microscopic analysis of alfalfa roots fixed 2 days after microtargeting with enod40 derivatives. (A and B) Transversal sections showing transient expression of an Mtenod40-uidA translational fusion in outer cortical cells of alfalfa roots. The colored GUS reaction product is indicated by arrows: (A) 80-μm section; (B) 8-μm section. (C to F) Cortical cell divisions 2 days after microtargeting. Note the difference in cell length of recently divided and nondivided cells (brackets). Nuclei are indicated by arrowheads. (C and D) Whole roots in phase contrast, showing cortical cell divisions. (E and F) Sections (8 μm) of cortical cell divisions as seen in bright-field mode (E) and fluorescence (F). (G) Early root primordium. Extensive concomitant divisions (within brackets) are found in the pericycle (p) and endodermis (e). Bars: 25 μm (A, E, and F), 15 μm (B), 20 μm (C and G), and 50 μm (D).
Then, the reporter gene was fused to the next AUG codon, M4, which is located 8 bp after the stop codon of sORF I within the M3 sORF (Fig. 1A) and codes for a 7-amino-acid peptide. Translation was detectable, albeit inefficient (Fig. 2A, construct 8). The next ATG, M5, the start of a 5-amino-acid sORF, also showed reduced translation, whereas M6, corresponding to the sORF-spanning region 2 (sORF II), was clearly translated (35%; Fig. 2A, constructs 9 to 11). To confirm that the stop codon of sORF I arrested protein translation, fusions downstream in the three reading frames were constructed. Translation was not detected in any case (Fig. 2A, construct 12). Fusions to AUG codons of other reading frames within sORF II (M7 and M8) or after sORF II (e.g., M9) also exhibited undetectable or very low levels (less than 1%) of translation (Fig. 2A, constructs 14 to 16).
Mutation of ATG codon M6 completely abolished GUS activity, indicating that sORF II translation started from this codon (Fig. 2A, construct 13). Interestingly, even between two Medicago species (M. sativa and M. truncatula), sORF II is not conserved in either size or amino acid sequence except for the amino acids encoded in the region of the so-called box II (Fig. 1). Moreover, AUG M6 only exists in enod40 genes of a few legume species and tobacco, and the nucleotide sequences of the different enod40 genes after box II are not conserved.
These results suggest that sORFs do not arrest 5′-to-3′ ribosome scanning and that reinitiation can occur along the enod40 transcript. Two sORFs spanning the conserved nucleotide regions of Mtenod40 can be efficiently translated in root tissues.
Activity of the different Mtenod40 regions depends on translation of sORFs.
Expression of enod40 in roots resulted in cortical cell divisions (9). In order to study the molecular mechanism of enod40 action, a microtargeting apparatus was used to introduce DNA in alfalfa root cells to monitor events related to enod40 expression in a defined root region. Two days after microtargeting, the bombarded area was inspected for cortical cell division. Particles were found only in superficial cell layers (epidermis and outermost cortex) and were used to localize the bombarded area (diameter, less than 0.5 mm). Cell divisions detected in the inner cortex below this area (Fig. 3), without divisions in the underlying pericycle and endodermis, were observed as either (i) foci of recently divided cells showing a dense cytoplasm and conspicuous nuclei (Fig. 3C) or (ii) foci of divided cells that had already started to elongate (Fig. 3D). Sectioning confirmed the presence of divided cells (Fig. 3E and F). Early root primordia (Fig. 3G) were distinguished from cortical cell divisions by the occurrence of concomitant, extensive divisions of the pericycle, i.e., below the endodermis, which is recognized by the presence of Casparian strips and a strong autofluorescence.
The enod40 activity of different constructs was monitored in a semiquantitative manner by counting the number of foci of dividing cortical cells. Transient expression of the Mtenod40 construct in alfalfa roots induced cortical cell divisions at high frequency, giving an fdiv value (cell division foci per root) of 0.51. This frequency is more than two times higher than those obtained in our previous experiments using conventional particle bombardment of whole roots from transgenic alfalfa plants carrying an Msenod12A promoter-uidA fusion (9). The empty vector had no effect (fdiv = 0.02; Table 1). In agreement with our previous experiments, constructs spanning either box I or II had similar activity (corresponding to Δ7 and Δ5, respectively; Fig. 1B), as both also induced divisions at high frequency (Table 1). Mutating the start codon of the 13-amino-acid peptide sequence (ATG changed to ACG; p35S-Δ7-ACG sORF I) abolished the cell division-inducing activity of Δ7 (fdiv = 0.03), suggesting that translation of sORF I is required for activity. Mutating the ATG codon of sORF II yielded a Δ5 derivative with highly reduced activity. Deletions containing the conserved nucleotide box II but not the entire sORF II did not show significant activity (Table 1, p35S-Mtenod40-Δ8 and p35S-Mtenod40-Δ4, respectively). In contrast, a short region spanning the unmodified sORF II retained biological activity, indicating that translation of this sORF might be responsible for the induction of cortical cell divisions elicited by Δ5 (Table 1, p35S-Mtenod40-Δ12). These values are significantly different, as demonstrated by a chi-square statistical test (P < 0.01).
TABLE 1.
Microtargeting of alfalfa seedlings with enod40 DNA constructs
| Construct | No. of focia | No. of roots examined | fdiv ± SEb |
|---|---|---|---|
| pDH51 | 1 | 64 | 0.02 ± 0.02 |
| p35S-Mtenod40 | 18 | 35 | 0.51 ± 0.09 |
| p35S-Mtenod40-Δ7 | 8 | 20 | 0.40 ± 0.11 |
| p35S-Mtenod40-Δ5 | 11 | 30 | 0.37 ± 0.09 |
| p35S-Mtenod40-Δ7-ACG sORF I | 1 | 38 | 0.03 ± 0.03 |
| p35S-Mtenod40-Δ5-ACG sORF II | 1 | 15 | 0.07 ± 0.07 |
| p35S-Mtenod40-Δ4 | 3 | 20 | 0.15 ± 0.08 |
| p35S-Mtenod40-Δ8 | 0 | 20 | 0.00 ± 0.00 |
| p35S-Mtenod40-Δ12 | 6 | 12 | 0.50 ± 0.15 |
| p35S-Mtenod40-ACG sORF I | 4 | 46 | 0.09 ± 0.04 |
| p35S-Msenod40-ACG sORF I | 1 | 9 | 0.11 ± 0.11 |
| p35S-Mtenod40-TCA sORF I | 2 | 33 | 0.06 ± 0.04 |
| p35S-Mtenod40-mod.sORF I | 8 | 27 | 0.30 ± 0.09 |
| p35S-SoyMtenod40 | 1 | 15 | 0.07 ± 0.07 |
| p35S-Mtenod40-ACG sORF II | 2 | 15 | 0.13 ± 0.09 |
| p35S-Mtenod40-ΔRNA | 2 | 15 | 0.13 ± 0.09 |
Number of cortical cell division foci observed 2 days after microtargeting.
Number of cortical cell division foci per bombarded root.
Thus, microtargeting of Mtenod40 and two nonoverlapping deleted derivatives induced a cell-specific growth response in alfalfa roots. The cortical cell division-inducing activity observed for enod40 deletions required the translation of two sORFs of 13 and 27 amino acids, sORFs I and II, respectively, spanning the conserved nucleotide boxes of the enod40 transcript.
Translation of sORF I and sORF II regulates Mtenod40 activity in alfalfa roots.
Neither in Northern blots nor in cDNA libraries have we identified Mtenod40 cDNAs corresponding only to Δ7 or Δ5 (not shown). Therefore, we decided to study the effects of sORF translation on the complete Mtenod40 transcript. Microtargeting of the Mtenod40 sequence with a mutated start codon for sORF I (p35S-Mtenod40-ACG sORF I and p35S-Msenod40-ACG sORF I; Table 1) had significantly reduced activity (fdiv = 0.09 and 0.11, respectively). This was surprising, since the activity of the 3′ region of Mtenod40 (see above) was expected to be retained in this derivative (only lacking sORF I). In addition, a construct in which the stop codon of the 13-amino-acid peptide-encoding sequence was mutated (p35S-Mtenod40-TCA sORF I), resulting in a longer sORF incorporating extra codons (up to 59 amino acids; asterisk in Fig. 1A), was also inactive (Table 1; fdiv = 0.06). The nucleotide changes assayed might disturb either translation of the peptide or the mRNA in this region.
To confirm that sORF I was responsible for activity, a mutant Mtenod40 sequence was created in which the 13-amino-acid peptide-encoding nucleotide sequence was modified while keeping the amino acid sequence intact (taking advantage of codon degeneracy). This yielded a construct (p35S-Mtenod40-mod.sORF I) with significantly high activity (fdiv = 0.30), confirming that the translation product of the 13-amino-acid sORF is crucial for enod40 activity. Finally, the Mtenod40 sORF I sequence was replaced by a 12-amino-acid-encoding sequence corresponding to the soybean Gmenod40 sORF I peptide (33). This p35S-SoyMtenod40 derivative was inactive (Table 1; fdiv = 0.07). This confirmed that the sORF I-derived peptide is essential for Mtenod40 activity.
These experiments also suggest that translation of sORF II may require translation of sORF I. To test this hypothesis, we assayed constructs in which uidA was fused to the ATG-M6 of sORF II with either a mutated ATG or TAA codon in sORF I (Fig. 2A, constructs 17 to 19; cf. construct 11). Mutating the initiating codon of sORF I increased the translational activity of sORF II, whereas increasing the size of sORF I dramatically reduced the capacity to reinitiate translation at sORF II. Hence, the level of sORF II translation does not correlate with the biological activity of the mutated Mtenod40 transcripts. Nonetheless, a point mutation of the ATG of sORF II abolishing its translation (Fig. 2A, construct 13) reduced the biological activity of Mtenod40 (Table 1; fdiv = 0.13). This indicates that translation of sORF II is important for cell division activity of the Mtenod40 transcript.
From these experiments, we concluded that translation of both sORF I and sORF II is required for full Mtenod40 activity. The effect of the 3′ Mtenod40 region in alfalfa roots depends on the presence and correct size and sequence of the 13-amino-acid peptide encoded by sORF I, even though deletions spanning either sORF I or sORF II are active. These results revealed a complex level of regulation acting on the Mtenod40 mRNA and are in line with the fact that nucleotide boxes I and II are highly conserved in enod40 genes from several species.
enod40 sORF-encoded oligopeptides require a minimum size to be detected by in vitro translation.
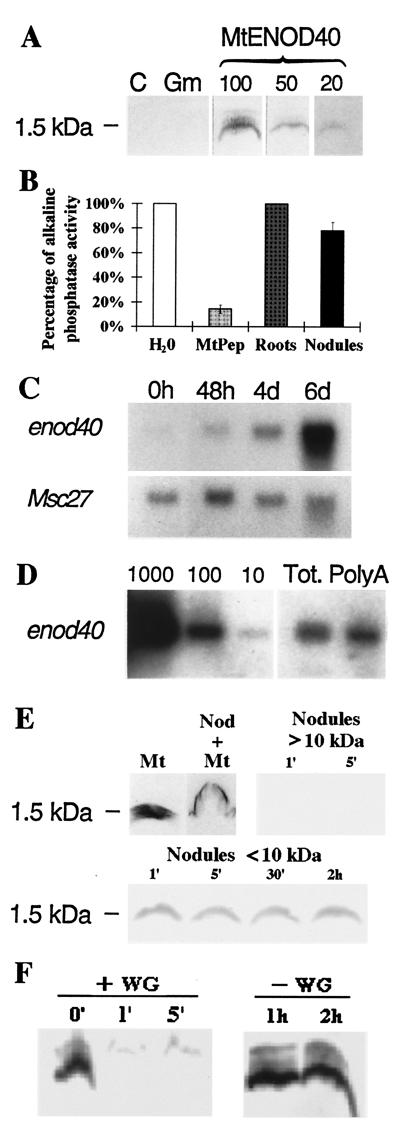
Having shown that the start codon of the Mtenod40 13-amino-acid sORF I can be recognized as a translation start site (in bombarded roots) and that sORF I is required for enod40 activity, we searched for the presence of the encoded peptide in planta by several methods based on immunodetection. Polyclonal antibodies against the 13-amino-acid MtENOD40 peptide coupled to ovalbumin were prepared and affinity purified using the synthetic peptide. In Western blots using high-resolution gels optimized for detection of small peptides, these antibodies specifically recognized as little as 20 ng of the synthetic MtENOD40 13-amino-acid peptide, giving a band at 1.5 kDa (Fig. 4A), whereas the preimmune serum did not react with this sample. In ELISA, as little as 1 ng of synthetic peptide could be detected (data not shown). Then, we assayed total extracts and subcellular fractions of roots and mature nodules using both Western blots (70 μg of total protein was loaded per lane) and ELISA (up to 600 μg of total protein was loaded per microtiter well). Moreover, ELISA was also used to test fractions obtained by separating up to 600 μg of protein extract from mature nodules through HPLC. Similarly, several subcellular fractions of root extracts (corresponding to soluble, nuclear, and microsomal fractions) were tested at different time points after inoculation of M. sativa with S. meliloti. Even though weak signals could be detected at higher molecular weights, no signal corresponding to a small oligopeptide was detected either during early nodule development or in mature nodules by any method using the different extraction procedures (data not shown). However, immunocompetition experiments revealed the presence of small amounts of an antigen related to the 13-amino-acid peptide in nodules (Fig. 4B). Interestingly, quantification of enod40 transcripts by Northern blotting indicated that they constitute approximately 0.5% of the total amount of mRNA in the nodule, accumulating rapidly during early nodule development (Fig. 4C).
FIG. 4.
Analysis of enod40 gene products and their stability. (A) Western blot of synthesized peptides showing the specificity of anti-ENOD40 13-amino-acid peptide antibodies. Lane C, 100 ng of a control peptide with a rearranged 13-amino-acid sequence; lane Gm, 100 ng of the Gmenod40 12-amino-acid peptide (31); lanes MtENOD40, 100, 50, or 20 ng of the Mtenod40 13-amino-acid peptide as marked. (B) ELISA immunocompetition assays. Root and nodule soluble extracts were used against 10 ng of bound Mtenod40 peptide. Antibodies were added alone (water control) in the presence of 500 ng of synthetic peptide (MtPep) or 500 μg of root or nodule extract. (C) Northern blot showing hybridization of an Mtenod40 probe to RNA isolated from S. meliloti-inoculated alfalfa roots during nodule development. Time after inoculation is indicated (hours or days [d]). The constitutively expressed Msc27 was used as a control for RNA loading. (D) Northern quantification of the Mtenod40 transcript amount in nodules. Lane Tot, 5 μg of total nodule RNA (approximately 2% corresponds to mRNA); lane PolyA, 200 ng of nodule poly(A)-containing RNA; 1,000, 100, and 10 pg of in vitro-transcribed Mtenod40 mRNA were used for calibration. Quantification of the hybridization signals indicates that Mtenod40 signals correspond to 50 and 80 pg for total and poly(A) RNA, respectively. (E and F) Western blots showing MtENOD40 13-amino-acid peptide stability in nodule extracts (E) and in wheat germ in vitro translation extracts (F). Mt, MtENOD40 peptide without extract; Nod+Mt, nodule extract denatured with 1% SDS before adding the peptide; nodule fractions enriched in proteins of >10 or <10 kDa incubated with the peptides for the indicated time periods (minutes). +WG and −WG, with and without wheat germ extracts for the indicated time periods (minutes or hours), respectively.
Thus, while enod40 is a very abundant mRNA in the nodule, the oligopeptide is either present at a very low concentration or unstable under our extraction conditions. To test the latter possibility, we added up to 2 μg of the synthetic peptide to nodule extracts, and the presence of the peptide was tested at different time points by Western blotting. The immunological signal had already disappeared after 1 min of incubation (Fig. 4E). Nevertheless, the peptide was stable in a soluble fraction inactivated by SDS treatment or fractionated to retain constituents of less than 10 kDa (Fig. 4E). These data indicate that oligopeptides may be highly unstable in plant extracts, due either to degradation or to binding to a component that masks it very rapidly from the antibodies.
We then tested the in vitro translation of various amounts of purified Mtenod40 RNA (transcribed in vitro) using both wheat germ and reticulocyte extracts (Fig. 5). No significant signal was observed in several experiments (Fig. 5A, constructs 1 and 2) using either Msenod40 or Mtenod40 RNAs. The occasionally translated short peptides (such as those depicted in Fig. 5B, lane 2) showed no correlation with the synthesized peptide using HPLC or immunoprecipitation (data not shown; see below). In addition, no in vitro translation products were observed using deletion derivatives of enod40 (Fig. 5, constructs 9 and 10). We tested the stability of the putative oligopeptide encoded by sORF I by adding the synthetic 13-amino-acid peptide to the extracts used for in vitro translation. The immunological signal disappeared again very rapidly (less than 1 min) (Fig. 4F). It is likely that the short unstable enod40 peptides need to be coupled to or protected by a large protein or subcellular structure to avoid rapid degradation. An enod40 derivative containing a point mutation in the stop codon of sORF I that should give translation of a 59-amino-acid peptide was then tested (Fig. 1A, asterisk). A clear band of the expected size was observed on gels after in vitro translation (Fig. 5, constructs 3 and 5). This in vitro translation product was not detected when the start codon of sORF I was mutated (Fig. 5, construct 4), further confirming that it corresponds to the mutated sORF I. Moreover, constructs with point mutations encoding the predicted peptides of 39 and 22 amino acids, respectively (Fig. 5, constructs 6 and 7), were tested. These transcripts are almost identical to Mtenod40, suggesting that the ATG of sORF I is indeed recognized but that peptides of less than 39 amino acids are very unstable and therefore cannot be detected under conditions for in vitro translation. This also suggests a correlation between the minimum size of a sORF whose product can be detected by in vitro translation and, as mentioned in the previous section, the maximum size of an upstream sORF that allows further 3′ translation (Fig. 2A, construct 19). As expected, small peptides could not be detected using pMtenod40-modified sORF I or pMtenod40-ACG sORF I (Fig. 5, constructs 12 and 13). The latter one occasionally showed short translated peptides similar to pMtenod40, confirming that these are not sORF I related. Finally, the presence of a poly(A) tail in the enod40 mRNA was required for detection of the in vitro translation products (Fig. 5, construct 11).
FIG. 5.
In vitro translation of Mtenod40 and Msenod40 RNA using wheat germ extracts. (A) Predicted peptides after in vitro translation of different Mtenod40 mRNA derivatives. Detection of an in vitro translation product of the expected size is denoted by +. Solid boxes indicate sORF I and sORF II present in the Mtenod40 and Msenod40 genes. TAG and TGA in bold type indicate the stop codon for a modified sORF I peptide after creation of a point mutation in the stop codon for sORF I (TAA to TCA). ACG in bold type indicates a point mutation changing the ATG start codon of sORF I to ACG. Predicted peptide values are in kilodaltons. (B) Radioactive products detected after in vitro translation of the different enod40 constructs, numbered as in panel A. Lane C, control translation reaction without mRNA.
To explain the presence of two biologically active regions in the enod40 transcript, it was previously hypothesized that the 3′ region might inhibit translation of sORF I (42). We tested this hypothesis by two different approaches. First, we introduced the 3′ region (Δ5) immediately downstream of the uidA sequence in the in-frame translational fusion with sORF I (pTra40M2-Δ5). Translation was reduced by half compared to pTra40M2 (Fig. 2B). Introducing this 3′ sequence in an inverted position (pTra40M2-inv.Δ5) yielded translation at the same level as that obtained without it. Second, in vitro translation of the 59-amino-acid sORF I Mtenod40 derivative (with a mutated stop codon) in the presence or absence of the 3′ Mtenod40 Δ5 sequence (Fig. 5, constructs 3 and 8) showed no significant effect of this 3′ region on sORF I translation.
Thus, the ATG codon of sORF I is recognized as a translation start site in roots and during in vitro translation. Detection of peptide products corresponding to an encoded sORF by in vitro translation requires a minimum size. The MtENOD40 oligopeptide(s) might be very difficult to detect due to high instability, even in plant tissues where transcripts accumulate abundantly. Nevertheless, epitopes related to the sORF I-encoded peptide may exist in nodule extracts. The 3′ enod40 region did not show any significant effect per se on translation of the 5′ sORF I.
Deletion of an Mtenod40 inter-sORF region spanning a predicted RNA structure affects biological activity.
Translation of Mtenod40 sORFs is necessary for full biological activity, although translation of either sORF is sufficient in the deletion- containing mRNAs. Hence, our results suggest that a novel type of regulation of sORF activity may exist in the whole transcript that is lacking in the deletion derivatives. We therefore decided to analyze the Mtenod40 mRNA in a genomic context. The sequence of a 3-kb Mtenod40 genomic region (containing a 1-kb upstream and a 1.3-kb downstream region) was determined, and its GC percentage was analyzed. The Mtenod40 mRNA separates from the isochore of the Medicago genome (38% GC 4) from the beginning of the transcript (Fig. 6B), suggesting a larger functional Mtenod40 region than that corresponding to the sORFs. In our previous work, we proposed that the Mtenod40 transcript codes for a highly structured RNA, based on calculations of the free energy of folding (12). By applying the same computer program using small (around 40 bp) scanning windows, we could now predict the presence of a highly structured RNA region which is located between the two sORFs of the Mtenod40 transcript (Fig. 6C, first panel; nς > 5; this number illustrates that this structure has a very low probability of appearing randomly on the same nucleotide sequence; for details see reference 12). This structure lies on the functional enod40 region predicted by the isochore analysis of the nucleotide sequence to lie between the two conserved nucleotide boxes. Therefore, a deletion was made to modify this “inter-sORF” region (Fig. 1, ΔRNA) without affecting the coding sequences of the sORFs. This Mtenod40 derivative showed a significant reduction in biological activity (Table 1, fdiv = 0.13) after microtargeting, indicating that the RNA region between sORF I and sORF II is also required for gene function. To show whether an RNA structure may be conserved, the presence of RNA structures was sought in various enod40 genes (Fig. 6C, panels 2 to 4). Interestingly, in a relatively homologous position (between the two conserved nucleotide boxes), highly probable predicted structures were found in enod40 genes from white clover, soybean, and rice (see Discussion).
FIG. 6.
Structural analysis of the Mtenod40 sequence. (A) Schematic representation of the Mtenod40 genomic sequence (accession number X80263). The cDNA sequence is represented by a striped box; sORF I and sORF II are indicated. (B) Analysis of the GC content of the Mtenod40 gene. The sequence upstream of the transcription start site shows an average GC content of 40% (dashed line), which is equivalent to the isochore of the Medicago genome. However, the cDNA sequence has an average GC content of 50% (dashed line). (C) Stability analysis of the enod40 genes. enod40 transcripts were scanned for predicted RNA structures (windows from 30 to 300 bp; increment, 2 bp), and the number of standard deviations (nς: value indicating the dispersion of the free energy of folding for each window, as described in reference 12) was calculated. Selected scans corresponding to specific window sizes (around 40 bp) are depicted for four enod40 transcripts: Mtenod40 (accession number X80264) for M. truncatula; Trenod40 for Trifolium repens (accession number AJ000268); Gmenod40 for Glycine max (accession number X69154), and Osenod40 for Oryza sativa (accession number AB024054; nucleotides 2200 to 2642 corresponding to transcript 22). Regions containing highly stable predicted RNA structures measured by a high nς are indicated with an arrowhead. Bars indicate the position of conserved nucleotide sequences 1 and 2 in the different transcripts.
We then tested whether this deletion of the inter-ORF region of the Mtenod40 transcript would inactivate translation of the sORFs. A translational fusion to sORF II in which this RNA region was deleted was used to bombard roots. No effect on translation of sORF II was detected in comparison to the fusion containing this RNA region (Fig. 2B, construct 20) (cf. construct 11). To analyze the effect of this region on sORF I translation, we introduced a point mutation in the sORF I ATG in a corresponding construct. Inactivation of sORF I affected sORF II translation as reported for the complete enod40 transcript (Fig. 2B, construct 21; cf. construct 17), indicating that sORF I is likely translated.
Therefore, an RNA region present between the Mtenod40 sORFs regulates the elicitation of cortical cell division, most likely without perturbing translation of the sORFs. These results reveal a new level of regulation acting on enod40 activity.
DISCUSSION
Peptides encoded in sORFs may act as signals during the induction of cortical cell divisions in roots.
Here we demonstrated that enod40-induced cell division in the alfalfa root inner cortex depends on the translation of two sORFs. Our assay was based on the introduction of Mtenod40 by microtargeting in the region of emerging root hairs where rhizobia and Nod factors induce cortical cell division. Division frequencies in our assay were comparable to those obtained by microtargeting chitin oligosaccharides or Nod factors (26, 36). Both the need to transduce a secondary signal from superficial cell layers where particles land into responsive cells in the inner cortex and the instability of the enod40 gene products are expected to lower this value. In order to analyze the effects of the bombarded constructs, we studied responses induced only 2 days after bombardment. Longer times may introduce further variables (e.g., light conditions or hormonal changes during development). Apparently, enod40 is not an inducer of cell division per se, but other factors likely present only in the inner cortex are required to complete cell cycle activation. Stable constitutive expression of enod40 in transgenic plants resulted in a large proportion of dividing root cortical cells when grown under nitrogen-limited conditions (9) as well as accelerated nodulation (10), further suggesting that our transient assay is related to the biological activity of enod40 in the root cortex.
enod40 is unusual in that it contains many sORFs, and the two peptides whose translation controls biological activity are encoded as such, in contrast to other eukaryotic small peptides, which are produced by cleavage of larger precursors containing signal sequences or related features (plant examples include systemin [32] and phytosulfokines [27]). Several reports suggest that plants do make use of peptides as signals in development, since both peptidic signals and putative receptors for them have been found (reviewed in reference 3). Moreover, mutations in these receptors influenced plant differentiation (2, 41). In our biological assay, however, we could not detect an effect of the synthetic peptide corresponding to sORF I (our unpublished results). The peptide may require certain modifications to become active, as has been shown for the phytosulfokine peptide growth factor (27). sORF-encoded small peptides may be able to diffuse out of the cell to act extracellularly, or alternatively, they may have intracellular targets that could be reached directly after translation. This is the case for the 5-amino-acid-encoding sORF in the 23S rRNA in E. coli, where the pentapeptide likely is produced and immediately binds its intracellular target, the rRNA (39). Several examples of the cis effects of upstream sORFs on 3′ translation mediated through binding to ribosomes have also been reported (25, 29) in eukaryotic cells. In these cases a 3′ long ORF codes for the protein having biological activity, in contrast to enod40 genes, where only sORFs are found all along the transcript.
sORF-encoded peptides can be translated in plant tissues.
We have previously proposed that enod40 may code for a nontranslatable RNA (12). This was based on the fact that it lacked long ORFs, the computer-assisted prediction that it forms highly stable secondary structures, and the fact that it did not produce detectable in vitro translation products, properties shared with biologically active RNAs. In addition, the enod40 RNA did not copurify with polysomes (1). In this work, however, using selected point mutants, we demonstrated that translation, size, and correct amino acid sequence of the 13-amino-acid sORF I as well as translation of sORF II are necessary for the biological activity of enod40. This supports the hypothesis that sORF-encoded peptides are biologically active, though we cannot exclude that translation from the enod40 RNA may alter either the secondary structure or its decay or its capacity to interact with other proteins to form a ribonucleoprotein. sORF-encoded peptides may exert critical functions in sORF-RNAs through a variety of mechanisms (15, 25, 29, 43).
Localization of the peptide in the cell might help us in determining such a mechanism. Immunocompetition experiments indicated the presence of a domain related to the enod40 sORF I-encoded peptide in alfalfa nodule extracts, in agreement with previous experiments done in soybean (42). However, by using Western analysis or direct immunological approaches in alfalfa plants, we could not detect this enod40 gene product even in young nodules, in which the transcript is very abundant. Oligopeptides are rapidly degraded in nodules and in vitro translation extracts, suggesting that immunological approaches may not be adequate to detect them biochemically. The high level of mRNA available for translation in the dividing cells of the nodule primordium may ensure continuous production of the oligopeptides in the cell via monosomes or a transient association with polysomes. Alternatively, the encoded peptides may be integrated into a larger protein complex (e.g., by coupling to an acceptor protein or a downstream sORF peptide) or into a structure (e.g., a ribonucleoprotein particle during translation), which could explain the presence of epitopes related to the enod40 sORF I peptide in nodule extracts.
The recognition of the sORF I ATG was demonstrated both in vivo using reporter gene fusions and in vitro by translation of a larger peptide. The soybean sORF I was also shown to be translated in tobacco protoplasts using a green fluorescent protein reporter gene (42). Our translational analysis indicated that sORFs do not prevent reinitiation at downstream AUG codons in either differentiated epidermal and cortical cells or nondifferentiated cultured cells. The different Mtenod40 sORFs are therefore likely to be translated with variable efficiency. Several plant mRNAs contain upstream sORFs that reduce translation of ORFs further downstream (15, 25, 29, 43). This was also the case for sORF II translation, as shown here using point mutations of sORF I. Several mechanisms to translate consecutive or overlapping ORFs exist in plant viruses (reviewed in reference 15) and might also exist in plants, where a polycistronic mRNA has recently been found (16). Even though sORFs are not generally considered gene products, our work indicates that both sORFs are required for enod40 function, and hence it acts as a polycistronic transcript.
A puzzling result is that replacement of sORF I by the homologous sORF I of soybean yielded an inactive derivative. This revealed a certain level of “species specificity” for sORF I regulation of enod40 action in alfalfa roots. A specific amino acid sequence might be required for appropriate function, suggesting that even nonconserved amino acids of the small peptide may play roles in target recognition. Strict sequence specificity of upstream sORFs has also been shown in translational regulation in mammalian cells (29).
Two regions of Mtenod40 are functionally connected.
Our data indicate that the action of sORF II depends on the translation of sORF I and this regulation is lost in the deleted Mtenod40 derivative spanning the 3′ region (Δ5). In addition, this control is exerted not only at the level of translation, since either the altered size or the absence of sORF I had negative effects on enod40 biological activity but decreased or increased sORF II translation, respectively. In these experiments, we also cannot exclude that interactions of the bombarded constructs with the endogenous alfalfa enod40 gene occurred, affecting either its transcription or translation. However, no enod40 mutant legume is yet available to analyze such an effect.
It has been proposed that the sORF I-encoded ENOD40 peptide is the active gene product and that the 3′ region might control its translation (even in trans on the endogenous gene 42). However, we could not detect activity of the enod40 peptide in plant tissues, and the work based on a tobacco protoplast assay (42) could not be reproduced (35). This model cannot explain the effects of several point mutations that we detected as affecting Mtenod40 activity in alfalfa roots. These mutations may perturb either the transcript stability or the capacity of the RNA to interact with the ribosomes or other intracellular target, having an indirect effect on enod40 function. Hence, we think that the identified regions and sORFs are more likely required for determining the subcellular site where enod40 is translated. This regulation can be overcome when using the deleted versions spanning only the sORF regions. Interestingly, RNA signals present in the 3′ untranslated regions of several genes have been shown to be involved in localization of mRNA translation (30), a feature that may be essential when the encoded gene products are unstable (such as the enod40 oligopeptides). An alternative hypothesis is that sORF-mediated translational control of the transcript may also be exerted to allow stability or export of the RNA into the cytoplasm (as shown in animals [19]). Finally, the translation process per se might impose certain properties on the enod40 mRNA. Nevertheless, we think that our experiments using the complete transcript are likely to reflect appropriately enod40 gene regulation in vivo, since shorter forms of the enod40 transcript have not been detected in planta.
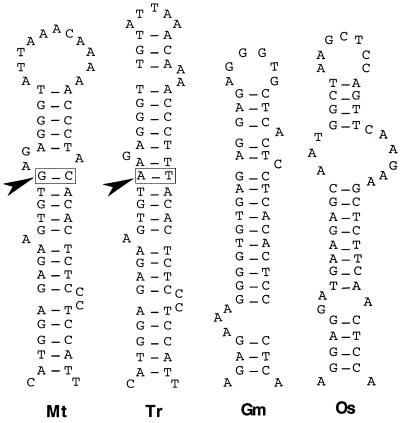
Microtargeting of the sORF I or sORF II region alone is sufficient for enod40 activity, whereas the inter-ORF region is inactive. This region, presenting predicted stable RNA structures, may determine transcript localization or stability. Alternatively, the critical parameter might be the correct spacing between sORFs that is also affected by the deletion of this region. Further studies on the involvement of RNA structures in gene function might await more sensitive methods to assay enod40 activity. A predicted highly structured RNA region related to that of Mtenod40 was detected in almost all enod40 cDNAs found in the databases except for the tobacco gene. In this case, only a PCR product has been cloned, and we cannot exclude that this RNA structure is not required in this nonlegume plant or that the cloned fragment corresponded to a nonfunctional gene. Despite the large sequence variation detected in the inter-ORF region between enod40 genes, it seems that a conserved stem structure could be detected even in the distantly related nonlegume rice plant. The predicted RNA structures were folded using an appropriate window according to the maximum nς value (Fig. 7). By comparing the two highly related sequences of Trifolium repens and M. truncatula, a compensatory mutation seemed to be present in the stem RNA region (arrow), further reinforcing the validity of the presence of an RNA structure in the enod40 transcripts. Hence, this inter-ORF region, even though probably not directly responsible for gene function, seems to be involved in enod40 gene regulation.
FIG. 7.
Proposed RNA structures from different enod40 transcripts. The regions presenting the highest nς in the different scans depicted in Fig. 6C were folded using MFOLD. A putative stem structure conserved in the enod40 genes of the different species is proposed. The squares (arrow) indicate the presence of a compensatory mutation in the Trenod40 and Mtenod40 genes. Mt, M. truncatula; Tr, T. repens; Gm, G. max; Os, O. sativa.
sORF translation and structured RNA signals might be important elements in the regulation of genes lacking long ORFs, particularly those having functions related to growth and differentiation, as shown here for enod40 regulation in nodule organogenesis. Further analysis of the mechanism of enod40 action in development [such as the identification of putative receptors or targets for the peptide(s) and/or structured RNA region, as well as their subcellular localization] may uncover novel means of translational control of the cell.
ACKNOWLEDGMENTS
We thank Elke Fenner for technical assistance, H. Küster for providing pGUS-INT, Y. D'Aubenton-Carafa and C. Thermes for RNA structure analysis and helpful discussions, and L. Troussard for sequencing help.
C.J. was supported by a postdoctoral fellowship from the Swedish Council for Forestry and Agricultural Research and an EEC PTP training fellowship, C. Sousa by a postdoctoral fellowship from the EEC (TMR Marie Curie training grant), C.C. by a fellowship from the Ministère Français de l'Enseignement Supérieur et de la Recherche, and H.M. by a short-term EMBO fellowship.
REFERENCES
- 1.Asad S, Fang Y, Wycoff K L, Hirsch A M. Isolation and characterization of cDNA and genomic clones of MsENOD40; transcripts are detected in meristematic cells of alfalfa. Protoplasma. 1994;183:10–23. [Google Scholar]
- 2.Becraft P W, Stinard P S, McCarty D R. CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science. 1996;273:1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- 3.Bisseling T. The role of plant peptides in intercellular signalling. Curr Opin Plant Biol. 1999;2:365–368. doi: 10.1016/s1369-5266(99)00006-0. [DOI] [PubMed] [Google Scholar]
- 4.Blondon F, Marie D, Brown S, Kondorosi A. Genome size and base composition in Medicago sativa and M. truncatula species. Genome. 1994;37:264–275. doi: 10.1139/g94-037. [DOI] [PubMed] [Google Scholar]
- 5.Bonneville J M, Sanfaçon H, Fütterer J, Hohn T. Post-transcriptional trans-activation in cauliflower mosaic virus. Cell. 1989;59:1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- 6.Bullock W O, Fernández J M, Short J M. XL-1 Blue: a high efficiency plasmid-transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 7.Burleigh S H, Harrison M J. A novel gene whose expression in Medicago truncatula roots is suppressed in response to colonization by vesicular-arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Mol Biol. 1997;34:199–208. doi: 10.1023/a:1005841119665. [DOI] [PubMed] [Google Scholar]
- 8.Cech T R, Bass B L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- 9.Charon C, Johansson C, Kondorosi E, Kondorosi A, Crespi M. enod40 induces dedifferentiation and division of root cortical cells in legumes. Proc Natl Acad Sci USA. 1997;94:8901–8906. doi: 10.1073/pnas.94.16.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charon C, Sousa C, Crespi M, Kondorosi A. Alteration of enod40 expression modifies Medicago truncatula root nodule development induced by Sinorhizobium meliloti. Plant Cell. 1999;11:1953–1965. doi: 10.1105/tpc.11.10.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corich V, Goormachtig S, Lievens S, van Montagu M, Holsters M. Patterns of ENOD40 gene expression in stem-borne nodules of Sesbania rostrata. Plant Mol Biol. 1998;37:67–76. doi: 10.1023/a:1005925607793. [DOI] [PubMed] [Google Scholar]
- 12.Crespi M D, Jurkevitch E, Poiret M, d'Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A. enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994;13:5099–5112. doi: 10.1002/j.1460-2075.1994.tb06839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Hirsch A M. Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol. 1998;116:53–68. doi: 10.1104/pp.116.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furini A, Koncz C, Salamini F, Bartels D. High level transcription of a member of a repeated gene family confers dehydration tolerance to callus tissue of Craterostigma plantagineum. EMBO J. 1997;16:3599–3608. doi: 10.1093/emboj/16.12.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fütterer J, Hohn T. Translation in plants—rules and exceptions. Plant Mol Biol. 1996;132:159–189. doi: 10.1007/BF00039382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Rios M, Fujita T, LaRosa P C, Locy R D, Clithero J M, Bressans R A, Csonka L N. Cloning of a polycistronic cDNA from tomato encoding γ-glutamyl phosphate reductase. Proc Natl Acad Sci USA. 1997;94:8249–8254. doi: 10.1073/pnas.94.15.8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 18.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 19.Hentze M, Kulozik A. A perfect message: RNA surveillance and non-sense mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 20.Herrero M, de Lorenzo V, Timmis K T. Transposon vectors containing non-antibiotic selection markers for cloning and stable chromosomal insertion of foreign DNA in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouchi H, Hata S. Isolation and characterization of novel cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- 22.Kouchi H, Takane K, So R, Ladha J, Reddy P. Rice ENOD40: isolation and expression analysis in rice and transgenic soybean root nodules. Plant J. 1999;18:121–129. doi: 10.1046/j.1365-313x.1999.00432.x. [DOI] [PubMed] [Google Scholar]
- 23.Leibovitch M P, Nguyen V C, Gross M S, Solhonne B, Leibovitch S A, Bernheim A. The human ASM (adult skeletal muscle) gene: expression and chromosomal assignment to 11p5*. Biochem Biophys Res Commun. 1991;180:1241–1250. doi: 10.1016/s0006-291x(05)81329-4. [DOI] [PubMed] [Google Scholar]
- 24.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 25.Lovett P S, Rogers E J. Ribosomal regulation by the nascent peptide. Microbiol Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathesius U, Schlaman H R M, Spaink H P, Weinman J J, Sautter C, Rolfe B G, Djordjevic M A. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 1998;14:23–34. doi: 10.1046/j.1365-313X.1998.00090.x. [DOI] [PubMed] [Google Scholar]
- 27.Matsubayashi Y, Sakagami Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA. 1996;93:7623–7627. doi: 10.1073/pnas.93.15.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minami E, Kouchi H, Cohn J R, Ogawa T, Stacey G. Expression of the early nodulin, ENOD40, in soybean roots in response to various lipo-chitin signal molecules. Plant J. 1996;10:23–32. doi: 10.1046/j.1365-313x.1996.10010023.x. [DOI] [PubMed] [Google Scholar]
- 29.Mize G J, Ruan H, Low J, Morris D. The inhibitory upstream open reading frame from mammalian S-adenosyl methionine decarboxylase mRNA has a strict sequence specificity in critical positions. J Biol Chem. 1998;273:32500–32505. doi: 10.1074/jbc.273.49.32500. [DOI] [PubMed] [Google Scholar]
- 30.Oleynikov Y, Singer R. RNA localization: different zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- 31.Olivas W M, Muhlrad D, Parker R. Analysis of the yeast genome: identification of new non-coding and small ORF-containing RNAs. Nucleic Acids Res. 1997;25:4619–4625. doi: 10.1093/nar/25.22.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearce G, Strydom D, Johnson S, Ryan C A. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253:895–898. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sautter C, Waldner H, Neuhaus-Url G, Galli A, Neuhaus G, Potrykus I. Micro-targeting: high efficiency gene transfer using a novel approach for the acceleration of micro-projectiles. Biotechnology. 1991;9:1080–1085. doi: 10.1038/nbt1991-1080. [DOI] [PubMed] [Google Scholar]
- 35.Schell J, Bisseling T, Dulz M, Franssen H, Fritze K, John M, Kleinow T, Lessnick A, Miklashevichs E, Pawlowski K, Rohrig H, van de Sande K, Schmidt J, Steinbiss H, Stoll M. Re-evaluation of phytohormone-independent division of tobacco protoplast-derived cells. Plant J. 1999;17:461–466. [Google Scholar]
- 36.Schlaman H R M, Gisel A A, Quaedvlieg N E M, Bloemberg G V, Lugtenberg B J J, Kijne J W, Potrykus I, Spaink H P, Sautter C. Chitin oligosaccharides can induce cortical cell division in roots of Vicia sativa when delivered by ballistic microtargeting. Development. 1997;124:4887–4895. doi: 10.1242/dev.124.23.4887. [DOI] [PubMed] [Google Scholar]
- 37.Schultze M, Kondorosi A. Regulation of symbiotic root nodule development. Annu Rev Genet. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- 38.Tam W, Ben-Yehuda D, Hayward W S. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol Cell Biol. 1997;17:1490–1502. doi: 10.1128/mcb.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenson T, DeBlasio A, Mankin A. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc Natl Acad Sci USA. 1996;93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teramoto H, Toyama T, Takeba G, Tsuji H. Noncoding RNA for CR20, a cytokinin-repressed gene of cucumber. Plant Mol Biol. 1996;32:797–808. doi: 10.1007/BF00020478. [DOI] [PubMed] [Google Scholar]
- 41.Torii K U, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier R F, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van de Sande K, Pawlowski K, Czaja I, Wieneke U, Schell J, Schmidt J, Walden R, Matvienko M, Wellink J, van Kammen A, Franssen H, Bisseling T. Modification of phytohormone response by a peptide encoded by ENOD40 of legumes and a nonlegume. Science. 1996;273:370–373. doi: 10.1126/science.273.5273.370. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Wessler S. Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell. 1998;10:1733–1745. doi: 10.1105/tpc.10.10.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamashita A, Watanabe Y, Nukina N, Yamamoto M. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell. 1998;95:115–123. doi: 10.1016/s0092-8674(00)81787-0. [DOI] [PubMed] [Google Scholar]
- 45.Yang W C, Katinakis P, Hendriks P, Smolders A, de Vries F, Spee J, van Kammen A, Bisseling T, Franssen H. Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 1993;3:573–585. doi: 10.1046/j.1365-313x.1993.03040573.x. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida H, Kumimoto H, Okamoto K. dutA RNA functions as an untranslatable RNA in the development of Dictyostelium discoideum. Nucleic Acids Res. 1994;22:41–46. doi: 10.1093/nar/22.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]