Abstract
Background:
Circular RNAs (CircRNAs) have been recently implicated in the progression of pancreatic cancer (PC).
Aims:
To investigate the involvement of CircCCT3 in PC and studying its interactions and functioning during the progression of PC in vitro and in vivo, using methods of molecular biology and bioinformatics.
Study Desig
n: Experimental study.
Methods:
The expressions of CircCCT3 and miR-613 in pancreatic carcinoma tissues and cell lines were evaluated by quantitative real-time polymerase chain reaction (PCR). The relationship between clinical pathologic features as well as the survival rate and CircCCT3 expression was analyzed with chi-square test and the Kaplan–Meier method. CCK-8, wound healing, transwell assays, and the fluorescein isothiocyanate-AnnexinV/propidium iodide (FITC-AnnexinV/PI) assay were used to assess cell proliferation, migration, invasion, and apoptosis after CircCCT3 overexpression or downregulation. The Dual-Luciferase reporter assay, RNA immunoprecipitation (RIP), RNA pull-down and fluorescence in situ hybridization (FISH) assays were performed to validate the potential interaction of CircCCT3, miR-613, and vascular endothelial growth factor (VEGFA). The nude mouse xenograft tumor assay was used to detect CircCCT3 effects on pancreatic tumorigenesis in vivo. Western blotting analysis was performed to examine the VEGFA and the vascular endothelial growth factor receptor 2 (VEGFR2) protein expressions following.
Results:
CircCCT3 expression was significantly increased in PC tissues (3.41 ± 0.57 vs. 1.00 ± 0.10, P < .01) and cell lines (Patu8988 2.57 ± 0.20; SW1990 2.88 ± 0.10; BxPC-3 2.45 ± 0.20; Panc02 2.99 ± 0.10 vs. H6c7 1.00 ± 0.10; all P < .001). CircCCT3 expression was negatively correlated with miR-613 expression. PC patients with high CircCCT3 expression exhibited significantly poorer overall survival rate than those patients with low CircCCT3 expression (P = .013). Moreover, it was found that CircCCT3 promoted cell proliferation, migration, and invasion, and inhibited cell apoptosis in PC cells. The CircCCT3 acted as a sponge for the miR-613 to facilitate VEGFA and VEGFR2 expression. si-CirCCT3 also inhibited tumor growth of PC in nude mice. si-CircCCT3 reduced VEGFA and VEGFR2 expression, whereas overexpression of CircCCT3 increased VEGFA and VEGFR2 expression.
Conclusion:
Increased CircCCT3 suggests a poor prognosis for PC patients and promotes the migration and invasion through targeting VEGFA/VEGFR2 signaling. CircCCT3 may serve as a potential and promising therapeutic target for PC treatment.
Keywords: CircCCT3, pancreatic cancer, miR-613, VEGFA/VEGFR2 signaling
Introduction
Pancreatic cancer (PC) is one of the most lethal malignancies and aggressive gastrointestinal tumors, remaining at the lowest 5-year relative survival rate of 9%.1 Since its aggressive nature, late diagnosis, and limitations of existing chemo and radiation therapies, PC has become the fourth leading cancer type in estimates of the cause of death, and has been predicted to become the second cause of cancer-related deaths in developed countries by 2030.2 Therefore, understanding the potential mechanism that promotes PC progression is vital for developing effective diagnostic techniques and therapeutic strategies.
Small non-coding RNAs including miRNA, lncRNA, and CircRNA, and others, have been the focus of many works in the recent 2 decades and their essential roles in various human diseases are already well identified.3 The research from these years has altered our perception of non-coding RNAs from “junk” transcriptional products to functional regulatory molecules that mediate cellular processes including chromatin remodeling, transcription, post-transcriptional modifications, and signal transduction.4 The non-coding RNAs networks could interact with different molecular targets to facilitate designated cell biological responses and fates.5 In particular, in the field of cancer, non-coding RNAs have been confirmed as both oncogenic drivers and (or) tumor suppressors in every major cancer type.6
Among non-coding RNAs, circular RNAs (CircRNAs) are defined as closed and single-stranded RNAs derived from back-spliced pre-mRNA7. Initially, CircRNAs, discovered 20 years ago, were believed to be by-products of aberrant splicing.8 Recent studies have shown that circRNAs exert various effects on normal metabolism and pathophysiological processes.9 Accruing evidence has shown that circRNAs function as modulators in cancer development, and they may be novel biomarkers and therapeutic targets.10 CircCCT3 (circBase ID: hsa_circ_0004680) is originated from exons 3-5 and introns 3-4 of chaperonin containing TCP1 subunit 3 (CCT3) by back-splicing,11 and is identified to be involved in the progression of hepatocellular carcinoma11 and colorectal cancer.12 So far, the role of CircCCT3 on PC progression has not been investigated.
The miRNAs are short non-coding RNAs that contain about 22 nucleotides.13 Emerging evidence has demonstrated that miRNAs are involved in cancer initiation and progression.13 Thus, miRNAs could be oncogenes or tumor suppressor genes mainly by regulating target genes that contribute to cellular processes such as cell cycle, mobility, and survival.14 miR-613 has been demonstrated to be downregulated in several types of cancers including renal cell carcinoma, glioma, breast cancer, and cervical cancer, implying that miR-613 may act as a tumor suppressor gene.15-18 However, its function and mechanism in PC remain unclear.
Vascular endothelial growth factor A (VEGFA) is one member of the VEGF family proteins.19 VEGFA is regarded as one of the most important in malignant diseases since it is the most predominant isoform secreted by human tumors20 including PC.21,22 Moreover, VEGFA/VEGFR2 signaling has been implicated in different cancer progression.23,24
Therefore, the scope of our study was to analyze the expression pattern of CircCCT3 in PC, explore the correlation of CircCCT3 with the PC phenotype, and investigate whether CircCCT3 leads to PC progression through targeting miR-613 and regulating VEGFA/VEGFR2.
Material and Methods
Tissue Specimens and Cell Culture
30 paired PC tissues and non-tumor adjacent tissues were collected from the Tianjin Baodi Hospital. Inclusion criteria: (1) There was no adjuvant therapy before surgery, including chemotherapy and radiotherapy. (2) PC pathology was confirmed by 2 experienced pathologists after surgery. (4) The patient could tolerate the operation, without evident heart, brain, or lung diseases. Exclusion criteria: (1) Preoperative chemotherapy, radiotherapy and other related adjuvant therapies. (2) Patients could not tolerate surgery. This investigation was approved by the Ethics Committee of our hospital. All patients agreed to the protocol and signed the informed consent. This investigation was conducted in accordance with the Declaration of Helsinki.
Human pancreatic duct epithelial cell (H6c7) and human PC cell lines (Patu8988, SW1990, BxPC-3, and Panc02) were obtained from ATCC (Manassas, VA). All cells were cultured in Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Invitrogen, Carlsbad, CA containing 10% fetal bovine serum (Gibco, Rockville, MD) and incubated in a humidified atmosphere with 5% CO2 at 37°C.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted by TRIzol (Invitrogen, Carlsbad) reagent, and cDNA was synthesized using a reverse transcription reaction kit (TaKaRa, Komatsu, Japan) following the manufacturer’s instructions. qRT-PCR was performed by SYBR Premix DimerEraser (TaKaRa) on a LightCycler 480 PCR System (Roche, Rotkreuz, Switzerland). The relative expression of RNAs was calculated by the 2−ΔΔCT method and normalized to U6 or glyceraldehyde phosphate dehydrogenase (GAPDH). Primers are listed as below: CircCCT3 Forward primer: GGACCCAGGATGAAGAGGTT; CircCCT3 Reverse primer: CATTGGGTCCAAAAGCATCT; miR-613 Forward primer: GTGAGTGCGTTTCCAAGTGT; miR-613 Reverse primer: GGGTCCCTTCACACTTGGAA; GAPDH: Forward primer: 5′-ACGCTGCATGTGTCCTTAG-3′; Reverse primer: 5′-GAGCCTCTTATAGCTGTTTG-3′; U6: Forward primer: 5′-CTCGCTTCGGCAGCACA-3′; Reverse primer: 5′-AACGCTTCACGAATTTGCGT-3′. Each sample for qRT-PCR was carried out 3 times and the average result was recorded.
CCK-8 Assay
For the CCK-8 assay, 2 × 105 cells in each well were inoculated in 96-well plates. The medium was used as blank control. At post-hypoxia reoxygenation, indicated cells in each well were added 10 μL of CCK-8 (Beyotime Institute of Biotechnology, Beijing, China) solution, and kept at 37°C for 2 hours. The OD value of 450 nm was detected. The final data were represented as cell viability.
Apoptosis Assay
For the apoptosis analysis, 1 × 106 cells were seeded in a 60 mm plate, treated with OE-CircCCT3 or CCT3 siRNA, and cultured for 24 hours. After harvesting, 1 × 105 cells were suspended in a 1x binding buffer provided with the Annexin V-FITC Apoptosis Detection kit II (BD Bioscience, San Jose, CA), then stained with FITC Annexin V (BD Bioscience) and PI (Sigma-Aldrich, St. Louis, MO). Fluorescence was detected with a BD Accuri C6 flow cytometer (BD Bioscience), and the data were analyzed with the BD Accuri C6 software (BD Bioscience).
Wound-Healing Assay
Cells were plated into 6-well plates and grown to nearly 90% confluence. The same size scratch was made through the cell monolayer using a 200 μL disposable pipette tip. After washing with PBS, fresh culture medium was added, and the cells were incubated at 37°C under an atmosphere with 5% CO2. Wound closure was imaged at 0 and 48 hours.
Bioinformatic Prediction
Circular RNA Interactome software (https://circinterac tome.nia.nih.gov) was used to predict CircCCT3-miRNA interactions, while the mRNA targets of miR-613 were predicted by TargetScan software (http://www.targetscan.org).
Luciferase Reporter Assay
First, a luciferase reporter vector (pGL3-Firefly_Luciferase-Renilla-Luciferase) with VEGFA 3’ UTR or CircCCT3 was built, and the mutant vectors were constructed by GeneChem (Shanghai, PR China). A luciferase vector and miR-613 mimic or mimic-negative control were co-transfected into cells and incubated for 24 hours. A Dual-Luciferase Reporter assay detection kit (Promega, WI) was used to detect firefly and Renilla luciferase activities, which were measured on a Fluoroskan Ascent device (Thermo Fisher Scientific, USA).
Fluorescence In Situ Hybridization
Cy3-CircCCT3 and fluorescein isothiocyanate (FITC)-miR-613 probes were synthesized and obtained from RiboBio (Guangzhou, PR China). Hybridization assays were performed using a FISH Detection Kit (RiboBio, PR China). All images were captured by a confocal microscope (FV1000; Olympus, Tokyo, Japan).
RIP Assay
A RIP kit (Millipore, Billerica, MA, USA) was purchased for performing the RIP assay. PC cells were collected, and lysed by RIP lysis buffer. Then, the cell lysates were incubated with anti-argonaute-2 (Ago2) or immunoglobulin G (IgG; negative control) antibody in RIP buffer at 4°C, overnight. Co-precipitated RNA was isolated after digestion with 150 μL protease K. The expression of CircCCT3 and miR-613 was detected.
Pull-down Assay with Biotinylated CircCCT3 and miR-613
Probe Panc02 cells (1 × 107) were obtained, lysed, and sonicated as indicated. To generate the probe-coated beads, C-1 magnetic beads (Life Technologies) were co-incubated with the CircCCT3 probe for 2.5 hours at 25°C. Then, the CircCCT3 probe or oligo probe was co-incubated with cell lysates at 4° overnight. RNA was eluted and extracted by wash buffer and used for qRT-PCR. The CircCCT3 probe with biotinylation was synthesized and purchased from RiboBio (Guangzhou, PR China).The miR-613 was also tested using the same procedure.
Western Blotting
Cells were lysed with precooled radio-immunoprecipitation assay lysis buffer supplemented with protease inhibitor (Beyotime Institute of Biotechnology, Shanghai, China) for 30 minutes on ice. The supernatant was collected after centrifugation at 14 000 rpm at 4°C for 20 minutes. The protein concentration was determined using a bicinchoninic acid (BCA) protein concentration determination kit (RTP7102; Real-Times Biotechnology Co. Ltd., Beijing, China). The samples (20 μg) were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membranes. A GAPDH antibody was used as an internal reference. The membranes were washed with TBST and incubated with goat anti-mouse/rabbit antibody. Color development was performed using the chemiluminescence detection method, and images of protein bands were obtained for analyses. The primary and secondary antibodies were as follows: anti-GAPDH monoclonal antibody (Cell Signaling Technology, Danvers, MA,USA; 5174S, diluted 1/1000), anti-VEGFA (Abcam, Cambridge, UK; ab214424 diluted 1/1000), anti-VEGFR2 (Abcam; ab134191, diluted 1/1000), goat anti-mouse IgG (ProteinTech; SA00001-1, diluted 1/3000), and goat anti-rabbit IgG (ProteinTech; SA00001-2, diluted 1/3000). Each western blot was repeated at least 3 times.
Xenograft Tumor Model
Animal experiments were conducted with the permission of the animal care and use committee of the local hospital and performed in accordance with the guidelines of the National Animal Care and Ethics Institution. The Panc02 cell line stably transfected with sh-CircCCT3 or sh-NC was established. Afterwards, the Panc02 cells stably transfected with sh-CircCCT3 or sh-NC were subcutaneously inoculated into the right flank of BALB/c male nude mice (Model Animal Research Center Of Nanjing University, Nanjing, China) in the sh-NC group and sh-CircCCT3 group (n = 6 for each group). The volume of tumors was recorded every week. After inoculation for 28 days, these nude mice were euthanized and the tumors were dissected and weighed. The expression of CircCCT3 was detected by qRT-PCR.
Statistical Analysis
The normality distribution of the numeric parameters was tested by the Kolmogorov–Smirnov test. The data were normally distributed after the test, and all data were described as mean ± SD and analyzed with GraphPad Prism 7.0 software (GraphPad Prism, San Diego, CA). One-way analysis of vairance with a Bonferroni correction or the Student’s t-test was used to analyze the differences between groups. The association between CircCCT3 expression and the clinicopathological features was evaluated using chi-square test. Survival data were analyzed using the Kaplan–Meier method and the log-rank test. The univariate and multivariate analyses were analyzed using Cox proportional hazards models. The correlation of CircCCT3 expression with miR-613 expression was carried out using Spearman’s method. P < .05 was regarded as statistical significance.
Results
CircCCT3 Expression is Increased in Clinical PC Tissues
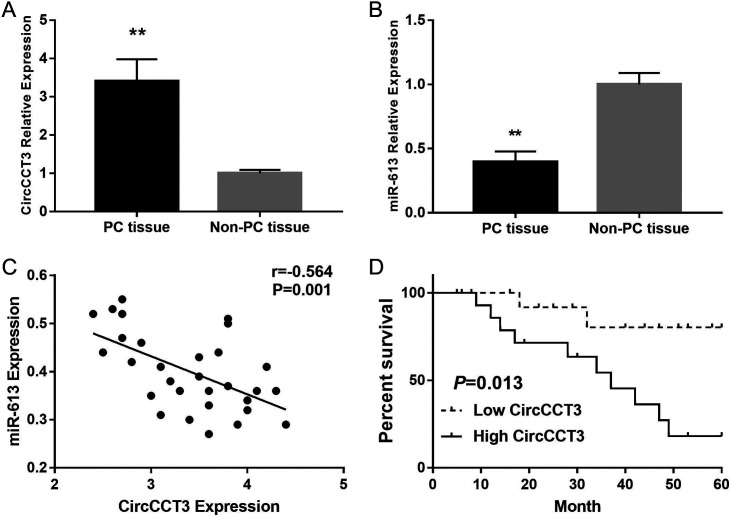
To investigate whether CircCCT3 played a role on PC metastasis and progression, the expression level of CircCCT3 was tested in PC and non-PC tissue. We observed that CircCCT3 levels in human PC tumors were significantly higher compared with adjacent non-PC tissues (P < .001) (Figure 1A), whereas miR-613 expression in PC tissue was markedly decreased compared to adjacent non-PC tissue. The higher levels of CircCCT3 in PC patients correlated with vascular invasion (P = .011), peritoneal metastasis (P = .013), lymph node metastasis (P = .020), and clinical progression (P = .009) (Table 1). Further, the Cox multivariate survival analysis revealed that high CircCCT3 expression was an independent prognostic factor for poor survival of PC patients (hazard ratio [HR] = 2.94, 95% confidence interval [CI] 1.85-6.16, P = .007) (Table 2). In addition, CircCCT3 expression was negatively correlated with miR-613 expression in PC tissue. PC patients with high CircCCT3 expression (n = 15) exhibited significantly poorer overall survival rate than those patients with low CircCCT3 expression (n = 15), as defined by the log-rank test (P = .013) (Figure 1 D).These results implicated that CircCCT3 may serve as an oncogene in PC.
FIG. 1.
(A-D). Data are shown as mean ± SD. (A). Expression levels of CircCCT3 in PC samples and adjacent non-PC tissue. (B). Expression levels of miR-613 in PC samples and adjacent non-PC tissue. (C) Correlation of CircCCT3 expression with miR-613 expression in PC. (D) Pancreatic carcinoma patients with high CircCCT3 expression (n = 15) exhibited significantly poorer overall survival rate than those patients with low CircCCT3 expression (n = 15), as defined by log-rank test (P = .013).
Table 1.
CircCCT3 Expression and its Correlation With Progression of PC
| Characteristics | Case (No.) | Expression of CircCCT3 | P | |
|---|---|---|---|---|
| High | Low | |||
| Age | ||||
| <50 | 8 | 5 | 3 | .409 |
| ≥50 | 22 | 10 | 12 | |
| Gender | ||||
| Male | 16 | 9 | 7 | .538 |
| Female | 14 | 6 | 8 | |
| Location | ||||
| Head | 11 | 6 | 5 | .705 |
| Body/tail | 19 | 9 | 10 | |
| Tumor size (cm) | ||||
| <4 | 20 | 12 | 8 | .245 |
| ≥4 | 10 | 3 | 7 | |
| Tumor differentiation | ||||
| High/medium | 12 | 7 | 5 | .176 |
| Low | 18 | 8 | 10 | |
| Vascular invasion | ||||
| Absent | 15 | 11 | 4 | .011* |
| Present | 15 | 4 | 11 | |
| Peritoneal metastasis | ||||
| No | 22 | 8 | 14 | .013* |
| Yes | 8 | 7 | 1 | |
| Lymph node metastasis | ||||
| Absent | 10 | 2 | 8 | .020* |
| Present | 20 | 13 | 7 | |
| Clinical stage | ||||
| I/II | 13 | 3 | 10 | .009** |
| III/IV | 17 | 12 | 5 | |
** P < .01, * P < .05.
Table 2.
Univariate and Multivariate Overall Survival Analysis of Prognostic Factors for PC Patients (n = 60)
| Clinicopathologic Parameters | Overall Survival | |||||
|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||||
| HR | 95% CI | P | HR | 95% CI | P | |
| Age (<50 years vs. ≥50 years) | 0.91 | 0.66-2.31 | .567 | |||
| Gender (male vs. female) | 1.08 | 0.75-2.04 | .622 | |||
| Tumor size (cm) (<4 vs. ≥4) | 1.13 | 0.49-1.87 | .255 | |||
| Location (head vs. body/tail) | 2.02 | 0.83-4.01 | .119 | |||
| Tumor differentiation (high/medium vs. low) | 2.25 | 0.97-4.33 | .085 | |||
| Vascular invasion (absent vs. present) | 3.14 | 1.98-5.88 | .011* | 1.47 | 0.68-2.83 | .167 |
| Peritoneal metastasis (yes vs. no) | 3.47 | 1.78-6.92 | .025* | 1.98 | 0.86-3.99 | .130 |
| Lymph node metastasis (absent vs. present) | 3.78 | 1.83-6.88 | .021* | 2.49 | 1.12-4.76 | .041* |
| Clinical stage (I/II vs. III/IV) | 5.01 | 2.08-8.45 | <.001** | 3.41 | 1.36-8.66 | .003** |
| CircCCT3 expression (low vs. high)* | 4.36 | 1.95-7.77 | <.001*** | 2.94 | 1.85-6.16 | .007** |
* P < .05; ** P < .01; *** P < .001 using median CircCCT3 values as cutoff.
CircCCT3 Expression was Upregulated in the PC Cell Line
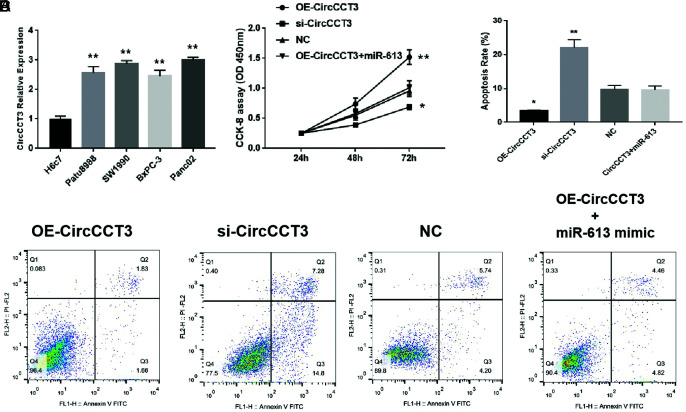
The CircCCT3 levels in different cell lines were then tested, and we found that CircCCT3 expressions in the normal cell line H6c7 were significantly lower than in PC tumor cell lines (Patu8988, SW1990, BxPC-3 and Panc02) (**P<.01) (Figure 1A). To explore the effects of CircCCT3 on the biological function of Panc02 cells, we first explored cell proliferation via cell counting Kit-8 (CCK-8) assays and apoptosis through the FITC-AnnexinV/PI assay. These results showed that overexpression of CircCCT3 increased cell proliferation of Panc02 cells compared with NC (P < .01), while silencing CircCCT3-suppressed cell proliferation (P < .05) (Figure 2B). In contrast, the OE-CircCCT3 adding miR-613 mimic reversed the effect of the OE-CCT3 by decreasing cell growth (Figure 2B).We also performed the FITC-AnnexinV/PI assay to evaluate the potential function of CircCCT3 on cell apoptosis. We found that the cell apoptosis rate was significantly decreased in the OE-CircCCT3 group (P < .05), while in the si-CircCCT3 group, the cell apoptosis rate was increased compared with the negative control group (P < .01). OE-CircCCT3 adding miR-613 had a similar apoptosis rate in comparison with the negative control group.
FIG. 2.
(A-D). Data are shown as mean ± SD. (A) Comparison of CircCCT3 levels between the normal cell line H6c7 and different tumor cell lines. (B) Proliferation rate following OE-CircCCT3 or si-CircCCT3 transfection. (C) Comparison of apoptosis rate following OE-CircCCT3 or si-CircCCT3 transfection. (D) Representative figures of cell apoptosis following OE-CircCCT3 or si-CircCCT3 transfection.
CircCCT3 Promotes the Migration and Invasion of PC Both In Vitro and In Vivo
In order to evaluate the potential role of CircCCT3 on the metastasis of PC, OE-CircCCT3 or CircCCT3 siRNA, and CircCCT3 siRNA+miR-613 were introduced into Panc02 cells.
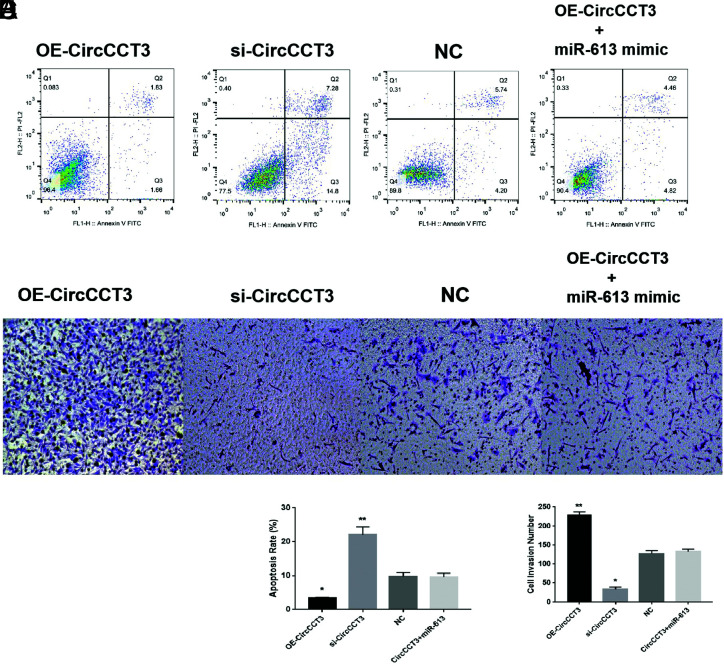
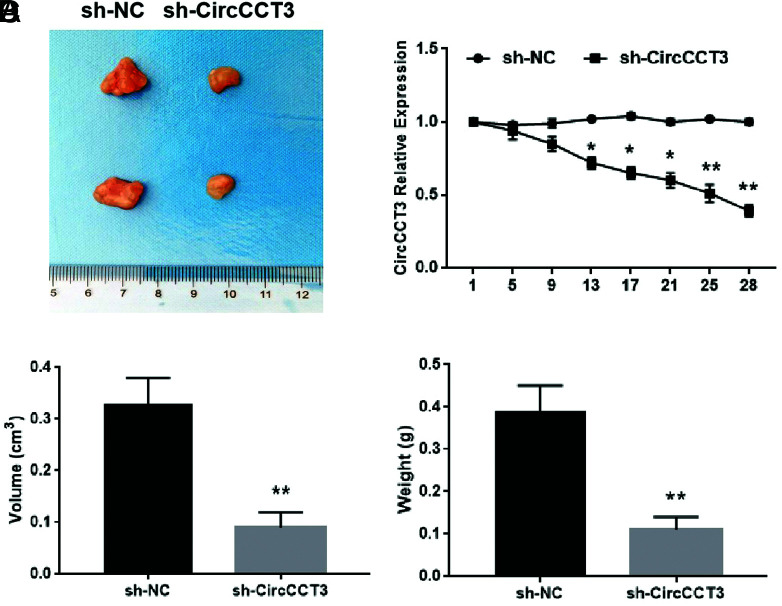
Subsequently, the wound-healing assay was conducted, and data showed that overexpression of CircCCT3 significantly increased Panc02 cell migration in vitro (Figure 2A and B, P < .05). On the other hand, the migratory ability of Panc02 cells was significantly inhibited by CircCCT3 siRNA (Figure 3A and B P < .05). Co-transfection of OE-CircCCT3 and the miR-613 mimic exhibited the similar effects compared with NC (Figure 3A and B). In addition, the percentages of invasion of CircCCT3-overexpressing Panc02 cells were significantly lower than those of negative control cells, whereas si-CircCCT3-transfecting Panc02 had markedly decreased invasion numbers compared to negative control cells (Figure 3C and D). Co-transfection of OE-CircCCT3 and miR-613 mimic exhibited similar effects compared with the negative control (Figure 3C and D). In short, these results demonstrated that CircCCT3 overexpression promotes the migratory and invasion capability of PC cells, and these effects could be reversed by adding the miR-613 mimic. Finally, in the xenograft tumor formation assay, we found that sh-CirCCT3 inhibited tumor growth of PC compared with control at 4 weeks (Figure 4A and B). The tumor volume and tumor weight in the sh-CirCCT3 group were both significantly lower than in the sh-CircNC group (Figure 4C and D).
FIG. 3.
(A-D). Data are shown as mean ± SD. (A and B) Wound-healing assay for Panc02 cell migration after 48h following OE-CirCCT3 or OE- CirCCT3 (*P < .05,**P < .01). (C and D) Transwell Invasion assay for Panc02 cells invasion after 48 hours using in Panc02 cells following OE-CirCCT3 or OE- CirCCT3 (*P < .05,**P < .01 vs. NC).
FIG. 4.
(A-D). Data are shown as mean ± SD. sh-CirCCT3 suppressed tumor growth of PC. (A) Representative photographs of PC tumor tissue in sh-CirCCT3 and sh-CircNC groups. (B) Expressions of CircCCT3 during the tumor growth between si-CirCCT3 and si-CircNC groups. (C) Comparison of tumor volume between the sh-CircCCT3 group and the sh-CircNC group at day 24 (**P < .01) (n = 6 for each group). (D) Comparison of tumor weight between the sh-CircCCT3 group and the sh-CircNC group tumor volume at day 24 (**P < .01) (n = 6 for each group).
CircCCT3 Abolishes the Interaction of miR-613 with the Targets of VEGFA
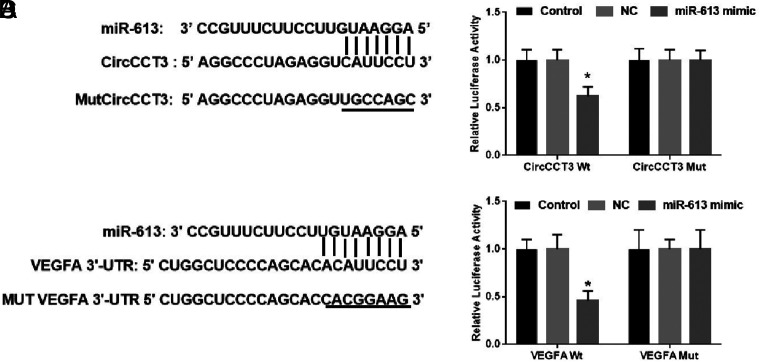
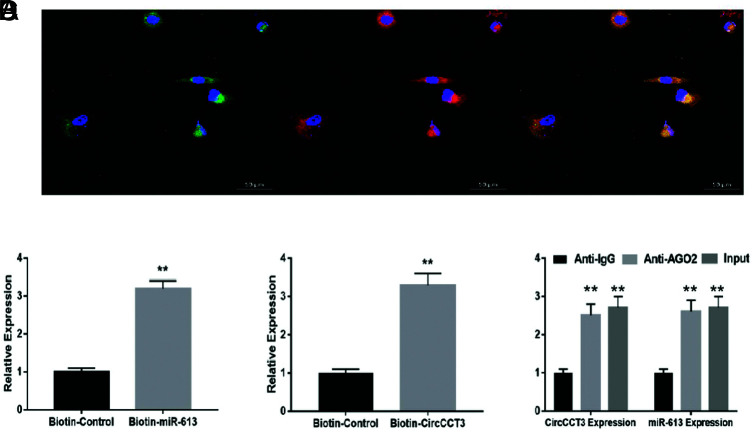
The predicted complementary binding sites at the 3’-UTR are shown in Figure 5A. The bioinformatics analysis also showed that the 3’-UTR of VEGFA were potential target genes of miR-613 (Figure 5A). As shown next, the luciferase assay confirmed that CircCCT3 was also a target of miR-613 (Figure 5C). Luciferase activity assay showed that the miR-613 mimic led to a notable decrease in luciferase activity in the CircCCT3-WT reporter, compared with the mimic-negative control group, whereas the miR-613 mimic had no obvious effect on luciferase activity in the CircCCT3-MUT reporter (Figure 5B). Similarly, the miR-613 mimic contributed to the significant reduction of luciferase activity in the VEGFA-WT reporter in comparison with the mimic-negative control group, whereas the miR-613 mimic had no obvious effect on luciferase activity in the VEGFA-MUT reporter (Figure 5D). In order to further identify the interaction between CircCCT3 and miR-613 in PC cells, we next carried out FISH, RNA pull-down, and RIP assays. FISH technology demonstrated that CircCCT3 (red fluorescence) and miR-613 (green fluorescence) could be visualized in both Panc02 and BxPC3 cells, and colocalization was also observed (Figure 6A). The RIP assays disclosed that CircCCT3 and miR-613 expressions were substantially enriched by Ago2 antibody compared with control IgG antibody (Figure 6B). The biotin-coupled probe pull-down assay was then performed and the results showed miR-613 detected in the CircCCT3 pulled-down pellet compared with the control group (Figure 6C). Also, CircCCT3 was detected in the miR-613 pulled-down pellet compared with the control group (Figure 6D).
FIG. 5.
(A-D). (A) Bioinformatics analysis of matching sequence of miR-613 within CircCCT3. MUT CircCCT3 is the mutation of the match sequence of CircCCT3 with miR-613. (B) The luciferase reporter assay revealed that miR-613 binds to the CircCCT3, not MUT CircCCT3. (C). Bioinformatics analysis of matching sequence of miR-613 within 3'-UTR of VEGFA. MUT VEGFA 3'-UTR is the mutation of the match sequence of 3'-UTR of VEGFA with miR-613. (D) The luciferase reporter assay revealed that miR-613 binds to the 3'-UTR of WT VEGFA, not MUT VEGFA. Relative luciferase activity was quantified and the data were demonstrated as mean ± SD. *P < .05 vs. respective NC groups; NC, negative control; WT, wild-type; MUT, mutant.
FIG. 6.
(A-D). Data are shown as mean ± SD. (A) CircCCT3 and miR-613 were colocalized in Panc02 cells by FISH using confocal microscope. CircCCT3 was stained red, miR-613 was stained green, nuclei were stained blue (DAPI) and overlapped expression was mixed (Scale bar, 20 μm). (B) The biotin-coupled probe pull-down assay was performed and the results showed that miR-613 was detected in the CircCCT3 pulled-down pellet compared with the control group. (C) CircCCT3 was detected in the biotin-miR-613 vector compared with the control group. (D) Relative CircCCT3 and miR-613 expressions presented as fold enrichment in Ago2 relative to normal IgG immunoprecipitates. RIP assays disclosed that CircCCT3 and miR-613 expressions were substantially enriched by Ago2 antibody compared with control IgG antibody (P < .01vs. control).
CircCCT3 Activated VEGFA/VEGFR2 Signaling to Facilitate PC Progression
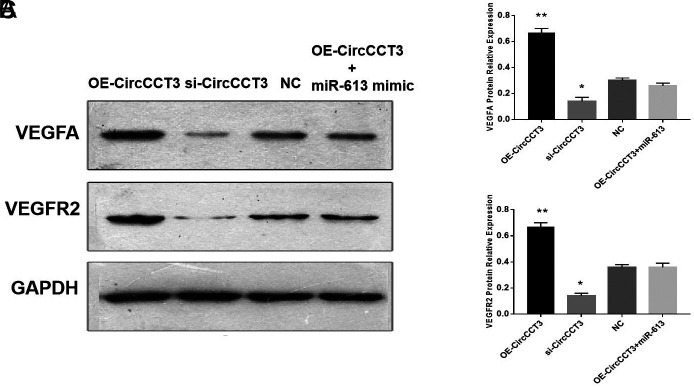
To explore the role of CircCCT3 on the VEGFA/VEGFR2 axis, the western blot analysis was performed, demonstrating that VEGFA and VEGFR2 expressions were both increased by transfecting OE-CircCCT3 compared with negative control, whereas these proteins were significantly downregulated by transfecting si-CircCCT3 in Panc02 cells (Figure 7. A-C.). In addition, VEGFA and VEGFR2 expressions in co-transfection of the OE-CircCCT3 and miR-613 mimic groups were not significantly different from the negative control groups. This finding suggested that CircCCT3 could regulate the VEGFA/VEGFR2 axis in PC.
FIG. 7.
(A-C). Data are shown as mean ± SD. VEFGA and VEGFR2 protein expressions detected through western blot analysis. (A) Representative bands of VEGFA and VEGFR2. (B) Relative expressions of VEGFA proteins following OE-CircCCT3 or si-CircCCT3 transfection. (C) Relative expressions of VEGFR2 proteins following OE-CircCCT3 or si-CircCCT3 transfection.
Discussion
The current study investigated the potential role of CircCCT3 in PC development. We demonstrated for the first time that CircCCT3 was upregulated in PC and predicted poor prognosis in PC patients. PC patients with high CircCCT3 expression exhibited significantly poorer overall survival rate than those patients with low CircCCT3 expression. Functionally, we found that CircCCT3 knockdown markedly inhibited PC cell proliferation, migration, and invasion in vitro and decreased tumor growth in vivo. Using bioinformatics prediction, luciferase activity assay, RIP, RNA pull-down, and FISH, we discovered that miR-613 binds with CircCCT3 in PC cells. Finally, we identified CircCCT3 as an miR-613 sponge that upregulates the expression of VEGFA and VEGFR2 to promote PC progression.
Accumulating evidence suggests that CircRNAs may be used as tumor biomarkers and regulators because of their crucial involvement in various biological processes. Mechanistically, most identified circRNAs are mainly localized in the cytoplasm of the cell,25 indicative of their roles in post-transcriptional regulation.26 The competing endogenous RNA (ceRNA) hypothesis indicates that circRNAs harbor microRNA response elements that bind miRNAs to reverse-regulate the activity of the miRNAs,27 thus attenuating the inhibitory effect on their target molecules. Mounting evidence has confirmed that some circRNAs can repress miRNA function and modulate target gene expression to play a tumor suppressor or oncogenic role by acting as miRNA sponges in different cancers including PC.28 For example, CircRNA_000864 represses migration and invasion in PC cells by targeting miR-361-3p.29 Hsa_circ_0000069 knockdown could suppress tumorigenesis and malignant transformation via inhibition of STIL in PC.30 hsa_circRNA_001587 upregulates SLC4A4 expression to inhibit migration, invasion, and angiogenesis of PC cells via binding to microRNA-223.31 circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in PC.32
Angiogenesis is the formation of new vessels from existing capillaries, which is also an especially important process for tumor development and metastasis. Among many factors involved in the regulation of complex angiogenesis, VEGFA is a predominant stimulant.33
The VEGFA/VEGFR2 axis serves as crucial signaling in angiogenesis and metastasis of tumor progression.34 Continuous VEGF expression through VEGFR2 leads to the development and maintenance of a vascular network that promotes tumor growth and metastasis.34 Drugs targeting the VEGF pathway have been studied, as single agents or in combination with chemotherapy, to suppress the VEGF signaling pathway.35 In our study, we found that higher levels of CircCCT3 are correlated with vascular invasion, implicating CircCCT3 involvement in the pathological process of angiogenesis in PC. We also discovered that overexpression of CircCCT3 increased VEGF and VEFGR2 protein expressions, suggesting CircCCT3 could promote VEGF/VEFGR2 signaling to facilitate angiogenesis in PC. One previous study has demonstrated that CircCCT3 could facilitate the progression of hepatocellular carcinoma by targeting miR-1287-5p,36 indicating that CircCCT3 maybe also deregulated in other gastrointestinal cancers. Further investigations of CircCCT3 expression in the colon cancer, gastric cancer and are worthy of study.
Our study has limitations. First, only 30 pairs of PC tissues were analyzed in this study due to the limited number of available PC samples. Second, all the patient samples are collected from only 1 Chinese hospital. A larger number of PC samples should be tested at multiple centers and from other countries to further confirm the conclusions of this study. Third, we look forward to further research to explore why and how CircCCT3 is upregulated in PC, which will provide a deeper understanding of the molecular mechanism. Finally, we did not consider the presence of hereditary PC (e.g. BRCA1, BRCA2, or PALLD mutations) or family history when enrolling PC patients.
In conclusion, our study first revealed the novel function of CircCCT3 in PC progression. We illustrated that CircCCT3-mediated miR-613 and VEGFA/VEGFR2 signaling promote PC development and progression, providing a novel therapeutic target for PC therapy.
Footnotes
Ethics Committee Approval: Ethics committee approval was received for this study from the Ethical Committee of Tianjin Baodi Hospital (Ethical Number: 20200012).
Patient Consent for Publication: Written informed consent was obtained from all patients.
Data-sharing Statement: The data that support the findings of this study are available from the corresponding author upon reasonable request.
Preprint History: Previously posted to Research Square as a preprint on March 4, 2021 (DOI: 10.21203/rs.3.rs-268983/v1).
Author Contributions: Concept - L-P.H.; Design - L-P.H.; Supervision - L-P.H., L-B.L.; Materials - L-Y.Y.; Data Collection and/or Processing - X-B.M.; Analysis and/or Interpretation - E-K.H., C-Q.H.; Literature Review - L-B.L.; Writing - X-B.M., C-Q.H., L-B.L.
Conflict of interest: The authors have no conflicts of interest to declare.
Funding: The authors declared that this study had received no financial support.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.. 10.3322/caac.21551) [DOI] [PubMed] [Google Scholar]
- 2. Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921.. 10.1158/0008-5472.CAN-14-0155) [DOI] [PubMed] [Google Scholar]
- 3. Clerget G, Abel Y, Rederstorff M. Small non-coding RNAs: a quick look in the rearview mirror.Methods Mol Biol. 2015;1296:3–9.. 10.1007/978-1-4939-2547-6_1) [DOI] [PubMed] [Google Scholar]
- 4. Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review) [review]. Oncol Rep. 2017;37(1):3–9.. 10.3892/or.2016.5236) [DOI] [PubMed] [Google Scholar]
- 5. Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer.Biochim Biophys Acta Rev Cancer. 2021;1875(1):188491. 10.1016/j.bbcan.2020.188491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38(5):485–491.. 10.1093/carcin/bgx026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen LL, Yang L. Regulation of circRNA biogenesis.RNA Biol. 2015;12(4):381–388.. 10.1080/15476286.2015.1020271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. 10.1186/s12943-017-0663-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases .Oncotarget. 2017;8(42):73271–73281.. 10.18632/oncotarget.19154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer.J Cancer. 2019;10(21):5015–5021.. 10.7150/jca.30828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lv B, Zhu W, Feng C. Coptisine blocks secretion of exosomal CircCCT3 from cancer-associated fibroblasts to reprogram glucose metabolism in hepatocellular carcinoma.DNA Cell Biol. 2020. 10.1089/dna.2020.6058) [DOI] [PubMed] [Google Scholar]
- 12. Li W, Xu Y, Wang X, et al. CircCCT3 modulates vascular endothelial growth factor A and Wnt signaling to enhance colorectal cancer metastasis Through sponging miR-613. DNA Cell Biol. 2020;39(1):118–125.. 10.1089/dna.2019.5139) [DOI] [PubMed] [Google Scholar]
- 13. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222.. 10.1038/nrd.2016.246) [DOI] [PubMed] [Google Scholar]
- 14. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20(1):5–20.. 10.1038/s41580-018-0059-1) [DOI] [PubMed] [Google Scholar]
- 15. Song H, Nan Y, Wang X, et al. MicroRNA-613 inhibits proliferation and invasion of renal cell carcinoma cells through targeting FZD7. Mol Med Rep. 2017;16(4):4279–4286.. 10.3892/mmr.2017.7076) [DOI] [PubMed] [Google Scholar]
- 16. Yu X, Wang W. Tumor suppressor microRNA-613 inhibits glioma cell proliferation, invasion and angiogenesis by targeting vascular endothelial growth factor A. Mol Med Rep. 2017;16(5):6729–6735.. 10.3892/mmr.2017.7422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu C, Jiang Y, Han B. miR-613 suppresses chemoresistance and stemness in triple-negative breast cancer by targeting FAM83A.Cancer Manag Res. 2020;12:12623–12633.. 10.2147/CMAR.S276316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ji H, Hu NJ. MiR-613 blocked the progression of cervical cancer by targeting LETM1. Eur Rev Med Pharmacol Sci. 2020;24(12):6576–6582.. 10.26355/eurrev_202006_21642) [DOI] [PubMed] [Google Scholar]
- 19. Sargent KM, Clopton DT, Lu N, Pohlmeier WE, Cupp AS. VEGFA splicing: divergent isoforms regulate spermatogonial stem cell maintenance. Cell Tissue Res. 2016;363(1):31–45.. 10.1007/s00441-015-2297-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reinmuth N, Parikh AA, Ahmad SA, et al. Biology of angiogenesis in tumors of the gastrointestinal tract. Microsc Res Tech. 2003;60(2):199–207.. 10.1002/jemt.10258) [DOI] [PubMed] [Google Scholar]
- 21. Itakura J, Ishiwata T, Shen B, Kornmann M, Korc M. Concomitant over-expression of vascular endothelial growth factor and its receptors in pancreatic cancer. Int J Cancer. 2000;85(1):27–34.. [DOI] [PubMed] [Google Scholar]
- 22. Seo Y, Baba H, Fukuda T, Takashima M, Sugimachi K. High expression of vascular endothelial growth factor is associated with liver metastasis and a poor prognosis for patients with ductal pancreatic adenocarcinoma. Cancer. 2000;88(10):2239–2245.. [DOI] [PubMed] [Google Scholar]
- 23. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317.. 10.1016/j.canlet.2017.06.027) [DOI] [PubMed] [Google Scholar]
- 24. Zhang Q, Lu S, Li T. et al. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J Exp Clin Cancer Res. 2019;38(1):173. 10.1186/s13046-019-1156-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9(3):e1471. 10.1002/wrna.1471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLOS ONE. 2012;7(2):e30733. 10.1371/journal.pone.0030733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58. 10.1186/s12943-017-0630-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rong Z, Xu J, Shi S, et al. Circular RNA in pancreatic cancer: a novel avenue for the roles of diagnosis and treatment. Theranostics. 2021;11(6):2755–2769.. 10.7150/thno.56174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang L, Han J, Yu H, et al. CircRNA_000864 Upregulates B-cell Translocation Gene 2 Expression and Represses Migration and Invasion in Pancreatic Cancer Cells by Binding to miR-361-3p. Front Oncol. 2020;10:547942. 10.3389/fonc.2020.547942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ye Z, Zhu Z, Xie J, et al. Hsa_circ_0000069 Knockdown Inhibits tumorigenesis and exosomes with Downregulated hsa_circ_0000069 Suppress Malignant Transformation via Inhibition of STIL in Pancreatic Cancer. Int J Nanomedicine. 2020;15:9859–9873.. 10.2147/IJN.S279258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X, Tan P, Zhuang Y, Du L. hsa_circRNA_001587 upregulates SLC4A4 expression to inhibit migration, invasion, and angiogenesis of pancreatic cancer cells via binding to microRNA-223. Am J Physiol Gastrointest Liver Physiol. 2020;319(6):G703–G717.. 10.1152/ajpgi.00118.2020) [DOI] [PubMed] [Google Scholar]
- 32. Kong Y, Li Y, Luo Y, et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol Cancer. 2020;19(1):82. 10.1186/s12943-020-01205-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Claesson-Welsh L, Welsh M.VEGFA and tumour angiogenesis. J Intern Med. 2013;273(2):114–127.. 10.1111/joim.12019) [DOI] [PubMed] [Google Scholar]
- 34. Wang Y, Zhang F, Wang J, et al. lncRNA LOC100132354 promotes angiogenesis through VEGFA/VEGFR2 signaling pathway in lung adenocarcinoma. Cancer Manag Res. 2018;10:4257–4266.. 10.2147/CMAR.S177327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mastrella G, Hou M, Li M, et al. Targeting APLN/APLNR improves antiangiogenic efficiency and blunts proinvasive side effects of VEGFA/VEGFR2 blockade in glioblastoma. Cancer Res. 2019;79(9):2298–2313.. 10.1158/0008-5472.CAN-18-0881) [DOI] [PubMed] [Google Scholar]
- 35. Fontanella C, Ongaro E, Bolzonello S, et al. Clinical advances in the development of novel VEGFR2 inhibitors. Ann Transl Med. 2014;2(12):123. 10.3978/j.issn.2305-5839.2014.08.14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin W, Zhang T, Ding G, et al. Circular RNA circCCT3 promotes hepatocellular carcinoma progression by regulating the miR12875p/TEAD1/PTCH1/LOX axis. Mol Med Rep. 2021;23(5):375. 10.3892/mmr.2021.12014) [DOI] [PMC free article] [PubMed] [Google Scholar]



 Content of this journal is licensed under a
Content of this journal is licensed under a