Abstract
Background
Neurofibromatosis type 1 (NF1) is a neurocutaneous disorder that results in a predisposition to the growth of multiple tumors in the central nervous system, the peripheral nervous system, and the skin. The clinical manifestations of neurofibromatosis are associated with loss of neurofibromin expression which causes the upregulation of the RAS pathway. Although neurofibromatosis type 1 can be diagnosed based on the National Institutes of Health criteria, sometimes the diagnosis is difficult, in cases where the characteristic features do not develop. Moreover, other RAS-related disorders may present with significantly overlapping clinical features.
Aims
To determine the clinical and molecular genetic characteristics of Turkish patients with neurofibromatosis type 1.
Study design
Cross-sectional study.
Methods
For the genetic analysis of 27 Turkish families clinically diagnosed with NF1 between 1990 and 2019, we used a multi-step process consisting of next-generation sequencing, multiplex ligation-dependent probe amplification, and array-comparative genomic hybridization.
Results
In this study, we identified 11 novel and 11 previously reported single-nucleotide variants in 22 families. Whole gene deletions were detected by multiplex ligation-dependent probe amplification analysis in 3 families. Of those, array comparative genomic hybridization analysis defined a 17q11.2 deletion in 4 patients from 2 families and 1.2-Mb involving 1 unrelated patient. All patients with a deletion had facial dysmorphism, suggesting a peculiar phenotype in this group. We could not find any pathogenic variant in the 2 families that met the National Institutes of Health criteria.
Conclusion
The novel pathogenic variants identified in this study broaden the spectrum of pathogenic variants in NF1 and provide better clinical characterization of NF1. RNA-seq experiments are recommended in patients who meet the National Institutes of Health diagnostic criteria for NF but have not identified any variants in next-generation sequencing, multiplex ligation-dependent probe amplification, or array-comparative genomic hybridization analysis.
Keywords: NF1 gene, genetic diagnosis, novel pathogenic variant, NF1 microdeletion syndrome
Introduction
Neurofibromatosis type 1 (NF1) is a neurocutaneous disorder that results in a predisposition to the growth of multiple tumors in the CNS, including the brain, spinal cord, the peripheral nervous system, and the skin.1,2 It is related to heterozygous germline pathogenic variants in the NF1 gene which causes activation of the RAS/MAPK pathway (mitogen-activated protein kinases), predisposing patients to developing multiple tumor types and other organ system anomalies.3 The pathogenic variant can affect cognition and behavior, causing anxiety and attention deficit disorders as well as learning disabilities. It can lead to café-au-lait spots, which are brown birthmarks on the skin. Axillary and inguinal freckling is a very characteristic pattern and involves the armpits and groins. Lisch nodules are small benign hamartomas of the iris. Scoliosis, sphenoid wing dysplasia, and tibial bowing are the most common skeletal manifestations of this disorder.4 Other manifestations, such as vascular abnormalities, vitamin D deficiency, and growth delay are less common.5 The prevalence of the disease is estimated to be about 1 in 3000.3 It is one of the most common genetic disorders and there is no predilection for any specific geographic location or ethnic or cultural group. Although NF1 is usually diagnosed based on diagnostic criteria, clinical diagnosis may be challenging in individuals with atypical findings or in children at an early age, in whom the characteristic features have not developed fully yet.6 In addition, members of RAS-related disorders may present with overlapping clinical findings, which can make diagnosis difficult, especially in the prenatal period.7,8 Therefore, due to the complexity of clinical diagnosis, understanding the mechanism of molecular pathogenesis in NF1 would be useful to characterize this disorder. The pathogenic variants identified in NF1 mostly lead to decreased protein levels or a truncated protein.1 Other pathogenic variants include single or multiple exon deletions/duplications and larger microdeletions involving NF1 and neighboring genes, which affect 4% of the patients.2 Therefore, we suggest that a multi-stage variant detection procedure that involves target sequencing of NF1 with next-generation sequencing (NGS), multiplex ligation-dependent probe amplification (MLPA) analysis, and array comparative genomic hybridization (a-CGH) is essential for the identification of NF1 variants and the classification of patients. Such a strategy will provide not only follow-up opportunities and early detection of benign and malignant tumors but also appropriate genetic counseling, including prenatal or pre-implantation genetic diagnosis.
Material and Methods
Patients
This study included 27 unrelated Turkish families that had at least 1 patient clinically diagnosed with or suspected to have NF1 based on the NIH criteria, between June 1990 and 2019 at the İstanbul University, İstanbul Faculty of Medicine (NIH, 1988). The study was approved by the İstanbul Medical Faculty Ethics Committee (June 30 2017, 825 issues) and informed written consent was obtained from each patient. After the patients’approval, a physical examination was performed. For each family, a pedigree involving at least 3 generations was drawn and all the available medical records (radiographic images, photographs, laboratory test results, etc.) from each family were obtained.
Methods
Sample Preparations
A targeted-panel gene test was designed for the coding exons of the NF1 gene (NM_001042492.3) in 70 amplicons with 100 coverage. DNA isolations were performed by using the commercially available DNA Isolation Kit for Mammalian Blood (Roche11667327001/Magnapure Large Volume kit).
Next-Generation Sequencing
The DNA Library was prepared using the Ion Ampliseq Library Kit 2.0 (Thermo Fisher Scientific) according to the manufacturer’s instructions. After primer sequences were partially digested, adapters and barcodes (Ion ExpressTM Barcode) (Thermo Fisher Scientific) were ligated to the amplicon using the Ion Xpress Barcode Adapters Kit (Life Technologies) procedure. Following equalization of barcoded libraries using the Ion Library Equalizer Kit (Thermo Fisher Scientific), the emulsion-based PCR process was applied with the PGM Template Hi-Q™ View kit on the Ion OneTouch 2 Instrument (Thermo Fisher Scientific). The enrichment of template-positive Ion Sphere™Particles was performed with Ion PGM™ template preparation kits using the Ion OneTouch™ ES Instrument (Thermo Fisher Scientific). Sequencing of enriched particles was performed by Ion PGM Hi-Q View Sequencing Kit, according to the manufacturer’s instructions (Thermo Fisher Scientific).
Data Analysis
For reference genome alignment, base calling, and filtering of weak signal reads, we used the IT platform-specific pipeline software Torrent Suite (v 4.2) with the plug-in ‘variant caller’ program (Life Technologies). Raw data were processed and aligned to the hg19 human reference genome (GRCh37) using Torrent Suite Software (v5) (Thermo Fisher Scientific) for a reliable variant calling. For annotation of the variants, Ion Reporter (v 4.0) software was used. Integrated Genomics Viewer (IGV) (http://software.broadinstitute.org/software/igv/) was used to see the coverage and quality of the variants. A cut-off of 100X coverage was applied to all analyses. In silico analysis programs (MutationTaster, PolyPhen and SIFT) as well as Combined Annotation-Dependent Depletion (CADD) (https://cadd.gs.washington.edu) were used for the prediction of the disease-causing status of novel variants.9,10,11 Variant frequency observed from the Genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org/about).12 The phenotypic impact was searched from ClinVar13 and a literature search. The guidelines of the American College of Medical Genetics and Genomics (ACMG) were used for variant classification.14
Sanger Sequencing
The confirmation and segregation analysis of the candidate pathogenic variants found with NGS were carried out with PCR followed by Sanger sequencing. Polymerase chain reaction (PCR) (Thermo Scientific) was performed only for exons with pathogenic variants for the NF1 gene. PCR products were sequenced utilizing the BigDye Terminator 3.1 Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. Sequence analyses were performed using ABI3500 Sequencing System (Applied Biosystems).
Multiplex Ligation-dependent Probe Amplification
NF1-MLPA reaction kits (SALSA P081/P082) (MRC-Holland, Amsterdam, The Netherlands) targeting whole exons in NF1 were used according to the manufacturer`s protocol. Fragment analyses were conducted utilizing the Genetic Analyzer 3730 capillary electrophoresis system (Applied Biosystems, CA, USA). Data analysis was performed using Coffalyser.Net software).15
Array Comparative Genomic Hybridization
The a-CGH analysis was performed by using Agilent SurePrint G3 CGH+SNP Microarray Kit (4x180K) (Agilent Technologies, Inc., Santa Clara, CA, USA). Data analysis was performed using Agilent Genomics Workbench (v5.0.2.5). Genomic linear positions were given relative to hg19. The deleted genomic regions of the patients were evaluated using some specific databases such as Online Mendelian Inheritance in Man (https://www.omim.org), The Database of Genomic Variants (DGV, http://dgv.tcag.ca/dgv/app/home), The DatabasE of genomiC varIations and Phenotype in Humans using Ensembl Resources (DECIPHER, https://decipher.sanger.ac.uk), and DISEASES (https://diseases.jensenlab.org/Search).
Results
Clinical Findings
Overall, 44 individuals from 27 families who had a clinical diagnosis or were suspected of NF1 based on NIH criteria were included in our study group (Table 1). Cafe-au-lait spots were the most common skin finding, which was present in 38 patients. Lisch nodules were diagnosed in 8 patients. Thirty-one patients had axillary and inguinal freckling. Optic pathway gliomas (astrocytomas of the optic pathway) were present in 3 patients. Neurofibromas of a variety of different types, either cutaneous neurofibromas, plexiform neurofibromas or malignant peripheral nerve sheath tumors were detected in 39, 9, and 1 patient(s) respectively. Distinctive bone abnormalities such as scoliosis and local bony overgrowths were identified in 6 and 1 patient(s), respectively.
Table 1.
Demographics, Clinical Findings and NF1 Mutational Spectrum of the Probands from NF1 Families by the NGS Study
| NIH Criteria | Molecular Results | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family | Case | Gender/Age | CAL | NF | PNF | AF/IF | OG | LN | SD/TBA | FH | Total | Other | Exon | Chromosomal Location | dbSNP ID | Nucleotide | Protein | Variant Affect | Ref | HGMD | ACMG | CADD | SIFT | PolyPhen | varSEAK Online | Mutation Taster |
| Family 1 | A1-II-10 | M/50 | + | + | - | - | - | - | - | + | 3 | - | 38 | 17:29654545 | rs1555533555 | c.5297C>A | p.ser1766* | Nonsense | 19 | CM080450 | PVS1 | 26.2 | Nap | Nap | _ | Disease-causing |
| A1-III- 1 | M/28 | + | - | - | + | - | - | - | + | 3 | - | |||||||||||||||
| A1-III- 2 | F/26 | + | + | + | - | - | - | - | + | 4 | - | |||||||||||||||
| A1-III- 3 | F/23 | + | + | - | - | - | - | + | + | 4 | Exostoses in juxta-epiphyseal regions of long bones | |||||||||||||||
| A1-III- 4 | M/20 | + | + | - | + | - | - | - | + | 4 | - | |||||||||||||||
| Family 2 | A2-II-7 | M/66 | - | + | - | + | - | - | - | + | 3 | - | 36 | 17:29592245 | : rs1295045178 | c.4725-2 A>C | - | Splicing | 1 | CS1311527 | PVS1 | 36 | Nap | Nap | Pathogenic (class 5) | Disease-causing |
| A2-III-3 | F/34 | + | + | - | + | - | + | + | 5 | MPNST, Breast cancer, lung cancer | ||||||||||||||||
| Family 4 | A4-II-11 | M/49 | + | + | + | + | - | - | - | + | 5 | Hypertension, scoliosis | 21 | 17:29556436 | rs786201823 | 2802_2803insT | p. N935* | Nonsense | Novel | NA | PVS1 | NA | Nap | Nap | - | Prediction disease-causing |
| A4-III-1 | F/18 | + | - | - | + | - | - | - | + | 3 | Temporal bossing, periorbital swelling, left ptosis, facial asymmetry, missing and decayed tooth, narrow and high palate, macrocephaly | |||||||||||||||
| A4-III-4 | F/9 | + | - | - | + | - | - | - | + | 3 | Scoliosis | |||||||||||||||
| Family 5 | A5-IV-8 | M/44 | + | + | + | + | - | - | - | + | 5 | Scoliosis, learning disability, Vertebral fusion between C5 and T1 (operated) | 11 | 17:29528504 | rs267606603 | c.1260+1G>A | - | Splicing | 26 | CS064442 | PVS1 | 34 | Nap | Nap | Pathogenic (class 5) | Disease-causing |
| A5-V-1 | F/18 | + | + | - | + | - | - | - | + | 4 | Ventriculomegaly | |||||||||||||||
| Family 6 | A6-IV-3 | F/49 | + | + | - | + | - | - | - | + | 4 | Macrocephaly, scoliosis | 53 | 17:29683984-29683987 | rs1135402907 | c.7747_7748delAG | p. R2583Dfs*12 | Frameshift Deletion | 20 | CD041598 | PVS1 | NA | Nap | Nap | - | Prediction disease-causing |
| A6-IV-4 | F/27 | + | + | - | + | - | - | - | + | 4 | - | |||||||||||||||
| A6-III-3 | F/25 | + | + | + | + | - | - | - | + | 5 | - | |||||||||||||||
| Family 7 | A7-II-4 | M/41 | + | + | - | + | - | - | - | + | 4 | Macrocephaly | 37 | 17:29653003 | - | C.4998delT | p.Pro1667fs | Frameshift Deletion | Novel | NA | PVS1 | 28.4 | Nap | Nap | - | Prediction disease-causing |
| A7-III-1 | F/11 | - | + | - | + | + | + | - | + | 5 | Neurofibromas of the brain and cerebellum | |||||||||||||||
| A7-III-3 | F/6 | - | + | - | + | + | + | - | + | 5 | Neurofibromas of the brain and cerebellum | |||||||||||||||
| Family 8 | A8-III-2 | F/39 | + | + | - | + | - | - | - | + | 4 | Macrocephaly | 26 | 17:29559859- 17:29559862 | - | c.3456_3459 delACTC | p.Leu1153METfs*4 | Frameshift Deletion | Novel | NA | PVS1 | NA | Nap | Nap | - | Prediction disease-causing |
| A8-IV-1 | M/8 | + | + | - | - | - | - | - | + | 3 | - | |||||||||||||||
| Family 9 | A9-III-3 | F/29 | + | + | - | + | + | - | - | + | 5 | - | 14 | 17:29546023 | - | c.1555C>T | P.Gln519* | Nonsense | 17 | CM1820908 | PVS1 | 37 | Nap | NAP | - | |
| A9-IV-1 | F/5 | + | - | - | - | - | - | - | + | 2 | - | |||||||||||||||
| Family 10 | A10-II-3 | M/44 | - | + | - | + | - | - | - | + | 3 | High palate, large/anteriorly turned and up-lifted ears, flattening of the helix | 18 | 17:29553484 | - | c.2033dupc | p.ile679aspfs*21 | Frameshift Duplication | 4 | CI951961 | PVS1 | NA | Nap | NAP | - | Prediction disease-causing |
| A10-III-2 | F/13 | + | - | - | - | - | - | - | + | 2 | - | |||||||||||||||
| Family 12 | A12- III-1 | M/50 | + | + | - | + | - | - | - | - | 3 | High palate | - | - | - | - | - | - | - | - | - | - | - | - | ||
| Family 13 | A13- II-8 | F/44 | - | + | - | - | - | - | - | - | 1 | Multiple polyps in the gall bladder | 55 | 17:29685597 | - | c.8070C>G | p.Tyr2690* | Nonsense | Novel | NA | PVS1 | 31 | Nap | Nap | - | Prediction disease-causing |
| Family 14 | A14-II-3 | F/54 | + | + | - | + | - | - | - | - | 3 | - | 5 | 17:29496994 | - | C.564delA | P.Lys189Asnfs*2 | Frameshift Deletion | Novel | NA | PVS1 | 36 | Nap | Nap | - | Prediction disease-causing |
| Family 15 | A15-II-4 | M/57 | + | + | - | - | - | + | - | - | 3 | Pituitary adenoma | 8 | 17:29509541 | rs1567826623 | c.746T>C | P.Leu249Pro | Missense | Novel | NA | PS1 | 23.9 | Damaging | 0.96 | - | Disease-causing |
| Family 16 | A16- III-4 | F/24 | - | + | - | + | - | - | - | - | 2 | Epilepsy, psycho-motor developmental delay, pulmonary stenosis, osteoporosis, scoliosis, learning disability | 32 | 17:29585419 | - | c.4231C>T | p.leu1411phe | Missense | 7 | CM098465 | PS1 | 28.9. | Damaging | 0.999 | - | Disease-causing |
| Family 17 | A17- III-5 | F/48 | + | + | + | - | - | - | - | - | 3 | - | 16 | 17:29550496- 17:29550499 | - | c.1756_1759 delACTA | P.Thr586valfs*18 | Frameshift Deletion | 8 | CD982825 | PVS1 | NA | Nap | Nap | - | Prediction disease-causing |
| Family 18 | A18- III-4 | F/26 | + | + | - | + | - | - | - | - | 3 | Pectus carinatum, hamartoma (1x7 mm) in the corpus callosum, epilepsy | 47 | 17:29667526 | - | c.6925 dellT | P.ser2309Argfs*10 | Frameshift Deletion | Novel | NA | PVS1 | 23.7 | Nap | Nap | - | Prediction disease-causing |
| Family 20 | A20- III-9 | F/34 | + | + | - | - | + | - | - | 3 | Scoliosis | 38 | 17:29654624 & 17:29654625 | - | c.5375_5376insC | P.Leu1793Profs*4 | Frameshift Insertion | Novel | NA | PVS1 | NA | Nap | Nap | - | Prediction disease-causing | |
| Family 21 | A21-III-2 | F/35 | + | + | + | + | - | + | - | - | 5 | Macrocephaly, meningioma | 32 | 17:29586057 | rs786204157 | c.4340A>G | p.Gln1447Arg | Missense | 30 | CM133021 | PS1 | 24.2 | Damaging | 0.163 | - | Disease-causing |
| Family 22 | A22-III-1 | M/35 | + | + | - | - | - | - | - | - | 2 | - | 23 | 17:29528428 | rs876660782 | c.2991-1G>A | - | Splicing | 26 | CS961633 | PVS1 | 35 | - | Pathogenic (class 5) | Disease-causing | |
| Family 23 | A23-III-1 | M/20 | + | + | - | + | - | - | - | - | 3 | 34 | 17:29587508 | 4552delA | p.Ile1518fs | Frameshift Deletion | Novel | NA | PVS1 | 26.7 | Nap | Nap | - | Prediction disease-causing | ||
| Family 24 | A24- III-4 | F/25 | + | + | - | + | - | - | - | - | 3 | Hamartomatous, hyper-pigmented skin lesion | 19 | 17:29554235 | rs587781577 | c.2252-1G>A | - | Splicing | Novel | NA | PVS1 | 34 | Nap | Nap | Pathogenic (class 5) | Disease-causing |
| Family 25 | A25- III-6 | F/54 | + | + | + | + | - | - | - | - | 4 | - | 9 | 17:29527461 | - | c.910C>T | P.Arg304* | Nonsense | 26 | CM087437 | PVS1 | 35 | Nap | NAP | - | Disease-causing |
| Family 26 | A26-III-1 | F/18 | + | + | - | - | - | - | - | - | 2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Family 27 | A27- III-3 | M/24 | + | + | + | - | - | - | - | - | 3 | Hypertelorism, hypertension | 4 | 17:29490322 | - | c.407delA | P.Asn136Phefs*19 | Frameshift Deletion | Novel | NA | PVS1 | 23.4 | Nap | Nap | - | Prediction disease-causing |
| Total, N | 33 | 34 | 8 | 26 | 3 | 6 | 2 | 24 | ||||||||||||||||||
*CAL, cafe-au-lait spots; NF, neurofibromas; PNF, plexiform neurofibroma; AF, axillary freckling; IF, inguinal freckling; OG, optic glioma;LN, lisch nodules; MPNST, malignant peripheral nerve sheath tumors; FH, family history; PVS1, pathogenic very strong 1; PS1, pathogenic strong 1; Ref, references; NAp, not applicable; NA, not available.
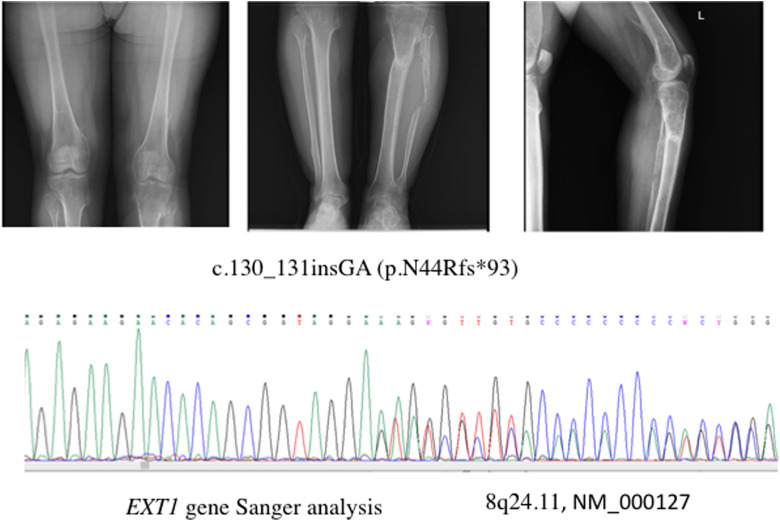
A 20-year-old female (A1-III-3) had a history of multiple exostoses for which she had been operated on previously (Figure 1A), Although there were 5 NF1 patients in this family, this finding was detected only in this patient. She was tested for exotosin-1 gene (EXT1) variants, based on a clinical diagnosis of multiple exostoses. We found a frameshift pathogenic variant [8q24.11, NM_000127, C.130_131insGA (p.N44Rfs*93)] in EXT1 by Sanger analysis (Figure 1B), which had not been reported before. The parents tested negative for this variant.
FIG. 1. A, B.
(A) Exostoses in the juxtaepiphyseal regions of long bones. (B) A pathogenic variant identified in the EXT1 gene with Sanger analysis in the (A1-III-3) case.
In our cohort, a female patient (A2-III-1) was diagnosed with breast cancer at the age of 35 years. The risk of breast cancer in NF1 is especially increased in patients younger than 50 years of age compared to the general population.16,17,18
The pathogenic variants of NF1 usually cause a similar clinical finding with other syndromes.7 Likewise in this study, a case (A16-III-4) with multiple café-au-lait spots, pulmonary stenosis, and intellectual difficulty was diagnosed with Watson syndrome.
NGS Analysis
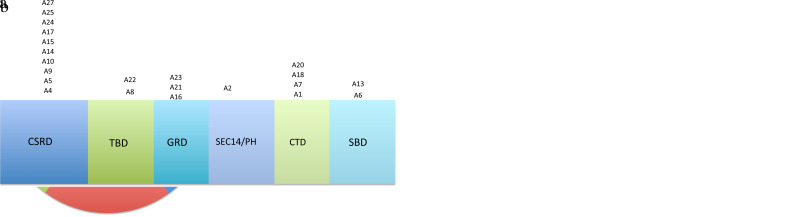
In the present study, we have identified NF1 pathogenic variants in 22 out of 27 families with targeted NGS analysis of NF1 (Table 1). There were 10 frameshifts, 5 nonsense, 4 splicing region and 3 missense pathogenic variants (Figure 2A). The frequency of truncated pathogenic variants was remarkably higher than that of other pathogenic variants (19/22; 86%). In this patient group, 9 pathogenic variants were familial (Figure 2B).
FIG. 2. A, B.
(A) The distribution of identified NF1 pathogenic variant types utilizing NGG. (B) Distribution of identified genetic variations of proband of NF1 family and possible defect on neurofibromin domains. *CSRD, cysteine-serine rich domain; TBD, tubulin-binding domain; GRD, GAP-related domain; SEC14/PH > SEC14 domain and pleckstrin homology (PH) domain; CTD > Carboxy-terminal domain-terminal domain; SBD, syndecan-binding domain (Frayling I et al, 2019).16
On comparing with the ClinVar, CLINVITAE, and Human Gene Mutation Database, we identified 11 pathogenic variants that were not previously reported in the literature. The novel pathogenic variants found in this study were checked both in the affected and healthy family members and 100 normal controls. None of the controls or the healthy family members were found to carry the pathogenic variant, confirming its pathogenicity.
Copy Number Variations (CNV) Analyses
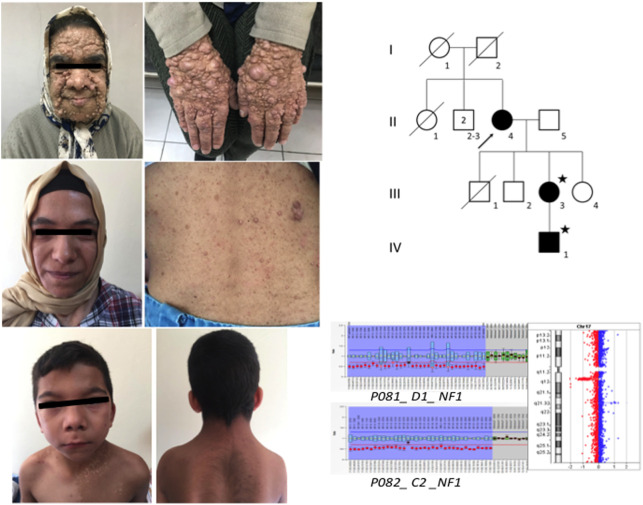
In 7 individuals from 3 unrelated families (A3, A11, A12, A19, A26) where NGS did not yield any pathogenic variants, NF1 rearrangements were analyzed with the MLPA approach. This approach detected heterozygous whole gene deletion in 5 patients (Table 2). Of those, 3 [(A11-III-4), (A11-IV-3), (A11-V-1)] belonged to the same family (Figure 3), whereas in the 2 others (A3 and A19 ), the deletion had occurred de novo (Table 2). The a-CGH analysis defined a 17q11.2 deletion of about 1.4 Mb in 4 of them; and a 1.2 Mb deletion in 1 patient (family A3). To evaluate the clinical significance of the deleted region, we searched OMIM, DGV, DISEASE, and DECIPHER databases. The genes CRLF3, ATAD5, TEFM, ADAP2, RNF135, OMG, EVI2B, EVI2A, RAB11FIP4 SUZ12, LRRC37B, and NF1 were found to be located in these regions.
Table 2.
Clinical and Genetic Features of NF1 Microdeletion Patients
| NIH Criteria | Molecular results | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family | Case | Gender/Age | CAL | NF | PNF | AF/IF | OG | LN | SD/TBA | FH | Total | Other | MLPA Results | A-CGH RESULTS | References | |
| Family 3 | A3-III-2 | F/18 | + | + | + | +/+ | - | + | - | - | 5 | Hypertelorism, macrocephaly, mild micrognathia, bilateral cubitus valgus, hyperextensibility, genu recurvatum, hamartoma of the splenium of the corpus callosum, ectopia of the cerebellar tonsils, hypothyroidism, and thyroiditis | Whole NF1 gene deletion | 17q11.2 deletion (1.2 Mb), Position (29124299_30326958). LOC107133515, SH3GL1P2, SUZ12P1, CRLF3, ATAD5, TEFM, ADAP2, RNF135, DPRXP4, MIR4733, NF1, OMG, EVI2B, EVI2A, RAB11FIP4, MIR4724, MIR193A, MIR4725, MIR365B | 28 | |
| Family 11 | A11- III-4 | F/66 | + | + | - | +/+ | - | + | - | + | 5 | Hypertelorism, macrocephaly, hyperparathyroidism, hypertension, Adrenal adenoma | Whole NF1 gene deletion | 17q11.2 deletion (1.4 Mb), Position (28941066_30367214). CRLF3, ATAD5, EFM, ADAP2, RNF135, NF1, OMG, EVI2B, EVI2A, RAB11FIP4, MIR193A, SUZ12, LRRC37B, LOC107133515, SH3GL1P2, SUZ12P1, DPRXP4, LOC107984974, LOC646030, MIR4733, MIR4724, MIR4725, MIR365B, COPRS, UTP6 | 2,29 | |
| A11- IV-3 | F/35 | + | + | - | +/+ | - | - | - | + | 4 | Hypertelorism | |||||
| A11-V-1 | M/8 | + | + | - | +/+ | - | - | - | + | 4 | Hypertelorism, Neurofibrosarcoma | |||||
| Family 19 | A19- III-5 | M/40 | + | + | - | +/+ | - | - | - | - | 3 | Glaucoma, neurofibrosarcoma, malignant mesenchymal tumor | Whole NF1 gene deletion | 17q11.2 deletion (1.4 Mb), Position (28941066_30367214). CRLF3, ATAD5,TEFM, ADAP2, RNF135, NF1, OMG, EVI2B, EVI2A, RAB11FIP4, MIR193A, SUZ12, LRRC37B, LOC107133515, SH3GL1P2, SUZ12P1, DPRXP4,LOC107984974, LOC646030, MIR4733, MIR4724, MIR4725, MIR365B, COPRS, UTP6 | 2,29 | |
| Total, N | 5 | 5 | 1 | 5 | 0 | 2 | 0 | 3 | ||||||||
*CAL, cafe-au-lait spots; NF, neurofibromas; PNF, plexiform neurofibroma; AF, axillary freckling;IF, inguinal freckling; OG, optic glioma; LN, lisch nodules; MPNST, malignant peripheral nerve sheath tumors; FH, family history.
FIG. 3.
Familial NF1 microdeletion in family A11.
With the molecular approach used in our study, we did not detect any pathogenic variants in 2 patients who met the NIH criteria for a diagnosis of NF1 (A12-III-1 and A26-III-1).
Discussion
NF1 is one of the most common genetic disorders with a broad spectrum of phenotypic manifestations that may progress throughout the life span of the patients.6 Although NF1 can be diagnosed readily based on criteria defined by NIH, diagnosis can sometimes be difficult, especially when a phenotype does not meet the diagnostic criteria of NIH. Therefore, the genetic analysis should be included in the diagnostic algorithm of this disorder. Loss of function of NF1 is responsible for the etiopathogenesis of clinical manifestations associated with this disorder.8 Most of the pathogenic variants detected in NF1 are single-base substitutions, insertions, or deletions.19,20,21 Other variants arise from multiple exon deletions or amplifications as well as microdeletions including NF1 and neighboring genes.22,23 Due to the numerous types of variants, it may not be possible to confirm the diagnosis with a single method.4,20,24 Therefore, a multi-step process is required for a comprehensive analysis of the gene. In this scope, we utilized a multi-step process consisting of NGS, MLPA, and a-CGH. In the first step, NGS yielded 22 pathogenic variants which were detected in 81% (22/27) of our patients. Additionally, in the (A1-III-3) patients from this study, a novel pathogenic variant in the EXT1 gene was found by NGS analysis. The EXT1 gene is associated with hereditary multiple exostoses (HME) disease. HME is a rare autosomal dominant disorder. The incidence is estimated to be about 1 in 50 000. The disease is characterized by multiple exostoses, skeletal deformities, and scoliosis. It is predominantly caused by a pathogenic variant in EXT1 and exostosin-2 (EXT2) genes, that encode heparan sulfate (HS) glycosyltransferases.25 Therefore, this patient (A1-III-3) had both NF1 and HME. This shows the significance of considering additional diagnoses.
In the second step, MLPA analyses detected a deletion in 5 patients, 3 of them being members of the same family (3/27; 11%). In the third step, the a-CGH study showed a large deletion involving NF1 and multiple genes in all these patients.
For 2 patients who completely met the clinical diagnostic criteria, no NF1 variants could be identified with this multi-step approach. Previous studies have reported NF1 pathogenic variants located in deep intronic or regulatory regions, which are not covered by NGS.26 Other possible explanations for the absence of pathogenic variants include somatic mosaicism or pathogenic variants of other genes in the RAS pathway which are known to be associated with overlapping clinical features.7,8 We suggest that this diagnostic gap may be filled by RNA-seq as well as whole-genome analysis which could provide information about other gene/genes and the possibility of a deep intronic or regulatory region variant that is associated with the phenotype.16
In patients whose NGS detected a pathogenic variant, there were no remarkable differences for the phenotype, which was in agreement with previous studies.6 The frequency of truncating pathogenic variants was remarkably higher than that of other types of pathogenic variants (19/22). Based on studies, the variants including frameshift (FS) and nonsense (nS) that result in a truncated protein and degradation via NMD (nonsense-mediated decay) by the early termination of mRNA cause non-expression of normal protein from one allele and dramatically reduce normal function.27 This mechanism results in the loss of the negative regulatory effect of the protein on the RAS/MAPK pathway. On the other hand, missense variants (MS) do not promote NMD, but result in the expression of the aberrant protein that decreases the normal function of normal allele with a dominant negative effect. In addition, the variants which were identified only in patients with clinical findings, but not in 100 healthy individuals, support the pathogenicity of these variants. We have planned further research including functional studies, especially for missense variants, that will support this data.
In patients in whom NGS did not yield any pathogenic variant, we carried out MLPA, which detected whole NF1 gene deletions in 5 patients from 3 families. Since MLPA cannot discriminate a whole gene deletion from a microdeletion, we applied a-CGH to see whether the deletion found in our patients included only NF1 gene or other additional genes in its neighborhood. In this stage, we identified a 1.4 Mb deletion in 4 patients and a 1.2 Mb deletion in 1 patient. Although clinical symptoms do not manifest fully before 8 years of life in most cases, patients with a large NF1 deletion present at an early age with a more a severe phenotype, including learning difficulty, dysmorphic features, and a higher frequency of tumors.28
The risk of plexiform neurofibroma at an early age is also increased.21 In our study group, the (A11-V-4) patient presented with neurofibrosarcoma before 8 years of age. In the present study, all patients with microdeletion had facial dysmorphism while none of the patients with other types of pathogenic variants displayed this finding.
The phenotypes of patients with NF1 microdeletion may be influenced by a loss of function of the genes CRLF3, ATAD5, TEFM, ADAP2, RNF135, OMG, EVI2B, EVI2A, RAB11FIP4 SUZ12, LRRC37B, and NF1 in the deleted region. Based on literature data, the dysmorphic facial features of patients in our study may be due to the haploinsufficiency of the RNF135 (MIM:611358) gene. Furthermore, the expression of only one copy of OMG (MIM: 164345) plays a significant role in intellectual disability traits in these patients.28,29 Similarly, COPRS is associated with learning disabilities in NF1 microdeletion patients and tumorigenesis.21 In addition, ADAP2 gene deletion (MIM: 608635) may lead to cardiovascular abnormalities. Likewise, the deletion of the SUZ12 and UTP6 genes, which are also expressed during human heart development, maybe related to cardiomyopathy.2,21,22 Although all these genes were deleted in our patients, only facial dysmorphism appeared to be a discriminating feature for microdeletion in our patient group.
The phenotypic variation of NF1 microdeletion syndrome depends on the extent of the chromosomal rearrangements and the genes that have been deleted.21 However, even in cases with the same deletion, phenotypic variations may be observed. It may be suggested that the NF1-related phenotypes may be influenced by additional mechanisms such as modifier genes and regulatory proteins that interact with the NF1 gene. Although the clinical features of NF1 microdeletion patients are generally more severe, we suggest that the presence of a mild clinical phenotype should not exclude the possibility of NF1 gene deletion in a patient.23 Despite the various types of pathogenic variants identified in this study, the evaluation of the correlation between the genotype–phenotype of these patients was not possible due to the small patient sample size. A systematic comparative study with more patients of the same age and with overlapping clinical manifestations may provide a more reliable definition of genotype–phenotype relations. Therefore, future WES/WGS studies may provide the variants in modifying genes which may help to explain genotype–phenotype correlations in NF1. Though prenatal diagnosis of NF1 is controversial, our three-step approach presented here seems to be more effective for symptomatic/presymptomatic diagnosis and genetic counseling for NF1.
Footnotes
Ethics Committee Approval: The study was approved by the İstanbul Medical Faculty Ethics Committee (June 30 2017, 825 issues).
Patient Consent for Publication: Informed written consent was obtained from each patient.
Data-sharing Statement: The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Author Contributions: Concept - S.S., K.C.; Design - S.S.; Supervision - S.S., K.C.; Data Collection and/or Processing - S.S.; Analysis and/or Interpretation - S.S.; Literature Review - S.S., K.C., T.K., Ş.P., Ş.Ö.; Writing - S.S.; Critical Review - K.C., T.K., Ş.P., Ş.Ö.
Acknowledgments: We thank Birsen Karaman, Cagrı Gulec, Sommayyeh Haydargholizadeh and Adem Nejefli from the İstanbul Faculty of Medicine, University of İstanbul and Gulcin Ozan, Burak Don, Ilknur Suer, Murat Kaya, Ayla Sahin and Harun Arslan from the Division of Medical Genetics, Department of Internal Medicine, İstanbul Faculty of Medicine, University of İstanbul, for their contribution in this study.
Conflict of Interest: The authors have declared that no conflicts of interest exist.
Funding: This research was supported by the Scientific Research Projects (BAP) of University of İstanbul ( Project No. TDK-2018-28151).
References
- 1. Sabbagh A, Pasmant E, Imbard A, et al. NF1 molecular characterization and neurofibromatosis type I genotype-phenotype correlation: the French experience. Hum Mutat. 2013;34(11):1510–1518.. 10.1002/humu.22392) [DOI] [PubMed] [Google Scholar]
- 2. Venturin M, Carra S, Gaudenzi G, et al. ADAP2 in heart development: a candidate gene for the occurrence of 132 cardiovascular malformations in NF1 microdeletion syndrome. J Med Genet. 2014;51(7):436–443.. 10.1136/jmedgenet-2013-102240) [DOI] [PubMed] [Google Scholar]
- 3. Pasmant E, Parfait B, Luscan A, et al. Neurofibromatosis type 1 molecular diagnosis: what can NGS do for you when you have a large gene with loss of function mutations? Eur J Hum Genet. 2015;23(5):596–601.. 10.1038/ejhg.2014.145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsipi M, Poulou M, Fylaktou I, et al. Phenotypic expression of a spectrum of neurofibromatosis type 1 (NF1) mutations identified through NGS and MLPA. J Neurol Sci. 2018;395:95–105.. 10.1016/j.jns.2018.10.006) [DOI] [PubMed] [Google Scholar]
- 5. Yoshida Y, Sato N, Furumura M, Nakayama J. Treatment of pigmented lesions of neurofibromatosis 1 with intense pulsed-radio frequency in combination with topical application of of vitamin D3 ointment. J Dermatol. 2007;34(4):227–230.. 10.1111/j.1346-8138.2007.00258.x) [DOI] [PubMed] [Google Scholar]
- 6. Barrea C, Vaessen S, Bulk S, Harvengt J, Misson JP. Phenotype–genotype correlation in children with neurofibromatosis type 1. Neuropediatrics. 2018;49(3):180–184.. 10.1055/s-0037-1620239) [DOI] [PubMed] [Google Scholar]
- 7. Nyström AM, Ekvall S, Allanson J, et al. Noonan syndrome and neurofibromatosis type I in a family with a novel mutation in NF1. Clin Genet. 2009;76(6):524–534.. 10.1111/j.1399-0004.2009.01233.x) [DOI] [PubMed] [Google Scholar]
- 8. Yap P, Super L, Qin J, et al. Congenital retroperitoneal teratoma in neurofibromatosis type 1. Pediatr Blood Cancer. 2016. ;63(4):706–708.. 10.1002/pbc.25812) [DOI] [PubMed] [Google Scholar]
- 9. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;76:7.20.1–7.20.41.. 10.1002/0471142905.hg0720s76) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081.. 10.1038/nprot.2009.86) [DOI] [PubMed] [Google Scholar]
- 11. Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–362.. 10.1038/nmeth.2890) [DOI] [PubMed] [Google Scholar]
- 12. Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443.. 10.1038/s41586-020-2308-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868.. 10.1093/nar/gkv1222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424.. 10.1038/gim.2015.30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coffa J, van den Berg J. Analysis of MLPA Data Using Novel Software Coffalyser.NET by MRC-Holland. Mod Approaches Qual Control. 2011;7:125–150.. 10.5772/21898) [DOI] [Google Scholar]
- 16. Frayling IM, Mautner V-F, van Minkelen R, et al. Breast cancer risk in neurofibromatosis type 1 is a function of the type of NF1 gene mutation: a new genotype-phenotype correlation. J Med Genet. 2019;56(4):209–219.. 10.1136/jmedgenet-2018-105599) [DOI] [PubMed] [Google Scholar]
- 17. Momozawa Y, Iwasaki Y, Parsons MT, et al. Germline pathogenic variants of 11 breast cancer genes in 7,051 Japanese patients and 11,241 controls. Nat Commun. 2018;9(1):4083. 10.1038/s41467-018-06581-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilson CH, Griffith CD, Shrimankar J, Douglas F. Gynaecomastia, neurofibromatosis and breast cancer. Breast. 2004;13(1):77–79.. 10.1016/j.breast.2003.08.002) [DOI] [PubMed] [Google Scholar]
- 19. Cassiman C, Casteels I, Jacob J, et al. Choroidal abnormalities in café-au-lait syndromes: a new differential diagnostic tool? Clin Genet. 2017;91(4):529–535.. 10.1111/cge.12873) [DOI] [PubMed] [Google Scholar]
- 20. De Luca AD, Schirinzi A, Buccino A, et al. Novel and recurrent mutations in the NF1 gene in Italian patients with neurofibromatosis type 1. Hum Mutat. 2004;23(6):629. 10.1002/humu.9245) [DOI] [PubMed] [Google Scholar]
- 21. Serra G, Antona V, Corsello G, et al. NF1 microdeletion syndrome: case report of two new patients. Ital J Pediatr. 2019;45(1):138. 10.1186/s13052-019-0718-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kehrer-Sawatzki H, Kluwe L, Sandig C, et al. High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene. Am J Hum Genet. 2004;75(3):410–423.. 10.1086/423624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mautner VF, Kluwe L, Friedrich RE, et al. Clinical characterisation of 29 neurofibromatosis type-1 patients with molecularly ascertained 1.4 Mb type-1 NF1 deletions. J Med Genet. 2010;47(9):623–630.. 10.1136/jmg.2009.075937) [DOI] [PubMed] [Google Scholar]
- 24. Ulusal SD, Gürkan H, Atlı E, et al. Genetic analyses of the NF1 gene in Turkish neurofibromatosis type I patients and definition of three novel variants. Balkan J Med Genet. 2017;20(1):13–20.. 10.1515/bjmg-2017-0008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsumoto K, Ogawa H, Nozawa S, Akiyama H. An analysis of osteoporosis in patients with hereditary multiple exostoses. Osteoporos Int. 2020;31(12):2355–2361.. 10.1007/s00198-020-05533-7) [DOI] [PubMed] [Google Scholar]
- 26. Xiong HY, Alipanahi B, Lee LJ, et al. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2014;347(6218):1254806. 10.1126/science.1254806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5(2):89–99.. 10.1038/nrm1310) [DOI] [PubMed] [Google Scholar]
- 28. Vogt J, Nguyen R, Kluwe L, et al. Delineation of the clinical phenotype associated with non-mosaic type-2 NF1 deletions: two case reports. J Med Case Rep. 2011;5:577. 10.1186/1752-1947-5-577) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Venturin M, Moncini S, Villa V, et al. Mutations and novel polymorphisms in coding regions and UTRs of CDK5R1 and OMG genes in patients with non-syndromic mental retardation. Neurogenetics. 2006;7(1):59–66.. 10.1007/s10048-005-0026-9) [DOI] [PubMed] [Google Scholar]
- 30. Ben-Shachar S, Constantini S, Hallevi H, et al. Increased rate of missense/in-frame mutations in individuals with NF1-related pulmonary stenosis: a novel genotype–phenotype correlation. Eur J Hum Genet. 2013;21(5):535–539.. 10.1038/ejhg.2012.221) [DOI] [PMC free article] [PubMed] [Google Scholar]



 Content of this journal is licensed under a
Content of this journal is licensed under a