Abstract
Background
Excessive inflammation has been implicated in the immunopathogenesis of coronavirus disease 2019 (COVID-19). In the current study, the involvement of S100 calcium binding protein S100A4, S100A9, and S100A10 in the inflammatory settings of COVID-19 patients were evaluated.
Methods
Peripheral blood samples were obtained from 65 COVID-19 subjects and 50 healthy controls. From the blood samples, RNA was extracted and cDNA was synthesized, and then the mRNA expression levels of S100A4, S100A9, and S100A10 were measured by Real-time PCR.
Results
The mRNA expression of S100A4 (fold change [FC] = 1.45, P = 0.0011), S100A9 (FC = 1.47, P = 0.0013), and S100A10 (FC = 1.35, P = 0.0053) was significantly upregulated in COVID-19 patients than controls. The mRNA expression of S100A4 (FC = 1.43, P = 0.0071), (FC = 1.66, P = 0.0001), and S100A10 (FC = 1.63, P = 0.0003) was significantly upregulated in the severe COVID-19 subjects than mild-to-moderate subjects. There was a significant positive correlation between mRNA expression of S100A4 (ρ = 0.49, P = 0.030), S100A9 (ρ = 0.55, P = 0.009), and S100A10 (ρ = 0.39, P = 0.040) and d-dimer in the COVID-19 patients. The AUC for S100A4, S100A9, and S100A10 mRNAs were 0.79 (95% CI 0.66–0.92, P = 0.004), 0.80 (95% CI 0.67–0.93, P = 0.002), and 0.71 (95% CI 0.56–0.85, P = 0.010), respectively.
Conclusions
S100A4, S100A9, and S100A10 play a role in the inflammatory conditions in COVID-19 patients and have potential in prognosis of severe form of COVID-19. Targeting these modules, hopefully, might confer a therapeutic tool in preventing sever symptoms in the COVID-19 patients.
Keywords: Coronavirus disease 2019, Inflammation, S100A4, S100A9, S100A10
Introduction
Coronavirus disease 2019 (COVID-19) pandemic resulted from Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to the emergence of a global challenge and a major health issue worldwide [1]. Angiotensin-converting enzyme 2 (ACE2) receptor, which is predominantly expressed on the alveolar epithelial cells of the lung, is the major receptor for S protein of SARS-CoV-2 that facilitate the virus entrance. Multiple organ involvement also occurs in the tissues expressing the ACE2 receptor, such as kidney and intestine [2–5]. Even though vast majority of the infected individuals experience a moderate form COVID-19 with little clinical symptoms, approximately 10–15% of subjects develop a severe illnesses needing hospitalization and supportive cares, and 5% of cases might need admission into intensive care units (ICU) [6]. During the severe forms of COVID-19, cytokine storm and cytokine release syndrome might culminate in acute respiratory distress syndrome (ARDS), sepsis, septic shock, multiorgan failure, and even death [7–9].
That notwithstanding, the main molecules involved in causing these severe symptoms have not yet been identified. Numerous studies have been performed on the molecular basis to identify the mechanisms involved in the development of acute inflammatory symptoms in patients with COVID-19 [10]. Acute phase proteins are one of the most important molecules that are produced during infectious and non-infectious injuries and lead to inflammation and overactivation of the immune system [11]. S100A calcium binding proteins play important roles during inflammatory processes and regulate different molecules and inflammatory signaling pathways leading to inflammation [12]. Among them, S100A4, S100A9, and S100A10 molecules are of special importance for causing inflammation that also have antimicrobial roles [13, 14]. The S100A9 molecule is highly expressed in immune inflammatory cells, such as neutrophils and monocytes [15]. This molecule is calcium-dependent and is involved in the production of cytokines, chemokines, and growth factors and plays a role in the infiltration of monocytes and neutrophils into the inflammatory sites [12]. The role of this molecule in causing neutrophil-induced inflammation in many diseases is well established [12]. As another molecule in this group, S100A10 plays an important role in causing inflammation by various mechanisms, such as increasing plasminogen production and subsequently activating the matrix metalloproteinase (MMP) 9 [16]. It seems that the inflammation caused by these molecules is mainly through the activation of type 1 helper (Th1) immune responses and plays a protective role in the Th2-mediated allergic airway inflammation [17]. Given that COVID-19-infected patients mainly suffer from Th1-mediated inflammation [18], these molecules are likely to play an important role in causing inflammatory complications in these patients. It was also observed that S100A8/A9 conferred a diagnostic biomarker value and could accurately identify COVID-19 patients who were subsequently admitted to ICU [19].
Nonetheless, there is insufficient data on the role of S100A4, S100A9, and S100A10 molecules in causing inflammation in patients infected with COVID-19. Therefore, the aim of this study was to evaluate the expression of S100A4, S100A9, and S100A10 molecules in patients with COVID-19 compared to healthy individuals. We evaluated the diagnostic value of these molecules in promoting severe forms of COVID-19. Hopefully, targeting these modules might confer a therapeutic tool in preventing sever complications in the COVID-19 patients.
Materials and methods
Study population
The study population in the present research comprised of 65 patients with COVID-19 referred to the Ali Ibn Abi Talib Hospital, Rafsanjan University of Medical Sciences, Rafsanjan, Iran. Diagnosis of COVID-19 was conducted based on the detection of SARS-CoV-2 genetic content in the nasopharyngeal swab samples by Real-Time PCR test, computerized tomography (CT) scan of chest for detection of COVID-19 patterns, and typical clinical manifestations of the patients. Patients who had respiratory failure, reduced oxygen saturation, and hospitalized in ICU were considered as sever COVID-19 subjects. As the control group, 50 healthy age- and sex-matched individuals were enrolled. None of the healthy controls were infected with other viruses and they did not have immune-related disorders, such as autoimmune diseases, allergy and cancer, or liver diseases. All patients and healthy controls had Iranian (Fars) ethnicity. The demographic data, laboratory indices, and clinical presentations of the study participants are listed in Table 1. The ethics committee of Rafsanjan University of Medical Sciences approved the protocol of this study (IR.RUMS.REC.1400.108) and all subjects signed a written informed consent to participate in this study. From all subjects, 5 ml of the peripheral blood was obtained in the EDTA-anticoagulated tubes using venipuncture.
Table 1.
Demographics and clinical presentations of COVID-19 patients
| Characteristic | Overall COVID-19 patients (N = 65) | Mild-to-moderate COVID-19 patients (N = 43) | Severe COVID-19 patients (N = 22) |
|---|---|---|---|
| Gender; male/female (N, %) | 36 (55.4%)/29 (44.6%) | 25 (58.2%)/18 (41.8%) | 11 (50%)/11(50%) |
| Smoker/non-smoker | 27 (41.5%)/38 (58.5%) | 15 (34.9%)/28 (65.2%) | 12 (54.6%)/10 (45.4%) |
| Age (year, mean ± SD) | 52.7 ± 11.3 | 51.1 ± 11.1 | 54.3 ± 11.5 |
| Duration of COVID-19 (Day) | 12.5 ± 2.2 | 12.4 ± 2.0 | 12.6 ± 2.4 |
| Oxygen saturation | 92.3 ± 6.3 | 91.8 ± 6.1 | 92.8 ± 6.5 |
| Systolic BP (mmHg) | 131.6 ± 21.7 | 128.5 ± 20.3 | 134.7 ± 23.1 |
| Diastolic BP (mmHg) | 72.2 ± 8.5 | 71.6 ± 8.2 | 72.8 ± 8.8 |
| WBC (cells/mm3) | 8154 ± 1739.4 | 8025 ± 1725 | 8283 ± 1753.8 |
| Neutrophil–lymphocyte ratio | 10.3 ± 7.4 | 10.1 ± 7.3 | 10.5 ± 7.5 |
| ALP (IU/L) | 218.3 ± 43.7 | 217.8 ± 43.1 | 218.8 ± 44.3 |
| AST (IU/L) | 27.3 ± 8.8 | 26.8 ± 8.5 | 27.8 ± 9.1 |
| ALT (IU/L) | 33.8 ± 6.2 | 33.3 ± 6.0 | 34.3 ± 6.4 |
| LDH (IU/L) | 398.3 ± 80.6 | 396.8 ± 78.9 | 399.8 ± 82.3 |
| CRP (mg/L) | 4.3 ± 1.8 | 3.4 ± 1.4 | 5.2 ± 2.2 |
| ESR (mm/h) | 18.3 ± 9.1 | 16.5 ± 8.7 | 20.1 ± 9.5 |
| BMI (kg/m2) | 28.7 ± 6.8 | 28.1 ± 6.6 | 29.3 ± 7.0 |
| Total cholesterol (mg/dl) | 206.7 ± 37.1 | 205.2 ± 35.9 | 208.2 ± 38.3 |
| TG (mg/dl) | 161.5 ± 58.3 | 160.2 ± 57.4 | 162.8 ± 59.2 |
| LDL (mg/dl) | 133.2 ± 38.1 | 131.8 ± 36.8 | 134.6 ± 39.4 |
| HDL (mg/dl) | 48.5 ± 13.8 | 48.1 ± 13.5 | 48.9 ± 14.1 |
| Creatinine (mg/dl) | 1.94 ± 0.55 | 1.74 ± 0.41 | 2.14 ± 0.69 |
| BUN (mg/dl) | 23.3 ± 14.8 | 22.8 ± 14.4 | 23.8 ± 15.2 |
| FBS (mg/dl) | 98.3 ± 27.6 | 98.9 ± 27.9 | 97.7 ± 27.3 |
| d-dimer (ng/ml) | 1.88 ± 0.14 | 1.74 ± 0.12 | 2.02 ± 0.16 |
| Cardiovascular diseases | 6 (9.23%) | 3 (7%) | 3 (13.6%) |
| Diabetes | 9 (13.8%) | 5 (11.6%) | 4 (18.2%) |
| Hypertension | 12 (18.5%) | 8 (18.6%) | 4 (18.2%) |
| Fever | 62 (95.4%) | 40 (93%) | 22 (100%) |
| Cough | 61 (93.8%) | 39 (90.7%) | 22 (100%) |
| Dyspnea | 59 (90.8%) | 38 (88.4%) | 21 (95.5%) |
| Sputum | 37 (56.9%) | 18 (41.9%) | 19 (86.36%) |
| Vomiting/diarrhea | 31 (47.7%) | 15 (34.9%) | 16 (72.8%) |
| Methylprednisolone use | 48 (73.8%) | 28 (65.1%) | 20 (90.9%) |
| Remdesivir use | 33 (50.8%) | 14 (32.5%) | 19 (86.3%) |
| Azithromycin use | 22 (33.8%) | 10 (23.25%) | 12 (54.5%) |
| Anticoagulation therapy | 16 (24.6%) | 6 (13.9%) | 10 (45.5%) |
COVID-19 coronavirus disease 2019; WBC white blood cell; CRP C-reactive protein; ALP alkaline phosphatase; AST aspartate aminotransferase; ALT alanine aminotransferase; LDH lactate dehydrogenase; ESR erythrocyte sedimentation rate; BMI body mass index; FBS fasting blood sugar; TG triglyceride; LDL low density lipoprotein; HDL high density lipoprotein; BUN blood urea nitrogen; OR odds ratio; CI confidence interval; SD standard deviation; BP blood pressure
RNA extraction, cDNA synthesis, and quantitative real-time PCR
RNA extraction from plasma was conducted using Trizol total RNA extraction kit (GeneAll, Korea) according to manufactures’ instructions. Determination of the relative absorbance ratio at A260/A280 and A260/A230 by a spectrophotometer (Nano Drop 2000, Thermo Scientific, USA) was exerted to assess the extracted RNA concentration and purity. Then, template RNA was reverse-transcribed by PrimeScript 1st strand cDNA Synthesis Kit (TAKARA, Japan) following the manufacturer’s guidelines using Thermal Cycler instrument (Eppendorf, Germany). Real-time mRNA expression of S100A4, S100A9, and S100A10 was conducted using TaqMan-based approach using ABI StepOnePlus real-time PCR System (Applied Biosystems, Foster City, CA, USA). The real-time analyses were conducted in triplicate order. The transcript level of β-Actin was measured as housekeeping gene to normalize the expression levels of target genes. The comparative threshold cycle method (2−∆∆ct) was exerted to measure the relative amounts of target genes in each sample.
Statistical analysis
The Statistical Package for the Social Sciences (SPSS, Chicago, IL, USA) software for windows version 23 was used to analysis of data. The Kolmogorov–Smirnov test was exerted to normality evaluation of Scale variables. Group comparisons of non-parametric variables were conducted via the Mann–Whitney U test. To determine the relationship between scale variables, Spearman’s correlations were used. The receiver operating characteristic (ROC) curves were measured and the area under the curve (AUC) was calculated for S100A4, S100A9, and S100A10 to determine the sensitivity and specificity of these molecules to predict severe form of COVID-19. For plotting the graphs, the GraphPad Prism version 8.00 for Windows (GraphPad Software, La Jolla, CA, USA) was applied. The study results were presented as mean ± Standard deviation (SD). P values < 0.05 were considered as statistically significant.
Results
Baseline data of the patients
The baseline characteristics of the study population in the study are listed in the Table 1. The COVID-19 group contained 65 patients, with a mean age of 52.7 ± 11.3 years old, involving 36 (55.4%) males and 29 (44.6%) females. In the control group, a total of 50 subjects containing 27 (54%) males and 23 (46%) females with a mean age of 50.4 ± 10.8 years old were included. The patient and control groups were age- and sex-matched. In the patients and control groups, 27 (41.5%) and 21 (42%) individuals were smokers, respectively.
Patient group consisted of 43 (66.2%) mild-to-moderate COVID-19 cases and 22 (33.8%) severe cases. No significant differences were observed in terms of clinicopathological manifestations between COVID-19 patients with mild-to-moderate and severe disease.
Expression level of S100A4, S100A9, and S100A10
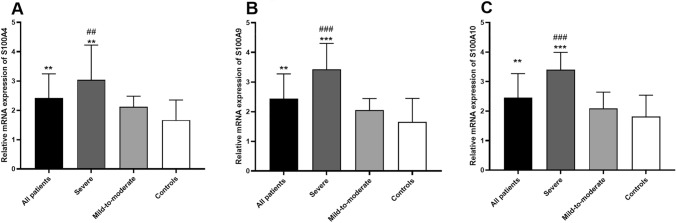
It was observed that the expression of S100A4 mRNA was significantly upregulated in the peripheral blood of COVID-19 patients in comparison to the healthy controls (fold change = 1.45, P = 0.0011; Fig. 1A). Moreover, a significant overexpression of S100A9 mRNA was detected in the peripheral blood samples from COVID-19 cases compared with healthy subjects (fold change = 1.47, P = 0.0013; Fig. 1B). In addition, mRNA of S100A10 was significantly upregulated in the peripheral blood of COVID-19 cases in comparison to the control group (fold change = 1.35, P = 0.0053; Fig. 1C).
Fig. 1.
Transcript level of S100A4 (A), S100A9 (B), and S100A10 (C) in the peripheral blood samples from the all COVID-19 cases, those with severe form of the diseases, cases with mild-to-moderate disease form, and healthy controls (* shows comparison with healthy control, # shows comparison with mild-to-moderate group; ** shows a P < 0.01, *** show a P < 0.001, ## shows a P < 0.01, ### show a P < 0.001)
The mRNA expression of S100A4 was significantly upregulated in the peripheral blood samples from severe COVID-19 subjects compared with mild-to-moderate subjects (fold change = 1.43, P = 0.0071; Fig. 1A). Additionally, a significant overexpression of S100A9 mRNA was detected in the peripheral blood samples from severe COVID-19 cases compared with mild-to-moderate COVID-19 patients (fold change = 1.66, P = 0.0001; Fig. 1B). Furthermore, mRNA of S100A10 was significantly overexpressed in the peripheral blood of severe COVID-19 cases in comparison to those with mild-to-moderate form of the disease (fold change = 1.63, P = 0.0003; Fig. 1C).
It was seen that the mRNA expression of S100A4 was significantly upregulated in the peripheral blood samples from severe COVID-19 subjects in comparison to the control group (fold change = 1.82, P = 0.0015; Fig. 1A). Moreover, a significant overexpression of S100A9 mRNA was detected in the peripheral blood samples from severe COVID-19 cases compared with healthy controls (fold change = 2.07, P = 0.0002; Fig. 1B). Also, S100A10 mRNA was significantly overexpressed in the peripheral blood of severe COVID-19 cases in comparison to healthy control group (fold change = 1.87, P = 0.0002; Fig. 1C).
No statistically significant differences were detected in the mRNA expression of S100A4, S100A9, and S100A10 between mild-to-moderate COVID-19 cases and healthy controls (Fig. 1).
Correlation analysis
The results of correlation analysis between clinicopathological characteristics of the patients with the mRNA expression of S100A4, S100A9, and S100A10 are shown in Table 2.
Table 2.
Correlation of S100A4, S100A9, and S100A10 with clinical presentations of the COVID-19 patients
| Item | S100A4 | S100A9 | S100A10 |
|---|---|---|---|
| Age | ρ = 0.25, P = 0.312 | ρ = 0.11, P = 0.490 | ρ = 0.08, P = 0.826 |
| Duration of COVID-19 | ρ = 0.24, P = 0.083 | ρ = 0.10, P = 0.231 | ρ = 0.16, P = 0.419 |
| Oxygen saturation | ρ = 0.43, P = 0.048 | ρ = 0.16, P = 0.345 | ρ = 0.42, P = 0.281 |
| Systolic BP | ρ = 0.17, P = 0.461 | ρ = 0.18, P = 0.520 | ρ = 0.21, P = 0.313 |
| Diastolic BP | ρ = 0.09, P = 0.309 | ρ = 0.08, P = 0.814 | ρ = 0.11, P = 0.636 |
| WBC | ρ = 0.51, P = 0.026 | ρ = 0.31, P = 0.068 | ρ = 0.31, P = 0.068 |
| Neutrophil–lymphocyte ratio | ρ = 0.54, P = 0.014 | ρ = 0.49, P = 0.028 | ρ = 0.15, P = 0.441 |
| ALP | ρ = 0.14, P = 0.416 | ρ = 0.13, P = 0.298 | ρ = 0.30, P = 0.480 |
| AST | ρ = 0.11, P = 0.216 | ρ = 0.10, P = 0.544 | ρ = 0.16, P = 0.203 |
| ALT | ρ = 0.15, P = 0.305 | ρ = 0.19, P = 0.651 | ρ = 0.20, P = 0.613 |
| LDH | ρ = 0.59, P = 0.011 | ρ = 0.61, P = 0.017 | ρ = 0.58, P = 0.038 |
| CRP | ρ = 0.71, P = 0.001 | ρ = 0.74, P = 0.006 | ρ = 0.68, P = 0.002 |
| ESR | ρ = 0.65, P = 0.018 | ρ = 0.70, P = 0.007 | ρ = 0.66, P = 0.010 |
| BMI | ρ = 0.32, P = 0.608 | ρ = 0.28, P = 0.216 | ρ = 0.22, P = 0.513 |
| Total cholesterol | ρ = 0.16, P = 0.617 | ρ = 0.19, P = 0.264 | ρ = 0.26, P = 0.610 |
| TG | ρ = 0.30, P = 0.144 | ρ = 0.36, P = 0.213 | ρ = 0.11, P = 0.394 |
| LDL | ρ = 0.23, P = 0.855 | ρ = 0.20, P = 0.462 | ρ = 0.19, P = 0.498 |
| HDL | ρ = 0.23, P = 0.411 | ρ = 0.26, P = 0.574 | ρ = 0.10, P = 0.207 |
| Creatinine | ρ = 0.24, P = 0.320 | ρ = 0.22, P = 0.506 | ρ = 0.31, P = 0.233 |
| BUN | ρ = 0.13, P = 0.511 | ρ = 0.10, P = 0.413 | ρ = 0.32, P = 0.084 |
| FBS | ρ = 0.15, P = 0.604 | ρ = 0.20, P = 0.210 | ρ = 0.19, P = 0.077 |
| D-dimer | ρ = 0.49, P = 0.030 | ρ = 0.55, P = 0.009 | ρ = 0.39, P = 0.040 |
Bold values show statistically significant correlations
COVID-19 coronavirus disease 2019; WBC white blood cell; CRP C-reactive protein; ALP alkaline phosphatase; AST aspartate aminotransferase; ALT alanine aminotransferase; LDH lactate dehydrogenase; ESR erythrocyte sedimentation rate; BMI body mass index; FBS fasting blood sugar; TG triglyceride; LDL low density lipoprotein; HDL high density lipoprotein; BUN blood urea nitrogen; OR odds ratio; CI confidence interval; BP blood pressure
It was seen that there was a significant positive correlation between percentage of oxygen saturation (ρ = 0.43, P = 0.048), WBC count (ρ = 0.51, P = 0.026), neutrophil–lymphocyte ratio (ρ = 0.54, P = 0.014), LDH (ρ = 0.59, P = 0.011), CRP (ρ = 0.71, P = 0.001), ESR (ρ = 0.65, P = 0.016), and d-dimer (ρ = 0.49, P = 0.030) in the COVID-19 patients with the mRNA expression of S100A4.
Neutrophil–lymphocyte ratio (ρ = 0.49, P = 0.028), LDH (ρ = 0.61, P = 0.017), CRP (ρ = 0.74, P = 0.006), ESR (ρ = 0.70, P = 0.007), and d-dimer (ρ = 0.55, P = 0.009) were correlated significantly with mRNA expression of S100A9 in the COVID-19 patients.
There was a significantly direct correlation of S100A10 mRNA expression with LDH (ρ = 0.58, P = 0.038), CRP (ρ = 0.68, P = 0.002), ESR (ρ = 0.66, P = 0.010), and d-dimer (ρ = 0.39, P = 0.040) in COVID-19 cases.
Potential of S100A molecules as biomarkers in COVID-19
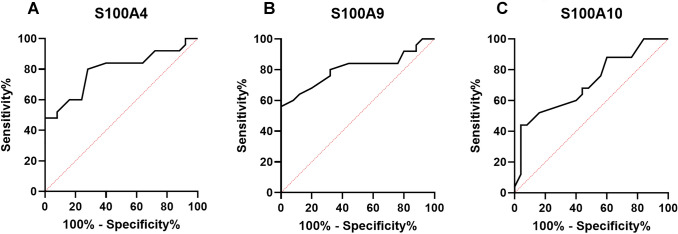
The ROC curves were plotted and the AUC was calculated for S100A4, S100A9, and S100A10 to determine the sensitivity and specificity of these molecules in distinguishing between severe form and mild-to-moderate form of COVID-19. It was observed that AUC for S100A4 mRNA was 0.79 (95% CI 0.66–0.92, P = 0.004; Fig. 2A), for S100A9 was 0.80 (95% CI 0.67–0.93, P = 0.002; Fig. 2B), and for S100A10 was 0.71 (95% CI 0.56–0.85, P = 0.010; Fig. 2C).
Fig. 2.
The ROC curves were plotted and the AUC was calculated for S100A4 (A), S100A9 (B), and S100A10 (C) to determine the sensitivity and specificity of these molecules in distinguishing between severe and mild-to-moderate forms of COVID-19. According to ROC cure analyses, AUC for S100A4 mRNA was 0.79 (95% CI 0.66–0.92, P = 0.004), for S100A9 was 0.80 (95% CI 0.67–0.93, P = 0.002), and for S100A10 was 0.71 (95% CI 0.56–0.85, P = 0.010)
Discussion
In this study, we intended to divulge the involvement of S100A4, S100A9, and S100A10 molecules in the inflammatory settings of COVID-19 patients. Then evaluated the diagnostic value of these molecules in promoting severe forms of COVID-19. Our experiments indicated upregulation of S100A4, S100A9, and S100A10 mRNAs in the peripheral blood samples from COVID-19 cases compared to healthy individuals. In addition, the mRNA expression of S100A4, S100A9, and S100A10 molecules conferred a value in discriminating severe form of COVID-19.
The coagulation system has been identified to be in a hyperactive status in most of theCOVID-19 cases with severe form of infection [20]. In fact, several patients show a condition that demonstrate a prothrombotic diathesis, such as increased levels of fibrin degradation products (d-dimer), higher levels of fibrinogen, and decreased levels of antithrombin [20, 21]. Additionally, d-dimer level was reported to be associated with disease severity and was indicated to be a valid prognostic biomarker for in-hospital death in subjects admitted due to COVID-19 [22]. In the hospitalized COVID-19 patients, an increased levels of neutrophil extracellular traps (NETs) were found in comparison to the healthy group [23]. Moreover, the remnants of NETs (such as myeloperoxidase-DNA complexes, cell-free DNA, and citrullinated histone H3) and neutrophil-derived S100A8/A9 heterodimer in the serum of patient were related to increased risk of dysregulated thrombotic events regardless of prophylactic anticoagulation treatment [24]. NETs are extracellular webs of chromatin and microbicidal proteins that are considered as an innate immune mechanism during infections. Nonetheless, NETs are also able to stimulate and perpetuate thrombosis [21]. In our investigation, we detected that S100A9 mRNA was upregulated in the COVID-19 patients compared to the control group as well as in the severe COVID-19 subjects compared to mild-to-moderate patients. Interestingly, there was a correlation between the mRNA levels of S100A9 and d-dimer levels in the COVID-19 patients. As a consequence, S100A9 might be involved in the coagulopathy in the COVID-19 patients. It should be noted that 16 (24.6%) cases in our study were using anticoagulants, but the mRNA expression of S100A4 was not different in those with and without anticoagulation therapy. Therefore, mechanistic inhibition of the coagulation pathway in the COVID-19 patients (probably through inhibition of S100A9) might confer better therapeutic tool in controlling coagulopathy in the COVID-19 patients.
Previous investigations have indicated that the cell-surface generation of plasmin is needed for macrophage recruitment, which is partially mediated via the plasmin-dependent activation of MMP-9 [25]. As a result, macrophage-generated plasmin might directly hydrolyze extracellular matrix (ECM) components as well as activate MMP-9, hence increasing further degradation of ECM. It was observed that S100A10 is involved in the macrophage plasmin generation that is needed for ECM hydrolysis and MMP-9 activation. Moreover, S100A10 deficiency was associated with decreased migration capacity of macrophages [16]. Here we detected increased levels of S100A10 in the peripheral blood samples from COVID-19 patients. On the other hand, increased levels of MMP-9 as well as molecules involved in the adhesion and infiltration of leukocytes were detected in the COVID-19 patients [26]. It seems that S100A10, by activation of pro-MMP-9, along with increased levels of adhesion molecules, such as Intercellular adhesion molecule 1 (ICAM-1), Vascular cell adhesion protein 1 (VCAM-1), and E-selectin plays a role in the recruitment of inflammatory cells (potentially to the lung tissue) and immunopathogenesis of COVID-19.
A study indicated that COVID-19 subjects hospitalized in general wards had significantly increased level of S100A8/A9 in comparison to healthy subjects [19]. Moreover, S100A8/A9 was increased in COVID-19 patients hospitalized in ICU compared with those without hospitalization in ICU, or in patients with fatal outcomes compared with the alive patients, implying that increased levels of S100A8/A9 was associated with high mortality rate in COVID-19 subjects. Additionally, levels of S100A8/A9 in serum during the admission time were correlated positively with peak CT score as well as demand for oxygen. Moreover, the peak level of d-dimer was increased as the serum S100A8/A9 elevated. On the other hand, the ratio of neutrophils to lymphocytes was positively correlated with the serum S100A8/A9 [19]. The serum S100A8/A9 was associated with more severe sepsis-related organ dysfunction. Also, the serum level of S100A8/A9 was associated with COVID-GRAM risk score, which is COVID-19 critical illness prediction tool and has been designed to predict occurrence of ICU admission, mechanical ventilation, or death in hospitalized patients with COVID-19 [27]. Our investigation indicated a correlation between mRNA expression of S100A4, S100A9, and S100A10 and LDH levels, implying to the involvement of these molecules in the severity of acute lung injury and ARDS. Moreover, we detected that mRNA expression of S100A4, S100A9, and S100A10 was correlated with neutrophils to lymphocytes ratio in the COVID-19 cases. This observation suggests that S100A4, S100A9, and S100A10 might be involved in the reduction of the peripheral blood lymphocytes in the COVID-19 patients. This might stem from higher infiltration of peripheral blood lymphocytes to the lung tissue, as S100A10 (in particular) might contributes to the infiltration of leukocytes in the COVID-19 patients.
It was observed that the AUCs for COVID-GRAM risk scores with S100A8/A9 was 0.810 for the prediction of ICU admission. For the prediction of subsequent death, the AUCs for COVID-GRAM risk scores by S100A8/A9 was 0.818. Additionally, higher S100A8/A9 level culminated in significant worse overall survival. The researchers classified the COVID-19 patients into low- or high-level groups according to the concentrations of S100A8/A9 at a cutoff of 6195 ng/ml, and into high-risk or non-high-risk groups according to COVID-GRAM risk scores. It was detected that the hazard ratio (HR) of high S100A8/A9 level was 13.32, which was higher than that of COVID-GRAM risk score (HR = 4.612). It was concluded that the concentrations of S100A8/A9 measured during hospital admission had better predictive value than COVID-GRAM risk scores for subsequent death in COVID-19 cases [19]. We also determined the ROC curves and measured the AUC was calculated for S100A4, S100A9, and S100A10 to determine the sensitivity and specificity of these molecules in distinguishing between severe form and mild-to-moderate form of COVID-19. It was observed that AUC for S100A4 mRNA was 0.79 (P = 0.004), for S100A8 was 0.80 (P = 0.002), and for S100A10 was 0.71 (P = 0.010). Therefore, evaluation of the serum levels of S100A4, S100A9, and S100A10 might be a promising approach in prognosis of the sever forms of COVID-19, ICU hospitalization, as well as the risk of mortality due to the infection.
Studies have reported that serum level of S100A8/A9 was correlated with both the severity of pathogen-associated tissue damage and excessive cytokine storm [28]. Chen et al. [19] assessed association between S100A8/A9 and cytokine storm in COVID-19. They observed a correlation between the serum levels of S100A8/A9 with the concentrations of a different spectrum of pro-inflammatory cytokines, most importantly IL-8, MCP-3, MCP-1, IL-1ra, β-NGF, IL-7, IL-10, RANTES, G-CSF, IL-1α, CTACK and IL-17A. Furthermore, myeloid chemokines IL-8, MCP-3 and MCP-1 were among the most significant mediators associated with levels of S100A8/A9. Our study also detected a significant correlation between mRNA expression of S100A4, S100A9, S100A10 with inflammatory indices (CRP, ESR) as well as increased leukocytosis in the COVID-19 patients. Therefore, S100A family molecules might regulate cytokine release syndrome and promote recruitment of monocytes and neutrophils to the involved sites in the COVID-19 patients.
In conclusion, our study found a significant association between S100A4, S100A9, S100A10 molecules and the inflammatory settings as well as severity of COVID-19 patients. Moreover, these molecules indicated a potential in the prognosis of the severity, hospitalization in ICU ward, and probably requirement for supportive cares (like mechanical ventilation) in the COVID-19 patients. In addition, due to the involvement of S100A10 in the coagulopathies, COVID-19 patients with higher levels of S100A10 might be demanding in receiving anticoagulation therapies. Nonetheless, further investigations are warranted to comprehensively clarify the implication of S100A4, S100A9, and S100A10 in the pathogenesis of COVID-19, which might open up new horizons in devising efficient prognostic and therapeutic tools for patients with COVID-19 infection.
Acknowledgements
The authors are grateful to the patients for their participation in this study.
Funding
This article was supported by a grant from Deputy of Research, Rafsanjan University of Medical Sciences.
Data availability
All data that support the conclusions of this manuscript are included within the article.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This study was approved by the ethics committee of Rafsanjan University of Medical Sciences and all individuals voluntarily signed a written informed consent form.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner AJ, Hiscox JA, Hooper NM. ACE2: from vasopeptidase to SARS virus receptor. Trends Pharmacol Sci. 2004;25:291–294. doi: 10.1016/j.tips.2004.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020;24:422–422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagheri-Hosseinabadi Z, Pirsadeghi A, Rahnama A, Bahrehmand F, Abbasifard M. Is there any relationship between serum zinc levels and angiotensin-converting enzyme 2 gene expression in patients with coronavirus disease 2019? Meta Gene. 2022;31:100991. doi: 10.1016/j.mgene.2021.100991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Organization WH. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected: interim guidance, 13 March 2020: World Health Organization, 2020.
- 7.Han S, Mallampalli RK. The acute respiratory distress syndrome: from mechanism to translation. J Immunol. 2015;194:855–860. doi: 10.4049/jimmunol.1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbasifard M, Khorramdelazad H. The bio-mission of interleukin-6 in the pathogenesis of COVID-19: a brief look at potential therapeutic tactics. Life Sci. 2020;257:118097. doi: 10.1016/j.lfs.2020.118097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagheri-Hosseinabadi Z, Ostad Ebrahimi H, Bahrehmand F, Taghipour G, Abbasifard M. The relationship between serum levels of interleukin-2 and IL-8 with circulating microRNA-10b in patients with COVID-19. Iran J Immunol. 2021;18:65–73. doi: 10.22034/iji.2021.88780.1904. [DOI] [PubMed] [Google Scholar]
- 10.Ebrahimi N, Aslani S, Babaie F, Hemmatzadeh M, Hosseinzadeh R, Joneidi Z, et al. Recent findings on the coronavirus disease 2019 (COVID-19); immunopathogenesis and immunotherapeutics. Int Immunopharmacol. 2020;89:107082. doi: 10.1016/j.intimp.2020.107082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruys E, Toussaint M, Niewold T, Koopmans S. Acute phase reaction and acute phase proteins. J Zhejiang Univ Sci B. 2005;6:1045. doi: 10.1631/jzus.2005.B1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong L, Lan W, Lim RR, Chaurasia SS. S100A proteins as molecular targets in the ocular surface inflammatory diseases. Ocul Surf. 2014;12:23–31. doi: 10.1016/j.jtos.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Ambartsumian N, Klingelhöfer J, Grigorian M. The multifaceted S100A4 protein in cancer and inflammation. In: Heizmann CW, editor. Calcium-binding proteins of the EF-hand superfamily. New York, NY: Humana Press; 2019. pp. 339–365. [DOI] [PubMed] [Google Scholar]
- 14.Lee T-H, Chang HS, Bae D-J, Song HJ, Kim M-S, Park JS, et al. Role of S100A9 in the development of neutrophilic inflammation in asthmatics and in a murine model. Clin Immunol. 2017;183:158–166. doi: 10.1016/j.clim.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in inflammation. Front Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connell PA, Surette AP, Liwski RS, Svenningsson P, Waisman DM. S100A10 regulates plasminogen-dependent macrophage invasion. Blood. 2010;116:1136–1146. doi: 10.1182/blood-2010-01-264754. [DOI] [PubMed] [Google Scholar]
- 17.Palmer LD, Maloney KN, Boyd KL, Goleniewska AK, Toki S, Maxwell CN, et al. The innate immune protein S100A9 protects from T-helper cell type 2–mediated allergic airway inflammation. Am J Respir Cell Mol Biol. 2019;61:459–468. doi: 10.1165/rcmb.2018-0217OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, et al. Concurrent human antibody and TH1 type T-cell responses elicited by a COVID-19 RNA vaccine. MedRxiv. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Long X, Xu Q, Tan J, Wang G, Cao Y, et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell Mol Immunol. 2020;17:992–994. doi: 10.1038/s41423-020-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabrizi ZA, Khosrojerdi A, Aslani S, Hemmatzadeh M, Babaie F, Bairami A, et al. Multi-facets of neutrophil extracellular trap in infectious diseases: moving beyond immunity. Microb Pathog. 2021;158:105066. doi: 10.1016/j.micpath.2021.105066. [DOI] [PubMed] [Google Scholar]
- 22.Yao Y, Cao J, Wang Q, Shi Q, Liu K, Luo Z, et al. d-dimer as a biomarker for disease severity and mortality in COVID-19 patients: a case control study. J Intensive Care. 2020;8:1–11. doi: 10.1186/s40560-020-00466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, et al. Neutrophil extracellular traps in COVID-19. JCI insight 2020; 5. [DOI] [PMC free article] [PubMed]
- 24.Zuo Y, Zuo M, Yalavarthi S, Gockman K, Madison JA, Shi H, et al. Neutrophil extracellular traps and thrombosis in COVID-19. J Thromb Thrombolysis. 2021;51:446–453. doi: 10.1007/s11239-020-02324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong Y, Hart E, Shchurin A, Hoover-Plow J. Inflammatory macrophage migration requires MMP-9 activation by plasminogen in mice. J Clin Investig. 2008;118:3012–3024. doi: 10.1172/JCI32750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammadhosayni M, Mohammadi FS, Ezzatifar F, Gorabi AM, Khosrojerdi A, Aslani S, et al. Matrix metalloproteinases are involved in the development of neurological complications in patients with coronavirus disease 2019. Int Immunopharmacol. 2021;100:108076. doi: 10.1016/j.intimp.2021.108076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang W, Liang H, Ou L, Chen B, Chen A, Li C, et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan JK, Roth J, Oppenheim JJ, Tracey KJ, Vogl T, Feldmann M, et al. Alarmins: awaiting a clinical response. J Clin Investig. 2012;122:2711–2719. doi: 10.1172/JCI62423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data that support the conclusions of this manuscript are included within the article.