Abstract
A variety of cancer entities are driven by KRAS mutations, which remain difficult to target clinically. Survival pathways, such as resistance to cell death, may represent a promising treatment approach in KRAS mutated cancers. Based on the frequently observed genomic deletions of BCL-2-related ovarian killer (BOK) in cancer patients, we explored the function of BOK in a mutant KrasG12D-driven murine model of lung cancer. Using KrasG12D/+ Bok−/− mice, we observed an overall tumor-promoting function of BOK in vivo. Specifically, loss of BOK reduced proliferation both in cell lines in vitro as well as in KrasG12D-driven tumor lesions in vivo. During tumor development in vivo, loss of BOK resulted in a lower tumor burden, with fewer, smaller, and less advanced tumors. Using KrasG12D/+ Tp53Δ/Δ Bok−/− mice, we identified that this phenotype was entirely dependent on the presence of functional p53. Furthermore, analysis of a human dataset of untreated early-stage lung tumors did not identify any common deletion of the BOK locus, independently of the TP53 status or the histopathological classification. Taken together our data indicate that BOK supports tumor progression in Kras-driven lung cancer.
Subject terms: Cancer models, Non-small-cell lung cancer
Introduction
Despite recent therapeutic advances, including the development of immune checkpoint inhibitors and other targeted therapies, lung cancer remains the leading cause of cancer-related deaths worldwide [1]. Lung adenocarcinoma (LUAD) is the most common subtype of lung cancer and is associated with significant rates of morbidity and mortality. About 33% of LUAD cases are associated with activating mutations in KRAS [2], which are difficult to target in a clinical setting. Furthermore, 46% of LUAD patients have somatic mutations in the tumor suppressor TP53––one of the most common events in cancer [2]. TP53 defends cells from acquired stress by triggering a multitude of anti-proliferative and stress-response signals such as senescence, cell cycle arrest, coordination of DNA damage repair, or apoptosis [3, 4].
Notably, defects in apoptosis can promote tumorigenesis per se through extending the life span of a cell, therefore increasing its chances of acquiring additional cancer-promoting mutations [5]. Furthermore, apoptosis inhibition enables cell resistance to stress caused by oncogenic alterations, nutrient deprivation, DNA damage, or the build-up of toxic components [6]. Under normal conditions, apoptosis is controlled by the BCL-2 protein family [7], which comprises both pro- and anti-apoptotic members. Apoptotic signals result in permeabilization of the mitochondrial outer membrane, caspase activation, and cell death [7]. Interestingly, several cancer entities have altered expression of BCL-2 family members [7–9].
The BCL-2 family member BOK (BCL-2-related ovarian killer) [10] is a multi-BH-domain family member with sequence and structural homology to pro-apoptotic BAX and BAK [11, 12]. BOK localizes primarily to the endoplasmic reticulum (ER) [13], where its activity is negatively regulated by ER-associated degradation [14]. Furthermore, BOK is a critical inducer of apoptosis in response to ER stress [15]. Notably, BOK has also been linked to the pathway of TP53-mediated cellular response to DNA damage, with Zhang et al. showing increased levels of damage in the absence of BOK [16, 17]. Recent data point to a role for BOK also in cell proliferation [18] and Srivastava et al. suggested that BOK interacts with the UMP synthetase enzyme (UMPS), aiding its function in nucleotide synthesis and thus promoting cellular proliferation [19].
While its function and purpose have remained in part elusive, Beroukhim et al. found that the genomic locus containing BOK amongst several other genes is frequently deleted across several cancer entities [20]. These data were initially interpreted as pointing to a previously unknown function of BOK as a tumor suppressor and thus underlining the need for further investigation.
Using an inducible model of Kras-driven murine lung cancer, we show that BOK plays a role in promoting tumor development, as Bok-proficient mice developed a larger number of more advanced lesions. This effect is mediated, at least in part, by defects in proliferation, and is entirely dependent on p53. Furthermore, we show that the BOK locus is not genetically deleted in human lung cancer naïve samples. Collectively, our data point towards a function of BOK in promoting the development of lung adenocarcinomas.
Results
Bok deletion reduces tumor burden
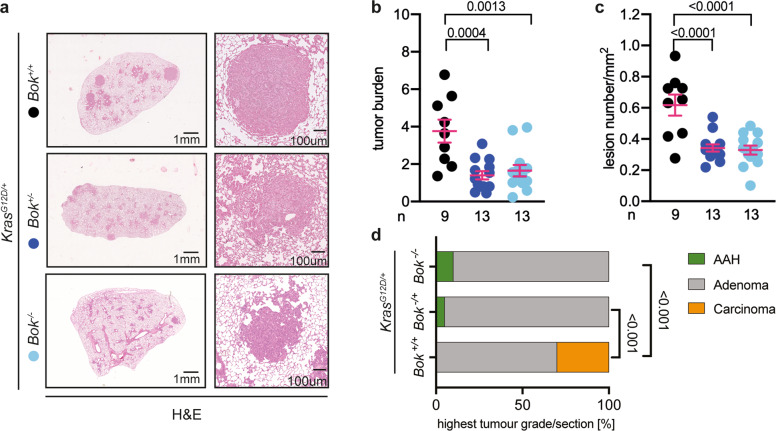
To analyze the role of BOK in lung cancer, we used mice harboring a lox-stop-lox-KrasG12D allele, in which the mutated Kras is expressed upon AdenoCre viral infection of the lungs [21]. Through this synchronized induction of tumor development, we compared the development of lung cancer between littermate mice with two copies (Bok+/+), one copy (Bok+/−) or none (Bok−/−) of the Bok gene. Intriguingly, KrasG12D/+Bok−/− and KrasG12D/+Bok+/− mice showed significantly lower tumor burden than the Bok-proficient animals (Fig. 1a, b). This was due to a decrease in overall lesion number (Fig. 1c). We then histologically graded the most advanced lesion present in the lungs of experimental mice [21]. Whereas Bok−/− and Bok−/+ mice more frequently presented with histologically well-differentiated lesions, Bok-proficient mice presented with more advanced lesions (Fig. 1d). Notably, most advanced lesions (carcinoma) were only observed in the Bok− proficient mice. These data indicate that loss of BOK leads to a lower tumor burden and lesion progression in Kras-driven lung cancer.
Fig. 1. Deletion of Bok reduces tumor burden.
a Representative images of H&E staining of lungs of the indicated genotypes 19 weeks after infection with 5 × 106 PFU of AdenoCre. Panels on the right are higher magnification. Scale bars represent 1 mm or 100 µm, respectively. b Quantification of the H&E staining expressed as the ratio between the area of the lesions over the total lung area. Data were analyzed by one-way ANOVA (p = 0.0002, F(2,32) = 10.93). Reported in the figure as p values from the post hoc analysis with Bonferroni correction. c Quantification of the H&E staining expressed as number of lesions per mm2. Data were analyzed by one-way ANOVA (p < 0.0001, F(2,32) = 15.51) and are reported in the figure as p values from the post hoc analysis with Bonferroni correction. d Percentage of slides that showed at least one lesion histologically graded as indicated by color coding from mice of the indicated genotypes at 19 weeks post-infection. Data were analyzed by chi-square test (p < 0.001). For the quantifications reported in panel (b–d), three sections per animal, separated by 100 µm, were analyzed. n = number of animals. Data in panel (b, c) are presented as dot plot (every dot represents one experimental animal) and report mean ± SEM. Color code as in panel (a).
Bok deletion decreases proliferation
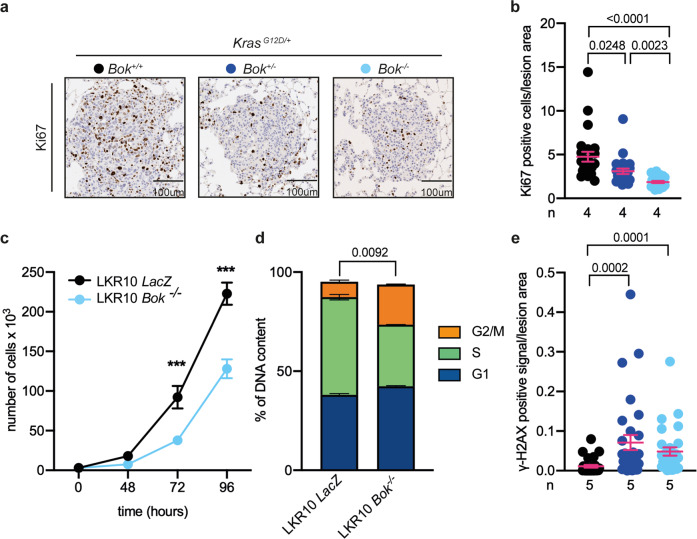
As the proliferative rate is an important characteristic of malignant lesions and BOK has been shown to affect proliferation [18, 19], we analyzed Ki67 in our setting, a well-established immunohistochemical marker of proliferation. Consistent with a prior publication on hepatocellular carcinoma [18], Bok+/+ lesions showed significantly more Ki67 positive cells than Bok−/− lesions (Fig. 2a, b). However, we did not observe any differences in the number apoptotic cells in Bok+/+, Bok+/− or Bok−/− lesions (Fig. S1).
Fig. 2. Bok-deficiency correlates with a lower proliferation rate and higher levels of DNA damage.
a, b Representative images and quantification of Ki67 staining. Data are reported as number of positive cells normalized to lesion area and analyzed by Kruskal–Wallis test (p < 0.0001, H(3) = 36.26). In panel (b), the p values from the post hoc analysis with Dunn´s correction are reported. c Live cell numbers evaluated as trypan blue exclusion over time. Data are presented as mean ± SEM and were analyzed by repeated measure two-way ANOVA (time, genotype, and interaction effects, p < 0.0001). Post hoc analysis with Bonferroni correction showed a significant difference between the genotypes at 72 h and 96 h (***p < 0.0001). d Cell cycle analysis performed with propidium Iodide (PI) staining. Data are presented as mean ± SEM and were analyzed by chi-square test (p = 0.0092, chi-square (2) = 9.373). e Quantification of γ-H2AX positive staining normalized to lesion area. Data were analyzed by Kruskal–Wallis test (p < 0.0001, H(3) = 22.34) and are reported in the figure as p values from the post hoc analysis with Dunn´s correction. For the quantification in panel (b) and (e), six lesions per slide/animal were evaluated and data are presented as dot plot and report mean ± SEM. For panel (c) and (d), each cell line was evaluated in at least three independent experiments.
To further explore the role of BOK, we used the mutant Kras-driven mouse lung adenocarcinoma cell line LKR10 [22] to generate Bok-deficient cells by CRISPR/Cas9 (LKR10 Bok−/− and, as control, LKR10 LacZ, Fig. S2a). Bok knockout had no effects on the protein expression of its close relatives BAK and BAX, or its well-established pro-survival interaction partner MCL-1 [10, 19] (Fig. S2b). As expected, we found that Bok-deficient cells proliferate slower than their counterparts (Fig. 2c). Furthermore, cell cycle analysis revealed that Bok-deficient cells show longer G2 duration (Fig. 2d), suggesting persistent DNA damage. Following this data, we analyzed the levels of phosphorylated histone γ-H2AX, an indicator of DNA double-strand breaks, in our experimental animals. In line with our hypothesis, we detected higher levels of γ-H2AX in Bok-deficient lesions (Fig. 2e). Taken together, these data show that BOK impacts on proliferation as well as on DNA damage repair response, which could explain the decreased tumor burden observed in the absence of BOK.
Functional Tp53 mediates the Bok− dependent phenotype
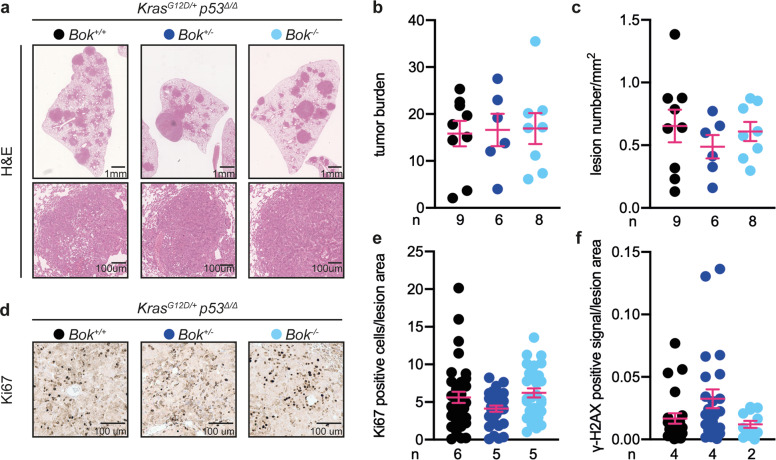
Since alterations in the TP53 tumor suppressor gene occur in about 50% of NSCLC cases [23], we investigated the contribution of functional p53 signaling to the effects of BOK in lung adenocarcinoma by crossing the KrasG12D Bok−/− mice in a Tp53fl/fl background. We generated mice in which the administration of Cre recombinase allowed for the simultaneous deletion of the tumor suppressor p53 as well as KrasG12D activation [23]. Notably, analysis of tumor burden showed no differences across genotypes (Fig. 3a–c), suggesting that the function of BOK is dependent on the presence of p53 in this model of tumorigenesis. Accordingly, proliferative rates and DNA damage levels were similar in Bok-deficient and -proficient mice (Fig. 3d–f). Knockout of p53 also rescued the defect in cell cycle progression in the Bok-deficient mouse lung adenocarcinoma cell line LKR10 (Fig. S3), supporting the findings in the KrasG12D/+ Tp53Δ/Δ mouse model.
Fig. 3. The effect of Bok in murine NSCLC is dependent on functional p53.
a Representative images of H&E staining of lungs of the indicated genotypes 13 weeks after infection with 5 × 106 PFU of AdenoCre. Lower panels are higher magnification of indicated areas. Scale bars represent 1 mm or 100 µm, respectively. b Quantification of the H&E staining expressed as the ratio between the area of the lesions over the total lung area. Data were analyzed by one-way ANOVA (p = 0.9654, F(2,20) = 0.0325). c Quantification of the H&E staining expressed as number of lesions per mm2. Data were analyzed by one-way ANOVA (p = 0.5803, F(2,20) = 0.5593). d, e Representative images, and quantification of Ki67 staining. Data are reported as number of positive cells normalized to lesion area. Data were analyzed by Kruskal–Wallis test (p = 0.1013, H(3) = 4.579). f Quantification of γ-H2AX staining. Data are reported as percentage of positive staining normalized to lesion area. Data were analyzed by Kruskal–Wallis test (p = 0.0783, H(3) = 5.094). For the quantifications reported in panel (b, c), three sections per animal, separated by 100 µm, were analyzed. For the quantification in panel (e) and (f), six lesion per slide/animal were evaluated. n = number of animals. Data in panel (b, c, e, and f) are presented as dot plot and report mean ± SEM.
Bok-deficiency sensitizes NSCLC cells against DNA damaging agents
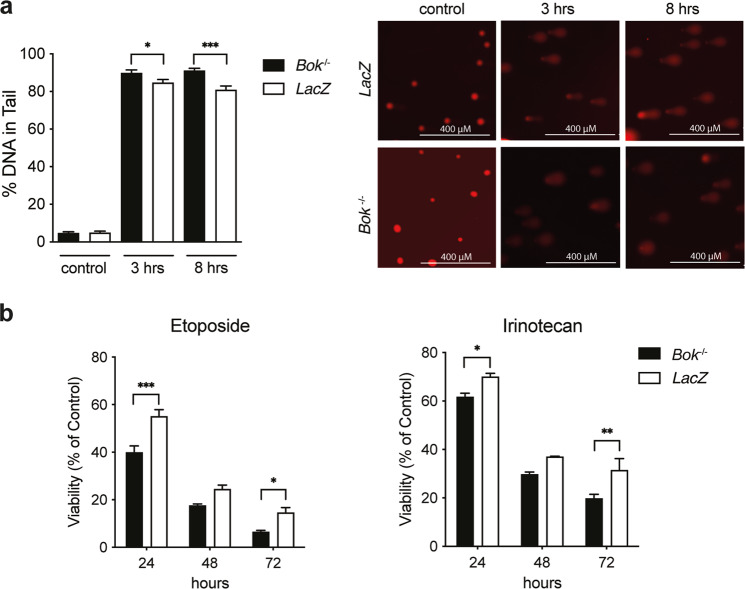
As our previous data suggested that Bok-deficient lung cancer cells have a defect in DNA repair leading to a p53-dependent arrest in G2 (Figs. 2 and S3), we wanted to determine if Bok-deficient cells are thus more sensitive to DNA damaging agents. First, to measure differences in DNA damage and DNA repair capacity in Bok-deficient versus Bok-proficient lung cancer cells upon induction of DNA damage, we performed comet assays in the LKR10 LacZ and Bok−/− cells treated with Etoposide. As expected, Etoposide-treated Bok-deficient cells had significantly higher levels of DNA damage (% DNA in tail) than the LacZ control cells, indicating that Bok-deficient cells have a decreased capacity to efficiently repair DNA strand brakes (Fig. 4a). Accordingly, LKR10 Bok−/− cells showed significantly increased sensitivity to DNA damaging standard chemotherapeutics such as Etoposide and Irinotecan (Fig. 4b). These results highlight the important role of BOK for DNA repair processes in lung cancer cells.
Fig. 4. Bok-deficiency sensitizes against DNA damaging agents.
a DNA damage in Etoposide-treated LKR10 LacZ and Bok−/− cells. Cells were treated with 6 µg/ml Etoposide for indicated times and the level of DNA damage (% DNA in tail) analyzed by alkaline comet assay. Percent tail in DNA (% DNA in tail as measured in comet assay) after exposure to Etoposide (left panel). Representative microscopic images of the comet assay (right panel). Data are means ± SEM of at least 25 cells per group analyzed across two slides. Statistical differences at the indicated times were determined by unpaired t-test with Welch correction. b Cell viability of LKR10 LacZ and Bok−/− cells treated with Etoposide (4 µg/ml) or Irinotecan (80 µM) for indicated times. Data are means ± SEM of three independent experiments. Significant differences from LacZ were revealed by two-way ANOVA, Bonferroni’s multiple comparison test (*: P < 0.05; **: P < 0.01; ***: P < 0.001).
The BOK locus remains unaffected in human lung cancer patients
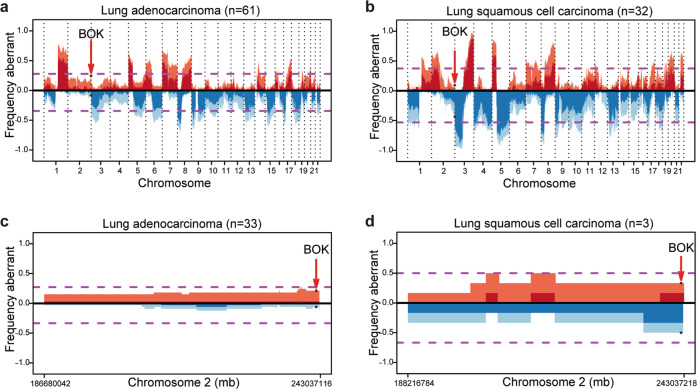
As stated before, BOK has been shown to be genomically deleted across several cancer entities [20]. To gain additional information specific to lung cancer, we interrogated an extended dataset of human patients from the TRACERx dataset [24]. The TRACERx prospective study was designed to analyze intra-tumor heterogeneity and the evolutionary trajectory of tumor cell genomes specifically in treatment-naïve lung squamous carcinoma (LUSC) and LUAD patients. Using the TRACERx data, we detected that BOK is not frequently deleted in either cohort (Fig. 5a, b). Our data in the mouse models suggest an interplay between BOK and TP53, therefore we re-run the same analysis in the TP53 wild type (WT) subpopulation. Again, both in LUSC and LUAD, we detected no significant BOK deletion (Fig. 5c, d).
Fig. 5. BOK is not subjected to copy number variation in lung cancer patients.
a, b Copy number aberrations (CNA) expressed as frequency in TRACERx LUAD (a, n = 61) and LUSC (b, n = 32) tumors. c, d CNA expressed as frequency in TRACERx LUAD grouped for TP53 wildtype (WT) in LUAD (c, n = 33) and LUSC (d, n = 3). Shading indicates clonal status. Dashed purple lines represent threshold for frequent gains and losses.
Taken together, our data suggest that, while BOK plays a role as tumor-promoting factor in lung cancer, and clearly does so in collaboration with TP53, it is not genomically deleted in lung cancer from patients in the TRACERx study.
Discussion
Despite the fact that the BCL-2 family member BOK was discovered more than 20 years ago, its function still remains ambiguous (reviewed in [25]). Using a constitutional Bok−/− knockout mouse model, we found that Bok-deficiency reduces tumor burden, by decreasing lesion number and histological grading. This phenotype was mainly driven by proliferation deficits both in vivo and in vitro models. Furthermore, these effects were entirely lost in the absence of functional p53, together suggesting a tumor promoting role for BOK.
Due to the structural homology of BOK with the apoptosis effectors BAX and BAK [11, 12] and the aggravated phenotype seen in BAX/BAK/BOK full body triple knockout mice [11], BOK is considered to be a pro-apoptotic member of the BCL-2 family. As such, its deletion should be an advantage in tumor survival and BOK should act as a tumor suppressor. However, recent publications also point towards cell death-independent functions of BOK, including a pro-proliferative role of BOK (first proposed by [26]). Bok−/− cell lines, including lung cancer cell lines, divided slower [19, 27] and Bok−/− hepatocellular carcinomas and cell lines showed lower proliferative indexes [18]. In line with these data, we showed that Bok-deficient lung cancer lesions have lower Ki67 levels and Bok-deficient lung cancer cell line proliferate slower.
It can be argued that apoptosis induction and proliferation inhibition are two sides of the same coin. It is known that apoptosis and cellular proliferation are linked by cell-cycle regulators and signaling pathways, affecting both processes [28]. Under physiologic conditions, apoptosis elicits a signaling cascade inducing cell proliferation in the neighboring cells [29]. Therefore, in our settings, the deletion of Bok may decrease proliferation in surrounding cancer cells, by repressing apoptosis in tumor cells.
Furthermore, the proliferative defect seen after downregulation of BOK in human HCT-116 colorectal carcinoma cells is dependent on increased cell cycle arrest, due to the higher levels of the cyclin kinase inhibitors p19INK4d and p21cip1 [18]. Along the same line, p53 was found to be upregulated in BOK-deficient cells [19]. These results are in line with our finding, showing that the absence of p53 eliminates any Bok-driven differences in vivo and in vitro. It could be argued that, in our mouse models, the differences are driven by a ceiling effect due to the different aggressiveness of the tumor models. However, while this could explain the tumor burden, it could not justify the effects on proliferation and DNA damage.
While Beroukhim et al. found the genomic region containing the BOK locus to be deleted in a wide-ranging screen of primary cancer tissue and established cell lines [20], we identified no recurrent deletion of the BOK locus in LUAD and LUSC treatment-naïve samples coming from the TRACERx dataset. These results are in line with our in vivo data, in which deletion of Bok impaired tumor proliferation. However, our experimental settings do not allow us to exclude a different BOK involvement at later time points. Moravcikova et al. showed detectable BOK mRNA and protein levels in biopsies from early and late-stage lung cancer patients as well as in various NSCLC cell lines, supporting our finding that the BOK gene is not frequently deleted in NSCLC [27]. Interestingly, the same study indicated that BOK may exert a tumor-suppressor-like function at later stages of disease. While our data suggest that BOK deletion does not represent a common event in early lung cancer as it interferes with tumor proliferation, it remains to be tested if and at what stage the proliferation defects seen upon loss of BOK are overcome by the numerous other transformative events in the course of tumor development.
Taken together, our data show that BOK promotes proliferation of tumor cells in mutant Kras-driven lung cancer and that, in contrast to common opinion, BOK genomic deletion does not represent a recurrent event in lung cancer.
Materials and methods
Mouse experiments
The previously described LSL-KrasG12D/+ mice [21] (re-derived on a C57BL/6 J background) were crossed with Bok−/− mice (generated on a C57BL/6 genetic background [30]) to generate the following genotypes: LSL-KrasG12D/+ Bok−/−, LSL-KrasG12D/+ Bok+/−, and LSL-KrasG12D/+ Bok+/+. In another set of experiments, these mice were backcrossed on a Tp53fl/fl background [31] to generate: LSL-KrasG12D/+;Bok−/−;Tp53fl/fl, LSL-KrasG12D/+;Bok−/+;Tp53fl/fl, and LSL-KrasG12D/+;Bok+/+;Tp53fl/fl animals. Mice from the same litters were used as age-matched control animals in all experiments. Mice were grouped according to their genotype. 6-8 week old mice were intranasally infected with 5*106 PFU of adenovirus expressing Cre recombinase, as previously described [32]. Mice were maintained in pathogen-free conditions with free access to food and water. Mice that developed a tumor-unrelated illness during the course of the experiment were euthanized and excluded from the study. All animal experiments were approved by and conduced in accordance with the District Government of Upper Bavaria (AZ: 55.2-1-54-2532-55-12).
Immunohistochemistry
FFPE lungs from experimental animals were cut into three-step sections at 100 µm intervals to yield slides of 2 µm thickness. One 2 µm slide from each step section was stained with Hematoxylin-Eosin (H&E) for tumor grading and quantitative tumor analysis. Specific immunostainings were performed using the Leica BOND RXm (Leica Biosystems, Nussloch, Germany), using a DAB-based detection system (BOND Dectection System, Leica Biosystem). The following primary antibodies were used: mouse monoclonal Ki-67 (clone B56, cat. #550609, BD Biosciences, NJ, USA), rabbit polyclonal cleaved caspase-3 (Asp175; Cell Signaling Technology, MA, USA), and rabbit monoclonal Phospho-Ser139-Histone H2AX (γ-H2AX clone 20E3, cat. #9718, Cell Signaling Technology, MA, USA). All stained sections were acquired with a SCN400 slide scanner (Leica Biosystem) and analyzed by experimenter blind to the genotype.
For tumor burden analysis, three slides per animal 100 µm apart were evaluated and the lobe and individual lesion areas were defined manually and measured using the Aperio ImageScope software (version 12.4, Leica Biosystems). The total area of the lesions was normalized over the total area of the lung for each slide. The average of three analyzed H&E slides was used for the reported data. For tumor grading, the slides were evaluated by a pathologist blind to the genotype and were classified as hyperplasia, atypical adenomatous hyperplasia (AAH), adenoma, and adenocarcinomas, as previously described [21, 32]. For the analysis of Ki67 and γ-H2AX immunostainings, the six most advanced lesions on each slide by software-based analysis using the Positive Pixel Count v9 algorithm available in the Aperio ImageScope software (version 12.4, Leica Biosystems) and the reported data were normalized to lesion area.
In vitro experiments
The Kras-driven Tp53 WT mouse lung adenocarcinoma cell line LKR10 [22] was used to target Bok using CRISPR/Cas9-mediated genome editing (targeting sequence: 5’-TCCCAGCGTATACCGGAACG-3’). Targeting guides were cloned in the px458 plasmid, which carries also a GFP expressing cassette (pSpCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang, Addgene plasmid # 48138; http://n2t.net/addgene:48138; RRID:Addgene_48138). LKR10 cells were transfected with the Bok-targeting plasmid and the LacZ control using Metafectene® (Biontex Laboratories, Germany). GFP positive cells were single sorted using BD FACSAria™ Fusion flow cytometer (BD Biosciences) to generate new cell lines, in which successful deletion of Bok was detected by Western blot. The newly generated LKR10 Bok−/− and LKR10 LacZ cells were maintained in RPMI medium 1640 (ThermoFisher Scientific, MA, USA) supplemented with 10% fetal bovine serum (FBS Good Forte, PAN-Biotech, Germany), 1% penicillin/streptomycin, and 2 mM L-Glutamine (ThermoFisher Scientific), at 37 °C in 5% CO2 an atmosphere. Cells were routinely checked for mycoplasma contamination. For knockout of Tp53 in the LKR10 Bok−/− cells (targeting sequence: 5′-GAAGTCACAGCACATGACGG-3′) an inducible CRISPR/Cas9 system was used as described by Aubrey et al. [33]. For proliferation analysis, 3,000 cells per line were plated in 96 well plates and were counted by Trypan Blue exclusion every 24 h. For cell cycle analysis, cells were collected and fixed in 70% ethanol, followed by Ribonuclease A treatment (RNAse A, ThermoFisher Scientific), and propidium iodide staining (PI, Sigma-Aldrich, MO, USA). Data were acquired by BD FACSCanto (BD Biosciences) and analyzed using the Watson pragmatic algorithm available in the FlowJo software (v. 9.7.6, BD Biosciences) or ModFit LT 5.0 (ModFit LT™). Cell viability in cells treated with chemotherapeutics was determined using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) according to the manufactures’ instructions. For Western blot analysis, the following antibodies were used: BOK (1:1000, Abcam, ab233072, clone BOK-R1-5-1); BOK (1:200, clone 1–5, provided by TK and already validated in [13]); p53 (1:1000, Cell Signaling, cat #32352, clone #D2H9O); β-actin (1:1000, Cell Signaling, cat #4970, clone #13E5).
Comet assay
Cells were seeded in 10 cm dishes at a density of 1.5 × 106 the day before the experiment and then treated with Etoposide (6 ug/ml) at 37 °C for the indicated times. The cells were harvested, resuspended in ice-cold PBS at 1.4 × 105, and then prepared using the Trevigen CometAssay Kit and its protocol. The cells were combined with low-melting agarose at a ratio of 1:10 and spread onto the CometSlides. Gelling was achieved at 4 °C in the dark for 10 min and the slides were then placed in 4 °C Lysis Solution overnight. After incubating the slides in alkaline unwinding solution for 20 min at room temperature (200 mM NaOH, 1 mM EDTA), electrophoresis took place at 4 °C and 1 V/cm in alkaline electrophoresis solution (200 mM NaOH, 1 mM EDTA) for 25 min. The slides were dried at 37 °C and DNA was stained in the dark for 15 min using propidium iodide (1 mg/ml stock solution) at a 1:1500 dilution. Slides were analyzed using the OpenComet software tool (https://cometbio.org/index.html).
TracerX analysis
The TRACERx study was conducted under the favorable opinion from the NRES Committee London––Camden & Islington Research Ethics Committee [24]. Informed consent was obtained from all subjects. BOK copy number alteration was evaluated using a similar approach to the one described in [32].
Statistical analysis
The details of experiments, including sample number, statistical tests used, number of experiments, and dispersion and precision measures, are stated in the figures or figure legends. The data analysis was performed with GraphPad Prism software and in the R statistical environment version ≥3.3.1. The appropriate statistical test was selected depending on data type and distribution. For all statistical tests statistical significance was determined if the p value was less than 0.05.
Supplementary information
Acknowledgements
We thank the members of the TRACERx consortium for participating in this study. The authors thank Bettina Flasch for excellent technical assistance. Grant support by Deutsche Krebshilfe for AM (Doktorandenstipendium 112066); the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation – FOR 2036, SFB 1335 (Project-ID 360372040)), German Consortium for Translational Cancer Research (DKTK) to KS and PJJ; Support by the Swiss National Science Foundation (project grant #31003A_173006) to TK; NM is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (Grant Number 211179/Z/18/Z), and also receives funding from Cancer Research UK Lung Cancer Center of Excellence, Rosetrees, and the NIHR BRC at University College London Hospitals.
Author contributions
MAD and PJJ designed and supervised the research; AM, EM, UH, MD, MD, AJ, KS, CB, LB and MAD performed experiments and/or analyzed data; MA and WW provided critical reagents and infrastructure; NM provided TRACERx data; TK had an advisory role and provided Bok−/− mice; AM, CB, MAD and PJJ wrote the paper; and all authors reviewed the data and approved the final version of the manuscript.
Competing interests
KS has received funding from Roche Pharma AG and is a member of the scientific advisory board of TRIMT GmbH. WW has attended Advisory Boards and served as a speaker for Roche, MSD, BMS, AstraZeneca, Pfizer, Merck, Lilly, Boehringer, Novartis, Takeda, Amgen, and Astellas. WW receives research funding from Roche, MSD, BMS, and Bruker. LB has had a consulting or advisory role, received honoraria, research funding, and/or travel/accommodation expenses from Abbvie, AstraZeneca, BMS, Boehringer Ingelheim, Eli Lilly, Janssen, MSD, Merck, Novartis, Pfizer, Roche, and Takeda. NM has stock options and has received consultancy fees from Achilles Therapeutics. PJJ has had a consulting or advisory role, received honoraria, research funding, and/or travel/accommodation expenses from Abbvie, Roche, BMS Celgene, Boehringer, Novartis, Pfizer, Pierre Fabre, and Servier. TK is a co-founder and shareholder of ATANIS Biotech AG, a company performing allergy diagnosis. All the other authors disclose no potential conflicts of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Michael A. Dengler, Philipp J. Jost.
Contributor Information
Michael A. Dengler, Email: michael.dengler@medunigraz.at
Philipp J. Jost, Email: philipp.jost@medunigraz.at
Supplementary information
The online version contains supplementary material available at 10.1038/s41388-021-02161-1.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janic A, Valente LJ, Wakefield MJ, Di Stefano L, Milla L, Wilcox S, et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med. 2018;24:947–53. doi: 10.1038/s41591-018-0043-5. [DOI] [PubMed] [Google Scholar]
- 4.Lieschke E, Wang Z, Kelly GL, Strasser A. Discussion of some ‘knowns’ and some ‘unknowns’ about the tumour suppressor p53. J Mol Cell Biol. 2019;11:212–23. doi: 10.1093/jmcb/mjy077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097–9. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 6.Ichim G, Tait SWG. A fate worse than death: apoptosis as an oncogenic process. Nat Rev Cancer. 2016;16:539–48. doi: 10.1038/nrc.2016.58. [DOI] [PubMed] [Google Scholar]
- 7.Cory S, Huang DCS, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 8.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, et al. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–9. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 9.Yip KW, Reed JC. Bcl-2 family proteins and cancer. Oncogene. 2008;27:6398–406. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 10.Hsu SY, Kaipia A, McGee E, Lomeli M, Hsueh AJ. Bok is a pro-apoptotic Bcl-2 protein with restricted expression in reproductive tissues and heterodimerizes with selective anti-apoptotic Bcl-2 family members. Proc Natl Acad Sci USA. 1997;94:12401–6. doi: 10.1073/pnas.94.23.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke FFS, Vanyai HK, Cowan AD, Delbridge ARD, Whitehead L, Grabow S, et al. Embryogenesis and adult life in the absence of intrinsic apoptosis effectors BAX, BAK, and BOK. Cell. 2018;173:1217–.e17. doi: 10.1016/j.cell.2018.04.036. [DOI] [PubMed] [Google Scholar]
- 12.Zheng JH, Grace CR, Guibao CD, McNamara DE, Llambi F, Wang Y-M, et al. Intrinsic instability of BOK enables membrane permeabilization in apoptosis. Cell Rep. 2018;23:2083–.e6. doi: 10.1016/j.celrep.2018.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Echeverry N, Bachmann D, Ke F, Strasser A, Simon HU, Kaufmann T. Intracellular localization of the BCL-2 family member BOK and functional implications. Cell Death Differ. 2013;20:785–99. doi: 10.1038/cdd.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llambi F, Wang Y-M, Victor B, Yang M, Schneider DM, Gingras S, et al. BOK is a non-canonical BCL-2 family effector of apoptosis regulated by ER-Associated degradation. Cell. 2016;165:421–33. doi: 10.1016/j.cell.2016.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpio MA, Michaud M, Zhou W, Fisher JK, Walensky LD, Katz SG. BCL-2 family member BOK promotes apoptosis in response to endoplasmic reticulum stress. Proc Natl Acad Sci USA. 2015;112:7201–6. doi: 10.1073/pnas.1421063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yakovlev AG, Di Giovanni S, Wang G, Liu W, Stoica B, Faden AI. BOK and NOXA are essential mediators of p53-dependent apoptosis. J Biol Chem. 2004;279:28367–74. doi: 10.1074/jbc.M313526200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang F, Ren L, Zhou S, Duan P, Xue J, Chen H, et al. Role of B-cell lymphoma 2 ovarian killer (BOK) in acute toxicity of human lung epithelial cells caused by cadmium chloride. Med Sci Monit Int Med J Exp Clin Res. 2019;25:5356–68. doi: 10.12659/MSM.913706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabachini T, Fernandez-Marrero Y, Montani M, Loforese G, Sladky V, He Z, et al. BOK promotes chemical-induced hepatocarcinogenesis in mice. Cell Death Differ. 2018;25:708–20. doi: 10.1038/s41418-017-0008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srivastava R, Cao Z, Nedeva C, Naim S, Bachmann D, Rabachini T, et al. BCL-2 family protein BOK is a positive regulator of uridine metabolism in mammals. Proc Natl Acad Sci USA. 2019;116:15469–74. doi: 10.1073/pnas.1904523116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 23.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–8. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 24.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–21. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 25.Naim S, Kaufmann T. The multifaceted roles of the BCL-2 family member BOK. Front Cell Dev Biol. 2020;8:574338. doi: 10.3389/fcell.2020.574338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray JE, Garcia J, Jurisicova A, Caniggia I. Mtd/Bok takes a swing: proapoptotic Mtd/Bok regulates trophoblast cell proliferation during human placental development and in preeclampsia. Cell Death Differ. 2010;17:846–59. doi: 10.1038/cdd.2009.167. [DOI] [PubMed] [Google Scholar]
- 27.Moravcikova E, Krepela E, Donnenberg VS, Donnenberg AD, Benkova K, Rabachini T, et al. BOK displays cell death-independent tumor suppressor activity in non-small-cell lung carcinoma. Int J Cancer. 2017;141:2050–61. doi: 10.1002/ijc.30906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alenzi FQB. Links between apoptosis, proliferation and the cell cycle. Br J Biomed Sci. 2004;61:99–102. doi: 10.1080/09674845.2004.11732652. [DOI] [PubMed] [Google Scholar]
- 29.Brock CK, Wallin ST, Ruiz OE, Samms KM, Mandal A, Sumner EA, et al. Stem cell proliferation is induced by apoptotic bodies from dying cells during epithelial tissue maintenance. Nat Commun. 2019;10:1044. doi: 10.1038/s41467-019-09010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ke F, Voss A, Kerr JB, O’Reilly LA, Tai L, Echeverry N, et al. BCL-2 family member BOK is widely expressed but its loss has only minimal impact in mice. Cell Death Differ. 2012;19:915–25. doi: 10.1038/cdd.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. doi: 10.1101/gad.14.8.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munkhbaatar E, Dietzen M, Agrawal D, Anton M, Jesinghaus M, Boxberg M, et al. MCL-1 gains occur with high frequency in lung adenocarcinoma and can be targeted therapeutically. Nat Commun. 2020;11:4527. doi: 10.1038/s41467-020-18372-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, Milla L, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–32. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.