Abstract
Inflammation impacts basal ganglia motor circuitry in association with psychomotor retardation, a key symptom of major depression (MD). We previously reported associations between circulating protein inflammatory biomarkers and psychomotor slowing as measured by neuropsychological tests probing psychomotor speed in patients with MD. To discover novel transcriptional signatures in peripheral blood immune cells related to psychomotor slowing, microarray data were analyzed in a primary cohort of 88 medically-stable, unmedicated, ambulatory MD patients. Results were confirmed and extended in a second cohort of 57 patients with treatment resistant depression (TRD) before and after anti-inflammatory challenge with the tumor necrosis factor antagonist infliximab versus placebo. Composite scores reflecting pure motor and cognitive-motor processing speed were linearly associated with 403 and 266 gene transcripts in each cohort, respectively (|R|>0.30, p<0.01), that were enriched for cytokine signaling and glycolysis-related pathways (p<0.05). Unsupervised clustering in the primary cohort revealed two psychomotor slowing-associated gene co-expression modules that were enriched for interferon, interleukin-6, aerobic glycolysis, and oxidative phosphorylation pathways (p<0.05, q<0.1). Transcripts were predominantly derived from monocytes, plasmacytoid dendritic cells, and natural killer cells (p’s<0.05). In infliximab-treated TRD patients with high plasma C-reactive protein concentrations (>5mg/L), two differential co-expression modules enriched for oxidative stress and mitochondrial degradation were associated with improvements in psychomotor reaction time (p<0.05). These results indicate that inflammatory signaling and associated metabolic reprogramming in peripheral blood immune cells are associated with systemic inflammation in depression and may affect relevant brain circuits to promote psychomotor slowing.
Introduction
Evidence suggests that inflammation contributes to symptoms of major depression (MD) that are common to other psychiatric illnesses including psychomotor retardation1, 2. Markers of inflammation, such as inflammatory cytokines and their soluble receptors, chemokines, and acute phase reactants like C-reactive protein (CRP), are reliably elevated in peripheral blood and cerebrospinal fluid of MD patients3–6. Moreover, administration of inflammatory stimuli (i.e., cytokines like interferon [IFN]-alpha or cytokine inducers like endotoxin and typhoid vaccination) decreases psychomotor speed in humans and locomotor activity in laboratory animals7–12. Regarding clinical relevance, both psychomotor slowing and inflammation have been independently associated with resistance to conventional antidepressant therapies13–15.
Psychomotor deficits in MD are thought to involve dopamine-rich regions of the basal ganglia, including dorsal striatum, which have been shown to be sensitive to the effects of inflammation. Indeed, administration of inflammatory stimuli alters neural activity and dopamine metabolism in dorsal striatal regions7, 16–18. In addition, we previously found that MD patients exhibit decreased functional connectivity between dorsal striatum and ventromedial prefrontal cortex (vmPFC) as well as presupplementary motor area as a function of increasing plasma CRP concentrations19. Moreover, decreased connectivity in these corticostriatal circuits correlated with both reduced psychomotor speed and increased concentrations of plasma inflammatory cytokines and their soluble receptors, particularly interleukin 6 (IL-6) and IL-1 receptor antagonist (IL-1RA).
We recently reported associations between plasma inflammatory markers and psychomotor slowing in a sample of medically-stable, medication-free patients with MD2. Patients were slower on all tasks relative to published normative standards. IL-6 was associated with increased movement latency on both the simple and choice movement time tasks from the Cambridge Neuropsychological Test Automated Battery (CANTAB). Monocyte chemoattractant protein 1 (MCP-1) predicted slower performance on both a pure motor Finger Tapping Test (FTT) and Digit Symbol Substitution Task (DSST) from the Wechsler Adult Intelligence Scale that measures psychomotor processing speed. IL-10, an anti-inflammatory cytokine, was associated with better performance on DSST. Finally, IL-6 was found to be associated with a psychomotor slowing factor created when all 7 tasks employed in the study were included in a principal component analysis (PCA).
High-throughput analysis of mRNA gene expression has been used to reveal a more nuanced picture of immune signaling pathways in MD that are difficult to measure at the protein level20, 21, and which may serve as clinically-relevant biomarkers for response to antidepressants or for patients who may benefit from anti-inflammatory therapies22–26. Indeed, we previously reported that inflammatory gene signatures were associated with symptoms of depression and fatigue in patients administered IFN-alpha for chronic hepatitis C virus27 and in cancer patients undergoing treatment28. Moreover, we found that antidepressant response to the tumor necrosis factor (TNF) antagonist infliximab was predicted by peripheral blood gene transcripts related not only to inflammation but also glucose and lipid metabolism in patients with treatment resistant depression (TRD)22. These findings are consistent with the change in utilization of energy sources (e.g., shifts away from oxidative phosphorylation [OXPHOS] toward glycolysis) that sustains activation of immune cells, as well as with evidence of insulin resistance that is associated with inflammation29, 30. Accordingly, we recently reported transcriptomic signatures in peripheral blood immune cells reflecting altered glucose metabolism (e.g., insulin signaling and resistance, PI3K/AKT signaling), glycolysis (hypoxia-inducible factor [HIF]-1, pentose phosphate metabolism), and low tyrosine metabolism selectively in MD patients with both higher levels of CRP and significant anhedonia31. We also observed that both inflammatory (e.g., TNF, NF-kB signaling) and metabolic (e.g. insulin, AKT) gene expression signatures in peripheral blood immune cells correlated with low functional connectivity in corticostriatal circuitry in MD, including circuits involving dorsal striatum, which have been associated with psychomotor processing speed32.
Nevertheless, cellular inflammatory and associated metabolic pathways in peripheral blood immune cells that may contribute to psychomotor slowing and lead to novel targets for therapeutic intervention have yet to be examined. To this end, we analyzed microarray data from a primary cohort of 88 medically stable, unmedicated MD patients2. Findings from the MD cohort were confirmed and extended by examining functional pathways before and after anti-inflammatory challenge with infliximab in an independent cohort of 57 TRD patients who exhibited high or low inflammation (as defined by plasma CRP >5 versus ≤5mg/L)22.
Methods
Participants
Primary MD cohort: We enrolled 88 participants with MD (ClinicalTrials.gov NCT01426997) excluded for medical conditions that might confound study interpretation and free of anti-inflammatory and psychotropic medications (see Supplementary Information [SI]). Psychomotor speed and protein inflammatory markers in these patients have been described previously2.
Secondary TRD cohort: Fifty-seven medically-stable, variably medicated participants with TRD were recruited from a study testing the efficacy of infliximab (ClinicalTrials.gov NCT00463580, see SI). Gene and protein signatures predicting antidepressant response to infliximab in these patients have been described22, 33. Patients received an infusion of infliximab (5 mg/kg) or placebo on three occasions (baseline, 2 weeks, and 6 weeks). Gene expression data were available at baseline (prior to infusion) as well as 6 hours, 24 hours, and 2 weeks following the first infusion of infliximab or placebo. Given that the Week 2 time point reflects both sustained anti-inflammatory effects of infliximab as well as its early antidepressant effects that were progressive over time and sustained for weeks after each infusion22, 33, change in gene expression from Baseline to Week 2 was examined herein as a predictor of subsequent change in psychomotor outcomes.
All procedures were approved a priori by the Institutional Review Board of Emory University. All participants provided written informed consent.
Psychomotor Assessments and Composite Factor
Psychomotor slowing was assessed on the same day of blood sampling between 3:00pm and 6:00pm using objective standardized measures of psychomotor speed that have been well-established by our group and others8, 10, 34–36 including: 1) Reaction and Movement Time Tasks of the CANTAB; 2) FTT; 3) DSST; and 4) Trail Making Test Part A (Trails A). See SI. Outliers in task data (≤1 per variable) were determined by Grubbs test37 and removed.
In the primary cohort, 88 of 93 subjects had outlier-free psychomotor data which were used for the composite psychomotor factor scores as a measure of psychomotor speed2. Ln-transformed scores on all tasks were entered into PCA (see SI). A single resulting Bartlett factor score reflecting contributions from all tasks was used to examine associations with gene expression38, 39.
In the TRD cohort, 57 of 58 subjects had outlier-free baseline data to create a composite psychomotor factor. Additionally, 50 subjects had complete psychomotor data at baseline and Week 8, along with gene expression data at baseline and Week 2. Change in psychomotor variables were calculated as Week 8 minus baseline measurements.
Blood Collection
Blood was collected at 9 am (±1 hour) on the morning of neurocognitive testing. Whole blood for gene expression was collected in Tempus Tubes (Applied Biosystems, Carlsbad, CA), then stored at −20 °C until RNA extraction by the Emory Cancer Genomics Core for microarray analysis. Additional blood was collected to measure plasma IL-6 and high sensitivity (hs)-CRP, which have been found to correlate with the composite psychomotor factor, to be elevated in MD patients6, 40, and/or to moderate antidepressant responses to infliximab2, 33 (see SI).
Microarray
RNA was extracted using Ambion Tempus RNA Spin kit (Thermo Fisher, Asheville, NC, USA) and RNA concentrations, A260/280 ratio and sample quality (Agilent Bioanalyzer 2100 RNA Nano assay, Agilent, Santa Clara, CA, USA) were determined. Samples were linearly amplified by TotalPrep-96 RNA Amplification Kit for Array Analysis (Illumina, San Diego CA, USA). After hybridization to Illumina Human HT-12 Expression BeadChips using the Whole-Genome Gene Expression Direct Hybridization Assay (Illumina), BeadChips were scanned on the Illumina HiScan to determine raw probe fluorescence intensity.
Gene-Psychomotor Linear Associations and Pathway Analyses
Raw expression data were exported from Illumina’s GenomeStudio and pre-processed in R. Following model-based background correction41, average signal intensity data were log2-transformed and robust spline normalized using lumi42, and technical batch effects of BeadChip barcode were corrected. 16,969 (MD) and 16,951 (TRD) probes passed filter criteria of Illumina probe detection p-value <0.01 in at least 10% of samples. Raw data are available as Gene Expression Omnibus series GSE135524 (MD) and GSE45468 (TRD). In both the MD cohort and the TRD cohort at baseline, gene probes associated with the composite psychomotor factor were identified based on a threshold of |Rpartial|>0.30 (after controlling for age, sex, race, body mass index [BMI], and education in general linear regression models43; power >0.80 to detect pre-specified effect size at α=0.05 with n=88) consistent with a biologically relevant, medium effect size44–49, combined with nominal p<0.0548–50, which together has been shown to be more reliable across pathway analyses than use of false discovery rate (FDR) alone47, 50, 51.
Functional annotation of resulting transcripts within curated pathways was assessed using WikiPathways and KEGG (Kyoto Encyclopedia Gene and Genome) databases as implemented in clusterProfiler52. An FDR significance threshold of q<0.153 was used herein for pathway analysis to ensure biologically meaningful pathway results21, 54–59. Nominal significance threshold was p<0.05.
Weighted Gene Co-Expression Network Analysis (WGCNA)
WGCNA60, an unsupervised hierarchical clustering method, was used to construct a gene co-expression network and identify clusters (i.e., “modules”) of highly inter-correlated genes associated with psychomotor slowing in patients with depression. Each gene co-expression module was summarized by its first principal component (called “module eigengene” [ME]), which was subsequently tested for association with phenotypic variables. Functional characterization of genes comprising modules of interest was made via pathway analysis as described above. Intra-modular connectivity analysis was conducted to identify the most highly-connected “hub” genes within a module (see SI; R code available upon request).
In the cross-sectional MD cohort, the gene co-expression module’s ME scores were correlated with the composite psychomotor factor and plasma IL-6. In the longitudinal TRD cohort, findings from the primary cohort were extended by testing the association between early gene expression change following a single infusion of infliximab or placebo (Week 2 post-infusion - baseline) and subsequent change in psychomotor variables (Week 8 - baseline) separately in patients with high and low inflammation (CRP >5mg/L and ≤5mg/L, respectively). A CRP cut-off of 5mg/L was used to index high inflammation because previous results in this TRD sample demonstrated that changes in depression scores (baseline to week 12) favored infliximab-treated versus placebo-treated patients at a baseline CRP concentration >5 mg/L33.
Transcript Origin Analysis
To identify the cellular source of differentially expressed genes, transcript origin analysis was conducted as previously published22, 27, 61 (see SI).
Statistical analysis
Demographic and clinical variables were characterized using descriptive statistics. Normality was assessed using the Shapiro test. Chi-square tests, linear models and Kruskal-Wallis rank tests were used to compare categorical, normal and non-normal/heteroskedastic continuous variables, respectively, between cohorts (see SI details). Analyses were conducted in R and IBM SPSS Statistics 27.0 (New York, NY, USA).
Results
Patient characteristics and psychomotor variables
Demographic, clinical, and psychomotor variables are shown in Table 1. The two cohorts were similar with regards to age, sex, BMI, plasma IL-6 concentrations, and depression severity per the Hamilton Depression Rating Scale (p’s>0.052). Of note, the TRD cohort had greater percentage of white race (X2(1,145)=16.286, p<0.001), college-educated subjects (X2(1,145)=11.22, p<0.001), and higher concentrations of plasma hs-CRP (H(1)=7.644, p<0.006) consistent with previous reports15, 62. One patient in the MD and nine in the TRD cohort were taking diabetes medication. Sensitivity analyses accounting for diabetes medication yielded consistent results as the primary analyses (see SI).
Table 1.
Demographic and clinical variables, and measures of psychomotor speed in the MD (N=88) and TRD cohorts (N=57).
| MD (N=88) Primary cohort |
TRD (N=57) Secondary cohort |
p-value | ||
|---|---|---|---|---|
| Age | Mean (SD) | 39.9 (11.6) | 42.8 (9.1) | 0.1161 |
| Sex | N (%) Female | 60 (68.2%) | 38 (66.7%) | 0.8492 |
| Race | N (%) White | 38 (43.2%) | 44 (77.2%) | < 0.0012 |
| BMI | Mean (SD) | 30.9 (7.4) | 31.7 (7.6) | 0.4921 |
| hsCRP (mg/L) | Median (IQR) | 1.5 (3.1) | 2.4 (6.1) | 0.0063 |
| IL-6 (pg/ml) | Median (IQR) | 1.2 (1) | 1.8 (1.4) | 0.0523 |
| HAMD | Median (IQR) | 23 (5) | 23 (5) | 0.1673 |
| Education | N (%) with college degree | 40 (45.5%) | 42 (73.7%) | < 0.0012 |
| TMT | Mean (SD) | 32.4 (13.4) | 32.4 (12.2) | 0.9751 |
| Factor loading | 0.60 | 0.52 | ||
| DSST | Mean (SD) | 53.5 (10.9) | 52.6 (10.1) | 0.5971 |
| Factor loading | −0.60 | −0.54 | ||
| FTT | Mean (SD) | 40.8 (9.4) | 41.0 (9.4) | 0.9431 |
| Factor loading | −0.47 | −0.45 | ||
| SRT | Mean (SD) | 364.3 (72.5) | 368.4 (87.1) | 0.7551 |
| Factor loading | 0.75 | 0.83 | ||
| SMT | Mean (SD) | 557.3 (189.2) | 513.7 (130.5) | 0.1311 |
| Factor loading | 0.59 | 0.77 | ||
| CRT | Mean (SD) | 383.4 (67.5) | 375.9 (66.0) | 0.5121 |
| Factor loading | 0.75 | 0.79 | ||
| CMT | Mean (SD) | 525.8 (130.7) | 501.3 (131.9) | 0.2751 |
| Factor loading | 0.69 | 0.79 |
Linear Model ANOVA,
Pearson’s Chi-squared test,
Kruskal-Wallis rank sum test. SD: standard deviation; BMI: body mass index; HAMD: Total score on 17-item Hamilton Depression Rating Scale; hsCRP: high-sensitivity C-reactive protein (mg/L); IQR: interquartile range; TMT: Trail making test A (s); DSST: Digit Symbol Substitution Task (# of filled boxes correct); FTT: Finger Tapping Task (mean # of taps by dominant hand); SRT: Simple Choice Reaction Time (ms); SMT: Simple Choice Movement Time (ms); CRT: Five Choice Reaction Time (ms); CMT: Five Choice Movement Time (ms). Adapted from Goldsmith, Haroon2.
In each cohort, PCA revealed one component comprising all seven psychomotor variables with individual loadings ranging from −0.60 to 0.79 in consistent directions (positive loadings for simple and choice reaction time [SRT, CRT], simple and choice movement time [SMT, CMT], Trails A; negative loadings for DSST and FTT). Higher composite scores indicated psychomotor slowing. All psychomotor variables, including the composite motor factor, were similar between cohorts (p’s>0.13).
Transcripts associated with psychomotor slowing enriched inflammatory and glucose metabolism pathways
In the primary cohort, 403 probes associated with the composite psychomotor factor. Functional annotation of genes revealed significant enrichment of 19 WikiPathways, including TNF-related weak inducer of apoptosis (TWEAK), IL-1, and the insulin signaling pathway (p<0.05 and q<0.1)(Fig 1A). Nineteen significant KEGG results included three viral infection pathways (p<0.05 and q<0.1). Other nominally-enriched KEGG pathways were related to immune response (TNF, NOD-like receptor signaling) and metabolism (sphingolipid, mammalian target of rapamycin [mTOR] signaling)(p’s<0.05). Confirmatory analysis in the secondary (TRD) cohort at baseline revealed 266 psychomotor factor-associated genes which nominally enriched WikiPathways’ Glycolysis and gluconeogenesis, KEGG’s HIF-1 signaling, and viral protein-cytokine interaction pathways (p’s<0.05)(Fig 1B). See Tables S1 and S2 for gene lists and pathway results from MD and TRD cohorts, respectively.
Fig 1. Gene transcripts associated with psychomotor slowing are enriched for immune and metabolic pathways.
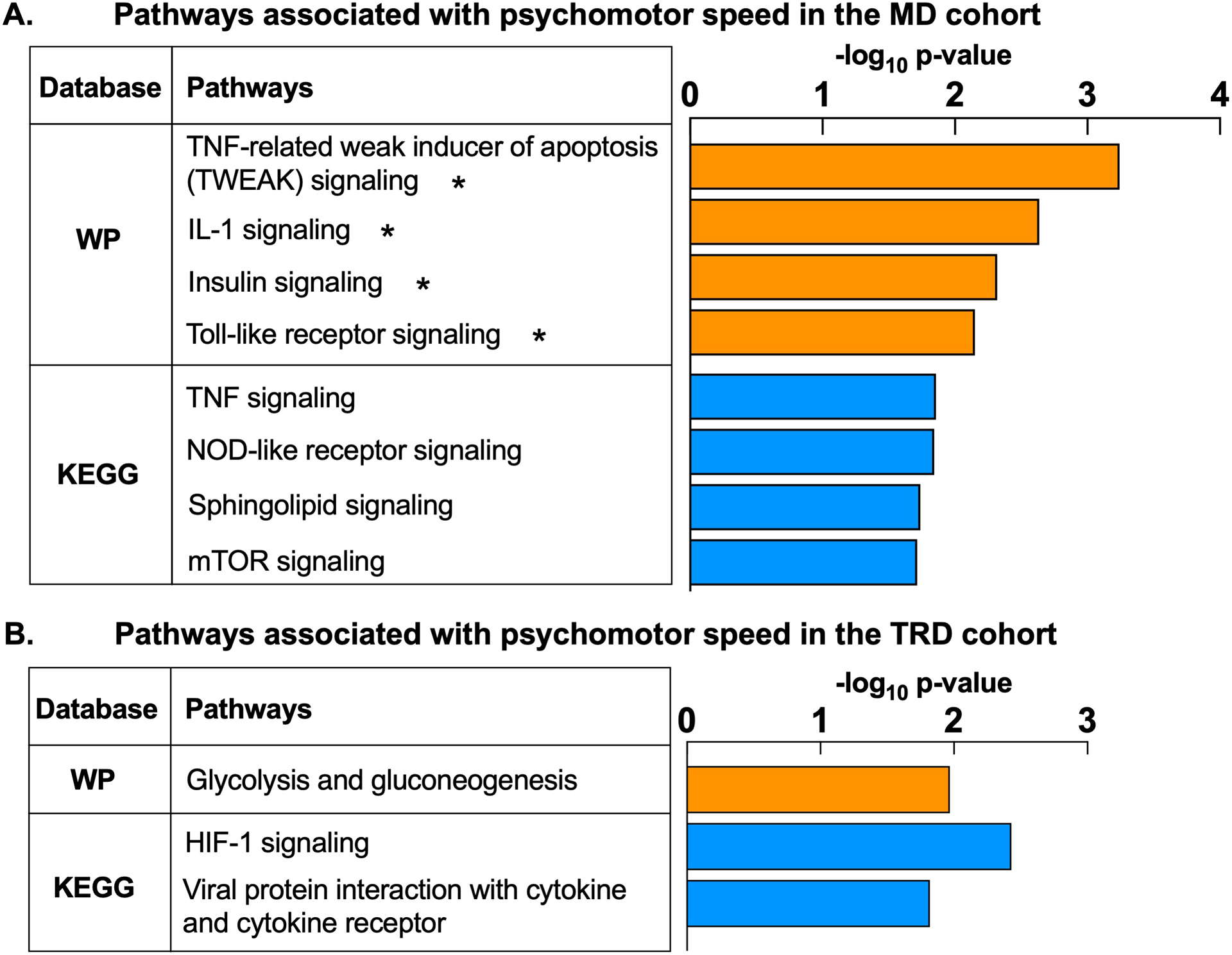
Composite psychomotor factor scores as calculated by Goldsmith, Haroon2 were used as the primary metric of psychomotor slowing in both the primary cohort of medically-stable, unmedicated patients with major depression (MD) (A) and a secondary cohort of medically-stable patients with treatment resistant depression (TRD) (B). The composite psychomotor factor reflects performance on 1) Finger Tapping Test, 2) Reaction Time Task from the Cambridge Neuropsychological Test Automated Battery (including simple and choice reaction and movement latency measurements), 3) Digit Symbol Substitution Task, and 4) Trail Making Test, with higher factor scores indicating increasing severity of psychomotor slowing. Association between composite psychomotor factor scores and whole blood gene expression was assessed in linear models adjusting for covariates. Pathway enrichment analysis using the WikiPathways and KEGG databases revealed that N=403 gene probes associated with the composite psychomotor factor in the MD cohort (n=88 patients) enriched immune (e.g. Tumor necrosis factor [TNF], TNF-related weak inducer of apoptosis [TWEAK], interleukin-1, toll-like and NOD-like receptor signaling) and metabolic (e.g. insulin, sphingolipid, mTOR signaling) pathways (all p<0.05). (A). Similarly, N=266 gene probes associated with the composite psychomotor factor in the TRD cohort (n=57 patients) enriched pathways related to inflammation (viral protein interaction with cytokine and cytokine receptor) and glucose metabolism (glycolysis and gluconeogenesis, hypoxia-inducible factor [HIF]-1 signaling) (all p<0.05) (B). WP, WikiPathways; KEGG, Kyoto Encyclopedia Gene and Genome; TNF, Tumor necrosis factor; IL-1, interleukin-1; NOD, Nucleotide-binding, oligomerization domain; mTOR, mammalian target of rapamycin; HIF-1, hypoxia-inducible factor-1. *pathways are significantly enriched at p<0.05 and q<0.1.
Co-expression analysis revealed inflammatory and metabolic shifts in peripheral blood immune cells in association with psychomotor slowing
In the MD cohort, covariate-adjusted gene expression data clustered into 13 co-expression modules, each arbitrarily assigned a color name (Fig 2A and Fig S1–2) and ranging in size from 38 to 4882 genes. Modules Red and Green (“MEred”, “MEgreen”) were of primary interest based on their positive association with psychomotor slowing (r=0.28, p=0.008 and r=0.23, p=0.032, respectively)(Fig 2A). Module Red comprised 254 highly intercorrelated gene probes which significantly enriched 24 WikiPathways and 19 KEGG pathways including Type I, II, III interferon, IL-6, NOD-like receptor, RIG-I-like receptor, Leptin signaling, Antigen processing and presentation, and Natural killer cell-mediated cytotoxicity (p<0.05 and q<0.1)(Fig 2B and Table S3). Consistent with the pathway results and previously demonstrated relationship between the composite psychomotor factor and IL-6 in this cohort2, MEred positively associated with plasma IL-6. Top 20 hub genes of Module Red were visualized as a two-dimensional network topology in Cytoscape v3.8.0 (Fig 2C), whereby 14 were involved in interferon signaling (shown in boxes). Genes comprising Module Red were estimated to be predominantly expressed by monocytes (z=6.45, p=5.72E-11), plasmacytoid dendritic cells (pDCs; z=3.28, p=0.0005), and natural killer cells (NKs; z=1.86, p=0.031).
Fig 2. Gene co-expression modules associated with psychomotor slowing in major depression (MD) are enriched for biological pathways related to immune response and metabolism.
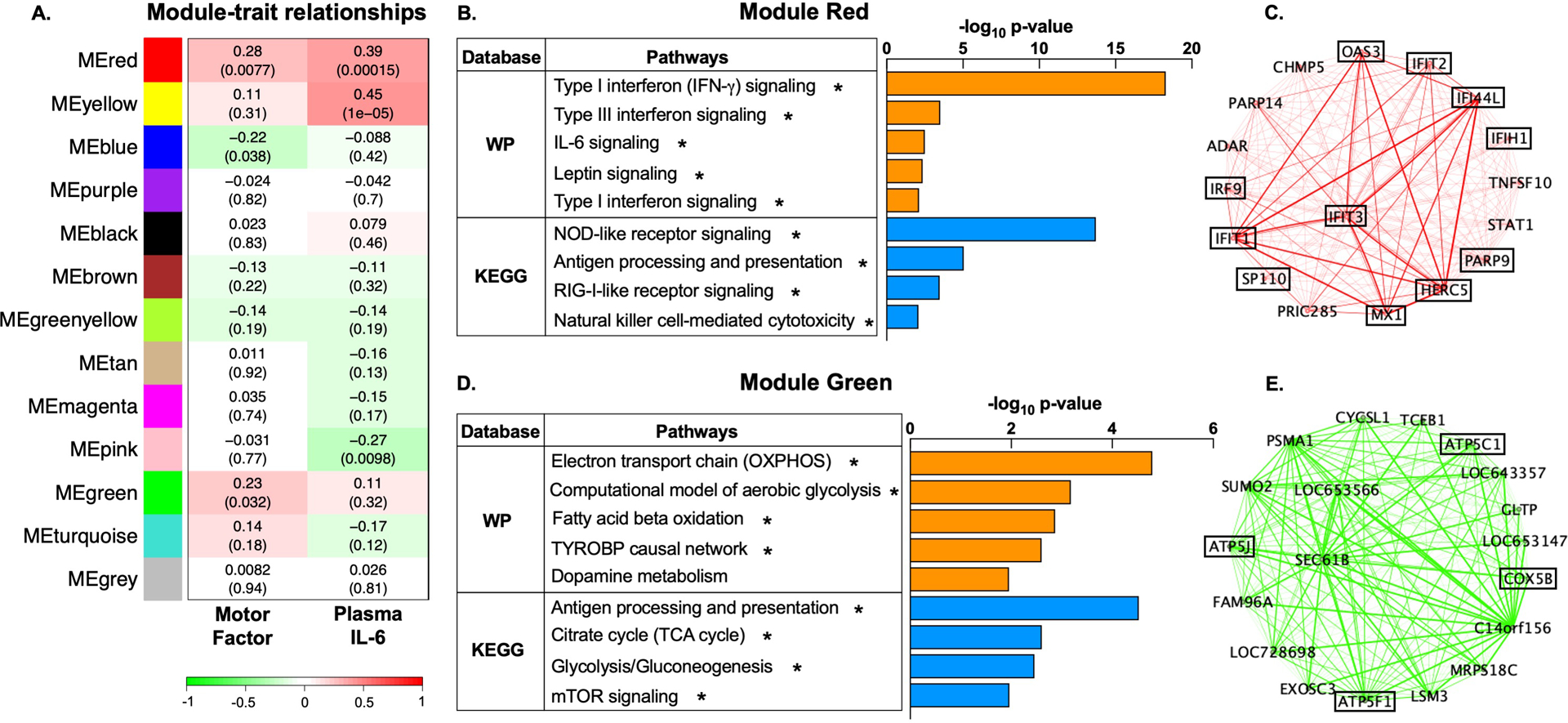
Using Weighted Gene Co-Expression Network Analysis (WGCNA), gene expression in the primary MD cohort (n=88 patients) was clustered into 13 co-expressed gene modules, each labeled by a distinct color name as shown (A). For each gene module, its constituent genes were summarized by the first principal component, referred to as “module eigengene” (ME). Analysis relating the MEs to phenotypic traits revealed that Modules Red and Green positively correlated with the composite psychomotor factor (r=0.28, p=0.008 and r=0.23, p=0.032, respectively) (A). Functional annotation of the gene transcripts in Module Red (n=254) within WikiPathways (WP) and KEGG databases revealed significant enrichment of immune pathways (interferon, IL-6, leptin, NOD-like receptor, RIG-I-like receptor signaling, antigen presentation, and natural killer cell-mediated toxicity) (all p<0.05) (B). To better visualize the gene co-expression patterns, we constructed a two-dimensional network topology in Cytoscape v3.8.0. Intramodular connectivity analysis revealed that the top 20 most connected “hub” genes within Module Red were primarily involved in interferon signaling (C, in boxes). Genes comprising Module Green (n=1382) significantly enriched WikiPathways and KEGG ontologies related to immune cell function (antigen processing and presentation, TYROBP causal network), glucose and lipid metabolism (glycolysis and gluconeogenesis, oxidative phosphorylation [OXPHOS], fatty acid beta oxidation, the citric acid [TCA] cycle, mammalian target of rapamycin (mTOR) signaling), and amino acid metabolism (dopamine metabolism pathway) (all p<0.05) (D). Several of the top 20 hub genes within Module Green were involved in mitochondrial respiration (E, in boxes). ME, module eigengene; IL-6, interleukin-6; WP, WikiPathways; KEGG, Kyoto Encyclopedia Gene and Genome; IFN, interferon; TYROBP, Transmembrane Immune Signaling Adaptor; NOD, Nucleotide-binding, oligomerization domain; RIG-I, retinoic-acid inducible gene; OXPHOS, oxidative phosphorylation; TCA, the citric acid. *pathways are significantly enriched at p<0.05 and q<0.1.
Module Green comprised 1382 gene probes which significantly enriched 11 WikiPathways and 54 KEGG pathways (Fig 2D). Top WikiPathways included Aerobic glycolysis, Electron transport chain (OXPHOS system in mitochondria), Fatty acid beta oxidation, and TYROBP causal network (p<0.05 and q<0.1). Nominally-significant results included dopamine metabolism (p<0.05). Significant KEGG results included Glycolysis/gluconeogenesis, Antigen processing and presentation, Citrate cycle (TCA cycle), and mTOR signaling (p<0.05 and q<0.1; Table S4). Top hub genes of Module Green included genes related to mitochondrial respiration and electron transport chain (ETC)(Fig 2E, in boxes). Similar to Module Red, genes comprising Module Green were predominantly derived from monocytes (z=15.52, p<1E-12), pDCs (z=11.98, p<1E-12), and NK cells (z=6.48, p=4.68E-11).
Gene modules co-regulated at Week 2 post-infliximab in association with improved reaction speed at Week 8 involved oxidative and mitochondrial stress pathways in patients with high inflammation
Separate WGCNA networks were constructed in TRD patients with high (CRP>5mg/L) and low (CRP≤5mg/L) inflammation who showed differential antidepressant response to infliximab33. Early change in post-infliximab or placebo gene expression (Week 2 - baseline) was covariate-adjusted and clustered into co-expression modules (representing genes co-regulated by treatment). Resulting MEs were tested for association with subsequent change (Week 8 - baseline) in SRT and CRT, which were found to be improved by infliximab selectively in patients with high inflammation (see SI).
Among TRD patients with high inflammation (n=20), 33 differential co-expression modules were detected, two of which (“MEblue” and “MEdarkred”) were associated with both treatment (infliximab versus placebo) and change in SRT or CRT (Fig 3A). Treatment with infliximab was associated with decreasing MEblue (r=−0.45, p=0.049). Lower values of MEblue corresponded to more negative change in CRT, indicating faster reaction speed at Week 8 versus baseline (r=0.46, p=0.04). Together this suggests that in patients with high inflammation, infliximab reduced expression of Module Blue genes at Week 2, which correlated with greater improvement in psychomotor reaction times at Week 8. Pathway analysis revealed enrichment of pathways related to oxidative stress and mitochondrial damage (ferroptosis, mitophagy) (p<0.05, q<0.1), immune function and cancer metabolism (p<0.05)(Fig 3B). Conversely, infliximab treatment positively correlated with MEdarkred (r=0.44, p=0.05). Larger values of MEdarkred (reflecting higher expression at Week 2) in turn correlated with more negative change in SRT at Week 8 (hence greater improvement)(r=−0.55, p=0.01). Genes within Module Darkred enriched broad immune-related pathways (p<0.05)(Fig 3C). See Table S5 for full results for Modules Blue and Darkred. Results for TRD patients with low inflammation (n=30) are provided in SI and Table S6.
Fig 3. Gene modules co-regulated by infliximab and associated with subsequent improvements in psychomotor speed in treatment resistant depression (TRD) are enriched for pathways related to oxidative stress, mitochondrial degradation, inflammation, and cancer metabolism.
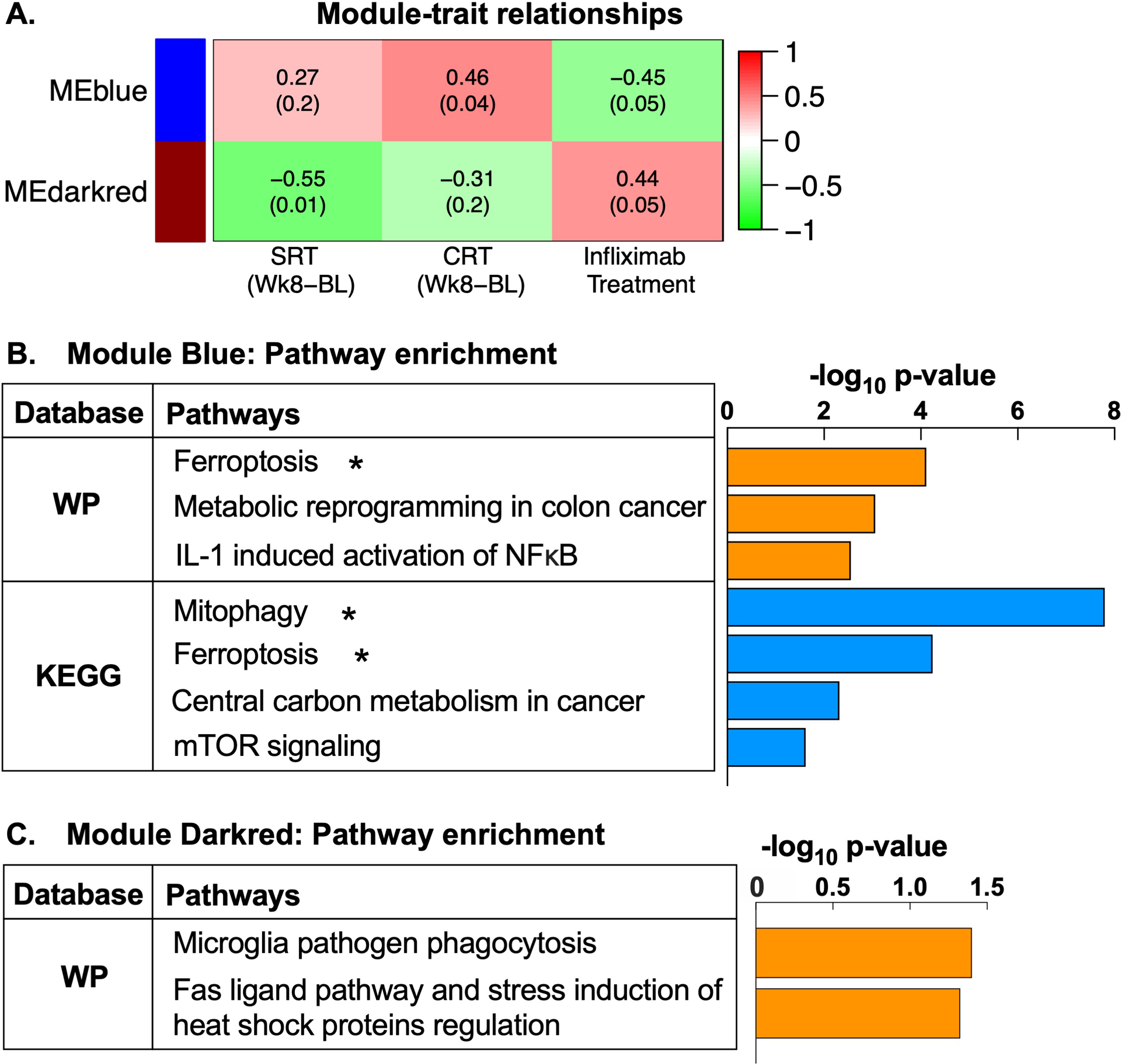
Among TRD patients with high inflammation (n=20 patients), change in gene expression following a single infusion of infliximab or placebo (Week 2 post-infusion - baseline) clustered into 33 differential gene co-expression modules, labeled by distinct color names, and constituent genes summarized by a module eigengene (ME). Of these, MEs of two gene modules, blue and darkred, were associated with both infliximab treatment and subsequent change in simple or choice reaction time (SRT and CRT) at Week 8 (A). Treatment with infliximab was associated with decreasing MEblue (r=−0.45, p=0.049), and lower values of MEblue corresponded to more negative values of change in CRT (indicating faster reaction speed at Week 8 relative to baseline) (r=0.46, p=0.04). Conversely, infliximab positively correlated with MEdarkred (r=0.44, p=0.05), and larger values of MEdarkred in turn correlated with more negative values of change in SRT (hence greater improvement) (r=−0.55, p=0.01). Functional annotation of the gene transcripts in Module Blue (n=1440) within WikiPathways and KEGG databases revealed significant enrichment of pathways related to oxidative stress and mitochondrial degradation (ferroptosis and mitophagy), as well as immune function (interleukin-1-induced activation of NF-kappa-B) and cancer metabolism (metabolic reprogramming in colon cancer, central carbon metabolism in cancer) (all p<0.05) (B). Gene transcripts within Module darkred (n=150) enriched immune pathways including microglia pathogen phagocytosis and a Fas ligand pathway (all p<0.05) (C). ME, module eigengene; SRT, simple reaction time; CRT, choice reaction time; WP, WikiPathways; KEGG, Kyoto Encyclopedia Gene and Genome; IL-1, interleukin-1; NFκB, nuclear factor kappa-B; mTOR, mammalian target of rapamycin. *pathways are significantly enriched at p<0.05 and q<0.1.
Discussion
We identified transcriptomic signatures of psychomotor slowing involving inflammatory and metabolic reprogramming consistent with activated peripheral blood immune cells in MD. Moreover, similar gene signatures were modified in association with psychomotor improvements following anti-inflammatory treatment with infliximab in TRD patients with high inflammation. Specifically, we uncovered concurrent changes in both inflammatory (cytokine and pattern recognition receptor signaling) and metabolic (insulin signaling, glycolysis, and OXPHOS) pathways that are characteristic of metabolic reprogramming in activated immune cells and particularly monocytes. Moreover, infliximab treatment in TRD patients with high inflammation led to differential co-expression modules related to oxidative and mitochondrial stress, inflammation, and energy metabolism at Week 2 in association with subsequent improvement in reaction speed at Week 8.
Enrichment of innate immune and cytokine signaling pathways has been previously demonstrated in transcriptomic studies of MD20, 21, 63. Herein, cytokine signaling pathways including IL-1 and TNF (TWEAK) were enriched in gene transcripts associated with psychomotor slowing. IL-1 has consistently been implicated in depression3, 6, 40. Notably, we previously found that baseline expression of TWEAK pathway genes predicted antidepressant response to infliximab in TRD22, and psychomotor retardation was one of the most responsive symptoms in patients with increased inflammation33. These results were augmented via co-expression analysis (e.g., Module Red) which uncovered several cytokine pathways including Type I, II, III interferon and IL-6 signaling, both of which have been implicated in MD21, 40. Moreover, pathways suggestive of immune cell activation, including retinoic-acid inducible gene-I (RIG-I)-like receptor, antigen processing and presentation, and NK cell-mediated cytotoxicity were also enriched. While increased circulating markers like CRP may reflect inflammatory activity in multiple peripheral tissues (immune, adipose, liver, etc.), as well as interactions with metabolic systems, little is known about the peripheral blood immune cell contribution to systemic inflammation in depression. It is, however, thought to involve myeloid cells and particularly monocytes64. Accordingly, results herein suggest that transcriptomic signatures related to motor slowing were likely the product of monocytes as well as pDCs and NKs, powerful producers of Type I and II IFNs, respectively65.
Consistent with the above, converging evidence from both cohorts indicated that glucose-related metabolic shifts in peripheral blood immune cells were related to psychomotor slowing. This included pathways for HIF-1, a transcription factor crucial for glycolysis66; mTOR, which upregulates glycolytic genes like glucose transporter 166; and insulin signaling, which has also been implicated in anhedonia symptoms in depression31. Results from individual gene-level linear associations appeared consistent with “Warburg effect” whereby activated immune cells shift from slower, energy-maximizing OXPHOS to faster but energy-inefficient glycolysis in order to rapidly generate energy29. Interestingly, co-expression analysis discovered that psychomotor slowing was associated with enrichment of both glycolysis and OXPHOS pathways, rather than a Warburg-like shift to aerobic glycolysis at the expense of mitochondrial respiration. Recent data indicate that metabolic requirements of immune cells vary substantially depending upon the activation stimulus and signaling cascade. When considering results from both pathway analyses and TOA on genes associated with motor slowing, IFN production from pDCs involves glycolysis and mTOR when pDCs are activated through toll-like receptors67, but relies upon OXPHOS during RIG-I-like receptor-mediated antiviral responses68. Furthermore, NK cell cytotoxicity and IFN-γ release69, 70, as well as monocytes activated by non-LPS stimuli71, depend on both increased OXPHOS and glycolysis. These results suggest that transcriptomic signatures of psychomotor slowing involve nuanced metabolic changes that reflect peripheral immune cell type and activation status. Together with our previous findings of transcriptomic alterations in glucose-related pathways (insulin, glycolysis) in MD patients with high inflammation and high anhedonia31, these results also suggest that bioenergetic shifts reflecting interactions between inflammation and metabolic disturbances may broadly associate with symptoms of depression relevant to energy expenditure, including both psychomotor slowing and anhedonia72.
While mitochondrial respiration deficits have been described in depression73, 74, the directionality of association between psychomotor slowing and mitochondrial respiration and OXPHOS observed herein is consistent with reports of upregulation of related genes and proteins in peripheral immune cells and brains of patients with depression and bipolar disorder75–78. The functional relevance of metabolism- and mitochondria-related gene modules to psychomotor slowing was uncovered using infliximab in our TRD cohort. In TRD patients with high baseline inflammation, infliximab improved reaction speed in association with early decreases in a co-expression module related to ferroptosis, reflecting oxidative stress-driven cell death, and mitophagy, a mitochondrial degradation process that promotes shifts to glycolysis in macrophages79. Consistently, normalization of mitochondrial antioxidant defense and mitochondrial respiration have been shown to associate with antidepressant response to ketamine80, 81 and lithium82. Infliximab also nominally impacted cancer metabolism pathways, which may reflect reversal of Warburg-like shifts in specific immune cell subsets. These results complement our previous findings indicating that genes related to cytokine signaling and metabolic reprograming were associated with antidepressant response to infliximab, including improvements in psychomotor slowing. Whereas genes associated with psychomotor slowing in the MD cohort reflected both metabolic as well as numerous inflammatory cytokine and viral signaling pathways, genes associated with psychomotor slowing in the higher inflammation TRD cohort predominantly reflected metabolic shifts toward glycolysis. Together with the above-mentioned effects of infliximab among TRD patients with CRP>5mg/L, these results suggest that immunometabolic gene expression signatures of psychomotor slowing are more specific to patients with higher inflammation.
Strengths of this study include medically-stable patients, who were free of psychotropic and anti-inflammatory medications in the primary cohort. In addition, psychomotor slowing was defined by a composite factor of extensively validated neurocognitive tasks that probed a range of aspects of psychomotor speed, previously established to be associated with plasma inflammatory markers in our MD cohort. Furthermore, to our knowledge, this is the first study to examine transcriptomic markers involved in the impact of an anti-inflammatory treatment on psychomotor symptoms in depression. While gene and psychomotor data were not available from the same post-infliximab timepoint in the TRD cohort, we were able to examine whether early transcriptomic changes predicted later psychomotor improvements. Moreover, although both cohorts were on average obese, BMI was controlled for in all transcriptomic analyses, suggesting that immunometabolic shifts in circulating inflammatory cells that characterize psychomotor slowing are relevant to patients with depression independent of BMI. Regarding limitations, no control group was included. Nevertheless, the study benefitted from published normative data for all tasks2, and MD/TRD subjects exhibited impaired performance83. A more lenient threshold of FDR <0.1 was used for pathway analysis to ensure biologically meaningful pathway results, consistent with similar analytic strategies in other bioinformatics studies including those in depression21, 54–59. However, converging pathway results from both linear associations at the individual gene-level and co-expression modules formed by unsupervised clustering suggest that the reported findings are robust. Finally, it should be noted that while TOA provides estimated cellular origins, single-cell profiling is required for a more refined insight into peripheral immune cell transcriptomic shifts underlying psychomotor symptoms.
In sum, these findings are consistent with the notion that activation of inflammatory (cytokine, antiviral) and metabolic pathways in peripheral blood immune cells contribute to elevated circulating inflammatory markers which are related to altered neurotransmitters and neurocircuits that drive motor deficits in psychiatric disorders like depression. Future work will determine precise immune cells and gene signatures that may serve as promising treatment targets for symptoms like psychomotor retardation in depressed patients with high inflammation or metabolic alterations.
Supplementary Material
Acknowledgements:
This study was supported by grants R01MH087604 (Miller), R01MH109637 (Felger), R01MH107033 (Haroon), R01MH112076 (Miller/Haroon), F32MH119750 (Bekhbat), and K23MH114037 (Goldsmith) from the National Institute of Mental Health; NARSAD Distinguished Investigator Grant (Miller) from the Brain and Behavioral Research Foundation. In addition, the study was supported in part by PHS Grants UL1TR000454, UL1TR002378, KL2TR000455, and TL1TR002382 from the Clinical and Translational Science Award program, by the NIH/NCI under award number P30CA138292, and the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities.
Footnotes
Financial Disclosure: All authors declare no conflicts of interest. In the past 12 months, Dr. Felger has consulted for Otsuka on a topic unrelated to this research.
Supplementary information is available at MP’s website.
References
- 1.Carvalho AF, Miskowiak KK, Hyphantis TN, Kohler CA, Alves GS, Bortolato B et al. Cognitive dysfunction in depression - pathophysiology and novel targets. CNS Neurol Disord Drug Targets 2014; 13(10): 1819–1835. [DOI] [PubMed] [Google Scholar]
- 2.Goldsmith DR, Haroon E, Woolwine BJ, Jung MY, Wommack EC, Harvey PD et al. Inflammatory markers are associated with decreased psychomotor speed in patients with major depressive disorder. Brain Behav Immun 2016; 56: 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldsmith DR, Rapaport MH, Miller BJ. A meta-analysis of blood cytokine network alterations in psychiatric patients: comparisons between schizophrenia, bipolar disorder and depression. Mol Psychiatry 2016; 21(12): 1696–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller AH, Raison CL. The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat Rev Immunol 2016; 16(1): 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zunszain PA, Hepgul N, Pariante CM. Inflammation and depression. Current topics in behavioral neurosciences 2013; 14: 135–151. [DOI] [PubMed] [Google Scholar]
- 6.Dowlati Y, Herrmann N, Swardfager W, Liu H, Sham L, Reim EK et al. A meta-analysis of cytokines in major depression. Biol Psychiatry 2010; 67(5): 446–457. [DOI] [PubMed] [Google Scholar]
- 7.Brydon L, Harrison NA, Walker C, Steptoe A, Critchley HD. Peripheral inflammation is associated with altered substantia nigra activity and psychomotor slowing in humans. Biol Psychiatry 2008; 63(11): 1022–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capuron L, Gumnick JF, Musselman DL, Lawson DH, Reemsnyder A, Nemeroff CB et al. Neurobehavioral effects of interferon-alpha in cancer patients: phenomenology and paroxetine responsiveness of symptom dimensions. Neuropsychopharmacology 2002; 26(5): 643–652. [DOI] [PubMed] [Google Scholar]
- 9.Haroon E, Felger JC, Woolwine BJ, Chen X, Parekh S, Spivey JR et al. Age-related increases in basal ganglia glutamate are associated with TNF, reduced motivation and decreased psychomotor speed during IFN-alpha treatment: Preliminary findings. Brain Behav Immun 2015; 46: 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majer M, Welberg LA, Capuron L, Pagnoni G, Raison CL, Miller AH. IFN-alpha-induced motor slowing is associated with increased depression and fatigue in patients with chronic hepatitis C. Brain Behav Immun 2008; 22(6): 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frenois F, Moreau M, O’Connor J, Lawson M, Micon C, Lestage J et al. Lipopolysaccharide induces delayed FosB/DeltaFosB immunostaining within the mouse extended amygdala, hippocampus and hypothalamus, that parallel the expression of depressive-like behavior. Psychoneuroendocrinology 2007; 32(5): 516–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lenczowski MJ, Bluthe RM, Roth J, Rees GS, Rushforth DA, van Dam AM et al. Central administration of rat IL-6 induces HPA activation and fever but not sickness behavior in rats. Am J Physiol 1999; 276(3): R652–658. [DOI] [PubMed] [Google Scholar]
- 13.Bruder GE, Alvarenga JE, Alschuler D, Abraham K, Keilp JG, Hellerstein DJ et al. Neurocognitive predictors of antidepressant clinical response. Journal of affective disorders 2014; 166: 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor BP, Bruder GE, Stewart JW, McGrath PJ, Halperin J, Ehrlichman H et al. Psychomotor slowing as a predictor of fluoxetine nonresponse in depressed outpatients. Am J Psychiatry 2006; 163(1): 73–78. [DOI] [PubMed] [Google Scholar]
- 15.Haroon E, Daguanno AW, Woolwine BJ, Goldsmith DR, Baer WM, Wommack EC et al. Antidepressant treatment resistance is associated with increased inflammatory markers in patients with major depressive disorder. Psychoneuroendocrinology 2018; 95: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capuron L, Pagnoni G, Drake DF, Woolwine BJ, Spivey JR, Crowe RJ et al. Dopaminergic mechanisms of reduced basal ganglia responses to hedonic reward during interferon alfa administration. Arch Gen Psychiatry 2012; 69(10): 1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenberger NI, Berkman ET, Inagaki TK, Rameson LT, Mashal NM, Irwin MR. Inflammation-induced anhedonia: endotoxin reduces ventral striatum responses to reward. Biol Psychiatry 2010; 68(8): 748–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison NA, Brydon L, Walker C, Gray MA, Steptoe A, Critchley HD. Inflammation causes mood changes through alterations in subgenual cingulate activity and mesolimbic connectivity. Biol Psychiatry 2009; 66(5): 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felger JC, Li Z, Haroon E, Woolwine BJ, Jung MY, Hu X et al. Inflammation is associated with decreased functional connectivity within corticostriatal reward circuitry in depression. Mol Psychiatry 2016; 21(10): 1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leday GGR, Vertes PE, Richardson S, Greene JR, Regan T, Khan S et al. Replicable and Coupled Changes in Innate and Adaptive Immune Gene Expression in Two Case-Control Studies of Blood Microarrays in Major Depressive Disorder. Biol Psychiatry 2018; 83(1): 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostafavi S, Battle A, Zhu X, Potash JB, Weissman MM, Shi J et al. Type I interferon signaling genes in recurrent major depression: increased expression detected by whole-blood RNA sequencing. Mol Psychiatry 2014; 19(12): 1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta D, Raison CL, Woolwine BJ, Haroon E, Binder EB, Miller AH et al. Transcriptional signatures related to glucose and lipid metabolism predict treatment response to the tumor necrosis factor antagonist infliximab in patients with treatment-resistant depression. Brain Behav Immun 2013; 31: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mamdani F, Berlim MT, Beaulieu MM, Labbe A, Merette C, Turecki G. Gene expression biomarkers of response to citalopram treatment in major depressive disorder. Transl Psychiatry 2011; 1: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guilloux JP, Bassi S, Ding Y, Walsh C, Turecki G, Tseng G et al. Testing the predictive value of peripheral gene expression for nonremission following citalopram treatment for major depression. Neuropsychopharmacology 2015; 40(3): 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cattaneo A, Gennarelli M, Uher R, Breen G, Farmer A, Aitchison KJ et al. Candidate genes expression profile associated with antidepressants response in the GENDEP study: differentiating between baseline ‘predictors’ and longitudinal ‘targets’. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 2013; 38(3): 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cattaneo A, Ferrari C, Uher R, Bocchio-Chiavetto L, Riva MA, Consortium MRCI et al. Absolute Measurements of Macrophage Migration Inhibitory Factor and Interleukin-1-beta mRNA Levels Accurately Predict Treatment Response in Depressed Patients. Int J Neuropsychopharmacol 2016; 19(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felger JC, Cole SW, Pace TW, Hu F, Woolwine BJ, Doho GH et al. Molecular signatures of peripheral blood mononuclear cells during chronic interferon-alpha treatment: relationship with depression and fatigue. Psychol Med 2012; 42(8): 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao C, Beitler JJ, Higgins KA, Conneely K, Dwivedi B, Felger J et al. Fatigue is associated with inflammation in patients with head and neck cancer before and after intensity-modulated radiation therapy. Brain Behav Immun 2016; 52: 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol 2016; 16(9): 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest 2006; 116(7): 1793–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bekhbat M, Treadway MT, Goldsmith DR, Woolwine BJ, Haroon E, Miller AH et al. Gene signatures in peripheral blood immune cells related to insulin resistance and low tyrosine metabolism define a sub-type of depression with high CRP and anhedonia. Brain Behav Immun 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldsmith DR, Bekhbat M, Le NA, Chen X, Woolwine BJ, Li Z et al. Protein and gene markers of metabolic dysfunction and inflammation together associate with functional connectivity in reward and motor circuits in depression. Brain Behav Immun 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raison CL, Rutherford RE, Woolwine BJ, Shuo C, Schettler P, Drake DF et al. A randomized controlled trial of the tumor necrosis factor antagonist infliximab for treatment-resistant depression: the role of baseline inflammatory biomarkers. JAMA Psychiatry 2013; 70(1): 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang HH, Lee IH, Gean PW, Lee SY, Chi MH, Yang YK et al. Treatment response and cognitive impairment in major depression: association with C-reactive protein. Brain Behav Immun 2012; 26(1): 90–95. [DOI] [PubMed] [Google Scholar]
- 35.Krogh J, Benros ME, Jorgensen MB, Vesterager L, Elfving B, Nordentoft M. The association between depressive symptoms, cognitive function, and inflammation in major depression. Brain Behav Immun 2014; 35: 70–76. [DOI] [PubMed] [Google Scholar]
- 36.Raison CL, Rye DB, Woolwine BJ, Vogt GJ, Bautista BM, Spivey JR et al. Chronic interferon-alpha administration disrupts sleep continuity and depth in patients with hepatitis C: association with fatigue, motor slowing, and increased evening cortisol. Biol Psychiatry 2010; 68(10): 942–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grubbs FE. Procedures for Detecting Outlying Observations in Samples. Technometrics 1969; 11(1): 1–21. [Google Scholar]
- 38.Bekhbat M, Chu K, Le NA, Woolwine BJ, Haroon E, Miller AH et al. Glucose and lipid-related biomarkers and the antidepressant response to infliximab in patients with treatment-resistant depression. Psychoneuroendocrinology 2018; 98: 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Felger JC, Haroon E, Patel TA, Goldsmith DR, Wommack EC, Woolwine BJ et al. What does plasma CRP tell us about peripheral and central inflammation in depression? Mol Psychiatry 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howren MB, Lamkin DM, Suls J. Associations of depression with C-reactive protein, IL-1, and IL-6: a meta-analysis. Psychosom Med 2009; 71(2): 171–186. [DOI] [PubMed] [Google Scholar]
- 41.Allen JD, Chen M, Xie Y. Model-Based Background Correction (MBCB): R Methods and GUI for Illumina Bead-array Data. J Cancer Sci Ther 2009; 1(1): 25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics 2008; 24(13): 1547–1548. [DOI] [PubMed] [Google Scholar]
- 43.Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics 2012; 28(9): 1280–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross KM, Carroll JE, Dunkel Schetter C, Hobel C, Cole SW. Pro-inflammatory immune cell gene expression during the third trimester of pregnancy is associated with shorter gestational length and lower birthweight. Am J Reprod Immunol 2019; 82(6): e13190. [DOI] [PubMed] [Google Scholar]
- 45.Miller GE, Chen E, Shalowitz MU, Story RE, Leigh AKK, Ham P et al. Divergent transcriptional profiles in pediatric asthma patients of low and high socioeconomic status. Pediatric pulmonology 2018; 53(6): 710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mellon SH, Wolkowitz OM, Schonemann MD, Epel ES, Rosser R, Burke HB et al. Alterations in leukocyte transcriptional control pathway activity associated with major depressive disorder and antidepressant treatment. Transl Psychiatry 2016; 6: e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole SW, Galic Z, Zack JA. Controlling false-negative errors in microarray differential expression analysis: a PRIM approach. Bioinformatics 2003; 19(14): 1808–1816. [DOI] [PubMed] [Google Scholar]
- 48.Han TJ, Felger JC, Lee A, Mister D, Miller AH, Torres MA. Association of childhood trauma with fatigue, depression, stress, and inflammation in breast cancer patients undergoing radiotherapy. Psychooncology 2016; 25(2): 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres MA, Pace TW, Liu T, Felger JC, Mister D, Doho GH et al. Predictors of depression in breast cancer patients treated with radiation: role of prior chemotherapy and nuclear factor kappa B. Cancer 2013; 119(11): 1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo L, Lobenhofer EK, Wang C, Shippy R, Harris SC, Zhang L et al. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nature biotechnology 2006; 24(9): 1162–1169. [DOI] [PubMed] [Google Scholar]
- 51.Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, Bao W et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol 2006; 24(9): 1140–1150. [DOI] [PubMed] [Google Scholar]
- 52.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16(5): 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 2003; 100(16): 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: two different approaches for Gene Ontology analysis. BMC Proc 2009; 3 Suppl 4: S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y, Han L, Yuan Y, Li J, Hei N, Liang H. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat Commun 2014; 5: 3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang D, Li Y, Xiao H, Liu Q, Zhang M, Zhu J et al. Gaining confidence in biological interpretation of the microarray data: the functional consistence of the significant GO categories. Bioinformatics 2008; 24(2): 265–271. [DOI] [PubMed] [Google Scholar]
- 57.Jansen R, Penninx BW, Madar V, Xia K, Milaneschi Y, Hottenga JJ et al. Gene expression in major depressive disorder. Mol Psychiatry 2016; 21(3): 339–347. [DOI] [PubMed] [Google Scholar]
- 58.de Kluiver H, Jansen R, Milaneschi Y, Penninx B. Involvement of inflammatory gene expression pathways in depressed patients with hyperphagia. Transl Psychiatry 2019; 9(1): 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y, Lutz PE, Wang YC, Ragoussis J, Turecki G. Global long non-coding RNA expression in the rostral anterior cingulate cortex of depressed suicides. Transl Psychiatry 2018; 8(1): 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008; 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci U S A 2011; 108(7): 3080–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chamberlain SR, Cavanagh J, de Boer P, Mondelli V, Jones DNC, Drevets WC et al. Treatment-resistant depression and peripheral C-reactive protein. Br J Psychiatry 2019; 214(1): 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hepgul N, Cattaneo A, Agarwal K, Baraldi S, Borsini A, Bufalino C et al. Transcriptomics in Interferon-alpha-Treated Patients Identifies Inflammation-, Neuroplasticity- and Oxidative Stress-Related Signatures as Predictors and Correlates of Depression. Neuropsychopharmacology 2016; 41(10): 2502–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J, Zhang L, Yu C, Yang XF, Wang H. Monocyte and macrophage differentiation: circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomark Res 2014; 2(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol 2011; 29: 163–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014; 345(6204): 1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nature immunology 2008; 9(10): 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fekete T, Suto MI, Bencze D, Mazlo A, Szabo A, Biro T et al. Human Plasmacytoid and Monocyte-Derived Dendritic Cells Display Distinct Metabolic Profile Upon RIG-I Activation. Front Immunol 2018; 9: 3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keating SE, Zaiatz-Bittencourt V, Loftus RM, Keane C, Brennan K, Finlay DK et al. Metabolic Reprogramming Supports IFN-gamma Production by CD56bright NK Cells. J Immunol 2016; 196(6): 2552–2560. [DOI] [PubMed] [Google Scholar]
- 70.Kumar A, Pyaram K, Yarosz EL, Hong H, Lyssiotis CA, Giri S et al. Enhanced oxidative phosphorylation in NKT cells is essential for their survival and function. Proc Natl Acad Sci U S A 2019; 116(15): 7439–7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lachmandas E, Boutens L, Ratter JM, Hijmans A, Hooiveld GJ, Joosten LA et al. Microbial stimulation of different Toll-like receptor signalling pathways induces diverse metabolic programmes in human monocytes. Nat Microbiol 2016; 2: 16246. [DOI] [PubMed] [Google Scholar]
- 72.Treadway MT, Cooper JA, Miller AH. Can’t or Won’t? Immunometabolic Constraints on Dopaminergic Drive. Trends Cogn Sci 2019; 23(5): 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allen J, Romay-Tallon R, Brymer KJ, Caruncho HJ, Kalynchuk LE. Mitochondria and Mood: Mitochondrial Dysfunction as a Key Player in the Manifestation of Depression. Front Neurosci 2018; 12: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karabatsiakis A, Bock C, Salinas-Manrique J, Kolassa S, Calzia E, Dietrich DE et al. Mitochondrial respiration in peripheral blood mononuclear cells correlates with depressive subsymptoms and severity of major depression. Transl Psychiatry 2014; 4: e397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beech RD, Lowthert L, Leffert JJ, Mason PN, Taylor MM, Umlauf S et al. Increased peripheral blood expression of electron transport chain genes in bipolar depression. Bipolar Disord 2010; 12(8): 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeng D, He S, Ma C, Wen Y, Xie Y, Zhao N et al. Co-Expression Network Analysis Revealed That the ATP5G1 Gene Is Associated With Major Depressive Disorder. Front Genet 2019; 10: 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Q, Dwivedi Y. Transcriptional profiling of mitochondria associated genes in prefrontal cortex of subjects with major depressive disorder. The World Journal of Biological Psychiatry 2016; 18(8): 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martins-de-Souza D, Guest PC, Harris LW, Vanattou-Saifoudine N, Webster MJ, Rahmoune H et al. Identification of proteomic signatures associated with depression and psychotic depression in post-mortem brains from major depression patients. Transl Psychiatry 2012; 2: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Esteban-Martinez L, Sierra-Filardi E, McGreal RS, Salazar-Roa M, Marino G, Seco E et al. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J 2017; 36(12): 1688–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weckmann K, Deery MJ, Howard JA, Feret R, Asara JM, Dethloff F et al. Ketamine’s antidepressant effect is mediated by energy metabolism and antioxidant defense system. Sci Rep 2017; 7(1): 15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weckmann K, Labermaier C, Asara JM, Muller MB, Turck CW. Time-dependent metabolomic profiling of Ketamine drug action reveals hippocampal pathway alterations and biomarker candidates. Transl Psychiatry 2014; 4: e481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stacey D, Schubert KO, Clark SR, Amare AT, Milanesi E, Maj C et al. A gene co-expression module implicating the mitochondrial electron transport chain is associated with long-term response to lithium treatment in bipolar affective disorder. Transl Psychiatry 2018; 8(1): 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sobin C, Sackeim HA. Psychomotor symptoms of depression. Am J Psychiatry 1997; 154(1): 4–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


