Abstract
Purpose:
One outcome of DNA damage from hydroxyl radical generated by ionizing radiation (IR) or by the Fenton reaction is oxidation of the nucleobases, especially guanine (G). While 8-oxo-7,8-dihydroguanine (OG) is a commonly studied oxidized lesion, several others are formed in high abundance, including 5-carboxamido-5-formamido-2-iminohydantoin (2Ih), a prevalent product in in vitro chemistry that is challenging to study from cellular sources. In this short review, we have a goal of explaining new insights into hydroxyl radical-induced oxidation chemistry of G in DNA and comparing it to endogenous DNA damage, as well as commenting on the biological outcomes of DNA base damage.
Conclusions:
Pathways of oxidation of G are discussed and a comparison is made between IR (hydroxyl radical chemistry) and endogenous oxidative stress that largely forms carbonate radical anion as a reactive intermediate. These pathways overlap with the formation of OG and 2Ih, but other guanine-derived lesions are more pathway specific. The biological consequences of guanine oxidation include both mutagenesis and epigenetics; a new mechanism of gene regulation via the base excision repair pathway is described for OG, whereas the impact of IR in forming guanine modifications may be to confound this process in addition to introduction of mutagenic sites.
Keywords: DNA damage, hydroxyl radical, guanine oxidation, base excision repair, gene expression
Purpose:
The purpose of this article is to present new insights into the chemistry of DNA damage to the heterocyclic base guanine (G) caused by hydroxyl radical generating sources, to compare this chemistry to that emanating from reactive oxygen species of endogenous origin, and to discuss briefly the consequences of these chemical modifications which include both mutagenic events and changes in gene expression. We focus on G because it is the heterocyclic base most susceptible to oxidation, and its modification has been demonstrated to have both mutagenic and epigenetic outcomes. The product 8-oxo-7,8-dihydroguanine, OG, is one of dozens of different oxidized bases generated in the process of oxidative DNA damage, and it is the focus here because of its abundance and it role in both mutagenesis and alteration of gene expression.
A second goal is to highlight how difficult it is to determine all the products of oxidative base damage, to quantify them and to find their locations in the genome. We and others find that G-rich regulatory regions of DNA, including gene promoters, are sensitive to oxidation and that the presence of lesions such as OG can therefore impact gene regulation (Amente et al. 2019; Ding et al. 2017; Fleming et al. 2017; Pan et al. 2016; Perillo et al. 2008). In brief, with this article we bring light to new findings concerning the role of hydroxyl radical in guanine oxidation, present a few mysteries regarding the identities of major oxidation products of guanine, and discuss the impact of guanine oxidation on gene expression via DNA repair.
Discussion:
Ionizing radiation (IR) is so termed because the photons or particles are of sufficient energy to eject electrons from the substrate creating ions from neutral components. In chemistry, any process that leads to loss of electrons is considered an oxidation. Ionization of water leads to hydroxyl radical formation plus an electron; the HO● so formed can add to any of the heterocyclic bases of DNA, or it can abstract a hydrogen atom (H●) from the base or the ribose (Pogozelski and Tullius 1998; Steenken 1989). IR can also directly impact DNA bases by loss of an electron and a proton generating, for example, guanine and adenine radicals, formally (G-H)● and (A-H)● (Greenberg 2021). The chemistry of guanine and adenine radicals is complex and depends upon the surrounding sequence and the presence of oxidants including O2 and reductants such as glutathione, ascorbate and urate. Many reviews detail these chemical pathways, a portion of which is shown in Figure 1 (Cadet et al. 2014; Greenberg 2021).
Figure 1.
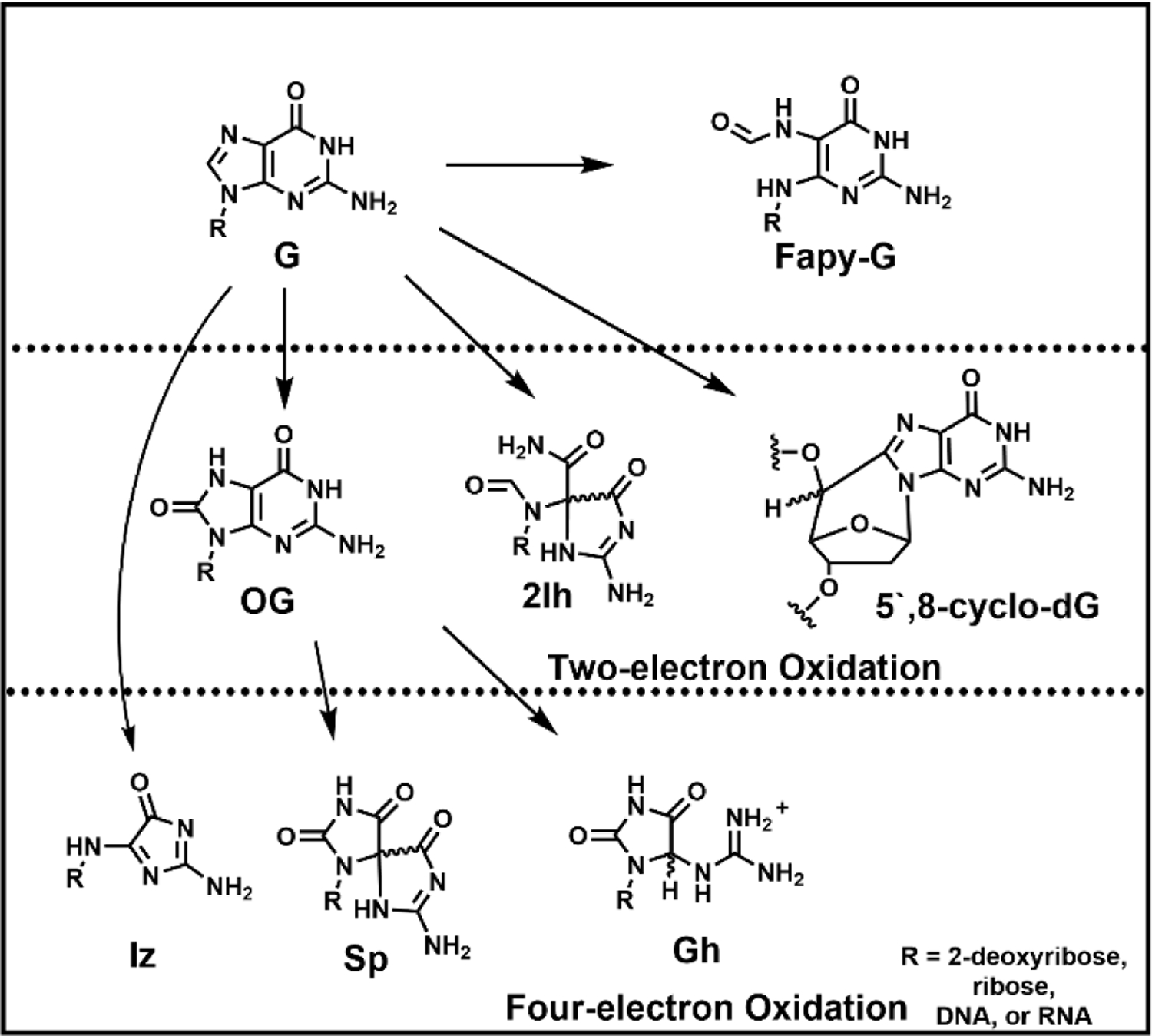
Overall pathways to products of G oxidation resulting from two- or four-electron oxidation processes.
Products of Guanosine Oxidation.
In Figure 1, we show the most commonly discussed structures that are derived from reactive oxygen species (ROS) attacking G, with OG being the major product in most studies (Cadet et al. 2008; Cadet et al. 2014; Fleming and Burrows 2017a; Neeley and Essigmann 2006). OG represents a formal two-electron oxidation of G, although the pathway by which it is formed typically involves unstable one-electron oxidized intermediates. To illustrate two very different mechanisms, we show in Figure 2 the reaction of G with hydroxyl radical vs. carbonate radical anion (Alshykhly et al. 2015b; Cadet et al. 2008; Crean et al. 2005); in the former, hydroxyl radical adds to the C8 position of G to form 8-hydroxy-G radical, (G-OH)●, which after the loss of one more electron and a proton leads to 8-hydroxyG and then OG after tautomerization. In the second pathway, loss of an electron to form G●+ occurs first, and attack of H2O at C8 leads to the same intermediate (G-OH) ●. Another proposed pathway to form G●+ via hydroxyl radical is addition of the radical across the C4-C5 double bond followed by elimination of HO- (Kumar et al. 2011). Thus, OG is the product formed from two very different types of ROS, the hydroxyl radical HO● and one-electron oxidants such as carbonate radical anion, CO3●─. Conditions under which these two radicals are formed will be discussed later.
Figure 2.
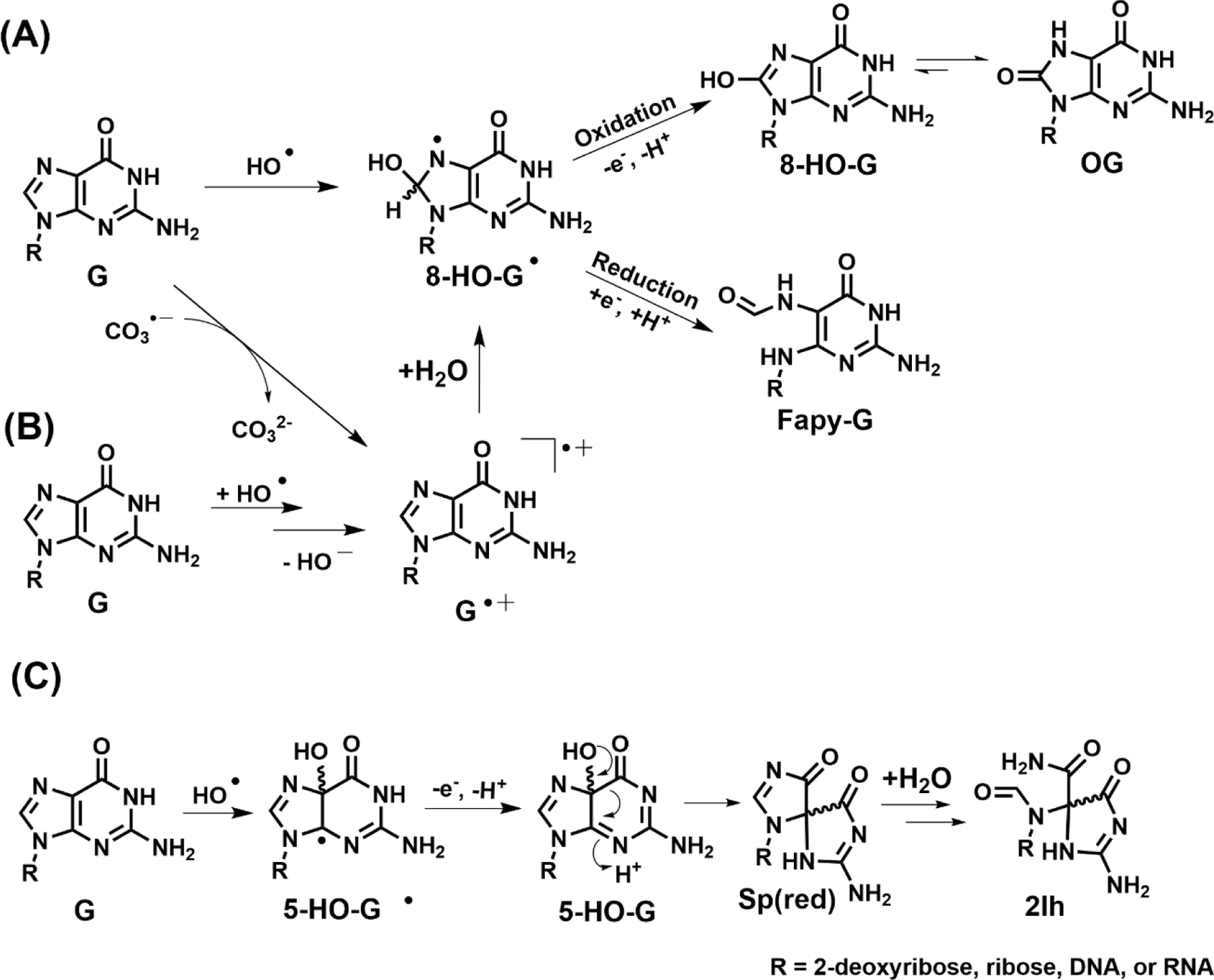
Proposed mechanisms for G oxidation at (A) C8 to yield OG or Fapy-G, (B) by hydroxy radical to yield G radical cation, or (C) at C5 to yield 2Ih.
Another potential outcome of the unstable intermediate (8-OH-G●) is one-electron reduction and protonation, which after ring opening to a more stable carbonyl compound would produce the formamidopyrimidine product derived from dG, or Fapy-G (Figure 1)(Cadet et al. 2008; Dizdaroglu 2015). This type of reduction can only occur under strongly anoxic conditions, achieved in very hypoxic tumors, or when ionizing radiation generates both HO● and H● that add sequentially to the G heterocycle without the radical intermediate (8-OH-G●) being intercepted by O2 or O2●─ (Douki et al. 1997). Fapy-G has been found in quantities nearly equal to that of OG under conditions of ionizing radiation (Frelon et al. 2000). Analysis of Fapy-G as a nucleoside is complicated by ribose ring opening and reclosure that can generate both α and β anomers at C1’ as well as both furanose (5-membered) and pyranose (6-membered) rings (Berger and Cadet 1985), as well as the glycosidic bond being labile toward hydrolysis to yield the free base. Thus, Fapy-G is a good example of a G lesion that has represented challenges for consistent detection and quantification between laboratories (Alshykhly et al. 2015b; Cui et al. 2013; Douki et al. 1997; Frelon et al. 2000; Swarts et al. 1996).
5-Hydroxy vs. 8-Hydroxy-dG.
We show in Figure 2A,B that three mechanisms could lead to 8-hydroxyguanosine which isomerizes to OG. Similarly, two analogous mechanisms, hydroxyl radical addition or hydration of G●+, could lead to hydroxylation of G at C5 rather than C8 (Fig. 2C) (Cadet et al. 2008; Cadet et al. 2014; Fleming and Burrows 2017a). The 5-hydroxy isomer is also an unstable intermediate; molecules of this sort are more stable as carbonyl compounds if possible, and in this case, a facile 1,2-acyl shift can occur to generate a new isomeric heterocycle, which after hydration and ring opening leads to 2Ih, first characterized by Louise Ball and coworkers (North Carolina) (Ye W et al. 2009). The rearrangement, proposed initially by Genevieve Pratviel (CNRS, Toulouse) by analogy to the same rearrangement reported in our laboratory in 2000 for 5,8-dihydroxyG, leads to a spirohydantoin structure, as shown in Figure 2C (Lapi et al. 2001; Luo et al. 2000; Vialas et al. 2000). Because this spirocycle is not oxidized at C8 and the 5-membered ring is no longer aromatic, it undergoes a slow addition of H2O at C8, followed by ring opening to a bis-formamido-2-iminohydantoin structure, 2Ih.
In vitro, the formation of 2Ih is on par with the yield of OG under conditions that mimic cellular systems, i.e., oxidation by transition metal-induced ROS or ionizing radiation in the presence of reductants such as ascorbate or thiols (Alshykhly et al. 2015b; Fleming et al. 2011). If cellular reductants are absent, the C5 position of G is subject to perhydroxylation (reaction with O2 or O2●─) rather than hydroxylation (reaction with H2O), and the final products are different—the imidazolone product Iz and subsequent degradation products are formed (Figure 1). Although we often find 2Ih to be as abundant as OG, it is severely understudied as a DNA lesion, likely because it is so difficult to detect. It lacks a UV chromophore and elutes with the void volume on a C18 HPLC column. The glycosidic bond in the 2Ih nucleoside is not very stable, and so it is perhaps unsurprising that this lesion has not been detected from cellular samples where considerable manipulation is required to identify these one-in-a-million G lesions. With no antibody available for 2Ih nor a reliable signature for sequencing, determining the sequence location of such lesions remains a challenge. Nevertheless, we and others have been able to isolate and characterize this lesion, both in the nucleoside and oligonucleotide contexts, in order to better define the likely chemistry and biochemistry that may occur in the cellular context (Alshykhly et al. 2015b, 2015a; Fleming et al. 2011; Rokhlenko et al. 2012; Vialas et al. 2000; Ye W et al. 2009).
Hyperoxidation products of G.
A curious feature of OG is its high sensitivity to further oxidation; whereas the one-electron redox potential of G is 1.29 V vs. NHE, the potential for OG is 700 mV lower, making OG thousands of times more reactive toward one-electron oxidants than G (Steenken et al. 2000). Indeed, one way to analyze OG by HPLC methods is through use of an electrochemical detector (Helbock et al. 1998). Studies with isotopically labeled H218O showed that OG●+ is susceptible to attack at C5, forming 5-hydroxy-OG, and as mentioned above, this species rearranges to a spirocyclic base if possible via an acyl shift of the C6 carbonyl to become bonded to C4 (Luo et al. 2000). This spirodihydantoin base, named Sp (Figure 1), is very chemically stable, although it can revert to 5-OH-OG at pH ≤ 3 (Ye Y et al. 2009). In duplex DNA where spirocyclization is sterically inhibited or at lower pH (e.g., < 6), the 5-OH-OG species instead becomes hydrated at C6, ring opens and decarboxylates to form 5-guanidinohydantoin, Gh (Figure 1) (Luo et al. 2001). Therefore, the final outcome of OG oxidation is principally a mixture of Sp and Gh, depending on single-stranded vs. duplex context (Fleming et al. 2012). The hydantoin structures, while initially controversial, have been thoroughly characterized including two crystal structures in collaboration with Sylvie Doublié (U. Vermont) (Aller et al. 2010; Eckenroth et al. 2014), and via extensive spectroscopic methods (Fleming et al. 2013). The hydantoins have also been detected in vivo in a mouse model of inflammation where levels of these lesions were 1–2 orders of magnitude lower than OG (Mangerich et al. 2012).
Notably, the hydantoin lesions Sp, Gh and 2Ih do not resemble G or OG in their base-pairing properties; that is, none of these is able to form a stable base pair with C (Alshykhly et al. 2015a; Fleming and Burrows 2017b). They are significantly destabilizing to duplex DNA, and these distortions have been addressed computationally by Suse Broyde (NYU) (Jia et al. 2005). When bypassed by polymerases, Sp and Gh lead to the misincorporation of both G and A opposite, i.e. G→C and G→T transversion mutations (Figure 3) (Kornyushyna and Burrows 2003). Not surprisingly, an in vivo mutagenesis assay indicated that the hydantoins are nearly 100% mutagenic if not repaired (Henderson et al. 2003). In contrast, OG is only about 2% mutagenic because it is able to base pair with C if it remains in the anti orientation, and if it does mispair with A in the syn orientation, there is a back-up mechanism for its repair via MUTYH (David et al. 2007; Neeley et al. 2007).
Figure 3.
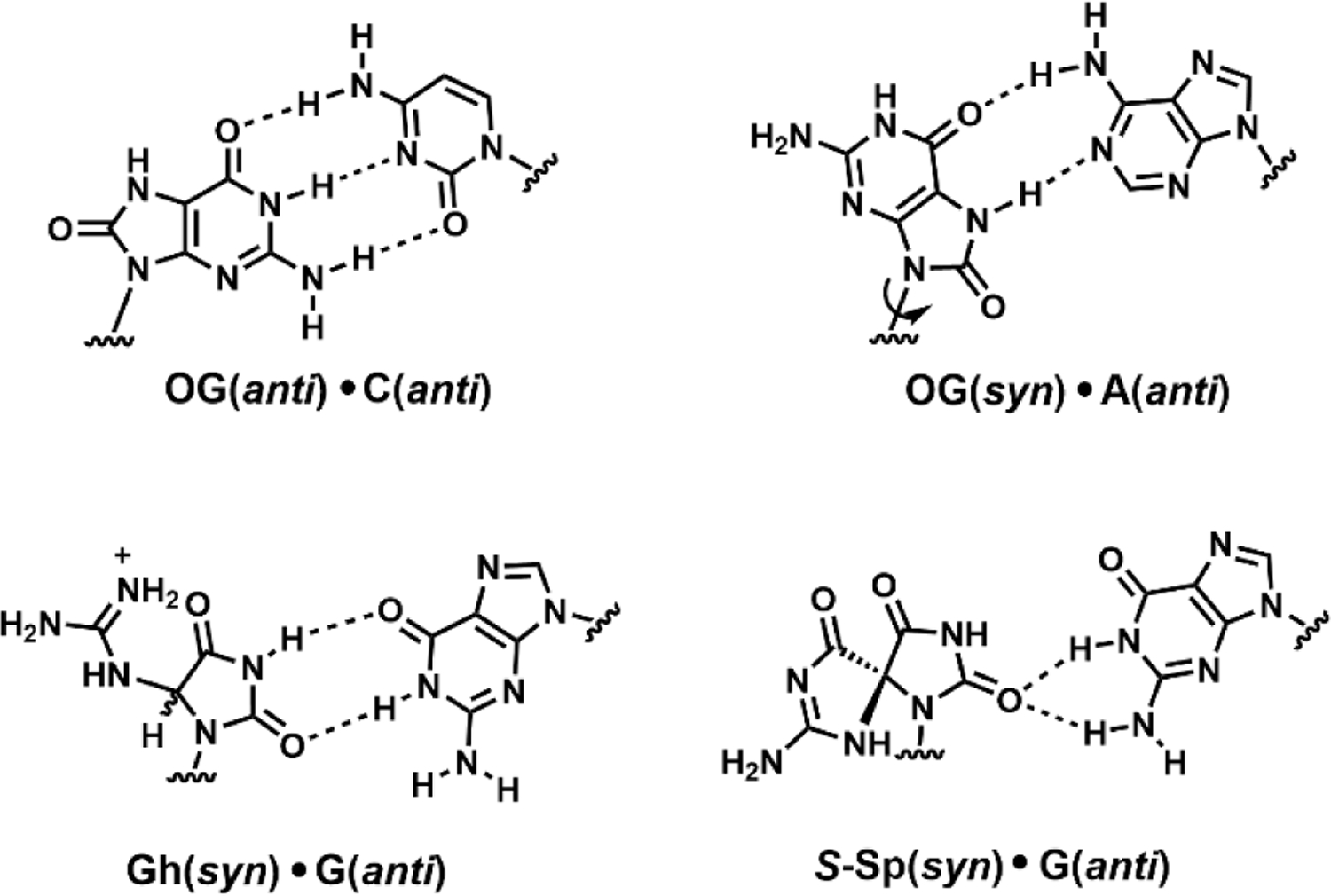
Base pairs formed by G oxidation products.
An argument for a significant role played by the hydantoins in cellular oxidative damage is the fact that they are by far the best substrates known for the NEIL repair enzymes (Fleming and Burrows 2017a; Krishnamurthy et al. 2008; Liu et al. 2010). Sheila David’s laboratory (UC-Davis) has shown that NEIL1, the human homolog of bacterial endonuclease VIII, recognizes and excises hydantoin bases with a strong preference over their originally discovered substrates such as thymine glycol, although the substrate specificity is dependent on native vs. edited versions of the glycosylase (Yeo et al. 2010). The hydantoins Sp and Gh are found to arise from endogenous oxidative stress whereas 2Ih can also be formed from radiation damage (Alshykhly et al. 2015b; Fleming and Burrows 2013). Thymine glycol (Tg) on the other hand is a product principally of radiation damage and metabolism because of the generation of the highly reactive hydroxyl radical which can hydroxylate any base (Cadet et al. 2008). It is fitting therefore that edited NEIL1, which is expected to increase during inflammation creating endogenous carbonate radical anion, has a heightened substrate preference for Sp and Gh compared to Tg.
The base excision repair (BER) pathway has evolved largely to deal with oxidative damage products in DNA but in fact, we expect a different spectrum of base oxidation products to be formed in DNA during radiation damage compared to endogenous oxidative stress because the ROS are somewhat different. It is somewhat surprising then that the BER system works as well as it does on radiation-damaged base lesions given that radiation was probably not a big evolutionary driver on early Earth. Perhaps this quandary is solved by noting that there is substantial cross-over among the damage products of endogenous and radiation-induced oxidation (Cadet et al. 2008; Cadet et al. 2014; Fleming and Burrows 2017a). Complicating this picture is the fact that radiation damage to tissues can then trigger subsequent inflammatory responses which result in a blend of multiple ROS (McKelvey et al. 2018). Therefore, the immediate vs. longer term DNA damage products may differ to some extent. A part of this picture is described next for two commonly generated ROS.
ROS: Hydroxyl radical vs. Carbonate Radical Anion.
Direct deposition of high energy into DNA can lead to frank strand breaks which are processed by homologous recombination and non-homologous end joining (Scully et al. 2019). Here we consider only the DNA nucleobase lesions that are generated by radiation vs. inflammation-induced ROS. Ionizing radiation events generating radiation tracks are known to damage DNA to yield complex products such as clustered lesions and strand breaks with dependency on the track structure; we suggest a few prior reports for readers interested in this topic (Georgakilas et al. 2013; Goodhead 1994; Sutherland et al. 2000). Hydroxyl radical (HO●) is typically blamed for indiscriminate DNA damage as a highly reactive radical that can add to all four bases and abstract hydrogen atoms from any accessible site in the ribose moiety, with base damage leading to mutations and sugar oxidation leading to strand breaks (Neeley and Essigmann 2006; Pogozelski and Tullius 1998). However, HO● may not be formed to the extent previously thought. Some of the pathways believed to form HO● actually form carbonate radical anion, CO3●─, instead due to the relatively high concentration of bicarbonate in cells, about 20 mM, and also to the presence of CO2 (~1 mM). For example, the inflammatory response generates nitric oxide (NO) and superoxide (O2●─) which combine to form peroxynitrite (ONOO─) (Lonkar and Dedon 2011). Early reports posited that ONOO─ decomposed to generate hydroxyl radical, but more recently it was shown that its preferred reaction is with CO2 leading to peroxynitrosocarbonate (ONOOCO2─), which decomposes to carbonate radical anion (CO3●─) and ●NO2 (Fig. 4) (Lonkar and Dedon 2011; Neeley and Essigmann 2006).
Figure 4.
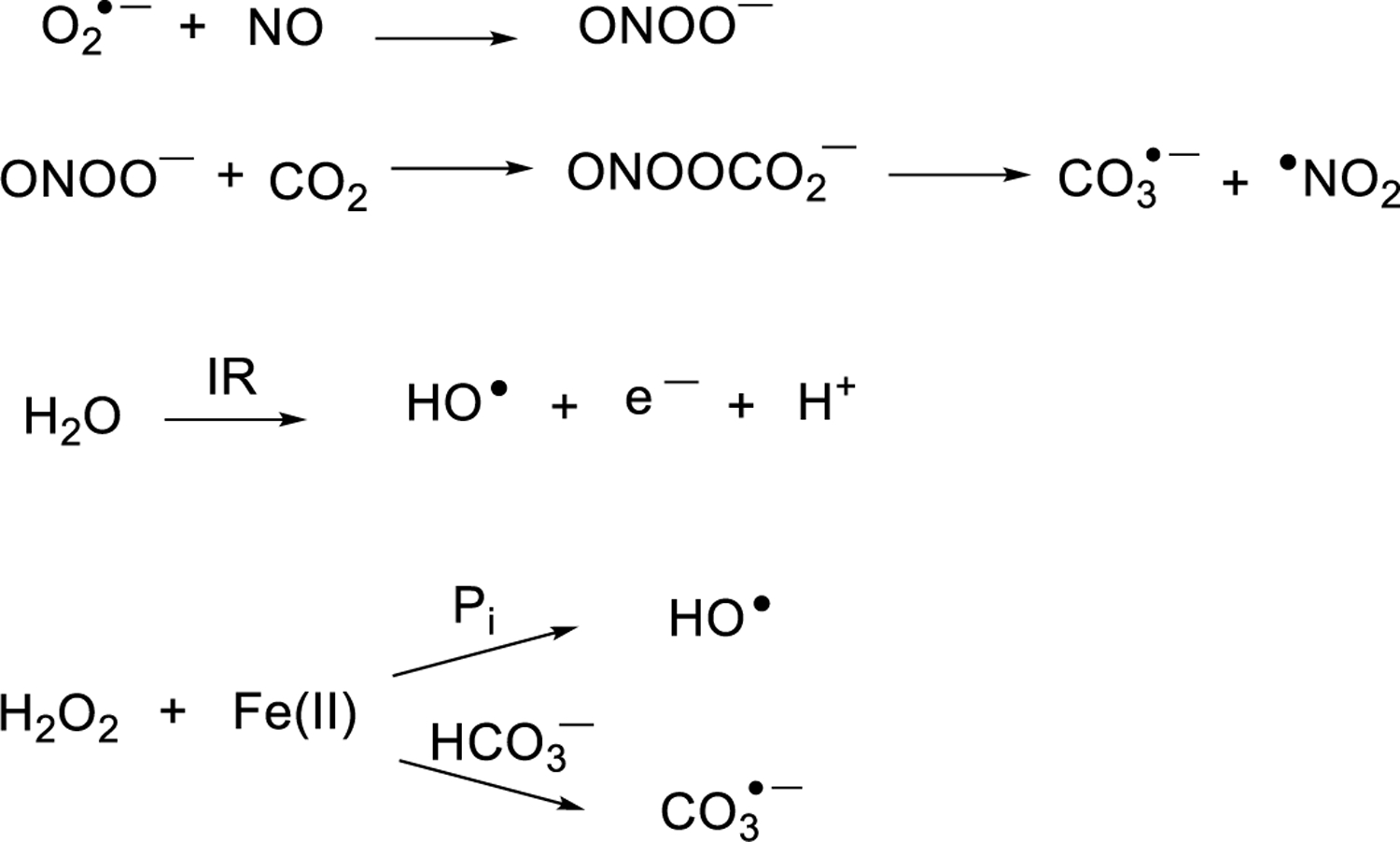
Pathways for formation of hydroxyl radical and carbonate radical anion.
The Fenton reaction is also thought of as a cellular source of hydroxyl radical because of the metabolic production of H2O2. In the presence of Fe(II), H2O2 will decompose to generate hydroxyl radical. This picture changed in 2019 when Meyerstein and coworkers reported that in the presence of physiological bicarbonate, which is normally around 20 mM but any amount above 500 μM will have the same impact, leads to an inner-sphere oxidation of iron-bound carbonate to CO3●─, circumventing formation of HO● (Fig 4) (Illes et al. 2019).
Why does the identity of the ROS matter? (1) Because the oxidation products are different. As already noted, HO● attacks any base or sugar forming adducts that include OG and Tg while CO3●─ is a one-electron oxidant that prefers to generate guanine radical cation, G●+. Some of the final outcomes are the same, for example, the formation of OG (Fig. 2). Further oxidation to Sp or Gh can also occur with CO3●─. Some products are different; notably, the less reactive CO3●─ does not react with A, C, and T to any major extent when G is abundant (Joffe et al. 2003). Also, hydroxyl radical attack at C5’ of a ribose abstraction a hydrogen atom and generating the C5’ radical can lead to the 5’,8-cyclo-dG adduct as signature of the involvement of hydroxyl radical chemistry (Chatgilialoglu et al. 2011).
To expand on the Meyerstein findings that hydroxyl radical is not formed in the presence of bicarbonate, we studied the Fe(II)-mediated oxidation products of guanosine in phosphate vs. bicarbonate buffers (Fleming and Burrows 2020a). When Fe(II) was complexed to either EDTA (as in classical Fenton chemistry) or to α-ketoglutarate (a common cellular ligand for iron) the outcome was the same; oxidation of G with H2O2 mediated by Fe(II) led to OG + 5’,8-cyclo-dG in phosphate buffer, but in contrast, the products were exclusively OG (and its further oxidation product Sp) in bicarbonate buffer (Figure 5). This tells us that endogenous Fenton chemistry generates principally CO3●─, not HO●. Preliminary studies on radiation-induced G oxidation indicate that some but not all of the HO● formed in IR is quenched in the presence of bicarbonate buffer, but more thorough studies are needed.
Figure 5.
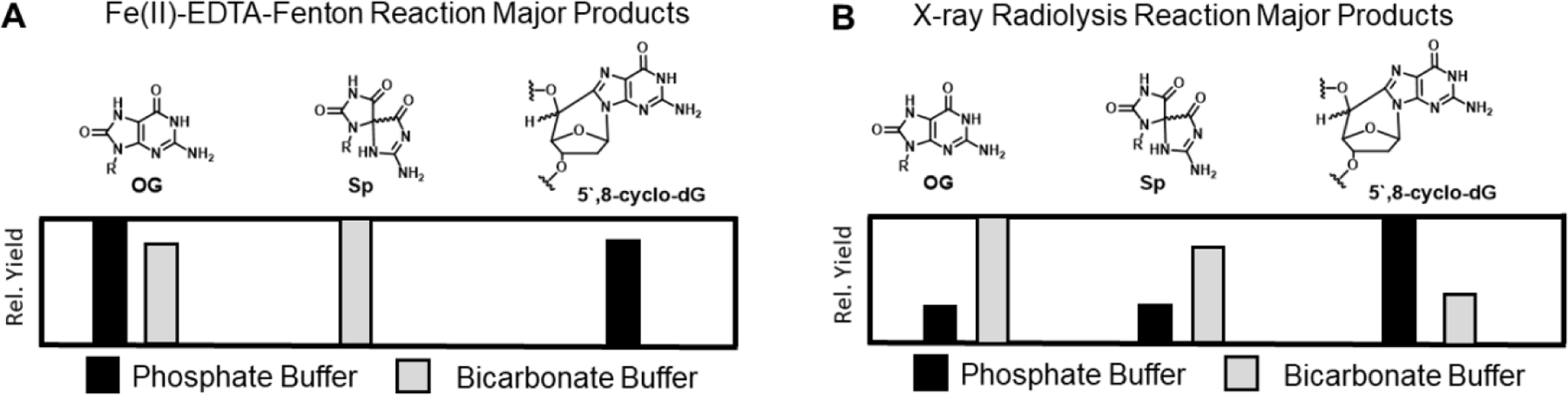
The buffer, phosphate vs. bicarbonate, impacts the ROS generated from the Fe(II)-EDTA Fenton reaction or X-ray radiolysis of water as reported by the major products detected from G oxidation. Relative yield refers to phosphate vs. bicarbonate buffer. Data in panel A were previously reported with complete details regarding the yields, as well as oxygen and iron ligand dependency of the reaction (Fleming and Burrows 2020a); panel B data are unpublished.
Why does the identity of the ROS matter? (2) Because the location of oxidation products can be different. IR-induced formation of neutral base radicals or hydroxyl radical adducts is fairly random in terms of the sites of attack in a DNA sequence because of the high reactivity of the species involved. In contrast, electron hole formation from removal of one electron by carbonate radical anion is strongly preferred at G to form G●+ (Rokhlenko et al. 2012), and furthermore, the electron hole can migrate in duplex DNA over hundreds or maybe thousands of nucleotides via charge transport to localize on the most stable site for oxidation before a slower second step that forms the final lesion (Tse et al. 2019), such as H2O attack and a further one-electron oxidation to form OG. In duplex DNA, the lowest redox potential is found at either OG (but these sites are rare) or at oligo-dG sites such as 5’-GGG-3’ or 5’-GGGG-3’ wherein the G●+ has been shown to prefer any of the underlined G’s, that is, all but the 3’-most G (Saito et al. 1995). This ‘damage-at-a-distance’ phenomenon has been intensely studied by Jacqueline Barton’s laboratory (Caltech), and it has been invoked as a signaling mechanism to home in on remote sequences for protein binding (Tse et al. 2019). Interestingly, these types of G-rich sequences are common in regulatory regions of DNA such as gene promoters, and when four or more GGG tracks are in close proximity these sequences can fold to G-quadruplexes (Fleming and Burrows 2020b). This suggests that G-quadruplex (G4)-forming sequences might be hotspots for oxidation and therefore sites of special BER activity.
About a decade ago, Susan Wallace (U. Vermont) suggested we synthesize part of the human telomere G-quadruplex sequence with hydantoin lesions at various positions in the sequence, and significantly, her laboratory then found that NEIL3 preferred to excise Sp or Gh from the G4 context (Zhou et al. 2013). These studies immediately pointed to a biological role for BER at the sites of G-quadruplexes during oxidative or radiation stress.
Biological Consequences of G Oxidation—Mutagenesis vs. Epigenetics.
The mutagenic properties of OG have been the subject of hundreds of papers in the literature in which the G→T transversion mutations observed from mispairing of OG in the syn conformation with A in the opposite strand lead ultimately to the replacement of OG with T at that position in the genome. The BER enzymes OGG1 and MUTYH are responsible for excising OG and the mispaired A, separately, from duplex DNA (David et al. 2007). However, OG is not “highly mutagenic” as often reported; Ogg1−/− mice live relatively normal lives (Klungland et al. 1999).
Another role for OG is to impact gene expression. The idea that OG might be epigenetic was discussed in 2013 by Phyllis Strauss (Northeastern U.) who performed studies with the CREB protein that controls 25% of the transcriptome (Moore et al. 2013). The CRE binding domain includes two Gs on each strand whose modification to OG resulted in diminished binding of CREB, thereby reducing gene induction.
In our work and others, a more surprising finding was that OG in promoter regions can actually increase transcription factor occupancy resulting in gene induction (Fleming et al. 2017; Pan et al. 2016; Pastukh et al. 2015; Perillo et al. 2008). We found that OG located in the loop of a promoter G4 could dramatically impact gene expression in a reporter plasmid. OG could be installed synthetically at specific sites or could be formed in cellulo by incubation with TNFα, which increases ONOO- formation (Fleming et al. 2017; Fleming, Zhu, Howpay Manage, et al. 2019). When OG was placed in the non-template strand just upstream of the transcription start site, gene expression actually increased. This increase was dependent upon the BER pathway, specifically the ability to form an abasic (AP) site, the folding of the strand into a G4, and the ability of AP-endonuclease to bind but not cleave the AP site in the G4 context (Figure 6). This mechanism is enabled by AP-endonuclease displacing OG-glycosylase 1 and preventing it from catalyzing lyase activity once the AP is formed (Vidal et al. 2001). The co-location of OG and a folded G4, both thought of as ‘DNA damage,’ had a strikingly positive effect, although the sign and magnitude of the effect was location dependent (Fleming, Zhu, Ding, et al. 2019). Thus, we agree that the term ‘epigenetic’ should be applied to OG in some circumstances, those being cases in which endogenous oxidative stress is funneling the electron hole to G-rich regulatory regions of the genome where G-quadruplexes serve as a sensor of oxidative stress, and the base modification impacts gene expression (Fleming and Burrows 2020b, 2021). In other locations, and in most instances of radiation-induced DNA damage, we would expect the chemical modification of DNA bases to be mutagenic (Fleming and Burrows 2020b).
Figure 6.
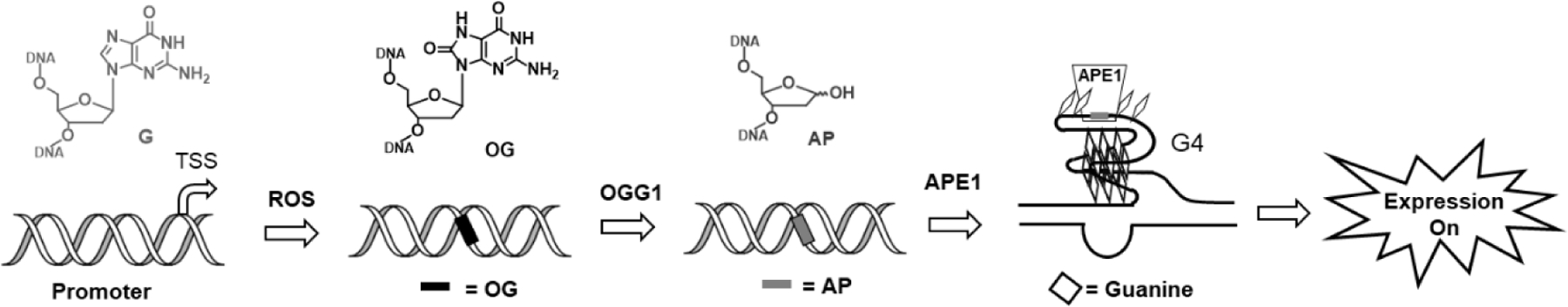
Gene expression is induced when G is oxidized to OG in a promoter G-quadruplex via BER.
Conclusions:
Many literature sources imply that the chemistry of oxidative DNA damage is very similar between radiation-induced vs. endogenous stress and point to hydroxyl radical as the primary culprit (Cooke et al. 2003; De Bont and van Larebeke 2004; Kowalska et al. 2020). We now understand that carbonate radical anion, a one-electron oxidant, is very important to guanine oxidation during endogenously generated oxidative stress (Fleming and Burrows 2020c). Even during the initial period of DNA damage invoked by IR, hydroxyl radical is not the lone damaging species; it is attenuated by the presence of physiological bicarbonate, thereby generating a mixture of products emanating from hydroxyl radical and from carbonate radical anion. A confounding fact in all of this is that OG is a product of both pathways, either from direct hydroxyl radical attack at C8 of G or from attack of H2O at C8 of G●+ (Figure 2). Therefore, we found that analysis of other products, such as 5’,8-cyclo-dG, is more diagnostic of the reactive species that is damaging DNA because this product arises only from radical formation at C5’ of the ribose and not from the one-electron pathway of CO3●─ (Fleming and Burrows 2020a).
Fortunately, base excision repair enzymes in the form of OGG1 + MUTYH and NEIL1/2/3 remove guanine oxidation products from the genome to avoid mutagenesis imposed by OG vs. the hydantoins Sp, Gh, and 2Ih (Fleming and Burrows 2017a). The observation that the BER pathway is involved in both up- and downregulation of gene expression brings many questions along with it, including the complete molecular picture of how BER enzymes operate on different lesions and different structural motifs including the presence of lesions in nucleosomal DNA, as is being investigated by Sarah Delaney (Brown) (Li and Delaney 2019).
In the future, we need a more complete atlas of radiation vs. endogenously-formed DNA modifications. For too long, our focus has been on low-hanging fruit such as OG, but the rarer lesions such as hydantoins could play significant roles. Important to the advancement of this field is improvements in DNA sequencing methodology to actually sequence for rare oxidative modifications at high resolution. Challenges in this area include finding strategies to pull-down only lesion-containing sequences, and then to solve the problem of amplification of lesion-containing DNA in such a way that information of the modified base is retained. Our laboratory has tackled some of these issues (Ding et al. 2017; Riedl et al. 2015), and advances have been made by Shana Sturla’s group (ETH-Zurich) (Wu et al. 2018).
Ample questions remain to be addressed by the next generation of researchers investigating the intersection of DNA damage, DNA repair and gene expression. Answers to these questions will aid in our understanding and treatment of aging, cancer, and neurological diseases as well as environmental exposure to radiation.
Acknowledgements:
We thank many coworkers and collaborators over the years for experimental work and critical insights. In particular, we thank the nine women mentioned in this paper for their inspirational accomplishments in the field. Work in our laboratory was funded by the U.S. National Institutes of Health via grants R01 CA090689 and R01 GM129267.
Biographical Notes:
Aaron M. Fleming, PhD, is a Research Associate Professor of Chemistry at the University of Utah. Cynthia J. Burrows, PhD, is Distinguished Professor of Chemistry and Thatcher Presidential Chair of Biological Chemistry at the University of Utah.
Footnotes
Disclosure Statement: The authors declare no competing interests.
References:
- Aller P, Ye Y, Wallace SS, Burrows CJ, Doublie S. 2010. Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin. Biochemistry 49(11):2502–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshykhly OR, Fleming AM, Burrows CJ. 2015a. Guanine oxidation product 5-carboxamido-5-formamido-2-iminohydantoin induces mutations when bypassed by DNA polymerases and is a substrate for base excision repair. Chem Res Toxicol 28(9):1861–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshykhly OR, Fleming AM, Burrows CJ. 2015b. 5-Carboxamido-5-formamido-2-iminohydantoin, in addition to 8-oxo-7,8-dihydroguanine, is the major product of the iron-Fenton or X-ray radiation-induced oxidation of guanine under aerobic reducing conditions in nucleoside and DNA contexts. J Org Chem 80(14):6996–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amente S, Di Palo G, Scala G, Castrignanò T, Gorini F, Cocozza S, Moresano A, Pucci P, Ma B, Stepanov I et al. 2019. Genome-wide mapping of 8-oxo-7,8-dihydro-2′-deoxyguanosine reveals accumulation of oxidatively-generated damage at DNA replication origins within transcribed long genes of mammalian cells. Nucleic Acids Res 47(1):221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M, Cadet J. 1985. Isolation and characterization of the radiation-induced degradation products of 2`-deoxyguanosine in oxygen-free aqueou solutions. Z Naturforsch [B] 40b:1519–1531. [Google Scholar]
- Cadet J, Douki T, Ravanat J-L. 2008. Oxidatively generated damage to the guanine moiety of DNA: Mechanistic aspects and formation in cells. Acc Chem Res 41(8):1075–1083. [DOI] [PubMed] [Google Scholar]
- Cadet J, Loft S, Olinski R, Evans MD, Bialkowski K, Wagner RJ, Dedon PC, Møller P, Greenberg MM, Cooke MS. 2012. Biologically relevant oxidants and terminology, classification and nomenclature of oxidatively generated damage to nucleobases and 2-deoxyribose in nucleic acids. Free Radic Res 46(4):367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Wagner RJ, Shafirovich V, Geacintov NE. 2014. One-electron oxidation reactions of purine and pyrimidine bases in cellular DNA. Int J Radiat Biol 90(6):423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatgilialoglu C, Ferreri C, Terzidis MA. 2011. Purine 5’,8-cyclonucleoside lesions: chemistry and biology. Chem Soc Rev 40(3):1368–1382. [DOI] [PubMed] [Google Scholar]
- Cho BP, Kadlubar FF, Culp SJ, Evans FE. 1990. 15N nuclear magnetic resonance studies on the tautomerism of 8-hydroxy-2’-deoxyguanosine, 8-hydroxyguanosine, and other C8-substituted guanine nucleosides. Chem Res Toxicol 3(5):445–452. [DOI] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. 2003. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J 17(10):1195–1214. [DOI] [PubMed] [Google Scholar]
- Cooke MS, Loft S, Olinski R, Evans MD, Bialkowski K, Wagner RJ, Dedon PC, Møller P, Greenberg MM, Cadet J. 2010. Recommendations for standardized description of and nomenclature concerning oxidatively damaged nucleobases in DNA. Chem Res Toxicol 23(4):705–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crean C, Geacintov NE, Shafirovich V. 2005. Oxidation of guanine and 8-oxo-7,8-dihydroguanine by carbonate radical anions: Insight from oxygen-18 labeling experiments. Angew Chem, Int Ed 44(32):5057–5060. [DOI] [PubMed] [Google Scholar]
- Cui L, Ye W, Prestwich EG, Wishnok JS, Taghizadeh K, Dedon PC, Tannenbaum SR. 2013. Comparative analysis of four oxidized guanine lesions from reactions of DNA with peroxynitrite, singlet oxygen, and gamma-radiation. Chem Res Toxicol 26(2):195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SS, O’Shea VL, Kundu S. 2007. Base-excision repair of oxidative DNA damage. Nature 447:941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N. 2004. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19(3):169–185. [DOI] [PubMed] [Google Scholar]
- Ding Y, Fleming AM, Burrows CJ. 2017. Sequencing the mouse genome for the oxidatively modified base 8-oxo-7,8-dihydroguanine by OG-Seq. J Am Chem Soc 139:2569–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M. 2015. Oxidatively induced DNA damage and its repair in cancer. Mutat Res Rev Mutat Res 763:212–245. [DOI] [PubMed] [Google Scholar]
- Douki T, Martini R, Ravanat JL, Turesky RJ, Cadet J. 1997. Measurement of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 8-oxo-7,8-dihydroguanine in isolated DNA exposed to gamma radiation in aqueous solution. Carcinogenesis 18(12):2385–2391. [DOI] [PubMed] [Google Scholar]
- Eckenroth BE, Fleming AM, Sweasy JB, Burrows CJ, Doublie S. 2014. Crystal structure of DNA polymerase beta with DNA containing the base lesion spiroiminodihydantoin in a templating position. Biochemistry 53(13):2075–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Muller JG, Ji I, Burrows CJ. 2011. Characterization of 2’-deoxyguanosine oxidation products observed in the Fenton-like system Cu(II)/H2O2/reductant in nucleoside and oligodeoxynucleotide contexts. Org Biomol Chem 9(9):3338–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Muller JG, Dlouhy AC, Burrows CJ. 2012. Context effects in the oxidation of 8-oxo-7,8-dihydro-2’-deoxyguanosine to hydantoin products: Electrostatics, base stacking, and base pairing. J Am Chem Soc 134(36):15091–15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Burrows CJ. 2013. G-Quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem Res Toxicol 26(4):593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Orendt AM, He Y, Zhu J, Dukor RK, Burrows CJ. 2013. Reconciliation of chemical, enzymatic, spectroscopic and computational data to assign the absolute configuration of the DNA base lesion spiroiminodihydantoin. J Am Chem Soc 135(48):18191–18204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Burrows CJ. 2017a. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic Biol Med 107:35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Burrows CJ. 2017b. 8-Oxo-7,8-dihydro-2’-deoxyguanosine and abasic site tandem lesions are oxidation prone yielding hydantoin products that strongly destabilize duplex DNA. Org Biomol Chem 15(39):8341–8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Ding Y, Burrows CJ. 2017. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc Natl Acad Sci USA 114:2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Zhu J, Ding Y, Burrows CJ. 2019. Location dependence of the transcriptional response of a potential G-quadruplex in gene promoters under oxidative stress. Nucleic Acids Res 47(10):5049–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Zhu J, Howpay Manage SA, Burrows CJ. 2019. Human NEIL3 gene expression is regulated by epigenetic-like oxidative DNA modification. J Am Chem Soc 141:11036–11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Burrows CJ. 2020a. Iron Fenton oxidation of 2’-deoxyguanosine in physiological bicarbonate buffer yields products consistent with the reactive oxygen species carbonate radical anion not the hydroxyl radical. Chem Commun 56(68):9779–9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Burrows CJ. 2020b. Interplay of guanine oxidation and G-quadruplex folding in gene promoters. J Am Chem Soc 142(3):1115–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Burrows CJ. 2020c. On the irrelevancy of hydroxyl radical to DNA damage from oxidative stress and implications for epigenetics. Chem Soc Rev 49(18):6524–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Burrows CJ. 2021. Oxidative stress-mediated epigenetic regulation by G-quadruplexes. NAR Cancer 3(3):doi: 10.1093/narcan/zcab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelon S, Douki T, Ravanat J-L, Pouget J-P, Tornabene C, Cadet J. 2000. High-performance liquid chromatography-tandem mass spectrometry measurement of radiation-induced base damage to isolated and cellular DNA. Chem Res Toxicol 13(10):1002–1010. [DOI] [PubMed] [Google Scholar]
- Georgakilas AG, O’Neill P, Stewart RD. 2013. Induction and repair of clustered DNA lesions: what do we know so far? Radiat Res 180(1):100–109. [DOI] [PubMed] [Google Scholar]
- Goodhead DT. 1994. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int J Radiat Biol 65(1):7–17. [DOI] [PubMed] [Google Scholar]
- Greenberg MM. 2021. Tandem and Clustered Lesions from Radicals in Nucleic Acids from a Single Initial Chemical Event. In: Dizdaroglu M, Lloyd RS, editors. DNA Damage, Repair and Disease Royal Society of Chemistry; p. 27–60. [Google Scholar]
- Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. 1998. DNA oxidation matters: The HPLC–electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc Nat Acad Sci USA 95(1):288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. 2003. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry 42:9257–9262. [DOI] [PubMed] [Google Scholar]
- Illes E, Mizrahi A, Marks V, Meyerstein D. 2019. Carbonate-radical-anions, and not hydroxyl radicals, are the products of the Fenton reaction in neutral solutions containing bicarbonate. Free Radic Biol Med 131:1–6. [DOI] [PubMed] [Google Scholar]
- Jaruga P, Speina E, Gackowski D, Tudek B, Olinski R. 2000. Endogenous oxidative DNA base modifications analysed with repair enzymes and GC/MS technique. Nucleic Acids Res 28(6):E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaruga P, Kirkali G, Dizdaroglu M. 2008. Measurement of formamidopyrimidines in DNA. Free Radic Biol Med 45(12):1601–1609. [DOI] [PubMed] [Google Scholar]
- Jia L, Shafirovich V, Shapiro R, Geacintov NE, Broyde S. 2005. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry 44:13342–13353. [DOI] [PubMed] [Google Scholar]
- Joffe A, Geacintov NE, Shafirovich V. 2003. DNA lesions derived from the site selective oxidation of guanine by carbonate radical anions. Chem Res Toxicol 16:1528–1538. [DOI] [PubMed] [Google Scholar]
- Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. 1999. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A 96(23):13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornyushyna O, Burrows CJ. 2003. Effect of the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin on proofreading by Escherichia coli DNA polymerase I (Klenow Fragment) in different sequence contexts. Biochemistry 42(44):13008–13018. [DOI] [PubMed] [Google Scholar]
- Kowalska M, Piekut T, Prendecki M, Sodel A, Kozubski W, Dorszewska J. 2020. Mitochondrial and nuclear DNA oxidative damage in physiological and pathological aging. DNA Cell Biol 39(8):1410–1420. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy N, Zhao X, Burrows CJ, David SS. 2008. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry 47(27):7137–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Pottiboyina V, Sevilla MD. 2011. Hydroxyl radical (OH•) reaction with guanine in an aqueous environment: A DFT study. J Phys Chem B 115(50):15129–15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapi A, Pratviel G, Meunier B. 2001. Guanine oxidation in double-stranded DNA by MnTMPyP/KHSO5: at least three independent reaction pathways. Met-Based Drugs 8(1):47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Delaney S. 2019. Challenges for base excision repair enzymes: Acquiring access to damaged DNA in chromatin. The Enzymes 45:27–57. [DOI] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. 2010. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A 107(11):4925–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonkar P, Dedon PC. 2011. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int J Cancer 128(9):1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Muller JG, Rachlin EM, Burrows CJ. 2000. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org Lett 2:613–616. [DOI] [PubMed] [Google Scholar]
- Luo W, Muller JG, Rachlin EM, Burrows CJ. 2001. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem Res Toxicol 14:927–938. [DOI] [PubMed] [Google Scholar]
- Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z et al. 2012. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA 109(27):E1820–E1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter B, Seiler CL, Murphy K, Ming X, Zhao J, Lindgren B, Jones R, Tretyakova N. 2018. Mapping three guanine oxidation products along DNA following exposure to three types of reactive oxygen species. Free Radic Biol Med 121:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKelvey KJ, Hudson AL, Back M, Eade T, Diakos CI. 2018. Radiation, inflammation and the immune response in cancer. Mamm Genome 29(11–12):843–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SP, Toomire KJ, Strauss PR. 2013. DNA modifications repaired by base excision repair are epigenetic. DNA Repair (Amst) 12(12):1152–1158. [DOI] [PubMed] [Google Scholar]
- Neeley WL, Essigmann JM. 2006. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol 19(4):491–505. [DOI] [PubMed] [Google Scholar]
- Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. 2007. DNA Polymerase V allows bypass of toxic guanine oxidation products in vivo. J Biol Chem 282(17):12741–12748. [DOI] [PubMed] [Google Scholar]
- Pan L, Zhu B, Hao W, Zeng X, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR et al. 2016. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase1-mediated epigenetic regulation of nuclear factor kappaB-driven gene expression. J Biol Chem 291(49):25553–25566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastukh V, Roberts JT, Clark DW, Bardwell GC, Patel M, Al-Mehdi AB, Borchert GM, Gillespie MN. 2015. An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am J Physiol Lung Cell Mol Physiol 309(11):L1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. 2008. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319(5860):202–206. [DOI] [PubMed] [Google Scholar]
- Pogozelski WK, Tullius TD. 1998. Oxidative strand scission of nucleic acids: Routes initiated by hydrogen abstraction from the sugar moiety. Chem Rev 98:1089–1108. [DOI] [PubMed] [Google Scholar]
- Riedl J, Ding Y, Fleming AM, Burrows CJ. 2015. Identification of DNA lesions using a third base pair for amplification and nanopore sequencing. Nat Commun 6:8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokhlenko Y, Geacintov NE, Shafirovich V. 2012. Lifetimes and reaction pathways of guanine radical cations and neutral guanine radicals in an oligonucleotide in aqueous solutions. J Am Chem Soc 134(10):4955–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I, Takayama M, Sugiyama H, Nakatani K, Tsuchida A, Yamamoto M. 1995. Photoinduced DNA cleavage via electron transfer: demonstration that guanine residues located 5’ to guanine are the most electron-donating sites. J Am Chem Soc 117:6406–6407. [Google Scholar]
- Scully R, Panday A, Elango R, Willis NA. 2019. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol 20(11):698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenken S. 1989. Purine bases, nucleosides, and nucleotides: aqueous solution redox chemistry and transformation reactions of their radical cations and e- and OH adducts. Chem Rev 89:503–520. [Google Scholar]
- Steenken S, Jovanovic SV, Bietti M, Bernhard K. 2000. The trap depth (in DNA) of 8-oxo-7,8-dihydro-2’deoxyguanosine as derived from electron-transfer equilibria in aqueous solution. J Am Chem Soc 122:2373–2374. [Google Scholar]
- Sutherland BM, Bennett PV, Sidorkina O, Laval J. 2000. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc Natl Acad Sci U S A 97(1):103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts SG, Smith GS, Miao L, Wheeler KT. 1996. Effects of formic acid hydrolysis on the quantitative analysis of radiation-induced DNA base damage products assayed by gas chromatography/mass spectrometry. Radiat Environ Biophys 35(1):41–53. [DOI] [PubMed] [Google Scholar]
- Tse ECM, Zwang TJ, Bedoya S, Barton JK. 2019. Effective distance for DNA-mediated charge transport between repair proteins. ACS Cent Sci 5(1):65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialas C, Claparols C, Pratviel G, Meunier B. 2000. Guanine oxidation in double-stranded DNA by Mn-TMPyP/KHSO5: 5,8-Dihydroxy-7,8-dihydroguanine residue as a key precursor of imidazolone and parabanic acid derivatives. J Am Chem Soc 122(10):2157–2167. [Google Scholar]
- Vidal AE, Hickson ID, Boiteux S, Radicella JP. 2001. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res 29(6):1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, McKeague M, Sturla SJ. 2018. Nucleotide-resolution genome-wide mapping of oxidative DNA damage by click-code-seq. J Am Chem Soc 140(31):9783–9787. [DOI] [PubMed] [Google Scholar]
- Ye W, Sangaiah R, Degen DE, Gold A, Jayaraj K, Koshlap KM, Boysen G, Williams J, Tomer KB, Mocanu V et al. 2009. Iminohydantoin lesion induced in DNA by peracids and other epoxidizing oxidants. J Am Chem Soc 131(17):6114–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Munk BH, Muller JG, Cogbill A, Burrows CJ, Schlegel HB. 2009. Mechanistic aspects of the formation of guanidinohydantoin from spiroiminodihydantoin under acidic conditions. Chem Res Toxicol 22(3):526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. 2010. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc Natl Acad Sci U S A 107(48):20715–20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. 2013. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem 288(38):27263–27272. [DOI] [PMC free article] [PubMed] [Google Scholar]


