Abstract
Well-orchestrated transcriptional programs in intestinal epithelial cells (IECs) are essential for maintenance of optimal mucosal barrier functions, whereas the contribution of elongation-related mechanisms to barrier function remains unknown. Here, a combination of genetic and genomic approaches defined a critical role of IEC-intrinsic negative elongation factor (NELF) complex in maintenance of epithelial homeostasis. By direct occupancy at endogenous gene loci, NELF sustained expression of a subset of genes related to junctional integrity. As a result, epithelial NELF deficiency results in subdued levels of these junction-related genes and excessive IEC necroptosis in vivo secondary to commensal microbial invasion. In a colitis model, NELF-deficient mice exhibited severely impaired barrier integrity characterized by increased intestinal permeability and significantly exacerbated intestinal inflammation with lethal consequences. Our findings reveal the protective function of the NELF complex against intestinal damage and inflammation and suggest that elongation represents a biologically important step in defining IEC transcriptome.
INTRODUCTION
The intestinal mucosal barrier constituted by specialized epithelial cells segregates mammalian hosts from the environmental microbes including pathogens and commensals1-5. In addition to creating the mucus layers, intestinal epithelial cells (IECs) constitute a continuous physical barrier that is connected by epithelial junctional complex formed by tight junctions, adherens junctions and desmosomes6. Adherens junctions are involved in control of cell–cell contacts7, while tight junctions seal adjacent epithelial cells to one another8. The major protein components of epithelial tight junctions include members of the claudin family, occludin, and junctional adhesion molecule-A8. Tight junctions are intercellular adhesion complexes that control intestinal permeability, allowing exchange of essential ions, nutrients, and water while guarding the epithelial surfaces against microbes9. Dysregulation of tight junctions is associated with increased intestinal permeability and inflammation8,10,11. Abnormal junctional integrity has been observed in patients with inflammatory bowel disease (IBD) and may contribute to IBD pathogenesis12,13. Thus, the gene expression of the major components of tight junctions needs to be tightly controlled to ensure proper barrier integrity9. However, the transcriptional regulatory mechanisms of controlling IEC junctional integrity has not been well understood.
Emerging evidence from advanced sequencing techniques reveals that in addition to widely appreciated RNA polymerase II (Pol II) recruitment and transcription initiation, Pol II promoter-proximal pausing and its release to productive elongation are key steps in a full-fledged transcription cycle14. Establishment of Pol II pausing and its release are tightly controlled by a number of positive and negative regulatory factors, including positive transcription elongation factor-b (P-TEFb), DRB sensitivity-inducing factor (DSIF), and negative elongation factor (NELF)15,16. NELF, a four-subunit complex that participates in the transcription elongation process, includes NELF-A, NELF-B, NELF-C, and NELF-E17. According to the canonical pause-release model, the four-subunit NELF complex colocalizes with the paused Pol II near the transcription start sites (TSSs), followed by signal-induced phosphorylation of Pol II and NELF as well as DSIF by the P-TEFb complex and subsequent dismissal of NELF from TSSs, which facilitates productive transcription elongation and thus gene expression16-18. According to the above paradigm, deletion of NELF would result in enhanced transcription of its target genes, which is indeed the case for a subset of NELF+ genes19,20. However, for a significant proportion (up to 23–86% depending on the experimental systems) of NELF+ genes19,21, NELF loss-of-function actually leads to reduced instead of enhanced transcription19,21-23. One plausible explanation for such findings is that NELF stabilizes paused Pol II in the pre-initiation complex by preventing premature promoter-proximal transcription termination17. Thus, the role of NELF in a given cell type in vivo could be complex, likely playing a positive or negative role in a gene-specific and context-specific manner.
The post-initiation mechanisms have been implicated in key biologic processes such as organism development and innate immunity14,19,24-31. Studies by us and others have shown that transcription elongation targeted by negative regulators including the glucocorticoid receptor and other transcription repressors represents a critical rate-limiting step in macrophage-mediated inflammatory responses19,30,32. Recently, we also have shown that NELF controls macrophage inflammatory transcriptome by counteracting an anti-inflammatory circuit21, resulting in prominent alterations of inflammatory responses in vivo, raising the possibility that NELF may act as a key regulator of inflammation in others tissues or cells such as IECs. In fact, studies on transcriptional mechanisms controlling IEC identities and barrier function have mainly focused on pre-initiation associated mechanisms such as those mediated by sequence-specific transcription factors33,34. Whether post-initiation mechanisms in the intestinal epithelium might influence the IEC identities and intestinal homeostasis remains unknown.
Here, using a combination of genetic and genomic approaches, we set to investigate the role of IEC-intrinsic NELF in the context of normal intestinal physiology. Comprehensive characterization of NELF target genes in primary IECs reveals that NELF is essential for expression of a subset of genes involved in maintaining IEC barrier function and intestinal homeostasis. Genetic disruption of Nelfb in IECs resulted in disrupted intestinal homeostasis, manifested as increased intestinal permeability, invasion of tissue by commensal bacteria and subsequent bacterium-dependent IEC necroptosis. Upon induction of colitis, NELF-deficient animals exhibit exacerbated intestinal damage and tissue inflammation. Together, our results uncover a critical role of IEC-intrinsic NELF in maintaining epithelial barrier integrity, thereby establishing a physiological role of NELF in controlling mammalian intestinal homeostasis.
RESULTS
Epithelial NELF deficiency renders mice highly susceptible to colitis
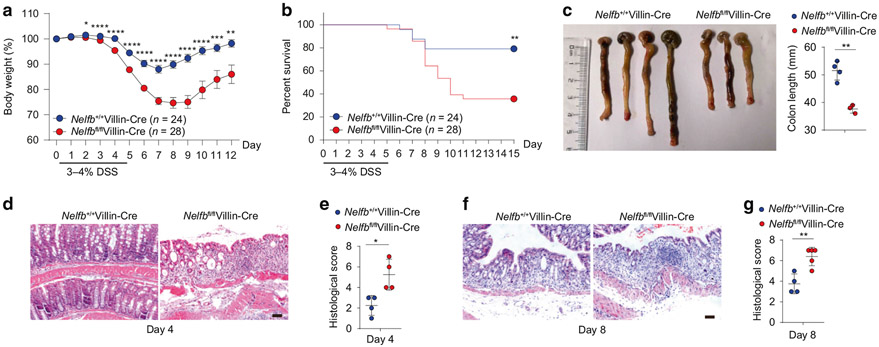
To investigate the in vivo functions of IEC-intrinsic NELF, we firstly generated mice lacking NELF-B specifically in IECs (Nelfbfl/flVillin-Cre). Consistent with the previous findings in other cell types21,35, ablation of Nelfb led to efficient depletion of NELF-B itself as well as diminished protein levels of additional NELF complex subunits such as NELF-E, confirming the validity of this genetic loss-of-function approach for studying the NELF complex in IECs (Fig. S1A-C). Housed in an SPF facility, Nelfbfl/flVillin-Cre mice appeared healthy and showed no overt developmental defects, evidenced by normal intestinal architecture and comparable IEC proliferation relative to WT control mice (Nelfb+/+Villin-Cre) (Fig. S1D, E). In addition, organoids grown from Nelfbfl/flVillin-Cre mice in vitro were indistinguishable in morphology and circularity from their WT counterparts (Fig. S1F). To investigate the role of IEC-intrinsic NELF in regulating intestinal barrier function, control mice and Nelfbfl/flVillin-Cre mice were co-housed to normalize for environmental factors prior to treatment with dextran sodium sulfate (DSS) for 5 days. Compared to DSS-treated control mice, Nelfbfl/flVillin-Cre animals demonstrated significantly exacerbated weight loss (Fig. 1a), high mortality (Fig. 1b), colonic shortening (Fig. 1c), and increased disease severity at day 4 and day 8 (Fig. 1d-g), indicating that IEC-intrinsic NELF plays an important role in limiting DSS-induced intestinal inflammation. The above results were further corroborated with littermate controls (Fig. S1G-I). Collectively, these data demonstrate that loss of NELF in IECs renders the mice highly susceptible to intestinal damage, implying a critical role of NELF complex in maintenance of intestinal homeostasis.
Fig. 1. High sensitivity to DSS-induced colitis in mice with NELF-B deletion in IECs.
Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice were treated with 3 or 4% DSS in drinking water for 5 days. a–c Body weights (a), survival (b) (n = 24–28) and colon length (c) (n = 3–4) of Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice after DSS treatment. d, e H&E staining (d) and histological scores (e) (n = 4) of colon sections from DSS treated mice at day 4. Scale bar, 50 μm. f, g H&E staining (f) and histological scores (g) (n = 4) of colon sections from DSS treated mice at day 8. Scale bar, 50 μm. Data are pooled from more than 3 (a, b) independent experiments and shown as mean ± SEM. Student’s t-test was performed; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Images represent at least two independent experiments (c, d, and f).
Epithelial NELF attenuates microbiota-dependent cell death in IECs
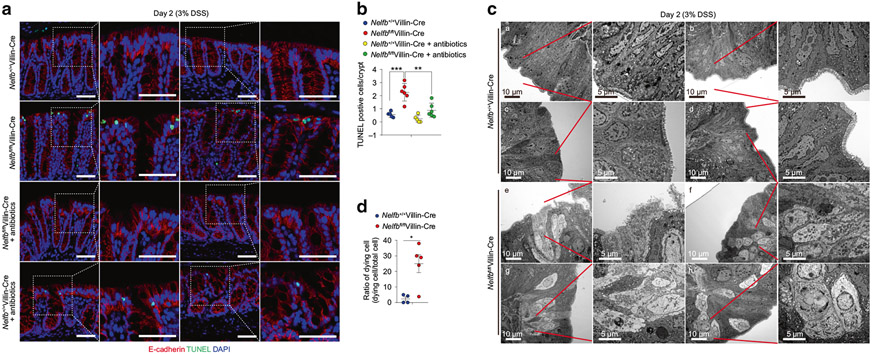
As above, the protective effects of NELF were evident as early as day 2 of post DSS challenge (Fig. 1a), raising the possibility that defects of barrier function might contribute to the increased susceptibility to DSS in Nelfbfl/flVillin-Cre mice. An elevated rate of IEC death is commonly associated with intestinal barrier breach observed both in IBD patients and in preclinical models of colitis36. To examine whether epithelial NELF regulated cell death, we performed TdT-mediated dUTP nick end labeling (TUNEL) staining analyses at day 2 of DSS treatment. Quantification of TUNEL+ cells in situ demonstrated that epithelial NELF deficiency significantly increased the number of dying IECs in colon and that such increase was reversed by antibiotics treatment (Fig. 2a, b), suggesting that NELF deficiency was associated with microbiota-dependent cell death. To characterize cell death at a fine resolution, colonic tissues were subjected to transmission electron microscopy (TEM) analyses, which revealed typical necrotic but not apoptotic features of dying IECs in Nelfbfl/flVillin-Cre mice including membrane destruction and loss of electron density accompanying disrupted cristae structures (Fig. 2c, d)37-39. Taken together, these data demonstrate that IEC-intrinsic NELF prevents epithelial microbiota-dependent necroptosis-like cell death to ameliorate experimental colitis.
Fig. 2. Epithelial NELF represses microbiota-dependent cell death in IECs before the onset of colitis.
Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice were pre-treated with water or antibiotics, and continued drink with 3% DSS for 2 days. a, b TUNEL analysis (a) and quantification (b) of cell death in Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice after DSS or DSS with antibiotics treatment (n = 4–6). Scale bars, 50 μm. c, d TEM analysis (c) and quantification (d) of IEC cell death in Nelfb+/+Villin-Cre (a–d) and Nelfbfl/flVillin-Cre (e–h) mice after DSS treatment (n = 4–5). Scale bars, 5 or 10 μm. Data are pooled from at least 2 (B and D) independent experiments and shown as mean ± SEM. Student’s t-test was performed; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Images (a, c) represent at least two independent experiments.
Epithelial NELF prevents IEC necroptosis
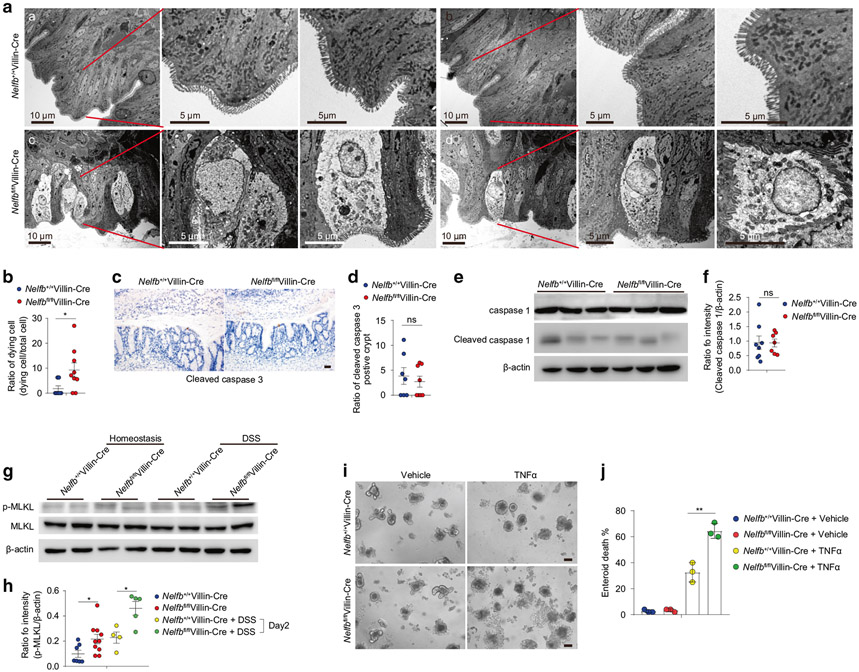
Given the appearance of necroptosis-like cell death associated with NELF deficiency at the early stage of colitis, we next determined whether such cell death could be observed at steady state. Although Nelfbfl/flVillin-Cre mice did not show increased intestinal permeability at steady state (Fig. S1J, K), these NELF-deficient mice did reveal increased frequency of IEC cell death in the colon, as indicated by TEM analysis which demonstrated typical necrotic features (Fig. 3a, b). Consistent with the morphological assessments of necroptotic rather than apoptotic or pyroptotic features, Nelfbfl/flVillin-Cre mice did not show increased levels of cleaved caspase-3 (Fig. 3c, d) and cleaved caspase-1 (Fig. 3e, f) but show increased activation of p-MLKL relative to WT controls (Fig. 3g, h). In line with increased activation of p-MLKL at steady state, Nelfbfl/flVillin-Cre mice also displayed significantly boosted activation of p-MLKL (Fig. 3g, h) and gene expression of Ripk3 (Fig. S2A, B) at day 2 of DSS treatment. To further confirm these phenotypes, we utilized the organoid system to determine whether loss of NELF would accelerate IEC death induced by TNF-α and found that NELF-deficient organoids exhibited increased cell death upon TNF-α stimulation (Fig. 3i, j), suggesting that regulation of necroptosis by epithelial NELF is an IEC-intrinsic phenomenon instead of the consequence secondary to excessive inflammation. Furthermore, we performed in vivo pharmacologic inhibition of necroptosis by treating mice with necrostatin-1 and found that necrostatin-1 administration relieved body weight loss and intestinal damage in DSS-treated Nelfbfl/flVillin-Cre mice (Fig. S2C-E), indicating that necroptosis played a key role in the exaggeration of mucosal injury in NELF knockout mice. Together, our data indicate that ablation of epithelial NELF sensitizes colonic IECs to necroptosis.
Fig. 3. Epithelial NELF inhibits IEC necroptosis.
a, b TEM analysis (a) and quantification (b) of IEC cell death in Nelfb+/+Villin-Cre (a, b) and Nelfbfl/flVillin-Cre (c, d) mice at stead state (n = 4–9). Scale bars, 5 or 10 μm. c, d Immunohistochemistry analysis (c) and quantification (d) of cleaved caspase 3 positive cell in colon section of Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice (n = 7). Scale bar, 50 μm. e, f Immunoblotting analysis (e) and quantification (f) of cleaved caspase 1 in colon tissues of Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice (n = 7–8). g, h Immunoblotting analysis (g) and quantification (h) of p-MLKL in colon tissues of Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice with or without DSS treatment (n = 4–10). i, j Organoids analysis (l) and quantification (j) of TNF-α induced cell death of Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice (n = 3). Scale bar, 50 μm. Data are pooled from at least two (b, d, f, h, and j) independent experiments and shown as mean ± SEM. Student’s t test was performed; *p ≤ 0.05; ns not significant (p > 0.05). Images (a, c, and i) represent at least two independent experiments.
Epithelial NELF deficiency leads to dysregulated IEC gene expression
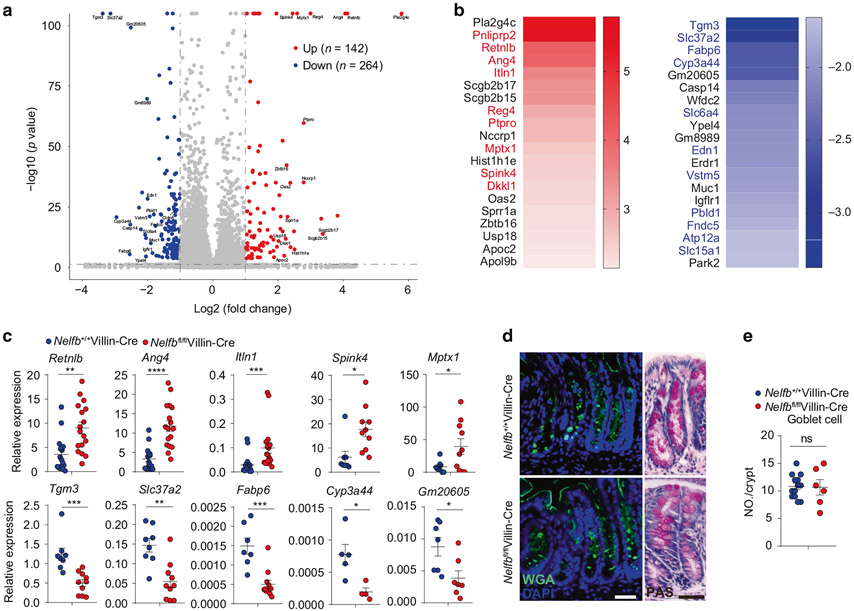
To better understand how NELF regulates epithelial function, we next carried out genome-wide transcriptional profiling of colonic tissues, which revealed that in vivo deletion of IEC-specific NELF resulted in substantial alterations in gene expression including upregulation of 142 genes and downregulation of 264 genes (Fig. 4a). Interestingly, a majority of the top upregulated genes are highly expressed in goblet cells (Fig. 4b), including Pnliprp2, Retnlb, Ang4, Itln1, Reg4, Ptpro, Mptx1, Spink4, and Dkkl1 according to the public single cell database Tabula Muris (https://tabula-muris.ds.czbiohub.org/)40. On the contrary, many of the top downregulated genes are highly expressed in enterocytes (Fig. 4b), including Tgm3, Slc37a2, Fabp6, Cyp3a44, Slc6a4, Edn1, Vstm5, Pbld1, Fndc5, and Atp12a according to Tabula Muris. Differential expression of representative genes in these two groups was confirmed by quantitative polymerase chain reaction (qPCR) analyses (Fig. 4c). Although the expression of a number of genes from specialized IECs were profoundly regulated by NELF, the IEC differentiation was not obviously affected as Nelfbfl/flVillin-Cre mice displayed a comparable number of goblet cells to control mice (Fig. 4d, e). In addition, we also examined the microbiota composition and found that loss of epithelial NELF results in minimal microbial changes in both of small intestine and colon (Fig. S2A-C), indicating a central role of NELF in regulation of IEC gene expression rather than luminal microbial ecology. Collectively, these analyses implicate a pivotal role for NELF in coordinating the expression of a subset of IEC-intrinsic genes without interfering with cellular development.
Fig. 4. Epithelial NELF deficiency leads to dysregulation of IEC gene expression without affecting cell differentiation.
a The volcano plot analysis of RNA-seq dataset showing the top upregulated and downregulated genes in colonic epithelia cell from Nelfbfl/flVillin-Cre mice versus Nelfb+/+Villin-Cre at stead state. b Heat map analysis of RNA-seq dataset showing the top 20 upregulated and downregulated genes in colonic epithelia cell from Nelfbfl/flVillin-Cre mice versus Nelfb+/+Villin-Cre. Red font indicates that upregulated genes are highly expressed in goblet cells and blue font indicates that downregulated genes are highly expressed in enterocytes. c qPCR analysis of the expression of representatively upregulated and downregulated genes from RNA-seq dataset in Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice (n = 5–17). d, e Immunofluorescent WGA and PAS staining (d) and quantification (e) analysis of the goblet cell in Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre (n = 6–13). Scale bars, 50 μm. Data are pooled from at least two (c, e) independent experiments and shown as mean ± SEM. Student’s t-test was performed; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; ns not significant (p > 0.05). Images (d) represent at least two independent experiments.
Epithelial NELF maintains the expression of cell junction-related genes
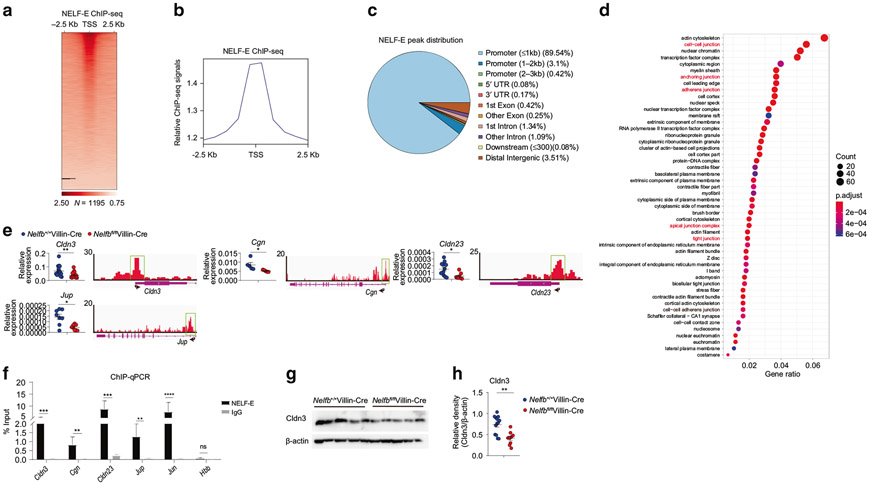
To further pinpoint NELF direct target genes that may regulate epithelial barrier function, we performed NELF-E chromatin immunoprecipitation in colonic IECs followed by high throughput sequencing (chromatin immunoprecipitation (ChIP)-seq) to assess genome-wide binding of NELF. Consistent with our previous results in macrophages21, NELF occupancy exhibited TSS-centric distribution with 89.5% of ChIP-seq peaks located near promoter regions (Figs. 5a-c and S4A-C). Intriguingly, the Gene Ontology (GO) analysis of peak annotation revealed an enrichment of genes whose function is related to epithelial junctional complexes including cell-cell junction, anchoring junction, adherens junction, tight junction (Fig. 5d), suggesting that NELF may directly regulate the expression of cell junction-related genes. Indeed, colonic tissue sampling from multiple independent experiments confirmed the ChIP-seq results that loss of epithelial NELF resulted in significantly decreased expression of cell junction-related genes Cldn3, Cgn, Cldn23, and Jup, all of which were direct NELF targets evidenced by robust NELF occupancy near their TSS regions (Fig. 5e). The presence of NELF complex at these tight junction genes in colonic IECs was confirmed by NELF-E chromatin immunoprecipitation at the individual gene loci, with Jun as a positive control gene and Hbb as a negative control gene respectively (Fig. 5f). The mRNA expression patterns were further corroborated by decreased protein levels of Cldn3, Cgn, and Cldn23 in colon from Nelfbfl/flVillin-Cre mice (Fig. 5g, h, and S5A-E). To test whether NELF has similar effects on regulation of tight junction gene expression in human, we next determined the gene expression of tight junction proteins in human IEC cell lines (LS174T) and found that knock-down of NELF-B by siRNA could reduce the expression of a couple of epithelial junction-related genes including CLDN3, CGN, CLDN23, and JUP (Fig. S5F), suggesting NELF may have similar effects on regulation of epithelial junction in human. Taken together, these results identify a unique role for epithelial NELF in directly promoting the expression of cell junction-related genes, implying a potential function of NELF in controlling intestinal junctional integrity.
Fig. 5. Epithelial NELF promotes the expression of a subset of cell junction-related genes with direct NELF binding to their promoter regions.
a Heat map of NELF-E ChIP-seq signals around TSS regions for colonic IEC-expressed genes in Nelfb+/+Villin-Cre mice at steady state. Each row indicates one gene sorted by the peak heights in Nelfb+/+Villin-Cre mice. The signals were normalized to input signals. The number of peaks in the whole genome is shown at the bottom. b Relative NELF-E ChIP-seq signals around TSS regions as (a) shown. c The genomic distribution of NELF-E ChIP-seq peaks. d GO analysis of NELF-E ChIP-seq peaks for cellular component classes and many cellular components (red font) are related to epithelia cell–cell junction. e qPCR analysis of epithelia cell junction-related genes expression in Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice (n = 6–12). Tracks of NELF-E ChIP-seq are shown for these genes. f The occupancies of NELF-E in the TSS region of cell junction-related genes were assessed by ChIP-qPCR in Nelfbfl/flVillin-Cre versus Nelfb+/+Villin-Cre mice. g, h Immunoblotting analysis (g) and quantification (h) of Cldn3 in colon tissues of Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice (n = 9–12). Data are pooled from at least two (e, f, and h) independent experiments and shown as mean ± SEM. Student’s t-test was performed; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; ns not significant (p > 0.05). Images (g) represent at least two independent experiments.
Epithelial NELF orchestrates barrier integrity to alleviate colitis
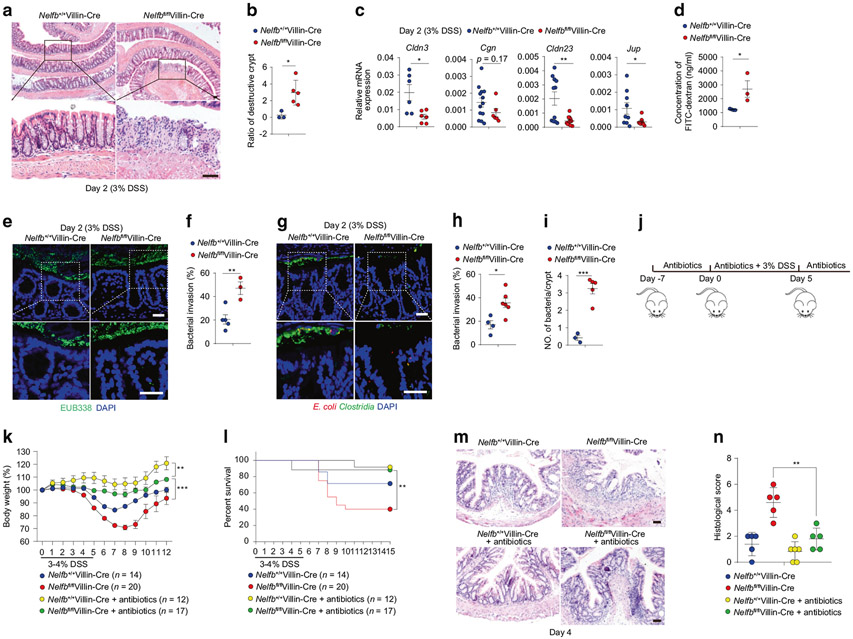
Abnormal epithelial junctional integrity is often associated with intestinal inflammation12. To further link necroptosis and dysregulation of junctional integrity to the phenotypes of increased susceptibility to intestinal damage in Nelfbfl/flVillin-Cre mice, we next focused our attention on the early stage of DSS-induced colitis. Consistent with the increased necroptosis, Nelfbfl/flVillin-Cre mice displayed increased destructive crypts in the distal colon as early as day 2 after DSS treatment (Fig. 6a, b), indicating an early breakdown of epithelial barrier integrity. Similar with the observations at steady state, Nelfbfl/flVillin-Cre mice exhibited reduced expression of cell junction-related genes (Cldn3, Cgn, Cldn23, and Jup) at day 2 of DSS treatment (Fig. 6c) and elevated gut permeability upon DSS treatment as indicated by increased plasma levels of FITC after oral administration of FITC-dextran (Fig. 6d), implicating compromised junctional integrity under conditions of intestinal damage. At day 2 of DSS treatment, increased bacterial invasion of colonic tissues in Nelfbfl/flVillin-Cre mice was visualized by fluorescence in situ hybridization (FISH) using a universal probe for bacterial 16S ribosomal RNA (Fig. 6e, f) and antibodies against E. coli and Clostridia (Fig. 6g-i), implying that the increased susceptibility to intestinal damage in Nelfbfl/flVillin-Cre mice was due to defects in barrier integrity and subsequent bacterial invasion. Consistent with the above hypothesis, depletion of microbiota by broad spectrum antibiotics along DSS-induced inflammation (Fig. 6j) markedly protected Nelfbfl/flVillin-Cre mice from weight loss and death (Fig. 6k, l) and diminished the differences in weight loss, survival rate and disease severity between Nelfbfl/flVillin-Cre and control mice (Fig. 6k-n). Collectively, these data demonstrate that epithelial NELF maintains barrier integrity to prevent bacterial invasion and to protect against intestinal damage.
Fig. 6. Epithelial NELF orchestrates barrier integrity before the onset of colitis.
Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice were treated with 3% DSS in drinking water for 2 days. a, b H&E staining (a) and histological scores (b) (n = 3–5) of colon sections from DSS treated mice at day 2. Scale bars, 50 μm. c qPCR analysis of cell junction-related genes in colon tissues from DSS treated mice at day 2 (n = 5–13). d Intestinal permeability analysis of Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice after DSS treatment at day 2 (n = 3). e, f FISH analysis (e) and quantification (f) of bacterial invasion in colon sections from DSS treated mice at day 2 (n = 3–5). Scale bars, 50 μm. g–i Immunofluorescent analysis (g) and quantification (h, i) of bacterial (E. coli and Clostridia). invasion in colon sections from DSS treated mice at day 2 (n = 3–6). Scale bars, 50 μm. j The experimental scheme for the procedure of antibiotics or/and DSS treatment. k, l Body weights (k) and survival (l) (n = 12–20) of Nelfb+/+Villin-Cre and Nelfbfl/flVillin-Cre mice. m, n H&E staining (m) and histological scores (m) (n = 5–6) of colon sections from antibiotics or/and DSS treated mice at day 4. Scale bars, 50 μm. All data are shown as mean ± SEM. Student’s t-test was performed; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; ns not significant (p > 0.05).
DISCUSSION
The cellular transcriptional network controls IEC identities and barrier function via sequence-specific transcription factors, DNA methylation, histone modifications, and chromatin accessibility33,34. However, little is known about how post-initiation mechanisms regulate the IEC identities and epithelial functions. In this study, we identify a previously unknown physiological function of IEC-intrinsic NELF complex in maintaining epithelial integrity by orchestrating expression of cell junction-associated genes and preventing epithelial necroptosis, thereby providing essential protection against commensal bacterial invasion in the context of intestinal inflammation. Consequently, NELF deficiency in IECs renders the mice highly susceptible to DSS-induced colitis (Fig. S4). DSS has been shown to make the inner colon mucus layer penetrable to bacteria41. Notably, in the absence of epithelial NELF, the intestinal epithelial barrier remained relatively intact without evident bacterial invasion under steady state. However, upon induction of intestinal damage with DSS, consequences of NELF deficiency such as impaired barrier function and thus increased bacterial invasion became apparent (Fig. S4), suggesting contributions of both genetic background (NELF deficiency) and environmental factors (DSS) to exacerbated intestinal inflammation. Therefore, NELF acts as a sentinel regulator guarding the intestinal epithelial surfaces against external stimuli such as commensal bacteria to maintain intestinal homeostasis.
Intact mucosal barrier function is indispensable for maintaining a healthy relationship between intestinal luminal contents and the host4,42. In fact, it has long been observed that patients with active Crohn’s disease showed increased intestinal permeability that may precede clinical relapses43,44. Moreover, newly discovered IBD risk loci are associated with genes encoding key regulators of barrier integrity such as C1orf106, RNF186, and HNF4A12,45. In our study, NELF-target genes encoding components and/or regulator of barrier function, such as tight junction proteins claudins, may underpin the observed phenotypes as well. Nevertheless, we cannot rule out the possibility that in addition to cell junction-related genes, other genes regulated by NELF may participate in the orchestration of mucosal barrier function that controls commensal microbial invasion by physical segregation or direct bacterial killing to restrain intestinal inflammation. Thus, it appears that concerted actions of multiple molecules and pathways are in place to sustain intestinal barrier integrity in NELF-sufficient animals, whose functional importance is manifested by NELF deficiency.
The transcriptional regulatory mechanisms not only shape the identity of the intestinal epithelium but also modulate the sensitivity of epithelium by responding to environmental cues33,34. Conventionally, as indicated by its name, NELF complex acts as a negative regulator of transcription, whose ablation is expected to result in potentiation of gene expression. However, recent studies with genetic and tissue-specific loss-of-function approaches in vivo have implicated NELF as a positive regulator for a number of genes22,23. In principle, two mechanisms could account for the positive role of NELF in controlling gene expression: direct action and indirect action. In the indirect manner, NELF still functions as a canonical transcription repressor to suppress negative factor(s) that dampen expression of gene X and thus the net effects of NELF on gene X is manifested as positive. One such example is the unexpected findings by us that in macrophages, NELF promotes instead of inhibits expression of inflammatory cytokines21. The pro-inflammatory effects of NELF are secondary to its canonical repressive function on genes encoding transcription factor AP-1 that is essential for production of a critical anti-inflammatory cytokine IL-10, illustrating a delicate NELF-centric transcriptional network fine-tuning macrophage inflammatory responses21. Intriguingly, the current study exemplifies the direct positive effects of NELF in IECs by binding and promoting expression of several cell junction-associated genes. The precise molecular details of NELF-mediated upregulation of transcription activities in IECs are of great interest and warrant future investigations. The plausible explanations include but are not limited to stabilization of pre-initiation complex and remodeling of TSS-surrounding regions17. It is also conceivable that the direct and indirect mechanisms by which NELF exerts its function are not mutually exclusive and may cooperate in a dynamic manner to achieve the optimal transcriptional outputs in a given tissue environment.
Dysfunction of intestinal barrier function due to tight junction disruption or abnormal cell death such as apoptosis and necroptosis is believed to exacerbate or directly trigger intestinal inflammation4,12,38,39. For instance, cell death often accelerates tissue inflammation due to disruption of barrier integrity38,39,46. These notions are supported by our observation of excessive necroptosis in NELF-B deficient IECs at the early time points of colitis (Fig. 2), suggesting necroptosis in IECs may further contribute to breakdown of barrier integrity, constituting an amplification step leading to the vicious and often deadly inflammatory cycle. As more and more factors are discovered to be involved in post-initiation and productive transcription elongation, the link between elongation mechanisms and human diseases is becoming increasingly recognized14,17,47 yet such connections in IECs remain elusive. Overall, our study provides evidence that NELF complex is essential for IECs to coordinate gene expression programs and maintain intestinal barrier functions, and highlights a potential link of post-initiation to the pathogenesis of IBDs. Nevertheless, the current study merely serves to unveil the vast complexity and biological significance of post-initiation regulation in IECs. Future investigations in this area are destined to be fertile for enriching our understanding of intestinal epithelial biology as well as for developing potential IBD therapeutic targets.
METHODS
Mice
The Villin-Cre mice were purchased from Jackson Laboratory and Nelfbfl/fl mice were generated48. To deplete Nelfb specifically in IECs, the Nelfbfl/flVillin-Cre mice were generated by crossing Nelfbfl/fl mice to Villin-Cre mice on the C57BL/6J background. Age and sex matched mice at 7–10 weeks of age were used for experiments. All the mice were maintained on specific pathogen-free (SPF) conditions on 12 h:12 h light/dark cycle. The littermates were separated after weaning and non-littermates were co-housed after weaning for 3 weeks and separated for more than 2 weeks before experiment. The laboratory animal facility of Tsinghua University has been accredited by AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care International) and all the animal protocols were approved by the IACUC (Institutional Animal Care and Use Committee) of Tsinghua University.
DSS-induced intestinal inflammation and microbiota depletion
Mice were treated with 3–4% DSS (MP Biomedicals, Santa Ana, CA) dissolved in drinking water for 2–5 days followed by normal drinking water until the end of the experiment. The body weight and dead mice were recorded daily. For some experiments with antibiotics, the intestinal microbiota was depleted by giving mice with combine antibiotics (neomycin sulfate, 1 g/l, Amresco; metronidazole, 1 g/l, Amresco; ampicillin, 1 g/l, Amresco; vancomycin, 0.5 g/l, Amresco; streptomycin 5 g/l, Amresco) for 7 days before and after DSS treatment until the end of the experiment.
Treatment with necrostatin-1
Mice were intraperitoneally injected with necrostatin-1 (MedChemExpress, USA) at the beginning of DSS treatment (3% DSS in drinking water for 5 days). During the DSS treatment, the Nelfbfl/flVillin-Cre mice received 10% DMSO or necrostatin-1 (4.5 mg/kg) twice daily. The body weight was recorded daily.
Quantitative PCR analysis of intestinal microbiota and 16S rDNA sequencing
Small intestine and colon feces were collected and bacterial DNA extraction as previously described49,50. Briefly, the fresh feces were flushed by 1× PBS, collected and weighed. Microbial DNA was extracted by Stool Genomic DNA Kit (CoWin Biosciences, China). The abundances of specific intestinal groups were determined by qPCR with FastSYBR mixture (CWBIO, China) and conserved specific 16S rDNA primers (Table S1).
The intestinal microbiota 16S rDNA sequencing and analysis were performed as previously described49,51. Briefly, the total bacteria 16S rDNA of distinct regions (16S V4) were amplified by specific barcoded primers through Phusion R High-Fidelity PCR Master Mix (New England Biolabs) and the products were purified with Qiagen Gel Extraction Kit (Qiagen, Germany). TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA) was used to generate libraries following the manufacturer’s protocol and the quality of libraries were measured by the Qubit@ 2.0 Fluorometer (Thermo Scientific, Waltham, MA) and Agilent Bioanalyzer 2100 system. Illumina HiSeq2500 platform was used to perform the sequencing. The 250 bp paired-end reads were generated and merged by FLASH. The sequences were analyzed by Gold database and UCHIME algorithm was used to clean the chimera sequences. More than 97% similarity were assigned to the same operational taxonomic units (OTUs). The representative sequences were annotated by GreenGene Database. The full 16S rDNA sequencing data have been deposited in NCBI’s Sequence Read Archive (SRA) and the access number is PRJNA700789.
Intestinal permeability assay
Mice starved for 24 h received either tap water or 3% DSS in drinking for 16 h before experiment. Then, the mice were given FITC-dextran (44 mg/100 g body weight; 4.4 kDa, sigma) by gavages. After 4h, the mice were sacrificed with carbon dioxide and fluorescence intensity of plasma samples was measured by spectrophotometer with an excitation of 485 nm and an emission wavelength of 528 nm.
Intestinal crypt isolation and organoid culture assay
The intestinal crypts were isolated and cultured as described previously52. Briefly, small intestine and colon were isolated and opened longitudinally. The intestine was cut into 3–5mm fragments and washed through vigorous shaking at least twice. The small intestinal tissue pieces were incubated in cold 1× PBS with 10mM EDTA for 1 h at 4 °C and the colonic tissue pieces were incubated on warm 1× PBS with 20mM EDTA for 35 min at 37 °C. Following the incubation, tissue pieces were transferred to 50 ml tube containing 15 ml cold 1× PBS with shaking. The villous materials and tissue fragments were discarded and the crypt was collected after centrifugation at 300×g for 2 min. The pellet was resuspended with 100 μl mixture medium (basement matrix gel:ENR medium = 1:1 (ENR mdium: DMEM/F12 (Gibco) with B27 supplement (1×, Gibco), N2 supplement (1×, Gibco), HEPES (Thermo Fisher), N-acetylcysteine (1 mM, Sigma), Penicillin/Streptomycin, GlutaMAX-I and growth factors including 50 ng/ml EGF (Peprotech), 100 ng/ml Noggin (Peprotech) and 500 ng/ml R-spondin (Peprotech); Wnt3a (Peprotech) was added in colonic crypt culture medium)) and seeded in a pre-warmed 96-well plate. The plate was incubated in cell incubator for 5 min and then overlaid with 200 μl ENR medium. The ENR medium was changed in every 2 days and then the organoids were transferred to the 24-well plate and overlaid with 500 μl ENR medium. The crypt growth was measured by microscope and the circularity was calculated by Image J as previously described53. The tumor necrosis factor-α (TNF-α) (100 ng/ml, Peprotech) was used to induce cell death of organoids for 24 h. The viable and dead organoids were observed and recorded by morphology through bright filed microscope.
Immunoblotting
The colon tissues were opened, washed by cold 1× PBS and snap-frozen immediately in liquid nitrogen, homogenized, and lysed by RIPA (Beyotime Biotechnology). The protein amounts were determined by the BCA Protein Assay (Thermo Fisher Scientific). The protein lysates were separated by the 10% SDS-PAGE gel and transferred to the PVDF membrane (Millipore). The membranes were blocked in the 5% skim milk/TBST for 1 h at room temperature, immunoblotted with the primary antibodies against NELF-B (Abcam, ab167401), NELF-E (Proteintech, 10705-1-AP), p-MLKL (Abcam, ab196436), Cldn3 (Affinity,DF7115), Cgn (Proteintech, 21369-1-AP), Cldn23 (Affinity, DF9395) and Cleaved Caspase 1 (Santa Cruz, sc-56036) dissolved in 2.5% BSA/TBST at 4 °C overnight. The primary antibody of β-actin was used as control. The membranes were washed by TBST for three times and then incubated with horseradish-peroxidase-conjugated secondary antibody for 1 h, visualized and captured by ECL Western Boltting Substrate (Thermo Fisher Scientific) and ImageQuant LAS 400 imaging system (GE Healthcare Life Sciences).
RNA extraction and quantitative RT-PCR
The colon tissue was opened and washed by cold 1× PBS and snap-frozen in liquid nitrogen and homogenized by liquid nitrogen. The total RNA was extracted by Total RNA Miniprep Purification Kit (GeneMarkbio) following the manufacturer’s protocols, and then was reverse-transcribed into cDNA by Reverse Transcriptase M-MLV (TakaRa). qPCR was performed with FastSYBR mixture (CoWin Biosciences) and specific primers (Table S1) on a qPCR system (StepOnePlus; Applied Biosystems). The comparative threshold cycle method and an internal control (Gapdh) were used to normalize the expression of target genes.
RNA sequencing and analysis
Total RNA was extracted from colonic tissues by Total RNA Miniprep Purification Kit (GeneMarkbio). After removal of rRNA, the RNA was converted into cDNA using random N6 primer followed by A-tailing and the cDNA was amplified. To generate libraries, the amplified products were separated into single strand DNA and cyclized. The RNA-seq libraries were subjected and sequenced with the pair-end option using BGISEQ-500 system at Beijing Genomics Institute (BGI), China. Bowtie 254 and HISAT55 were used to align the RNA-seq reads and the clean and unique mapped reads were preserved. The gene expression levels (FPKM) are quantified by RSEM56. The significant upregulated genes were defined with a p value <0.05 and (FPKM) fold change ≥2 and the significant downregulated genes were defined with a p value <0.05 and (FPKM+ 1) fold change ≤0.5. The RNA-seq data have been deposited in NCBI’s SRA and are accessible through the accession number PRJNA701356.
RNA-mediated interference
The siRNA against human NELFB and the negative control siRNA (nontargeting siRNA) were purchased from GenePharma Company. The siRNAs were transfected into human LS174T cells by INVI DNA RNA Transfection Reagent (Invigentech). After 48 h, the cells were collected for mRNA analysis.
Chromatin immunoprecipitation (ChIP) assay and ChIP-seq
The colonic tissues were opened and washed with cold 1× PBS for two times, and then transferred to the pre-warmed (37 °C) in 1× PBS with 20 mM EDTA and incubated at 37 °C for 35 min. The supernatant was removed and the sediment was resuspended with cold PBS and further vigorously shaken to release crypts. The contents were filtered through a 70mm cell strainer (BD Bioscience) and spun at 300× 3 g for 2 min to collect the crypts. The crypts were fixed in 1% methanol-free formaldehyde (Thermo Scientific) for 5 min at room temperature followed by quenching with 125mM glycine for another 5 min. The nuclear extracts from crypts were prepared as previously described21. The chromatin DNA was sonicated into 100–300 bp using a Bioruptor (Diagenode). For immuno-precipitations, the NELF-B (ab167401, Abcam) and IgG (2729, Cell Signaling Technology) were used to detect NELF-B and endogenous IgG. The purified immunoprecipitated DNA was subjected and analyzed by RT-PCR and the relative occupancies were normalized to input DNA. The primer sequences were listed in Table S1. For ChIP-seq, 10 ng of DNA was ligated to adapters and the 100–300 bp DNA fragments were purified to prepare the libraries by the NEBNext Ultra II Library Prep Kit for Illumina (NEB). The libraries were sequenced by HiSeq2500. FASTQ files were generated by CASAVA. ChIP-seq datasets were mapped to the mouse mm10 reference genome using Bowtie 254. Multiple mapped reads and PCR duplicates were removed. Monoclonal reads were then normalized by calculating the reads per kilobase of transcript per million mapped reads (RPKM). Enriched peaks were identified by MACS2 v.2.1.2 with default parameters57. The target genes of each peak were annotated by annotatePeak function in ChIPseeker package58. Heatmaps and profile plots for normalized ChIP-seq data were generated using deepTools v. 3.5.059. The genomic data have been deposited in NCBI’s SRA and are accessible through the accession number PRJNA701745.
Histological analysis, immunohistochemistry, and immunofluorescence staining
The small intestine and colon tissues were fixed with 4% paraformaldehyde at 4 °C for more than 4 h, paraffin embedded, and sectioned (5 μm). The sections were deparaffinized and rehydrated before staining with hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS). The PAS staining was performed by the PAS Stain Kit (Solarbio, G1285). The histological scores of H&E stained sections were used to assess the intestinal pathology, based on assigned scores of tissue damage (0 = normal; 1 = irregular crypts; 2 = mild to moderate crypt loss (10–50%); 3 = severe crypt loss (50–90%); 4 = complete crypt loss) and inflammation (0 = absent, 1 = mucosal, 2 = submucosal, 3 = transmural extending into muscularis and serosa and 4 = diffuse). More than six different locations on each sample were used to count the score for each mouse.
The sections were deparaffinized and rehydrated. Heat-induced epitope retrieval was performed by boiling for 15 min with the Improved Citrate Antigen Retrieval Solution (Beyotime) before incubation with 10% serum in room temperature for 1 h. Sections were stained with the primary antibodies (anti-Ki67 (Cell Signaling Technology, 1:200), anti-Cleaved caspase 3 (Cell Signaling Technology, 1:200), anti-Escherichia coli (Bio-Rad, 1:200), anti-Clostrida. spp (NOVUS, 1:200), anti-E-cadherin (Cell Signaling Technology, 1:200), Cgn (Proteintech, 1:100)) overnight at 4 °C and washed with TBS for several times. For immunohistochemistry, HRP-conjugated secondary antibodies (EasyBio, 1:50) were performed in room temperature for 1 h and then visualized with DAB kit (TIANGEN). For immunofluorescence, the sections were incubated with secondary antibodies conjugated with DyLight 594 (Invitrogen, 1:100) or Alexa Flour 488 (Cell Signaling Technology, 1:100, or WGA (Wheat Germ Agglutinin-Rhodamine, Vectorlabs, 1:100) in dark at room temperature for 1 h. Slides were washed again and incubated with DAPI (Beyotime Biotechnology) and mounting with Flioroshied Mounting Medium (Abcam).
Fluorescence in situ hybridization
The FISH analysis was performed as previously described51. Briefly, the colonic tissues were fixed with methanol-Carnoy’s fixative (60% methanol, 30% Chloroform, 10% glacial acetic acid) for 4 h at 4 °C. Paraffin-embedded sections were dewaxed and hydrated. The hybridization buffer (20mM Tris-HCl, pH = 7.4, 0.9M NaCl, 0.1% SDS, 20% formamide) and posthybridization wash buffer (20mM Tris-HCl, pH = 7.4, 0.9M NaCl, 0.5M EDTA, 0.1% SDS) were pre-warmed in a 50 °C incubator. The tissue sections were incubated with 500 ng Alexa Fluor 488-conjugated EUB338 probe (5′-GCTGCCTCCCGTAGGAGT-3′) to detect all bacteria at 50 °C for 3 h in dark and then washed with posthybridization wash buffer and water respectively for 5 min, followed by incubation with DAPI in PBS in the dark for 10 min at room temperature. The slides were mounted with Flioroshied Mounting Medium (Abcam).
TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay
The sections were deparaffinized and rehydrated before washing with PBS for two times and incubation with Permeabilization Buffer (Triton-100 dissolved 0.1% citrate sodium with final concentration of 0.1%) at room temperature for 8 min. The slides were washed with PBS for two times and incubated with 50 μl TdT Enzyme working solution (add 50 μl of Enzyme Solution to the remaining 450 μl Label Solution to obtain 500 μl TUNEL reaction mixture, Roche) to each section at 37 °C for 1 h. Then the slides were washed with PBS, stained by DAPI and mounted with Flioroshied Mounting Medium (Abcam).
Transmission electron microscope assay
The colon tissues were fixed in 0.1M sodium-cacodylate buffer (pH = 7.0) containing 2% paraformaldehyde and 2.5% glutaraldehyde at 4 °C for 24 h. The segments were rinsed three times for 10 min with 0.1M sodium-cacodylate buffer and then fixed with 1% cacodylate-buffered osmium tetroxide solution for second fixation at 4 °C for 1.5 h. After dehydration in ascending series of ethyl alcohol, the segments were embedded in epoxyresin. The segments were cut into ultrathin section by glass knife on microtome and stained with uranyl acetate. The sections were examined by transmission electron microscope (Hitachi, H-7650B).
Statistical analysis
GraphPad Prism software was used for data analysis. For graphs, data are shown as mean ± SEM. Statistical significance was determined using a two-tailed unpaired Student’s t-test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns = non-significant.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by National Natural Science Foundation of China (32030037, 31821003, and 31725010 to X.H., 81601443 to X.-K.G.) and National Key Research and Development Program of China (2020YFA0509100 to X.H.), funds from Tsinghua-Peking Center for Life Sciences (X.H., X.-K.G), and Institute for Immunology at Tsinghua University (X.H.). R.L. is supported by the NIH R01CA220578.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41385-021-00465-9.
REFERENCES
- 1.Peterson LW & Artis D Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol 14, 141–153 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV Epithelial cell contributions to intestinal immunity. Adv. Immunol 126, 129–172 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Martens EC, Neumann M & Desai MS Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol 16, 457–470 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Turner JR Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol 9, 799–809 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Caruso R, Lo BC & Nunez G Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol 20, 411–426 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Zihni C, Mills C, Matter K & Balda MS Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol 17, 564–580 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Ivanov AI & Naydenov NG Dynamics and regulation of epithelial adherens junctions: recent discoveries and controversies. Int. Rev. Cell Mol. Biol 303, 27–99 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Suzuki T Regulation of intestinal epithelial permeability by tight junctions. Cell Mol. Life Sci 70, 631–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramanan D & Cadwell K Intrinsic defense mechanisms of the intestinal epithelium. Cell Host Microbe 19, 434–441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lechuga S et al. Loss of β-cytoplasmic actin in the intestinal epithelium increases gut barrier permeability in vivo and exaggerates the severity of experimental colitis. Front. Cell Dev. Biol 8, 588836 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laukoetter MG et al. JAM-A regulates permeability and inflammation in the intestine in vivo. J. Exp. Med 204, 3067–3076 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham DB & Xavier RJ Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 578, 527–539 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mankertz J & Schulzke JD Altered permeability in inflammatory bowel disease: pathophysiology and clinical implications. Curr. Opin. Gastroenterol 23, 379–383 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Jonkers I & Lis JT Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol 16, 167–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwak H & Lis JT Control of transcriptional elongation. Annu. Rev. Genet 47, 483–508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adelman K & Lis JT Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet 13, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen FX, Smith ER & Shilatifard A Born to run: control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol 19, 464–478 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi Y et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Sacta MA et al. Gene-specific mechanisms direct glucocorticoid-receptor-driven repression of inflammatory response genes in macrophages. Elife 7, e34864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rawat P et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell 81, 1013–1026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu L et al. Negative elongation factor complex enables macrophage inflammatory responses by controlling anti-inflammatory gene expression. Nat. Commun 11, 2286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilchrist DA et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 22, 1921–1933 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Core LJ et al. Defining the status of RNA polymerase at promoters. Cell Rep. 2, 1025–1035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Lee C, Gilmour DS & Gergen JP Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev. 21, 1031–1036 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeitlinger J et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet 39, 1512–1516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagha M et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976–987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams LH et al. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol. Cell 58, 311–322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adelman K et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl Acad. Sci. USA 106, 18207–18212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hargreaves DC, Horng T & Medzhitov R Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138, 129–145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang Y et al. The transcriptional repressor Hes1 attenuates inflammation by regulating transcription elongation. Nat. Immunol 17, 930–937 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogatsky I & Adelman K Preparing the first responders: building the inflammatory transcriptome from the ground up. Mol. Cell 54, 245–254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupte R, Muse GW, Chinenov Y, Adelman K & Rogatsky I Glucocorticoid receptor represses proinflammatory genes at distinct steps of the transcription cycle. Proc. Natl Acad. Sci. USA 110, 14616–14621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heppert JK et al. Transcriptional programmes underlying cellular identity and microbial responsiveness in the intestinal epithelium. Nat. Rev. Gastroenterol. Hepatol 18, 7–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beumer J & Clevers H Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol 22, 39–53 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Sun J et al. Deregulation of cofactor of BRCA1 expression in breast cancer cells. J. Cell Biochem 103, 1798–1807 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patankar JV & Becker C Cell death in the gut epithelium and implications for chronic inflammation. Nat. Rev. Gastroenterol. Hepatol 17, 543–556 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Ueda H, Fujita R, Yoshida A, Matsunaga H & Ueda M Identification of prothymosin-alpha1, the necrosis-apoptosis switch molecule in cortical neuronal cultures. J. Cell Biol 176, 853–862 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunther C et al. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature 477, 335–339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welz PS et al. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 477, 330–334 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Tabula Muris C et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansson ME et al. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS ONE 5, e12238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Souza HS & Fiocchi C Immunopathogenesis of IBD: current state of the art. Nat. Rev. Gastroenterol. Hepatol 13, 13–27 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Wyatt J, Vogelsang H, Hubl W, Waldhoer T & Lochs H Intestinal permeability and the prediction of relapse in Crohn’s disease. Lancet 341, 1437–1439 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Bjarnason I, O’Morain C, Levi AJ & Peters TJ Absorption of 51chromium-labeled ethylenediaminetetraacetate in inflammatory bowel disease. Gastroenterology 85, 318–322 (1983). [PubMed] [Google Scholar]
- 45.de Lange KM et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet 49, 256–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi N et al. RIPK1 ensures intestinal homeostasis by protecting the epithelium against apoptosis. Nature 513, 95–99 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Lee TI & Young RA Transcriptional regulation and its misregulation in disease. Cell 152, 1237–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amleh A et al. Mouse cofactor of BRCA1 (Cobra1) is required for early embryogenesis. PLoS ONE 4, e5034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ou J, Liang S, Guo XK & Hu X alpha-Defensins promote bacteroides colonization on mucosal reservoir to prevent antibioticiInduced dysbiosis. Front. Immunol 11, 2065 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang S et al. Nutrient sensing by the intestinal epithelium orchestrates mucosal antimicrobial defense via translational control of Hes1. Cell Host Microbe 25, 706–718 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Guo XK, Ou J, Liang S, Zhou X & Hu X Epithelial Hes1 maintains gut homeostasis by preventing microbial dysbiosis. Mucosal Immunol. 11, 716–726 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Sato T et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 53.McCray T, Richards Z, Marsili J, Prins GS & Nonn L Handling and assessment of human primary prostate organoid culture. J. Vis. Exp 10.3791/59051 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D, Langmead B & Salzberg SL HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu G, Wang LG & He QY ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Ramirez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.