Abstract
The process of protein import into plastids has been studied extensively using isolated pea (Pisum sativum) chloroplasts. As a consequence, virtually all of the known components of the proteinaceous apparatus that mediates import were originally cloned from pea. With the recent completion of the Arabidopsis genome sequencing project, it is now possible to identify putative homologs of the import components in this species. Our analysis has revealed that Arabidopsis homologs with high sequence similarity exist for all of the pea import complex subunits, making Arabidopsis a valid model for further study of this system. Multiple homologs can be identified for over one-half of the components. In all but one case it is known that more than one of the putative isoforms for a particular subunit are expressed. Thus, it is possible that multiple types of import complexes are present within the same cell, each having a unique affinity for different chloroplastic precursor proteins, depending upon the exact mix of isoforms it contains. Sequence analysis of the putative Arabidopsis homologs for the chloroplast protein import apparatus has revealed many questions concerning subunit function and evolution. It should now be possible to use the genetic tools available in Arabidopsis, including the generation of knockout mutants and antisense technology, to address these questions and learn more about the molecular functions of each of the components during the import process.
The availability of the sequence for the entire genome of Arabidopsis allows a detailed analysis of all the genes involved in a particular biological process, regardless of the plant species in which the system was first identified. One such process is the import of cytoplasmically synthesized precursor proteins into chloroplasts. Most of the current information regarding this process, including the identification of components of the import apparatus that mediates it, has come from biochemical studies in pea (Pisum sativum; Fig. 1; Chen and Schnell, 1999; Keegstra and Cline, 1999; Keegstra and Froehlich, 1999; May and Soll, 1999; Schleiff and Soll, 2000). From these studies it has been determined that nuclear-encoded, chloroplast-localized enzymes are synthesized in the cytoplasm as precursors containing an N-terminal transit peptide not seen in the mature protein within the chloroplast (for review, see Bruce, 2000). A precursor protein initially interacts with a complex located within the outer membrane of the chloroplast envelope that consists of at least three subunits: translocon at the outer envelope membrane of chloroplasts (Toc) 159, Toc75, and Toc34 (Waegemann and Soll, 1991; Hirsch et al., 1994; Kessler et al., 1994; Perry and Keegstra, 1994; Schnell et al., 1994; Seedorf et al., 1995; Tranel et al., 1995). These early events involve the hydrolysis of GTP, presumably by Toc159 and Toc34, which are known to be GTP-binding proteins (Kessler et al., 1994; Seedorf et al., 1995). A recent report by Sohrt and Soll (2000) has also implicated a fourth component, Toc64, as being a member of the outer membrane import machinery. Hydrolysis of low concentrations of ATP in the cytoplasm or intermembrane space results in the irreversible association of precursor proteins with the translocation machinery of both the outer and inner envelope membranes (Olsen et al., 1989; Olsen and Keegstra, 1992). The import complex of the chloroplastic inner envelope membrane also consists of at least three subunits: translocon at the inner envelope membrane of chloroplasts (Tic) 110, Tic20, and Tic22 (Kessler and Blobel, 1996; Lübeck et al., 1996; Kouranov and Schnell, 1997; Kouranov et al., 1998). Two additional components, Tic55 and Tic40, have also been reported to be a part of this translocon, but their inclusion is more controversial (Wu et al., 1994; Ko et al., 1995; Caliebe et al., 1997; Stahl et al., 1999). Complete translocation of precursor proteins into the chloroplast interior is accomplished via the hydrolysis of ATP within the stroma (Theg et al., 1989). This ATP hydrolysis is presumably mediated by stromal molecular chaperones, at least one of which, heat shock protein (Hsp) 93 (a member of the Hsp100 family of molecular chaperones), has been found to interact with the import complex (Akita et al., 1997; Nielsen et al., 1997). As the precursor enters the chloroplast, the transit peptide is cleaved off by the stromal processing peptidase (SPP) and the mature protein begins the process of folding and assembly (Oblong and Lamppa, 1992; VanderVere et al., 1995; Richter and Lamppa, 1998).
Figure 1.
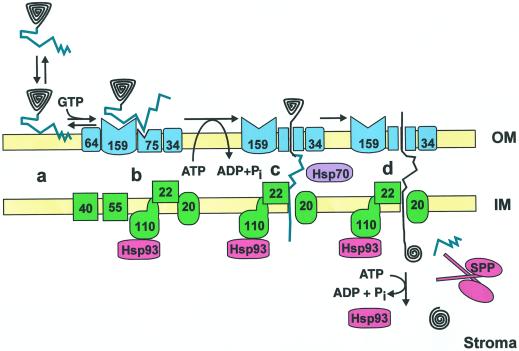
Current model describing the process of protein import into pea chloroplasts. Nuclear-encoded chloroplastic proteins are initially synthesized in the cytoplasm with a transit peptide (teal) that targets them to the plastid surface (a). In a process stimulated by GTP, the precursor protein associates with the components (blue) of the outer envelope membrane translocon (b). Hydrolysis of ATP in the cytoplasm and/or intermembrane space causes the precursor to interact with the components (green) of the inner membrane translocon as well (c). It is postulated that this step may be assisted by chaperones residing in the intermembrane space (purple). Hydrolysis of stromal ATP results in the complete translocation of the precursor protein into the chloroplast interior, where the transit peptide is removed (d). This final step is mediated at least in part by stromal factors (red). The numbers within the components of the outer and inner membrane translocons refer to the calculated molecular mass of each subunit. OM, Outer membrane; IM, inner membrane.
Although virtually all of the conclusions described above were derived from work done with pea chloroplasts, expressed sequence tags (ESTs) for homologs of the various import components can be identified in the databases for a variety of monocots and dicots, including maize, tomato, and Arabidopsis. More importantly, the recent completion of the Arabidopsis genome sequencing project (The Arabidopsis Genome Initiative, 2000) has made it possible to find, in this species, homologs of those components for which no ESTs exist. In addition to establishing the general significance of the components of the import apparatus, identification of Arabidopsis homologs for the subunits of the pea import complex will allow the use of this species to perform molecular work that is not practical and/or possible with pea, including isolation of “knockout” mutants and generation of transgenic plants expressing sense or antisense copies of the genes encoding one or more of these components.
In this paper, we analyze the Arabidopsis genomic, cDNA, and EST information currently available in GenBank concerning each of the known and putative subunits of the chloroplast protein import machinery. All of these components have homologs of high sequence identity within the Arabidopsis genome that are expressed and likely act as functional counterparts to the pea proteins. For several of these translocation components, multiple putative homologs are present in the Arabidopsis genome. However, in most cases, it is unclear whether all copies are expressed, or if they are, whether they are all acting as functional homologs within Arabidopsis chloroplasts. The information revealed by this analysis will allow important new questions to be raised, and further experimental work can then be designed to answer them in the near future.
RESULTS AND DISCUSSION
Outer Envelope Membrane Proteins
Toc159, a GTP-binding protein, is postulated to be the first subunit of the import complex with which an incoming precursor protein interacts, serving as the receptor for transit peptides (Waegemann and Soll, 1991; Hirsch et al., 1994; Kessler et al., 1994; Perry and Keegstra, 1994; Ma et al., 1996). There are three homologs of this protein in Arabidopsis (Table I) designated AtToc159, AtToc132, and AtToc120 based on their predicted molecular masses (Bauer et al., 2000). All three are expressed, as demonstrated by the presence of at least one Arabidopsis EST for each and by reverse transcriptase-PCR experiments (Bauer et al., 2000).
Table I.
Putative Arabidopsis homologs for the components of the pea chloroplast protein import machinery
| Pea Import Component | Arabidopsis Homologsa | Chromosomeb | GenBank Designationc | EST in Database | No. of Intronsd |
|---|---|---|---|---|---|
| Toc159 | AtToc159 | IV | T14P8.24 | Yes | 1 |
| AtToc132 | II | At2g16640 | Yes | 0 | |
| AtToc120 | III | MGL6.8 | Yes | 0 | |
| Toc75 | AtToc75-III | III | T6H20.230 | Yes | 6 |
| AtToc75-I | I | F10O5.4 | No | NDe | |
| AtToc75-IV | IV | At4g09080 | No | 5 | |
| Toc34 | AtToc34 | V | MUG13.14 | Yes | 6 |
| AtToc33 | I | T7I23.11 | Yes | 6 | |
| Toc64 | AtToc64-III | III | MEB5.17 | Yes | 12 |
| AtToc64-V | V | T5E8_220 | Yes | 12 | |
| AtToc64-I | I | F7G19.15 | Yes | NDe | |
| Tic110 | AtTic110 | I | F10K1.33 and F4H5.1f | Yes | 14 |
| Tic20 | AtTic20-I | I | F13M7.7 | Yes | NDe |
| AtTic20-IV | IV | F4C21.25 | Yes | 2 | |
| Tic22 | AtTic22-IV | IV | F17M5.110 | Yes | 7 |
| AtTic22-III | III | MYM9.5 | Yes | 7 | |
| Tic55 | AtTic55 | II | At2g24820 | Yes | 2 |
| Tic40 | AtTic40 | V | MTG13.6 | Yes | 13 |
| Hsp93gh | AtHsp93-V | V | K3K7.7 | Yes | 8 |
| AtHsp93-III | III | T21J18_140 | Yes | 8 | |
| SPP (CPE) | AtCPE | V | MDH9.8 | Yes | 23 |
| Hsp70g | AtHsp70-V | V | K9P8.5 | Yes | 7 |
| AtHsp70-IVa | IV | At4g24280 | Yes | 7 | |
| AtHsp70-IVb | IV | At4g37910 | Yes | 5 |
See “Materials and Methods” for an explanation of the criteria used in designating a sequence as a homolog of the pea import apparatus.
The chromosomal location for the genes encoding each of the putative Arabidopsis chloroplast protein import components is indicated.
The entries in this column refer to the bacterial artificial chromosome or P1 clone designation in GenBank for each of the putative homologs.
Gene structure predictions are based on the annotations given in the GenBank entry indicated in this table, unless otherwise noted.
ND, Not determined. The current annotations given in the database for these homologs are predicted to be incorrect because the regions of sequence identity to the pea proteins are very different from the predicted boundaries of the open reading frame.
This coding region is split between two bacterial artificial chromosomes, which overlap by 200 bp.
Arabidopsis homologs given for these components are only those predicted to have a chloroplastic targeting sequence (see “Materials and Methods” for an explanation of how this was determined.
Arabidopsis homologs given for this protein are only those predicted to belong to the ClpC class of the Hsp100 family of chaperones.
The pea Toc159 protein is composed of three domains: an N-terminal acidic region, a central domain encompassing the GTP-binding motifs, and a C-terminal domain containing the membrane-spanning regions (Chen et al., 2000a). AtToc159 shares approximately 48% identity with the pea protein, most of which is concentrated in the central and C-terminal domains (approximately 69% identity in these regions). Pea Toc159 and AtToc159 are highly acidic, especially in their N-terminal regions (Bölter et al., 1998a; Bauer et al., 2000; Chen et al., 2000a). Approximately 30% and 27%, respectively, of the amino acids in this domain are Asp or Glu (Table II). This is in contrast to the other members of the outer membrane import complex (Toc75, Toc34, and Toc64) in which the percentage of acidic residues ranges from 9% to 11% for the Arabidopsis isoforms. One of the defining features of transit peptides is that they lack acidic amino acids, resulting in an overall basic pI and net positive charge (Keegstra et al., 1989). Thus, it is interesting to speculate that the N-terminal acidic domain of Toc159, which is localized on the cytoplasmic face of the chloroplast, is involved in an electrostatic interaction with positively charged transit peptides, increasing the overall efficiency of precursor protein binding (Bölter et al., 1998a). This is similar to the situation described by the acid chain hypothesis for the early interaction of basic mitochondrial targeting sequences with their acidic receptors (Komiya et al., 1998).
Table II.
Comparison of the acidic properties of various members of the outer envelope membrane import complex
| Import Component | Percentage of Acidic Residues in Whole Proteina | pI of Whole Protein | Percentage of Acidic Residues in N-Terminal Domaina | pI of N-Terminal Domain |
|---|---|---|---|---|
| Pea Toc159 | 20 | 4.2 | 30 | 3.6 |
| AtToc159 | 19 | 4.3 | 27 | 3.8 |
| AtToc132 | 17 | 4.7 | 28 | 3.8 |
| AtToc120 | 16 | 4.9 | 26 | 3.9 |
| AtToc75-III | 10 | 8.8 | N/A | N/A |
| AtToc34 | 10 | 9.4 | N/A | N/A |
| AtToc64-III | 10 | 8.2 | N/A | N/A |
Acidic residues are aspartate (D) and glutamate (E).
AtToc132 and AtToc120 show less overall identity with pea Toc159 (approximately 37% and 39%, respectively), the majority of which is again concentrated in the central and C-terminal domains (approximately 50% for each). In addition, their levels of identity to AtToc159 are also relatively low (approximately 37% and 38%, respectively). On the other hand, the two proteins share approximately 70% amino acid identity with each other. This suggests that AtToc132 and AtToc120 share a common ancestor that diverged from AtToc159 before these two proteins diverged from one another.
AtToc132 and AtToc120 are also highly acidic in their N-terminal regions (approximately 28% and 26% acidic residues, respectively). In fact, this is the main feature shared between the Arabidopsis homologs at their N-termini. There is very little conservation of primary structure between the three proteins before the GTP-binding domain (Bauer et al., 2000). However, despite a maintenance of the overall percentage of acidic residues within the N-terminal domains, the pI of the N-termini and the whole proteins differs between the three isoforms (Table II). Thus, the question arises of whether these subtle changes in size and overall charge between the Arabidopsis Toc159 homologs reflect differences in the types of precursors with which these proteins interact (Bauer et al., 2000). It is interesting to note that mutant Arabidopsis plants that lack AtToc159 are still able to import some, but not all, chloroplastic proteins, suggesting that some other factor, perhaps AtToc132 and/or AtToc120, is substituting for AtToc159 in the import of some precursors (Bauer et al., 2000).
Toc75 has been shown to form a voltage-gated, peptide-sensitive channel in artificial lipid bilayers (Hinnah et al., 1997). Thus, it is hypothesized that this protein forms the channel through which precursor proteins cross the outer envelope membrane (Perry and Keegstra, 1994; Schnell et al., 1994; Tranel et al., 1995; Hinnah et al., 1997). Analysis of the Arabidopsis genome sequence reveals at least three coding regions that have strong similarity to pea TOC75: AtTOC75-III, AtTOC75-I, and AtTOC75-IV, named according to their chromosomal location. Only one of these genes, AtTOC75-III, is represented by an EST. More than 10 ESTs for this gene can be found, but none currently exist for the other two homologs. In addition, of the three, AtToc75-III shows the highest levels of identity with the pea protein (approximately 74%). As a consequence, it is likely that AtToc75-III is the major Toc75 isoform in Arabidopsis cells.
AtToc75-III and AtToc75-I are quite similar to one another in size and amino acid sequence, sharing >60% identity throughout their lengths. On the other hand, AtToc75-IV displays some striking differences from its two homologs. First of all, the protein encoded by AtTOC75-IV is much smaller at 407 amino acids in length versus 818 amino acids for the protein encoded by AtTOC75-III. Furthermore, the region of similarity between AtToc75-IV and the other two Arabidopsis homologs is confined to the C-termini of the larger proteins. It appears that AtTOC75-IV may represent just the last six exons of AtTOC75-III. In fact, this gene seems to be an extreme case of a more common phenomenon. For a few components, including Toc75 and Toc159, BLAST searches reveal several small regions with high levels of sequence similarity to these subunits throughout the genome. Although these putative open reading frames do show similarity to the import components outside of commonly found motifs (i.e. nucleotide-binding domains), the regions of similarity are not extensive. In general, they constitute less than one-quarter of the total length of the queried import component, not enough to really be considered a possible functional homolog. One possible explanation for the occurrence of these presumably unexpressed regions of similarity is that these short open reading frames are examples of the evolutionary process of exon shuffling in progress.
In the case of AtToc75-IV, the region of similarity extends for approximately 50% of the length of the larger Toc75 homologs. It is possible that this may be enough for the protein made by AtTOC75-IV to be functional. Future research should address this problem, but some observations suggest that it may indeed be needed in Arabidopsis cells. It is interesting to note that the levels of identity between this coding region and its “parent” are quite high at the amino acid level (approximately 65% with AtToc75-III) and at the nucleotide level. Moreover, the splicing pattern of AtTOC75-IV is identical to that seen in the 3′ region of AtTOC75-III, implying that selection pressure on AtTOC75-IV may still be relatively high.
Toc34, another GTP-binding protein of the translocation apparatus, is hypothesized to have a regulatory function during precursor import (Kessler et al., 1994; Seedorf et al., 1995; Kouranov and Schnell, 1997). This subunit has two homologs in Arabidopsis named AtToc34 and AtToc33 based on their predicted molecular masses (Jarvis et al., 1998). ESTs are present for both of these homologs within the Arabidopsis database, and their expression has been verified via northern and western-blot analysis (Jarvis et al., 1998; Gutensohn et al., 2000). It appears that the two proteins, which are >60% identical to each other and to the pea protein, can at least partially substitute for one another within plant cells. Arabidopsis mutants that lack AtToc33 display a delayed greening phenotype and reduced levels of chloroplast protein import early in their development, but are otherwise normal (Jarvis et al., 1998; Gutensohn et al., 2000).
The genes for AtToc34 and AtToc33 provide an example of the evolutionary process of gene duplication. Each coding region consists of six introns and seven exons; five of the seven exons are exactly the same size between the two genes. In addition, in every case, the exon-intron junctions occur at homologous positions within the sequences. Thus, it appears that these two coding regions have diverged from one another only relatively recently after the duplication of a common ancestral gene.
A fourth putative subunit of the outer envelope membrane import apparatus, Toc64, was recently isolated (Sohrt and Soll, 2000). The amino acid sequence for this component contains an amidase domain, but the protein itself has no measurable amidase activity (Sohrt and Soll, 2000). In addition, Toc64 contains three tetratricopeptide repeats (TPR), which are hypothesized to be involved in protein-protein interactions with cytosolic factors complexed with a precursor protein and/or with the precursor itself, perhaps serving as a docking site for the incoming protein (Sohrt and Soll, 2000). Within the Arabidopsis genome there are three coding regions that display extensive similarity with the pea protein outside of the amidase domain and/or the TPR motifs. These homologs have been designated AtToc64-III, AtToc64-V, and AtToc64-I. For all three isoforms, cognate ESTs have been isolated. However, only AtToc64-III and AtToc64-V contain regions similar to both the amidase domain and the TPR motifs of pea Toc64 (approximately 67% and 50% identical, respectively). Thus, although it is likely that the proteins encoded by AtTOC64-III and AtTOC64-V could serve as functional homologs of pea Toc64 within Arabidopsis cells, further experiments will need to be done to determine whether AtToc64-I, which lacks the TPR motifs, is playing a similar role.
Inner Envelope Membrane Proteins
The first component of the inner membrane import complex to be cloned and characterized was Tic110 (Kessler and Blobel, 1996; Lübeck et al., 1996). This subunit consists of a large globular domain localized in the chloroplast stroma and anchored to the envelope by a membrane-spanning α-helix at the N terminus (Kessler and Blobel, 1996; Jackson et al., 1998). Based on this topology it has been proposed that Tic110 acts as an anchor for stromal molecular chaperones involved in precursor protein import (Kessler and Blobel, 1996; Jackson et al., 1998). Preliminary evidence suggests that Tic110 may physically interact with at least one molecular chaperone (M. Akita and K. Keegstra, unpublished data). BLAST searches on the Arabidopsis genome sequence reveal only one coding region, AtTIC110, similar to the pea gene (Table I). The protein encoded by AtTIC110 is expressed and displays high levels of identity (approximately 68%) to pea Tic110. In addition, it appears to have the same overall structure as the pea protein, with a predicted transmembrane helix at the N terminus followed by a large hydrophilic domain. Thus, it is reasonable to conclude that AtTic110 acts as a functional homolog of pea Tic110 within Arabidopsis cells.
The gene structure for AtTIC110 is quite complicated; the coding region consists of 15 exons and 14 introns. Overall, the coding region is 5,261 bp in length, with 42% of this length comprising the introns. This complexity is in contrast to the genes encoding the Arabidopsis Toc159 isoforms. The coding regions for these proteins are also quite long, ranging from 3,270 bp (AtTOC120) to 4,595 bp (AtTOC159) in length. However, they contain only one small intron (83 bp; AtTOC159) or none at all (AtTOC132 and AtTOC120). This diversity in gene structure is seen for the other components of the import complex as well. The genes encoding the Arabidopsis homologs of Tic20 and Tic55 are relatively simple (two or fewer introns), whereas the genes for the remaining subunits are more complicated, containing between six and 23 introns (Table I).
Tic20, an integral protein of the inner envelope membrane, is believed to form at least a portion of the channel through which chloroplast precursors traverse the inner membrane (Kouranov and Schnell, 1997; Kouranov et al., 1998). The Arabidopsis genome contains two genes encoding proteins, AtTic20-I and AtTic20-IV (designated according to the chromosomal locations of the genes), that are similar to pea Tic20. Both of these genes have corresponding ESTs within the Arabidopsis database. AtTic20-I is highly similar to the pea protein, sharing >60% identity with pea Tic20. As a consequence, it is likely to act as the functional counterpart to the pea protein in Arabidopsis chloroplasts. On the other hand, AtTic20-IV is only approximately 33% identical to pea Tic20 and approximately 40% identical to AtTic20-I. Although these levels of identity are relatively high, it is quite low for this system; most of the putative Arabidopsis homologs for the other import components show much higher levels of identity to their pea counterparts and related Arabidopsis isoforms. Thus, it appears that these two Tic20 isoforms may have diverged from one another earlier in evolution than is the case for isoforms of some of the other subunits of the import complex.
BLAST searches for Arabidopsis homologs of pea Tic20 reveal a third putative isoform on chromosome II. However, this protein is much smaller (by approximately 70 amino acids) than the other two Arabidopsis homologs. More importantly, BLAST searches using this putative isoform as the query sequence fail to detect either AtTic20-I or AtTic20-IV. Thus, it was concluded that this coding region, despite sharing approximately 26% identity with pea Tic20 at the amino acid level, should not be considered an Arabidopsis homolog of the pea protein.
Tic22 is localized in the intermembrane space of the chloroplast envelope and appears to be peripherally associated with the inner envelope membrane (Kouranov and Schnell, 1997; Kouranov et al., 1998). Due to its localization, it has been proposed that Tic22 may be involved in the formation of contact sites between the import complexes of the outer and inner membranes (Kouranov and Schnell, 1997; Kouranov et al., 1998). Within the Arabidopsis genome there are at least two coding regions, AtTIC22-IV and AtTIC22-III, of high similarity to pea TIC22. These genes are expressed, as determined by the presence of several ESTs for each in the database. The encoded proteins share approximately 62% and 41% identity, respectively, with pea Tic22.
Tic55, an iron-sulfur protein believed to play a regulatory role during chloroplast protein import (Caliebe et al., 1997), and Tic40, which is proposed to recruit chaperones to the site of precursor protein import (Wu et al., 1994; Ko et al., 1995; Stahl et al., 1999), each have one clear homolog of high similarity in Arabidopsis. ESTs exist for both AtTic55 and AtTic40. The proteins display approximately 78% and 52% identity, respectively, with their pea counterparts. Thus, it is likely that they serve as functional homologs to the corresponding pea proteins.
Soluble Factors
It is thought that molecular chaperones within the chloroplast stroma provide the driving force, through the hydrolysis of ATP, for the translocation of precursor proteins into the chloroplast interior (Chen and Schnell, 1999; Keegstra and Cline, 1999; Keegstra and Froehlich, 1999). At the present time the best candidate for this role is Hsp93, a member of the Hsp100 family of chaperones that is consistently found in import complexes isolated from pea chloroplasts (Akita et al., 1997; Nielsen et al., 1997; Kouranov et al., 1998). This chaperone has at least two homologs (Table I) predicted to be present in Arabidopsis chloroplasts, AtHsp93-V (approximately 88% identity to pea Hsp93) and AtHsp93-III (approximately 83% identity to the pea protein; Nakabayashi et al., 1999). These two proteins, along with pea Hsp93, belong to the caseinolytic protease (Clp) C class of Hsp100 chaperones. Hsp100 proteins of other classes, specifically the ClpB and ClpD classes, that are predicted to be chloroplast-localized can also be detected in the Arabidopsis genome, as can potentially chloroplastic members of the Hsp70 and Hsp60 chaperone families. This diversity of stromally localized chaperones raises the question of whether Hsp93 is the only chaperone that interacts with the protein import complex or whether other types of chaperones could substitute for it in different species. Further work will be needed to confirm that the AtHsp93 homologs directly interact with the import complex in Arabidopsis chloroplasts as Hsp93 does in pea chloroplasts.
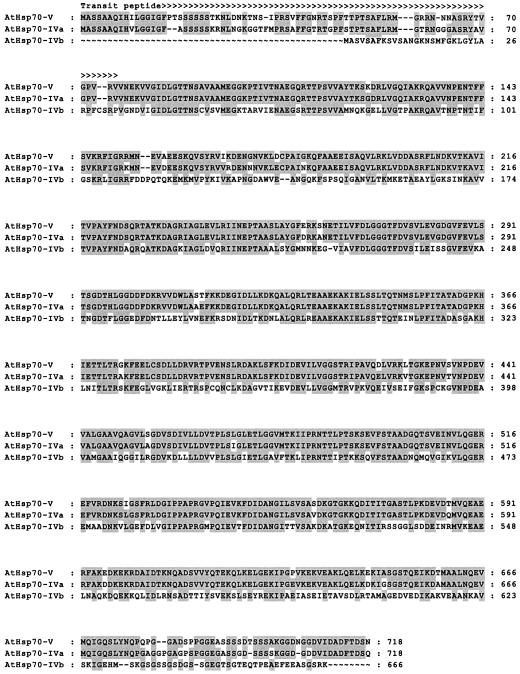
Although no stromal Hsp70 proteins have been found to interact with import complexes (Akita et al., 1997; Nielsen et al., 1997), there is evidence to suggest that Hsp70 molecules do bind to precursor proteins before and/or during envelope translocation (Schnell et al., 1994; Wu et al., 1994; Kourtz and Ko, 1997; Ivey et al., 2000; May and Soll, 2000). Furthermore, an outer membrane-associated Hsp70 protein, which faces the intermembrane space of the chloroplast envelope, is believed to interact with precursor proteins as they move between the outer and inner membrane translocons (Marshall et al., 1990; Schnell et al., 1994). Within the Arabidopsis genome there are several coding regions that encode proteins similar to known Hsp70 molecules from other species. These Arabidopsis Hsp70 proteins can be classified into one of four groups: proteins of approximately 650 residues that likely represent cytosolic Hsp70 molecules, proteins that are 668 or 669 residues long and contain an obvious signal peptide at their N-termini, molecules with clear chloroplastic (two proteins) or mitochondrial (one protein) targeting motifs, and proteins that do not fit into any of the previous three groups. Of the proteins within the last group, only one shows some characteristics of a chloroplast transit peptide at its N terminus. Sequence alignment between this protein and the two obvious chloroplast-targeted Hsp70 molecules is shown in Figure 2.
Figure 2.
Multiple sequence alignment for the putative chloroplast-localized Arabidopsis Hsp70 isoforms. Shaded residues designate sequence identities between two or more of the proteins. The predicted transit peptide is indicated (>). The predicted cleavage site is based on sequence identity to a pea chloroplastic Hsp70 (accession no. L03299) and has not been experimentally verified. The alignment was created using the PileUp program from the Wisconsin package of sequence analysis tools (Genetics Computer Group, Madison, WI).
The only known intermembrane space protein that has been cloned is Tic22 (Kouranov et al., 1998). An analysis of the transit peptide for pea Tic22 reveals that it has a relatively high incidence of acidic amino acids: three within the 50 residues of its length (Kouranov et al., 1998). AtTic22 has five acidic residues within the same region. The paradigm for chloroplast transit peptides is that they are deficient in acidic amino acids, having no more than two over their length (Keegstra et al., 1989). Thus, the transit peptides for pea and Arabidopsis Tic22 are somewhat unusual, and this fact may account for why these proteins are targeted to the intermembrane space of the chloroplast envelope rather than the stroma, although this has not been experimentally verified. We analyzed the transit peptides of the possible chloroplastic Hsp70 proteins to see if we could detect, based on what is observed from the transit peptide of Tic22, which one (or ones) might be targeted to the intermembrane space. However, all three of these proteins display a low incidence of acidic amino acids within their presumed transit peptides (Fig. 2). Thus, either the presence of acidic amino acids within the transit peptide is not the determining factor for intermembrane space targeting or Arabidopsis may not contain an intermembrane space-localized Hsp70 protein as has been suggested for pea (Marshall et al., 1990; Schnell et al., 1994). Further experimental work will be needed to differentiate between these possibilities.
The SPP (also known as the chloroplast processing enzyme [CPE]) is a metalloendopeptidase that cleaves transit peptides off precursor proteins as they enter the chloroplast stroma (Oblong and Lamppa, 1992; VanderVere et al., 1995; Richter and Lamppa, 1998). This component has one homolog in Arabidopsis, named AtCPE, which shares approximately 75% identity with the pea protein (Richter and Lamppa, 1998). The SPP currently is the only constituent of the import machinery whose molecular function has been studied in enough detail to be unequivocally assigned (Richter and Lamppa, 1998, 1999).
CONCLUSIONS
Analysis of the Arabidopsis sequence database has revealed that homologs of high sequence similarity can be found for each of the chloroplast protein import components that were originally identified in pea. This suggests that the protein import system is conserved between pea and Arabidopsis, making Arabidopsis a valid model for its study. It is likely that the import complex is conserved in other plant species as well. EST sequences similar to the known import components can be found in many species, including maize, soybean, and rice. In addition, antibody cross-reactivity studies on species as diverse as mosses and tomato have suggested that at least some of the subunits of the import machinery can be found in all chloroplast-containing eukaryotes (J. Davila-Aponte and K. Keegstra, unpublished data). Various lines of evidence have also indicated that cyanobacteria contain homologs of at least some of the import components (Bölter et al., 1998b; Reumann and Keegstra, 1999; Reumann et al., 1999). Thus, the chloroplast protein import system is likely to be conserved, at least in part, in all plant (and related) species.
For at least seven (Toc159, Toc75, Toc34, Toc64, Tic20, Tic22, and Hsp93) of the 11 known import components, multiple homologs can be found within the Arabidopsis genome. In all but one of these cases it is known that more than one of these homologs is expressed within Arabidopsis cells (Jarvis et al., 1998; Bauer et al., 2000; Gutensohn et al., 2000). This observation immediately suggests that multiple isoforms of the same subunit may be present in the same cells at the same time (Jarvis et al., 1998; Bauer et al., 2000; Chen et al., 2000b; Gutensohn et al., 2000). If this is the case, then one may imagine the existence of multiple types of import complexes within the chloroplast envelope, each with their own particular precursor specificity. For example, if all three Arabidopsis Toc159 homologs are expressed within the same cell, then the chloroplasts within that cell may have a mixture of import complexes: some containing AtToc159, others containing AtToc132, and still others containing AtToc120. However, because the stoichiometry of the subunits within the outer membrane translocon is not known, it is also possible that all three may exist within the same import complex. It is obvious that such questions cannot be answered by sequence analysis alone, and further experiments will be needed to address these issues.
The possibility of multiple isoforms for some of the protein import components within Arabidopsis chloroplasts also raises the question of whether the same situation is present in pea plants. Is Arabidopsis “unusual” in having multiple genes for at least some of the subunits of the import complex or is this the case in pea as well? So far, only one isoform has been identified for each component of the pea import apparatus. However, this fact does not mean that additional homologs do not exist within the pea genome. Since the pea import components were all initially isolated via biochemical means, it is possible that isoforms not present at high concentrations or at the particular stage of development studied would be missed. At this time there is not enough pea sequence information in GenBank to determine if multiple genes for the import components may indeed also be found in this species.
It is interesting to note that none of the coding regions for the Arabidopsis import components are found close to one another within the genome. Even for the components that have multiple putative isoforms, the genes encoding these proteins are located on separate chromosomes (see Table I). This is in contrast to the situation known for several other gene families (Lin et al., 1999; Mayer et al., 1999; The Arabidopsis Genome Initiative, 2000). Often, homologs of a particular coding region can be found nearby in the genome, if not in tandem (Lin et al., 1999; Mayer et al., 1999; The Arabidopsis Genome Initiative, 2000). In the case of the chloroplast protein import complex, however, the genes encoding the various subunits are found scattered throughout the genome. The explanation for this observation is not clear. Perhaps recombination in the areas immediately surrounding the genes for the import components is suppressed due to the essential nature of the import complex genes themselves or other genes in their local environment. Additional work will be needed to test this hypothesis.
It has been known for many years that the components of the pea chloroplast protein import complex show little sequence similarity to proteins of known function from other organisms (with the exception of the molecular chaperones and the SPP), including the subunits of the protein import systems of other organelles (Reumann and Keegstra, 1999; Reumann et al., 1999). Thus, it has not been possible to use information gained from the genetic study of other protein import systems to learn more about the functions of the individual subunits in the chloroplast import complex. Identification of the Arabidopsis homologs for the pea import components has now made it practical to analyze the functions of these proteins genetically, especially through the use of knockout mutants and antisense technology. Such experiments are already being carried out in several laboratories, and three reports have recently emerged from these investigations (Jarvis et al., 1998; Bauer et al., 2000; Gutensohn et al., 2000). The study of knockout mutants and antisense plants for each of the import components should lead to a better understanding of their molecular functions. Cross-complementation studies in knockout mutants will also be useful in determining whether the putative Arabidopsis import complex isoforms are the functional homologs of the corresponding pea proteins, as is predicted. However, it should be noted that since several of these proteins appear to have multiple isoforms within Arabidopsis cells, double and triple mutants may need to be constructed in some cases before component function can be analyzed in detail. Despite this limitation, the genetic study of chloroplast protein import in Arabidopsis should provide a great deal of information concerning this system in the coming years.
MATERIALS AND METHODS
All sequence comparisons were done using the BLASTN, BLASTP, and TBLASTN programs (versions 2.0) available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST; Altschul et al., 1990; Altschul et al., 1997). The weight matrix used was the blosum62 matrix, and no settings were changed from the default. The database searched was the Arabidopsis Database Project, found at The Arabidopsis Information Resource (http://www.Arabidopsis.org/blast), which contains genomic and EST sequences. This database was checked for the final time between October 30, 2000 and November 5, 2000, just before manuscript submission. During manuscript revision, a recheck of the database between January 11, 2001, and January 18, 2001, found no additional homologs.
A sequence was considered a homolog only if the following conditions were met, unless otherwise noted: (a) using the pea (Pisum sativum) sequence as the query, one of the BLAST programs used detected this sequence with an expect value of less than or equal to 0.0001; (b) using the putative Arabidopsis homolog as the query, one of the BLAST programs used detected the pea sequence and other Arabidopsis isoforms with an expect value of less than or equal to 0.0001; (c) the region of similarity between the pea protein and the putative Arabidopsis homolog extended for approximately 50% or more of the sequence lengths; (d) the region of similarity to the pea protein extended beyond common motifs (i.e. nucleotide-binding domains); and (e) the putative Arabidopsis homolog was not already annotated as being more similar to another protein of known function. Levels of identity between different amino acid sequences were calculated with the MegAlign program (Lipman-Pearson algorithm; ktuple = 2, gap penalty = 4, gap length penalty = 12) of the Lasergene software package (DNASTAR, Inc., Madison, WI). Predictions concerning chloroplast targeting were made using the TargetP program (version 1.01), available at http://www.cbs.dtu.dk/services/TargetP (Emanuelsson et al., 2000).
ACKNOWLEDGMENTS
We thank K. Bird, Dr. J. Froehlich, Dr. K. Inoue, and Dr. A. Sanderfoot for their helpful comments on this manuscript.
Footnotes
This work was supported in part by the Division of Energy Biosciences at the U.S. Department of Energy (grants to K.K.), by the Cell Biology Program at the National Science Foundation (to K.K.), and by the Graduate Fellowship Program at the National Science Foundation (to D.J.-C.).
LITERATURE CITED
- Akita M, Nielsen E, Keegstra K. Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J Cell Biol. 1997;136:983–994. doi: 10.1083/jcb.136.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- Bölter B, May T, Soll J. A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide. FEBS Lett. 1998a;441:59–62. doi: 10.1016/s0014-5793(98)01525-7. [DOI] [PubMed] [Google Scholar]
- Bölter B, Soll J, Schulz A, Hinnah S, Wagner R. Origin of a chloroplast protein importer. Proc Natl Acad Sci USA. 1998b;95:15831–15836. doi: 10.1073/pnas.95.26.15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce BD. Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 2000;10:440–447. doi: 10.1016/s0962-8924(00)01833-x. [DOI] [PubMed] [Google Scholar]
- Caliebe A, Grimm R, Kaiser G, Lübeck J, Soll J, Heins L. The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a mononuclear iron-binding protein. EMBO J. 1997;16:7342–7350. doi: 10.1093/emboj/16.24.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Chen X, Schnell DJ. Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol. 2000a;122:813–822. doi: 10.1104/pp.122.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Chen X, Schnell DJ. Mechanism of protein import across the chloroplast envelope. Biochem Soc Trans. 2000b;28:485–491. [PubMed] [Google Scholar]
- Chen X, Schnell DJ. Protein import into chloroplasts. Trends Cell Biol. 1999;9:222–227. doi: 10.1016/s0962-8924(99)01554-8. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Gutensohn M, Schulz B, Nicolay P, Flügge U-I. Functional analysis of the two Arabidopsis homologues of Toc34, a component of the chloroplast protein import apparatus. Plant J. 2000;23:771–783. doi: 10.1046/j.1365-313x.2000.00849.x. [DOI] [PubMed] [Google Scholar]
- Hinnah SC, Hill K, Wagner R, Schlicher T, Soll J. Reconstitution of a chloroplast protein import channel. EMBO J. 1997;16:7351–7360. doi: 10.1093/emboj/16.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Muckel E, Heemeyer F, von Heijne G, Soll J. A receptor component of the chloroplast protein translocation machinery. Science. 1994;266:1989–1992. doi: 10.1126/science.7801125. [DOI] [PubMed] [Google Scholar]
- Ivey RA, Subramanian C, Bruce BD. Identification of a Hsp70 recognition domain within the Rubisco small subunit transit peptide. Plant Physiol. 2000;122:1289–1299. doi: 10.1104/pp.122.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DT, Froehlich JE, Keegstra K. The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J Biol Chem. 1998;273:16583–16588. doi: 10.1074/jbc.273.26.16583. [DOI] [PubMed] [Google Scholar]
- Jarvis P, Chen L-J, Li H-M, Peto CA, Fankhauser C, Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Cline K. Protein import and routing systems of chloroplasts. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K, Froehlich JE. Protein import into chloroplasts. Curr Opin Plant Biol. 1999;2:471–476. doi: 10.1016/s1369-5266(99)00021-7. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Olsen LJ, Theg SM. Chloroplastic precursors and their transport across the envelope membranes. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:471–501. [Google Scholar]
- Kessler F, Blobel G. Interaction of the protein import and folding machineries in the chloroplast. Proc Natl Acad Sci USA. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F, Blobel G, Patel HA, Schnell DJ. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- Ko K, Budd D, Wu C, Seibert F, Kourtz L, Ko ZW. Isolation and characterization of a cDNA clone encoding a member of the Com44/Cim44 envelope components of the chloroplast protein import apparatus. J Biol Chem. 1995;270:28601–28608. doi: 10.1074/jbc.270.48.28601. [DOI] [PubMed] [Google Scholar]
- Komiya T, Rospert S, Koehler C, Looser R, Schatz G, Mihara K. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the “acid chain” hypothesis. EMBO J. 1998;17:3886–3898. doi: 10.1093/emboj/17.14.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Chen X, Fuks B, Schnell DJ. Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A, Schnell DJ. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J Cell Biol. 1997;139:1677–1685. doi: 10.1083/jcb.139.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtz L, Ko K. The early stage of chloroplast protein import involves Com70. J Biol Chem. 1997;272:2808–2813. doi: 10.1074/jbc.272.5.2808. [DOI] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito M-I, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Lübeck J, Soll J, Akita M, Nielsen E, Keegstra K. Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J. 1996;15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Kouranov A, LaSala SE, Schnell DJ. Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J Cell Biol. 1996;134:315–327. doi: 10.1083/jcb.134.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JS, DeRocher AE, Keegstra K, Vierling E. Identification of heat shock protein hsp70 homologues in chloroplasts. Proc Natl Acad Sci USA. 1990;87:374–378. doi: 10.1073/pnas.87.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T, Soll J. Chloroplast precursor protein translocon. FEBS Lett. 1999;452:52–56. doi: 10.1016/s0014-5793(99)00527-x. [DOI] [PubMed] [Google Scholar]
- May T, Soll J. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12:53–63. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K, Schüller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Düsterhöft A, Stiekema W, Entian K-D, Terryn N. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature. 1999;402:769–777. doi: 10.1038/47134. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Ito M, Kiyosue T, Shinozaki K, Watanabe A. Identification of clp genes expressed in senescing Arabidopsis leaves. Plant Cell Physiol. 1999;40:504–514. doi: 10.1093/oxfordjournals.pcp.a029571. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K. Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 1997;16:935–946. doi: 10.1093/emboj/16.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oblong JE, Lamppa GK. Identification of two structurally related proteins involved in proteolytic processing of precursors targeted to the chloroplast. EMBO J. 1992;11:4401–4409. doi: 10.1002/j.1460-2075.1992.tb05540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen LJ, Keegstra K. The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J Biol Chem. 1992;267:433–439. [PubMed] [Google Scholar]
- Olsen LJ, Theg SM, Selman BR, Keegstra K. ATP is required for the binding of precursor proteins to chloroplasts. J Biol Chem. 1989;264:6724–6729. [PubMed] [Google Scholar]
- Perry SE, Keegstra K. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 1994;6:93–105. doi: 10.1105/tpc.6.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Davila-Aponte J, Keegstra K. The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proc Natl Acad Sci USA. 1999;96:784–789. doi: 10.1073/pnas.96.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Keegstra K. The endosymbiotic origin of the protein import machinery of chloroplastic envelope membranes. Trends Plant Sci. 1999;4:302–307. doi: 10.1016/s1360-1385(99)01449-1. [DOI] [PubMed] [Google Scholar]
- Richter S, Lamppa GK. A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc Natl Acad Sci USA. 1998;95:7463–7468. doi: 10.1073/pnas.95.13.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Lamppa GK. Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J Cell Biol. 1999;147:33–43. doi: 10.1083/jcb.147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Soll J. Travelling of proteins through membranes: translocation into chloroplasts. Planta. 2000;211:449–456. doi: 10.1007/s004250000357. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Waegemann K, Soll J. A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 1995;7:401–411. doi: 10.1046/j.1365-313x.1995.7030401.x. [DOI] [PubMed] [Google Scholar]
- Sohrt K, Soll J. Toc64, a new component of the protein translocon of chloroplasts. J Cell Biol. 2000;148:1213–1221. doi: 10.1083/jcb.148.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl T, Glockmann C, Soll J, Heins L. Tic40, a new “old” subunit of the chloroplast protein import translocon. J Biol Chem. 1999;274:37467–37472. doi: 10.1074/jbc.274.52.37467. [DOI] [PubMed] [Google Scholar]
- Theg SM, Bauerle C, Olsen LJ, Selman BR, Keegstra K. Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J Biol Chem. 1989;264:6730–6736. [PubMed] [Google Scholar]
- Tranel PJ, Froehlich J, Goyal A, Keegstra K. Acomponent of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 1995;14:2436–2446. doi: 10.1002/j.1460-2075.1995.tb07241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderVere PS, Bennett TM, Oblong JE, Lamppa GK. A chloroplast processing enzyme involved in precursor maturation shares a zinc-binding motif with a recently recognized family of metalloendopeptidases. Proc Natl Acad Sci USA. 1995;92:7177–7181. doi: 10.1073/pnas.92.16.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K, Soll J. Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1991;1:149–158. [Google Scholar]
- Wu C, Seibert FS, Ko K. Identification of chloroplast envelope proteins in close physical proximity to a partially translocated chimeric precursor protein. J Biol Chem. 1994;269:32264–32271. [PubMed] [Google Scholar]