Abstract
The pandemic caused by the new coronavirus SARS-CoV-2 has made evident the need for broad-spectrum, efficient antiviral treatments to combat emerging and re-emerging viruses. Plitidepsin is an antitumor agent of marine origin that has also shown a potent pre-clinical efficacy against SARS-CoV-2. Plitidepsin targets the host protein eEF1A (eukaryotic translation elongation factor 1 alpha) and affects viral infection at an early, post-entry step. Because electron microscopy is a valuable tool to study virus-cell interactions and the mechanism of action of antiviral drugs, in this work we have used transmission electron microscopy (TEM) to evaluate the effects of plitidepsin in SARS-CoV-2 infection in cultured Vero E6 cells 24 and 48h post-infection. In the absence of plitidepsin, TEM morphological analysis showed double-membrane vesicles (DMVs), organelles that support coronavirus genome replication, single-membrane vesicles with viral particles, large vacuoles with groups of viruses and numerous extracellular virions attached to the plasma membrane. When treated with plitidepsin, no viral structures were found in SARS-CoV-2-infected Vero E6 cells. Immunogold detection of SARS-CoV-2 nucleocapsid (N) protein and double-stranded RNA (dsRNA) provided clear signals in cells infected in the absence of plitidepsin, but complete absence in cells infected and treated with plitidepsin. The present study shows that plitidepsin blocks the biogenesis of viral replication organelles and the morphogenesis of virus progeny. Electron microscopy morphological analysis coupled to immunogold labeling of SARS-CoV-2 products offers a unique approach to understand how antivirals such as plitidepsin work.
Keywords: Plitidepsin, SARS-CoV-2, Antiviral, Covid-19, Electron microscopy
1. Introduction
Infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) urgently needs effective antiviral treatments with a significant clinical benefit for patients. An increasing number of randomized clinical trials are starting to identify potent antivirals targeting the virus. Indeed, remdesivir, molnupiravir and nirmatrelvir have recently shown clinical benefits when administered early upon infection (Beigel et al., 2020; Garibaldi et al., 2021; Grein et al., 2020; Imran et al., 2021; Owen et al., 2021). However, coronaviruses have a reduced number of molecular druggable targets, and as new variants arise, these targets evolve and could eventually develop antiviral resistance. Monoclonal antibodies such as casirivimab-imdevimab have shown activity in clinical trials before the surge of omicron variant, but pre-clinical evidence suggest now that this combination lacks neutralization activity against omicron (Agarwal et al., 2020). Although Sotrovimab may retain activity, higher concentrations are needed for neutralization of omicron (Agarwal et al., 2020).
An interesting approach to overcome these limitations relies on the use of compounds against highly conserved cellular host factors required to complete the replication cycle of distinct types of viruses, which offer a common targeted solution to diverse viral threats. Furthermore, targeting host factors could offer a pan-antiviral strategy to combat not only viruses known at present, but also future pandemics to come (Baggen et al., 2021). Presently, there are only a limited number of approved drugs involved in targeting host factors at post-entry steps (Baggen et al., 2021). This approach is especially relevant for pan-antiviral solutions given that viruses may use alternative pathways to enter a cell, but will most likely converge at intracellular processes involving genome replication and protein production. One of these compounds is plitidepsin, which has shown a potent preclinical efficacy by targeting the host protein eEF1A (Losada et al., 2016; Rodon et al., 2021; White et al., 2021). In 2018, the Therapeutic Goods Administration (TGA; Australian Government) approved the combination of plitidepsin with dexamethasone for the treatment of patients with relapsed/refractory multiple myeloma (https://www.tga.gov.au/auspar-plitidepsin). The results of randomized phase I studies of plitidepsin in adults hospitalized with Covid-19 have been recently published (Varona et al., 2022) and plitidepsin has been successfully used as a rescue treatment for prolonged viral SARS-CoV-2 replication in a patient with chronic lymphocytic leukemia (Guisado-Vasco et al., 2022). Currently, plitidepsin is being evaluated in a phase 3, multicenter, randomized, controlled trial to determine the efficacy and safety of two dose levels of plitidepsin versus control in adult patients requiring hospitalization for management of moderate coronavirus infectious disease 2019 (COVID-19) (ClinicalTrials.gov Identifier: NCT04784559). eEF1A is necessary to transport aminoacyl-tRNAs to the A site of the ribosome during protein translation, but is also implicated in other activities (Mateyak and Kinzy, 2010) such as inhibition of apoptosis (Sun et al., 2014), proteasome degradation (Hotokezaka et al., 2002), and actin bundling and cytoskeleton reorganization (Edmonds et al., 1998) among other non-canonical functions. Also, eEF1A is implicated in the replication of distinct viruses, including coronaviruses (Zhang et al., 2014), and was identified as a potential SARS-CoV-2 interacting protein in one of the first screenings performed to identify novel targets (Gordon et al., 2020).
Upon SARS-CoV-2 infection, plitidepsin inhibits nucleocapsid viral protein expression and viral induced cytopathic effect in vitro (Rodon et al., 2021; White et al., 2021). In addition, it also reduces genomic and subgenomic RNA expression (White et al., 2021). In contrast to Remdesivir, the in vitro inhibitory activity of plitidepsin is enhanced when the drug is added several hours post-infection (White et al., 2021). Current models of SARS-CoV-2 replication propose that upon viral fusion, non-structural viral proteins form a replication–transcription complex that is continuous with the ER and has a double membrane vesicle (DMV) morphology that shelters the viral genome replication (Baggen et al., 2021; Wolff et al., 2020a). A negative RNA strand is used as a template for the generation of positive strands that are translated and incorporated into nascent viruses (Baggen et al., 2021; Wolff et al., 2020a). Discontinued transcription of positive RNA strands produce negative subgenomic RNAs, which are then used as templates for positive subgenomic RNA generation that codify for structural and accessory proteins (Baggen et al., 2021; Wolff et al., 2020a). Translation of viral proteins is facilitated by mRNA export via molecular pores located in the DMV that enable viral protein production in the cytoplasm (Wolff et al., 2020b). Yet, how plitidepsin exerts its intracellular antiviral activity and influences the formation of viral replication DMV remains unknown.
Here we aimed to explore the antiviral effect of plitidepsin at the cellular level to understand its impact on SARS-CoV-2 replication and DMV formation. Using transmission electron microscopy (TEM), we recapitulated the infectious SARS-CoV-2 cycle in Vero E6 cells and observed a lack of viral DMVs in plitidepsin-treated cells. Complementary immunohistochemistry analyses using nucleocapsid and dsRNA immunogold labeling unambiguously confirmed the lack of viral replication in plitidepsin treated cells.
2. Material & methods
Biosafety Approval. The biologic biosafety committee of the Research Institute Germans Trias i Pujol approved the execution of SARS-CoV-2 experiments at the BSL3 laboratory of the Center for Bioimaging and Comparative Medicine, CMCiB (CSB-20-015-M3).
Materials. Plitidepsin was synthesized at PharmaMar, S.A. (Colmenar Viejo, Madrid, Spain).
Cell culture, viral isolation and titration. Vero E6 cells (ATCC CRL-1586) were cultured in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen), 100 U/mL penicillin, 100 μg/mL streptomycin (all from Invitrogen).
SARS-CoV-2 D614G was isolated from a nasopharyngeal swab collected in March 2020 in Spain in Vero E6 cells as previously described in detail (Rodon et al., 2021). The virus was propagated for two passages and a virus stock was prepared collecting the supernatant from Vero E6 cell and sequenced as described elsewhere (Rodon et al., 2021). Genomic sequence was deposited at GISAID repository (http://gisaid.org) with accession ID EPI_ISL_510689. Viral stock was titrated in 10-fold serial dilutions on Vero E6 cells to calculate the TCID50 per mL.
The following SARS-CoV-2 variants with deposited genomic sequence at the GISAID repository were also tested: Delta or B.1.617.2 (originally detected in India; EPI_ISL_3342900) isolated in Spain in February 2021, and Omicron or B.1.1.529 isolated in Spain in December 2021 (originally detected in South Africa; EPI_ISL_8151031). Genomic sequencing was performed from viral supernatant by using standard ARTIC v3 or v4 based protocols followed by Illumina sequencing (dx.doi.org/10.17504/protocols.io.bhjgj4jw). Viral stocks were obtained as previously described and titrated in 1/3 serial dilutions on Vero E6 cells to achieve a 50% of cytopathic effect three days post infection as described in the antiviral activity section.
Antiviral Activity. Increasing concentrations of plitidepsin were added to Vero E6 cells and SARS-CoV-2 viruses were added subsequently as described in (Rodon et al., 2021). Assay was set up to achieve a 50% of cytopathic effect three days post infection. Cytopathic or cytotoxic effects of viruses or drugs were measured using the CellTiter-Glo luminescent cell viability assay (Promega). Luminescence was measured in a Fluoroskan Ascent FL luminometer (ThermoFisher Scientific). Response curves were adjusted to a non-linear fit regression model, calculated with a four-parameter logistic curve with variable slope to calculate the IC50. Cells not exposed to the virus were used as negative controls of infection, and were set as 100% of viability to normalize data and calculate the percentage of cytopathic effect. Analyses were generated with the GraphPad Prism v9 Software (GraphPad Software, San Diego,CA, USA).
Cellular infection. Vero E6 cells were infected with SARS-CoV-2 at a multiplicity of infection MOI of 0.02 for 24 and 48 hours (h) in the presence or absence of 0.2 and 0.05 μM of plitidepsin added at the time of infection. These two concentrations are close to the IC90 and IC50 of plitidepsin, respectively, determined by the SARS-CoV-2 induced cytopathic effect on Vero E6 cells (Rodon et al., 2021). As negative controls, Vero E6 cells were treated with or without plitidepsin for the same time but without virus. At each studied time point, cell monolayers were chemically fixed using two different protocols depending on the type of analysis (morphology or immunohistochemistry).
Morphology Analysis. Cell monolayers were fixed with 4% paraformaldehyde and 1% glutaraldehyde in phosphate buffered saline (PBS) for 2h at room temperature (RT). Cells were removed from the plates in the fixative, pelleted by centrifugation and washed three times with PBS. Post-fixation of cell pellets was done on ice with 1% osmiumtetroxide +0.8% potassium ferrocyanide in water. Afterwards the pellets were dehydrated on ice with increasing concentrations of acetone and processed for embedding in the epoxy resin EML-812 (Taab Laboratories), as previously described (Tenorio et al., 2018). Sections of cells in epoxy resins have high contrast and optimal morphological details. Infiltration with epoxy resin was performed at RT. All samples were polymerized at 60 °C for 48h. Ultrathin sections (50–70 nm) were cut with a Leica UC6 microtome and placed on uncoated 300 mesh copper grids. Sections were contrasted with 4% uranyl acetate and Reynold's lead citrate. Images were taken with a Tecnai G2 TEM operated at 120 kV with a Ceta camera or with a Jeol 1400 operated at 120 kV with a Gatan Rio camera. At least 50 cells per condition were studied by TEM.
Immunohistochemistry. Cells were fixed with 4% paraformaldehyde and 0.1% glutaraldehyde in PBS for 1h at RT and removed from the plates in the fixative, pelleted by centrifugation and washed three times with PBS. Cell pellets were incubated 30 min with 2% uranyl acetate in water at RT, gradually dehydrated on ice with ethanol and infiltrated at RT with LR-White acrylic resin as previously described (de Castro Martin et al., 2017). Sections of cells in acrylic resins have low contrast but an optimal preservation of protein epitopes, which is fundamental for immunolabeling studies. After polymerization at 60 °C for 48h, the samples were sectioned using a Leica UC6 ultramicrotome. Ultrathin sections were collected on copper grids with a carbon-coated Formvar film. For immunogold labeling, unspecific binding on sections was blocked by incubation with Tris-buffered gelatin (TBG) for 5 min. N protein was labeled with the rabbit anti-SARS-CoV-2 nucleocapsid antibody (GXT 135357 GeneTex) diluted 1/50 in TBG for 1h. Double stranded RNA (dsRNA), an intermediate of viral replication, was labeled with the mouse monoclonal anti-dsRNA J2 antibody (10010200, English and Scientific Consulting Kft, SCICONS) diluted 1/30 in TBG for 1h. After three washes with PBS and incubation with TBG for 5 min, grids were labeled with a secondary antibody, corresponding to the species of the first antibody, conjugated with 10-nm colloidal gold particles (BB international) in TBG for 30 min. Sections were washed three times with PBS and five times with milliQ water and stained with saturated uranyl acetate for 20 min. After washing three times with milliQ water, grids were allowed to dry at RT. Images were taken with a Jeol 1400 TEM operated at 80 kV with a Gatan One view camera or with a Jeol 1011 TEM operated at 100 kV with a Gatan ES1000W camera. At least 50 cells per condition were studied by TEM.
3. Results
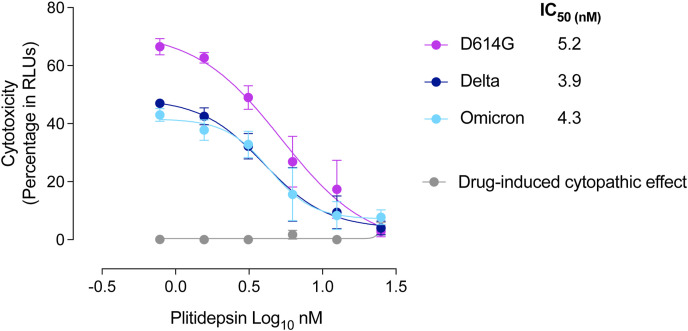
To understand how plitidepsin exerts its mechanism of action, we studied how plitidepsin interferes with viral replication of three SARS-CoV-2 variants (D614G, Delta and Omicron) and observed a dose response reduction of the viral-induced cytopathic effect three days post-infection on Vero E6 cells (Fig. 1 ).
Fig. 1.
Viral induced cytopathic effect on Vero E6 cells exposed to a fixed concentration of different SARS-CoV-2 VOC (D614G, Delta and Omicron variants) in the presence of increasing concentrations of plitidepsin. Non-linear fit to a variable response curve showing mean values and S.E.M from two representative experiments with four replicates are shown (red lines), excluding data from drug concentrations with associated toxicity. The IC50 value for each VOC is indicated. Cytotoxic effect on Vero E6 cells exposed to increasing concentrations of plitidepsin in the absence of virus is also shown (grey lines) in relative light units (RLUs).
We then followed SARS-CoV-2 D614G variant replication cycle using electron microscopy.
We found no replication organelles nor viral particles in cells infected for 24h at an MOI of 0.02, although compared to control cells, the viral infection induced some stress in cells, which showed mitochondria with swollen cristae and Golgi with swollen cisternae (not shown).
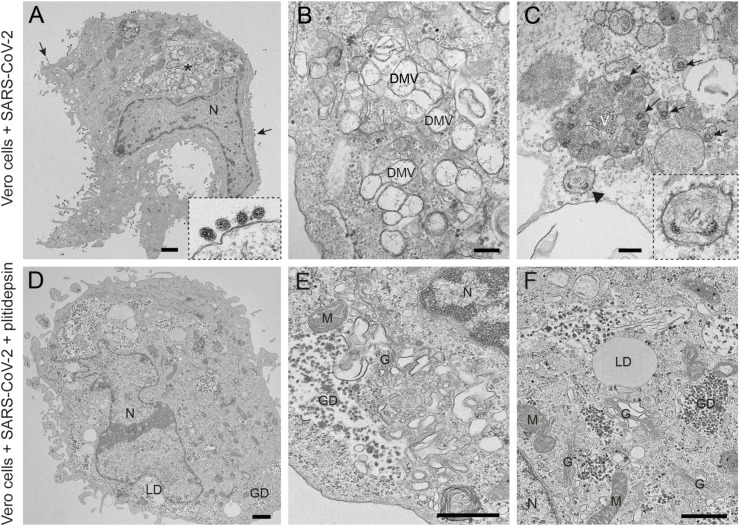
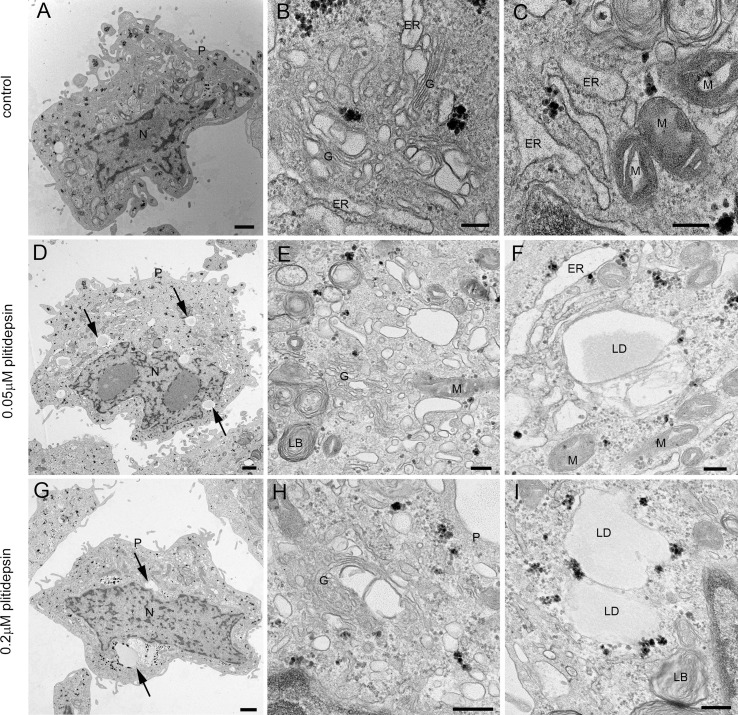
In Vero E6 cells infected for 48h at an MOI of 0.02 with the D614G variant, clusters of characteristic DMVs, which are the replication organelles of SARS-CoV-2 (Ogando et al., 2020), were present in the cytoplasm and occupied a significant area of the cytosol (Fig. 2 A and B). DMVs often had an electron lucent content with fibrillar material (Fig. 2B). Intracellular viral particles and extracellular virions attached to the plasma membrane were abundant (Fig. 2A and C). On average, in ultrathin sections of infected cells we found 30 intracellular viral particles and 73 extracellular particles attached to the plasma membrane per cell. Single or very few viral particles were present inside small vesicles with an electron lucent lumen while larger vacuoles with numerous viral particles were often observed (Fig. 2C). These vacuoles that contained internal membranes and/or amorphous electron dense material might represent late endosomes/lysosomes. We seldom observed budding of viral particles into the endoplasmic reticulum (ER) (Fig. 2C).
Fig. 2.
Transmission electron microscopy of Vero E6 cells infected with SARS-CoV-2 and effects of plitidepsin. (A) to (C) ultrathin sections of cells infected at an MOI of 0.02 and 48 h post-inoculation. (A) Cell containing a viral replication organelle (asterisk) made of a collection of double-membrane vesicles (DMVs) near the nucleus (N). Numerous viral particles are seen on the cell surface (arrows and inset in A). (B) DMVs have a round shape and fibrillar content. (C) Individual intracellular viral particles (arrows) are seen inside single-membrane vesicles and a large group of viruses inside a vacuole (V). Budding of viral particles in RER membranes (arrowhead and inset). (D) to (F) Ultrathin sections of Vero E6 cells infected with SARS-CoV-2 and treated with 0.2 μM plitidepsin. Low (D) and high (E and F) magnification views show that these cells contain lipid droplets (LD), large glycogen deposits (GD) and an altered Golgi complex (G) but no viral structures. M, mitochondrion. Scale bars, 1 μm in A and D; 200 nm in B, C, E and F.
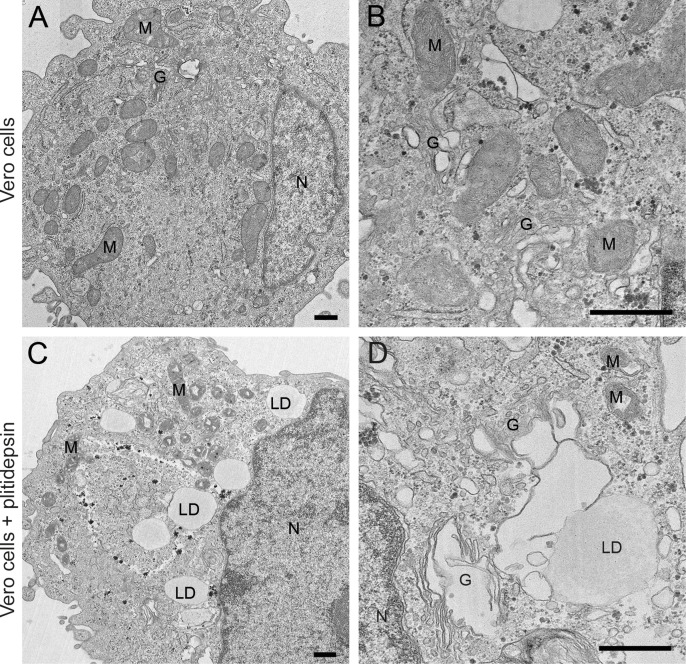
When 0.2 μM of plitidepsin was added to Vero E6 cells, the drug had a profound impact on viral replication 48h later (Fig. 2D–F). We did not observe any DMVs in the cytoplasm, nor viral particles inside cells or at the plasma membrane (Fig. 2D). We found clusters of potential single membrane vesicles present in the cytoplasm, which are formed in early stages of SARS-CoV-2 infection (Eymieux et al., 2021a), that could however also reflect parts of a swollen Golgi (Fig. 2E). Lipid droplets (LDs) and glycogen granules were abundant (Fig. 2F), something also observed in mock-infected cells treated with 0.2 μM of plitidepsin (Fig. S1). This effect was observed in cells incubated with a lower concentration of plitidepsin, that is 0.05 μM, only less pronounced (Fig. S2).
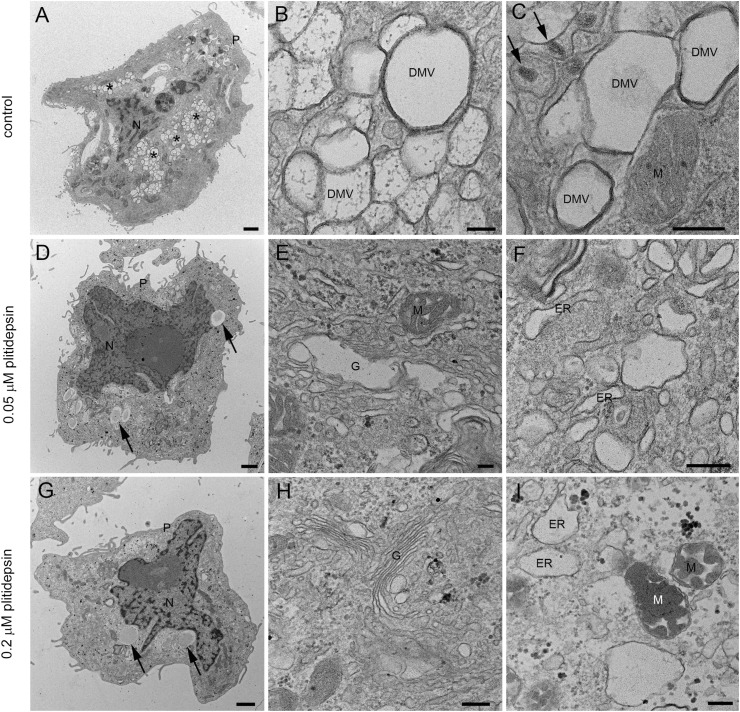
We then studied in detail the effects of 0.05 or 0.2 μM of plitidepsin on the assembly of SARS-CoV-2 replication organelles or DMVs. Of note, these two concentrations are close to the IC50 and IC90 estimated for SARS-CoV-2 induced cytopathic effect on Vero E6 cells (Rodon et al., 2021). When plitidepsin was added to Vero E6 cells at the same time of viral infection (MOI of 0.02 for 48h), both concentrations had a deep impact on the biogenesis of DMVs (Fig. 3 ). Cells infected without plitidepsin contained numerous, typical DMVs with fibrillar content surrounded by viral particles inside single-membrane vesicles (Fig. 3A–C) but when the drug was added no DMVs were found (Fig. 3D–I). Already at the low concentration, no DMVs were found in the cytosol (Fig. 3D–F). Single membrane vesicles were present but they did not contain fibrillar content in their lumen as opposed to infected cells, and this could reflect a part of a swollen Golgi or ER (Fig. 3F and I). These morphological analyses on plitidepsin-treated cells failed to identify any structures implicated in SARS-CoV-2 replication that were unambiguously detected in untreated infected cells (Fig. 2A–C and 3A-C).
Fig. 3.
Transmission electron microscopy of Vero E6 cells infected with SARS-CoV-2 in the absence or presence of two doses of plitidepsin. (A) to (C) Cells infected in the absence of plitidepsin. Low (A) and high (B and C) magnification of ultrathin sections of cells infected 48 h at an MOI of 0.02. (A) Groups of characteristic double membrane vesicles (DMVs; asterisks) occupy large areas of infected cells. (B) Typical DMVs exhibit electron dense membranes and sometimes a fibrillar content. (C) Single membrane vesicles with viruses (arrows) are often found in the close vicinity of DMVs. (D) to (F) Low (D) and high (E) and (F) magnification of Vero E6 cells infected with SARS-CoV-2 48 h at an MOI of 0.02 and treated with 0.05 μM plitidepsin. Cells contain lipid droplets (arrows in D), swollen Golgi stacks (G), mitochondria with swollen cristae (M) and swollen endoplasmic reticulum (ER) cisternae. No viral structures are seen. (G) to (I) Cells infected with SARS-CoV-2 48 h at an MOI of 0.02 and treated with 0.2 μM plitidepsin. Cells contain lipid droplets (arrows in G), swollen ER and altered mitochondria (I). N, nucleus; P, plasma membrane. Scale bars, 1 μm in A, D and G; 200 nm in B, C, E, F, H and I.
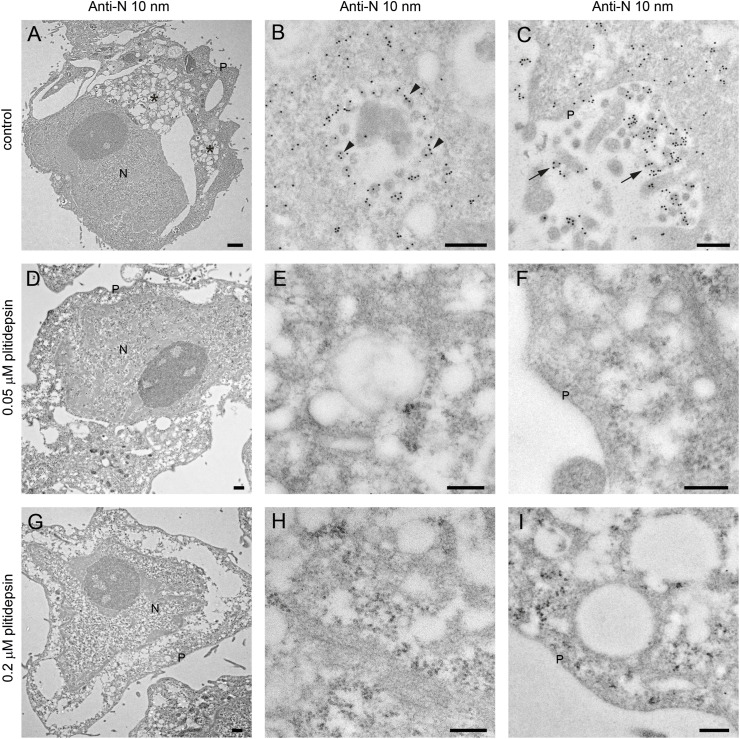
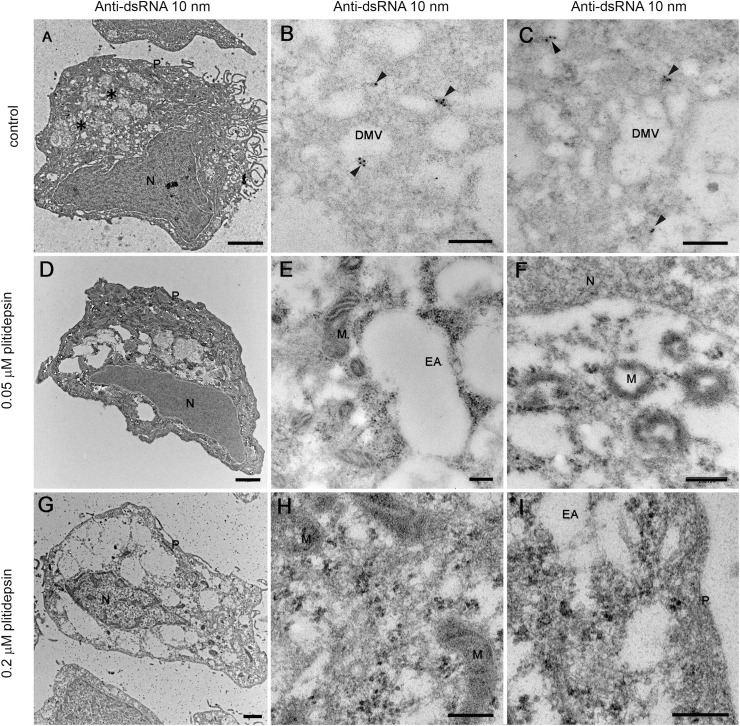
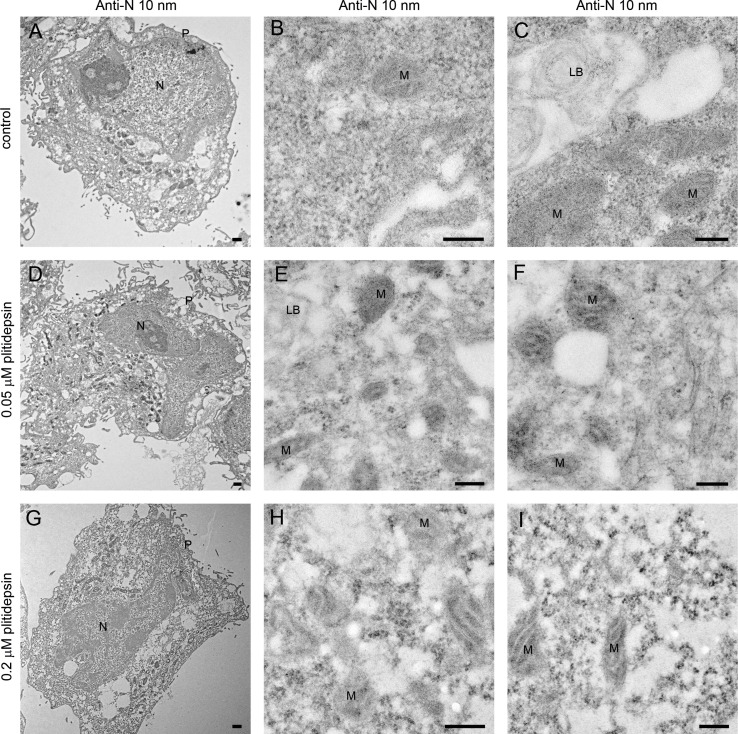
The absence of structures implied in viral replication was confirmed by immunodetection of two key products of SARS-CoV-2: the nucleocapsid and double stranded RNA (dsRNA). The nucleocapsid participates in replication and translation of viral RNA and maintains the structure of ribonucleoprotein complex, whereas the dsRNA is synthesized by the viral polymerase in the host cell, once viral replication has started in DMVs. For both targets, no signal by immunogold labeling was detected in non-infected cells, either non treated with plitidepsin or incubated with low or high concentration of plitidepsin (Figs. S3 and S4). However, virus-infected cells showed a strong labeling for the nucleocapsid in the cytosol and in viral particles that accumulated intra- as well as extracellularly (Fig. 4 A–C). Coherent with the morphological observations, after application of plitidepsin at 0.05 μM, no signal was detected by antibody labelling for nucleocapsid (Fig. 4D–F). The absence of labelling was also found for the high concentration of plitidepsin (0.2 μM) (Fig. 4G–I). In SARS-CoV-2 infected cells without plitidepsin, the labeling for dsRNA was restricted to the lumen of large electron lucent vesicles, which represent the DMVs (Fig. 5 A–C). Due to the low contrast of ultrathin sections of cells embedded in the LR-White acrylic resin, the fibrillar content in the lumen of DMVs is not visible in these samples but the labeling of dsRNA marked them unequivocally as DMVs. After application of plitidepsin at 0.05 or 0.2 μM, no signal for dsRNA was detected by immunogold labelling (Fig. 5D–I). Overall, immunodetection of viral proteins and genetic material confirmed that the morphological analysis performed corresponded to viral replication structures and that plitidepsin completely blocked the assembly of the viral replication organelles or DMVs and the accumulation or viral nucleocapsid protein and dsRNA. Taken together these results provide evidence that plitidepsin inhibits intracellular viral replication and abrogates the formation of DMVs.
Fig. 4.
Immunogold detection of SARS-CoV-2 nucleocapsid protein in infected cells in the absence or presence of two doses of plitidepsin. Ultrathin sections of cells were incubated with anti-N primary antibody followed by a secondary antibody conjugated with 10 nm colloidal gold particles and visualized by TEM. (A) to (C) Ultrathin sections of cells infected for 48 h with SARS-CoV-2 at an MOI of 0.02. (A) Low magnification image of an infected cell with characteristic DMVs (asterisks) near the nucleus (N). (B) High magnification image of a group of intracellular viruses (arrowheads) inside a vacuole. Viral particles and areas of cytosol are labeled. (C) Labeled extracellular virions (arrows) on the cell surface. (D) to (F) Cells infected 48 h with SARS-CoV-2 at an MOI of 0.02 and treated with 0.05 μM plitidepsin. Low (D) and high (E) and (F) magnification images of cells show no labeling. (G) to (I) Cells infected 48 h with SARS-CoV-2 at an MOI of 0.02 and treated with 0.2 μM plitidepsin. Low (G) and high (H) and (I) magnification images of cells show no labeling. P, plasma membrane. Scale bars, 0.5 μm in A, D and G; 200 nm in B, C, E, F, H and I.
Fig. 5.
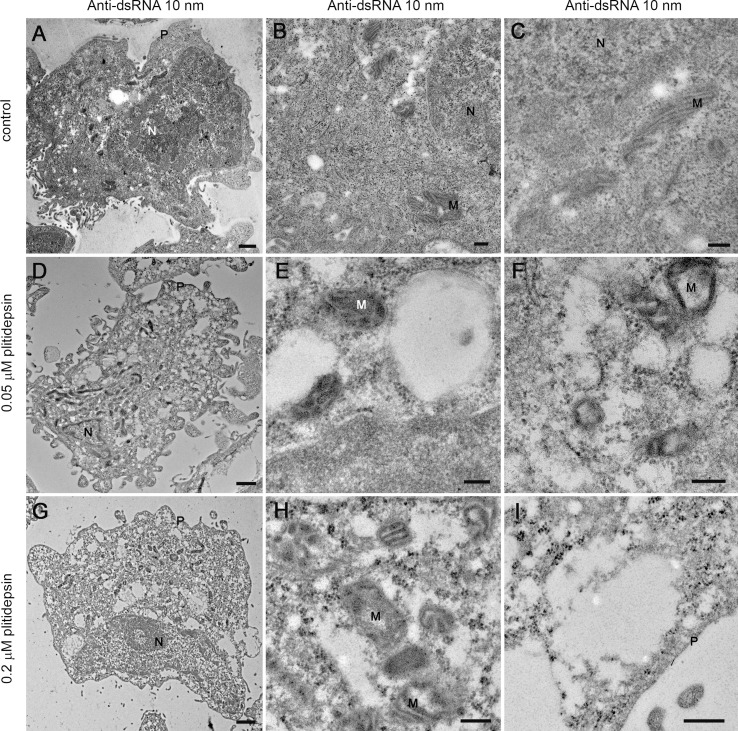
Immunogold labelling of SARS-CoV-2 dsRNA in infected cells in the absence or presence of two doses of plitidepsin. Ultrathin sections of cells were incubated with anti-dsRNA primary antibody followed by a secondary antibody conjugated with 10 nm colloidal gold particles and visualized by TEM. (A) to (C) Ultrathin sections of cells infected 48 h with SARS-CoV-2 at an MOI of 0.02. (A) Low magnification image of an infected cell with characteristic DMVs (asterisks) near the nucleus (N). (B) and (C) High magnification images of labeled DMVs (arrowheads). (D) to (F) Cells infected 48 h with SARS-CoV-2 at an MOI of 0.02 and treated with 0.05 μM plitidepsin. Low (D) and high (E) and (F) magnification images of cells show no labeling. (G) to (I) Cells infected 48 h with SARS-CoV-2 at an MOI of 0.02 and treated with 0.2 μM plitidepsin. Low (G) and high (H) and (I) magnification images of cells show no labeling. N, nucleus; EA, empty areas; M, mitochondrion. Scale bars, 1 μm in A, D and G; 200 nm in B, C, E, F, H and I.
4. Discussion
The TEM analysis of SARS-CoV-2-infected Vero E6 cells performed herein reveals the formation of DMV structures characteristic of the viral replication organelle, and the accumulation of intracellular viral particles in small vesicles and large vacuoles as well as extracellular virions at the plasma membrane. These structures have been described before as characteristic features of SARS-CoV-2 infection in cell culture (Eymieux et al., 2021a, 2021b; Ogando et al., 2020). In this study, infected cells displayed a clear specific labelling for both the nucleocapsid viral protein and the dsRNA viral replication intermediate. Specific labelling of two key viral products is highly relevant in a context in which TEM analysis of SARS-CoV-2-infected samples has led to the misidentification of viral particles and to ambiguous results (Dittmayer et al., 2020; Miller and Goldsmith, 2020). Together, the morphological analysis along with the immunogold detection of two complementary SARS-CoV-2 products performed herein provided conclusive evidence that the subcellular structures detected in infected cells represented viral replication organelles and virus progeny. In the present set of experiments, plitidepsin abrogated the formation of DMVs and the detection of nucleocapsid and dsRNA viral products in SARS-CoV-2-infected Vero E6 cells. Since plitidepsin interferes with the host factor eEF1A implicated in RNA translation (Losada et al., 2016), it is worthwhile to know the impact of this mechanism on the formation of DMVs.
The antiviral effect of plitidepsin against SARS-CoV-2 is mediated throughout inhibition of the host protein eEF1A. siRNA silencing of eEFA1A in host cells induces a significant reduction in the nucleocapsid protein levels, as well as a reduction in the viral RNA, which demonstrates a direct involvement of eEF1A in the viral replication (Zhang et al., 2014). In addition, the exposure of plitidepsin to cells at the moment of SARS-CoV-2 infection reduced the viral replication and transcription, which induced a reduction in the accumulation of genomic RNA and sub-genomic N RNA viral levels, respectively (White et al., 2021). Due to the known high sensitivity of coronavirus to translation inhibitors (Bojkova et al., 2020; van den Worm et al., 2011), especially at an early stage of infection, a plitidepsin-mediated inhibition of SARS-CoV-2 protein translation process could represent one possible mechanism, which could lead to the abrogation of DMV formation. The results gathered from the present experiments demonstrate that a concentration as low as 50 nM of plitidepsin, completely abolished the formation of DMVs, and the synthesis of N protein and viral dsRNA in SARS-CoV-2 infected Vero E6 cells both at 24 and 48 h post-inoculation. The plitidepsin-mediated anti-SARS-CoV-2 activity at such a low concentration may be the result of a highly specific inhibition of genomic RNA viral translation. This specific antiviral effect is explained by the lack of plitidepsin-mediated protein synthesis inhibition in eukaryotic cells at high concentrations (e.g., 450 nM or higher), far above those used herein (Losada et al., 2016). Inhibition of the viral protein translation may affect several essential proteins, such as non-structural proteins, whose predicted transmembrane domains are key for DMV formation during the replication of distinct coronaviruses (Angelini et al., 2013; Oudshoorn et al., 2017; Wolff et al., 2020a). It is tempting to speculate that the lack of translation of these viral proteins could directly abrogate the formation of these replication–transcription complexes necessary for DMV formation. This possible mechanism is supported by prior observations where plitidepsin treatment decreased nucleocapsid content and reduced subgenomic RNA detection (Rodon et al., 2021; White et al., 2021). An alternative mechanism has been recently proposed with eEF1A binding to membranes and thereby recruiting a subset of proteins related to DMV formation (Carriles et al., 2021). It is worth highlighting that the activity of plitidepsin against SARS-CoV-2 is mediated by targeting the host factor eEF1A rather than by exerting a direct action against a viral protein. In contrast to those monoclonal antibodies that have lost activity with the surge of omicron variant (Agarwal et al., 2020), the mechanism of action of plitidepsin remains active against omicron as we have demonstrated herein, and in agreement with a recent publication (Varona et al., 2022). Plitidipsin is therefore a one of a kind compound that exerts a potent antiviral activity against different SARS-CoV-2 variants by targeting a cellular host factor.
In the present study, TEM morphological analysis coupled to immunogold labeling of SARS-CoV-2 products has demonstrated to be a valuable tool to better understand the antiviral effects, as previously reported for other viruses (García-Serradilla and Risco, 2021; Sachse et al., 2019). Remarkably, the plitidepsin-induced complete blockade of the assembly of viral structures detected here by electron microscopy is rather unique. In the presence of non-toxic, high concentrations of other antiviral drugs, SARS-CoV-2 often assembles small amounts of DMVs and viral particles (Izquierdo-Useros, Cerón and Risco, unpublished results). The generated insights warrant new experiments that aim to better understand the mechanism of plitidepsin-induced antiviral activity. This knowledge will be crucial to identify the mechanism of action for promising compounds that interfere with host factors whose implication in key biological processes can be applied as a pan-antiviral strategies.
Financial support
This research was funded by Pharma Mar, which commercializes Aplidin/Plitidepsin. The authors also acknowledge the crowdfunding initiative #Yomecorono (https://www.yomecorono.com) and the CBIG consortium supported by Grifols. N.I–U. is supported by grant PID2020-117145RB-I00 from the Spanish Ministry of Science and Innovation. C.R. is supported by grant RTI2018-094445-B-I00 (MCI/AEI/FEDER, UE) from the Spanish Ministry of Science and Innovation.
Data availability
Data related to this work is available from corresponding authors upon reasonable request.
Declaration of competing interest
P.A., J.V-A and N.I–U. are inventors in a patent application related to Aplidin/Plitidepsin (EP20382821.5). Unrelated to the submitted work, N.I–U. reports institutional grants from Grifols, Dentaid, Hipra and Palobiofarma. The authors declare that no other competing financial interests exist. A.L., P.A. and C.C. are PharmaMar S.A. employees or PharmaMar S.A. shareholders or both.
Acknowledgements
We acknowledge J. Díaz Pedroza from the CMCiB for his constant help at the BSL3 facility, I. Blanco, E. Martro, and V. Saludes from the Microbiology department of HUGTIP for providing key samples and outstanding help to sequence all VOC, and C. Andres, A. Antón and T. Pumarola from Respiratory Virus Unit, Microbiology Department, Vall d’Hebron Institut de Recerca (VHIR), Vall d’Hebron Hospital Universitari for providing key samples. We are grateful to M. Noguera and M. Parera from IrsiCaixa for their VOC sequencing support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105270.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
Effects of plitidepsin on the morphology of mock-infected Vero E6 cells. (A) and (B) Low and high magnification images of control Vero E6 cells with normal nucleus (N), Golgi complex (G) and mitochondria (M). (C) and (D) Low and high magnification images, respectively, of Vero E6 cells treated 48 h with 0.2 μM plitidepsin. Cells show lipid droplets (LD), altered mitochondria (M), glycogen deposits (GD) and swollen Golgi (G). Scale bars, 500 nm.
figs2.
Dose-dependent effects of plitidepsin in mock-infected Vero E6 cells. (A) to (C) control cells. Low (A) and high (B) and (D) magnification images, respectively, of untreated cells. (D) to (F) Vero E6 cells incubated 48 h with 0.05 μM plitidepsin. Low (D) and high (E) and (F) magnification images, respectively. Arrows in (D) point to lipid droplets. (G) to (I) Vero E6 cells incubated 48 h with 0.2 μM plitidepsin. Low (G) and high (H) and (I) magnification images, respectively. N, nucleus; P, plasma membrane; G, Golgi complex; ER, endoplasmic reticulum; LB, lamellar body; GD, glycogen deposits; LD, lipid droplet. Scale bars, 1 μm in A, D and G; 200 nm in B, C, E, F, H and I.
figs3.
Immunogold cytochemical control of anti-nucleocapsid (N) antibody in non-infected Vero E6 cells, with and without plitidepsin. Ultrathin sections of cells were incubated with anti-N primary antibody followed by a secondary antibody conjugated with 10 nm colloidal gold particles and visualized by TEM. (A) to (C) control, non-infected Vero E6 cells in the absence of plitidepsin. Low (A) and high (B) and (C) maginification images. (D) to (F) Vero E6 cells treated 48 h with 0.05 μM plitidepsin. Low (D) and high (E) and (F) magnification images. (G) to (I) Vero E6 cells treated 48 h with 0.2 μM plitidepsin. Low (G) and high (H) and (I) magnification images. Cells present no unspecific background. N, nucleus; P, plasma membrane; M, mitochondrion; LB, lamellar body. Scale bars, 0.5 μm in A, D and G; 200 nm in B, C, E, F, H and I.
figs4.
Immunogold cytochemical control of anti-dsRNA antibody in non-infected Vero E6 cells, with and without plitidepsin. Ultrathin sections of cells were incubated with anti-dsRNA primary antibody followed by a secondary antibody conjugated with 10 nm colloidal gold particles and visualized by TEM. Low (A) and high (B) and (C) magnification images. (D) to (F) Vero E6 cells treated 48 h with 0.05 μM plitidepsin. Low (D) and high (E) and (F) magnification images. (G) to (I) Vero E6 cells treated 48 h with 0.2 μM plitidepsin. Low (G) and high (H) and (I) magnification images. Cells present no unspecific background. N, nucleus; P, plasma membrane; M, mitochondrion. Scale bars, 1 μm in A, D and G; 200 nm in B, C, E, F, H and I.
References
- Agarwal A., Rochwerg B., Lamontagne F., Siemieniuk R.A., Agoritsas T., Askie L., Lytvyn L., Leo Y.-S., Macdonald H., Zeng L., Amin W., Barragan F.A., Bausch F.J., Burhan E., Calfee C.S., Cecconi M., Chanda D., Dat V.Q., De Sutter A., Du B., Geduld H., Gee P., Harley N., Hashmi M., Hunt B., Jehan F., Kabra S.K., Kanda S., Kim Y.-J., Kissoon N., Krishna S., Kuppalli K., Kwizera A., Lisboa T., Mahaka I., Manai H., Mino G., Nsutebu E., Preller J., Pshenichnaya N., Qadir N., Sabzwari S., Sarin R., Shankar-Hari M., Sharland M., Shen Y., Ranganathan S.S., Souza J.P., Stegemann M., Swanstrom R., Ugarte S., Venkatapuram S., Vuyiseka D., Wijewickrama A., Maguire B., Zeraatkar D., Bartoszko J.J., Ge L., Brignardello-Petersen R., Owen A., Guyatt G., Diaz J., Kawano-Dourado L., Jacobs M., Vandvik P.O. A living WHO guideline on drugs for covid-19. BMJ. 2020;m3379 doi: 10.1136/bmj.m3379. [DOI] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. e00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggen J., Vanstreels E., Jansen S., Daelemans D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021 doi: 10.1038/s41564-021-00958-0. [DOI] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S. Remdesivir for the treatment of covid-19 — final report. N. Engl. J. Med. 2020;14 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PubMed] [Google Scholar]
- Carriles A.A., Mills A., Muñoz-Alonso M.-J., Gutiérrez D., Domínguez J.M., Hermoso J.A., Gago F. Structural cues for understanding eEF1A2 moonlighting. Chembiochem. 2021;22:374–391. doi: 10.1002/cbic.202000516. [DOI] [PubMed] [Google Scholar]
- de Castro Martin I.F., Fournier G., Sachse M., Pizarro-Cerda J., Risco C., Naffakh N. Influenza virus genome reaches the plasma membrane via a modified endoplasmic reticulum and Rab11-dependent vesicles. Nat. Commun. 2017;8:1396. doi: 10.1038/s41467-017-01557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmayer C., Meinhardt J., Radbruch H., Radke J., Heppner B.I., Heppner F.L., Stenzel W., Holland G., Laue M. Why misinterpretation of electron micrographs in SARS-CoV-2-infected tissue goes viral. Lancet. 2020;396:e64–e65. doi: 10.1016/S0140-6736(20)32079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds B.T., Bell A., Wyckoff J., Condeelis J., Leyh T.S. The effect of F-actin on the binding and hydrolysis of guanine nucleotide by Dictyostelium elongation factor 1A. J. Biol. Chem. 1998;273:10288–10295. doi: 10.1074/jbc.273.17.10288. [DOI] [PubMed] [Google Scholar]
- Eymieux S., Rouillé Y., Terrier O., Seron K., Blanchard E., Rosa-Calatrava M., Dubuisson J., Belouzard S., Roingeard P. Ultrastructural modifications induced by SARS-CoV-2 in Vero cells: a kinetic analysis of viral factory formation, viral particle morphogenesis and virion release. Cell. Mol. Life Sci. 2021;78:3565–3576. doi: 10.1007/s00018-020-03745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymieux S., Uzbekov R., Rouillé Y., Blanchard E., Hourioux C., Dubuisson J., Belouzard S., Roingeard P. Secretory vesicles are the principal means of SARS-CoV-2 egress. Cells. 2021;10:2047. doi: 10.3390/cells10082047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Serradilla M., Risco C. Light and electron microscopy imaging unveils new aspects of the antiviral capacity of silver nanoparticles in bunyavirus-infected cells. Virus Res. 2021;16 doi: 10.1016/j.virusres.2021.198444. [DOI] [PubMed] [Google Scholar]
- Garibaldi B.T., Wang K., Robinson M.L., Zeger S.L., Bandeen-Roche K., Wang M.-C., Alexander G.C., Gupta A., Bollinger R., Xu Y. Comparison of time to clinical improvement with vs without remdesivir treatment in hospitalized patients with COVID-19. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X., Nicastri E., Oda R., Yo K., Quiros-Roldan E., Studemeister A., Redinski J., Ahmed S., Bernett J., Chelliah D., Chen D., Chihara S., Cohen S.H., Cunningham J., D'Arminio Monforte A., Ismail S., Kato H., Lapadula G., L'Her E., Maeno T., Majumder S., Massari M., Mora-Rillo M., Mutoh Y., Nguyen D., Verweij E., Zoufaly A., Osinusi A.O., DeZure A., Zhao Y., Zhong L., Chokkalingam A., Elboudwarej E., Telep L., Timbs L., Henne I., Sellers S., Cao H., Tan S.K., Winterbourne L., Desai P., Mera R., Gaggar A., Myers R.P., Brainard D.M., Childs R., Flanigan T. Compassionate use of remdesivir for patients with severe covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2007016. NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisado-Vasco P., Carralón-González M.M., Aguareles-Gorines J., Martí-Ballesteros E.M., Sánchez-Manzano M.D., Carnevali-Ruiz D., García-Coca M., Barrena-Puertas R., de Viedma R.G., Luque-Pinilla J.M., Sotres-Fernandez G., Fernández-Sousa J.M., Luepke-Estefan X.E., López-Martín J.A., Jimeno J.M. Plitidepsin as a successful rescue treatment for prolonged viral SARS-CoV-2 replication in a patient with previous anti-CD20 monoclonal antibody-mediated B cell depletion and chronic lymphocytic leukemia. J. Hematol. Oncol.J Hematol Oncol. 2022;15:4. doi: 10.1186/s13045-021-01220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotokezaka Y., Többen U., Hotokezaka H., van Leyen K., Beatrix B., Smith D.H., Nakamura T., Wiedmann M. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J. Biol. Chem. 2002;277:18545–18551. doi: 10.1074/jbc.M201022200. [DOI] [PubMed] [Google Scholar]
- Imran Mohd, Kumar Arora M., Asdaq S.M.B., Khan S.A., Alaqel S.I., Alshammari M.K., Alshehri M.M., Alshrari A.S., Mateq Ali A., Al-shammeri A.M., Alhazmi B.D., Harshan A.A., Alam MdT., Abida A. Discovery, development, and patent trends on molnupiravir: a prospective oral treatment for COVID-19. Molecules. 2021;26:5795. doi: 10.3390/molecules26195795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Muñoz-Alonso M.J., García C., Sánchez-Murcia P.A., Martínez-Leal J.F., Domínguez J.M., Lillo M.P., Gago F., Galmarini C.M. Translation elongation factor eEF1A2 is a novel anticancer target for the marine natural product plitidepsin. Sci. Rep. 2016;6 doi: 10.1038/srep35100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak M.K., Kinzy T.G. eEF1A: thinking outside the ribosome. J. Biol. Chem. 2010;285:21209–21213. doi: 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.E., Goldsmith C.S. Caution in identifying coronaviruses by electron microscopy. J. Am. Soc. Nephrol. 2020;31:2223–2224. doi: 10.1681/ASN.2020050755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Bárcena M., Sidorov I., Snijder E.J. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudshoorn D., Rijs K., Limpens R.W.A.L., Groen K., Koster A.J., Snijder E.J., Kikkert M., Bárcena M. Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio. 2017;8 doi: 10.1128/mBio.01658-17. mBio.01658-17, e01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;8 doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Rodon J., Muñoz-Basagoiti J., Perez-Zsolt D., Noguera-Julian M., Paredes R., Mateu L., Quiñones C., Perez C., Erkizia I., Blanco I., Valencia A., Guallar V., Carrillo J., Blanco J., Segalés J., Clotet B., Vergara-Alert J., Izquierdo-Useros N. Identification of plitidepsin as potent inhibitor of SARS-CoV-2-induced cytopathic effect after a drug repurposing screen. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.646676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse M., Fernández de Castro I., Tenorio R., Risco C. Advances in Virus Research. Elsevier; 2019. The viral replication organelles within cells studied by electron microscopy; pp. 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Du C., Wang B., Zhang Y., Liu X., Ren G. Up-regulation of eEF1A2 promotes proliferation and inhibits apoptosis in prostate cancer. Biochem. Biophys. Res. Commun. 2014;450:1–6. doi: 10.1016/j.bbrc.2014.05.045. [DOI] [PubMed] [Google Scholar]
- Tenorio R., de Castro I.F., Knowlton J.J., Zamora P.F., Lee C.H., Mainou B.A., Dermody T.S., Risco C. Reovirus σNS and μNS proteins remodel the endoplasmic reticulum to build replication neo-organelles. mBio. 2018;9:15. doi: 10.1128/mBio.01253-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Worm S.H.E., Knoops K., Zevenhoven-Dobbe J.C., Beugeling C., van der Meer Y., Mommaas A.M., Snijder E.J. Development and RNA-synthesizing activity of coronavirus replication structures in the absence of protein synthesis. J. Virol. 2011;85:5669–5673. doi: 10.1128/JVI.00403-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varona J.F., Landete P., Lopez-Martin J.A., Estrada V., Paredes R., Guisado-Vasco P., Fernandez de Orueta L., Torralba M., Fortun J., Vates R., Barberan J., Clotet B., Ancochea J., Carnevali D., Cabello N., Porras L., Gijon P., Monereo A., Abad D., Zuñiga S., Sola I., Rodon J., Vergara-Alert J., Izquierdo-Useros N., Fudio S., Pontes M.J., de Rivas B., Giron de Velasco P., Nieto A., Gomez J., Aviles P., Lubomirov R., Belgrano A., Sopesen B., White K.M., Rosales R., Yildiz S., Reuschl A.-K., Thorne L.G., Jolly C., Towers G.J., Zuliani-Alvarez L., Bouhaddou M., Obernier K., McGovern B.L., Rodriguez M.L., Enjuanes L., Fernandez-Sousa J.M., Krogan N.J., Jimeno J.M., Garcia-Sastre A. Preclinical and randomized phase I studies of plitidepsin in adults hospitalized with COVID-19. Life Sci. Alliance. 2022;5 doi: 10.26508/lsa.202101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.M., Rosales R., Yildiz S., Kehrer T., Miorin L., Moreno E., Jangra S., Uccellini M.B., Rathnasinghe R., Coughlan L., Martinez-Romero C., Batra J., Rojc A., Bouhaddou M., Fabius J.M., Obernier K., Dejosez M., Guillén M.J., Losada A., Avilés P., Schotsaert M., Zwaka T., Vignuzzi M., Shokat K.M., Krogan N.J., García-Sastre A. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science. 2021;371:926–931. doi: 10.1126/science.abf4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., Melia C.E., Snijder E.J., Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28:1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., Zheng S., Koster A.J., Snijder E.J., Bárcena M. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;5 doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi H., Chen J., Shi D., Li C., Feng L. EF1A interacting with nucleocapsid protein of transmissible gastroenteritis coronavirus and plays a role in virus replication. Vet. Microbiol. 2014;172:443–448. doi: 10.1016/j.vetmic.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data related to this work is available from corresponding authors upon reasonable request.