Abstract
Jasmonate ZIM-domain (JAZ) proteins are the key repressors of the jasmonic acid (JA) signal transduction pathway and play a crucial role in stress-related defense, phytohormone crosstalk and modulation of the growth-defense tradeoff. In this study, the sorghum genome was analyzed through genome-wide comparison and domain scan analysis, which led to the identification of 18 sorghum JAZ (SbJAZ) genes. All SbJAZ proteins possess the conserved TIFY and Jas domains and they formed a phylogenetic tree with five clusters related to the orthologs of other plant species. Similarly, evolutionary analysis indicated the duplication events as a major force of expansion of the SbJAZ genes and there was strong neutral and purifying selection going on. In silico analysis of the promoter region of the SbJAZ genes indicates that SbJAZ5, SbJAZ6, SbJAZ13, SbJAZ16 and SbJAZ17 are rich in stress-related cis-elements. In addition, expression profiling of the SbJAZ genes in response to phytohormones treatment (JA, ET, ABA, GA) and sugarcane aphid (SCA) was performed in two recombinant inbred lines (RILs) of sorghum, resistant (RIL 521) and susceptible (RIL 609) to SCA. Taken together, data generated from phytohormone expression and in silico analysis suggests the putative role of SbJAZ9 in JA-ABA crosstalk and SbJAZ16 in JA-ABA and JA-GA crosstalk to regulate certain physiological processes. Notably, upregulation of SbJAZ1, SbJAZ5, SbJAZ13 and SbJAZ16 in resistant RIL during JA treatment and SCA infestation suggests putative functions in stress-related defense and to balance the plant defense to promote growth. Overall, this report provides valuable insight into the organization and functional characterization of the sorghum JAZ gene family.
Subject terms: Genetics, Molecular biology, Plant sciences
Introduction
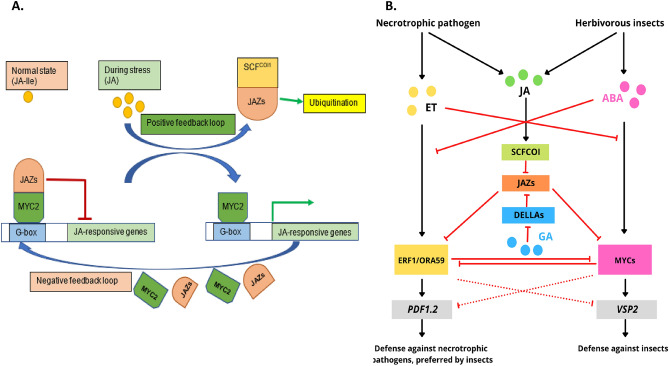
Jasmonates (JAs), a group of jasmonic acid and its bioactive derivatives, offer a critical role in plant resilience during biotic and abiotic stresses. JAs play a central role in activating plant responses to herbivory, pathogens invasion, UV radiation and ozone stress1–4. In addition, JAs also play a role in development processes like flower initiation and plant morphogenesis5–7. In general, JAs response promotes defense and reproduction by reducing growth processes like photosynthesis and cell division1,8. Such contrasting functions suggest a broader role of JAs in balancing defense and growth to optimize plant fitness9. JA signaling pathway is a complex process and consists of three distinct processes: biosynthesis, JA signal transduction and downstream gene activation. During JA signal transduction, SCF-type E3 ubiquitin ligase (SCFCOI1) receptor complex, Jasmonate-ZIM-domain (JAZ) and bHLH (basic helix-loop-helix) transcription factor MYC2 interact with each other to repress or derepress JA-responsive gene expression9–11 (Fig. 1A). Besides that, JAZ also plays a central role in the crosstalk between phytohormones signaling cascades of JA, abscisic acid (ABA), ethylene (ET) and gibberellins (GA) in response to stress12,13 (Fig. 1B). During herbivory, JA and ABA act synergistically on the expression of the MYC branch which results in the activation of defense-responsive genes14,15. In summary, JAZ plays a crucial role in the JA signaling pathway and are central mediators of the signal transduction pathways to activate defense genes.
Figure 1.
Proposed model for JA perception via COI1-JAZ co-receptor complex (A) and crosstalk between phytohormones in the defense signaling pathway (B) (adapted from Wasternack and Hause2; Pieterse et al.13). (A) During stress and wounding JAZ are recruited by the SCFCOI1 and MYC2 are released thus activating JA-defense genes (positive feedback loop). The newly synthesized JAZ genes during defense response binds to the MYC2 thus repressing the JA-defense genes (negative feedback loop). (B) Herbivorous insects induce JA- and ABA-dependent signaling pathways while necrotrophic pathogens induce JA- and ET-dependent signaling pathways. Solid lines are established interactions, dashed lines are hypothesized interactions, arrows indicate positive effects, and red inhibition lines represent negative effects. JA, Jasmonic acid; JAZ, JASMONATE ZIM DOMAIN; SCFCOI1, SCF-type E3 ubiquitin ligase; MYC2, bHLHzip transcription factor MYC2; ERF1, Ethylene Response Factor 1; ORA59, octadecanoid-responsive Arabidopsis AP2/ERF-domain protein 59; VSP2, VEGETATIVE STORAGE PROTEIN2; PDF1.2, PLANT DEFENSIN1.2.
The JAZ protein family belongs to the TIFY superfamily, which also includes three other subfamilies: ZIM-like (ZML), TIFY and PEAPOD (PPD) families16,17. All of the sub-families contain TIFY domains, with one or two different additional domains. The JAZ protein family consists of two highly conserved functional domains, TIFY (also called ZIM) and Jas (also called CCT_2)18. The TIFY domain (PF06200) usually consists of 28 amino acids with a consensus sequence “TIF[F/Y]XG”16, at the N-terminal of the JAZ protein sequences. Similarly, Jas domain (PF09425) consists of “SLX2FX2KRX2RX5PY” at the C-terminal of the JAZ protein sequence18. Both highly conserved domains of JAZ play irreplaceable roles in the JA signaling pathway. The TIFY domain mediates the interaction between JAZ proteins and the NINJA-TPL complex and both jointly inhibit the JA signaling19. The Jas domains mediate the interaction of JAZ with other genes, such as COI1 and MYC, resulting in the inhibition of transcriptional activities20–22. In addition, a previous study indicated that a lack of Jas domain in JAZ reduced the sensitivity of plant resistance to herbivory9.
JAZ proteins were first reported in 200718,23 and several studies afterward have well-characterized their functions in Arabidopsis thaliana. In A. thaliana there are 12 JAZ genes9. During mechanical wounding and Spodoptera exigua herbivory, 11 of the 12 Arabidopsis JAZ (AtJAZ) were induced significantly9,24,25. Among them AtJAZ1, AtJAZ2, AtJAZ5, AtJAZ6, AtJAZ9, AtJAZ10 and AtJAZ12 showed a relatively higher expression in S. exigua damaged leaves9. This event shows the negative feedback loop in which newly synthesized JAZ can repress the activity of MYC2 thus repressing the defense genes18,23 (Fig. 1A). Guo et al.26 reported that an Arabidopsis mutant (defective in ten AtJAZ genes) exhibited robust resistance to insect herbivores and fungal pathogens at expense of slow vegetative growth and poor reproductive performance. This observation suggests the positive feedback loop since a decrease in JAZ amplifies the plant capacity to release the MYC2 and thus expressing the defense genes9 (Fig. 1A). The increase of JAZ in the negative feedback loop is to trade-off growth and defense because overactivation of JA defense response leads to elevated carbon starvation, near loss of seed production and lethality in extreme conditions26. Chung et al.9 reported that wound-induced synthesis of JAs triggers the degradation of JAZ and subsequent expression of JAZ gene response in both positive and negative feedback loops. Taken together, JAZ proteins play an important role during biotic stress responses in plants by regulating defense to balance growth.
Sorghum [Sorghum bicolor (L.) Moench] is an important cereal crop in the world for the food, feed and biofuel industry. It ranks fifth in terms of both production and area planted among cereal crops in the world. Sorghum production was declining in years of 2015/2016 and 2016/2017 by 21.6% and 19.5%, respectively27. The loss in production is attributed to abiotic and biotic factors, among them, sugarcane aphid (SCA) (Melanaphis sacchari) is one of them. SCA outbreak in sorghum occurred in Texas in 2013 and after that it has seriously threatened the sorghum production in more than 20 US states28. SCA, a phloem-feeding insect, can attack sorghum at all developmental stages resulting in a significant yield loss and reduced quality of grain29. Further, SCA produces honeydew that ultimately reduces photosynthetic area, reduces the seed set and hinders the harvesting process30,31. All these have significantly increased the management cost for sorghum growers, yet genetic and molecular mechanisms behind sorghum plant responses to SCA are still unclear. For instance, to develop a resistant variety against SCA, identification of defense-genes and -pathways are crucial.
JAZ is one of the important defense gene families and it has been identified and functionally characterized in various plant species, including A. thalina9, Malus domesticus32, Camellia sinensis33, Brassica oleracea34, Petunia35 and Triticum aestivum36. Yet, identification and functional characterization of JAZ has not been reported in sorghum to date. In our study, we performed comparative genomics, domain-scan and phylogenetic analysis to identify and see the evolutionary relationship of the sorghum JAZ gene family. A detailed in silico analysis of promoter regions was conducted to identify the stress-related cis-acting elements and transcription factors (TFs). In addition, two Recombinant Inbred lines (RILs), resistant (RIL 521) and susceptible (RIL 609) to SCA, were used for expression profiling. The expression profiling of the sorghum JAZ gene in response to SCA infestation and four phytohormone treatments (JA, GA, ET, ABA) were investigated in both RILs. Our analysis provides insight into the biological function of sorghum JAZ genes and elucidates its possible role in phytohormone crosstalk and restraining a defense to balance the growth.
Materials and methods
Sequence acquisition of JAZ genes
To identify JAZ genes in sorghum, a single two-step approach was used. First, all the Arabidopsis JAZ proteins were used to perform a BLAST search among the whole genome sequences of sorghum. The protein sequences identified from the searches with E-value < 10−5 were pooled, and redundant sequences (identical sequences from multiple matching) were removed. Secondly, the acquired sorghum proteins were checked for the presence of TIFY and Jas domains using the InterProScan program (http://www.ebi.ac.uk/Tools/InterProScan/) and the Pfam database (http://pfam.xfam.org/) was used to confirm their presence37. The identified sorghum JAZ genomic sequence and coding sequence (CDS) were retrieved from Phytozome (https://phytozome-next.jgi.doe.gov/). These sequences were analyzed for exon/intron organization using the gene structure display (GSDS) tool38. The physical and chemical parameters of each sorghum JAZ (SbJAZ) protein were predicted using the ExPasy program (https://web.expasy.org/protparam/) and subcellular localization were predicted through WoLF PSORT online program (https://wolfpsort.hgc.jp/). The chromosomal position of SbJAZ genes obtained from the Phytozome were used to create the genetic map using chromoMap package in R39.
Phylogenetic analysis of plant JAZ
To establish the evolutionary relationship of SbJAZ proteins, a total of 106 JAZ proteins from seven representative plants, including a bryophyte (Physcomitrella patens), lycopodiophyte (Selaginella moellendorffii), monocots (O. sativa and S. bicolor) and eudicots (A. thaliana, Brassica oleracea and Camelia sinensis), were used for the phylogenetic analysis. The protein sequences of these seven plants were downloaded from NCBI and Phytozome33. A phylogenetic tree was constructed using the maximum likelihood method with Poisson correction model using 1000 bootstrap values40,41 in the MEGAX program42. To better understand the evolutionary relations, sorghum JAZ genes were compared to those homologs in rice for their chromosomal location and genomic structure.
Sequence analysis and identification of conserved motifs
Structural motif annotation for all 18 JAZ proteins were performed using the MEME program (http://meme-suite.org/tools/meme). We also manually checked the alignments of all SbJAZ proteins to identify the conserved sequences and pivotal amino acids. The online tool weblogo43 was used to generate the sequence logos of conserved regions present in SbJAZ proteins. Furthermore, the two conserved domains, TIFY and Jas domains, of sorghum were aligned with the domain of O. sativa and A. thaliana to observe the cross-species conservation of these domains.
Gene duplication and calculation of dN/dS ratio
The two types of gene duplication, segmental and tandem duplication are well known. Segmental gene pairs consist of 90% sequence similarity, while tandem consists of five or fewer genes within the 100 kb region44,45. After the identification of duplicated gene pairs, CDS sequences of each JAZ genes were used to calculate the synonymous (dS) and nonsynonymous (dN) rate of substitution using MEGA X according to Han and Luthe46. First, the coding sequences were aligned by Clustal W, then dN and dS was estimated using Nei-Gojbori substitution model47. Next, to calculate the p-value of the selection, codon-based Z-test was performed on each pair of sequences. With the p-value (< 0.05 are considered significant at the 5% level) of each pair of sequences, neutral evolution (dN = dS), positive selection (dN > dS) or purifying selection (dN < dS) were tested on each pair of sequences.
Prediction of cis‑regulatory elements and transcription factor networks
About 2000 bp promoter sequences upstream to ATG for all SbJAZ genes obtained from the Phytozome were analyzed using the PlantCARE promoter analysis tool48. Then, the identified various cis-elements were used to construct the heatmap through the gplots package in R software49. Further, transcription factor network prediction was performed as described by Wang et al.50 with minor modifications. The promoter sequence used earlier was submitted to the Plant Transcriptional Regulatory Map (PTRM) (http://planttfdb.gao-lab.org/prediction.php) to predict the transcription factors. These transcription factors were subjected to Cytoscape 3.8.2 to visualize the transcription factor regulatory network51. Furthermore, KEGG and GO analyses were conducted using the PTRM and DAVID Bioinformatics resources 6.8 (https://david.ncifcrf.gov/).
Sorghum plant growth and treatments
Sorghum seeds from parental lines (Tx2783 and BTx623) and the recombinant inbred lines (RILs), RIL 521 and RIL 609 developed from the parents, were grown in greenhouse at constant temperature (28 ± 2 °C) and 60% relative humidity under constant photoperiod of 14 h-light/10 h-dark. The seeds of the two parental lines (Tx2783 and BTx623) and Tx7000 were available to public use, which were originally obtained from the Germplasm Resources Information Network (GRIN, https://www.ars-grin.gov/) in the U.S. and the recombinant inbred lines (RIL 521 and RIL 609) were produced in our lab. All plant materials used in this study comply with local and national guidelines. Sugarcane aphid colonies were cultured on susceptible sorghum line (Tx7000). Sorghum seedlings of the four lines of 10–12 days old (2–3 leaf stage) were infested with 20 adults of apterous sugarcane aphid to the adaxial surface of the first true leaf. Each infested plant and the control plants (not infested with aphids) were covered, respectively, with a transparent cylindrical cage with nylon mesh on the top. To evaluate the differential responses to aphid in the four lines (Tx2783, BTx623, RIL 521 and RIL 609), the aphids on each plant were counted and recorded at 1, 3, 6 and 9 days post infestation (dpi) from ten independent plants of each infested lines. For phytohormones treatment, two RILs were sprayed separately with MeJA (100 μM), ABA (100 μM), ET (100 μM), SA (100 μM) and sterile distilled water (control) until runoff. Samples, first two true leaves and stem of the seedlings, were harvested from each treatment (MeJA, ABA, ET, GA and control) at 6 h after spraying. For SCA infestation analysis, samples (first two true leaves and stem below the second leaf) were collected from the two RILs infested with sugarcane aphids and without (control) at 0, 6-, 24- and 48-h post infestation (hpi). Each sample harvested had three biological replicates for each time point and were frozen immediately in liquid nitrogen and stored at − 80 °C. The control samples were collected at each time point to eliminate the circadian rhythm effect on gene expression.
RNA extraction and quantitative real-time PCR analysis
Quantitative RT-PCR (qRT-PCR) was used to estimate the relative expression of sorghum JAZ in response to aphid infestation and phytohormone treatment. A Trizol reagent (Invitrogen, Carlsbad, CA) was used to extract the total RNA from each sample and then it was treated with DNase (Turbo DNA-free kit, Thermo Fisher Scientific, Waltham, MA). A total of 2.5 μg of RNA was reverse-transcribed using the GoScript reverse transcriptase kit (Promega, Madison, WI) and the resulted cDNA was diluted four-fold before using for the qRT-PCR reaction. Primers were designed using the IDT DNA program (https://www.idtdna.com/PrimerQuest/Home/Index), which are listed in Table S1. A sorghum α-Tubulin gene (Sobic.001G107200) was used as the internal control as described previously, and this gene showed a stable expression throughout various treatments in sorghum52. qRT-PCR was performed on a Bio-Rad iCycler thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using the iTaq™ universal SYBR® green supermix (Bio-Rad Laboratories, Inc.). The qRT-PCR reaction was performed in a volume of 10 μl, containing 1 μl of cDNA, 0.4 μl (10 μM) each of the reverse and forward primers, 5 μl of SYBR green master mix and 3.2 μl of ddH2O under the following conditions: one cycle at 95 °C for 3 m, 40 cycles at 95 °C for 10 s and 55 °C for 30 s, followed by one cycle each of one min at 95 °C and 55 °C. The final melting curve was 81 cycles at 55 °C for 30 s.
Statistical analysis
The relative expression level of each gene was calculated using the 2−ΔΔCt method53 and the data presented are the averages of three biological and two technical replicates. For the aphid count data, ANOVA and Tukey test was used to estimate the significant difference. For expression analysis during phytohormones and SCA infestation, Student’s t-test was used to estimate the significant difference (*P < 0.05, **P < 0.01 and ***P < 0.001).
Results
Identification of the JAZ gene family in sorghum
To identify the SbJAZ gene family a single two-step approach was used, homology search and proteome scan for the presence of “TIFY” and “Jas” domains. From the first approach, 26 SbJAZ genes were identified. The second approach narrowed the SbJAZ to eighteen (Table 1). The decrease in the JAZ genes is due to the missing of one of the domains, or the addition of extra domains (VEFS and GATA) (Table S2). The true JAZ sequence should contain both TIFY and Jas domains34,35.
Table 1.
Structural features of the JAZ genes in sorghum.
| Name | Gene ID | Chromosome | Exon_count | Orientation | Location coordinates | Genomic (bp) | CDS (bp) | ORF (aa) | PI | MW (kDa) | GRAVY |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SbJAZ1 | Sobic.001G259300 | 1 | 3 | Forward | 31035454..31036798 | 1345 | 681 | 226 | 9.06 | 24.29 | −0.538 |
| SbJAZ2 | Sobic.001G259400 | 1 | 1 | Forward | 31205564..31206743 | 1180 | 507 | 168 | 9.92 | 17.34 | −0.089 |
| SbJAZ3 | Sobic.001G259600 | 1 | 1 | Forward | 31327155..31328316 | 1162 | 528 | 175 | 9.29 | 17.97 | 0.006 |
| SbJAZ4 | Sobic.001G259700 | 1 | 1 | Forward | 31345064..31346119 | 1056 | 666 | 221 | 9.81 | 22.72 | −0.316 |
| SbJAZ5 | Sobic.001G259900 | 1 | 1 | Forward | 31385111..31385784 | 674 | 561 | 186 | 8.42 | 19.58 | −0.152 |
| SbJAZ6 | Sobic.001G276900 | 1 | 2 | Forward | 54019381..54020155 | 775 | 675 | 224 | 8.32 | 24.09 | −0.462 |
| SbJAZ7 | Sobic.001G343900 | 1 | 5 | Forward | 63205787..63208276 | 2490 | 687 | 228 | 8.98 | 24.05 | −0.445 |
| SbJAZ8 | Sobic.001G482600 | 1 | 1 | Reverse | 75415876..75416893 | 1018 | 585 | 194 | 8.78 | 19.94 | −0.153 |
| SbJAZ9 | Sobic.001G482700 | 1 | 3 | Reverse | 75423136..75425280 | 2145 | 606 | 201 | 7.70 | 21.14 | −0.372 |
| SbJAZ10 | Sobic.001G482800 | 1 | 1 | Forward | 75429377..75432763 | 3387 | 543 | 180 | 9.33 | 18.87 | −0.274 |
| SbJAZ11 | Sobic.002G036100 | 2 | 2 | Reverse | 3392335..3395495 | 3161 | 729 | 242 | 9.33 | 24.82 | −0.517 |
| SbJAZ12 | Sobic.002G196200 | 2 | 7 | Reverse | 58508296..58511272 | 2977 | 1227 | 408 | 9.83 | 42.80 | −0.338 |
| SbJAZ13 | Sobic.002G214800 | 2 | 5 | Reverse | 60694460..60696521 | 2062 | 666 | 221 | 9.30 | 23.38 | −0.476 |
| SbJAZ14 | Sobic.002G374100 | 2 | 5 | Forward | 73193557..73195985 | 2429 | 708 | 235 | 9.25 | 24.92 | −0.37 |
| SbJAZ15 | Sobic.003G410300 | 3 | 4 | Forward | 71777501..71782156 | 4656 | 303 | 100 | 9.16 | 10.60 | −0.559 |
| SbJAZ16 | Sobic.006G056400 | 6 | 5 | Forward | 40020080..40027257 | 7178 | 537 | 178 | 9.81 | 18.84 | −0.341 |
| SbJAZ17 | Sobic.006G244800 | 6 | 5 | Forward | 58463788..58467690 | 3903 | 660 | 219 | 7.71 | 23.16 | −0.47 |
| SbJAZ18 | Sobic.007G132700 | 7 | 7 | Reverse | 55342328..55345325 | 2998 | 1287 | 428 | 9.55 | 45.30 | −0.34 |
JAZ genes having only TIFY or CCT domain are listed in Supplementary Table S2.
CDS coding DNA sequence, PI isoelectric point, MW molecular weight.
Table 1 summarizes the characteristics of the SbJAZ gene family, including Gene IDs and gene features. In this study, gene name was assigned from JAZ1 to JAZ18 according to their positions in the chromosome. At the genomic level SbJAZ sequences ranges from 674 to 7178 bp and coding DNA sequences (CDS) range from 303 to 1287 bp. The length of SbJAZ proteins varied between 100 and 428 aa and the predicted molecular weights range from 10.60 to 45.30 kDa. The PI ranges from 7.7 to 9.92, indicating that all the sorghum JAZ proteins were basic. The grand average of hydropathicity (GRAVY) values of all SbJAZ proteins, except SbJAZ3, is less than 0, implying that they all are hydrophilic proteins. Further detailed information about the instability index, aliphatic index, and subcellular localization of all the SbJAZ proteins are listed in Table S3.
Phylogenetic and structural analysis of sorghum JAZ genes
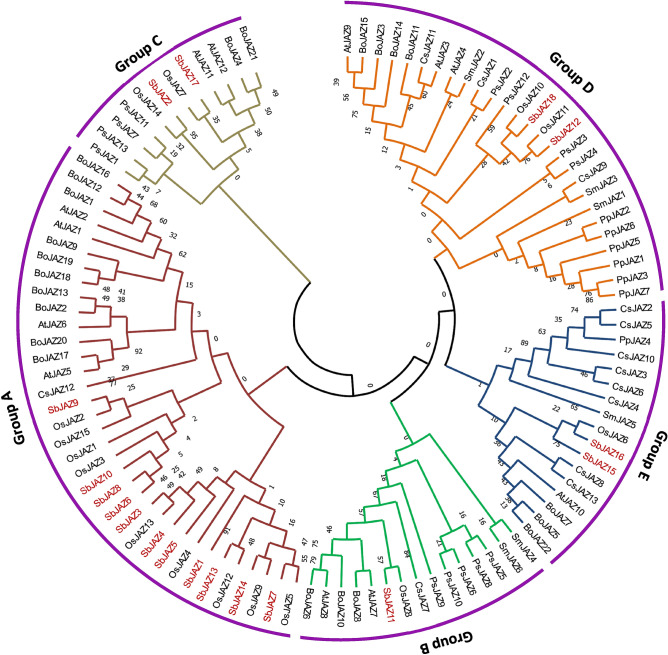
To determine the evolutionary relationship of sorghum JAZ, a total of 106 JAZ proteins from eight representative plant species, including a bryophyte (Physcomitrella patens, 7), lycopodiophyte (Selaginella moellendorffii, 6), gymnosperms (Picea sitchensis, 13), monocots (O. sativa (15) and S. bicolor) and eudicots (A. thaliana (12), B. oleracea (22) and C. sinensis (13) were used for the phylogenetic analysis33 (Table S4). The JAZ proteins were divided into five groups (Groups A–E), which is in line with previous classifications (Fig. 2)35. Amongst five groups, Group A was the largest one, with three sub-groups followed by Group D and Group E with two sub-groups in each. The sorghum JAZ protein family were distributed in all the groups: 11 in group A (SbJAZ1, SbJAZ3-10, SbJAZ13-14), two in each of the group C (SbJAZ2 and SbJAZ17), D (SabJAZ12 and SbJAZ18) and E (SbJAZ15-16), and the remaining one in group B (SbJAZ11).
Figure 2.
Phylogenetic analysis of the JAZ gene family. The circular tree represents the evolutionary relationship between the sorghum JAZ protein family with those in bryophyte, lycopodiophyte, gymnosperms, monocots and eudicots. The tree was generated by MEGAX using the Maximum Likelihood method based on the Poisson correction model using 1000 bootstrap values40,41. The analysis involved 106 amino acid sequences and they are divided into five classes. Species abbreviations used for phylogeny are as follows. At: Arabidopsis thaliana, Bo: Brassicae oleracea; Cs: Camellia sinensis; Os: Oryza sativa; Pp: Physcomitrella patens; Ps: Picea sitchensis; Sb: Sorghum bicolor and Sm: Selaginella moellendorffii.
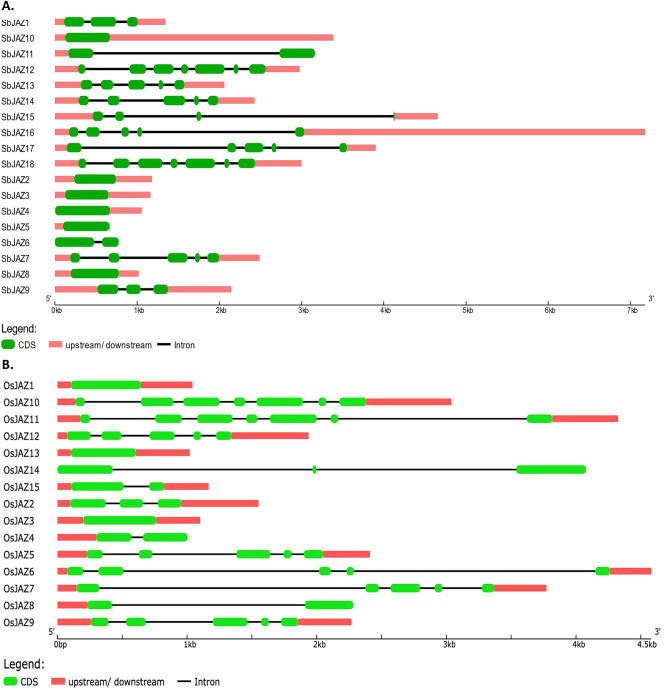
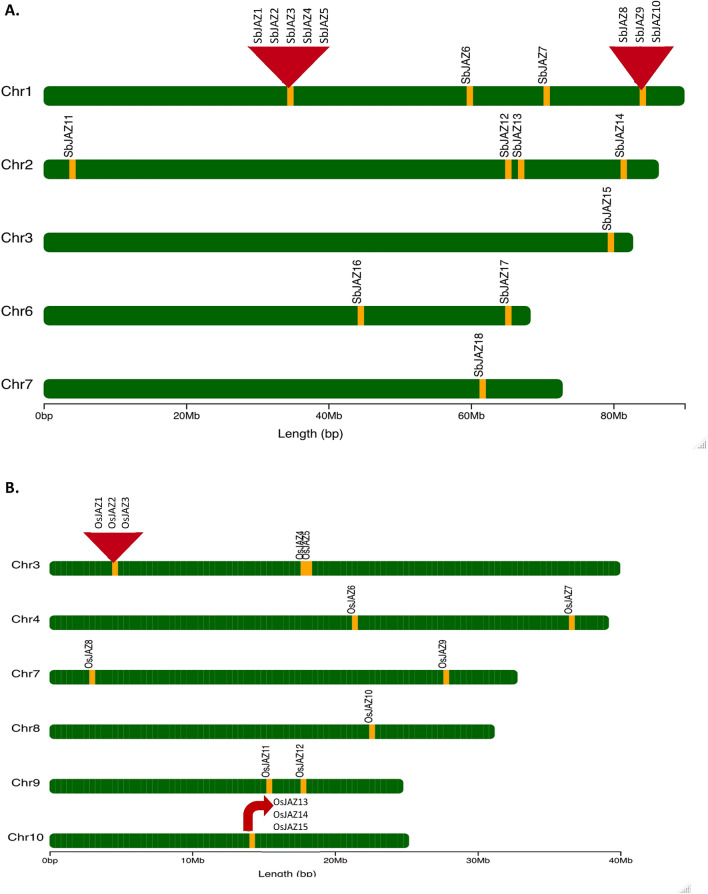
To study the structural diversity of the SbJAZ genes, intron/exon organization of coding sequence of each gene were constructed (Fig. 3). The 18 SbJAZ genes are mapped to five chromosomes (Chr 1, 2, 3, 6 and 7) out of the ten sorghum chromosomes (Fig. 4A), with ten residing on Chr 1, four on Chr 2, two on Chr 6 and one on each of Chr 3 and 7. All sorghum JAZs are consisted of 5′ and 3′ untranslated regions (UTRs) except SbJAZ6 and SbJAZ11. Similarly, all sorghum JAZ have exon number ranging from 1–7 and intron numbers ranging from 0–6 (Fig. 3). The divergence in gene structure could also support the clustering of SbJAZ proteins in the phylogenetic tree (Fig. 2)54. The cross-species comparison with the 15 OsJAZs showed that O. sativa has only two genes with single exons in comparison to six in SbJAZs (Fig. 3). In terms of JAZs gene distribution, O. sativa has more uniform distribution of JAZs in chromosomes with highest in Chr3 (five JAZ genes), compared to ten SbJAZ genes on Chr 1 of sorghum (Fig. 4).
Figure 3.
The schematic genomic structure for each SbJAZ gene (A) and OsJAZ (B) was generated using gene structure display server (GSDS). The size of coding DNA sequence (CDS), introns and untranslated regions are proportional to their sequence lengths.
Figure 4.
Genome mapping of the 18 SbJAZ genes on S. bicolor chromosomes (A) and 15 OsJAZ genes on O. sativa chromosomes (B). Gene locations and chromosome size can be measured using the scale on the bottom of the figure.
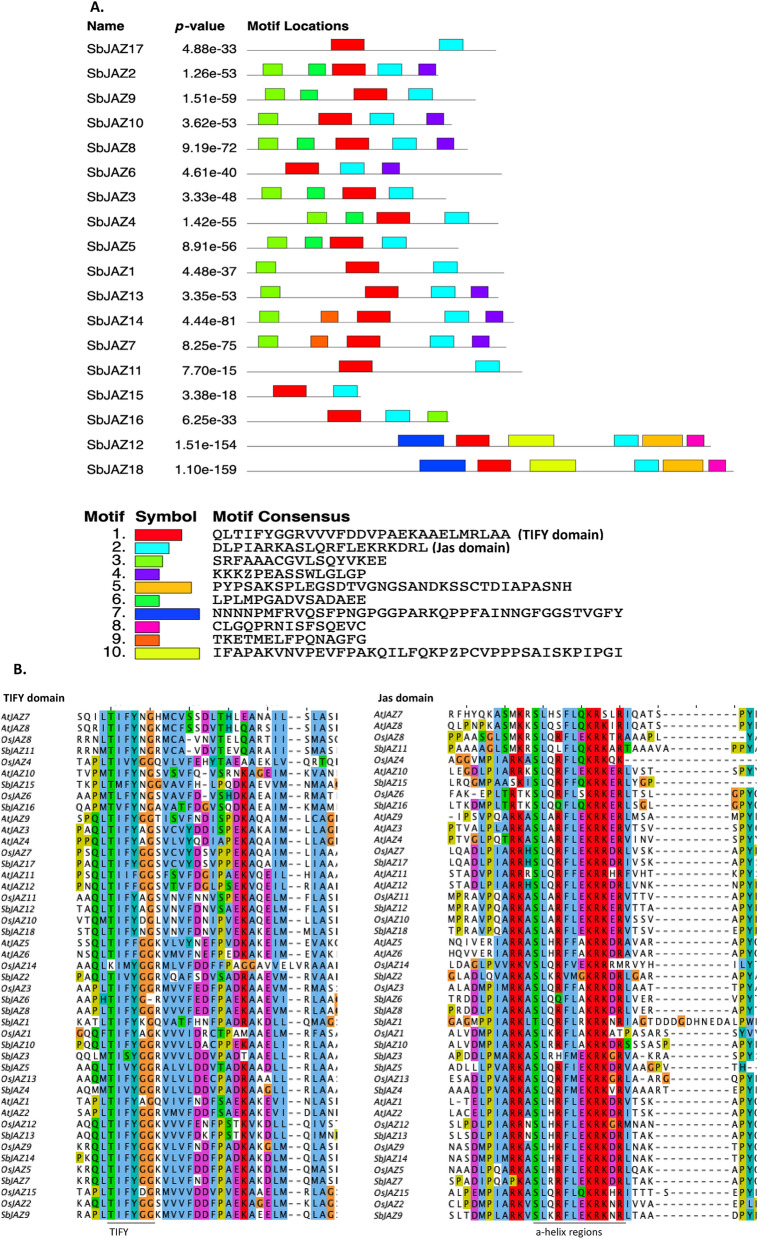
Conserved motif analysis in sorghum JAZ proteins
A total of ten distinct motifs in the SbJAZ protein family were identified through MEME suite (Fig. 5A). Among them two motifs, TIFY (red) and Jas domain (sky blue), were conserved in all the SbJAZ proteins (Fig. 5A). These two motifs and their locations vary among the sub-groups, but are present in all members of the JAZ family. To identify the conserved residues in those two domains, all 18 SbJAZ protein sequences were subjected to weblogo. Here, the TIFY domain has core “TI[F/V]YXG” motif and Jas domain has “SLX2FX2KRX2RX7PY”. Both of these domains are the defining trait of JAZ proteins, and are well conserved in sorghum JAZ. The sequence logo for TIFY and Jas domains generated from SbJAZ members are similar to the HMM profile generated from Pfam database (Fig. S1) which suggests these residues are conserved throughout the species. Further cross-species alignment of TIFY and Jas domain showed that these domains are well conserved in the monocots and dicots with some minor changes in few genes (Fig. 5B). The Jas domain motif of some of the proteins (OsJAZ4, SbJAZ15, SbJAZ1, SbJAZ5, OsJAZ2, OsJAZ14, OsJAZ8) lack the PY motif at the end. Previous studies have showed that the PY motif is not required for the ligand-dependent COI1-JAZ interaction, but the a-helix region of Jas domain is the one to bind the COI1 and JA hormone, which is conserved throughout the species55. Similarly, TIFY domain is highly conserved in all of the genes which acts as a repressor for JA signaling19.
Figure 5.
(A) Schematic representation of the conserved motifs in sorghum JAZ proteins. Conserved motifs and their alignment among JAZ proteins was elucidated by using MEME tool. Most of the proteins possess similar domains revealing their conserveness. Each colored box represents a motif in the protein, with the consensus motif sequence below in the index. (B) Cross-species comparison of TIFY and Jas domains revealed the sequence conservation of these domains.
Gene duplication and estimation of dN/dS values
In general, there are two major events in gene duplication, tandem or segmental, leading to an increase in number of genes in a family. The results of phylogenetic analysis showed that none of the SbJAZ gene pairs has 90% similarity between each other to be defined as segmental repeats56. Based on chromosomal location, six gene pairs were identified in Chr 1 as tandem duplication (Table 2). These pairs might have originated from two separate self-duplication events in Chr 1, first among SbJAZ3, SbJAZ4 and SbJAZ5 and second among SbJAZ8, SbJAZ9 and SbJAZ10. To further explore the driving force of SbJAZ gene evolution, dN/dS ratio and Z-test were calculated. The ratio of 1 indicates neutral selection, ratio > 1 indicates positive selection and ratio < 1 indicates purifying selection. From the Table 2, The dN/dS ratios of all six gene pairs were below 1, but the p-value showed significant purifying selection for SbJAZ8-9 gene pairs only, so other gene pairs occurred as the neutral selection.
Table 2.
Analysis of tandem duplication events of sorghum JAZ gene pairs.
| Gene pairs | dN | dS | dN/dS | p-value |
|---|---|---|---|---|
| SbJAZ3-SbJAZ4 | 0.434 | 0.465 | 0.934 | 0.282 |
| SbJAZ3-SbJAZ5 | 0.381 | 0.397 | 0.960 | 0.383 |
| SbJAZ4-SbJAZ5 | 0.344 | 0.414 | 0.830 | 0.082 |
| SbJAZ8-SbJAZ9 | 0.377 | 0.474 | 0.795 | 0.027* |
| SbJAZ8-SbJAZ10 | 0.341 | 0.354 | 0.961 | 0.392 |
| SbJAZ9-SbJAZ10 | 0.413 | 0.485 | 0.852 | 0.088 |
dN nonsynonymous substitutions rate, dS synonymous substitutions rate. To be considered purifying selection dN/ < 1 and p-value for the Z-test should be below 0.05 (*P < 0.05).
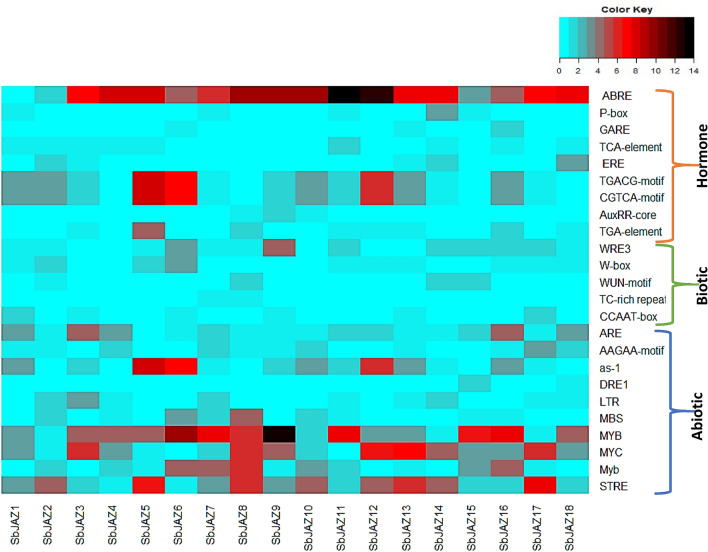
Identification of cis-elements within the SbJAZ genes
The cis-elements present upstream of a gene play a critical role in regulatory function involved in plant stress response and growth57. An in silico analysis of the 2000 bp upstream of the SbJAZ genes were conducted through the Plant Care database. The summary of the cis-elements was plotted in heatmap (Fig. 6) and according to their functions, the cis-elements were grouped into categories such as abiotic, biotic and hormone responsive. Almost all the SbJAZ genes possess plant hormone-responsive elements, ABA (ABRE), GA (P-BOX and GARE), SA (TCA-element), AUX (TGA-element, AuxRR-core) and JA (TGACG-motif and CGTXA-motif) (Table S5). Among them, ABA and JA responsive motifs were present abundantly in the SbJAZ genes. These results suggest that the SbJAZ family may be associated with the complex hormone regulatory network. Similarly, abiotic stress-responsive elements, such as STRE, Myb, MYC, MYB, MBS and as-1 were also present abundantly. Among them, the two cis-elements MBS and Myb were the drought-inducible motif. Meanwhile, some biotic-responsive elements, like WRE3, W-box, WUN and CCAAT-box, were present in a small number in some SbJAZ genes (Fig. 6 and Table S5). These motifs WUN and CCAAT-box are stress and defense inducible elements. Notably among 18 sorghum JAZ genes promoters, SbJAZ5, SbJAZ6, SbJAZ8, SbJAZ13, SbJAZ16 and SbJAZ17 are rich in cis-elements.
Figure 6.
Heatmap representing the distribution of cis-elements in the promoter regions (2000 bp upstream) of sorghum JAZ genes. The scale (color key) represents the number of cis-elements in each SbJAZ genes (1: sky blue, 7: red, 14: black).
Identification of transcription factor regulatory network of the SbJAZ genes
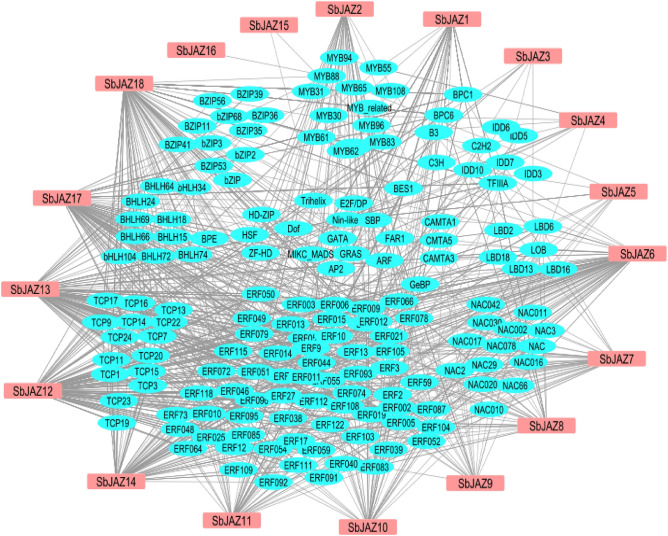
To explore the potential transcriptional network of SbJAZ genes, 2000 bp upstream sequences from the 18 SbJAZ genes were analyzed through the PTRM website. The results showed 211 transcription factors (TFs) participated in the regulation of sorghum JAZ genes (Table S6). These TFs were used to construct a transcription factor regulatory network (Fig. 7). The TFs network shows that the SbJAZ family were possibly regulated by the following seven transcription factor families, ERF, TCP, bHLH, MYB, C2H2, LBD and NAC. Among them, members of the ERF family were the most abundant, followed by TCP, NAC and bHLH.
Figure 7.
The putative transcription factor regulatory network of the SbJAZ genes. The network was constructed using the PTRM tool and Cystoscope.
Furthermore, to understand the biological functions of the 18 SbJAZ genes, GO and KEGG enrichment analyses were conducted. The results (Table 3) showed almost all the SbJAZ genes except SbJAZ8 were related to the plant responses to defense (GO:0031347), wounding (GO:0009611) and the jasmonic acid signaling pathway (GO:2000022) (Table 3). Moreover, these genes were also enriched with transcription corepressor activity (GO:0003714) and negative regulation of nucleic acid transcription (GO:1903507). Interestingly, the KEGG pathway analysis indicated that all the SbJAZ genes were regulated only in the JA signal transduction pathway (map04075) (Fig. S2).
Table 3.
Gene ontology of the sorghum JAZ genes.
| Gene ontology | Process | Functions associated with GO | Present in all SbJAZ genes except |
|---|---|---|---|
| GO:1903507 | BP | Negative regulation of nucleic acid-templated transcription | SbJAZ8 |
| GO:2000022 | BP | Regulation of jasmonic acid mediated signaling pathway | SbJAZ8 |
| GO:0031347 | BP | Regulation of defense response | SbJAZ8 |
| GO:0009611 | BP | Response to wounding | SbJAZ8 |
| GO:0005634 | CC | Nucleus | SbJAZ8 |
| GO:0003714 | MF | Transcription corepressor activity | SbJAZ8 |
BP biological process, CC cellular component, MF molecular function.
Differential response between resistant and susceptible lines
The two parental lines (Tx2783 and BTx623) and two RILs (RIL 521, SCA-resistant and RIL 609 SCA-susceptible) infested with SCA showed differential responses to aphid infestation. The Tx2783 is a resistant and BTx623 a susceptible genotype against SCA infestation58,59. Following aphid infestation, Tx2783 and RIL 521 lines showed adverse effect on aphid development and fecundity in comparison to the susceptible genotype. The number of aphids per plant was counted and based on that the rate of aphid regeneration was significantly reduced on Tx2783 and RIL 521 lines from early dpi (1- and 3-dpi) to late (6- and 9-dpi) (Fig. S3 and Table S7). Similarly, less SCA damage was noted on Tx2783 and RIL 521 lines in compared to the susceptible genotype.
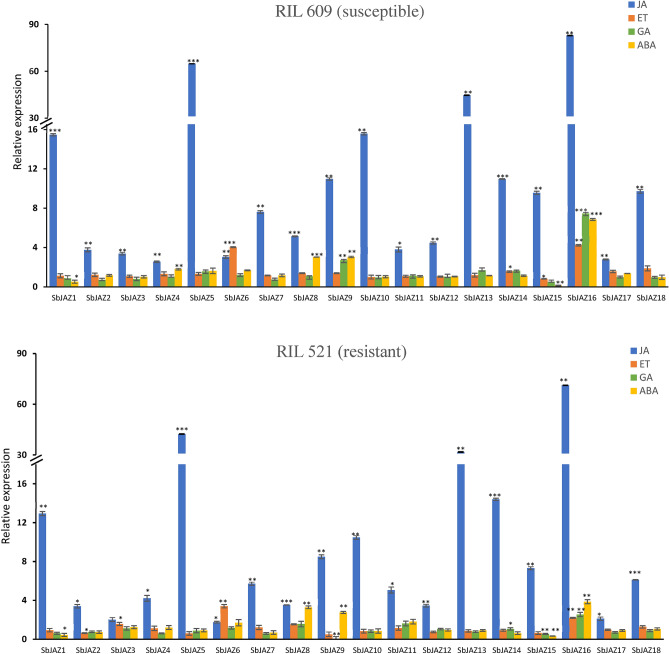
Expression patterns of SbJAZ genes after exogenous phytohormone treatments
Here, qRT-PCR was conducted to evaluate the response of sorghum JAZ genes in both resistant (RIL 521) and susceptible (RIL 609) RILs after phytohormones (JA, ET, GA and ABA) treatments. Following treatment with MeJA, almost all the JAZ genes in both RILs were upregulated (Fig. 8). Among 18 SbJAZ genes, three of them were upregulated more than 30-fold, SbJAZ5, SbJAZ13 and SbJAZ16, four of them were upregulated more than eightfold, SbJAZ1, SbJAZ9, SBJAZ10 and SbJAZ14 while the other eleven SbJAZ genes were upregulated between two to eight-fold (Fig. 8). In contrast, other three phytohormones, treatments with ET, GA and ABA didn’t show consistent upregulation in expression of sorghum JAZ genes as that induced by MeJA. After ABA treatments, SbJAZ8, SbJAZ9 and SbJAZ16 showed upregulation of more than three-fold while SbJAZ15 showed downregulation. Similarly, after ET treatment, SbJAZ6 and SbJAZ16 showed more than four-fold upregulation in both RILs. Among all SbJAZ genes, SbJAZ16 showed concurrent upregulation in all four phytohormone treatments.
Figure 8.
Expression pattern of 18 SbJAZ genes in response to the treatments with phytohormones (JA, ET, GA, and ABA) in resistant (RIL 521) and susceptible (RIL 609) RILs to SCA. qRT-PCR was used to determine the relative expression of each SbJAZ gene, and the relative expression was estimated using the 2−ΔΔCt method. Error bars in each bar represent the ± standard error (n = 3) and asterisks indicate significant differences between the control and phytohormone treated samples, *P < 0.05, **P < 0.01, ***P < 0.001. The bars without asterisk, are non-significant (P > 0.05).
Expression patterns of SbJAZ genes after SCA infestation
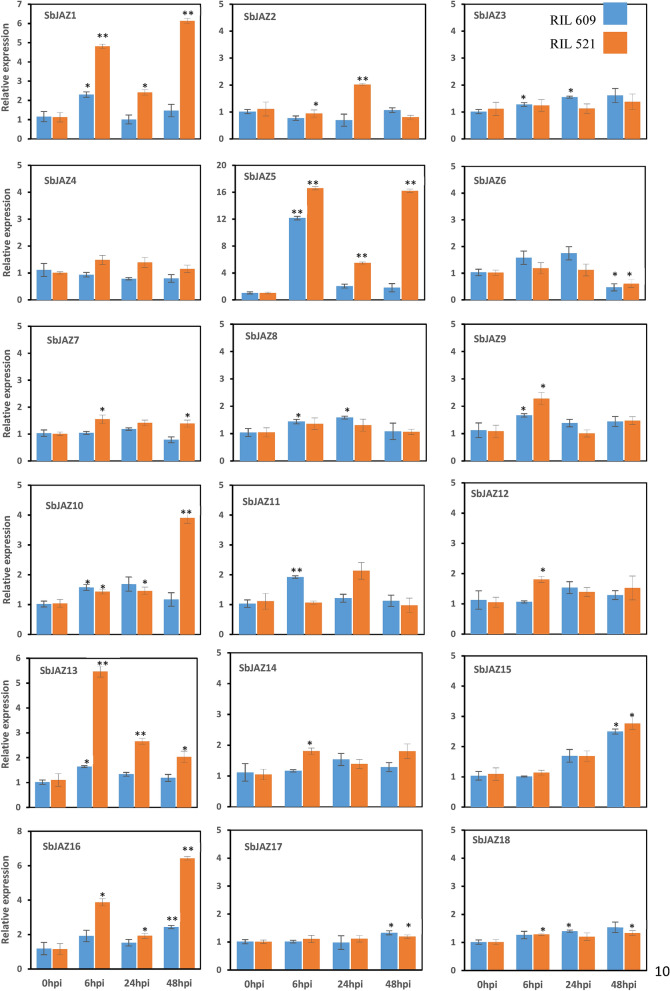
The qRT-PCR was conducted to observe the expression patterns of sorghum JAZ genes in both RILs after SCA infestation. The expression pattern was observed in all four-time points, 0, 6, 24 and 48 hpi. Most of the sorghum JAZ genes, except SbJAZ3, SbJAZ6, SbJAZ8, SbJAZ17 and SbJAZ18, at different time points (except 0 hpi) showed significantly higher expression in the RIL 521 in comparison to RIL 609 (Fig. 9). Among 18 SbJAZ genes, SbJAZ1, SbJAZ5, SbJAZ13 and SbJAZ16 showed more than five-fold upregulation in one or more of the time points in RIL 521. SbJAZ5 showed the highest upregulation among these four SbJAZ, around 16-fold in both 6 hpi and 48 hpi following SCA infestation and only five-fold in 24 hpi. Similar pattern of up and downregulation was observed in SbJAZ1 and SbJAZ16. These up and downregulation of JAZ genes at different time points suggest there are continuous interactions going on between the plant and aphid to dominate each other. Three other genes, SbJAZ9, SbJAZ10, SbJAZ15, showed 2 to fivefold upregulation in resistant lines at different time points. Interestingly, SbJAZ6 showed downregulation in both the RILs during SCA infestation at 2 dpi.
Figure 9.
Expression pattern of 18 SbJAZ genes in response to sugarcane aphid infestation in in resistant (RIL 521) and susceptible (RIL 609) RILs to SCA. qRT-PCR was used to determine the relative expression of each SbJAZ gene, and the relative expression was estimated using the 2−ΔΔCt method. Error bars in each bar represent the ± standard error (n = 3) and asterisks indicate significant differences between the control and phytohormone treated samples, *P < 0.05, **P < 0.01. The bars without asterisk, are non-significant (P > 0.05).
Discussion
JAZ proteins participate in stress response and play a central mediator role in the phytohormone signaling pathways to activate defense genes. Besides that, JAZ proteins balance growth and defense response by avoiding the detrimental effects of hyper-immunity caused by JA defense26. To date, 12 JAZ genes have been reported and studied in A. thaliana, 18 in M. domesticus32, 13 in C. sinensis36 and 14 in T. aestivum36. However, the study of the sorghum JAZ gene family and its functional role has not been reported. In this study, we carried out genome-wide analysis and expression profiling of the SbJAZ gene family and identified its functional role in plant development and defense.
Structural characteristics of sorghum JAZ genes
All JAZ genes contain of TIFY domain at N-terminal and Jas domains at C-terminal, which are characteristic features of JAZ. The TIFY domains mediate the interactions with NINJA, whereas Jas domain mediates the interactions with COI and MYC. The sorghum genome possesses a total of 18 JAZ genes that contain both domains (Table 1). The sorghum JAZ number is similar to Hevea brasiliensis (18)60, M. domestica (18), O. sativa (15) and B. oleracea (22), but higher than in P. patens (7) and S. moellendorfii (6). The higher number of JAZ family members indicates the expansion event in seed plants33. Interestingly, all 18 JAZ were basic in nature with PI value ranging from 7.70–9.92, suggesting that individual SbJAZ proteins may require different ionic strength for their optimal activity44. The sorghum JAZ genes consist of introns and exons ranging from 0–6 and 1–7, respectively. Similar number of introns (0–7) were noted in JAZ gene family of other plants such as rice and wheat61. Most of the dicots have at least one intron in JAZ genes, but the introns were lacking in monocots: rice, wheat, maize and sorghum36,62,63. The reason behind lacking introns in monocots might reflect the evolutionary difference from dicots35. The study suggests that fewer introns within a gene family will make plants more responsive to environmental stresses64,65. Therefore, sorghum JAZ, SbJAZ1-6 and SbJAZ8-10 may respond quickly to environmental stresses, for which further verification is needed.
Evolutionary relationship of the JAZ protein family
In this study, a total of 106 JAZ proteins from eight representative plants including a bryophyte lycopodiophyte, gymnosperms, dicots and monocots were used for phylogenetic analysis. These eight plant species belong to four plant terrestrial groups, which include the earliest land plants (bryophyte) to recently evolved C4 plants (sorghum). These JAZ family members are supposed to be originated from terrestrial plants because they were not found in algal genomes33. All the 106 JAZ proteins were clustered into 5 distinct groups (A–E) (Fig. 2). Within these groups, we found some lineage-specific JAZ sub-families. In group D, there was a separate clade only for bryophyte, and a clade for gymnosperms in Group B and C. Similarly, Group A showed exclusive clusters of monocots and dicots, which suggest that these genes might have evolved after the separation of monocots and dicots66. The grass family has accumulated many JAZ genes though duplication and transposon insertion46. The phylogeny and cross-species comparison with O. sativa (15 JAZs) revealed that a higher number in sorghum (18 JAZs) may be related to this extra duplication event in SbJAZ1-5. The phylogenetic tree (Fig. 2) forms a clade of SbJAZ2 with OsJAZ14 and SbJAZ3-5 with OsJAZ13, which suggests they are orthologs. Phylogenetic tree also reveals that the extra duplication events are SbJAZ4-5, which is further supported by their genomic structure as three SbJAZ3-5 had one exon which is similar to one exon of OsJAZ13 (Fig. 3).
Gene duplication event undergoes through either neutral, purifying and positive selection. In the SbJAZ gene family, evidence of neutral and purifying selection was obtained based on dN/dS and codon-based Z test (Table 2). Among six gene-pairs, five of them showed a neutral selection, indicating that the duplicated genes in these groups were not in strong selection pressure and many mutations evolved and remain in the population. These gene members could have developed more precise or new functions during the evolution46. One reason for most of the sorghum gene pairs showing neutral selection probably is that most of these pairs belong to tandem duplications, which tends to have a larger dN/dS ratio67. Interestingly, one of the gene pair (SbJAZ8-9) showed a purifying selection, indicating that they are evolving slowly at the protein level and suggests the stability of SbJAZ protein family during evolutionary process. Similar results of both purifying and neutral selection were reported in maize JAZs46.
Putative functions of the sorghum JAZ genes
The TFs are central regulators of the gene expression as it modulates the gene expression by binding to local and distal cis-acting elements of neighboring gene under different stresses68. The TFs regulatory network shows the sorghum JAZ genes are rich in ERF, TCP, NAC bHLH and MYB families (Fig. 7). All five TFs are induced during both biotic and abiotic stresses. Among them, NAC is one of the largest plant-specific TFs and acts via an ABA-dependent as well as independent pathway and play a vital role in both abiotic and biotic stress69. A previous study reported that NAC TFs was induced in sorghum during greenbug infestation70. Similarly, MYB TFs of sorghum and maize were also induced during fungal pathogen71 and leaf blight pathogen ingress72. The ERFs TFs play a vital role in ABA-independent pathways and are involved in both biotic and abiotic stress (drought and salt stress)68. The VaERF in grapes showed higher expression in response to Botrytis cinerea infection73. The other TFs, bHLH are induced during drought, osmotic stress and salt stress74. The sorghum JAZ genes rich with these TFs suggest the potential role of these genes being activated under biotic and abiotic stresses.
Expression profiling of sorghum JAZ during phytohormone treatment
Phytohormone signaling pathways are involved during plant-pest/pathogen interaction and several studies showed the repressor role of JAZ in defense signaling and in crosstalk between JA and other hormones12. For the expression analysis, RILs were used because one can phenotype multiple individuals from each RIL while reducing the individual, environmental and measurement variation75. The phytohormone treatment results (Fig. 8) showed that most of the sorghum JAZ genes were significantly upregulated in both the RIL 521 and RIL 609. During MeJA treatment, most of the SbJAZ genes were significantly upregulated, and in ABA treatments, SbJAZ8, SbJAZ9 and SbJAZ16 showed significant upregulation. Similarly, SbJAZ6 and SbJAZ16 were induced by the ET treatment and SbJAZ16 also showed significant upregulation during GA treatment. Among them, SbJAZ16 was upregulated during all the four phytohormone treatments. A similar kind of upregulation was reported in B. oleracea34 and C. sinensis76 during different phytohormone treatments. B. oleracea treated with MeJA showed significantly higher upregulation in eight BoJAZ genes out of 18, while when treated with ET, only two of them showed significant upregulation34. By combining the cis-elements results of this upregulated genes (Fig. 6), we found SbJAZ8 and SbJAZ9 were rich in ABA-responsive elements and SbJAZ16 were rich in the JA-, ABA- and GA-responsive elements. In contrast, both the SbJAZ6 and SbJAZ16 lacked the EA-responsive elements. Interestingly, all SbJAZ promoter regions have JA-responsive elements except SbJAZ4, SbJAZ8, SbJAZ15 and SbJAZ18. By combining the results of cis-elements and expression analysis we speculated that two SbJAZ genes (SbJAZ9 and SbJAZ16) were involved in JA-ABA crosstalk and SbJAZ16 in JA-GA crosstalk. Similar research conducted by Wang et al.36 in JAZ genes of T. aestivum indicated the crosstalk between JA-GA and JA-ABA with expression profiling of phytohormones treatment. They also reported the upregulation in a JAZ gene although they lack the respective cis-elements. In contrast, some of the SbJAZ genes contain abundant ABA (SbJAZ11-12) and MeJA (SbJAZ6 and SbJAZ12) responsive cis elements but showed low gene expression during their respective hormone treatment33. Similar low expression pattern was reported for CsJAZ genes during ABA and MeJA treatment though they were rich in the respective cis elements. One possible reason is that gene expression is not only determined by the presence of relevant cis elements but also by other physiological processes76. Similarly, besides promoter regions there are also several non-coding sequences in the front of the gene, which can either induce or suppress transcription of the gene. The JA-GA crosstalk is to promote plant growth and defense. Similarly, JA-ABA crosstalk acts synergistically to activate the defense-responsive genes against herbivory (Fig. 1B). Besides that, JA-ABA crosstalk also provides tolerance against salinity and drought and low temperature36. These results indicate that the SbJAZ genes can respond to JA, ABA and GA signaling, suggesting their role in the crosstalk between these phytohormones to promote defense gene activation34.
In both A. thaliana and T. aestivum, eight JAZ genes out of 12 and 21, respectively, were responsive to MeJA9,24,36. Similarly, in C. sinensis eight JAZ genes were significantly upregulated when treated with MeJA, among them four increased more than 30-fold76. The upregulation of JAZ genes induced by MeJA is controlled by short transcriptional cascades77. The upregulation of SbJAZ in both susceptible and resistant RILs during MeJA treatment implies the existence of a negative feedback loop to minimize the detrimental effects during hyper-immunity. In a negative feedback (Fig. 1A) loop, the newly synthesized JAZ repressor dampens the JA defense response by inhibiting the activity of MYC2 TFs9. Plant immune responses mediated by JA-Ile are metabolically costly and often linked to stunted growth26. Similarly, suppressed JAZ proteins further exacerbate the growth, nearly abolish the seed production and cause tissue death under extreme conditions26. The upregulation of SbJAZ genes in both RILs suggests its functions in minimizing the effects of hyper-immunity, promoting the growth and maintaining reproductive success.
Expression and functions of SbJAZ genes in response to SCA infestation
JAZ are key repressors in the JA signal transduction pathway and play a crucial role in stress-related defense and restraining a defense to balance growth. The SCA infestation expression analysis (Fig. 9) indicated that out of 18 SbJAZ genes, four of them (SbJAZ1, SbJAZ5, SbJAZ13 and SbJAZ16) were significantly upregulated in RIL 521 (resistant) in comparison to un-infested and RIL 609 (susceptible). This significantly higher expression of SbJAZ genes in resistant lines further supports the involvement of the JA pathway in host plant defense against SCA. Previous research showed significant upregulation of the lipoxygenase (LOX) genes in sorghum resistant lines during SCA infestation as the LOX gene is a marker gene for JA biosynthesis pathway78. All three SbJAZ genes (SbJAZ1, SbJAZ5, SbJAZ13) are in Group A clustered with four orthologous genes in A. thaliana (JAZ1, JAZ2, JAZ5, JAZ6) and B. oleracea (JAZ12, JAZ18, JAZ19, JAZ20), respectively, in a phylogenetic tree (Fig. 2). The remaining SbJAZ16 is clustered with AtJAZ10 and BoJAZ7 in Group E. All the five Arabidopsis JAZ were induced to high levels following mechanical wounding and Spodoptera exigua herbivory8,9,25. Similarly, all the five BoJAZ genes were significantly induced in resistant lines when infested with Plasmodiophora brassicae34. The BoJAZ7 clustered with SbJAZ16 is also significantly induced by Xanthomonas campestris and Fusarium oxysporum inoculation34. Four SbJAZ genes were also significantly increased during MeJA treatment in resistant lines (Fig. 8). Wound-induced expression of JAZ genes has been reported in Populus79, Solanum lycopersicom80 and Arabidopsis9, indicating that this phenomenon is conserved in the plant kingdom. The increased expression of JAZ proteins in resistant sorghum suggests a negative feedback loop (Fig. 1A) where the newly synthesized JAZ repressor dampens the JA response by inhibiting the activity of MYC2 TFs9. Overactivation of JA defense leads to carbon starvation, near loss of seed production and plant lethality (under extreme conditions)26. Interestingly, SbJAZ6 showed downregulation in the resistant line during SCA infestation. A similar result was reported in BoJAZ17 which is close to SbJAZ6 during F. oxysporum inoculation34. The inhibition of SbJAZ6 indicates the positive feedback loop (Fig. 1A) where the decrease in JAZ amplifies the plant capacity to release the MYC2 and thus expressing the defense genes9. Similar up and downregulation of the JAZ genes were noted in B. juncea during P. brassicae inoculation81. Previous research has also suggested the role of JAZ proteins in regulating plant processes that may confer resistance to insect herbivores through production of glucosinolate-based defenses9,82. In short, expression of JAZ genes helps in the regulation of plant defense, promotes growth and ensures reproductive success by restraining the immune response.
As shown in Fig. 6, the promoter regions of the five sorghum genes (SbJAZ1, SbJAZ5, SbJAZ6, SbJAZ13 and SbJAZ16) are rich in phytohormone motifs (ABA- and JA-responsive elements) and abiotic stress motifs (MYB, STRE). Among the five genes, SbJAZ5 consists of more motifs related to phytohormones and abiotic stresses, likely supported by its upregulation during MeJA treatment and SCA infestation in the resistant line. These results imply that these cis-elements present upstream of the gene regulates its expression under stresses. In the future, RNAi and CRISPR/Cas9 technology can be used on the potential SbJAZ genes from this study to develop a resistant cultivar against SCA. Similarly, yeast two-hybrid assay can be used to verify the functions of the potential SbJAZ genes by looking its interaction with MYC2 TFs. The role of SbJAZ during SCA infestation can also be validated by measuring the endogenous JA content and secondary metabolites during SCA infestation.
Conclusion
For the first time, this study has identified and characterized 18 JAZ genes from the sorghum genome through bioinformatic analysis and expression profiling. An in silico protein analysis showed the conserved TIFY and Jas domains in all SbJAZ genes, implying both the critical structural features and the conserved functions. Simultaneously, in silico analysis of the promoter region of SbJAZ genes revealed that the six sorghum JAZ genes were rich in cis-elements and TFs. These cis-elements and TFs are responsive to a variety of stresses and are verified in other plant species. The JAZ genes play an important role in phytohormone crosstalk to activate defense genes and are irreplaceable during wound and stress response. The expression profiling of phytohormone treatments in resistant (RIL 521) and susceptible (RIL 609) RILs to SCA has shown the possible role of SbJAZ9 in JA-ABA crosstalk and SbJAZ16 in both JA-ABA and JA-GA crosstalk to regulate certain physiological processes in plants. Notably, during JA treatment and SCA infestation, four SbJAZ genes (SbJAZ1, SbJAZ5, SbJAZ13 and SbJAZ16) showed strong expression in resistant RIL, implying their potential roles in stress response and regulating plant defense to balance the growth. Overall, these findings provide an insight into the important functions of the JAZ genes in host plant defense and genetic resources for genetic engineering in sorghum.
Supplementary Information
Acknowledgements
We thank Ms. Angela L. Phillips for her excellent technical assistance, and Dr. Jian Huang for his development of the sugarcane aphid resistance RIL population. This research was supported by the funding from the United States Department of Agriculture—Agricultural Research Service, CRIS project number: 3072-21000-009-00D (YH). USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Author contributions
Y.H. and K.S. designed the research and Y.H. supervised the entire study. K.S. performed genome analysis and all experiments, both K.S. and Y.H. interpreted the data, contributed to the discussion of the results and wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-07181-9.
References
- 1.Devoto A, Turner JG. Jasmonate-regulated Arabidopsis stress signalling network. Physiol. Plant. 2005;123:161–172. doi: 10.1111/j.1399-3054.2004.00418.x. [DOI] [Google Scholar]
- 2.Wasternack C, Hause B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013;111:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Browse J, Howe GA. New weapons and a rapid response against insect attack. Plant Physiol. 2008;146:832–838. doi: 10.1104/pp.107.115683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe GA, Jander G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 5.Boter M, et al. FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell. 2015;27:3160–3174. doi: 10.1105/tpc.15.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wasternack C. A plant’s balance of growth and defense—revisited. New Phytol. 2017;215:1291–1294. doi: 10.1111/nph.14720. [DOI] [PubMed] [Google Scholar]
- 7.Yu X, et al. The Jasmonate ZIM-domain protein gene SlJAZ2 regulates plant morphology and accelerates flower initiation in Solanum lycopersicum plants. Plant Sci. 2018;267:65–73. doi: 10.1016/j.plantsci.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung HS, et al. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146:952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katsir L, Chung HS, Koo AJ, Howe GA. Jasmonate signaling: A conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 2008;11:428–435. doi: 10.1016/j.pbi.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chini A, Boter M, Solano R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009;276:4682–4692. doi: 10.1111/j.1742-4658.2009.07194.x. [DOI] [PubMed] [Google Scholar]
- 12.Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 13.Shan X, Yan J, Xie D. Comparison of phytohormone signaling mechanisms. Curr. Opin. Plant Biol. 2012;15:84–91. doi: 10.1016/j.pbi.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Abe M, Shibaoka H, Yamane H, Takahashi N. Cell cycle-dependent disruption of microtubules by methyl jasmonate in tobacco BY-2 cells. Protoplasma. 1990;156:1–8. doi: 10.1007/BF01666500. [DOI] [Google Scholar]
- 15.Anderson JP, et al. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16:3460–3479. doi: 10.1105/tpc.104.025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. The tify family previously known as ZIM. Trends Plant Sci. 2007;12:239–244. doi: 10.1016/j.tplants.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Bai Y, Meng Y, Huang D, Qi Y, Chen M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics. 2011;98:128–136. doi: 10.1016/j.ygeno.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 19.Geerinck J, Pauwels L, De Jaeger G, Goossens A. Dissection of the one-MegaDalton JAZ1 protein complex. Plant Signal. Behav. 2010;5:1039–1041. doi: 10.4161/psb.5.8.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melotto M, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine-and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55:979–988. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011;62:2143–2154. doi: 10.1093/jxb/erq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández-Calvo P, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thines B, et al. JAZ repressor proteins are targets of the SCF COI1 complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 24.Chung HS, Niu Y, Browse J, Howe GA. Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry. 2009;70:1547–1559. doi: 10.1016/j.phytochem.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S-J, Huang Y, Ayoubi P. Identification of expression profiles of sorghum genes in response to greenbug phloem-feeding using cDNA subtraction and microarray analysis. Planta. 2006;223:932–947. doi: 10.1007/s00425-005-0148-1. [DOI] [PubMed] [Google Scholar]
- 26.Guo Q, et al. JAZ repressors of metabolic defense promote growth and reproductive fitness in Arabidopsis. Proc. Natl. Acad. Sci. 2018;115:E10768–E10777. doi: 10.1073/pnas.1811919115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.USDA, N. Quick stats USDA National Agricultural Statistics Service. Accessed 3 May 2019. https://quickstats.nass.usda.gov/ (Washington, DC, 2019).
- 28.Bowling RD, et al. Sugarcane aphid (Hemiptera: Aphididae): A new pest on sorghum in North America. J. Integr. Pest Manag. 2016;7:12. doi: 10.1093/jipm/pmw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh B, Padmaja P, Seetharama N. Biology and management of the sugarcane aphid, Melanaphis sacchari (Zehntner) (Homoptera: Aphididae), in sorghum: A review. Crop Prot. 2004;23:739–755. doi: 10.1016/j.cropro.2004.01.004. [DOI] [Google Scholar]
- 30.Narayana D. Screening for aphids and sooty molds in sorghum. Sorghum Newsl. 1975;18:21–22. [Google Scholar]
- 31.Carena M, Glogoza P. Resistance of maize to the corn leaf aphid: A review. Maydica. 2004;49:241–254. [Google Scholar]
- 32.Li X, et al. Genome-wide identification and analysis of the apple (Malus × domestica Borkh.) TIFY gene family. Tree Genet. Genomes. 2015;11:808. doi: 10.1007/s11295-014-0808-z. [DOI] [Google Scholar]
- 33.Zheng Y, et al. Genome-wide and expression pattern analysis of JAZ family involved in stress responses and postharvest processing treatments in Camellia sinensis. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, et al. Genome-wide identification, expression profile of the TIFY gene family in Brassica oleracea var. capitata, and their divergent response to various pathogen infections and phytohormone treatments. Genes. 2020;11:127. doi: 10.3390/genes11020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian S, et al. Genome-wide identification and characterization of JAZ protein family in two Petunia progenitors. Plants. 2019;8:203. doi: 10.3390/plants8070203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, et al. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.) BMC Genom. 2017;18:152. doi: 10.1186/s12864-017-3582-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quevillon E, et al. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu B, et al. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anand, L. chromoMap: An R package for interactive visualization and annotation of chromosomes. BioRxiv, 605600 (2019). [DOI] [PMC free article] [PubMed]
- 40.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 41.Zuckerkandl E, Pauling L. Evolving Genes and Proteins. Elsevier; 1965. Evolutionary divergence and convergence in proteins; pp. 97–166. [Google Scholar]
- 42.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Upadhyay RK, Mattoo AK. Genome-wide identification of tomato (Solanum lycopersicum L.) lipoxygenases coupled with expression profiles during plant development and in response to methyl-jasmonate and wounding. J. Plant Physiol. 2018;231:318–328. doi: 10.1016/j.jplph.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:1–21. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han Y, Luthe D. Identification and evolution analysis of the JAZ gene family in maize. BMC Genom. 2021;22:1–21. doi: 10.1186/s12864-021-07522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 48.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Team, R. C. R: A language and environment for statistical computing (2013).
- 50.Wang P, et al. Genome-wide identification of WOX genes and their expression patterns under different hormone and abiotic stress treatments in tea plant (Camellia sinensis) Trees. 2019;33:1129–1142. doi: 10.1007/s00468-019-01847-0. [DOI] [Google Scholar]
- 51.Kohl M, Wiese S, Warscheid B. Data Mining in Proteomics. Springer; 2011. Cytoscape: Software for visualization and analysis of biological networks; pp. 291–303. [DOI] [PubMed] [Google Scholar]
- 52.Scully ED, et al. Overexpression of SbMyb60 impacts phenylpropanoid biosynthesis and alters secondary cell wall composition in Sorghum bicolor. Plant J. 2016;85:378–395. doi: 10.1111/tpj.13112. [DOI] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Sagnard F, et al. Genetic diversity, structure, gene flow and evolutionary relationships within the Sorghum bicolor wild–weedy–crop complex in a western African region. Theor. Appl. Genet. 2011;123:1231. doi: 10.1007/s00122-011-1662-0. [DOI] [PubMed] [Google Scholar]
- 55.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharp AJ, et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu Z, Ricci WA, Schmitz RJ, Zhang X. Identification of cis-regulatory elements by chromatin structure. Curr. Opin. Plant Biol. 2018;42:90–94. doi: 10.1016/j.pbi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 58.Pant S, Huang Y. Elevated production of reactive oxygen species is related to host plant resistance to sugarcane aphid in sorghum. Plant Signal. Behav. 2021;16:1849523. doi: 10.1080/15592324.2020.1849523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paudyal S, et al. Categories of resistance to sugarcane aphid (Hemiptera: Aphididae) among sorghum genotypes. J. Econ. Entomol. 2019;112:1932–1940. doi: 10.1093/jee/toz077. [DOI] [PubMed] [Google Scholar]
- 60.Chao J, et al. Genome-wide identification and characterization of the JAZ gene family in rubber tree (Hevea brasiliensis) Front. Genet. 2019;10:372. doi: 10.3389/fgene.2019.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duan L, Mu X, Li W. Molecular evolution of transcriptional repressor JAZ protein family in jasmonic acid signaling pathway. Chin. Bull. Bot. 2013;48:623. [Google Scholar]
- 62.Zhang F, et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature. 2015;525:269–273. doi: 10.1038/nature14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ye H, Du H, Tang N, Li X, Xiong L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009;71:291–305. doi: 10.1007/s11103-009-9524-8. [DOI] [PubMed] [Google Scholar]
- 64.Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24:375–378. doi: 10.1016/j.tig.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Figueroa P, Browse J. Male sterility in Arabidopsis induced by overexpression of a MYC 5-SRDX chimeric repressor. Plant J. 2015;81:849–860. doi: 10.1111/tpj.12776. [DOI] [PubMed] [Google Scholar]
- 66.Chen Z, et al. The lipoxygenase gene family in poplar: Identification, classification, and expression in response to MeJA treatment. PLoS One. 2015;10:e0125526. doi: 10.1371/journal.pone.0125526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W-H, Graur D. Fundamentals of Molecular Evolution. Sinauer Ass. Inc.; 1991. [Google Scholar]
- 68.Baillo EH, Kimotho RN, Zhang Z, Xu P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes. 2019;10:771. doi: 10.3390/genes10100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakashima K, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, H. & Huang, Y. Genome-wide survey and characterization of greenbug induced nac transcription factors in sorghum [Sorghum bicolor (L.) Moench]. Plant Anim. Genom. Conf. (PAG XXI), P0191 (2013).
- 71.Baldoni E, Genga A, Cominelli E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015;16:15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ibraheem F, Gaffoor I, Tan Q, Shyu C-R, Chopra S. A sorghum MYB transcription factor induces 3-deoxyanthocyanidins and enhances resistance against leaf blights in maize. Molecules. 2015;20:2388–2404. doi: 10.3390/molecules20022388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu Y, et al. Genome-wide identification and expression analysis reveal the potential function of ethylene responsive factor gene family in response to Botrytis cinerea infection and ovule development in grapes (Vitis vinifera L.) Plant Biol. 2019;21:571–584. doi: 10.1111/plb.12943. [DOI] [PubMed] [Google Scholar]
- 74.Di C, et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014;80:848–861. doi: 10.1111/tpj.12679. [DOI] [PubMed] [Google Scholar]
- 75.Broman KW. The genomes of recombinant inbred lines. Genetics. 2005;169:1133–1146. doi: 10.1534/genetics.104.035212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen J, et al. Genome-wide analysis reveals stress and hormone responsive patterns of JAZ Family genes in Camellia sinensis. Int. J. Mol. Sci. 2020;21:2433. doi: 10.3390/ijms21072433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hickman R, et al. Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell. 2017;29:2086–2105. doi: 10.1105/tpc.16.00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shrestha K, Pant S, Huang Y. Genome-wide identification and classification of Lipoxygenase gene family and their roles in sorghum-aphid interaction. Plant Mol. Biol. 2021;105:527–541. doi: 10.1007/s11103-020-01107-7. [DOI] [PubMed] [Google Scholar]
- 79.Major IT, Constabel CP. Molecular analysis of poplar defense against herbivory: Comparison of wound-and insect elicitor-induced gene expression. New Phytol. 2006;172:617–635. doi: 10.1111/j.1469-8137.2006.01877.x. [DOI] [PubMed] [Google Scholar]
- 80.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cai Z, Chen Y, Liao J, Wang D. Genome-wide identification and expression analysis of jasmonate ZIM domain gene family in tuber mustard (Brassica juncea var. tumida) PLoS One. 2020;15:e0234738. doi: 10.1371/journal.pone.0234738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mewis I, et al. Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry. 2006;67:2450–2462. doi: 10.1016/j.phytochem.2006.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.