Abstract
Glucose homeostasis, regulated by glucose-dependent insulinotropic polypeptide (GIP), glucagon-like peptide-1 (GLP-1) and glucagon (GCG) is critical to human health. Several multi-targeting agonists at GIPR, GLP-1R or GCGR, developed to maximize metabolic benefits with reduced side-effects, are in clinical trials to treat type 2 diabetes and obesity. To elucidate the molecular mechanisms by which tirzepatide, a GIPR/GLP-1R dual agonist, and peptide 20, a GIPR/GLP-1R/GCGR triagonist, manifest their multiplexed pharmacological actions over monoagonists such as semaglutide, we determine cryo-electron microscopy structures of tirzepatide-bound GIPR and GLP-1R as well as peptide 20-bound GIPR, GLP-1R and GCGR. The structures reveal both common and unique features for the dual and triple agonism by illustrating key interactions of clinical relevance at the near-atomic level. Retention of glucagon function is required to achieve such an advantage over GLP-1 monotherapy. Our findings provide valuable insights into the structural basis of functional versatility of tirzepatide and peptide 20.
Subject terms: Hormone receptors, Cryoelectron microscopy, G protein-coupled receptors, Receptor pharmacology
Multi-targeting agonists at GIPR, GLP-1R or GCGR are pursued vigorously. Here, the authors report cryo-EM structures of tirzepatide-bound GIPR and GLP-1R, peptide 20-bound GIPR, GLP-1R and GCGR, revealing the molecular basis of their multiplexed pharmacological actions.
Introduction
Glucose-dependent insulinotropic polypeptide (also known as gastric inhibitory peptide, GIP), glucagon-like peptide-1 (GLP-1) and glucagon (GCG) are peptide hormones responsible for glucose homeostasis1,2. Their cognate receptors, GIPR, GLP-1R and GCGR, belong to class B1 G protein-coupled receptor (GPCR) family. Successful application of various GLP-1 mimetics to treat type 2 diabetes mellitus (T2DM) and obesity highlights the clinical value of this group of drug targets3. However, development of GIPR- and GCGR-based therapeutics has encountered drawbacks due to the complexity of physiology associated with GIP and GCG4–6. For example, GIP stimulates insulin secretion but also increases GCG levels7,8, while the latter has a parallel role in elevating energy expenditure and blood glucose9.
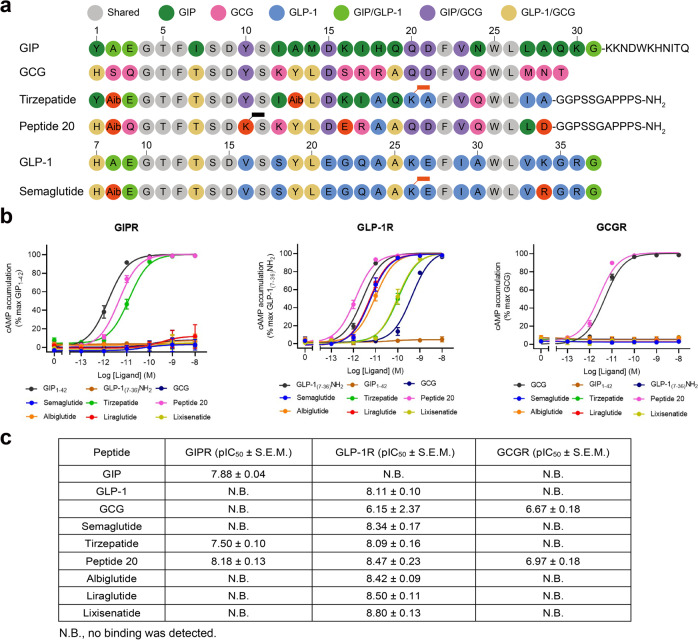
It was reported that the weight loss property of most GLP-1 analogs, except for semaglutide administered subcutaneously once weekly of smoother pharmacokinetics, is hampered by the therapeutic window because of the dose-dependent side-effects10,11. Chimeric peptides consisting of amino acids from GIP and GLP-1 were then designed to maximize their metabolic benefits12. Additional consideration was given to GCG for its role in energy expenditure13. Therefore, multi-targeting or unimolecular peptides possessing combinatorial agonism at GIPR, GLP-1R and GCGR have been extensively explored and more than a dozen peptides including two GIPR/GLP-1R dual agonists, ten GLP-1R/GCGR dual agonists and five GIPR/GLP-1R/GCGR triagonists have entered into clinical development (Supplementary Fig. 1a and Supplementary Table 1)14. Of them, two pioneered multi-targeting agonists, tirzepatide (LY3298176) and peptide 20 (MAR423) have attracted significant attention from both academic and industrial communities (Fig. 1a). Tirzepatide is an investigational once-weekly GIPR/GLP-1R dual agonist15 with a profound therapeutic superiority in reducing blood glucose and body weight beyond several approved drugs such as semaglutide16 and dulaglutide17 in multiple head-to-head clinical trials. Peptide 20, a GIPR/GLP-1R/GCGR triagonist (phase 1 clinical trials completed)18–20 with balanced potency at the three receptors, is evolved from a GLP-1R/GCGR dual agonist21 through iterative sequence refinement and modification (Supplementary Fig. 1b)22. It reversed glucose dysregulation without detrimental effects on metabolically healthy animals and reduced body weight, lowered fasting blood glucose, decreased glycosylated hemoglobin (HbA1C), improved glucose tolerance, and protected pancreatic islet architecture in diabetic fatty Zucker rats22–24.
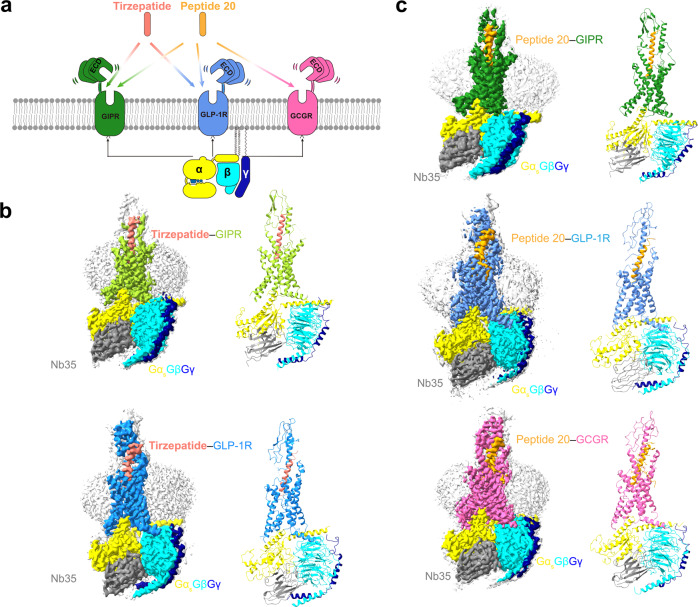
Fig. 1. Cryo-EM structures of tirzepatide and peptide 20-bound GIPR, GLP-1R and GCGR in complex with Gs.
a Multi-targeting peptides tirzepatide and peptide 20 possess distinct combinatorial agonism at GIPR, GLP-1R and GCGR. b Cryo-EM maps (left) and structural models (right) of tirzepatide-bound GIPR (top) and GLP-1R (bottom) in complex with Gs. The sharpened cryo-EM density map at the 0.243 threshold shown as light gray surface indicates a micelle diameter of 10 nm. The colored cryo-EM density map is shown at the 0.424 threshold. The tirzepatide is shown in salmon, GIPR in yellow green, GLP-1R in dodger blue, Gαs in yellow, Gβ subunit in cyan, Gγ subunit in navy blue and Nb35 in gray. c Cryo-EM maps (left) and structural models (right) of peptide 20-bound GIPR (top), GLP-1R (middle) and GCGR (bottom) in complex with Gs. The sharpened cryo-EM density map at the 0.228 threshold shown as light gray surface indicates a micelle diameter of 11 nm. The colored cryo-EM density map is shown at the 0.576 threshold. The peptide 20 is shown in orange, GIPR in forest green, GLP-1R in blue, GCGR in hot pink, Gαs in yellow, Gβ subunit in cyan, Gγ subunit in navy blue and Nb35 in gray.
The aim of this work is to understand molecular mechanisms of the dual and triple agonism conferred by tirzepatide and peptide 20. Thus, we determine five cryo-electron microscopy (cryo-EM) structures, including GIPR and GLP-1R bound with tirzepatide and GIPR, GLP-1R and GCGR bound with peptide 20, all in complex with Gs proteins at global resolutions of 3.4 Å, 3.4 Å, 3.1 Å, 3.0 Å and 3.5 Å, respectively. Integrated with pharmacological and clinical data, this work reveals the structural basis of peptide recognition by each receptor and provides important information for the design of better drugs through combinatorial agonism.
Results
Overall structure
The tirzepatide–GIPR–Gs, tirzepatide–GLP-1R–Gs, peptide 20–GIPR–Gs, peptide 20–GLP-1R–Gs and peptide 20–GCGR–Gs structures were determined by the single-particle cryo-EM approach with overall resolutions of 3.4 Å, 3.4 Å, 3.1 Å, 3.0 Å, and 3.5 Å, respectively (Fig. 1b, c, Supplementary Figs. 2–7, Supplementary Tables 2, 3). Apart from the α-helical domain of Gαs, the presence of bound tirzepatide and peptide 20, individual receptor and heterotrimeric Gs in respective complex was clearly visible in all five EM maps, thereby allowing unambiguous modeling of the secondary structure and side chain orientation of all major components of the complexes (Supplementary Fig. 6).
Tirzepatide has two non-coded amino acid residues at positions 2 and 13 (Aib, α-aminoisobutyric acid), and is acylated on K20P (P indicates that the residue belongs to the peptide) with a γGlu-2×OEG linker and C18 fatty diacid moiety. The first 30 and 29 amino acids of tirzepatide were modeled for the tirzepatide–GIPR–Gs and tirzepatide–GLP-1R–Gs complexes, respectively.
Peptide 20 contains two modifications: A2P with Aib and K10P that is covalently attached by a 16-carbon acyl chain (palmitoyl; 16:0) via a gamma carboxylate (γE spacer)22. The γE spacer and palmitic acid (C16:0) were well resolved in the final models of peptide 20–GLP-1R–Gs and peptide 20–GCGR–Gs, while only the γE spacer was modeled for peptide 20–GIPR–Gs with high-resolution features. The first 30, 29, and 28 amino acids of peptide 20 were modeled for the peptide 20–GIPR–Gs, peptide 20–GLP-1R–Gs and peptide 20–GCGR–Gs complexes, respectively.
As shown in Fig. 2a, the tirzepatide–GIPR–Gs and peptide 20–GIPR–Gs complex structures closely resembled that of the GIP–GIPR–Gs complex25 with Cα root mean square deviation (RMSD) values of 0.5 and 0.4 Å, respectively. Notable conformational differences were observed in the positions of peptide C-terminal half and the surrounding extracellular loop 1 (ECL1) and extracellular domain (ECD), indicative of GIPR-associated ligand specificity. Through two mutations (M14PL and H18PA), the dense contacts between ECL1 (residues 194 to 211) and GIP were disrupted by peptide 20, as seen from the buried surface area that decreased from 406 Å2 for GIP to 278 Å2 for peptide 20. Consequently, ECL1 adopted a more relaxed conformation, making peptide 20 straighter by shifting its tip toward the transmembrane domain (TMD) core by 4.2 Å (measured by the Cα of L27P). Similar movement was also seen for the C-terminal half of tirzepatide (2.1 Å measured by the Cα of I27P). As far as the N terminus is concerned, GIP and tirzepatide were stabilized by multiple strong contacts with the TMD core through a common N terminus (Y1P-A/Aib2P-E3P), while that of peptide 20 (H1P-Aib2P-Q3P) formed weaker interactions with the TMD core by abolishing the hydrogen bond with Q2243.37b (class B GPCR numbering in superscript)26, salt bridge with R1832.60b and hydrophobic contacts with V2273.40b (Fig. 2b). Such deficiency of peptide 20 was rescued by the introduction of T7P (hydrogen bond with R1902.67b), lipidated K10P and Y13P that contributed additional contacts with GIPR not observed in GIP25. The hydrogen bond between T7P and R1902.67b was also found in the tirzepatide–GIPR–Gs complex.
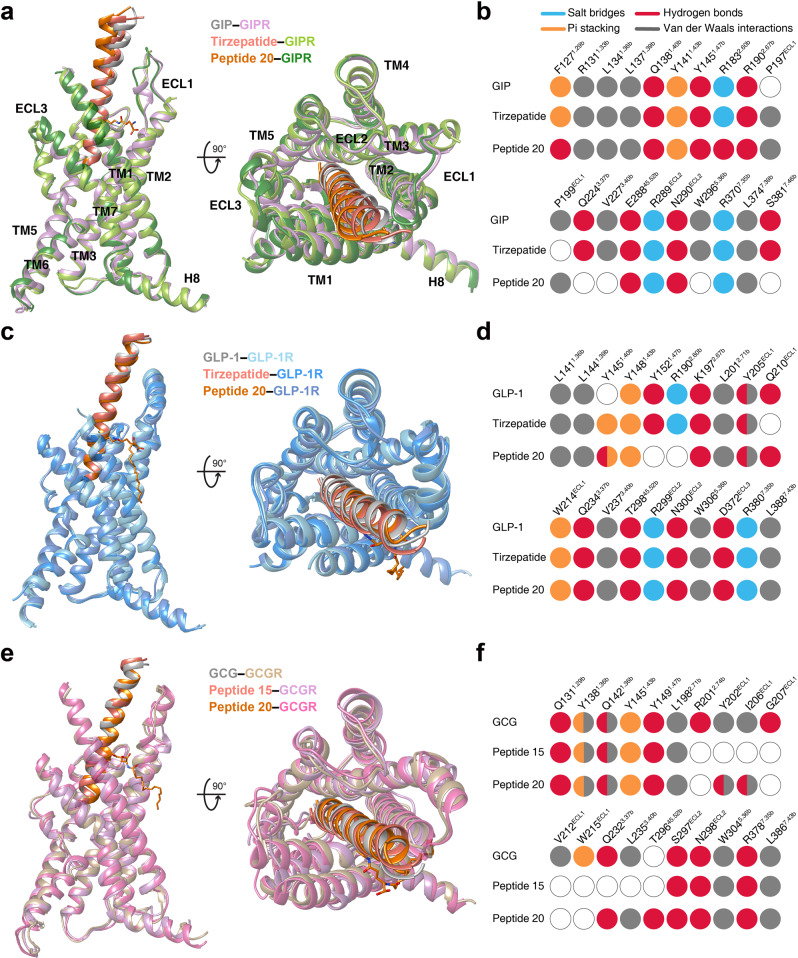
Fig. 2. Structural comparison of GIPR, GLP-1R and GCGR bound by mono-, dual and triple agonists.
a Structural comparison of GIP–GIPR–Gs25, tirzepatide–GIPR–Gs and peptide 20–GIPR–Gs. Receptor ECD and G protein are omitted for clarity. b Comparison of residue interactions employed by GIPR to recognize GIP, tirzepatide and peptide 20, described by fingerprint strings encoding different interaction types of the surrounding residues in each peptide. Color codes are listed on the top panel. Residues that show no interaction with ligands are displayed as white circles. c Structural comparison of GLP-1–GLP-1R–Gs27, tirzepatide–GLP-1R–Gs and peptide 20–GLP-1R–Gs. Receptor ECD and G protein are omitted for clarity. d Comparison of residue interactions that GLP-1R employed to recognize GLP-1, tirzepatide and peptide 20, described by fingerprint strings encoding different interaction types of the surrounding residues in each peptide. e Structural comparison of GCG–GCGR–Gs4, peptide 15–GCGR–Gs28 and peptide 20–GCGR–Gs. Receptor ECD and G protein are omitted for clarity. f Comparison of residue interactions that GCGR employed to recognize GCG, peptide 15 and peptide 20, described by fingerprint strings encoding different interaction types of the surrounding residues in each peptide.
The structures of tirzepatide- and peptide 20-bound GLP-1R are highly similar to that bound by GLP-127, with Cα RMSD of 0.8 Å and 0.7 Å, respectively (Fig. 2c). The bound peptides (GLP-1, tirzepatide and peptide 20) overlapped well and penetrated into the receptor TMD core by an identical angle and orientation, thereby exploiting a similar ligand recognition pattern for most residues except for a few positions that have distinct amino acids (Fig. 2c, Supplementary Tables 4, 5). The substitution (Y10P in tirzepatide) and modification (lipidated K10P in peptide 20) stabilized the binding of dual and triple agonists by newly-formed interactions with residues surrounding the TM1-TM2 cleft, a phenomenon unseen in the case of GLP-127. Meanwhile, some favorable interactions in GLP-1 recognition were absent for both tirzepatide (Y13PA decreased the hydrophobic interactions with TM1, E21PA broke the hydrogen bond with Q210ECL1) and peptide 20 (E3PQ eliminated the salt bridge with R1902.60b) (Fig. 2d). Interestingly, the residues at multiple positions (12, 16, 17, 20, 21, 24 and 28) of the multi-targeting agonists are highly solvent-accessible and of limited contact with GLP-1R, allowing them to employ distinct amino acids from GLP-1 without altering GLP-1R signaling profiles. As a comparison, superimposing either GIP or GCG with GLP-1 analogs suggest that they have potential steric clashes with ECL1 of GLP-1R via H18P of GIP and R18P of GCG. Two residues with shorter side chains (I7P and A13P) in GIP further weakened its binding to GLP-1R, consistent with the distinct cross-reactivity features of GIP and GCG with GLP-1R5,6.
Superimposing the structures of GCGR–Gs bound by GCG4, peptide 15 (GLP-1R and GCGR dual agonist)28 and peptide 20 reveals that these three peptides adopt a similar binding pose: a single continuous helix that penetrates into the TMD core through their N-terminal halves (residues 1 to 15), while the C-terminal halves (residues 16 to 30) are recognized by the ECD, ECL1 and TM1 (Fig. 2e). Given that both peptide 15 and peptide 20 are modified forms of GCG (differed by 7 residues), ligand recognition patterns are highly conserved across the three peptides except for a few positions. For example, by choosing alanine at position 18 instead of arginine in GCG, peptide 20 lost the cation-pi stacking with W215ECL1 and hydrogen bond with Q204ECL1, thereby allowing its outward movement toward ECL1 and leading to the formation of another hydrogen bond (D21P-I206ECL1) (Fig. 2f). Probably due to the lack of complementary interacting residues, superimposing GIP or GLP-1 to GCG bound to GCGR shows that the aligned peptides reduced favorable contacts with GCGR compared to GCG by removing one hydrogen bond (Y10P(GCG)/Y10P(GIP)/V16P(GLP-1)-Q1421.40b(GCGR)) and pi-pi stacking (Y13P(GCG)/A13P(GIP)/Y19P(GLP-1)-Y1381.36b(GCGR)) and by repulsing the interaction between Y1P(GIP) and I2353.40b(GCGR). These observations receive the support of our current and previous functional data showing that both GIP and GLP-1 were unable to activate GCGR (Supplementary Table 6)5,6.
Collectively, the binding mode comparison of the three peptides bound by the same receptor demonstrates common structural features in ligand recognition and distinct conformational adaptability of GIPR, GLP-1R and GCGR in response to different agonist stimulation.
Recognition of tirzepatide
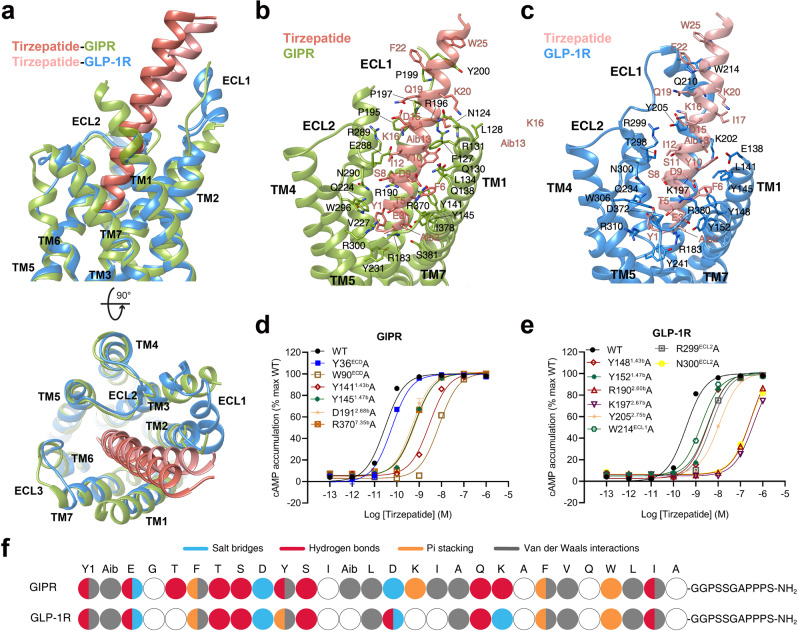
The tirzepatide–GIPR–Gs and tirzepatide–GLP-1R–Gs exhibit a similar peptide-receptor binding interface, where distinct structural features were observed at ECL1, ECL3 and the extracellular tips of TM1 and TM3 (Fig. 3a). GIPR-bound tirzepatide is rotated by 8.3° compared to that in complex with GLP-1R, such a movement shifted its C terminus toward the TMD core by 5.2 Å (measured by the Cα of I27P). The N-terminal region of tirzepatide (residues 1 to 10) in GIPR and GLP-1R overlapped well with the formation of a network of extensive interactions with multiple conserved residues (Y1.43b, Y1.47b, R190/K1972.67b, Q3.37b, V3.40b, N290/N300ECL2, R7.35b and I378/L3887.43b) (Fig. 3b–e, Supplementary Tables 4, 7). Notably, the inward movement of GIPR R3005.40b contributed one hydrogen bond with T5P (Fig. 3b, f). The middle region of tirzepatide in GLP-1R was stabilized by the peptide-ECD-ECL1-ECL2 interface through both a polar network (T29845.52b-S11P-Y205ECL1-R299ECL1-D15P-L32ECD-S31ECD-Q19P) and a complementary nonpolar network with ECD (L32, V36, W39 and Y88), ECL1 (W214), TM1 (L1411.36b) and TM2 (L2012.71b) via S11P, Aib13P, F22P, W25P and L26P (Fig. 3c). As a comparison, the ECL1 of GIPR partially unwound with the presence of three proline residues (P195ECL1, P197ECL1 and P199ECL1), resulting in reduced interactions between ECL1 and tirzepatide compared to that in GLP-1R (Fig. 3b). However, the α-helical extension in TM1 of GIPR provides additional residues for tirzepatide recognition including one hydrogen bond (Y10P and Q1381.40b) and a stacking interaction (K16P and F1271.29b). These observations receive support of our mutagenesis studies by measuring both cAMP responses and receptor binding affinities (Fig. 3d, e and Supplementary Table 7). The acylation on K20P by γGlu-2×OEG linker and C18 fatty diacid moiety that enables enhanced binding to plasma albumin and extended the peptide half-life in vivo29 were not resolved in both structures, indicating a high conformational flexibility, in line with the recently published cryo-EM structure of semaglutide-bound GLP-1R30 and our molecular dynamics (MD) simulation results (Supplementary Fig. 8a–c). Consistently, the non-acylated tirzepatide maintained high affinity and potency to both GLP-1R and GIPR (Supplementary Fig. 2f, g). Besides, GIPR has a relatively larger conformational difference between tirzepatide-bound and non-acylated tirzepatide-bound structures in most regions compared with GLP-1R (Supplementary Fig. 7c).
Fig. 3. Molecular recognition of tirzepatide by GIPR and GLP-1R.
a Structural comparison of tirzepatide–GIPR–Gs and tirzepatide–GLP-1R–Gs. Receptor ECD and G protein are omitted for clarity. b Interactions between tirzepatide (salmon) and the TMD of GIPR (yellow green). Residues involved in interactions are shown as sticks. c Interactions between tirzepatide (light salmon) and the TMD of GLP-1R (dodger blue). Residues involved in interactions are shown as sticks. d–e Effects of receptor mutations on tirzepatide-induced cAMP accumulation. Data shown are means ± S.E.M. of three independent experiments (n = 3) performed in quadruplicate. f The peptide recognition modes are described by fingerprint strings encoding different interaction types of the surrounding residues in each receptor. Residues that show no interaction with receptors are displayed as white circles. Color codes are listed on the top panel. WT, wild-type. Source data are provided as a Source Data file.
Peptide 20 recognition
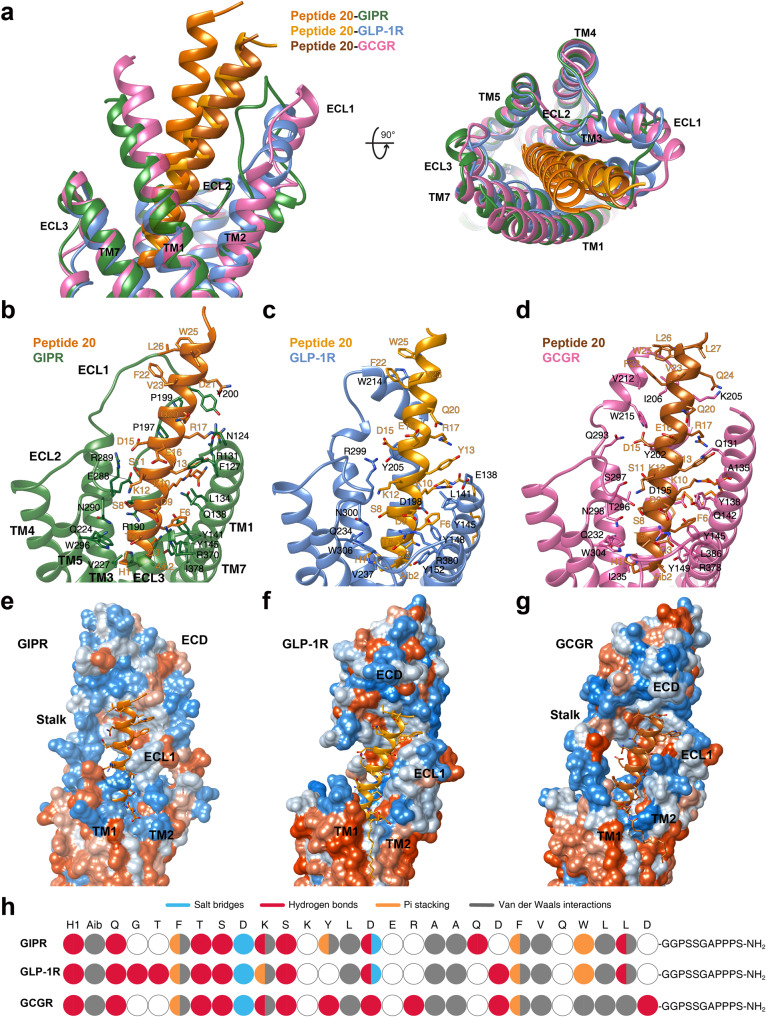
Superimposition of the TMDs of GIPR, GLP-1R and GCGR bound by peptide 20 shows that the three receptors employed conserved residues in the lower half of the TMD pocket to recognize the well-overlapped peptide N-terminal region (residues 1 to 11), while the peptide C terminus, engaged by ECL1, the N-terminal α-helix of ECD and the extracellular tip of TM1, displays receptor-specific positions and orientations (Fig. 4). Accompanying the inward movement of GIPR ECL1 by 6.4 Å relative to that of GCGR (measured by Cα of G202ECL1 in GIPR and G207ECL1 in GCGR), the C terminus of peptide 20 bound by GIPR shifted toward the TMD core by 8.1 Å (measured by Cα of L27P) and consequently pushed the extracellular tip of TM1 towards TM7 by 2.8 Å (measured by Cα of the residues at 1.29b). ECL1 and ECD of the three receptors coincidently constructed a complementary binding groove for the entrance of the C terminus of peptide 20 through multiple hydrophobic residues (A19P, F22P, V23P, W25P, L26P and L27P). However, several additional interactions were observed in GLP-1R (S11P-Y205ECL1 and D21P-Q210ECL1) and GCGR (D15P-Y202ECL1 and D21P-I206ECL1), but not in GIPR (Fig. 4b–h, Supplementary Table 5). Measurements of cAMP responses and ligand binding affinities in mutated GIP, GLP-1 and GCG receptors yielded similar results (Supplementary Fig. 9 and Supplementary Tables 8, 9).
Fig. 4. Molecular recognition of peptide 20 by GIPR, GLP-1R and GCGR.
a Structural comparison of peptide 20–GIPR–Gs, peptide 20–GLP-1R–Gs and peptide 20–GCGR–Gs. Receptor ECD and G protein are omitted for clarity. b–d Interactions between peptide 20 and the TMDs of GIPR (forest green), GLP-1R (blue), and GCGR (hot pink). Residues involved in interactions are shown as sticks. e–g Surface representations of the receptor for each of the peptide-receptor complex, with the peptides shown as ribbon and sticks. The receptor is shown in surface representation and colored from dodger blue for the most hydrophilic region, to white, to orange red for the most hydrophobic region. h The peptide recognition modes are described by fingerprint strings encoding different interaction types of the surrounding residues in each receptor. Color codes are listed on the top panel. Residues that show no interaction with receptors are displayed as white circles.
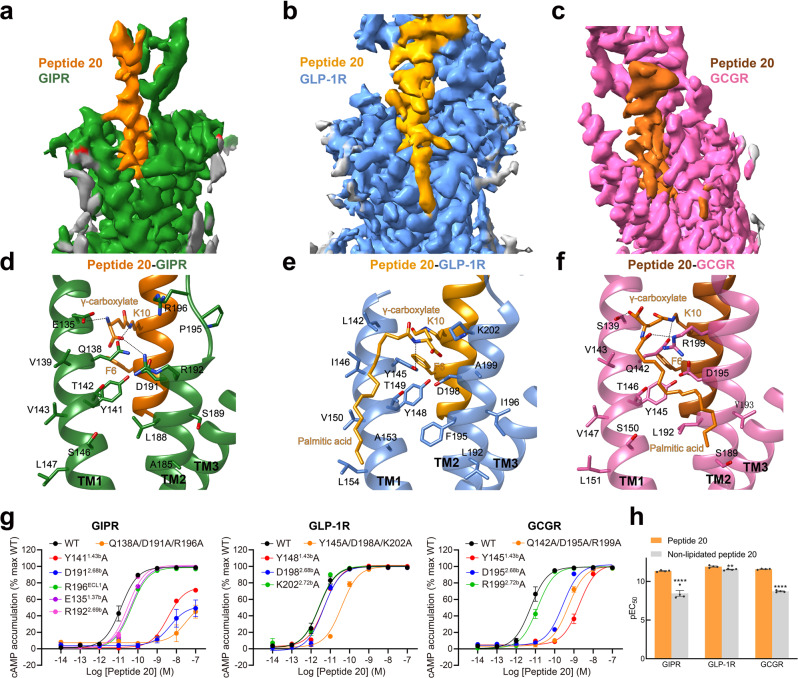
Notably, strong cryo-EM densities were observed in the crevices between TM1 and TM2 of the three complexes (Fig. 5a–c). They were connected to the side chain end of K10P of peptide 20, allowing unambiguous assignment of the binding sites of lipidated K10P with a 16-carbon palmitic acid through a γ-carboxylate spacer (Fig. 5d–f). Such a modification on K10P greatly stabilized the peptide binding through extensive contacts with both receptors and lipid membrane. For GCGR, the lipidated K10P contributed three hydrogen bonds (with S1391.37b, Q1421.40b and R1992.72b), extensive hydrophobic contacts with V1431.41b, T1461.44b, L1922.65b, V1932.66b, and membrane lipids where the 16-carbon palmitic chain surrounded (Fig. 5d–f). Removal of these contacts by GCGR triple mutant (Q142A + D195A + R199A) markedly reduced peptide 20 potency by 93-fold (Fig. 5g). For GLP-1R, the γ-carboxylate spacer formed two hydrogen bonds (with Y1451.40b and D1982.68b), and the 16-carbon palmitic chain terminus dropped down along TM1 with the formation of multiple strong hydrophobic interactions with I1461.41b, T1491.44b, V1501.45b, A1531.48b and L1541.49b. Similar phenomenon was also observed in GIPR. Consistently, regardless of the initial positions of palmitate either located in the membrane as seen in the cryo-EM structure or placed outside of the membrane, our MD simulations found that the γ-carboxylate spacer stably inserted into the TM1-TM2 cleft and the 16-carbon palmitic chain is deeply buried in the receptor-lipid interface, contributing multiple strong contacts to stabilize the complexes (Supplementary Fig. 10). The importance of K10P lipidation receives the support of our structure-activity relationship study where peptide 20 without K10P lipidation reduced the receptor-mediated cAMP accumulation by 1,288-fold and 759-fold for GIPR and GCGR in the presence of 0.1% bovine serum albumin (BSA), respectively, but inappreciably influenced that of GLP-1R (Fig. 5h, Supplementary Table 10). Similar phenomenon was observed in the presence of 0.1% casein. These results suggest that specific modification of peptide is equally significant to sequence optimization in terms of demonstration of a desired polypharmacology of a unimolecular dual or triple agonist.
Fig. 5. Structural and functional feature of lipidated K10P of peptide 20.
a–c Close-up views of the crevices between TM1 and TM2 displayed by cryo-EM maps of peptide 20-bound GIPR a, GLP-1R b, and GCGR c. Continuous electron densities connected to K10 in peptide 20 were observed in the three peptide 20-bound receptor–Gs complexes. d–f Interactions between lipidated K10P and the TM1-TM2 crevice of GIPR d, GLP-1R e, and GCGR f, with interacting residues shown in sticks. Hydrogen bonds are shown with dashed lines. g Effects of receptor mutations on peptide 20-induced cAMP accumulation. Data shown are means ± S.E.M. of at least three independent experiments (n = 3–9) performed in quadruplicate. h Effects of K10 lipidation on peptide 20-induced cAMP accumulation. The bar graph represents the average pEC50 (that is, −logEC50) and data are presented as means ± S.E.M. of four independent experiments (n = 4) performed in quadruplicate. Statistically significant differences were determined with a two-tailed Student’s t test. **P < 0.01 and ****P < 0.0001. WT, wild-type. Source data are provided as a Source Data file.
Receptor activation
Despite the existence of unique structural features among the ligand-binding pockets of GIPR, GLP-1R and GCGR, both tirzepatide and peptide 20 triggered receptor conformational changes similar to that induced by GLP-1 or GCG4,27 and distinct from the inactive or apo GLP-1R and GCGR structures (Supplementary Fig. 11)31,32. Compared to the inactive GCGR, the extracellular tip of TM7 in peptide 20-bound GCGR moved outward by 5.1 Å (measured by Cα atom of L3777.34b) and the α-helical structure of the extracellular half of TM6 was partially unwounded. In the intracellular side, a sharp kink located in the conserved Pro6.47b-X-X-Gly6.50b motif pivoted the intracellular tip of TM6 to move outwards by 19.3 Å (measured by Cα atom of K3446.35b), slightly higher than that seen with the GCG–GCGR–Gs (17.7 Å)4. This, in conjunction with the movement of TM5 towards TM6, opened up the cytoplasmic face of GCGR to accommodate G protein coupling. Similar conformational change was also observed in the tirzepatide–GIPR–Gs, tirzepatide–GLP-1R–Gs, peptide 20–GIPR–Gs and peptide 20–GLP-1R–Gs complexes, compared to peptide-free apo GLP-1R structure31. At the residue level, signaling initiation by either peptide 20, tirzepatide or endogenous peptide hormones rendered a common arrangement of residue contacts for the three receptors, including the reorganization of the central polar network that located just below the peptide binding site, opening of the hydrophobic packing to favor the formation of the TM6 kink at the PXXG motif and the rearrangement of two polar networks (HETX motif and TM2-6-7-helix 8) at the cytoplasmic face33,34.
G protein coupling
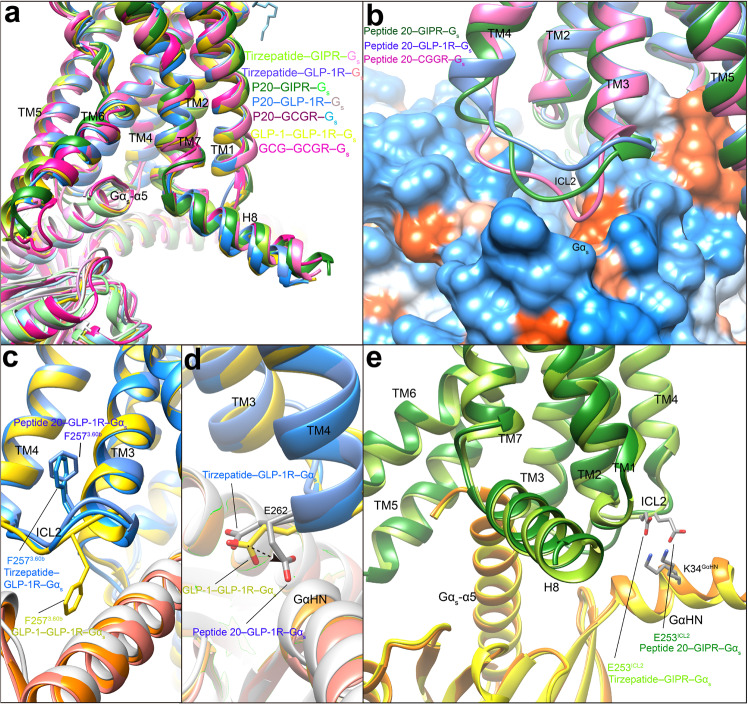
Comparison of the two tirzepatide- and three peptide 20-bound GPCR–Gs complex structures with that of other class B1 GPCR family members reveals a high similarity in the G protein binding interface, suggesting a common mechanism for Gs engagement4,33,35–38 (Fig. 6a). These complexes are anchored by the α5 helix of Gαs, which fits to the cytoplasmic cavity formed by TMs 2, 3, 5, 6, 7 and intracellular loop 1 (ICL1). Besides, H8 contributes several polar interactions with the Gβ subunit. There are some receptor- and ligand-specific structural features displayed by ICL2. For peptide 20-bound GCGR, its ICL2 moved downward and made extensive polar and nonpolar contacts with the binding groove formed by the αN helix, β1 strand and α5 helix of Gαs, resulting in an ICL2–Gαs interface area of 799 Å2, significantly larger than that of GLP-1R (396 Å2) or GIPR (416 Å2) (Fig. 6b). Different from the dipped down side chain conformation observed in GLP-1-bound GLP-1R27, F2573.60b in the peptide 20–GLP-1R–Gs complex rotated its side chain upwards (Fig. 6c). Furthermore, E262ICL2 was reoriented ~90° from an outside facing position to a position pointing to Gαs, thus introducing a hydrogen bond with Q35GαHN (GαHN means the αN helix of Gα) (Fig. 6d). Similar G protein interface was also observed in the tirzepatide-bound GLP-1R except for the orientation of E262ICL2 that is closer to that of GLP-1. In the case of peptide 20- and tirzepatide-bound GIPR complexes, the side chain of E253ICL2 contributed one salt bridge with K34GαHN, not observed in the peptide 20-bound GLP-1R and GCGR complexes (Fig. 6e).
Fig. 6. G protein coupling of multi-targeting agonist-bound GIPR, GLP-1R and GCGR.
a Comparison of G protein coupling among GIPR, GLP-1R and GCGR4, 25, 27. The Gαs α5-helix of the Gαs Ras-like domain inserts into an intracellular crevice of receptor’s TMD. The receptors and G proteins are colored as the labels. b Comparison of ICL2 conformation in the peptide 20-bound GIPR, GCGR and GLP-1R. c Comparison of F2573.60b conformation in the GLP-1R bound by GLP-1, tirzepatide and peptide 20. d Comparison of E262ICL2 conformation in the GLP-1R bound by GLP-1, tirzepatide and peptide 20. e Comparison of E253ICL2 conformation in the GIPR bound by tirzepatide and peptide 20. Residues involved in interactions are shown as sticks. Polar interactions are shown as black dashed lines.
Efficacy superiority
The superior therapeutic efficacy of tirzepatide over approved selective GLP-1 analogs was reported recently17,39, whereas the outcome of clinical trials on peptide 20 is not available in the literature. The five high-resolution cryo-EM structures reported here, together with abundant structural and pharmacological data of monospecific peptides documented previously4,25,27,30,40, provide us an excellent opportunity to analyze the molecular basis of the superior clinical efficacy presented by multi-targeting agonists.
Semaglutide and tirzepatide share two common substitutions, one is Aib at the second N-terminal residue and the other is acylated lysine at the 20th N-terminal residue by C18 diacid via a γGlu-2×OEG linker, to reduce degradation by dipeptidyl peptidase-4 (DPP-4) and to prolong their half-lives by enhanced binding to plasma albumin (Fig. 7a)41. Besides, there is only one residue in semaglutide (R34P) that is different from GLP-1 but does neither form any interaction with GLP-1R30 nor affect receptor binding and signaling29. However, tirzepatide has 14 unique amino acids (engineered from the GIP sequence) and an amidated exenatide-like C terminus as opposed to GLP-1 which allow the peptide to possess a GIPR binding ability equivalent to GIP(1–42) and to steadily interact with GLP-1R with a reduced potency compared to GLP-127 (Fig. 2a-d). Like GLP-1, semaglutide is not able to bind or activate GIPR. These findings were confirmed by GIPR or GLP-1R mediated cAMP accumulation assays (Fig. 7b–c)39. Of note is that tirzepatide was previously reported to cause biased signaling at GLP-1R in favor of cAMP response over β-arrestin recruitment in the presence of 0.1% casein39, which was confirmed in the present study showing that non-acylated tirzepatide elicited better cAMP responses (EC50 = 0.90 pM for 0.1% casein and 4.66 pM for 0.1% BSA) than that of tirzepatide (EC50 = 3.05 pM for 0.1% casein and 103.10 pM for 0.1% BSA) (Supplementary Table 11) highlighting the impact of acylation on their pharmacological properties. The combined activation of GIPR and GLP-1R by tirzepatide not only improved both glucose-dependent insulin secretion and glucose tolerance in mice15, but also showed significantly better efficacy than semaglutide and dulaglutide with regard to glucose control and weight loss16,17.
Fig. 7. Structure-basis of receptor selectivity demonstrated by tirzepatide, peptide 20 and GLP-1 analogs.
a Amino acid sequences of endogenous agonists, multi-targeting agonists and approved GLP-1 analogs including semaglutide. Residues are colored according to sequence conservation among GIP, GLP-1 and GCG. Aib, aminoisobutyric acid. Semaglutide and tirzepatide are conjugated by a C20 fatty diacid moiety via a linker connected to the lysine residue at position 20, while peptide 20 is covalently attached by a 16-carbon acyl chain (palmitoyl; 16:0) via a γ-carboxylate spacer at K10P. b Receptor signaling profiles of endogenous agonists, multi-targeting agonists and approved drug GLP-1 analogs including semaglutide. Data shown are means ± S.E.M. of four independent experiments (n = 4) performed in quadruplicate. Source data are provided as a Source Data file. c Receptor binding profiles of endogenous agonists, multi-targeting agonists and approved GLP-1 analogs. Data shown are means ± S.E.M. of three independent experiments (n = 3) performed in duplicate.
It is known that peptide 20 potently reversed metabolic disorders in rodent models of obesity and diabetes, characteristic of increased energy expenditure and elevated circulating FGF21 levels as a result of GCGR agonism22,23. Meanwhile, peptide 20 causes a biased signaling at GIPR and GCGR in favor of cAMP accumulation over β-arrestin 2 recruitment, with no significant biased influence on GLP-1R signaling5,6. Both the N terminus and C terminus of peptide 20 play important roles in its multiplexed pharmacological actions. The N terminus (the first 11 residues) that is highly conserved across GIP, GLP-1 and GCG interacts with the lower half of the TMD pocket of the three receptors consisting of conserved residues such as L/Y1.36b (hydrophobic with K10P), Q/Y1.40b (hydrogen bond with K10P), Y1.43b (stacking with F6P), Y1.47b (hydrogen bond with Q3P), Q3.37b (hydrogen bond with H1P), ECL2 (hydrogen bond with S8P), R7.35b (salt bridge with D9P), I/L7.43b (hydrophobic with Aib2P) and L7.43b (hydrophobic with F6P) (Figs. 2, 4b–d, 7a)42,43. A similar approach was applied to the design of peptide 20’s C terminus that occupies the hydrophobic binding groove of ECD, with residues (A19P, F22P, V23P, W25P, L26P and L27P) adopted from GIP, GLP-1 and GCG (Figs. 4e–g, 7a)42,43. To accommodate the upper half of the TMD pocket formed by ECL1 and the extracellular tips of TM1 and TM2 that diversified in both sequence and conformation across the three receptors, peptide 20 employs distinct residues to recognize specific region of a given receptor (Fig. 4h). For GIPR whose ECL1 was loosely compacted by peptide 20, three residues (Y13P-L14P-D15P) strengthened the peptide-binding interface by forming a hydrogen bond with F1271.29b and a salt bridge with R289ECL2, significantly stronger than that observed in GLP-1R and GCGR. Alternatively, another three residues (D21P-F22P-W25P) compacted well with the ordered ECL1 of GLP-1R via a hydrogen bond with Q210ECL1 and packing with W214ECL1. Two hydrogen bonds (D15P-Y2022.75b and R17P-Y2022.75b) were only seen in GCGR.
The most impressive structural feature of peptide 20 is the lipidated K10P by a 16-carbon palmitic acid through a γ-carboxylate spacer, which perfectly inserted into the TM1-TM2 crevice and made extensive contacts with both receptors and lipid membrane to stabilize the binding poses (Fig. 5). These observations suggest a combined mechanism of action for peptide 20 that uses conserved residues for ligand recognition and specific residues to induce conformational changes unique to each receptor, leading to a highly potent and balanced multi-targeting agonist for GIPR, GLP-1R and GCGR with a cAMP signaling profile similar to that of GIP, GLP-1 and GCG (Fig. 7b)22.
Discussion
Due to the central roles exerted by the three metabolically related peptide hormone receptors (GIPR, GLP-1R and GCGR) in the management of T2DM and obesity, the concept of combinatorial agonism or polypharmacology to synergize metabolic actions and maximize therapeutic benefits has been explored in the past decade with remarkable preclinical and clinical achievements. The 3-dimensional structures of GCGR, GLP-1R and GIPR solved previously helped us better understand the molecular basis of ligand recognition and receptor activation of these important class B1 GPCRs25,32,44–46. In this paper, we report five cryo-EM structures of two well-recognized multi-targeting agonists (tirzepatide and peptide 20) in complex with individual receptors and Gs proteins. The structural basis of the superior clinical efficacy of tirzepatide relative to monospecific agonists such as semaglutide and the triagonist peptide 20 is elucidated. Our results provide a near-atomic level visualization of the molecular action of multi-targeting agonists on three cognate receptors and offer valuable information for the design of better drugs to combat metabolic diseases.
Superimpositions of the two tirzepatide- and three peptide 20-bound structures to the three receptors bound by the endogenous ligands (GIP, GLP-1 and GCG) showed that the five peptides all adopt a single continuous helix, with the well-overlapped N terminus penetrating to the TMD core stabilized by conserved interactions, while the C terminus anchors the ECD, ECL1 and ECL2 in a receptor- and ligand-specific manner. With the presence of three proline residues (P195ECL1, P197ECL1 and P199ECL1), the ECL1 of GIPR presents a notable conformational adaptability in recognition of different agonists, a phenomenon that was not seen with that of GLP-1R and GCGR as their binding pockets exhibit less flexibility when recognizing the peptides through a combination of common segment that contributes to conserved interactions and distinct sequences that govern receptor selectivity. The distinct sequences that tirzepatide and peptide 20 employed, respectively, to recognize GIPR or GLP-1R are obviously different: the former was primarily based on the GIP sequence with engineered GLP-1 activity15, whereas the latter was derived from a GLP-1R/GCGR dual agonist in conjunction with GIP agonism22. Such a sequence and receptor binding divergence may consequently alter pharmacological and clinical outcomes. Clearly, distinct sequence and structural features of tirzepatide and peptide 20 allow them to exert combinatorial agonism at two or more receptors at the same time thereby maximize the benefit of polypharmacology and minimize the limitation of mono-targeting.
Both GIP and GLP-1 are released upon nutrient ingestion to promote insulin secretion by pancreatic β-cells. However, they have opposed effects on circulating GCG levels7,16. GIPR activation also has different roles in lipid metabolism from that of GLP-147. Maintenance of GCG action might be a key to the superior therapeutic efficacy of tirzepatide16,17,48. Structurally, the binding of tirzepatide to GIPR reshaped the ECL1 conformation relative to that of GIP, but made no change in the GLP-1R structure. As far as peptide 20 is concerned, the peptide binding pocket of both GLP-1R and GCGR closely resembled that of GLP-1 and GCG bound structures, where notable conformational change was only observed in the ECL1 of GIPR. These differences in structural plasticity or rigidity among the three receptors give clues to further optimize multi-targeting agonists using complementary amino acids to target common regions of individual receptors and distinct sequences to confer receptor selectivity.
Unlike tirzepatide that retains GCG function via counteracting that of GLP-1 through activation of GIPR, causing a glucagonotropic action by stimulating GCG release in states of hypoglycemia7,49, peptide 20 is capable of activating GCGR directly. Consistent with the effects of GCGR in increasing lipolysis and thermogenesis besides elevating blood glucose levels, preclinical studies have found that peptide 20 improved energy metabolism and hepatic lipid handling without exacerbating preexisting hyperglycemia22. Peptide 20 was developed through a series of optimizing processes based on GCGR agonism in diet-induced obese mice, concluding that the ideal metabolic benefits of triagonism predominantly depend on fine-tuning the GCG component22. The structures reveal that lipidation at K10 of peptide 20 allows the hydrophobic acyl tail to interact with the TMD region of all three receptors, providing a fresh clue for peptidic ligand design. Previous studies on optimizing the peptide sequences towards potent dual or triple agonists provide important information on the combinatorial agonism. For example, the subtle modification on the third N-terminal residue of peptide 20 showed diversified effects on its triagonism, highlighting the complexity of peptide-induced signal transduction. From the perspective of precision medicines, combinatorial agonism might be precisely designed to reflect pharmacological profiles of individual receptors such that diabetic patients at different disease stages could be prescribed with different multi-targeting agonists to take personalized therapeutic advantages.
Methods
Cell lines
Spodoptera frugiperda 9 (Sf9) (Invitrogen) and High Five™ insect cells (Expression Systems) were cultured in ESF 921 serum-free medium (Expression Systems) at 27 °C and 120 rpm. Human embryonic kidney 293 cells containing SV40 large T-antigen (HEK293T) were cultured in DMEM (Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco), 1 mM sodium pyruvate (Gibco) and 100 units/mL penicillin and 100 μg/mL streptomycin at 37 °C in 5% CO2. Chinese hamster ovary (CHO-K1) cells were cultured in F-12 (Gibco) containing 10% FBS, 100 units/mL penicillin and 100 μg/mL streptomycin at 37 °C in 5% CO2. For cAMP and receptor expression assays, HEK293T cells were seeded into 6-well cell culture plates at a density of 7 × 105 cells per well. After overnight incubation, cells were transfected with GIPR, GLP-1R or GCGR construct using Lipofectamine 2000 or Lipofectamine 3000 transfection reagent (Invitrogen). For whole-cell binding assay, CHO-K1 cells were seeded into 96-well fibronectin-treated cell culture plates at a density of 3 × 104 cells per well. After overnight incubation, cells were transfected with GIPR, GLP-1R or GCGR construct using Lipofectamine 2000 transfection reagent (Invitrogen). Following 24 h culturing, the transfected cells were ready for use.
Construct
The human GIPR DNA (Genewiz) with one mutation (T345F) was cloned into the pFastBac vector (Invitrogen) with its native signal peptide replaced by the haemagglutinin (HA) signal peptide. A BRIL fusion protein was added at the N-terminal of the ECD with a TEV protease site and 2GSA linker between them. C-terminal 45 amino acids (Q422-C466) of the receptor were truncated. LgBiT was added at the end of helix 8 with a 15-amino acid (15AA) polypeptide linker in between, followed by a TEV protease cleavage site and an OMBP-MBP tag. A dominant-negative bovine Gαs (DNGαs) construct with 9 mutations (S54N, G226A, E268A, N271K, K274D, R280K, T284D, I285T and A366S)50,51 was used to help stabilize the tirzepatide–GIPR–Gs complex. Meanwhile, a DNGαs construct with 8 mutations (S54N, G226A, E268A, N271K, K274D, R280K, T284D and I285T) was used to help stabilize the peptide 20–GIPR–Gs complex38,51. Rat Gβ1 was cloned with a C-terminal SmBiT34 (peptide 86 or HiBiT, Promega) connected with a 15AA polypeptide linker. The modified rat Gβ1 and bovine Gγ2 were both cloned into a pFastBac vector. The construct and various mutants of human GIPR were cloned into pcDNA3.1 vector for cAMP accumulation and whole-cell binding assays.
The human GLP-1R was modified with its native signal sequence (M1-P23) replaced by the HA signal peptide to facilitate receptor expression. To obtain a GLP-1R–Gs complex with good homogeneity and stability, we used the NanoBiT tethering strategy, in which the C terminus of GLP-1R was directly attached to LgBiT subunit followed by a TEV protease cleavage site and a double MBP tag. Rat Gβ1 was the same as the construct used in the GIPR structure determination. The Gαs (DNGαs with 9 mutations) used to stabilize the tirzepatide–GLP-1R–Gs complex was the same as that employed for the tirzepatide–GIPR–Gs complex. A dominant-negative human Gαs (DNGαs) with 8 mutations (S54N, G226A, E268A, N271K, K274D, R280K, T284D, and I285T) was generated as previously described to limit G protein dissociation51. The constructs were cloned into both pcDNA3.1 and pFastBac vectors for functional assays in mammalian cells and protein expression in insect cells, respectively. Other constructs including the full-length and various mutants of human GLP-1R were cloned into pcDNA3.1 vector for cAMP accumulation and whole-cell binding assays.
The human GCGR gene was cloned into pFastBac1 vector with GP64 promoter at the N terminus to enhance the protein yield. Forty-five residues (H433-F477) were truncated at the C terminus to improve the thermostability and an affinity tag, HPC4 tag, was added to the C terminus (GP64-HA-GCGR-GSGS linker-HPC4). Gαs (DNGαs with 8 mutations) was modified as above to stabilize the interaction with βγ subunits. The rat Gβ1 and bovine Gγ2 were used in the structure determination.
Additionally, we used an engineered Gs (mini-Gs) protein to stabilize the non-acylated tirzepatide (the side chain was removed at C20) bound GIPR or GLP-1R, as described previously52.
Protein expression
Baculoviruses containing the above complex constructs were prepared by the Bac-to-Bac system (Invitrogen). For the tirzepatide–GIPR–Gs and non-acylated tirzepatide–GIPR–mini-Gs complexes, GIPR and DNGαs or mini-Gs heterotrimer were co-expressed in High Five™ cells. Briefly, insect cells were grown in ESF 921 culture medium (Expression Systems) to a density of 3.2 × 106 cells/mL. The cells were then infected with BRIL-TEV-2GSA-GIPR(22-421)T345F-15AA-LgBiT-TEV-OMBP-MBP, DNGαs or mini-Gs, Gβ1-peptide 86 and Gγ2, respectively, at a ratio of 1:4:4:4. For the peptide 20–GIPR–Gs complex, GIPR and Gs heterotrimer were co-expressed in High Five™ cells grown in ESF 921 culture medium (Expression Systems) to a density of 3.2 × 106 cells/mL. The cells were then infected with BRIL-TEV-2GSA-GIPR(22-421)T345F-15AA-LgBiT-TEV-OMBP-MBP, DNGαs, Gβ1-peptide 86 and Gγ2, respectively, at a ratio of 1:3:3:3. After 48 h incubation at 27 °C, the cells were collected by centrifugation and stored at −80 °C until use.
The GLP-1R-LgBiT-2MBP, DNGαs or mini-Gs, Gβ1-peptide 86 and Gγ2 were co-expressed at multiplicity of infection (MOI) ratio of 1:1:1:1 by infecting Sf9 cells at a density of 3.0 × 106 cells/mL. Other operations are the same as GIPR.
The GCGR construct, DNGαs and Gβ1 and Gγ2 were co-expressed in High Five™ cells and infected with four separate baculoviruses at a ratio of 4:1:1:1. Other operations are the same as GIPR.
Nb35 expression and purification
Nanobody 35 (Nb35) with a 6× his tag at the C-terminal was expressed in the periplasm of E. coli BL21 (DE3) cells. Briefly, Nb35 target gene was transformed in the bacterium and amplified in TB culture medium with 100 μg/mL ampicillin, 2 mM MgCl2, 0.1% (w/v) glucose at 37 °C, 180 rpm. When OD600 reached 0.7–1.2, 1 mM IPTG was added to induce expression followed by overnight incubation at 28 °C. The cell pellet was then collected under 4 °C and stored at −80 °C. Nb35 was purified by size-exclusion chromatography (SEC) using a HiLoad 16/600 Superdex 75 column (GE Healthcare) with running buffer containing 20 mM HEPES, 100 mM NaCl, pH 7.4. Fractions of Nb35 were concentrated to ~2 mg/mL and quickly frozen in the liquid nitrogen with 10% glycerol and stored in −80 °C.
Complex formation and purification
Tirzepatide, non-acylated tirzepatide and peptide 20 were chemically synthesized in GL Biochem with a purity of 95.89%, 95.56% and 96.46%, respectively. For the tirzepatide–GIPR–Gs complex, cell pellets were lysed in a buffer containing 20 mM HEPES, 100 mM NaCl, pH 7.4, 10 mM MgCl2, 1 mM MnCl2 and 10% glycerol supplemented with protease inhibitor cocktail, EDTA-free (TragetMol). Cell membranes were then collected by ultracentrifugation at 4 °C, 90,000 g for 35 min. A buffer consisting of 20 mM HEPES, 100 mM NaCl, pH 7.4, 10 mM MgCl2, 1 mM MnCl2 and 10% glycerol was used to re-suspend the collected membranes. To assemble the GIPR–Gs complex, 15 μM tirzepatide (GL Biochem) was added to the preparation accompanied by 100 μM TCEP, 25 mU/mL apyrase (Sigma-Aldrich), 20 μg/mL Nb35 and 100 U salt active nuclease (Sigma-Aldrich) supplemented with protease inhibitor cocktail for 1.5 h incubation at room temperature (RT). The membrane was then solubilized with 0.5% (w/v) lauryl maltose neopentylglycol (LMNG, Anatrace) and 0.1% (w/v) cholesterol hemisuccinate (CHS, Anatrace) with additional 2 μM tirzepatide for 3 h at 4 °C. The supernatant was isolated by centrifugation at 90,000 g for 35 min and the solubilized complex was incubated with amylose resin (NEB) for 2.5 h at 4 °C. The resin was collected by centrifugation at 550 g and loaded onto a gravity flow column. The resin in the column was first washed with 5 column volumes (CVs) of buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 10% (v/v) glycerol, 5 mM MgCl2, 1 mM MnCl2, 25 μM TCEP, 5 μM tirzepatide, 0.1% (w/v) LMNG and 0.02% (w/v) CHS. After this, the resin was further washed with 25 CVs of buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 10% (v/v) glycerol, 5 mM MgCl2, 1 mM MnCl2, 25 μM TCEP, 5 μM tirzepatide, 0.03% (w/v) LMNG, 0.01% (w/v) glyco-diosgenin (GDN, Anatrace) and 0.008% (w/v) CHS. The protein was then incubated with a buffer consisting of 20 mM HEPES, pH 7.4, 100 mM NaCl, 10% (v/v) glycerol, 5 mM MgCl2, 1 mM MnCl2, 25 μM TCEP, 50 μM tirzepatide, 20 μg/mL Nb35, 0.03% (w/v) LMNG, 0.01% (w/v) GDN, 0.008% (w/v) CHS and 30 μg/mL His-tagged TEV protease on the column overnight at 4 °C. The flow through was collected and concentrated to 500 μL using a 100 kDa filter (Merck Millipore). SEC was performed by loading the protein onto Superose 6 Increase 10/300GL (GE Healthcare) column with running buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 10 mM MgCl2, 100 μM TCEP, 5 μM tirzepatide, 0.00075% (w/v) LMNG, 0.00025% (w/v) GDN, 0.0002% (w/v) CHS and 0.00025% digitonin (Anatrace). The tirzepatide–GIPR–Gs complexes were collected and concentrated for cryo-EM analysis.
For the non-acylated tirzepatide–GIPR–mini-Gs complex, the operations of the purification were the same as the tirzepatide–GIPR–Gs complex, except that the peptide was replaced by the non-acylated tirzepatide. The complex samples were concentrated to 14–16 mg/mL for cryo-EM analysis.
For the tirzepatide–GLP-1R–Gs complex, cells were suspended in 20 mM HEPES, pH 7.4, 100 mM NaCl and 10% (v/v) glycerol in the presence of protease inhibitor cocktail. Complex was formed by adding 10 mM MgCl2, 1 mM MnCl2, 50 mU/mL apyrase, 30 μM tirzepatide, 100 μM TCEP and 10 μg/mL Nb35 to the cell lysate and incubated at RT for 1.5 h. Cell membranes were solubilized by adding 0.5% (w/v) LMNG supplemented with 0.1% (w/v) CHS at 4 °C for 2 h, followed by centrifugation at 65,000 g for 30 min at 4 °C. The supernatant was taken to bind with amylose resin for 2 h at 4 °C. After packing, the column was washed with buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 10% (v/v) glycerol, 5 μM tirzepatide, 25 μM TCEP, 5 mM MgCl2, 1 mM MnCl2, 0.1% (w/v) LMNG and 0.02% (w/v) CHS first (10 CVs), and then with decreased concentrations of detergents, 0.03% (w/v) LMNG, 0.01% (w/v) GDN and 0.006% (w/v) CHS (20 CVs). TEV enzyme was added to the resin and kept at 4 °C overnight to remove the OMBP-MBP tag. The complex was eluted from the resin and concentrated to 500 μL using a 100 kDa MWCO Amicon Ultra Centrifugal Filter. SEC was carried out by loading the protein sample to Superdex 200 Increase 10/300GL (GE Healthcare) to obtain the monomer complex. The column was pre-equilibrated with 20 mM HEPES, pH 7.4, 100 mM NaCl, 5 μM tirzepatide, 100 μM TCEP, 2 mM MgCl2, 0.00075% (w/v) LMNG, 0.00025% (w/v) GDN, 0.00015% (w/v) CHS and 0.00025% digitonin.
For the non-acylated tirzepatide–GLP-1R–mini-Gs complex, the operations of the purification were the same as the peptide 20–GLP-1R–Gs complex, except that the peptide was replaced by the non-acylated tirzepatide, and the detergent of SEC running buffer was changed to 0.01% digitonin. The complex samples were concentrated to 16-18 mg/mL for cryo-EM analysis.
For the peptide 20–GIPR–Gs complex, the operations of the purification was the same as the tirzepatide–GIPR–Gs complex, except that the peptide was replaced by the peptide 20. The complex samples were concentrated to 5–6 mg/mL for cryo-EM analysis.
For the peptide 20–GLP-1R–Gs complex, cell pellets were thawed and lysed in a buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 10% (v/v) glycerol, 10 mM MgCl2, 1 mM MnCl2 and 100 μM TCEP supplemented with EDTA-free protease inhibitor cocktail by dounce homogenization. The complex formation was initiated by the addition of 20 μM peptide 20, 10 μg/mL Nb35 and 25 mU/mL apyrase. After 1.5 h incubation at RT, the membrane was solubilized in the buffer above supplemented with 0.5% (w/v) LMNG and 0.1% (w/v) CHS for 2 h at 4 °C. The supernatant was isolated by centrifugation at 65,000 g for 30 min and incubated with amylose resin for 2 h at 4 °C. The resin was then collected by centrifugation at 500 g for 10 min and washed in gravity flow column with 5 CVs of buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 10% (v/v) glycerol, 5 mM MgCl2, 1 mM MnCl2, 25 μM TCEP, 0.1% (w/v) LMNG, 0.02% (w/v) CHS and 5 μM peptide 20, followed by washing with 15 CVs of buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 10% (v/v) glycerol, 5 mM MgCl2, 1 mM MnCl2, 25 μM TCEP, 0.03% (w/v) LMNG, 0.01% (w/v) GDN, 0.008% (w/v) CHS and 5 μM peptide 20. The protein was then incubated overnight with TEV protease on the column to remove the C-terminal 2MBP-tag in the buffer above at 4 °C. The flow through was collected next day and concentrated with a 100 kDa molecular weight cut-off concentrator. The concentrated product was loaded onto a Superdex 200 increase 10/300 GL column with SEC running buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 100 μM TCEP, 2 μM peptide 20, 0.00075% LMNG, 0.00025% GDN and 0.0002% (w/v) CHS. The fractions for monomeric complex were collected and concentrated to 15-20 mg/mL for cryo-EM examination.
For the peptide 20–GCGR–Gs complex, cell pellets were resuspended in 20 mM HEPES, pH 7.4, 50 mM NaCl, 2 mM MgCl2 with protease inhibitor cocktail, EDTA-free, 5 µM peptide 20, 10 μg/mL Nb35 and 25 mU/mL apyrase. The suspension was incubated at RT for 2 h to promote the formation of complexes. Membranes were collected by centrifugation (30,000 rpm) at 4 °C for 30 min, and solubilized in 0.5% (w/v) LMNG, 0.1% (w/v) CHS, 10 µM peptide 20, 2 mM MgCl2, 100 U salt active nuclease and 25 mU/mL apyrase for 2.5 h at 4 °C. Supernatant was collected by centrifugation at 30,000 rpm for 30 min. The GCGR complex was incubated overnight with anti-HPC4 affinity resin in the presence of 2 mM CaCl2, washed with 20 CVs of 20 mM HEPES, pH 7.4, 100 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 5 μM peptide 20, 0.02% (w/v) LMNG and 0.004% (w/v) CHS, and eluted with 5 CVs of buffer by adding 6 mM EDTA and 5 µM peptide 20. The complexes were concentrated by a molecular weight cut-off concentrator and separated by SEC on a Superose 6 Increase 10/300GL column with running buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 2 mM MgCl2, 0.01% (w/v) LMNG, 0.002% (w/v) CHS and 5 μM peptide 20. The complex samples were concentrated to 12–14 mg/mL for cryo-EM analysis.
Structure determination
To prepare high-quality human GIPR–Gs complexes, the receptor’s C-terminal forty-five amino acids (Q422-C466) were truncated, and the NanoBiT tethering strategy was applied25,37,38,53. To enhance the receptor’s expression, a BRIL fusion protein and an optimized maltose binding protein-maltose binding protein tag (OMBP-MBP)54 were added to the N and C termini of the receptor to facilitate the receptor stability and expression (Supplementary Fig. 2a). To solve the tirzepatide–GIPR–Gs complex structure, we introduced one mutation (T345F) to stabilize complex assembly (Supplementary Fig. 3a). This mutation did not affect ligand binding and signaling properties as verified by both cAMP accumulation and receptor binding assays (Supplementary Fig. 2d).
The tirzepatide–GLP-1R–Gs complex was prepared using the same NanoBiT technique to achieve good homogeneity and stability as described previously46 (Supplementary Fig. 2b). Large-scale purification was performed and the complexes were collected by SEC for cryo-EM studies, with all components of the complex identified in SDS-PAGE of the SEC peak (Supplementary Fig. 3b). Activation of the modified GIPR and GLP-1R constructs by tirzepatide were confirmed by cAMP accumulation and receptor binding assays, showing similar responses to those of the wild-type (WT) receptors (Supplementary Fig. 3e–h). Acylated and non-acylated tirzepatide displayed reduced potencies in eliciting GIPR- or GLP-1R-mediated cAMP responses (Supplementary Fig. 2f, g).
Identical GIPR and GLP-1R constructs were used for the complex structure with peptide 20. Large-scale purification was conducted and the peptide 20–GIPR/GLP-1R–Gs complexes were collected by SEC for cryo-EM studies, with all components of the complex identified in SDS-PAGE of the SEC peak (Supplementary Fig. 4a, b). Activation of the modified GIPR and GLP-1R constructs by peptide 20 were confirmed by cAMP accumulation assays, showing similar responses to those of the WT (Supplementary Fig. 4d, e). To obtain the peptide 20–GCGR–Gs complexes, 45 residues (H433-F477) were truncated at the C terminus of the receptor followed by a HPC4 tag28 (Supplementary Fig. 2c). We used DNGαs34,51 and Nb35 that binds across the Gα:Gβ interface55 to enhance protein stability. Purified complex was resolved as a monodisperse peak on SEC, with all components of the complex identified in SDS-PAGE of the SEC peak (Supplementary Fig. 4c). The modified GCGR construct had a lower potency than that of the WT but did not significantly affect the binding affinity and cAMP signaling of GCG (Supplementary Fig. 4f).
Data acquisition and image processing
The purified tirzepatide–GIPR–Gs–Nb35 complex at a concentration of 18-20 mg/mL was mixed with 100 μM tirzepatide at 4 °C and applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, Au 300 mesh) that were subsequently vitrified by plunging into liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific). A Titan Krios equipped with a Gatan K3 Summit direct electron detector and serial EM3.7 were used to acquire cryo-EM images. The microscope was operated at 300 kV accelerating voltage, at a nominal magnification of 46,685× in counting mode, corresponding to a pixel size of 1.071 Å. Totally, 5,434 movies were obtained with a defocus range of -1.2 to -2.2 μm. An accumulated dose of 80 electrons per Å2 was fractionated into a movie stack of 36 frames.
The purified tirzepatide–GLP-1R–Gs–Nb35 complex (3 μL at about 20 mg/mL) was applied to a glow-discharged holey carbon grid (Quantifoil R1.2/1.3) and blotted subsequently. Sample-coated grids were vitrified by plunging into liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific). Automatic data collection was performed using serial EM3.7 on a Titan Krios equipped with a Gatan K3 Summit direct electron detector. The microscope was operated at 300 kV accelerating voltage, at a nominal magnification of 46,685× in counting mode, corresponding to a pixel size of 1.071 Å. A total of 9,309 movies were obtained with a defocus ranging from -1.2 to -2.2 μm. An accumulated dose of 80 electrons per Å2 was fractionated into a movie stack of 45 frames.
The purified peptide 20–GIPR–Gs–Nb35 complex at a concentration of 5–6 mg/mL was mixed with 100 μM peptide 20 at 4 °C and applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, Au 300 mesh) that were subsequently vitrified by plunging into liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific). A Titan Krios equipped with a Gatan K3 Summit direct electron detector and serial EM3.7 were used to acquire cryo-EM images. The microscope was operated at 300 kV accelerating voltage, at a nominal magnification of 46,685× in counting mode, corresponding to a pixel size of 1.071 Å. Totally, 3,948 movies were obtained with a defocus range of -1.2 to -2.2 μm. An accumulated dose of 80 electrons per Å2 was fractionated into a movie stack of 36 frames.
The purified peptide 20–GLP-1R–Gs–Nb35 complex (3.5 μL) was applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, 300 mesh), and subsequently vitrified using a Vitrobot Mark IV (ThermoFisher Scientific) set at 100% humidity and 4 °C. Cryo-EM images were collected using serial EM3.7 on a Titan Krios microscope (FEI) equipped with Gatan energy filter and K3 direct electron detector. The microscope was operated at 300 kV accelerating voltage and a calibrated magnification of 46,685× in counting mode, corresponding to a pixel size of 1.071 Å. The total exposure time was set to 7.2 s with intermediate frames recorded every 0.2 s, resulting in an accumulated dose of 80 electrons per Å2 with a defocus range of -1.2 to -2.2 μm. Totally, 4,778 images were collected and used for data processing.
The purified peptide 20–GCGR–Gs–Nb35 complex at a concentration of 12–14 mg/mL was mixed with 100 μM peptide 20 at 4 °C and applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, Au 300 mesh) that were subsequently vitrified by plunging into liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific). A Titan Krios equipped with a Gatan K3 Summit direct electron detector and serial EM3.7 were used to acquire cryo-EM images. The microscope was operated at 300 kV accelerating voltage, at a nominal magnification of 46,685× in counting mode, corresponding to a pixel size of 1.071 Å. Totally, 4,620 movies were obtained with a defocus range of -1.2 to -2.2 μm. An accumulated dose of 80 electrons per Å2 was fractionated into a movie stack of 36 frames.
The purified non-acylated tirzepatide–GIPR–mini-Gs–Nb35 complex at a concentration of 14-16 mg/mL was mixed with 100 μM non-acylated tirzepatide at 4 °C and applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, Au 300 mesh) that were subsequently vitrified by plunging into liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific). A Titan Krios equipped with a Gatan K3 Summit direct electron detector and serial EM3.7 were used to acquire cryo-EM images. The microscope was operated at 300 kV accelerating voltage, at a nominal magnification of 46,685× in counting mode, corresponding to a pixel size of 1.071 Å. Totally, 8,159 movies were obtained with a defocus range of -1.2 to -2.2 μm. An accumulated dose of 80 electrons per Å2 was fractionated into a movie stack of 36 frames.
The purified non-acylated tirzepatide–GLP-1R–mini-Gs–Nb35 complex (3.5 μL) was applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3, 300 mesh), and subsequently vitrified using a Vitrobot Mark IV (ThermoFisher Scientific) set at 100% humidity and 4 °C. Cryo-EM images were collected using serial EM3.7 on a Titan Krios microscope (FEI) equipped with Gatan energy filter and K3 direct electron detector. The microscope was operated at 300 kV accelerating voltage and a calibrated magnification of 46,685× in counting mode, corresponding to a pixel size of 1.071 Å. The total exposure time was set to 7.2 s with intermediate frames recorded every 0.2 s, resulting in an accumulated dose of 80 electrons per Å2 with a defocus range of -1.2 to -2.2 μm. Totally, 4,778 images were collected and used for data processing.
Dose-fractionated image stacks were subjected to beam-induced motion correction using MotionCor2.156. A sum of all frames, filtered according to the exposure dose, in each image stack was used for further processing. Contrast transfer function parameters for each micrograph were determined by Gctf v1.0657. Automated particle selection and data processing were performed using RELION-3.0 beta258.
For the dataset of the tirzepatide–GIPR–Gs–Nb35 complex, automated particle selection yielded 4,260,187 particles, which were subjected to reference-free 2D classification, producing 1,771,599 particles with well-defined averages. This subset of particle projections was subjected to a round of 3D classification resulting in one well-defined subset with 870,227 projections. Further 3D classification focusing on the alignment on the whole complex produced one high-quality subset accounting for 511,557 particles. These particles were subsequently subjected to CTF refinement and Bayesian polishing, which generated a map with an indicated global resolution of 3.4 Å.
For the dataset of the tirzepatide–GLP-1R–Gs–Nb35 complex, automated particle selection yielded 4,213,140 particles, which were subjected to reference-free 2D classification, producing 668,880 particles with well-defined averages. This subset of particle projections was subjected to a round of 3D classification resulting in one well-defined subset with 296,989 projections. Further 3D classification focusing on the alignment on the whole complex produced one high-quality subset accounting for 125,391 particles. These particles were subsequently subjected to CTF refinement and Bayesian polishing, which generated a map with an indicated global resolution of 3.4 Å.
For the dataset of the peptide 20–GIPR–Gs–Nb35 complex, automated particle selection yielded 5,322,921 particles. The particles were extracted on a binned dataset with a pixel size of 2.142 Å and were subjected to reference-free 2D classification, producing 4,334,371 particles with well-defined averages. This subset of particle projections was subjected to a round of 3D classification resulting in one well-defined subset with 1,876,783 projections. Further 3D classifications focusing on the alignment on the whole complex and the receptor produced one high-quality subset accounting for 255,256 particles. These particles were subsequently subjected to CTF refinement and Bayesian polishing, which generated a map with an indicated global resolution of 3.1 Å.
For the dataset of the peptide 20–GLP-1R–Gs–Nb35 complex, automated particle selection yielded 4,124,536 particles, which were subjected to reference-free 2D classification, producing 2,354,838 particles with well-defined averages. This subset of particle projections was subjected to a round of 3D classification resulting in one well-defined subset with 1,523,580 projections. Further 3D classifications focusing on the alignment on the whole complex and the receptor produced one high-quality subset accounting for 241,786 particles. These particles were subsequently subjected to CTF refinement and Bayesian polishing, which generated a map with an indicated global resolution of 3.0 Å.
For the dataset of the peptide 20–GCGR–Gs–Nb35 complex, automated particle selection yielded 3,931,945 particles, which were subjected to reference-free 2D classification, producing 917,065 particles with well-defined averages. This subset of particle projections was subjected to a round of 3D classification resulting in one well-defined subset with 578,668 projections. Further 3D classification focusing on the alignment on the whole complex produced one high-quality subset accounting for 383,657 particles. These particles were subsequently subjected to CTF refinement and Bayesian polishing, which generated a map with an indicated global resolution of 3.5 Å.
For the dataset of the non-acylated tirzepatide–GIPR–mini-Gs–Nb35 complex, automated particle selection yielded 7,204,521 particles, which were subjected to reference-free 2D classification, producing 2,718,249 particles with well-defined averages. This subset of particle projections was subjected to a round of 3D classification resulting in one well-defined subset with 2,102,580 projections. Further 3D classification focusing on the alignment on the whole complex produced one high-quality subset accounting for 1,251,553 particles. These particles were subsequently subjected to CTF refinement and Bayesian polishing, which generated a map with an indicated global resolution of 3.2 Å.
For the dataset of the non-acylated tirzepatide–GLP-1R–mini-Gs–Nb35 complex, automated particle selection yielded 5,985,110 particles, which were subjected to reference-free 2D classification, producing 1,723,671 particles with well-defined averages. This subset of particle projections was subjected to a round of 3D classification resulting in one well-defined subset with 906,824 projections. Further 3D classification focusing on the alignment on the whole complex produced one high-quality subset accounting for 452,921 particles. These particles were subsequently subjected to CTF refinement and Bayesian polishing, which generated a map with an indicated global resolution of 3.0 Å.
Model building and refinement
The models of the tirzepatide–GIPR–Gs complex and peptide 20–GIPR–Gs complex were built using the cryo-EM structure of the GIP–GIPR–Gs complex (PDB code: 7DTY)25 as the starting point. The models of the tirzepatide–GLP-1R–Gs complex and peptide 20–GLP-1R–Gs complex were built using the cryo-EM structure of the GLP-1–GLP-1R–Gs complex (PDB code: 6×18)27 as the starting point. The model of the peptide 20–GCGR–Gs complex was built using the cryo-EM structure of the GCG–GCGR–Gs complex (PDB code: 6LMK)4 as the starting point. The models were docked into the EM density maps using Chimera v1.1559, followed by iterative manual adjustment and rebuilding in COOT 0.9.660. Real space refinement was performed using Phenix 1.1661. The model statistics were validated with MolProbity v4.262. The final refinement statistics are provided in Supplementary Table 2. Structural figures were prepared in UCSF Chimera v1.15 and PyMOL 2.1 (https://pymol.org/2/).
cAMP accumulation assay
For GIPR, GLP-1R and GCGR, multi-targeting agonist stimulated cAMP accumulation was measured by a LANCE Ultra cAMP kit (PerkinElmer). After 24 h culture, the transfected cells were seeded into 384-well microtiter plates at a density of 3,000 cells per well in HBSS supplemented with 5 mM HEPES, 0.1% (w/v) BSA or 0.1% (w/v) casein and 0.5 mM 3-isobutyl-1-methylxanthine. The cells were stimulated with different concentrations of peptide agonists for 40 min at RT. Eu-cAMP tracer and ULightTM-anti-cAMP were then diluted by cAMP detection buffer and added to the plates separately to terminate the reaction. Plates were incubated at RT for 1 h and the fluorescence intensity measured at 620 nm and 650 nm by an EnVision multilabel plate reader (PerkinElmer).
Whole-cell binding assay
For GIPR, CHO-K1 cells were cultured in F-12 medium with 10% FBS and seeded at a density of 30,000 cells/well in Isoplate-96 plates (PerkinElmer). The WT or mutant GIPR was transiently transfected using Lipofectamine 2000 transfection reagent as previous described25. They were then washed twice using F-12 with 0.1% BSA or 0.1% casein, 33 mM HEPES, and incubated for 2 h at 37 °C. For homogeneous binding, cells were incubated in binding buffer with a constant concentration of 125I-GIP1-42 (30 pM, PerkinElmer) and increasing concentrations of unlabeled peptide agonists (3.57 pM to 5 μM) at RT for 3 h. Following incubation, cells were washed three times with ice-cold PBS and lysed by addition of 50 μL lysis buffer (PBS supplemented with 20 mM Tris-HCl, 1% Triton X-100, pH 7.4). Fifty µL of scintillation cocktail (OptiPhase SuperMix, PerkinElmer) were added and the plates were subsequently counted for radioactivity (counts per minute, CPM) in a MicroBeta2 microplate counter (PerkinElmer).
For GLP-1R and GCGR, CHO-K1 cells (30,000 cells/well) were seeded into Isoplate-96 plates and incubated for 24 h at 37 °C in 5% CO2. The WT or mutant constructs were transiently transfected using Lipofectamine 2000 transfection reagent as previous described25. They were then washed twice using F-12 with 0.1% BSA or 0.1% casein, 33 mM HEPES, and incubated for 2 h at 37 °C. The medium was removed and 125I-GLP-1(7-36)NH2 (60 pM) or 125I-GCG (40 pM) (PerkinElmer) and increasing concentrations unlabeled peptide agonists were added for overnight incubation at 4 °C. Cells were washed three times with ice-cold PBS and lysed in PBS with 1% Triton X-100, 20 mM Tris-HCl. After addition of scintillation cocktail (PerkinElmer), radioactivity (CPM) was counted on a MicroBeta2 microplate counter (PerkinElmer). Data were normalized to the WT response and analyzed using three-parameter logistic equation.
Receptor expression
Cell surface expression of GIPR, GLP-1R and GCGR were determined by flow cytometry 24 h post-transfection in HEK293T cells. Briefly, approximately 2 × 105 cells were blocked with PBS containing 5% BSA (w/v) at RT for 15 min. After that, cells expressing GIPR and GLP-1R were incubated with 1:300 anti-Flag primary antibody (diluted with PBS containing 5% BSA, Sigma), and those expressing GCGR were incubated with 1:50 anti-GCGR antibody (diluted with PBS containing 5% BSA, Abcam) at RT for 1 h. The cells were then washed three times with PBS containing 1% BSA (w/v) followed by 1 h incubation with 1:1,000 anti-mouse Alexa Fluor 488 conjugated secondary antibody (diluted with PBS containing 5% BSA, Invitrogen) at RT in the dark. After washing three times, cells were resuspended in 200 μL PBS containing 1% BSA for detection by NovoCyte (Agilent) utilizing laser excitation and emission wavelengths of 488 nm and 530 nm, respectively. For each sample, 20,000 cellular events were collected, and the total fluorescence intensity of positive expression cell population was calculated by NovoExpress 1.2.1. Data were normalized to the WT receptor.
β-arrestin 2 recruitment
HEK293T cells (3.2 × 106 cells/10 cm plate) were grown for 24 h before transiently transfection with 15 μg plasmid containing GIPR, GLP-1R or GCGR tagged with Rluc8 and β-arrestin 2 with a Venus-tag at the N terminus at a ratio of 1:9 by Lipofectamine 3000 transfection reagent. After cells were grown for 24 h, transiently transfected cells were then seeded onto poly-D-lysine coated 96-well culture plates (50,000 cells/well) in DMEM with 10% FBS. Cells were grown for 20 h before incubation in assay buffer (HBSS supplemented with 10 mM HEPES, 0.1% BSA or 0.1% casein, pH 7.4) for 30 min at 37 °C. Coelentrazine-h (Yeasen Biotech) was added to a final concentration of 5 μM for 5 min before bioluminescence resonance energy transfer (BRET) readings were made using an EnVision plate reader (PerkinElmer). BRET baseline measurements were collected for 15 cycles prior to ligand addition. After which, BRET was measured for 55 cycles. The BRET signal (ratio of 535 nm over 470 nm emission) was corrected to the baseline and then vehicle-treated condition to determine ligand-induced changes in BRET response. Concentration-response values were obtained from the area-under-the-curve (AUC) of the responses elicited by GIP1-42, GLP-1(7-36)NH2, glucagon, tirzepatide, non-acylated tirzepatide, peptide 20 or non-lipidated peptide 20.
Molecular dynamics simulation
Molecular dynamics (MD) simulation was performed by Gromacs 2020.163. The peptide-receptor complexes were prepared by the Protein Preparation Wizard (Schrodinger 2017-4) with G protein and Nb35 removed. The receptors were capped with acetyl and methylamide, and the titratable residues were left in their dominant state at pH 7.0. The complexes were embedded in a bilayer composed of 195–200 POPC lipids and solvated with 0.15 M NaCl in explicit TIP3P waters using CHARMM-GUI Membrane Builder v3.2.264. The CHARMM36-CAMP force filed65 was adopted for protein, peptides, lipids and salt ions. The 16-carbon acyl chain (palmitoyl; 16:0) covalently attached to the side chain amine of Lys10 in peptide 20 through a γ-carboxylate spacer and the γGlu-2×OEG linker, and C18 fatty diacid moiety that was acylated on Lys20 in tirzepatide were modeled with the CHARMM CGenFF small-molecule force field 66, program version 1.0.0. The Particle Mesh Ewald (PME) method was used to treat all electrostatic interactions beyond a cut-off of 10 Å and the bonds involving hydrogen atoms were constrained using LINCS algorithm67. The complex system was first relaxed using the steepest descent energy minimization, followed by slow heating of the system to 310 K with restraints. The restraints were reduced gradually over 50 ns. Finally, restrain-free production run was carried out for each simulation, with a time step of 2 fs in the NPT ensemble at 310 K and 1 bar using the Nose-Hoover thermostat and the semi-isotropic Parrinello-Rahman barostat68, respectively.
Statistical analysis
All functional data were presented as means ± standard error of the mean (S.E.M.). Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software). Concentration-response curves were evaluated with a three-parameter logistic equation. The significance was determined with either two-tailed Student’s t-test or one-way ANOVA. Significant difference is accepted at P < 0.05.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We are grateful to Ms. Jiahui Yan, Drs. Fan Wu and Fulai Zhou for technical assistance and to Drs. Raymond C. Stevens, Radostin Danev, Denise Wootten and Patrick M. Sexton for valuable interactions. This work was partially supported by National Natural Science Foundation of China 81872915 (M.-W.W.), 82073904 (M.-W.W.), 32071203 (L.H.Z), 81922071 (Y.Z.), 81773792 (D.Y.), 81973373 (D.Y.) and 21704064 (Q.T.Z.); National Science and Technology Major Project of China – Key New Drug Creation and Manufacturing Program 2018ZX09735–001 (M.-W.W.) and 2018ZX09711002–002–005 (D.Y.); the National Key Basic Research Program of China 2018YFA0507000 (M.-W.W.) and 2019YFA0508800 (Y.Z.); Ministry of Science and Technology of China 2018YFA0507002 (H.E.X.); Shanghai Municipal Science and Technology Major Project 2019SHZDZX02 (H.E.X.); the Strategic Priority Research Program of Chinese Academy of Sciences XDB37030103 (H.E.X.); Novo Nordisk-CAS Research Fund grant NNCAS-2017–1-CC (D.Y.); Zhejiang Province Science Fund for Distinguished Young Scholars LR19H310001 (Y.Z.); Fundamental Research Funds for Central Universities 2019XZZX001-01-06 (Y.Z.); Shanghai Science and Technology Development Funds 18ZR1447800 (L.H.Z.) and 18431907100 (M.-W.W.); The Young Innovator Association of CAS 2018325 (L.H.Z.) and SA-SIBS Scholarship Program (L.H.Z. and D.Y.). The cryo-EM data were collected at Cryo-Electron Microscopy Research Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences.
Source data
Author contributions
F.H.Z., Z.T.C., K.N.H., and C.Z. designed expression constructs, purified the receptor complexes, screened the specimen, prepared the final samples for negative staining, collected cryo-EM data and participated in manuscript preparation. X.Y.Z., A.Y.L. and T.X. conducted map calculation and participated in figure preparation; Q.Q.M., M.W., L.-N.C. and L.H.Z. built the models of the complexes and carried out structural analyses; Q.T.Z. and W.F. conducted MD simulations, comparative structural analysis and figure preparation; A.T.D. and Y.C. performed ligand binding and signaling experiments under the supervision of D.H.Y.; R.L.C. and P.Y.X. participated in method development; Y.Z. and B.W. assisted in structural studies on GLP-1R and GCGR; H.E.X. and M.-W.W. initiated the project; Q.T.Z., L.H.Z., H.E.X. and M.-W.W. supervised the studies, analyzed the data and wrote the manuscript with inputs from all co-authors.
Peer review
Peer review information
Nature Communications thanks Christopher Reynolds and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
The raw data underlying Figs. 3d, e, 5g, h, 7b and Supplementary Figs. 2d–g, 3a–l, 4a–f, 9a, b are provided as a Source Data file. The atomic coordinates and electron microscopy maps have been deposited in the Protein Data Bank (PDB) under accession codes: 7FIY (tirzepatide–GIPR–Gs complex), 7VAB (non-acylated tirzepatide–GIPR–Gs complex), 7FIM (tirzepatide–GLP-1R–Gs complex), 7VBI (non-acylated tirzepatide–GLP-1R–Gs complex), 7FIN (peptide 20–GIPR–Gs complex), 7VBH (peptide 20–GLP-1R–Gs complex) and 7V35 (peptide 20–GCGR–Gs complex), as well as Electron Microscopy Data Bank (EMDB) under accession codes: EMD-31606 (tirzepatide–GIPR–Gs complex), EMD-31836 (non-acylated tirzepatide–GIPR–Gs complex), EMD-31603 (tirzepatide–GLP-1R–Gs complex), EMD-31880 (non-acylated tirzepatide–GLP-1R–Gs complex), EMD-31604 (peptide 20–GIPR–Gs complex), EMD-31879 (peptide 20–GLP-1R–Gs complex) and EMD-31676 (peptide 20–GCGR–Gs complex), respectively. All relevant data are available from the authors and/or included in the manuscript or supplemental data. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fenghui Zhao, Qingtong Zhou, Zhaotong Cong, Kaini Hang, Xinyu Zou.
Contributor Information
Dehua Yang, Email: dhyang@simm.ac.cn.
Lihua Zhao, Email: zhaolihuawendy@simm.ac.cn.
H. Eric Xu, Email: eric.xu@simm.ac.cn.
Ming-Wei Wang, Email: mwwang@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-28683-0.
References
- 1.Wootten D, Miller LJ, Koole C, Christopoulos A, Sexton PM. Allostery and biased agonism at class B G protein-coupled receptors. Chem. Rev. 2017;117:111–138. doi: 10.1021/acs.chemrev.6b00049. [DOI] [PubMed] [Google Scholar]
- 2.Hollenstein K, et al. Insights into the structure of class B GPCRs. Trends Pharm. Sci. 2014;35:12–22. doi: 10.1016/j.tips.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stemmer K, Finan B, DiMarchi RD, Tschop MH, Muller TD. Insights into incretin-based therapies for treatment of diabetic dyslipidemia. Adv. Drug Deliv. Rev. 2020;159:34–53. doi: 10.1016/j.addr.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Qiao A, et al. Structural basis of Gs and Gi recognition by the human glucagon receptor. Science. 2020;367:1346–1352. doi: 10.1126/science.aaz5346. [DOI] [PubMed] [Google Scholar]
- 5.Yuliantie E, et al. Pharmacological characterization of mono-, dual- and tri-peptidic agonists at GIP and GLP-1 receptors. Biochem. Pharm. 2020;177:114001. doi: 10.1016/j.bcp.2020.114001. [DOI] [PubMed] [Google Scholar]
- 6.Darbalaei S, et al. Evaluation of biased agonism mediated by dual agonists of the GLP-1 and glucagon receptors. Biochem. Pharm. 2020;180:114150. doi: 10.1016/j.bcp.2020.114150. [DOI] [PubMed] [Google Scholar]
- 7.Christensen M, Vedtofte L, Holst JJ, Vilsboll T, Knop FK. Glucose-dependent insulinotropic polypeptide: a bifunctional glucose-dependent regulator of glucagon and insulin secretion in humans. Diabetes. 2011;60:3103–3109. doi: 10.2337/db11-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gault VA, Flatt PR, O’Harte FPM. Glucose-dependent insulinotropic polypeptide analogues and their therapeutic potential for the treatment of obesity-diabetes. Biochem Biophys. Res. Commun. 2003;308:207–213. doi: 10.1016/s0006-291x(03)01361-5. [DOI] [PubMed] [Google Scholar]
- 9.Campbell JE, Drucker DJ. Islet alpha cells and glucagon–critical regulators of energy homeostasis. Nat. Rev. Endocrinol. 2015;11:329–338. doi: 10.1038/nrendo.2015.51. [DOI] [PubMed] [Google Scholar]
- 10.Brandt SJ, Gotz A, Tschop MH, Muller TD. Gut hormone polyagonists for the treatment of type 2 diabetes. Peptides. 2018;100:190–201. doi: 10.1016/j.peptides.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilding JPH, et al. Once-Weekly Semaglutide in Adults with Overweight or Obesity. N. Engl. J. Med. 2021;384:989. doi: 10.1056/NEJMoa2032183. [DOI] [PubMed] [Google Scholar]
- 12.Frias JP, et al. The sustained effects of a dual GIP/GLP-1 receptor agonist, NNC0090-2746, in patients with type 2 diabetes. Cell Metab. 2017;26:343–352 e342. doi: 10.1016/j.cmet.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Brandt SJ, Muller TD, DiMarchi RD, Tschop MH, Stemmer K. Peptide-based multi-agonists: a new paradigm in metabolic pharmacology. J. Intern Med. 2018;284:581–602. doi: 10.1111/joim.12837. [DOI] [PubMed] [Google Scholar]
- 14.Yang D, et al. G protein-coupled receptors: structure- and function-based drug discovery. Signal Transduct. Target Ther. 2021;6:7. doi: 10.1038/s41392-020-00435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coskun T, et al. LY3298176, a novel dual GIP and GLP-1 receptor agonist for the treatment of type 2 diabetes mellitus: from discovery to clinical proof of concept. Mol. Metab. 2018;18:3–14. doi: 10.1016/j.molmet.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frias JP, et al. Tirzepatide versus semaglutide once weekly in patients with type 2 diabetes. N. Engl. J. Med. 2021;385:503–515. doi: 10.1056/NEJMoa2107519. [DOI] [PubMed] [Google Scholar]
- 17.Frias JP, et al. Efficacy and safety of LY3298176, a novel dual GIP and GLP-1 receptor agonist, in patients with type 2 diabetes: a randomised, placebo-controlled and active comparator-controlled phase 2 trial. Lancet. 2018;392:2180–2193. doi: 10.1016/S0140-6736(18)32260-8. [DOI] [PubMed] [Google Scholar]
- 18.Alexiadou K, Anyiam O, Tan T. Cracking the combination: gut hormones for the treatment of obesity and diabetes. J. Neuroendocrinol. 2019;31:e12664. doi: 10.1111/jne.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ClinicalTrials.gov. Investigation of Single Ascending Doses of NNC9204-1706 in Male Subjects Being Overweight or With Obesity, https://clinicaltrials.gov/ct2/show/NCT03095807 (2018).
- 20.ClinicalTrials.gov. Research Study of a New Medicine (NNC9204-1706) in People With Overweight or Obesity, https://clinicaltrials.gov/ct2/show/NCT03661879 (2019).
- 21.Day JW, et al. A new glucagon and GLP-1 co-agonist eliminates obesity in rodents. Nat. Chem. Biol. 2009;5:749–757. doi: 10.1038/nchembio.209. [DOI] [PubMed] [Google Scholar]
- 22.Finan B, et al. A rationally designed monomeric peptide triagonist corrects obesity and diabetes in rodents. Nat. Med. 2015;21:27–36. doi: 10.1038/nm.3761. [DOI] [PubMed] [Google Scholar]
- 23.Tschop MH, et al. Unimolecular polypharmacy for treatment of diabetes and obesity. Cell Metab. 2016;24:51–62. doi: 10.1016/j.cmet.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 24.Jall S, et al. Monomeric GLP-1/GIP/glucagon triagonism corrects obesity, hepatosteatosis, and dyslipidemia in female mice. Mol. Metab. 2017;6:440–446. doi: 10.1016/j.molmet.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao F, et al. Structural insights into hormone recognition by the human glucose-dependent insulinotropic polypeptide receptor. Elife. 2021;10:e68719. doi: 10.7554/eLife.68719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wootten D, Simms J, Miller LJ, Christopoulos A, Sexton PM. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl Acad. Sci. USA. 2013;110:5211–5216. doi: 10.1073/pnas.1221585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, et al. Differential GLP-1R binding and activation by peptide and non-peptide agonists. Mol. Cell. 2020;80:485–500 e487. doi: 10.1016/j.molcel.2020.09.020. [DOI] [PubMed] [Google Scholar]
- 28.Chang R, et al. Cryo-electron microscopy structure of the glucagon receptor with a dual-agonist peptide. J. Biol. Chem. 2020;295:9313–9325. doi: 10.1074/jbc.RA120.013793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau J, et al. Discovery of the Once-Weekly Glucagon-Like Peptide-1 (GLP-1) Analogue Semaglutide. J. Med Chem. 2015;58:7370–7380. doi: 10.1021/acs.jmedchem.5b00726. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Structure and dynamics of semaglutide- and taspoglutide-bound GLP-1R-Gs complexes. Cell Rep. 2021;36:109374. doi: 10.1016/j.celrep.2021.109374. [DOI] [PubMed] [Google Scholar]
- 31.Wu F, et al. Full-length human GLP-1 receptor structure without orthosteric ligands. Nat. Commun. 2020;11:1272. doi: 10.1038/s41467-020-14934-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, et al. Structure of the full-length glucagon class B G-protein-coupled receptor. Nature. 2017;546:259–264. doi: 10.1038/nature22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature. 2017;546:248–253. doi: 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang YL, et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex. Nature. 2018;555:121–125. doi: 10.1038/nature25773. [DOI] [PubMed] [Google Scholar]
- 35.Zhao LH, et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science. 2019;364:148–153. doi: 10.1126/science.aav7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilger, D. et al. Structural insights into differences in G protein activation by family A and family B GPCRs. Science369, 10.1126/science.aba3373 (2020). [DOI] [PMC free article] [PubMed]
- 37.Sun W, et al. A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor. Cell Res. 2020;30:1098–1108. doi: 10.1038/s41422-020-00442-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou F, et al. Structural basis for activation of the growth hormone-releasing hormone receptor. Nat. Commun. 2020;11:5205. doi: 10.1038/s41467-020-18945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willard, F. S. et al. Tirzepatide is an imbalanced and biased dual GIP and GLP-1 receptor agonist. JCI Insight5, 10.1172/jci.insight.140532 (2020). [DOI] [PMC free article] [PubMed]
- 40.Novikoff A, et al. Spatiotemporal GLP-1 and GIP receptor signaling and trafficking/recycling dynamics induced by selected receptor mono- and dual-agonists. Mol. Metab. 2021;49:101181. doi: 10.1016/j.molmet.2021.101181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pabreja K, Mohd MA, Koole C, Wootten D, Furness SG. Molecular mechanisms underlying physiological and receptor pleiotropic effects mediated by GLP-1R activation. Br. J. Pharm. 2014;171:1114–1128. doi: 10.1111/bph.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoare SR. Mechanisms of peptide and nonpeptide ligand binding to Class B G-protein-coupled receptors. Drug Disco. Today. 2005;10:417–427. doi: 10.1016/S1359-6446(05)03370-2. [DOI] [PubMed] [Google Scholar]
- 43.Zhao LH, et al. Differential requirement of the extracellular domain in activation of class B G Protein-coupled Receptors. J. Biol. Chem. 2016;291:15119–15130. doi: 10.1074/jbc.M116.726620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siu FY, et al. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013;499:444–449. doi: 10.1038/nature12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song G, et al. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature. 2017;546:312–315. doi: 10.1038/nature22378. [DOI] [PubMed] [Google Scholar]
- 46.Cong Z, et al. Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor. Nat. Commun. 2021;12:3763. doi: 10.1038/s41467-021-24058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samms RJ, Coghlan MP, Sloop KW. How may GIP enhance the therapeutic efficacy of GLP-1? Trends Endocrinol. Metab. 2020;31:410–421. doi: 10.1016/j.tem.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Tuttle KR. Breaking new ground with incretin therapy in diabetes. N. Engl. J. Med. 2021;385:560–561. doi: 10.1056/NEJMe2109957. [DOI] [PubMed] [Google Scholar]
- 49.Skow MA, Bergmann NC, Knop FK. Diabetes and obesity treatment based on dual incretin receptor activation: ‘twincretins’. Diabetes Obes. Metab. 2016;18:847–854. doi: 10.1111/dom.12685. [DOI] [PubMed] [Google Scholar]
- 50.Dong M, et al. Structure and dynamics of the active Gs-coupled human secretin receptor. Nat. Commun. 2020;11:4137. doi: 10.1038/s41467-020-17791-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang YL, et al. Dominant negative G proteins enhance formation and purification of agonist-GPCR-G protein complexes for structure determination. ACS Pharm. Transl. Sci. 2018;1:12–20. doi: 10.1021/acsptsci.8b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou F, et al. Molecular basis of ligand recognition and activation of human V2 vasopressin receptor. Cell Res. 2021;31:929–931. doi: 10.1038/s41422-021-00480-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duan J, et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat. Commun. 2020;11:4121. doi: 10.1038/s41467-020-17933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang Y, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 61.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abraham MJ, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 64.Wu EL, et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guvench O, et al. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 2011;7:3162–3180. doi: 10.1021/ct200328p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu W, He X, Vanommeslaeghe K, MacKerell AD., Jr Extension of the CHARMM General Force Field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 2012;33:2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 68.Aoki KM, Yonezawa F. Constant-pressure molecular-dynamics simulations of the crystal-smectic transition in systems of soft parallel spherocylinders. Phys. Rev. A. 1992;46:6541–6549. doi: 10.1103/physreva.46.6541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data underlying Figs. 3d, e, 5g, h, 7b and Supplementary Figs. 2d–g, 3a–l, 4a–f, 9a, b are provided as a Source Data file. The atomic coordinates and electron microscopy maps have been deposited in the Protein Data Bank (PDB) under accession codes: 7FIY (tirzepatide–GIPR–Gs complex), 7VAB (non-acylated tirzepatide–GIPR–Gs complex), 7FIM (tirzepatide–GLP-1R–Gs complex), 7VBI (non-acylated tirzepatide–GLP-1R–Gs complex), 7FIN (peptide 20–GIPR–Gs complex), 7VBH (peptide 20–GLP-1R–Gs complex) and 7V35 (peptide 20–GCGR–Gs complex), as well as Electron Microscopy Data Bank (EMDB) under accession codes: EMD-31606 (tirzepatide–GIPR–Gs complex), EMD-31836 (non-acylated tirzepatide–GIPR–Gs complex), EMD-31603 (tirzepatide–GLP-1R–Gs complex), EMD-31880 (non-acylated tirzepatide–GLP-1R–Gs complex), EMD-31604 (peptide 20–GIPR–Gs complex), EMD-31879 (peptide 20–GLP-1R–Gs complex) and EMD-31676 (peptide 20–GCGR–Gs complex), respectively. All relevant data are available from the authors and/or included in the manuscript or supplemental data. Source data are provided with this paper.