Abstract
Background & Aims
TOB1 is an anti-proliferative protein of Tob/BTG family and typically involved in the tumorigenesis and T cell activation. Although TOB1 is associated with T helper 17 cell–related autoimmunity, its role in modulating T cell–mediated immune responses in IBD remains poorly understood. Here, we explored its expression and the underlying mechanisms involved in the pathogenesis of inflammatory bowel disease (IBD).
Methods
TOB1 and ID2 expression in IBD patients was examined by quantitative real time polymerase chain reaction and immunohistochemistry. IBD CD4+ T cells were transfected with lentivirus expressing TOB1, ID2, TOB1 short hairpin RNA and ID2 short hairpin RNA, respectively, and Tob1–/–CD4+ T cells were transfected with lentivirus expressing Id2. Experimental colitis was established in Tob1–/– mice by trinitrobenzene sulfonic acid enema and in Rag1–/– mice reconstituted with Tob1–/–CD45RBhighCD4+ T cells to further explore the role of Tob1 in intestinal mucosal inflammation. Splenic CD4+ T cells of Tob1–/– mice were sorted to determine transcriptome differences by RNA sequencing.
Results
TOB1 expression was decreased in inflamed mucosa and peripheral blood CD4+ T cells of IBD patients compared with healthy subjects. Overexpression of TOB1 downregulated IBD CD4+ T cells to differentiate into Th1/Th17 cells compared with control subjects. Severe colitis was observed in Tob1–/– mice through trinitrobenzene sulfonic acid enema or in Rag1–/– mice reconstituted with Tob1–/–CD45RBhighCD4+ T cells, compared with control animals. RNA sequencing analysis revealed ID2 as functional target of TOB1 to inhibit IBD CD4+ T cell differentiation into Th1/Th17 cells. Mechanistically, TOB1 was associated with Smad4/5 to induce ID2 expression and restrain Th1/Th17 cell differentiation.
Conclusions
TOB1 restrains intestinal mucosal inflammation through suppressing Th1/Th17 cell–mediated immune responses via the Smad4/5-ID2 pathway. It may serve as a novel therapeutic target for treatment of human IBD.
Keywords: TOB1, Inflammatory Bowel Disease, CD4+ T Cells, ID2, Mucosal Inflammation
Abbreviations used in this paper: A-CD, active Crohn’s disease; A-UC, active ulcerative colitis; CD, Crohn’s disease; CDAI, Crohn’s disease activity index; ELISA, enzyme-linked immunosorbent assay; HC, healthy control; IBD, inflammatory bowel disease; IFN, interferon; IFX, infliximab; IL, interleukin; LP, lamina propria; LPMC, lamina propria mononuclear cell; LV-ID2, lentivirus-expressing ID2; LV-NC, negative control lentiviral vector; LV-shID2, lentivirus-expressing ID2 short hairpin RNA; LV-shSmad4, lentivirus-expressing short hairpin Smad4; LV-shTOB1, lentivirus-expressing TOB1 short hairpin RNA; LV-TOB1, lentivirus-expressing TOB1; mAb, monoclonal antibody; MLN, mesenteric lymph node; mRNA, messenger RNA; MS, multiple sclerosis; PB, peripheral blood; PBMC, peripheral blood mononuclear cell; PCR, polymerase chain reaction; qRT-PCR, quantitative real-time polymerase chain reaction; R-CD, Crohn’s disease in remission; R-UC, ulcerative colitis in remission; RNA-seq, RNA sequencing; shRNA, short hairpin RNA; shSmad4, short hairpin Smad4; Th, T helper; TNF, tumor necrosis factor; Treg, regulator T; UC, ulcerative colitis; WT, wild-type
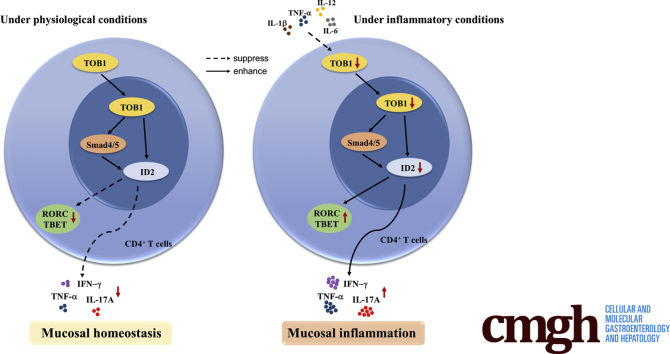
Graphical abstract
Summary.
TOB1 is downregulated in inflamed mucosa of inflammatory bowel disease patients and induces the expression of ID2 in CD4+ T cells by Smad4/5, contributing to the restraint of mucosal inflammation through preventing excessive T helper 1/T helper 17 cell–mediated immune responses in intestinal mucosa.
Inflammatory bowel diseases (IBDs) are chronic inflammatory conditions with a lifelong relapsing-remitting course in the gastrointestinal tract, resulting from an aberrant CD4+ T cell–mediated immune responses to the luminal microbiota in genetically susceptible hosts.1, 2, 3, 4 Crohn’s disease (CD) and ulcerative colitis (UC) are 2 major forms of IBD, with distinct clinical manifestations and endoscopic and histological features. Increasing lines of evidence have demonstrated that aberrant activation of T helper 1 (Th1), Th2, and Th17 cells and defects of regulatory T cells are involved in the pathogenesis of IBD characterized by increased expression of proinflammatory cytokines (eg, interferon γ [IFN-γ], tumor necrosis factor α [TNF-α], interleukin [IL]-17A).5, 6, 7, 8, 9, 10 However, the underlying mechanisms whereby the effector CD4+ T cells are regulated in inflamed mucosa of IBD are not fully understood.
TOB1, originally defined by its interaction with the receptor tyrosine kinase ErbB-211, belongs to the Tob/BTG antiproliferative family.12,13 The Tob/BTG family proteins show high structural similarities in their N-terminal 120-amino-acid region, namely Tob/BTG homology domain, which likely accounts for their function in preventing cell cycle progression and suppressing cell proliferation.13,14 Owing to the ability to inhibit cell growth, TOB1 has been shown to be involved in the occurrence and progression of a variety of tumors, including lung cancer, papillary carcinoma of thyroid, breast cancer, and gastric cancer.15, 16, 17, 18, 19 Moreover, mice with Tob deficiency are predisposed to spontaneously developing tumors in multiple organs (eg, lung, liver, and lymph node), suggesting that Tob1 functions as an important suppressor during tumorigenesis.20 In addition, Tob1 represses the osteoblast proliferation and bone formation through binding Smad proteins and negatively regulating the BMP/Smad signaling.21
Previous studies have also emphasized the crucial roles of TOB1 in regulating T cell activation and immune-mediated disorders.14,22 Recently, decreased TOB1 expression in CD4+ T cells was strongly associated with the progression of multiple sclerosis (MS).23 Interestingly, Tob1–/– mice develop more severe inflammation in the central nervous system during the induction of experimental autoimmune encephalomyelitis, a murine model mimicking MS.24 These results imply that TOB1 acts as a negative regulator of T cell activation and may play a protective role in the pathogenesis of T cell–mediated autoimmunity.
In this study, we identified TOB1 as a crucial regulator in controlling Th1/Th17 cell–mediated intestinal inflammation of IBD and found that TOB1 expression was downregulated in inflamed mucosa and peripheral blood (PB) CD4+ T cells of active IBD patients compared with that in IBD patients in remission and healthy control (HC) subjects. Transfection with lentivirus-expressing TOB1 (LV-TOB1) could suppress IBD PB-CD4+ T cells to differentiate into Th1 and Th17 cells. Severe colitis was observed in Tob1–/– mice through trinitrobenzene sulfonic acid (TNBS) enema or in Rag1–/– mice reconstituted with Tob1–/–CD45RBhighCD4+ T cells. RNA sequencing (RNA-seq) analysis revealed that ID2 acted as the functional target of TOB1 to inhibit IBD CD4+ T cell differentiation into Th1/Th17 cells. Importantly, we verified that TOB1 negatively regulated IBD CD4+ T cell differentiation into Th1/Th17 cells through the Smad4/5-ID2 signaling pathway.
Results
TOB1 Is Downregulated in Active IBD Patients and Refers to the Disease Activities
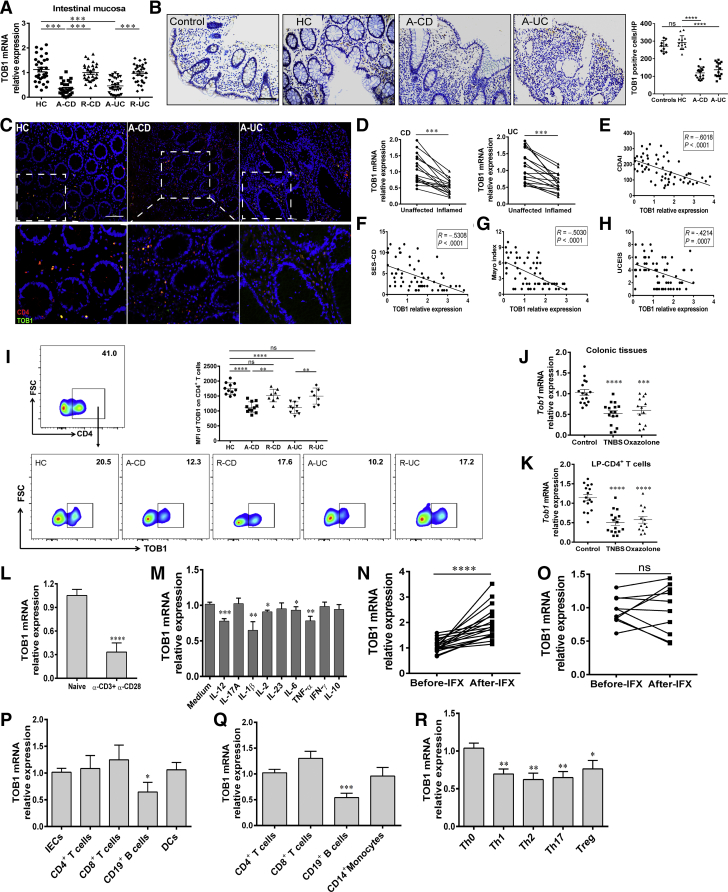
Given that TOB1 plays a crucial role in regulating immune responses and the pathogenesis of MS14,24 and that TOB1 is decreased in inflamed tissues of UC patients as reported previously,25 we sought to investigate whether TOB1 is associated with the pathogenesis of IBD. We found that the expression of TOB1 messenger RNA (mRNA) was downregulated in inflamed mucosa of active CD and UC patients compared with HC subjects (Figure 1A). Consistently, TOB1+ cells were decreased in inflamed mucosa of IBD patients compared with those in normal mucosa or non-IBD inflammatory control subjects (eg, celiac disease, Clostridium difficile infectious colitis, and ischemic enteritis), while no difference in TOB1+ cells was observed between healthy subjects and non-IBD inflammatory control subjects (Figure 1B). Similar trend of TOB1+CD4+ cells was observed in inflamed mucosa of IBD patients compared with those in healthy donors (Figure 1C). A notable decrease of TOB1 expression was also seen in inflamed intestinal mucosa of CD and UC patients, respectively, when compared with that in the unaffected tissues from the same patients (Figure 1D). Furthermore, we analyzed the correlation between TOB1 expression and disease severity of IBD. We found that the levels of TOB1 mRNA in inflamed colon tissues were inversely correlated with CDAI and Simple Endoscopic Score for Crohn’s Disease in CD patients, and the Mayo index and Ulcerative Colitis Endoscopic Index of Severity in UC patients, respectively (Figure 1E–H). Quantitative polymerase chain reaction (PCR) and flow cytometry analysis further revealed that expression of TOB1 was reduced in PB-CD4+ T cells from active IBD patients compared with control subjects (Figure 1I).
Figure 1.
TOB1 expression is decreased in active IBD patients. (A) Colon biopsies were collected from 29 patients with A-CD, 29 patients with R-CD, 31 patients with A-UC, 30 patients with R-UC, and 31 HC subjects, respectively, and TOB1 expression was analyzed by qRT-PCR. (B) Immunohistochemistry (IHC) analysis of TOB1 in representative sections from colon mucosa of non-IBD inflammatory control subjects (n = 9, including 3 patients with celiac disease, 3 patients with Clostridium difficile infectious colitis, and 3 patients with ischemic enteritis), HC subjects (n = 13), patients with A-CD (n = 17), and patients with A-UC (n = 16), respectively. Scale bars = 100 μm. (C) Immunofluorescence double staining showed co-localization of TOB1 with CD4+ cells in HC, A-CD, and A-UC samples, respectively. Scale bars = 100 μm. (D) TOB1 expression in inflamed and unaffected colon tissues from the same patients with active CD (n = 16) and active UC (n = 17), respectively, was examined by qRT-PCR. (E–H) TOB1 expression in intestinal mucosa of CD patients was observed to be negatively associated with CDAI and Simple Endoscopic Score for Crohn’s Disease (R = –0.6018, P < .0001; and R = –0.5308, P < .0001, respectively) and the correlation analysis between Mayo score, Ulcerative Colitis Endoscopic Index of Severity, and TOB1 expression in intestinal mucosa of UC patients (R = –0.5030, P < .0001; and R = –0.4214, P = .0007, respectively). (I) PB-CD4+ T cells were obtained from HC subjects (n = 11), patients with A-CD (n = 11), patients with R-CD (n = 8), patients with A-UC (n = 10), and patients with R-UC (n = 7), respectively. TOB1 expression was analyzed by flow cytometry, and MFIs (Median Fluorescence Intensity) of TOB1 expression were shown in each group. (J and K) Colon tissues and LP-CD4+ T cells were collected from TNBS- (n = 16), oxazolone-induced (n = 13) colitis mice and WT control animals (n = 16), respectively, and Tob1 expression was examined by qRT-PCR. (L) Naïve PB-CD4+ T cells were sorted from 13 healthy donors and cultured with plate-bound anti-CD3 mAb (5 μg/mL) and soluble anti-CD28 mAb (2 μg/mL) for 24 hours, and qRT-PCR was performed to analyze TOB1 expression. ∗∗P < .01 vs naïve CD4+ T cells. (M) PB-CD4+ T cells (5 × 105/mL) from healthy donors (n = 10) were stimulated with various cytokines (10 ng/mL) for 48 hours, and TOB1 expression was analyzed by qRT-PCR. Gene expression was normalized to GAPDH in each group. (N and O) 29 patients with active CD were receiving anti-TNF mAb (ie, infliximab, IFX) treatment at weeks 0, 2, and 6, and intestinal mucosal biopsies were collected from these patients at week 14 after an induction therapy, (N) 19 patients achieved clinical remission, and (O) 10 patients failed to IFX therapy. (P) Expression of TOB1 mRNA in intestinal mucosal biopsies was analyzed by qRT-PCR. LP-CD4+, CD8+ T, CD19+ B cells, dendritic cells (DCs), and intestinal epithelial cells (IECs) were isolated from normal colon tissues of 6 patients who underwent colectomy for colon cancer, and the mRNA levels of TOB1 were analyzed by qRT-PCR. ∗P < .05 vs IECs. (Q) CD4+, CD8+ T, CD19+ B cells, and CD14+ monocytes were isolated from PB of 8 healthy donors by immunomagnetic positive selection, and TOB1 expression was determined by qRT-PCR. ∗∗∗P < .001 vs CD4+ T cells. (R) PB-CD4+ T cells were isolated from 6 healthy donors and cultured with plate-bound anti-CD3 mAb (5 μg/mL) and soluble anti-CD28 mAb (2 μg/mL) under Th0, Th1, Th2, Th17, and Treg cell–polarizing conditions, respectively, for 5 days, and qRT-PCR was performed to analyze TOB1 expression in these CD4+ T cells. ∗P < .05, and ∗∗P < .01 vs Th0 cells. Spearman rank correlation was used to analyze the correlation between TOB1 expression and disease activities of IBD patients. Statistical analysis was performed with paired 2-sided Student’s t tests or analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
To further corroborate our preceding findings, we induced experimental colitis models in wild-type (WT) C57BL/6 mice by TNBS and oxazolone enema, respectively. We observed that Tob1 expression was significantly reduced in the inflamed colon tissues and lamina propria (LP)-CD4+ T cells of TNBS- or oxazolone-induced colitis mice compared with that of ethanol-treated control animals (Figure 1J and K). Collectively, these data indicate that TOB1 may play a role in the pathogenesis of IBD and serve as a biomarker for evaluating disease severity.
Previous study has demonstrated that TOB1 is highly expressed in unstimulated PB-CD4+ T cells but is downregulated during activation.22 Consistently, we also observed that TOB1 mRNA was decreased in activated CD4+ T cells compared with naïve CD4+ T cells in vitro (Figure 1L). Because a variety of proinflammatory cytokines are present in inflamed mucosa of IBD patients, we then explored whether those cytokines could modulate TOB1 expression in CD4+ T cells. We found that TNF-α, IL-1β, IL-2, IL-6, and IL-12 could suppress TOB1 expression in CD4+ T cells (Figure 1M). To further clarify if TNF-α, a critical proinflammatory cytokine present in human IBD, is involved in the downregulation of TOB1 in active IBD patients, we measured the alternations of TOB1 mRNA expression in intestinal mucosa of active CD patients who received an induction therapy with anti-TNF-α mAb (ie, infliximab [IFX]) and found that TOB1 mRNA increased in intestinal mucosa of patients with active CD in the response group (CDAI <150) at week 14 after an induction therapy with IFX compared with that before IFX therapy, but it did not change in gut mucosa of CD patients in nonresponse group (Figure 1N and O). Altogether, these data suggest that proinflammatory cytokines (eg, TNF-α, IL-1β, IL-2, IL-6, and IL-12) enable to restrain TOB1 expression, and that blockage of TNF-α could enhance TOB1 expression in intestinal mucosa.
We further found that the levels of TOB1 mRNA in normal colon tissues were comparable among intestinal epithelial cells and different immune cell subsets, albeit at a relatively lower level in B cells (Figure 1P). Likewise, similar results were also observed in immune cells from PB of healthy volunteers (Figure 1Q). A previous study reported that TOB1 expression is increased in Th17 cells (CD161+IFN-γ–IL-17A+CD4+ cells) compared with Th1 cells (CD161–IL-17A–IFN-γ+CD4+ cells) after being stimulated in vitro with anti-CD3/CD28 for 7 days.26 To delineate the expression of TOB1 in CD4+ T cells under differential conditions, we sorted naïve CD4+ T cells from healthy donors, cultured them in vitro under different Th-polarizing conditions as indicated for 5 days, and found that TOB1 was decreased in different activated CD4+ T cells including Th1, Th2, Th17, and regulator T (Treg) cells (Figure 1R), which was consistent with the previous work.22 Taken together, these data indicate that TOB1 is ubiquitously expressed in different immune cells of LP mononuclear cells (LPMCs) and PB mononuclear cells (PBMCs) and that its decreased expression may contribute to the development of IBD.
Forced TOB1 Expression in CD4+ T Cells Restricts Excessive Th1/Th17 Cell Immune Responses in IBD Patients
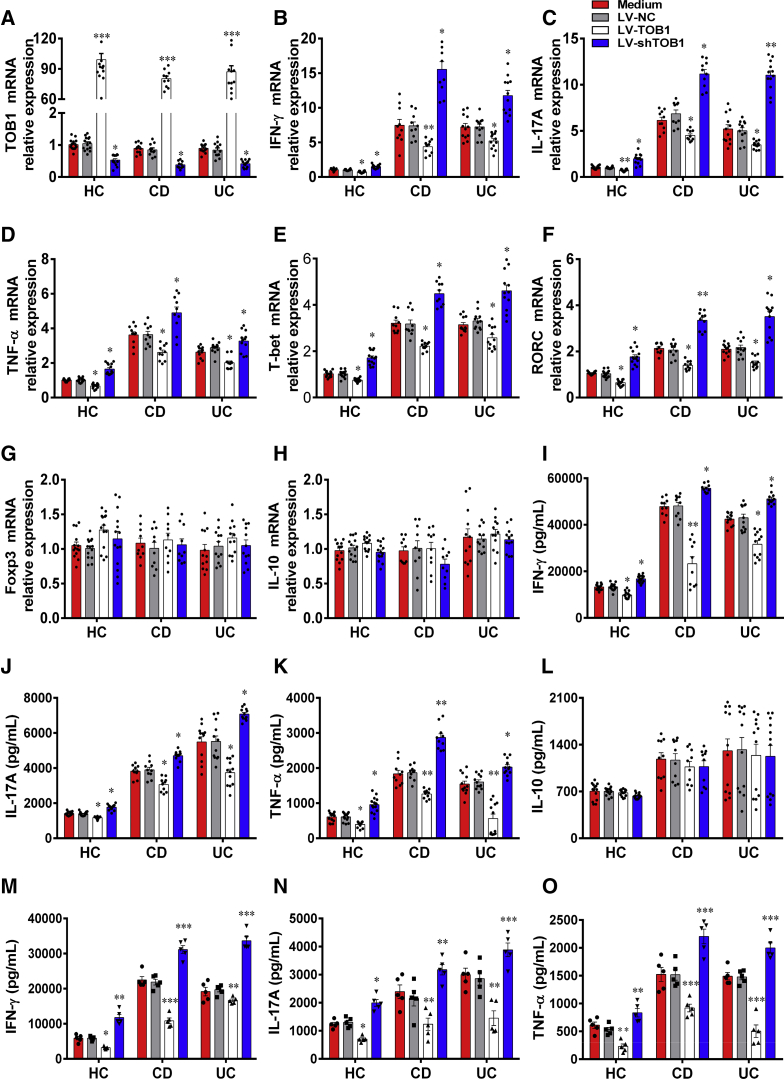
The decreased expression of TOB1 in IBD patients promoted us to investigate whether TOB1 affects IBD CD4+ T cell differentiation. To this end, we isolated PB-CD4+ T cells from active IBD patients and healthy subjects and transfected them with LV-TOB1, lentivirus-expressing TOB1 short hairpin RNA (shRNA) (LV-shTOB1), and negative control lentiviral vector (LV-NC), respectively. We found that the expression of TOB1 mRNA was significantly higher in LV-TOB1–transfected CD4+ T cells than in LV-NC–transfected CD4+ T cells, while LV-shTOB1 transfection strongly inhibited TOB1 expression in CD4+ T cells (Figure 2A). We then examined the expression of cytokines and related transcription factors in these transfected CD4+ T cells. Interestingly, we found that LV-TOB1–transfected IBD CD4+ T cells expressed lower levels of IFN-γ, IL-17A, TNF-α, T-bet, and RORC compared with control subjects, whereas the inhibition of TOB1 in IBD CD4+ T cells resulted in an opposite effect (Figure 2B–F). In addition, both IL-10 and Foxp3 expression was not affected in CD4+ T cells by LV-TOB1 or LV-shTOB1 transfection (Figure 2G and H). These findings were further confirmed by enzyme-linked immunosorbent assay (ELISA), showing that the amounts of signature cytokines (eg, IFN-γ, IL-17A, and TNF-α) were significantly decreased in culture supernatants of LV-TOB1–transfected CD4+ T cells compared with those in LV-NC–transfected CD4+ T cells, whereas IL-10 production was not altered when CD4+ T cells were transfected with LV-TOB1 or LV-shTOB1 (Figure 2I–L). In addition, we further isolated LP-CD4+ T cells from active IBD patients and healthy subjects and transfected them with LV-TOB1, LV-shTOB1, and LV-NC, respectively. These transfected LP-CD4+ T cells were cultured for 48 hours under stimulation with immobilized anti-CD3 and anti-CD28 monoclonal antibodies (mAbs) in vitro. The expression of IFN-γ, IL-17A, and TNF-α was then assessed in culture supernatants by ELISA, and we found that these proinflammatory cytokines were markedly decreased in culture supernatants of LV-TOB1–transfected LP-CD4+ T cells but were significantly increased in LV-shTOB1–transfected LP-CD4+ T cells compared with those in LV-NC–transfected LP-CD4+ T cells (Figure 2M–O). Taken together, these data indicate that TOB1 deficiency in CD4+ T cells results in augmenting Th1 and Th17 cell immune responses in IBD patients, while forced TOB1 expression in CD4+ T cells could reverse such an effect.
Figure 2.
Forced expression of TOB1 in IBD CD4+T cells restricts excessive Th1/Th17 cell immune responses. PB-CD4+ T cells from patients with A-CD (n = 10), patients with A-UC (n = 12), and HC subjects (n = 14) were transfected with LV-TOB1, LV-shTOB1, and LV-NC, respectively, and then cultured under stimulation with plate-bound anti-CD3 mAb (5 μg/mL) and soluble anti-CD28 mAb (2 μg/mL) for 5 days. (A) The transfected CD4+ T cells were harvested on day 5, and the transfection efficiencies of LV-TOB1 and LV-shTOB1 were confirmed by qRT-PCR, and (B-H) qRT-PCR was performed to determine the mRNA expression of IFN-γ, IL-17A, TNF-α, T-bet, RORC, Foxp3, and IL-10, respectively, in these CD4+ T cells. (I-L) The culture supernatants of these transfected CD4+ T cells were also collected for detecting the levels of IFN-γ, IL-17A, TNF-α, and IL-10, respectively, using ELISA (M-O). LP-CD4+ T cells isolated from inflamed colon tissues of A-CD patients (n = 5), A-UC patients (n = 5) and normal colon tissues of 5 patients who underwent colectomy for colon cancer, respectively, were transfected with LV-TOB1, LV-shTOB1, and LV-NC, respectively, and then cultured under stimulation with plate-bound anti-CD3 mAb (5 μg/mL) and soluble anti-CD28 mAb (2 μg/mL) for 48 hours. IFN-γ, IL-17A, and TNF-α were detected by ELISA in the culture supernatants of these transfected LP-CD4+ T cells. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 vs LV-NC from the same group.
Tob1 Deficiency Exacerbates TNBS-Induced Colitis in Mice
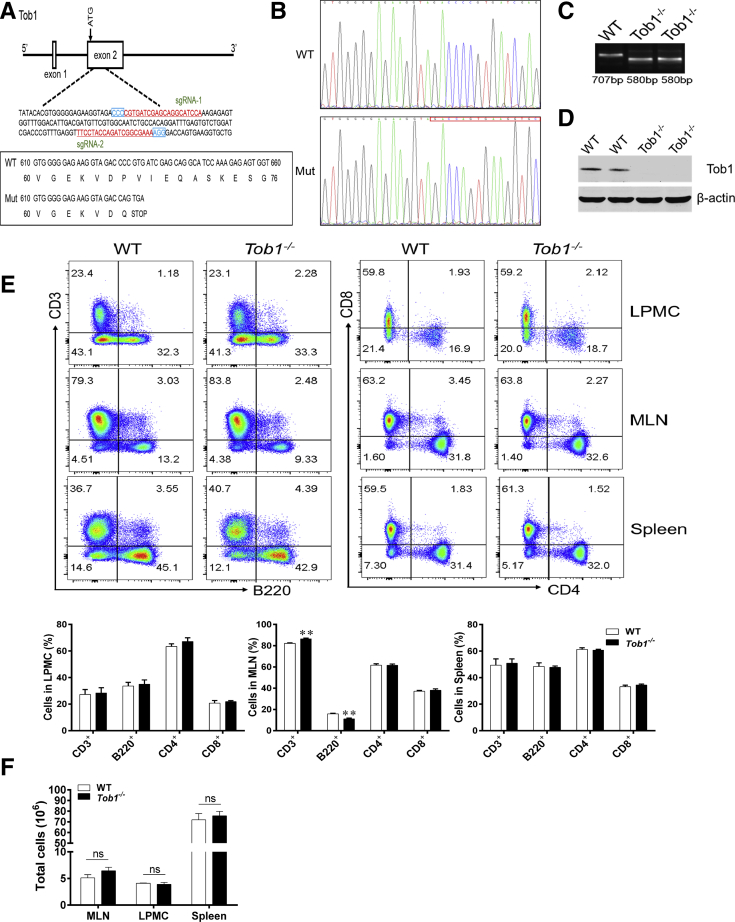
To explore potential roles of Tob1 in the pathogenesis of colitis in vivo, we generated Tob1–/– mice by the CRISPR/Cas9 technique (Figure 3A–C). Western blotting analysis confirmed the absence of TOB1 protein in the colon tissues of Tob1–/– mice (Figure 3D). Tob1–/– mice were viable and appeared apparently healthy over the duration of experiments. By flow cytometric analysis, we found that Tob1 deficiency had no appreciable influence on the frequencies of B, CD4+, and CD8+ T cells in spleen and LPMCs, although Tob1–/– mice showed slightly higher percentage of CD3+ T cells and lower percentage of B cells in the mesenteric lymph node (MLN) compared with their WT littermates (Figure 3E). Moreover, the total number of immune cells in MLN, LPMC, and spleen was not affected by Tob1 deficiency (Figure 3F). Therefore, these data indicate that Tob1 deficiency does not affect immune cell proliferation under physiological conditions.
Figure 3.
The generation and genotyping of Tob1–/–mice. (A) Tob1–/– mice were generated using the CRISPR/Cas9 technique. The 2 single guide RNAs targeting the WT and Tob1 sequence are underlined and highlighted in red, and the protospacer adjacent motif sequences are labeled in blue. The reading frames of WT and mutant Tob1 sequence are listed below. The 127-bp deletion in Tob1 sequence results in an early stop codon TGA. (B and C) Target loci of Tob1 were amplified using genomic DNA templates from mouse tails of WT and Tob1–/– mice. PCR products of the targeted fragment were subjected to (B) sequencing and (C) agarose gel electrophoresis, indicating the 127 missing bp in the Tob1–/– mice. (D) Protein levels of Tob1 in the colon tissues of WT littermates and Tob1–/– mice were detected by Western blotting, and β-actin was used as endogenous reference gene. (E) LPMCs and single-cell suspension of MLNs and spleens were obtained from 6- to 8-week-old WT littermates (n = 4) and Tob1–/– mice (n = 4), respectively, and then stained with fluorochrome-conjugated anti-CD3, anti-CD4, anti-CD8, and anti-B220 mAbs, respectively. The frequencies of B220+ B cells, CD3+ T cells (gated on total cells), and CD4+ and CD8+ T cells (gated on CD3+ T cells) in LPMCs, MLNs, and spleens, respectively, were analyzed by flow cytometry. Percentages of these cells in LPMCs, MLNs, and spleens were shown in the bar chart, respectively. (F) The total cells in MLN, LPMC, and spleen were calculated and are presented in the bar chart. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗∗P < .01 vs WT control animals from the same group.
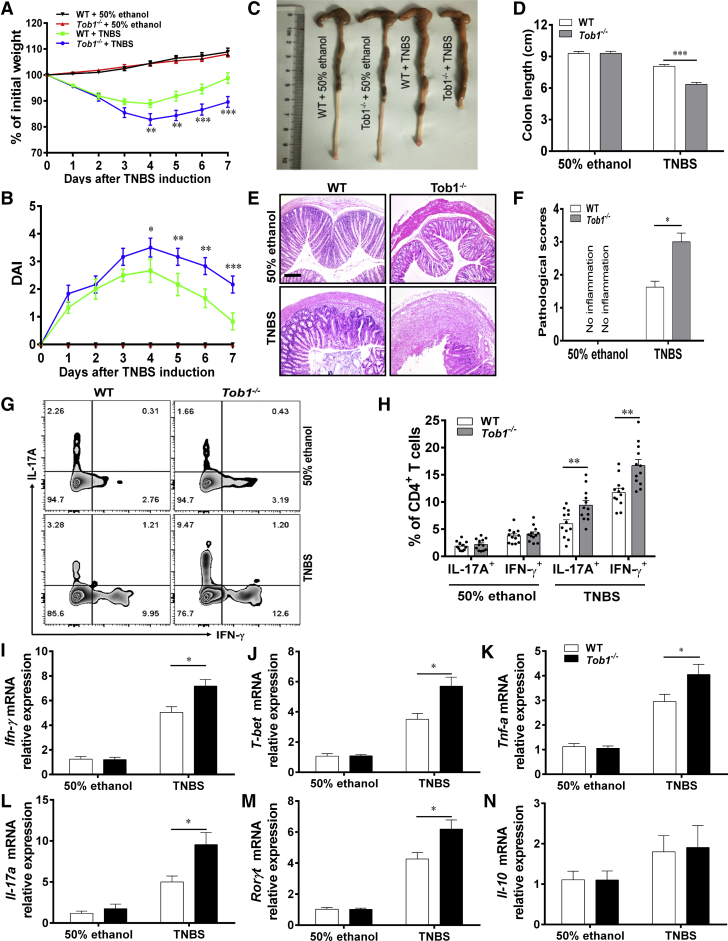
To determine whether Tob1 deficiency affects the development of colitis, we established TNBS-induced acute colitis model in Tob1–/– mice and WT littermates. We found that Tob1–/– mice showed enhanced disease severity of colitis compared with WT littermates, as exemplified by an excessive weight loss, shortened colon length, increased lymphocyte infiltration, and higher levels of pathological scores after TNBS treatment (Figure 4A–F). On the basis of our preceding observation in IBD patients, we asked whether the loss of Tob1 favored Th1 and Th17 cell differentiation in inflamed colon of colitic mice. As shown in Figure 4G and H, LP-CD4+ T cells from the colon tissues of TNBS-induced colitic Tob1–/– mice displayed higher frequencies of Th1 and Th17 cells compared with those from WT littermates, as determined by intracellular cytokine staining analysis. In agreement, quantitative real-time PCR (qRT-PCR) analysis also revealed that the mRNA levels of Ifn-γ, T-bet, Il-17a, Rorγt, and Tnf-α were significantly increased in the colon tissues of TNBS-induced colitic Tob1–/– mice compared with those of WT control animals (Figure 4I–M). However, no significant difference in expression of Il-10 between WT littermates and Tob1–/– mice was seen upon TNBS exposure (Figure 4N).
Figure 4.
Tob1 deficiency exacerbates TNBS-induced colitis in mice. The TNBS-induced colitis model was established in WT littermates (n = 12) and Tob1–/– mice (n = 12). (A) The changes of body weight in WT littermates and Tob1–/– mice were recorded every day over the duration of this experiment. (B) The scores of disease activity index (DAI) were calculated daily after TNBS administration. (C) WT littermates and Tob1–/– mice were sacrificed on day 7 after TNBS induction, and gross morphology of colons is shown. (D) The colon length of WT littermates and Tob1–/– mice is shown in the bar chart. (E) Representative colon sections were stained with hematoxylin and eosin. Scale bars = 200 μm. (F) Pathological scores of the colon tissues were calculated and shown as indicated. (G and H) Intracellular expression of IFN-γ and IL-17A in LP-CD4+ T cells was analyzed by flow cytometry after stimulation with PMA and ionomycin for 5 hours, the percentages of IFN-γ+CD4+ and IL-17A+CD4+ T cells are shown in the bar chart. (I–N) Colon tissues were collected from colitic mice and control animals, and the expression of Ifn-γ, T-bet, Tnf-α, Il-17a, Rorγt, and Il-10 mRNA was examined by qRT-PCR. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001 vs TNBS-treated WT group. Data are representative of 3 independent experiments.
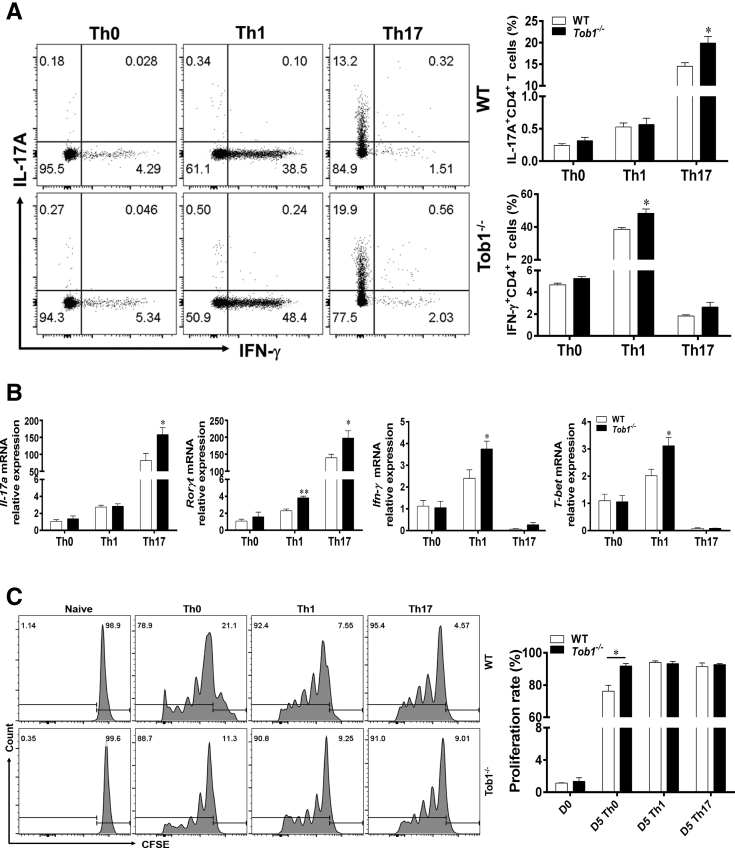
To further verify these findings, we differentiated splenic CD4+ T cells derived from Tob1–/– mice and WT littermates under Th0-, Th1-, and Th17-polarizing conditions in vitro. As shown in Figure 5A, Tob1–/–CD4+ T cells generated more IFN-γ and IL-17A under Th1- and Th17-polarizing conditions, respectively, compared with WT littermates. These results were further confirmed by qRT-PCR analysis, showing that the expression of Ifn-γ/T-bet and Il-17a/Rorγt mRNA was also enhanced in Tob1–/–CD4+ T cells in comparison with that in control animals under the same conditions (Figure 5B). Meanwhile, we detected the proliferation of splenic CD4+ T cells from both WT littermates and Tob1–/– mice under Th0-, Th1-, and Th17-polarizing conditions, respectively, in vitro, and found that the proliferation of Tob1–/–CD4+ T cells was increased under the Th0 condition, but did not alter under Th1 and Th17 conditions compared with WT control animals (Figure 5C). Taken together, our data indicate that Tob1 is required for controlling excessive TNBS-induced colitis in mice.
Figure 5.
Tob1 deficiency facilitates Th1 and Th17 cell differentiation in vitro. Naïve CD4+ T cells were isolated from spleens of WT littermates (n = 4) or Tob1–/– mice (n = 4) using anti-mouse CD4 magnetic beads, and cultured with plate-bound anti-CD3 mAb (5 μg/mL) and soluble anti-CD28 mAb (2 μg/mL) under Th0, Th1, and Th17 cell–polarizing conditions, respectively. (A and B) The polarizing CD4+ T cells were harvested on day 5, and intracellular expression of IFN-γ and IL-17A was analyzed by flow cytometry and qRT-PCR after stimulation with PMA and ionomycin. Percentages of IFN-γ+CD4+ and IL-17A+CD4+ T cells are shown in the bar chart. (C) Splenic CD4+ T cells of WT and Tob1–/– mice (n = 4) were labeled with CFSE (2 μM) and stimulated with plate-bound anti-CD3 (5 μg/mL) and soluble anti-CD28 (2 μg/mL) under Th0, Th1, and Th17 cell–polarizing conditions, respectively. The cell proliferation rates were then measured by flow cytometry on day 5. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗P < .05 vs WT control animals from the same group. Data are representative of 3 independent experiments.
Tob1–/–CD45RBhighCD4+ T Cells Induce Exaggerated Intestinal Mucosal Inflammation in Rag1–/– Mice
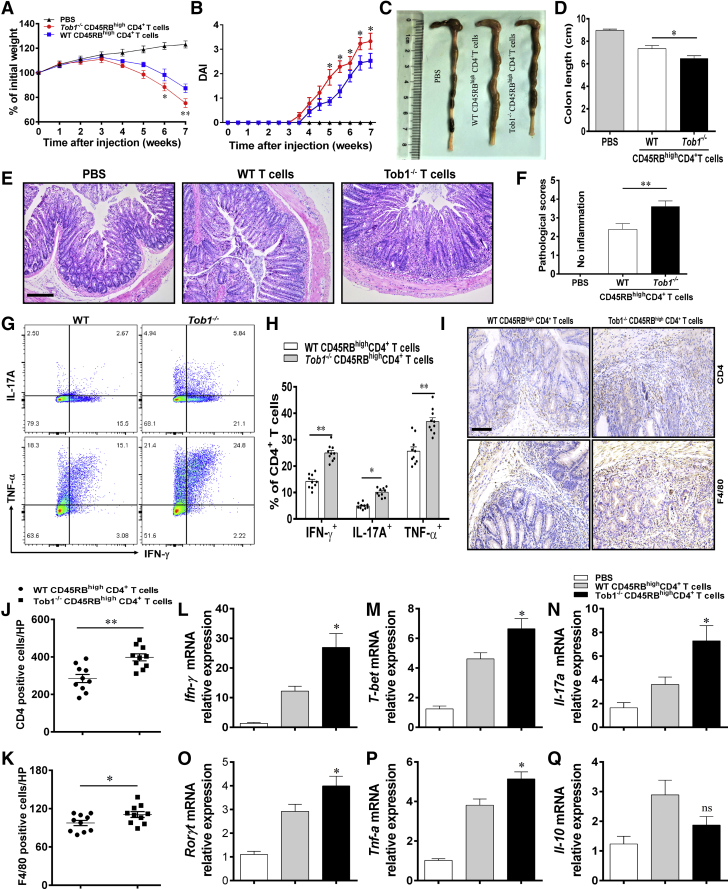
With these data in hand, we further investigated whether the loss of Tob1 in CD4+ T cells preferentially induced chronic mucosal inflammation in vivo. To address this, we established a chronic colitis model in Rag1–/– mice by intraperitoneal injection of CD45RBhighCD4+ T cells from WT littermates or Tob1–/– mice according to our previous report.27 As shown in Figure 6A–F, we found that Rag1–/– mice reconstituted with Tob1–/–CD45RBhighCD4+ T cells appeared with more severe loss of body weight and shortened colon length compared with the recipient mice reconstituted with WT CD45RBhighCD4+ T cells. Flow cytometric analysis further revealed that the lack of Tob1 in CD4+ T cells gave rise to enhanced Th1/Th17 cell immune responses in Tob1–/–CD45RBhighCD4+ T cell–reconstituted Rag1–/– mice, as evidenced by higher proportions of IFN-γ+CD4+, IL-17A+CD4+, and TNF-α+CD4+T cells in LPMCs (Figure 6G and H). In agreement, Rag1–/– mice reconstituted with Tob1–/–CD45RBhighCD4+ T cells appeared an increase of inflammatory cell (eg, CD4+ T cells and F4/80+ macrophages) infiltrations in inflamed colonic tissues compared with the recipient mice reconstituted with WT CD45RBhighCD4+ T cells (Figure 6I–K). Meanwhile, the mRNA levels of Ifn-γ, T-bet, Il-17a, Rorγt, and Tnf-α were elevated in the colon tissues from Tob1–/–CD45RBhighCD4+ T cell–reconstituted Rag1–/– mice compared with those from control animals, whereas no statistical difference was seen in the expression of Il-10 mRNA between these 2 groups (Figure 6L–Q). Collectively, these data imply that loss of Tob1 in CD4+ T cells contributes to an enhanced susceptibility to intestinal mucosal inflammation via augmenting Th1/Th17 cell immune responses.
Figure 6.
Tob1–/–CD45RBhighCD4+T cells induce exaggerated intestinal mucosal inflammation in Rag1–/–mice. Naïve CD25–CD45RBhighCD4+ T cells were sorted from spleen of WT littermates and Tob1–/– mice (n = 5) and injected intraperitoneally into 8-week-old Rag1–/– mice (5 × 105 cells/mouse and n = 10, respectively). (A) Changes of body weight in Rag1–/– mice reconstituted intraperitoneally with WT and Tob1–/–CD45RBhighCD4+ T cells, respectively, or injected intraperitoneally with phosphate-buffered saline as negative control, were monitored weekly after T cell transfer. (B) The scores of disease activity index (DAI) were calculated daily after T cell adoptive transfer. (C and D) Mice were sacrificed on week 7 after T cell transfer, and gross morphology of colons is shown. (E and F) Pathological scores of the colon tissues were calculated and are shown as indicated, and representative colon sections were stained with hematoxylin and eosin. Scale bars = 100 μm. (G) LPMCs were isolated from the colons of Rag1–/– mice transferred with WT and Tob1–/–CD45RBhighCD4+ T cells, respectively, and intracellular expression of IFN-γ and IL-17A in LP-CD4+ T cells was analyzed by flow cytometry. (H) Percentages of IFN-γ+CD4+ and IL-17A+CD4+ T cells are shown in the bar chart. (I–K) Representative images for the detection of CD4+ T cells and F4/80+ macrophages in colon tissues 7 weeks after T cell transfer in Rag1–/– mice by immunohistochemistry. Scale bar = 100 μm. (L–Q) Colon tissues were collected from these recipient mice 7 weeks after T cell transfer, and the expression of Ifn-γ, T-bet, Il-17a, Rorγt, Tnf-α, and Il-10 mRNA was examined by qRT-PCR. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗P < .05 and ∗∗P < .01 vs Rag1–/– mice transferred with WT CD45RBhighCD4+ T cells.
Tob1 Suppresses Th1 and Th17 Cell Differentiation in the Smad4/5-Id2-Dependent Pathway
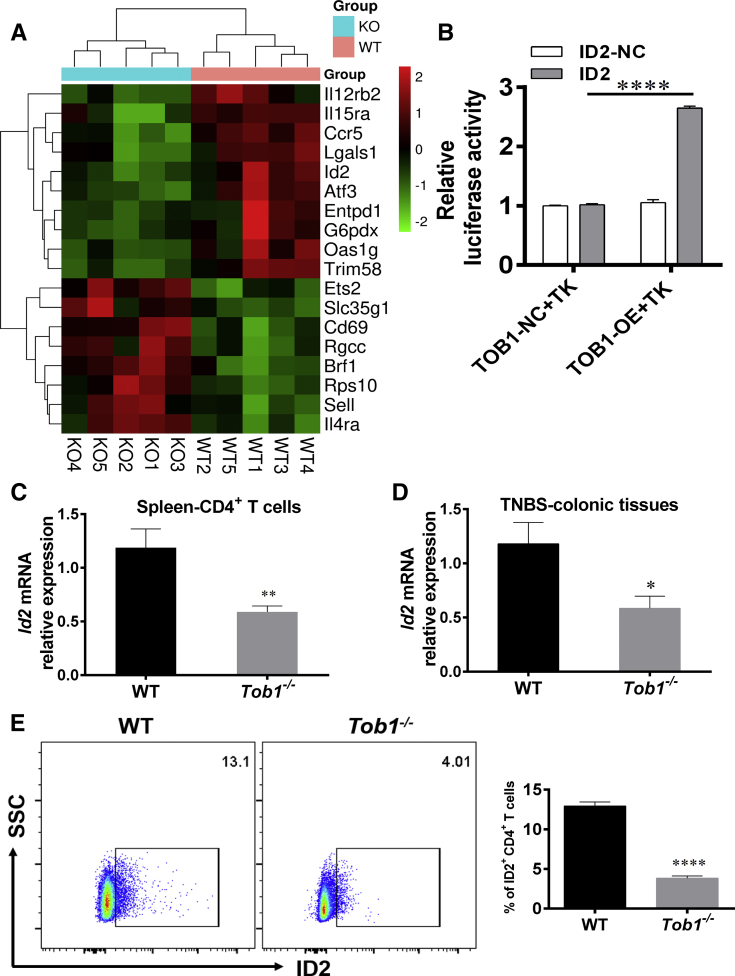
To further clarify the underlying mechanisms whereby Tob1 restrains colitis in mice and modulates CD4+ T cell differentiation, we sorted splenic CD4+ T cells of Tob1–/– mice and WT littermates for RNA-seq. We observed that an Id2 was markedly downregulated in Tob1–/–CD4+ T cells compared with that in WT control animals (Figure 7A). Id2 is a transcription factor of the HLH family and target of BMP signals in various types of cells,28 and upregulated expression of Id2 via the BMP-responsive Smad suppresses cell differentiation.29 A previous study demonstrated that Tob enhances Smad DNA binding by associating with Smad2 and Smad4, and augments Smad-mediated inhibition of IL-2 transcription.22 We further utilized luciferase reporter assays including ID2 or control vectors and found that 293T cells transfected with TOB1-overexpressing plasmid increased the relative luciferase activity of ID2 (Figure 7B), proving that TOB1 induces ID2 expression. Consistently, we observed that Id2 was reduced in splenic CD4+ T cells and inflamed colon tissues of Tob1–/– mice (Figure 7C–E), suggesting that Tob1 plays a crucial role in modulating CD4+ T cell differentiation in an Id2-dependent manner.
Figure 7.
Id2 is downregulated in splenic CD4+cells of Tob1–/–mice. (A) Compared with WT littermates (n = 5), RNA-seq of splenic Tob1–/–CD4+ T cells showed that the mRNA levels of several anti-inflammatory genes (eg, Il12rb2, Il15ra, Ccr5, Lgals1, Id2, Atf3, Entpd1, G6pdx, Oas1g, and Trim58) were decreased in Tob1–/– mice (n = 5). (B) Luciferase reporter assays using vectors constituted with ID2 promoter or control in the presence of pGMLR-TK and TOB1-overexpressing vector or control. (C and D) Id2 expression was reduced in splenic CD4+ T cells and colon tissues from TNBS-induced colitis of Tob1–/– mice (n = 10) compared with WT littermates (n = 10). (E) Id2 expression was decreased in splenic CD4+ T cells of Tob1–/– mice. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001 vs control animals.
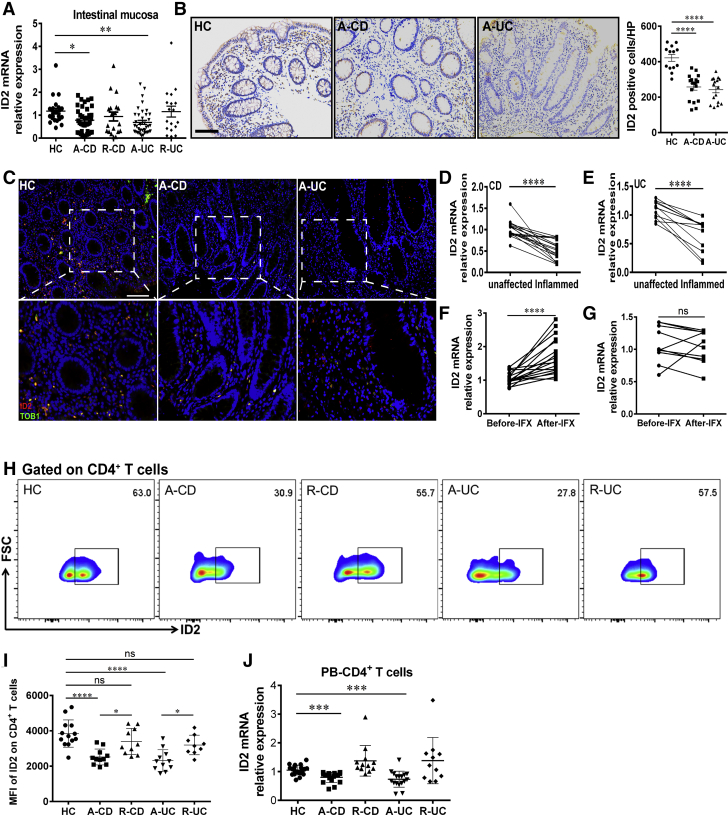
The decreased expression of Id2 in Tob1–/– mice promoted us to investigate whether ID2 is associated with the development of human IBD. As shown in Figure 8A–C, we found that the levels of ID2 mRNA and the frequencies of ID2+TOB1+ cells were downregulated in inflamed mucosa of active CD and UC patients compared with those in healthy donors. Moreover, a decrease of ID2 expression was also observed in inflamed mucosa of CD or UC patients when compared with that in the unaffected colon tissues from the same patients (Figure 8D and E). Interestingly, ID2 expression was increased in intestinal mucosa of CD patients in the response group (CDAI <150) at week 14 after an induction therapy with IFX compared with that before therapy, but it did not alter in CD patients in the nonresponse group (Figure 8F and G). Consistently, ID2 was observed to be downregulated in PB-CD4+ T cells from active IBD patients by contrast to healthy subjects (Figure 8H–J). Taken together, these results suggest that ID2 may play a role in the pathogenesis of IBD.
Figure 8.
Expression of ID2 is decreased in active IBD patients. (A) Colon biopsies were collected from patients with A-CD (n = 39), patients with R-CD (n = 19), patients with A-UC (n = 39), patients with R-UC (n = 18), and HC subjects (n = 23), respectively, and ID2 expression was analyzed by qRT-PCR. (B) Immunohistochemistry (IHC) analysis of ID2 in representative sections from normal colon of HC subjects (n = 13) and inflamed colons of patients with A-CD (n = 17) and patients with A-UC (n = 16), respectively. Scale bars = 100 μm. (C) Immunofluorescence double staining for co-localization of ID2 with TOB1 cells was shown in normal colon of HC and inflamed colon from active CD and UC patients, respectively. Scale bars = 100 μm. (D and E) ID2 expression in inflamed and unaffected colon tissues from the same patients with A-CD (n = 18) and patients with A-UC (n = 14) was examined by qRT-PCR. (F and G) Twenty-nine patients with active CD were receiving IFX treatment at weeks 0, 2, and 6, and intestinal mucosal biopsies were collected from these patients at week 14 after the first infusion. (F) Nineteen patients achieved clinical remission and (G) 10 patients failed to response to IFX therapy before and 14 weeks after the first infusion. Expression of ID2 mRNA in intestinal mucosa of these patients was analyzed by qRT-PCR. (H and I) PB-CD4+ T cells were obtained from HC subjects (n = 13), patients with A-CD (n = 11), patients with R-CD (n = 10), patients with A-UC (n = 11), and patients with R-UC (n = 9), respectively, and ID2 expression was determined by flow cytometry. (J) PB-CD4+ T cells were isolated from patients with A-CD (n = 17), patients with R-CD (n = 12), patients with A-UC (n = 17), and patients with R-UC (n = 11), and HC subjects (n = 20), respectively, and qRT-PCR was performed to determine ID2 expression in PB-CD4+ T cells. Statistical analysis was performed with paired 2-sided Student’s t tests or analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
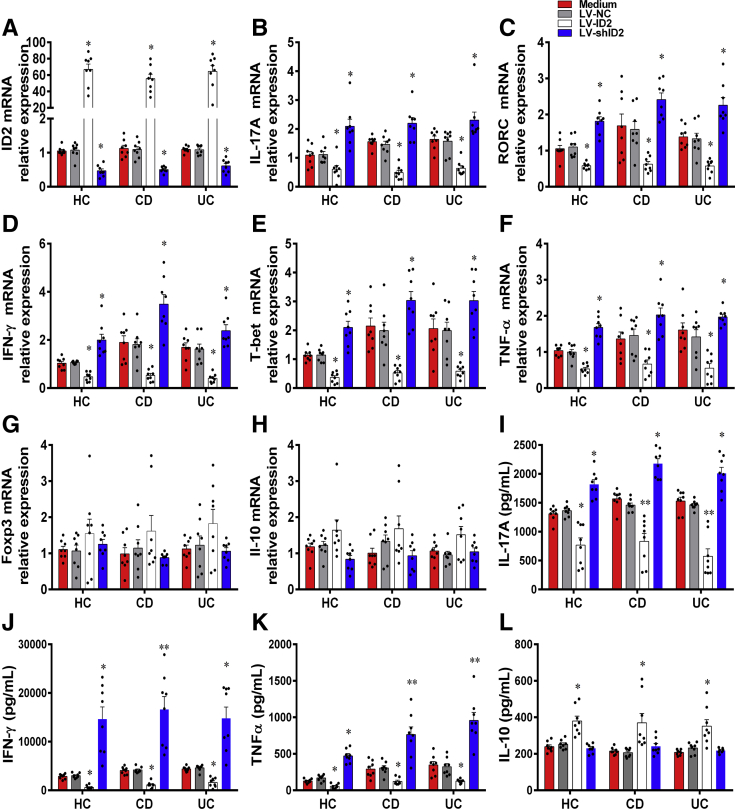
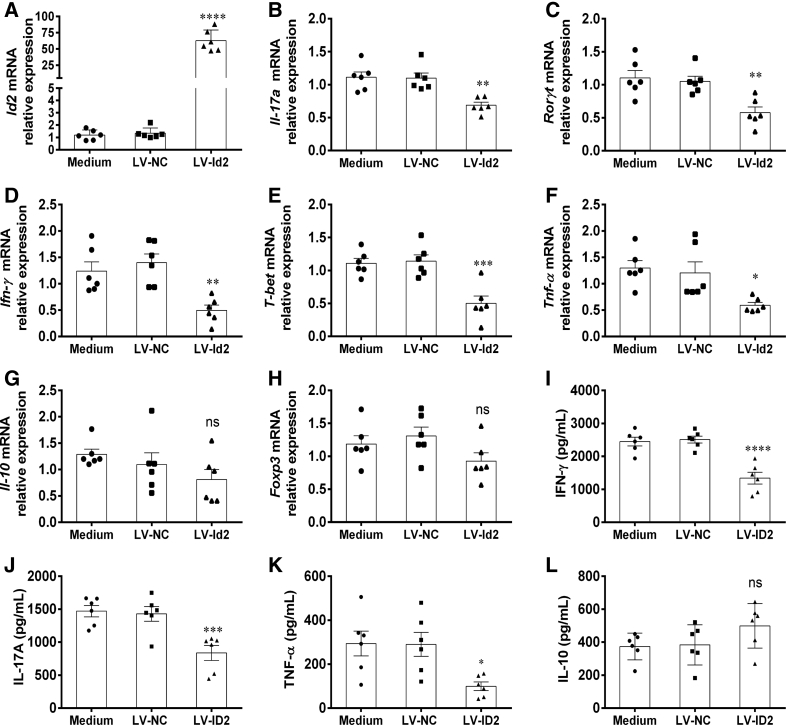
Furthermore, we also isolated CD4+ T cells from active IBD patients and healthy individuals, and then transfected them with LV-expressing ID2 (LV-ID2), LV-expressing ID2 shRNA (LV-shID2), and LV-NC, respectively. We observed that the expression of ID2 was higher in LV-ID2–transfected CD4+ T cells than that in LV-NC–transfected CD4+ T cells, while LV-shID2 transfection strongly inhibited ID2 expression in CD4+ T cells (Figure 9A). We then found that LV-ID2–transfected IBD CD4+ T cells expressed lower levels of IL-17A, RORC, IFN-γ, T-bet, and TNF-α compared with control subjects, whereas the inhibition of ID2 expression in IBD CD4+ T cells had an opposite effect (Figure 9B–F). However, the expression of IL-10 and Foxp3 was not altered in IBD CD4+ T cells by LV-ID2 transfection (Figure 9G and H). These findings were further confirmed by ELISA, showing that the amounts of signature cytokines (eg, IFN-γ, IL-17A, and TNF-α) were decreased, while IL-10 was increased in culture supernatants of LV-ID2–transfected CD4+ T cells in comparison with those in LV-NC–transfected CD4+ T cells (Figure 9I–L). In line with these results, overexpression Id2 in Tob1–/–CD4+ T cells resulted in lowered expression of Th1/Th17-related cytokines (eg, Il-17a, Rorγt, Ifn-γ, T-bet, and Tnf-α) (Figure 10A–F), while no change in Il-10 and Foxp3 expression was seen (Figure 10G and H). Moreover, the amounts of signature cytokines (eg, IFN-γ, IL-17A, and TNF-α) were decreased, whereas IL-10 appeared to increase but did not reach a statistical difference in culture supernatants of LV-Id2–transfected Tob1–/–CD4+ T cells in contrast to control animals (Figure 10I–L). Collectively, these data indicate that ID2 deficiency in CD4+ T cells augments Th1 and Th17 cell immune responses and that forced ID2 expression in CD4+ T cells restrains this effect.
Figure 9.
Enhanced expression of ID2 in IBD CD4+T cells confines Th1/Th17 cell immune responses. PB-CD4+ T cells were harvested from patients with A-CD (n = 8), patients with A-UC (n = 8), and HC subjects (n = 8), transfected with LV-ID2, LV-shID2, and LV-NC, respectively, and then cultured under stimulation with plate-bound anti-human CD3 mAb (5 μg/mL) and soluble anti-human CD28 mAb (2 μg/mL) for 5 days. (A) The transfected CD4+ T cells were harvested on day 5, and transfection efficiencies of LV-ID2 and LV-shID2 were confirmed by qRT-PCR. (B–H) qRT-PCR was performed to examine the mRNA expression of IFN-γ, IL-17A, TNF-α, T-bet, RORC, Foxp3, and IL-10 in these CD4+ T cells. (I–L) The culture supernatants of these transfected CD4+ T cells were also collected for detecting the levels of IFN-γ, IL-17A, TNF-α and IL-10 using ELISA. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗P < .05, and ∗∗P < .01 vs LV-NC from the same group.
Figure 10.
Enhanced expression of ID2 in Tob1–/–CD4+T cells restrains Th1/Th17 cell immune responses. Splenic CD4+ T cells were harvested from Tob1–/– mice (n = 6), transfected with LV-Id2 and LV-NC, respectively, and then cultured under stimulation with plate-bound anti-CD3 mAb (5 μg/mL) and soluble anti-CD28 mAb (2 μg/mL) for 5 days. (A) The transfection efficiency of LV-Id2 was confirmed by qRT-PCR, and (B–H) the mRNA expression of Il-17a, Rorγt, Ifn-γ, T-bet, Tnf-α, Il-10, and Foxp3 in these CD4+ T cells was detected by qRT-PCR. (I–L) The culture supernatants of these transfected CD4+ T cells were also collected for detecting the levels of IFN-γ, IL-17A, TNF-α, and IL-10, respectively, by ELISA. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001 vs LV-NC from the same group.
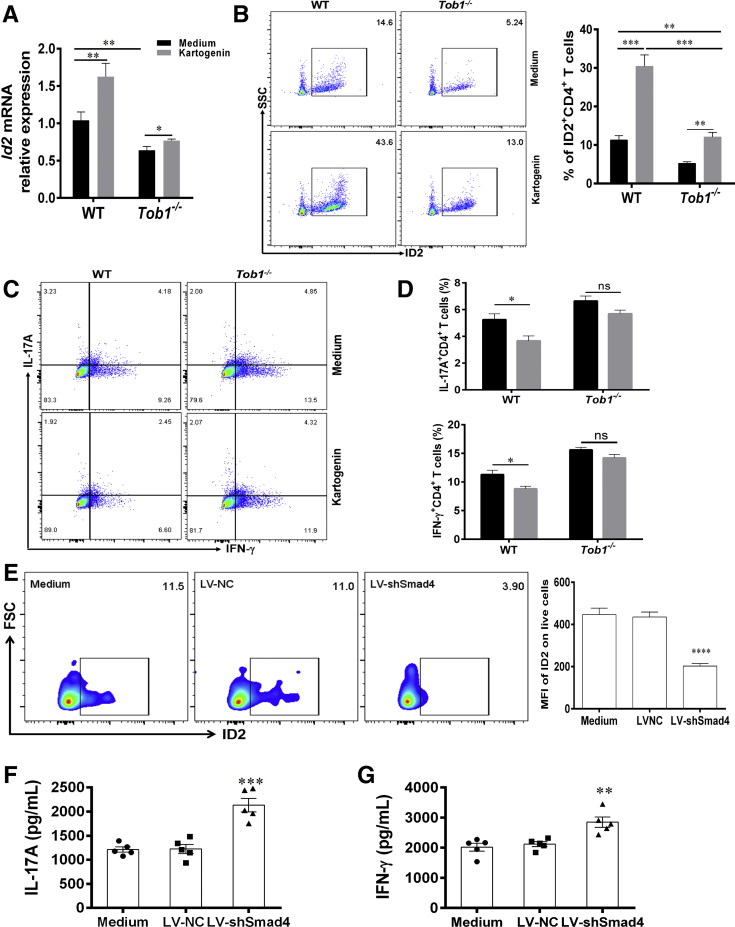
Additionally, we also utilized kartogenin, an agonist of Smad4/Smad5, to stimulate splenic CD4+ T cells of WT mice in vitro and found that it powerfully induced the expression of Id2 (Figure 11A and B), thus leading to inhibiting the protein expression of IL-17A and IFN-γ (Figure 11C and D). Moreover, we also transfected splenic CD4+ T cells with LV-expressing short hairpin Smad4 (LV-shSmad4) and determined whether it could modulate Id2 expression and the role in regulating the CD4+ T cell differentiation. These transfected CD4+ T cells were cultured for 5 days under stimulation with immobilized anti-CD3 and anti-CD28 mAbs in vitro. The levels of ID2 and Smad4 were reduced in LV-shSmad4–transfected CD4+ T cells compared with control animals (Figure 11E). Importantly, we found that the levels of IL-17A and IFN-γ were higher in culture supernatants than control animals (Figure 11F and G). Taken together, these data suggest that Tob1 is associated with Smad4/5 to upregulate the expression of Id2 and further restrain Th1/Th17 cell immune responses.
Figure 11.
Tob1 is associated with Smad4/5 to induce Id2 expression and restrain Th1/Th17 cell differentiation. (A–D) Splenic CD4+ T cells of WT littermates and Tob1–/– mice (n = 6, respectively) stimulated with kartogenin (1 μM, an agonist of Smad4/5) and plate-bound anti-CD3 mAb (5 μg/mL) and soluble anti-CD28 mAb (2 μg/mL) for 5 days. (A, B) Id2 expression was upregulated in splenic CD4+ T cells of WT littermates and Tob1–/– mice. (C and D) Intracellular expression of IFN-γ and IL-17A in these CD4+ T cells was analyzed by flow cytometry. (E–G) Splenic CD4+ T cells from WT littermates (n = 5) were transfected with LV-shSmad4 and LV-NC, respectively, and then cultured under stimulation with plate-bound anti-CD3 mAb (5 μg/mL) and soluble anti-CD28 mAb (2 μg/mL) for 5 days. (F and G) The expression of ID2 was detected by flow cytometry (E). The culture supernatants of these transfected CD4+ T cells were collected for detecting the levels of IL-17A and IFN-γ by using ELISA. Statistical analysis was performed with unpaired 2-sided Student’s t tests. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Discussion
Aberrant adaptive immune response mediated by CD4+ T cells, especially by proinflammatory Th1 and Th17 cells, has been regarded as one of the main drivers of intestinal mucosal inflammation in IBD patients. Seeking for crucial regulators of T cell activation and differentiation is thought to be of great significance for the clinical treatment of IBD. In the present study, we demonstrated that TOB1 plays a key protective role in the pathogenesis of IBD by preventing excessive Th1/Th17 cell–mediated immune responses. TOB1 has been reported to involve in the development of various tumors (eg, lung cancer, papillary carcinoma of thyroid, and gastric cancer), as evidenced by decreased TOB1 expression or higher level of TOB1 phosphorylation, which elucidates diminished antiproliferative activity.16,17,19,30 Moreover, reduced expression of TOB1 mRNA is also observed in PBMCs of MS patients and spinal cords of mice with experimental autoimmune encephalomyelitis, indicating that TOB1 is related to the pathogenesis of immune-related diseases.23,31 However, whether TOB1 is implicated in the progression of IBD remains unexplored. In this report, we observed that the levels of TOB1 were remarkably downregulated in the inflamed mucosa and PB-CD4+ T cells of patients with active CD and UC, and that TOB1 expression in the colon tissues was negatively associated with disease activity of IBD patients. Moreover, we observed that the polarized Th1, Th2, Th17, and Treg cells displayed lower levels of TOB1 mRNA than Th0 cells. Because a previous study demonstrated that IL-1β downregulates TOB1 expression in endometriotic stromal cells,32 we speculate that excessive CD4+ T cell activation and the presence of a variety of proinflammatory cytokines (eg, TNF-α, IL-1β, IL-6, IL-12) are responsible for the decreased TOB1 expression in the inflamed mucosa of patients with IBD.
TOB1 has already been demonstrated to negatively regulate T cell activation or proliferation by means of multiple mechanisms. TOB1 directly inhibits transcription of IL-2 gene through interacting with Smad2/4.22 In addition, TOB1 also interacts with iPABP to suppress the translation of IL-2 mRNA in anergic T cells.33 Currently, the function of TOB1 in modulating the differentiation of CD4+ T cells in IBD patients is not fully elucidated. Here, we found that LV-TOB1–transfected PB-CD4+ T cells displayed a damaged ability for the differentiation toward Th1/Th17 cells, while the elimination of TOB1 in PB-CD4+ T cells from IBD patients and healthy individuals resulted in increased expression level of Th1/Th17 cell–related cytokines and their transcription factors. Conversely, transfection with either LV-TOB1 or LV-shTOB1 appeared not to affect the expression of Foxp3 and IL-10 in CD4+ T cells cultured under Th0 conditions. Taken together, these results indicate that decreased TOB1 expression in inflamed tissues of IBD patients may augment Th1/Th17 cell–mediated mucosal immune responses, thus aggravating intestinal inflammation of IBD.
The importance of TOB1 in controlling intestinal inflammation was further evidenced by the study of Tob1–/– mice. In our study, we found that severe colitis was present in both Tob1–/– mice induced by rectal administration of TNBS and Tob1–/–CD45RBhighCD4+ T cell–reconstituted Rag1–/– mice, suggesting that Tob1 deficiency augments the mucosal immune responses mediated by Th1/Th17 cells in intestinal mucosa. Importantly, we ascertained the effects of TOB1 on Th1 and Th17 cell differentiation in vitro. These results were consistent with a previous report showing that Tob1–/–CD4+ T cells activated by WT-APCs (Antigen Presenting Cells) and MOG35-55 preferentially differentiate into Th1/Th17 cells in vitro.24
To clarify the underlying mechanisms whereby Tob1 restrains Th1/Th17 cell differentiation, we sorted splenic CD4+ T cells from both Tob1–/– mice and WT littermates for RNA-seq and found that expression of Id2 was significantly decreased in Tob1–/–CD4+ T cells than in WT cells. ID2 as an important molecule of signaling pathways and is involved in the development and cycle of cells and tumorigenesis.29 Moreover, ID2 is a part of the HLH transcription factors family and target of BMP signals in various types of cells.28 Importantly, upregulated expression of Id2 via BMP-responsive Smad suppresses cell differentiation.29 Recent study reported that Id2 and Id3 expression in Treg cells is required for restraining the development of fatal inflammatory disease.34 However, whether ID2 is related to the development of IBD is still elusive. In this study, we observed that ID2 was decreased in inflamed mucosa and PB-CD4+ T cells in active IBD patients and that overexpression of TOB1 promoted the relative luciferase activity of ID2. Importantly, LV-ID2–transfected IBD CD4+ T cells and Tob1–/–CD4+ T cells failed to differentiate into Th1 and Th17 cells, while suppression of ID2 expression resulted in facilitating IBD PB-CD4+ T cells to differentiate into Th1 and Th17 cells, suggesting a critical role of ID2 in modulating IBD CD4+ T cell differentiation. Additionally, a previous study has also demonstrated that Id2 could enhance Th1 cell differentiation but suppress T follicular helper cell differentiation through blocking E2A expression in the context to infection with Toxoplasma gondii.35 We suppose that this discrepancy may arise from the different status of CD4+ T cells and disease conditions. Besides, we also discovered that LV-shSmad4–transfected CD4+ T cells constrained ID2 expression. Kartogenin, an agonist of Smad4/Smad5, was found to promote the expression of Id2 in splenic CD4+ T cells of WT mice. Collectively, these data illustrate that Tob1 is associated with Smad4/5 to upregulate the expression of Id2.
In conclusion, our study proves that TOB1 expression is downregulated in IBD patients and that TOB1 induces the expression of ID2 by Smad4/5, thus contributing to the restraint of mucosal inflammation through preventing excessive Th1/Th17 cell–mediated immune responses in intestinal mucosa. Therefore, our data shed some light on a crucial role of TOB1 in the pathogenesis of IBD, and provide a novel therapeutic approach as to the supplementation or forced expression of TOB1 in the treatment of IBD.
Materials And Methods
Subjects
All patients and healthy individuals were recruited from Center for IBD Research, the Shanghai Tenth People’s Hospital of Tongji University (Shanghai, China), from January 2017 to November 2021. Among them, colon biopsies were collected from 46 patients with active CD (A-CD), 29 patients with CD in remission (R-CD), 47 patients with active UC (A-UC), 30 patients with UC in remission (R-UC), 44 HC subjects, and 9 non-IBD inflammatory control subjects (3 patients with celiac disease, 3 patients with Clostridium difficile infectious colitis, and 3 patients with ischemic enteritis), respectively. PB samples (10 mL) were obtained from 35 patients with A-CD, 32 patients with R-CD, 33 patients with A-UC, 32 patients with R-UC, and 43 HC subject, respectively. Table 1 summarizes the clinical features of IBD patients and healthy volunteers. Our study was approved by the Institutional Review Board for Clinical Research of the Shanghai Tenth People's Hospital of Tongji University (SHSY-IEC-4.0/19-52/01). Written informed consent was obtained from all participants before study.
Table 1.
Clinical Characteristics of IBD Patients and HC Subjects
| Biopsy Samples |
Blood Samples |
|||||
|---|---|---|---|---|---|---|
| HC Subjects | Patients With CD (A/R) | Patients With UC (A/R) | HC Subjects | Patients With CD (A/R) | Patients With UC (A/R) | |
| Number of patients | 44 | 75 (46/29) | 77 (47/30) | 43 | 67 (35/32) | 65 (33/32) |
| Sex | ||||||
| Male | 20 | 35 (21/14) | 38 (25/13) | 21 | 30 (14/16) | 32 (15/17) |
| Female | 24 | 40 (25/15) | 39 (22/17) | 22 | 37 (21/16) | 33 (18/15) |
| Age, y | 33.2 ± 8.5 | 31.5 ± 8.7 | 35.1 ± 12.3 | 31.2 ± 7.2 | 31.8 ± 7.1 | 34.8 ± 9.6 |
| Disease duration, mo | 42.7 ± 13.6 | 44.8 ± 19.5 | 36.8 ± 15.5 | 41.4 ± 15.3 | ||
| Disease location (CD)a | ||||||
| L1 | 21 (13/8) | 9 (5/4) | ||||
| L2 | 9 (4/5) | 10 (4/6) | ||||
| L3 | 45 (29/16) | 48 (26/22) | ||||
| L4 | 0 | 0 | ||||
| Disease extent (UC)a | ||||||
| E1 | 10 (6/4) | 9 (5/4) | ||||
| E2 | 31 (17/14) | 27 (15/12) | ||||
| E3 | 36 (24/12) | 29 (17/12) | ||||
| Current therapy | ||||||
| Mesalamine | 22 (13/9) | 29 (18/11) | 27 (15/12) | 30 (17/13) | ||
| Biologics | 35 (25/10) | 12 (10/2) | 31 (22/11) | 15 (10/5) | ||
| Azathioprine | 21 (14/7) | 27 (17/10) | 17 (12/5) | 24 (15/9) | ||
| Methotrexate | 8 (5/3) | 0 | 8 (6/2) | 0 | ||
| Glucocorticoids | 19 (19/0) | 31 (28/3) | 16 (11/5) | 27 (23/4) | ||
| CDAI (mean ± SEM) | 228.3 ± 49.4/102.5 ± 27.8 | 211.9 ± 35.7/90.7 ± 20.4 | ||||
| Mayo Score (mean ± SEM) | 6.8±1.9/1.7±0.5 | 5.2 ± 1.4/1.4 ± 0.5 | ||||
Values are n or mean ± SEM, unless otherwise indicated.
A/R, active/remission; CD, Crohn’s disease; CDAI, Crohn’s disease activity index; HC, healthy control; IBD, inflammatory bowel disease; UC, ulcerative colitis.
According to the Montreal classification system.
Mice
Six-week-old male Rag1–/– mice on C57BL/6 background were purchased from the Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China). All Rag1–/–, Tob1–/– and WT mice were raised and bred in the specific pathogen-free animal facility of the Tongji University. Mice at 8–10 weeks of age were used for experiments in this study. All animal experiments were performed following the Guide for the Care and Use of Laboratory Animals, with the approval of the Institutional Animal Care and Use Committee of Tongji University (SHDSYY-2018-3912).
Generation of Tob1–/–Mice
Tob1–/– mice on C57BL/6 background were generated at the Shanghai Key Laboratory of Regulatory Biology (Shanghai, China) using the CRISPR/Cas9 technique as described previously.36 In brief, 2 sequences in exon 2 of Tob1 gene were selected to design and synthesize single guide RNAs: 5′-CCCCGTGATCGAGCAGGCATCCA-3′, and 5′-TTCCTACCAGATCGGCGAAAAGG-3′. The mixture of in vitro transcribed Cas9 mRNA and Tob1 single guide RNAs was microinjected into the cytoplasm of zygotes obtained from superovulated female C57BL/B6 mice. These injected zygotes were then transferred into pseudopregnant recipient to bring forth founder mice. To identify the genotypes of offspring, the genomic DNA from mouse tails was amplified by PCR (sense primer: 5′-TTTTCTGGTGGAGGAGTTG-3′; anti-sense primer: 5′-CAGGAGGTCGTTCACATTTA-3′), and PCR products were sent for sequencing. The founder mouse carrying a 127-bp deletion in the Tob1 genomic DNA sequence was selected to mate with WT C57BL/6 mice to generate heterozygous Tob1+/-. Heterozygous Tob1+/- mice were then intercrossed to acquire homozygous Tob1–/– mice and WT littermates to further research.
Quantitative Real-Time PCR
Total RNA was extracted by using TRIzol (Invitrogen, Carlsbad, CA, USA), and reversely transcribed into complementary DNA using 5×All-in-one RT MasterMix kit (Applied Biological Materials, Richmond, Canada) following the manufacturer’s instructions. qRT-PCR was performed as described previously.37 Relative expression of genes was normalized to GAPDH, and reckoned by the 2-ΔΔCt algorithm.
Lentivirus-Mediated CD4+ T Cell Transfection
LV-TOB1 (transcript ID: NM_0057494, CDS: 444-1481) or LV-shTOB1 (target sequence: 5′-GCAGTATTCTAACCAGCAATT-3′) and LV-ID2 (transcript ID: NM_002166, CDS: 51-452) or LV-shID2 (target sequence: 5′-CCCACTATTGTCAGCCTGCAT-3′) were constructed in Hanbio Tech (Shanghai, China). PBMC were separated from EDTA anticoagulated PB of IBD patients and healthy donors by Ficoll-Paque PLUS (GE Healthcare, Uppsala, Sweden) gradient centrifugation.38 CD4+ T cells were then isolated from PBMCs using anti-human CD4 magnetic beads (BD Biosciences, San Diego, CA, USA) and cultured in complete RPMI-1640 medium under stimulation with immobilized anti-human CD3 mAb (5 μg/mL) and soluble anti-human CD28 mAb (2 μg/mL) (eBioscience, San Diego, CA) for 48 hours.39 These preactivated CD4+ T cells were reseeded in 24-well plates at a density of 1 × 105/well, and added with LV-TOB1, LV-shTOB1, LV-ID2, LV-shID2, and LV-NC, respectively (multiplicity of infection = 180), followed by centrifugation at 800 g for 2 hours at 37°C. After that, the 24-well plates containing CD4+ T cells and lentivirus were transferred into incubator of 37°C, and cultured for another 5 hours. Subsequently, these transfected CD4+ T cells were cultured with immobilized anti-human CD3 mAb (5 μg/mL) and soluble anti-human CD28 mAb (2 μg/mL) in complete RPMI-1640 medium for 5 days. Finally, these cells were harvested, and that qRT-PCR was executed to determine the transfection efficiency and levels of various cytokines and related transcription factors. In addition, cytokines (eg, IFN-γ, IL-17A, TNF-α, and IL-10) released into the supernatant were also detected by ELISA according to the manufacturer’s instructions.
TNBS-Induced Colitis in Mice
TNBS-induced colitis in mice was conducted as described previously.40,41 In brief, male Tob1–/– mice and WT littermates were mildly anesthetized by intraperitoneal injection of 1.25% pentobarbital sodium after fasting for 24 hours. TNBS mixture (3.75 mg TNBS dissolved in 150 μL 50% ethanol per mouse) (Sigma-Aldrich, St. Louis, MO, USA) was then slowly administered into the rectum of anesthetized mice through a polyethylene catheter. These mice were maintained in a perpendicular position for 2 minutes to assure the diffuse distribution of TNBS mixture in colon. As control animals, Tob1–/– mice and WT littermates were treated with 50% ethanol alone using the same protocol. The changes of weight and fecal consistency were monitored daily after TNBS induction. Day 7 post-TNBS/ethanol administration, mice were sacrificed. Colonic tissues were collected for hematoxylin and eosin staining, RNA analysis and isolation of LPMC from all mice. The histological grading of TNBS colitis was scored from 0 to 4 as described previously.
T Cell Adoptive Transfer Colitis Model in Rag1–/– Mice
The chronic colitis model was established according to our previous report.27 Briefly, splenic CD4+ T cells were separated from Tob1–/– mice and WT littermates using anti-mouse CD4 magnetic beads (BD Biosciences). After surface staining, naïve CD25–CD45RBhighCD4+ T cells of WT littermates and Tob1–/– mice were sorted from splenic CD4+ T cells on a BD FACSAria II Flow Cytometer and then transferred intraperitoneally into 8-week-old Rag1–/– mice (5 × 105 cells/mouse), respectively. These recipient Rag1–/– mice were monitored weekly for clinical characteristics such as weight loss and diarrhea. These recipients were sacrificed after 7 weeks of T cell adoptive transfer. Then, the severity of colitis was assessed according to the changes of clinical characteristics. Colon tissues were collected for hematoxylin and eosin staining, RNA analysis, and LPMC isolation, respectively.
Separation of LPMCs
LPMCs were isolated following the way as detailed in our previous study.42 Concisely, the washed colon pieces were incubated in phosphate-buffered saline containing 1 mM EDTA and 5% fetal bovine serum at 37°C to eliminate intestinal epithelial cells for 2 × 20 minutes. After that, the remaining colon tissues were minced, and digested in 10 mL 5% fetal bovine serum-RPMI-collagenase A (1 mg/mL; Sigma-Aldrich) at 37ºC for 30 minutes. LPMCs were then harvested from the interphase by density gradient centrifugation for further assays.
Flow Cytometric Analysis
For cell surface staining, LPMCs and single-cell suspension of spleen and MLN were obtained from Tob1–/– mice and WT littermates, first incubated with Fc Block (BD Biosciences), and then stained with fluorochrome-conjugated mAbs against CD3, CD4, CD8, and B220, respectively, for 30 minutes at 4°C.43 Concomitantly, the Live/Dead Fixable Dead Cell stain kits (Invitrogen, Eugene, OR) was used to exclude dead cells. For intracellular staining of TOB1, PBMCs were harvested and performed surface staining, followed by fixation and permeabilization for 30 minutes at 4°C. After 3 washes, PBMCs were performed with anti-TOB1 mAbs for 30 minutes at 4°C, then 3 washes, and incubated with donkey anti-mouse IgG (Alexa Fluor 488) for 30 minutes at 4°C. For intracellular cytokine staining, CD4+ T cells or LPMCs were treated with PMA (50 ng/mL; Sigma-Aldrich) and ionomycin (750 ng/mL; Sigma-Aldrich) for 5 hours at 37°C, along with the stimulation of brefeldin A (3 μg/mL; eBioscience) for the last 3 hours. Subsequently, these cells were harvested and processed for surface staining, followed by fixation and permeabilization for 30 minutes at 4°C. After 3 washes, intracellular staining was performed with fluorochrome-conjugated anti-ID2, anti-IFN-γ, anti-IL-17A, anti-TNF-α, anti-IL-10, and anti-Foxp3 mAbs. All stained samples were analyzed on a BD FACSCanto II Flow Cytometer. All data were processed using FlowJo software (Version 10.0.7; Tree Star; Ashland, OR).
Statistical Analysis
All data were analyzed using Prism 6.0 (GraphPad Software; San Diego, CA, USA). Statistical comparisons between different groups were performed using unpaired or paired Student’s t tests, and 1-way analysis of variance. Spearman rank correlation was used to analyze the correlation between TOB1 expression and disease activities of IBD patients. Data were shown as mean ± SEM, and P < .05 was considered as statistical significance.
Acknowledgments
CRediT authorship contributions
Ritian Lin, MD (Data curation: Lead; Formal analysis: Lead; Resources: Equal; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Caiyun Ma, MD, PhD (Conceptualization: Equal; Data curation: Lead; Formal analysis: Equal; Validation: Equal; Visualization: Equal; Writing – original draft: Equal)
Leilei Fang, MD, PhD (Data curation: Equal; Formal analysis: Equal; Fundingacquisition: Equal)
Chunjin Xu, MD, PhD (Data curation: Equal; Formal analysis: Equal; Resources: Equal)
Cui Zhang, MD, PhD (Data curation: Equal; Formal analysis: Equal; Resources: Equal)
Xiaohan Wu, MD (Data curation: Equal; Formal analysis: Equal; Resources: Equal)
Wei Wu, MD, PhD (Data curation: Equal; Resources: Equal; Writing – review & editing: Supporting)
Ruixin Zhu, MD, PhD (Data curation: Supporting; Writing – review & editing: Supporting)
Yingzi Cong, PhD (Conceptualization: Equal)
Zhanju Liu, MD, PhD (Conceptualization: Lead; Funding acquisition: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by grants from the National Key R&D Program of China (2018YFC1705400) and the National Natural Science Foundation of China (81630017, 91942312, 81800487, 82100550).
References
- 1.Chang J.T. Pathophysiology of inflammatory bowel diseases. N Engl J Med. 2020;383:2652–2664. doi: 10.1056/NEJMra2002697. [DOI] [PubMed] [Google Scholar]
- 2.Graham D.B., Xavier R.J. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578:527–539. doi: 10.1038/s41586-020-2025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun M., He C., Cong Y., Liu Z. Regulatory immune cells in regulation of intestinal inflammatory response to microbiota. Mucosal Immunol. 2015;8:969–978. doi: 10.1038/mi.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maloy K.J., Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 5.Park J.H., Peyrin-Biroulet L., Eisenhut M., Shin J.I. IBD immunopathogenesis: A comprehensive review of inflammatory molecules. Autoimmun Rev. 2017;16:416–426. doi: 10.1016/j.autrev.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Friedrich M., Pohin M., Powrie F. Cytokine networks in the pathophysiology of inflammatory bowel disease. Immunity. 2019;50:992–1006. doi: 10.1016/j.immuni.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 7.Strober W., Fuss I.J. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1756–1767. doi: 10.1053/j.gastro.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cader M.Z., Kaser A. Recent advances in inflammatory bowel disease: mucosal immune cells in intestinal inflammation. Gut. 2013;62:1653–1664. doi: 10.1136/gutjnl-2012-303955. [DOI] [PubMed] [Google Scholar]
- 9.Clough J.N., Omer O.S., Tasker S., Lord G.M., Irving P.M. Regulatory T-cell therapy in Crohn's disease: challenges and advances. Gut. 2020;69:942–952. doi: 10.1136/gutjnl-2019-319850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt H., Neurath M.F., Atreya R. Role of the IL23/IL17 pathway in Crohn's Disease. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.622934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuda S., Kawamura-Tsuzuku J., Ohsugi M., Yoshida M., Emi M., Nakamura Y., Onda M., Yoshida Y., Nishiyama A., Yamamoto T. Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene. 1996;12:705–713. [PubMed] [Google Scholar]
- 12.Matsuda S., Rouault J., Magaud J., Berthet C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001;497:67–72. doi: 10.1016/s0014-5793(01)02436-x. [DOI] [PubMed] [Google Scholar]
- 13.Winkler G.S. The mammalian anti-proliferative BTG/Tob protein family. J Cell Physiol. 2010;222:66–72. doi: 10.1002/jcp.21919. [DOI] [PubMed] [Google Scholar]
- 14.Baranzini S.E. The role of antiproliferative gene Tob1 in the immune system. Clin Exp Neuroimmunol. 2014;5:132–136. doi: 10.1111/cen3.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H.S., Kundu J., Kim R.N., Shin Y.K. Transducer of ERBB2.1 (TOB1) as a tumor suppressor: a mechanistic perspective. Int J Mol Sci. 2015;16:29815–29828. doi: 10.3390/ijms161226203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwanaga K., Sueoka N., Sato A., Sakuragi T., Sakao Y., Tominaga M., Suzuki T., Yoshida Y., J K.T., Yamamoto T., Hayashi S., Nagasawa K., Sueoka E. Alteration of expression or phosphorylation status of tob, a novel tumor suppressor gene product, is an early event in lung cancer. Cancer Lett. 2003;202:71–79. doi: 10.1016/j.canlet.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Ito Y., Suzuki T., Yoshida H., Tomoda C., Uruno T., Takamura Y., Miya A., Kobayashi K., Matsuzuka F., Kuma K., Yamamoto T., Miyauchi A. Phosphorylation and inactivation of Tob contributes to the progression of papillary carcinoma of the thyroid. Cancer Lett. 2005;220:237–242. doi: 10.1016/j.canlet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 18.O'Malley S., Su H., Zhang T., Ng C., Ge H., Tang C.K. TOB suppresses breast cancer tumorigenesis. Int J Cancer. 2009;125:1805–1813. doi: 10.1002/ijc.24490. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S.Q., Sun K.K., Wu X.Y., Zhong N., Zhao H., Li D.C. Clinicopathological significance of cytoplasmic transducer of ErbB2. 1 expression in gastric cancer. Mol Med Rep. 2015;12:1177–1182. doi: 10.3892/mmr.2015.3470. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida Y., Nakamura T., Komoda M., Satoh H., Suzuki T., Tsuzuku J.K., Miyasaka T., Yoshida E.H., Umemori H., Kunisaki R.K., Tani K., Ishii S., Mori S., Suganuma M., Noda T., Yamamoto T. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 2003;17:1201–1206. doi: 10.1101/gad.1088003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida Y., Tanaka S., Umemori H., Minowa O., Usui M., Ikematsu N., Hosoda E., Imamura T., Kuno J., Yamashita T., Miyazono K., Noda M., Noda T., Yamamoto T. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell. 2000;103:1085–1097. doi: 10.1016/s0092-8674(00)00211-7. [DOI] [PubMed] [Google Scholar]
- 22.Tzachanis D., Freeman G.J., Hirano N., van Puijenbroek A.A., Delfs M.W., Berezovskaya A., Nadler L.M., Boussiotis V.A. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat Immunol. 2001;2:1174–1182. doi: 10.1038/ni730. [DOI] [PubMed] [Google Scholar]
- 23.Corvol J.C., Pelletier D., Henry R.G., Caillier S.J., Wang J., Pappas D., Casazza S., Okuda D.T., Hauser S.L., Oksenberg J.R., Baranzini S.E. Abrogation of T cell quiescence characterizes patients at high risk for multiple sclerosis after the initial neurological event. Proc Natl Acad Sci U S A. 2008;105:11839–11844. doi: 10.1073/pnas.0805065105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulze-Topphoff U., Casazza S., Varrin-Doyer M., Pekarek K., Sobel R.A., Hauser S.L., Oksenberg J.R., Zamvil S.S., Baranzini S.E. Tob1 plays a critical role in the activation of encephalitogenic T cells in CNS autoimmunity. J Exp Med. 2013;210:1301–1309. doi: 10.1084/jem.20121611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fonseca-Camarillo G., Furuzawa-Carballeda J., Priego-Ranero A.A., Martinez-Benitez B., Barreto-Zuniga R., Yamamoto-Furusho J.K. Expression of TOB/BTG family members in patients with inflammatory bowel disease. Scand J Immunol. 2021;93 doi: 10.1111/sji.13004. [DOI] [PubMed] [Google Scholar]
- 26.Santarlasci V., Maggi L., Mazzoni A., Capone M., Querci V., Rossi M.C., Beltrame L., Cavalieri D., De Palma R., Liotta F., Cosmi L., Maggi E., Romagnani S., Annunziato F. IL-4-induced gene 1 maintains high Tob1 expression that contributes to TCR unresponsiveness in human T helper 17 cells. Eur J Immunol. 2014;44:654–661. doi: 10.1002/eji.201344047. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z., Geboes K., Colpaert S., Overbergh L., Mathieu C., Heremans H., de Boer M., Boon L., D'Haens G., Rutgeerts P., Ceuppens J.L. Prevention of experimental colitis in SCID mice reconstituted with CD45RBhigh CD4+ T cells by blocking the CD40-CD154 interactions. J Immunol. 2000;164:6005–6014. doi: 10.4049/jimmunol.164.11.6005. [DOI] [PubMed] [Google Scholar]
- 28.Perk J., Iavarone A., Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 29.Ruzinova M.B., Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki T., J K.T., Ajima R., Nakamura T., Yoshida Y., Yamamoto T. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes Dev. 2002;16:1356–1370. doi: 10.1101/gad.962802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Q., Gao Q., Zhang Y., Li Z., Mei X. MicroRNA-590 promotes pathogenic Th17 cell differentiation through targeting Tob1 and is associated with multiple sclerosis. Biochem Biophys Res Commun. 2017;493:901–908. doi: 10.1016/j.bbrc.2017.09.123. [DOI] [PubMed] [Google Scholar]
- 32.Lebovic D.I., Baldocchi R.A., Mueller M.D., Taylor R.N. Altered expression of a cell-cycle suppressor gene, Tob-1, in endometriotic cells by cDNA array analyses. Fertil Steril. 2002;78:849–854. doi: 10.1016/s0015-0282(02)03319-8. [DOI] [PubMed] [Google Scholar]
- 33.Okochi K., Suzuki T., Inoue J., Matsuda S., Yamamoto T. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells. 2005;10:151–163. doi: 10.1111/j.1365-2443.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- 34.Miyazaki M., Miyazaki K., Chen S., Itoi M., Miller M., Lu L.F., Varki N., Chang A.N., Broide D.H., Murre C. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat Immunol. 2014;15:767–776. doi: 10.1038/ni.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw L.A., Belanger S., Omilusik K.D., Cho S., Scott-Browne J.P., Nance J.P., Goulding J., Lasorella A., Lu L.F., Crotty S., Goldrath A.W. Id2 reinforces TH1 differentiation and inhibits E2A to repress TFH differentiation. Nat Immunol. 2016;17:834–843. doi: 10.1038/ni.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao Y., Guan Y., Wang L., Qiu Z., Liu M., Chen Y., Wu L., Li Y., Ma X., Liu M., Li D. CRISPR/Cas-mediated genome editing in the rat via direct injection of one-cell embryos. Nat Protoc. 2014;9:2493–2512. doi: 10.1038/nprot.2014.171. [DOI] [PubMed] [Google Scholar]
- 37.Li G., Lin J., Zhang C., Gao H., Lu H., Gao X., Zhu R., Li Z., Li M., Liu Z. Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes. 2021;13 doi: 10.1080/19490976.2021.1968257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu H., Lin J., Xu C., Sun M., Zuo K., Zhang X., Li M., Huang H., Li Z., Wu W., Feng B., Liu Z. Cyclosporine modulates neutrophil functions via the SIRT6-HIF-1alpha-glycolysis axis to alleviate severe ulcerative colitis. Clin Transl Med. 2021;11 doi: 10.1002/ctm2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou G., Wu W., Yu L., Yu T., Yang W., Wang P., Zhang X., Cong Y., Liu Z. Tripartite motif-containing (TRIM) 21 negatively regulates intestinal mucosal inflammation through inhibiting TH1/TH17 cell differentiation in patients with inflammatory bowel diseases. J Allergy Clin Immunol. 2018;142:1218–1228.e12. doi: 10.1016/j.jaci.2017.09.038. [DOI] [PubMed] [Google Scholar]
- 40.Yang W., Zhou G., Yu T., Chen L., Yu L., Guo Y., Cong Y., Liu Z. Critical role of ROCK2 activity in facilitating mucosal CD4(+) T cell activation in inflammatory bowel disease. J Autoimmun. 2018;89:125–138. doi: 10.1016/j.jaut.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 41.Wirtz S., Popp V., Kindermann M., Gerlach K., Weigmann B., Fichtner-Feigl S., Neurath M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12:1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- 42.He C., Shi Y., Wu R., Sun M., Fang L., Wu W., Liu C., Tang M., Li Z., Wang P., Cong Y., Liu Z. miR-301a promotes intestinal mucosal inflammation through induction of IL-17A and TNF-alpha in IBD. Gut. 2016;65:1938–1950. doi: 10.1136/gutjnl-2015-309389. [DOI] [PubMed] [Google Scholar]
- 43.Osorio-Barrios F., Navarro G., Campos J., Ugalde V., Prado C., Raich I., Contreras F., Lopez E., Espinoza A., Lladser A., Franco R., Pacheco R. The heteromeric complex formed by dopamine receptor D5 and CCR9 leads the gut homing of CD4(+) T cells upon inflammation. Cell Mol Gastroenterol Hepatol. 2021;12:489–506. doi: 10.1016/j.jcmgh.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]