Summary
Myeloid cells, such as macrophages, are critical components of the inflammatory response, in which infected, injured, or necrotic tissues are targeted for resolution. A key aspect of the inflammatory response is migration of myeloid cells to target tissues. Although studying immune cell migration in mammalian models in vivo is challenging, zebrafish are more tractable owing to their optical transparency and rapidity in generating transgenic lines. Here, we present a tailfin injury assay protocol for quantifying immune infiltration at injury sites.
For complete details on the use and execution of this profile, please refer to Anderson-Baucum et al. (2021) and Kulkarni et al. (2021).
Subject areas: Cell Biology, Immunology, Microscopy, Model Organisms, Antibody
Graphical abstract
Highlights
-
•
Migration is a function of immune cells that enable their homing to sites of injury
-
•
A protocol for zebrafish tailfin injury and visualization of immune cell migration
-
•
Use of transgenic zebrafish lines with fluorescently-labeled immune cells
Myeloid cells, such as macrophages, are critical components of the inflammatory response, in which infected, injured, or necrotic tissues are targeted for resolution. A key aspect of the inflammatory response is migration of myeloid cells to target tissues. Although studying immune cell migration in mammalian models in vivo is challenging, zebrafish are more tractable owing to their optical transparency and rapidity in generating transgenic lines. Here, we present a tailfin injury assay protocol for quantifying immune infiltration at injury sites.
Before you begin
Immune cell migration to the site of injury or cellular damage is a characteristic of initiation as well as resolution of inflammation (Luster et al., 2005). Innate immune cells like neutrophils and macrophages are among the first cells to respond to any kind of tissue damage and inflammation. These myeloid immune cells perform phagocytosis of pathogens or apoptotic cells and then promote tissue remodeling (Silva and Correia-Neves, 2012). However, in conditions of maladaptive inflammation, these cells can induce tissue damage primarily through their proinflammatory activities. This deleterious behavior of myeloid cells is a characteristic of several diseases including sepsis, neurodegenerative disorders, arthritis, obesity, type1 and type2 diabetes (Ardura et al., 2019). Here we describe a detailed protocol to study immune cell migration in vivo using the zebrafish (Danio rerio) animal model.
Note: This protocol is described for performing studies in the larval stage zebrafish (3–4 days post-fertilization). For performing studies with older zebrafish, appropriate approvals from the ethics committee are necessary.
Note: This protocol describes the use of a transgenic zebrafish (Danio rerio) line expressing green fluorescent protein (GFP) under the macrophage specific mpeg (macrophage expressed gene) promoter. Other fluorescent reporters can be used.
Note: All procedures described in this protocol have been approved by the University of Chicago’s Institutional Animal Care and Use Committee. Experiments performed following this protocol should be approved by the user’s Institutional Animal Care and Use Committee.
Zebrafish husbandry and maintenance
Zebrafish are kept in a circulating system that continuously filters and aerates the system water to maintain the water quality required for a healthy aquatic environment. The circulating system also helps to filter excess food and fish excreta. Different companies provide zebrafish systems but we use systems from Tecniplast in our University fish facility. The room temperature or the tank temperature is generally maintained between 26°C-28.5°C and the lighting conditions are 14:10 h (light: dark). Multiple lines of fish (e.g., transgenic, mutant, wild type) can also be housed on the same system. A set of different kinds of filters are used in the system. In our system, water from all the tanks passes through a coarse filter pad, a fine particulate canister filter, biological filter , active carbon absorption filter and UV disinfection filter before being circulated back into the tank. Reverse osmosis purified water is used to top off water losses in the zebrafish system. The pH of the system water should be checked daily and maintained between 6.8 and 7.5. pH generally falls due to wastes excreted by the zebrafish, and to decomposition of uneaten food. When necessary, sodium bicarbonate should be used to increase the pH.
Prepare PTU-egg water to block pigmentation
-
1.
Prepare egg-water by adding 0.3 g/L of ‘Instant Ocean’ sea salt and 0.5 mM CaSO4 to distilled water.
-
2.
Add 0.003% 1-phenyl-2-thiourea.
Note: PTU dissolution takes time. It is advisable to add PTU to egg water and stir it for 18 h before treating the embryos.
Prepare PEM buffer
-
3.
Add 0.21 M PIPES, 1 mM EGTA, and 2 mM MgSO4 to ddH2O.
-
4.
Adjust pH to 7.0.
Collect and prepare staining reagents
-
5.
PBS – Phosphate Buffered Saline pH 7.4 (1×)
-
6.
PW – PBS + 0.1% Tween-20
-
7.
PWD – PBS + Tween-20 + DMSO
-
8.
PWDT – PBS + Tween-20 + DMSO + Triton-X100
-
9.
PWDB – PBS + Tween-20 + DMSO + BSA
Immobilize the zebrafish with tricaine for tailfin injury
-
10.
Prepare stock solution of tricaine by adding 400 mg of tricaine powder and 2.1 mL 1 M Tris (pH -9.0) to ∼97.9 mL ddH2O.
-
11.
Adjust pH to 7.
-
12.
Add 400 μL of stock tricaine solution in 10 mL PTU-egg water in a 100 mm petri dish.
Sort transgene-positive embryos
-
13.
Using an epifluorescence dissecting scope (i.e., Olympus SZX2-ILLTS) sort transgene-positive (e.g., GFP-positive macrophages) embryos
Key resources table
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GFP (1:500 dilution) | Aves labs | Cat# GFP-1020 ; RRID: AB_10000240 |
| TO-PRO3 (1:500 dilution) | Thermo Fisher Scientific | Cat# T3605 |
| anti-chicken secondary antibody | Molecular probes | Cat# A-11040; RRID: AB_2534097 |
| Chemicals, peptides, and recombinant proteins | ||
| 1-Phenyl-2-thiourea (PTU) | Acros organics | Cat# 207250250 |
| Methylene blue | Fisher Scientific | Cat# M291-25 |
| Tricaine-S | Syndel | Cat# 200-266 |
| Instant Ocean Sea Salt | Instant Ocean | Cat# SS15-10 |
| Formaldehyde | Fisher Scientific | Cat# 04042-500 |
| Bovine serum Albumin (BSA) | Sigma-Aldrich | Cat# A8806 |
| Phosphate Buffered Saline (pH, 7.4) | Thermo Fisher Scientific | Cat# 10010023 |
| Dimethyl sulfoxide (DMSO) | Fisher Scientific | Cat# D128-500 |
| Triton-X100 | Sigma-Aldrich | Cat# T8787 |
| PIPES | Sigma-Aldrich | Cat# P6757-500 |
| EGTA | Fisher Scientific | Cat# 02783-100 |
| MgSO4 | Fisher Scientific | Cat# BP213-1 |
| Tween-20 | Sigma-Aldrich | Cat# P7949 |
| Vectashield | Vector Laboratories, Inc. | Cat# H1000 |
| Experimental models: Organisms/strains | ||
| Tg(mpeg:GFP) zebrafish | Zebrafish International Resource Center | Cat# ZL9940 |
| Other | ||
| Fluorescence microscope | Olympus | Model :SZX2-ILLTS |
| Confocal Microscope | Nikon | Model: Eclipse Ti2 |
| Corning™ Untreated Culture Dishes | Fisher Scientific | Cat# 08-772-32 |
| Disposable safety scalpels (Curved) | Fine Science Tools | Cat# 10000-15 |
Materials and equipment
Preparation of PEM buffer:
| Reagent | Final concentration | Amount |
|---|---|---|
| PIPES | 0.21 M | 63.5 g |
| EGTA | 1 mM | 0.38 g |
| MgSO4 | 2 mM | 0.24 g |
| Adjust pH-7.0 | n/a | Variable |
| ddH2O | n/a | Variable |
| Total | n/a | 1,000 mL |
Note: Store at 4°C.
Preparation of PWD (wash buffer):
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 1× | 494.5 mL |
| Tween-20 | 0.1% | 0.5 mL |
| DMSO | 1% | 5 mL |
| Total | n/a | 500 mL |
Note: Store at 22°C–28°C.
Preparation of PWDT (permeabilization buffer):
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 1X | 49.45 mL |
| Tween-20 | 0.1% | 0.05 mL |
| DMSO | 1% | 0.5 mL |
| TritonX-100 | 0.3% | 0.15 mL |
| Total | n/a | 50 mL |
Note: Store at 22°C–28°C.
Preparation of PWDB (blocking buffer):
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 1× | 49.45 mL |
| Tween-20 | 0.1% | 0.05 mL |
| DMSO | 1% | 0.5 mL |
| BSA | 4% | 2 g |
| Total | n/a | 50 mL |
Note: Store at 4°C. PWDB needs to be prepared freshly every time.
Step-by-step method details
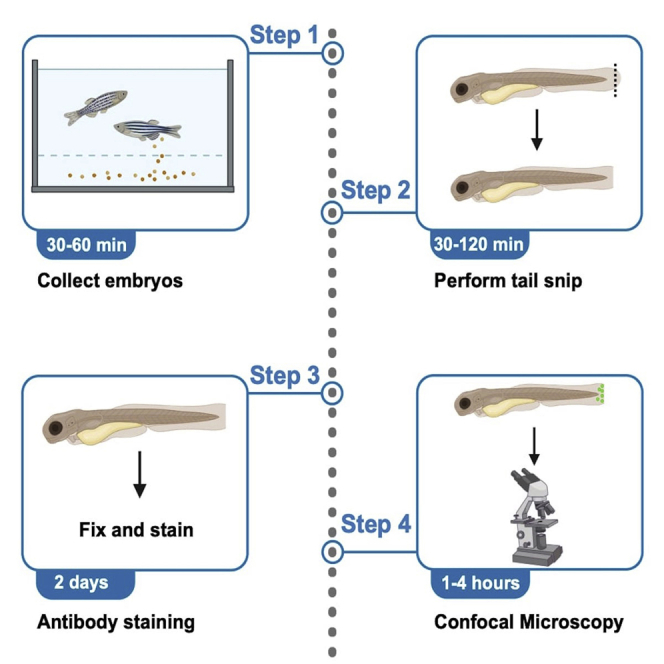
Collect the zebrafish embryos
-
1.
Cross the transgenic zebrafish (with labeled immune cells – e.g., Tg(mpeg:GFP) a day prior to the collection of embryos.
Note: Zebrafish eggs are collected from timed pairwise spawning. One female and one male are placed together in non-circulating 1-liter mesh-bottomed spawning tanks the afternoon/evening before collection. Tanks are set before the dark period of the light cycle begins, but after the day's feedings have been completed.
-
2.
Next day, collect the embryos in 100 mm petri dish in egg-water supplemented with methylene blue (100 μL in 1 L) and incubate at 37°C for 6 h.
Note: Eggs are generally spawned shortly after the daylight period of the light cycle begins, but several hours should be allotted for disinterested zebrafish. Once eggs are apparent in the bottom of the tank, the adult zebrafish are returned to their home tanks, and the spawning tank water is poured through a fine metal mesh strainer to capture eggs. Eggs are maintained at 28°C in ∼20 ml of system water in 100 mm plastic petri plates until several rounds of cleavage have occurred, so that unfertilized eggs can be identified and removed. Fertilized eggs are sorted into fresh petri plates at a density of <50 embryos, and dishes are filled with approximately 20 ml of fresh egg water.
-
3.
Change to PTU-egg-water to prevent pigmentation of the fish.
-
4.
At 3 day post-fertilization (dpf), transfer the larvae to a new petri dish containing PTU-egg-water that has been supplemented with 0.01% Tricaine to immobilize the larvae.
Note: Use non-tissue culture (TC)-treated petri dish, as larvae can stick to the TC-treated dish causing unexpected injuries.
-
5.
Sort transgenic larvae by epifluorescence using a dissecting microscope.
Tailfin injury
Timing: 30 min–2 h
In this step, we perform the tailfin cut to induce an inflammatory injury environment, which promotes immune cell migration to the injury site.
-
6.
Align the larvae horizontally and dissect the distal tip of the tailfin with a sharp scalpel.
-
7.
Transfer the larvae to fresh PTU-egg-water to wash out tricaine and revive.
-
8.
Recover the larvae in a 28.5°C incubator for 6 h post-injury.
-
9.
Following the recovery period, transfer the larvae to a freshly prepared 3% Formaldehyde solution in PEM buffer, and incubate it for 16–18 h at 4°C.
Note: Ideal time for this fixation is 16–18 h. An incubation longer than 18 h will result in brittle larvae, making subsequent processing more difficult.
Note: As an alternative, larvae can be utilized for live imaging studies. Instead of transferring to PTU-egg water for recovery, transfer up to 6 larvae to 3% molten low-melting temperature agarose prepared in egg water + 0.01% Tricaine. Larvae can be then transferred to coverslip-bottom 35 mm petri dishes (Mattek) and imaged on a confocal microscope for a time course of up to 12 h.
Immunostaining
Timing: 2 days
Here we describe the protocol for immunostaining of the larvae to facilitate visualization of the fluorescently labeled migrating immune cells.
-
10.
Wash the larvae twice with PBS
Note: Wash step involves incubating the larvae with indicated reagent (PBS here) on an orbital shaker for 5 min per wash.
-
11.
Permeabilize with PWDT for 20 min at 22°C–28°C.
Note: Do not permeabilize for more than 20 min. It can lead to degradation and inconsistent staining of the larvae.
-
12.
Wash once with PWD.
-
13.
Block non-specific antibody binding with PWDB (contains 4% BSA) for 30 min at 22°C–28°C.
-
14.
Wash once with PWD.
-
15.
Immunostain with primary antibodies by incubation at 4°C for 18 h in 50–100 μL of a 1:250 dilution of Chicken anti-GFP antibody.
-
16.
Wash three times with PWD.
-
17.
Immunostain with secondary antibodies by incubation at 4°C for 18 h in 250 μL of a 1:500 dilution of desired AlexaFluor labeled antibody.
Note: Secondary antibody staining may alternatively be carried out for 2–4 h at 22°C–28°C, but better and more consistent results are obtained with 18 h staining.
-
18.
Wash three times with PWD.
-
19.
Mount the larvae on glass slides in an anti-fade mounting solution e.g.,.Vectashield (Vector Labs Cat# H-1000).
Imaging and quantification
Timing: 1–4 h
-
20.
Mount the slides on the confocal microscope
-
21.
Image the larvae using the appropriate laser wavelengths at 20×. If necessary to capture all labeled cells in a thick sample, collect a Z-stack by imaging multiple Z planes with an optical section depth of 1 μm.
-
22.
Manually quantify the number of immune cells located distal to the notochord and through to the injury site.
-
23.
Perform appropriate statistical analysis to analyze the differences in immune cell migration.
Expected outcomes
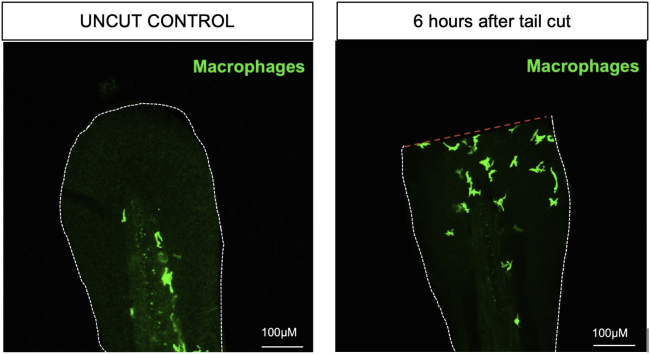
In the control zebrafish larvae that were not subjected to tail slice, there should be zero to one macrophage present in the region distal to the notochord. On the other hand, in the larvae subjected to tail snip, within 6 h post-injury we typically observe 8–12 macrophages at the injury site (Figure 1).
Figure 1.
Tailfin injury assay
Representative images of 3 dpf uninjured (left) and injured Tg(mpeg:GFP) zebrafish tails (right) that have been immunostained for GFP (macrophages, green). White dotted line shows the outline of tailfin and red dotted line indicates the site of tailfin injury. Scale bar indicates 100μm.
Limitations
This protocol relies on transgenic zebrafish in which the immune cells are tagged with a fluorescent marker such as GFP. Hence, the overexpression of these tags need to be performed using robust, cell-specific promoters to facilitate image analysis. Secondly, the intensity of the fluorescence differs based on the homozygosity or heterozygosity of the gene expression. Therefore, not every clutch of embryos will exhibit identical fluorescence intensity characteristics. Weak fluorescence can be enhanced by antibody-based staining.
Troubleshooting
Problem 1
Larval death post-tailfin injury (Refer to step-by-step methods details section: tailfin injury)-
Potential solution
One of the common issues with the tailfin injury assay is the death of the larvae after the injury. It is critical to cut the distal end of the zebrafish tailfin. Be careful with the tail snip; do not accidentally snip the end of the notochord as it is fatal for the fish.
Problem 2
Variable fluorescence intensities (Refer to step-by-step methods details section: imaging and quantification)
Potential solution
Although most of the fluorescent tags are sufficiently intense, it is recommended to immunostain for the fluorescent tags to enhance the brightness. It is also essential to titrate the antibody concentrations to optimize immunostaining specificity and intensity.
Problem 3
Absence of fluorescence (Refer to step-by-step methods details section: immunostaining) - Sometimes, despite crossing appropriate transgenic fish, there is absence of fluorescence.
Potential solution
The larvae can be immunostained with antibodies against proteins such as L-plastin to stain for the leukocytes.
Problem 4
Non-specific injuries (Refer to step-by-step methods details section: tailfin injury)- - Occasionally, the larval zebrafish can be mildly injured by handling, or sticking to the plastic dish. This injury will attract macrophages and may affect the number of cells attracted to the tailfin incision.
Potential solution
Use conditioned plates, handle with care, exclude damaged larvae from the analysis.
Problem 5
Pigmentation of zebrafish (Refer to before you begin section: Prepare PTU-egg water)- : Despite adding PTU to egg-water, the zebrafish can still be completely or partially pigmented. This will add to imaging difficulties.
Potential solution
Prepare fresh PTU-egg water. PTU remains efficacious in solution for 1–3 weeks.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Raghavendra Mirmira (mirmira@uchicago.edu)
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This research was supported by the NIH grants R01 DK060581 (to R.G.M.), R01 DK 105588 (to R.G.M.), and a DeVault fellowship (to A.K.). Research core services were provided by NIH grants P30 DK097512 (to Indiana University) and P30 DK020595 (to University of Chicago).
Author contributions
A.K., R.M.A., and R.G.M. conceived the study, developed the methodology, analyzed the data, wrote the original draft, and approved and edited the final draft.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Ryan Anderson, Email: ryananderson@uchicago.edu.
Raghavendra G. Mirmira, Email: mirmira@uchicago.edu.
Data and code availability
The datasets supporting the current study are available from the corresponding author on request.
References
- Anderson-Baucum E., Piñeros A.R., Kulkarni A., Webb-Robertson B.-J., Maier B., Anderson R.M., Wu W., Tersey S.A., Mastracci T.L., Casimiro I., et al. Deoxyhypusine synthase promotes a pro-inflammatory macrophage phenotype. Cell Metab. 2021;33:1883–1893.e7. doi: 10.1016/j.cmet.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardura J.A., Rackov G., Izquierdo E., Alonso V., Gortazar A.R., Escribese M.M. Targeting macrophages: friends or foes in disease? Front. Pharmacol. 2019;10:1255. doi: 10.3389/fphar.2019.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A., Pineros A.R., Walsh M.A., Casimiro I., Ibrahim S., Hernandez-Perez M., Orr K.S., Glenn L., Nadler J.L., Morris M.A., et al. 12-Lipoxygenase governs the innate immune pathogenesis of islet inflammation and autoimmune diabetes. JCI Insight. 2021;6:147812. doi: 10.1172/jci.insight.147812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luster A.D., Alon R., von Andrian U.H. Immune cell migration in inflammation: present and future therapeutic targets. Nat. Immunol. 2005;6:1182–1190. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- Silva M., Correia-Neves M. Neutrophils and macrophages: the main partners of phagocyte cell systems. Front. Immunol. 2012;3:174. doi: 10.3389/fimmu.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the current study are available from the corresponding author on request.