ABSTRACT
INO80 is the catalytic subunit of the INO80-chromatin remodeling complex that is involved in DNA replication, repair and transcription regulation. Ino80 deficiency in murine spermatocytes (Ino80cKO) results in pachytene arrest of spermatocytes due to incomplete synapsis and aberrant DNA double-strand break repair, which leads to apoptosis. RNA-seq on Ino80cKO spermatocytes revealed major changes in transcription, indicating that an aberrant transcription program arises upon INO80 depletion. In Ino80WT spermatocytes, genome-wide analysis showed that INO80-binding sites were mostly promoter proximal and necessary for the regulation of spermatogenic gene expression, primarily of premeiotic and meiotic genes. Furthermore, most of the genes poised for activity, as well as those genes that are active, shared INO80 binding. In Ino80cKO spermatocytes, most poised genes demonstrated de-repression due to reduced H3K27me3 enrichment and, in turn, showed increased expression levels. INO80 interacts with the core PRC2 complex member SUZ12 and promotes its recruitment. Furthermore, INO80 mediates H2A.Z incorporation at the poised promoters, which was reduced in Ino80cKO spermatocytes. Taken together, INO80 is emerging as a major regulator of the meiotic transcription program by mediating poised chromatin establishment through SUZ12 binding.
KEY WORDS: INO80, Poised chromatin, Chromatin remodeler, Germ cells, Polycomb subunits, Spermatogenesis
Summary: INO80 is required for meiotic progression in spermatocytes, which involves global changes in transcription. These changes are facilitated by INO80 in coordination with H2A.Z and SUZ12 to establish poised chromatin.
INTRODUCTION
Mammalian gametogenesis is dependent on the accumulation of cellular and epigenetic changes that are required to generate haploid gametes from diploid cells through meiosis. In males, meiotic prophase I involves the synapsis and recombination of homologous chromosomes (Handel and Schimenti, 2010), which is accompanied by dynamic changes in gene expression patterns to transit from somatic to germline-specific gene signatures (Sasaki and Matsui, 2008). Germ cells also undergo major changes in chromatin structure and histone modifications, which in turn allow changes in the cellular macromolecular assembly for execution of synapsis and recombination (Kota and Feil, 2010).
Histone modifications are integral to changes in chromatin dynamics. H3K9 methyltransferases, such as suppressor of variegation 3-9 H1 or H2 (SUV39-H1 or -H2) and euchromatic histone lysine N-methyltransferase 2 (G9a) (EHMT2), play crucial roles in synapsis (Tachibana et al., 2007; Takada et al., 2011). H3K27 methyltransferase activity of polycomb repressive complex 2 (PRC2) modulates the epigenome in spermatocytes and is responsible for the establishment and maintenance of bivalent chromatin (Hasegawa et al., 2015; Maezawa et al., 2018; Mu et al., 2014). PRC2 is also required for proper synapsis and double strand break repair (Mu et al., 2014).
As germ cells begin meiosis, they undergo epigenetic suppression of genes that are important for somatic developmental pathways. However, these genes remain in what is known as a poised state for activation at later developmental stages because they carry bivalent domains that are characterized by the co-occurrence of activating H3K4me3 and repressive H3K27me3 modifications at somatic gene promoters (Bernstein et al., 2006; Brykczynska et al., 2010; Hammoud et al., 2009, 2014; Sin et al., 2015). Trithorax complexes (e.g. MLL2, SET1A and SET1B) and polycomb group complexes (PRC2) are the main methyltransferases that catalyze the deposition of H3K4me3 and H3K27me3, respectively, at these sites (Piunti and Shilatifard, 2016). However, the regulatory mechanism of bivalency at poised chromatin in germ cells is not well understood.
Chromatin reorganization is actively facilitated by ATP-dependent chromatin remodeling complexes, which regulates the accessibility of co-factors and processes such as transcription and DNA damage repair. Several chromatin remodelers are known to play important roles in germ cell development (Dowdle et al., 2013; Imai et al., 2020; Kim et al., 2012; Li et al., 2014; Serber et al., 2016; Spruce et al., 2020; Wang et al., 2012). For example, the SWI/SNF catalytic subunit BRG1 promotes synapsis and DNA damage repair in germ cells, and is necessary for spermatogenesis (Kim et al., 2012). In contrast, imitation switch (ISWI) and chromodomain helicase DNA-binding (CHD) complexes are required for subsequent processes such as spermiogenesis and fertilization (Dowdle et al., 2013; Li et al., 2014). For INO80, our laboratory has previously shown that germ cell-specific depletion of INO80 causes a meiotic arrest in spermatocytes and results in infertility due to aberrant synapsis and incomplete DNA double strand break repair (Serber et al., 2016).
The INO80 complex is a highly conserved chromatin remodeler that binds nucleosome-free regions around promoter and transcriptional start sites, and organizes chromatin architecture by repositioning nucleosomes (Clapier and Cairns, 2009; Yen et al., 2013). INO80 also has a broad effect on promoters, which facilitates both transcriptional activation and repression, as shown in yeast, Drosophila and human (Cao et al., 2015; Chen et al., 2011; Klopf et al., 2017; Neuman et al., 2014; Poli et al., 2017; Yen et al., 2013). In ES cells, INO80 is required for pluripotency, reprogramming and regulation of transcription by facilitating the recruitment of co-factors and RNA polymerase II (RNAPII) to the promoters of pluripotency network genes (Wang et al., 2014; Zhou et al., 2016). INO80 also facilitates the exchange and turnover of the histone variant H2A.Z (Alatwi and Downs, 2015; Brahma et al., 2017; Papamichos-Chronakis et al., 2011; Ranjan et al., 2013; Yen et al., 2013). Bivalent promoters are also highly enriched with H2A.Z, which facilitates access of regulatory complexes to chromatin in ES cells (Hu et al., 2013; Ku et al., 2012). Chromatin remodeler complex members, such as BRG1, SMARCD1 and ARID1A of the esBAF complex, and CHD3/4 of the NuRD complex, were demonstrated to be involved in the regulation of bivalency (Alajem et al., 2015; Ho et al., 2009; Lei et al., 2015; Reynolds et al., 2012). Furthermore, BRG1 maintains poised promoters in spermatocytes, which is mediated by PRC1 member SCML2 (Menon et al., 2019).
In this study, we set out to determine how INO80 regulates the transcription program of meiotic spermatocytes. We present evidence that INO80 regulates transcription in meiotic spermatocytes, and that it is required to repress somatic gene expression in these cells. INO80 promotes both H2A.Z occupancy and H3K27me3 occupancy at poised promoters. Additionally, the repression of somatic genes is regulated by INO80-mediated recruitment of the PRC2 member SUZ12 at their promoters to maintain H3K27me3 levels and therefore bivalency. Taken together, these findings establish a crucial role for INO80-mediated regulation of meiotic transcription, and in the maintenance of poised genes through PRC2 recruitment.
RESULTS
INO80 occupies promoter regions with transcriptionally active and poised chromatin
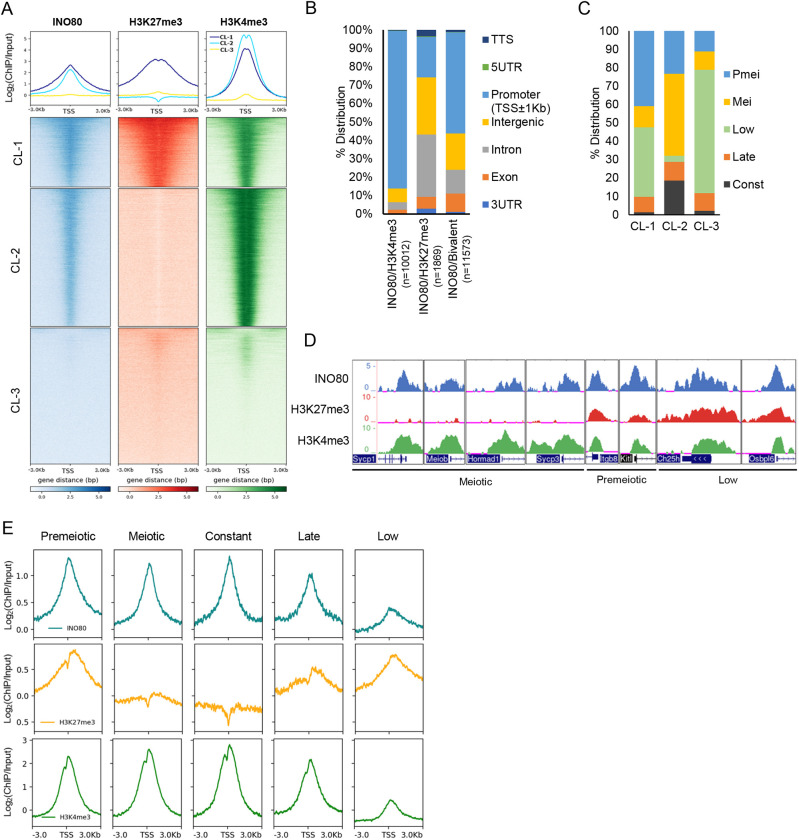
To determine how chromatin remodeler INO80 regulates meiotic progression by modulating chromatin activity in germ cells, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) in Ino80WT postnatal day (P) 18 spermatocytes. INO80 binding was most highly enriched around promoter proximal regions, with a lower occupancy at intron and intergenic regions (Fig. S1A). Some INO80 binding was also observed around 3′UTR and transcriptional start sites (TSSs) (Fig. S1A). INO80 peaks were distributed genome wide (Table S1) throughout all chromosomes (Fig. S1B). The extensive promoter occupancy of INO80 (Fig. S1A) prompted us to investigate the level of promoter activity of all genes based on active and repressive histone modifications. We compared the distribution of INO80-binding sites at all promoters with the activating and repressive histone modifications, such as histone H3 lysine 4 trimethyl (H3K4me3) and histone H3 lysine 27 trimethyl (H3K27me3), respectively. The comparison was made at a similar developmental timepoint (P17) (Mu et al., 2014). K-means clustering of these datasets based on INO80 occupancy around the TSS of all genes revealed three different clusters (Fig. 1A). INO80 enrichment was moderate-to-high in cluster (CL) 1 and CL2, whereas CL1 demonstrated slightly wider and more enriched INO80 occupancy relative to CL2. Additionally, CL1 also demonstrated robust binding of both H3K27me3 and H3K4me3, whereas H3K4me3 enrichment was higher in CL2 with no detectable H3K27me3 levels. CL3 was devoid of INO80 binding and displayed either no, or very low levels of, H3K4me3 and H3K27me3 (Fig. 1A). The co-occurrence of H3K27me3 and H3K4me3 in CL1 is indicative of bivalent chromatin, which is ‘poised’ and can be activated when necessary (Bernstein et al., 2006). Among all INO80-binding sites associated with promoters, 41% displayed both H3K27me3 and H3K4me3 (bivalent), and 56% were marked with H3K4me3 only, while only about 3% of INO80 peaks featured H3K27me3 only (Fig. S1C).
Fig. 1.
Genome-wide occupancy of INO80, H3K27me3 and H3K4me3. (A) K-Means clustering of INO80, H3K27me3 and H3K4me3 occupancy at the TSS±3 kb for all UCSC Refseq genes. Three clusters (CL) were identified with metaplots (top panels) illustrating enrichment of INO80, H3K27me3 and H3K4me3 in CL1 (blue), CL2 (cyan) and CL3 (yellow). (B) Genome-wide distribution of the INO80 peaks that co-occur with either H3K27me3 or H3K4me3, or with both modifications. (C) Representation of spermatogenic gene cohorts from each of the three clusters (CL1-CL3). (D) Genomic tracks exhibiting enrichment of INO80, H3K27me3 and H3K4me3 at representative meiotic (Sycp1, Sycp3, Hormad1 and Meiob), premeiotic (Itgb8 and Kitl) and somatic (Ch25h and Osbpl6) genes. (E) Metaplots showing relative enrichment of INO80, H3K27me3 and H3K4me3 at the TSS±3 kb region of the genes from the five meiotic temporal expression gene cohorts.
Genome-wide annotation of the INO80 peaks revealed that most peaks shared with bivalent modifications were near the TSS (Fig. S1D), associated primarily with promoters and with gene bodies, including exons, introns and some intergenic regions (Fig. 1B). Similarly, the majority (86%) of INO80-binding sites shared only with H3K4me3 were observed at the promoter/TSS regions, as well as some intergenic areas (Fig. 1B, Fig. S1E). However, a small proportion of INO80-binding sites with only H3K27me3 modification were distributed throughout a larger area, around promoters, gene bodies and intergenic regions (Fig. 1B, Fig. S1F).
In addition to the distribution of these marks, we explored the functional aspect of INO80 binding at either poised or active genes, and their role in the first wave of spermatocyte development. To determine how INO80 binding, in collaboration with these histone modifications might regulate gene expression in early spermatogenesis, we used previously determined temporal gene expression profiles of the postnatal developing testis (Margolin et al., 2014). The temporal cohorts include genes expressed: before meiosis at P6 (premeiotic); during early meiosis from P8 to P20 (meiotic); constantly from the premeiotic stage to adult stage from P6-P38 (constant); only at adult stage (P38) (late); and at very low levels throughout all the stages from P6 to P38 (low). Overall, the largest proportion (59%) of INO80-bound genes was present in the premeiotic and low cohorts, whereas 24% belong to the meiotic cohort (Fig. S1G). Among the clusters, 79% of CL1 comprised INO80-bound premeiotic and low genes, while INO80-bound meiotic genes represented 45% of CL2 (Fig. 1C). CL3 was primarily enriched with genes belonging to the low cohort (Fig. 1C). The bivalent chromatin associated with INO80 targets in CL1 indicates that the genes within the premeiotic and low cohorts are poised, whereas meiotic genes marked by H3K4me3 in CL2 are active.
An examination of representative INO80 target genes from the meiotic, premeiotic and low cohorts reveals the presence of H3K4me3 only at the active meiotic promoters (e.g. Sycp1, Sycp3, Hormad1 and Meiob). In contrast, repressed premeiotic (Kitl and Itgb6) and somatic promoters (Ch25h and Osbpl6) display bivalent modifications (Fig. 1D). Overall, premeiotic and late cohorts demonstrated high enrichment of both H3K4me3 and H3K27me3, along with high enrichment of INO80 (Fig. 1E). Meiotic and constant cohorts demonstrated only H3K4me3 and INO80 enrichment, whereas the low cohort displayed low INO80 and H3K4me3 enrichment with high H3K27me3 (Fig. 1E). Together, these data suggest a role for INO80 in gene regulation, possibly by regulating histone modification and therefore promoter activity of either repressed (premeiotic, late and low) or active (meiotic and constant) genes in developing spermatocytes.
INO80 regulates gene expression in developing spermatocytes
Ino80 deletion in spermatocytes results in meiotic arrest in late zygotene/early pachytene stage spermatocytes in adult testis (Serber et al., 2016). Because previous studies in other cell types suggested a role for INO80 regulation of transcription (Poli et al., 2017), we investigated a possible role for INO80 in the spermatogenic transcription program. We used spermatocytes from a germ cell-specific Stra8-Cre-driven INO80 knockout mouse model (Ino80cKO) and its wild-type littermates (Ino80WT) to determine the effect of Ino80 deletion on transcription in these cells. Stra8-driven Cre expression in murine testis begins postnatally with detectable CRE recombinase protein expression at P7 in spermatogonia and pre-leptotene spermatocytes (Sadate-Ngatchou et al., 2008). This expression allowed us to introduce Ino80 deletion at the beginning of the first wave of spermatocyte development. We examined comparable spermatocyte populations, where INO80 downregulation did not result in overt changes in cell populations. Testes missing INO80 were similar in size to INO80 heterozygous (Ino80Het) and Ino80WT at P18, whereas, at P30, Ino80cKO testis volume was reduced (Fig. S2A). At P18, late zygotene and pachytene spermatocytes showed robust INO80 expression (Fig. S2C). In contrast, INO80 expression in Ino80cKO testis was not detectable in spermatocytes (Fig. S2C), and overall INO80 protein expression was reduced in a dose-dependent fashion in both Ino80Het and Ino80cKO testes when compared with Ino80WT (Fig. S2B). We examined whether there was widespread cell death in Ino80cKO testis using TUNEL assays and found a lack of apoptosis until P21. However, by P30, the presence of apoptotic cells significantly increased in the Ino80cKO testis (Fig. S2D).
Meiotic substages were also similar at P18 between Ino80WT and Ino80cKO testes (Fig. S3A). In addition, no significant changes in overall meiotic substage-specific gene-expression patterns between Ino80WT and Ino80cKO testis were observed up to the early pachytene stage (Fig. S3B). However, the significant reduction in diplotene spermatocyte expression patterns observed in Ino80cKO testis (Fig. S3B) is indicative of aberrant gene regulation leading to meiotic arrest prior to diplonema.
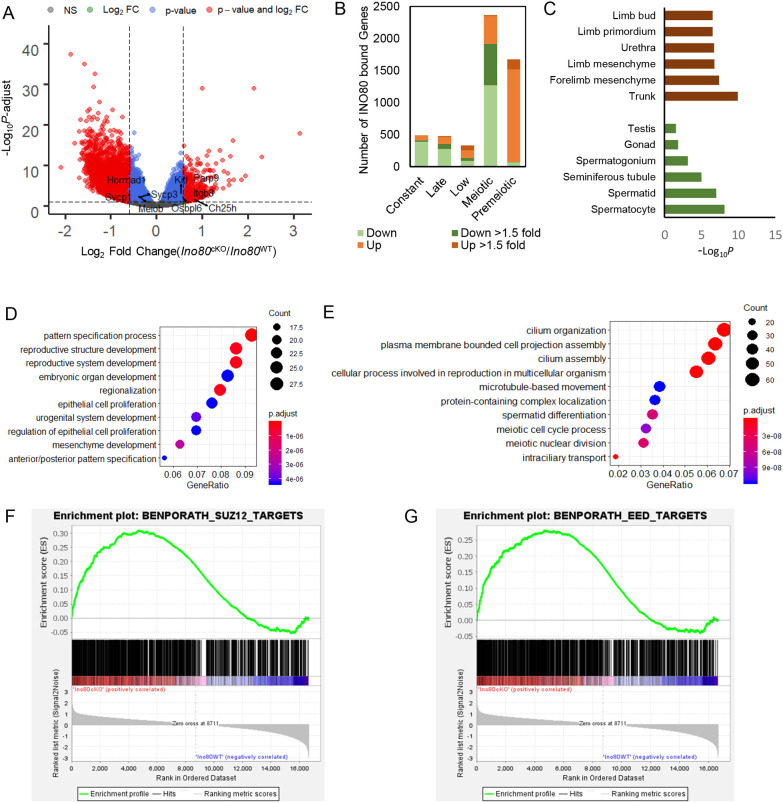
To determine how INO80 affects the meiotic transcription program, we performed mRNA seq from spermatocytes isolated from Ino80WT and Ino80cKO testes on P18. Differential expression analysis (Table S2) revealed upregulation of 3549 genes (P-adjust<0.05) and concomitant downregulation of 4103 genes (P-adjust<0.05) in Ino80cKO spermatocytes compared with Ino80WT (Fig. 2A). Among the misregulated genes, 2155 genes were downregulated and 632 genes were upregulated at 1.5-fold or higher. Among all the differentially expressed genes (DEG) (P-adjust<0.05), 62% of the downregulated and 71% of the upregulated DEGs were bound by INO80 at promoter/TSS regions. Although the pattern of INO80-binding around promoters was largely similar between up- and downregulated DEGs, upregulated DEGs showed a slight increase in overall INO80 enrichment compared with downregulated DEGs (Fig. S4B). Interestingly, most of the significantly upregulated genes in the INO80 mutant background belong to premeiotic and low cohorts, whereas most of those genes that were downregulated were present in the meiotic cohort of genes, with some from late and constant cohorts (Fig. 2B). Overall, a relatively low number of genes from constant, late and low cohorts were significantly changed in the mutant background when compared with premeiotic and meiotic cohorts (Fig. 2B), suggesting the importance of INO80 for regulating premeiotic and meiotic genes in spermatocytes.
Fig. 2.
INO80 regulates the transcription program in spermatocytes. (A) Volcano plot illustrating altered gene transcription in Ino80cKO spermatocytes compared with Ino80WT spermatocytes. Gray dots represent P-adjust>0.05; blue dots represent P-adjust<0.05 and fold change<1.5-fold; red dots represent P-adjust<0.05 and fold change>1.5-fold (n=5). P-adjust was derived by Benjamini–Hochberg method. (B) The number of differentially expressed (P-adjust<0.05) genes from each temporal expression cohort that also interact with INO80 at promoter proximal regions. (C) Enrichment of the anatomical terms (EMAPA) in the upregulated (brown) and downregulated (green) genes due to Ino80 deletion in spermatocytes. (D,E) Gene ontology biological pathways that were enriched in the differentially expressed genes (P-adjust<0.05), either upregulated (D) or downregulated (E) more than 1.5-fold. The size of the circles represents gene count and the color represents the adjusted P-value (Benjamini–Hochberg) for each pathway. (F,G) Gene set enrichment analysis depicting enrichment of SUZ12 (F) and EED (G) targets in the significantly upregulated genes due to Ino80 deletion in spermatocytes.
Gene enrichment analysis was performed in the mutant background to understand the biological nature of genes that are mis-expressed 1.5× in either direction. The upregulated genes were somatic in nature and related to structures such as embryo mesenchyme, trunk and limb (Fig. 2C), and functions related to pattern specification, reproductive structure and embryonic organ development (Fig. 2D). In contrast, the downregulated genes, which are primarily related to male gonad (Fig. 2C), cause male infertility, and abnormal spermatogenesis and sperm development in the mutants. The upregulated genes were also enriched in PRC2 subunits SUZ12 (Fig. 2F) and EED targets (Fig. 2G), which suggests a possible association between INO80-mediated gene repression and the actions of PRC2.
INO80 regulates the establishment of poised chromatin in spermatocytes
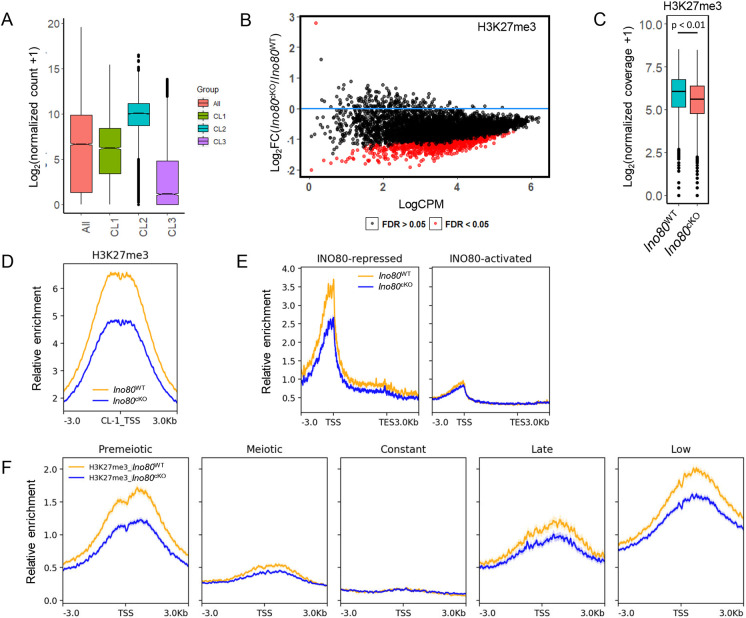
The observed change in the overall transcription program of developing spermatocytes prompted us to determine the mechanism by which INO80 regulates spermatocyte transcription. As INO80-bound promoters share either active H3K4me3 or repressive bivalent modifications, we determined the average level of gene expression from these clusters. The average expression of genes with bivalent promoters (CL1) was lower than the CL2 genes in Ino80WT spermatocytes (Fig. 3A). Additionally, the overall expression level of the genes in CL3, which were devoid of INO80, was much lower than the average gene expression level (Fig. 3A). Next, among the DEGs, CL1 genes demonstrated an overall increase in the average change of expression compared with the average of all genes. However, despite the presence of the majority of the downregulated DEGs, the mean change was negligible for genes in CL2 (Fig. S4A). Furthermore, we determined that the INO80-bound genes that were upregulated in Ino80cKO were highly enriched in bivalent promoters and therefore repressed in Ino80WT spermatocytes. In contrast, all the INO80-bound downregulated gene promoters were devoid of any H3K27me3 modifications and only displayed active H3K4me3 modifications and therefore were active in Ino80WT spermatocytes (Fig. S4B).
Fig. 3.
INO80 regulates establishment of histone H3 lysine 27 trimethylation. (A) The average gene expression level of all expressed genes and genes that belong to each of the three gene clusters in Ino80WT, based on the presence of either H3K4me3 or bivalent marks along with INO80. Lower and upper limits of the box represent first and third quartiles, midline represents the median, whiskers denote lower and upper limit of the dataset, and black dots represent outliers. (B) Differential analysis of H3K27me3 binding at bivalent domains (CL1) bound to INO80. Red dots represent promoters that have a significant (FDR<0.05) change in H3K27me3 enrichment in Ino80cKO compared with Ino80WT. Black dots represent FDR>0.05. FDR was derived by the Benjamini–Hochberg method (n=3). (C) Comparison of normalized coverage for H3K27me3 between Ino80cKO and Ino80WT at bivalent INO80-interacting regions (CL1). P<0.01, as calculated by Wilcoxon signed rank test (two-tailed). Lower and upper limits of the box represent first and third quartiles, midline represents the median, whiskers denote lower and upper limit of the dataset, and black dots represent outliers. (D) Metaplots showing relative enrichment for H3K27me3 between Ino80cKO and Ino80WT for CL1. Yellow, Ino80WT; blue, Ino80cKO. Plots are centered at the TSS. (E) Metaplots showing relative enrichment of H3K27me3 between Ino80cKO and Ino80WT 3 kb upstream and downstream of the promoter and gene body (represented as a 5 kb region) for either INO80-repressed or INO80-activated genes in Ino80WT. Yellow, Ino80WT; blue, Ino80cKO. (F) Relative enrichment of H3K27me3 between Ino80cKO and Ino80WT in each of the five temporal gene expression cohorts. Plots are centered at the TSS. Yellow, Ino80WT; blue, Ino80cKO.
To determine whether INO80 regulates the establishment of H3K4me3 and H3K27me3 modifications at poised promoters, genome-wide distribution of these modifications was examined either in the presence (Ino80WT) or absence of INO80 (Ino80cKO) in P18 spermatocytes. No detectable difference in the occurrence of H3K4me3 was observed between Ino80WT and Ino80cKO spermatocytes, either at the INO80-bivalent CL1 (Fig. S4C) or at INO80-repressed and INO80-activated genes (Fig. S4D). These data suggest that INO80 does not involve H3K4 methylation to regulate gene expression. In contrast, analysis of differential occupancy for H3K27me3 modifications revealed that many INO80-bound bivalent promoters exhibited a decrease (FDR<0.05) in H3K27me3 occupancy (Fig. 3B) along with a significant overall decrease in H3K27me3 enrichment at these sites (Fig. 3C,D). Change was also apparent at INO80-repressed genes (Fig. 3E), whereas no difference was observed for INO80-activated genes (Fig. 3E). Furthermore, among the temporal gene expression cohorts in Ino80cKO spermatocytes, H3K27me3 was reduced at TSSs associated with premeiotic, late and low groups (Fig. 3F). These data suggest that de-repression of these INO80-repressed genes is likely due to loss of H3K27me3.
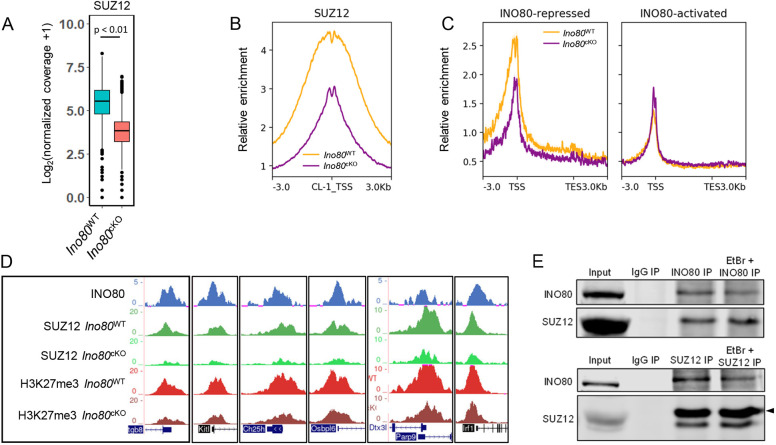
PRC2 is the only histone methyltransferase complex responsible for the establishment and maintenance of H3K27me3 modifications. As the genes encoding PRC2 complex members such as Ezh2, Suz12 and Eed displayed INO80 occupancy at their promoter (Fig. S5A), we determined the protein expression from these genes, which remained relatively unchanged between Ino80WT and Ino80cKO (Fig. S5B) at P18. To determine how INO80 regulates the establishment of H3K27me3 at bivalent promoters, we studied the PRC2 subunit SUZ12 occupancy in the presence or absence of INO80. CUT and RUN was performed to determine global SUZ12 binding affinity between Ino80WT and Ino80cKO spermatocytes from P18 testis. Analysis of differential occupancy revealed significant decrease in SUZ12 enrichment at INO80-bound bivalent sites (Fig. 4A,B). Additionally, INO80-repressed genes showed an overall decrease in SUZ12 enrichment, whereas no detectable changes were observed at the INO80-activated genes (Fig. 4C). This was further demonstrated by changes at representative genes from the premeiotic cohort, such as Kitl and Itgb8, in addition to transcription factor genes, such as Parp9 and Irf1, from the premeiotic cohort and somatic genes, such as Ch25h and Osbpl6, from the low cohort (Fig. 4D). These genes were upregulated in Ino80cKO (Fig. S4E) and demonstrated reduced enrichment of SUZ12 and H3K27me3 at their promoters (Fig. 4D).
Fig. 4.
INO80 promotes SUZ12-chromatin interaction. (A) Comparison of normalized coverage for SUZ12 between Ino80cKO and Ino80WT at bivalent INO80-interacting regions. P<0.01, as calculated by Wilcoxon signed rank test (two-tailed). Lower and upper limits of the box represent first and third quartiles, midline represents the median, whiskers denote lower and upper limit of the dataset, and black dots represent outliers. (B) Metaplot showing relative enrichment for SUZ12 between Ino80cKO and Ino80WT at CL1. Yellow, Ino80WT; purple, Ino80cKO. (C) Metaplots showing relative enrichment of SUZ12 between Ino80cKO and Ino80WT 3 kb upstream and downstream of the promoter and gene body (represented as 5 kb regions) in either INO80-repressed or INO80-activated genes in Ino80WT. Yellow, Ino80WT; purple, Ino80cKO. (D) Genomic tracks showing comparative enrichment of SUZ12 and H3K27me3 at representative genes. (E) Immunoblot showing the presence of SUZ12 in INO80 immunoprecipitated Ino80WT spermatocyte homogenate (top) and the presence of INO80 in SUZ12 immunoprecipitated Ino80WT spermatocyte homogenate (bottom), in either the presence or absence of ethidium bromide (EtBr). Arrowhead indicates the SUZ12 band in the bottom panel.
To determine whether INO80 interacts with SUZ12 to regulate H3K27 methylation, we performed immunoaffinity precipitation (IP) of INO80 from P18 wild-type spermatocytes. The co-IP of SUZ12 with INO80 suggests that INO80 physically interacts with SUZ12 to facilitate its chromatin interaction (Fig. 4E). We also performed reverse IP of SUZ12 and successfully determined the presence of INO80, corroborating the interaction (Fig. 4E). Furthermore, a similar immunodetection of both INO80 and SUZ12 in either SUZ12 IP or INO80 IP, respectively, occurred in the presence of ethidium bromide (Lai and Herr, 1992), suggesting that the observed interaction was specific and not due to indirect DNA-mediated artifacts.
INO80 regulates H2A.Z incorporation at poised promoters in spermatocytes
As a chromatin remodeler, INO80 is capable of nucleosome repositioning and exchange. Therefore, we examined whether changes in chromatin accessibility due to INO80 activity led to increased PRC2 binding and activity resulting in establishment of H3K27me3 at poised promoters. We found no significant differences in chromatin accessibility between Ino80WT and Ino80cKO spermatocytes from P18 testis either at INO80-bound bivalent regions (CL1) (Fig. S6A,B) or at CL2 (Fig. S6C), leading to the possibility that INO80 may affect the establishment of poised chromatin by its other effector H2A.Z.
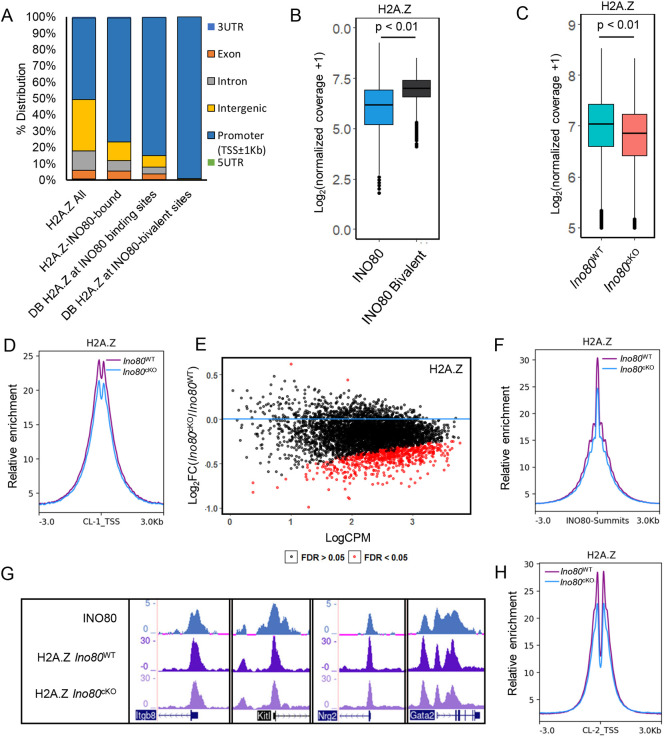
INO80 has been reported to play an important role in H2A.Z exchange at chromatin (Brahma et al., 2017; Papamichos-Chronakis et al., 2011). To determine whether INO80 mediates H2A.Z incorporation at the bivalent promoters, we performed ChIP-seq for H2A.Z in Ino80WT and Ino80cKO spermatocytes from P18 testis. The majority of H2A.Z peaks (Table S3) were distributed at promoter regions (Fig. 5A). The abundance of H2A.Z signal was significantly higher (P<0.01) at INO80-bound bivalent sites compared with the average signal at all INO80-binding sites (Fig. 5B). When comparing Ino80WT and Ino80cKO spermatocytes, a significant (FDR<0.05) decrease in H2A.Z occupancy was detected globally at bivalent promoters associated with INO80 binding (Fig. 5C,D), as well as many individual promoters (FDR<0.05) among them (Fig. 5E) at all INO80 peaks (Fig. 5F, Fig. S7A,B). Decreases in H2A.Z enrichment at all INO80 peaks were also apparent in Ino80cKO (Fig. 5F, Fig. S7A,B), suggesting a role for INO80 in promoting H2A.Z incorporation into developing spermatocytes. These decreases are further demonstrated in a few representative INO80-bound bivalent genes, such as Itgb8, Kitl, Nrg2 and Gata2 (Fig. 5G). Interestingly, among INO80-bound and differentially enriched H2A.Z sites between Ino80WT and Ino80cKO spermatocytes, 85% were located at promoters (Fig. 5A). Additionally, almost all (>99%) of the binding sites near the INO80-bivalent sites (TSS±1Kb) were promoter bound (Fig. 5A), suggesting that INO80 regulates H2A.Z incorporation at the bivalent promoters in developing spermatocytes. Among the promoters with H3K27me3, 84% showed H2A.Z occupancy in Ino80WT (Fig. S7C). Furthermore, when we compared fold changes in either H2A.Z occupancy or H3K27me3 occupancy at INO80-bound bivalent regions between Ino80cKO and Ino80WT spermatocytes, 83% of the change was concordant (Fig. S7D), suggesting a possible relationship between H2A.Z occupancy and H3K27me3 establishment at these regions. However, the reduction in H2A.Z levels was not limited to the INO80-bound bivalent promoters. Reduction was also observed in H3K27me3-free promoters in CL2 (Fig. 5H). This result was further demonstrated by reduced H2A.Z levels at representative genes with INO80 occupancy but devoid of either H3K27me3 or SUZ12 (Fig. S7E), suggesting that INO80-mediated H2A.Z incorporation is not dependent upon H3K27me3 establishment in spermatocytes.
Fig. 5.
INO80 regulates H2A.Z occupancy genome wide. (A) Relative distribution of genomic features at (1) all H2A.Z peaks, (2) those that also interact with INO80, (3) differentially accessible H2A.Z binding sites at INO80 peaks or (4) differentially accessible H2A.Z binding sites at INO80/bivalent regions. (B) Normalized coverage of H2A.Z at either all INO80-binding sites or INO80 interacting bivalent sites. P<0.01, as calculated by Wilcoxon rank sum test (two-tailed). Lower and upper limits of the box represent first and third quartiles, midline represents the median, whiskers denote lower and upper limit of the dataset, and black dots represent outliers. (C) Comparison of normalized coverage for H2A.Z between Ino80cKO and Ino80WT at bivalent INO80 interacting regions. P<0.01, as calculated by Wilcoxon signed rank test (two-tailed). Lower and upper limits of the box represent first and third quartiles, midline represents the median, whiskers denote lower and upper limit of the dataset, and black dots represent outliers. (D) Relative enrichment of H2A.Z between Ino80cKO and Ino80WT at INO80 interacting bivalent TSS±3 kb (CL1). Purple, Ino80WT; blue, Ino80cKO. (E) Dot plot showing differential binding analysis of H2A.Z binding at bivalent domains bound to INO80. Red dots represent promoters that have a significant (FDR<0.05) change in H2A.Z enrichment in Ino80cKO when compared with Ino80WT. Black dots represent FDR>0.05. FDR was derived by Benjamini–Hochberg method (n=2). (F) Relative enrichment of H2A.Z between Ino80cKO and Ino80WT at all INO80 summits±3 kb. Purple, Ino80WT; blue, Ino80cKO. (G) Genomic tracks showing comparative enrichment of INO80 and H2A.Z at representative genes with bivalent promoters. (H) Relative enrichment of H2A.Z between Ino80cKO and Ino80WT at INO80 interacting promoters with H3K4me3 only (CL2). Purple, Ino80WT; blue, Ino80cKO.
DISCUSSION
In this study, we have demonstrated the function of INO80 in regulating spermatogenic transcription to promote meiosis progression, and in repressing somatic gene expression in developing spermatocytes. We also showed that INO80 interacts with the PRC2 subunit SUZ12, and, in the absence of INO80, maintenance of bivalent promoters was perturbed due to a reduction in H3K27me3 modification. Furthermore, we found that INO80 promotes H2A.Z incorporation at these poised sites, possibly facilitating PRC2 binding and activity to introduce repressive H3K27me3 modifications.
Previous studies have shown that INO80 is associated with promoters/TSSs and is functionally correlated with changes in gene expression in several organisms (Cao et al., 2015; Chen et al., 2011; Klopf et al., 2017; Neuman et al., 2014; Yen et al., 2013). Our observed INO80 chromatin interaction in prepubertal murine spermatocytes is consistent with these observations. INO80 appears to regulate spermatogenic gene expression in a dichotomous way by promoting the expression of meiotic genes and silencing genes needed for somatic developmental pathways. The extensive association of INO80 with almost all active and poised chromatin in these cells also suggests a broad role for INO80 in regulating spermatogenic gene expression.
The presence of poised chromatin in developing germ cells is important for recovering totipotency following maturation and fertilization (Lesch and Page, 2014). Our study presents a novel cooperation between INO80 and PRC2 in the regulation of repression in poised chromatin during mammalian meiosis. Here, we propose that INO80 maintains bivalency by enabling H3K27 trimethylation at gene promoters through recruitment of SUZ12, which is known to work as a scaffold for PRC2 complex formation and also to facilitate chromatin binding of PRC2 (Højfeldt et al., 2018; Laugesen et al., 2019). This novel interaction of INO80 is in agreement with other studies showing that the INO80 complex can interact with other factors to regulate transcription, such as the transcription factor MBF in yeast, to facilitate the induction of gene transcription in a H2A.Z-dependent manner (Knezevic et al., 2018). INO80 also interacts with RBP1 and PAF1 in order to evict RNA polymerase II during stress in fission yeast (Lafon et al., 2015; Poli et al., 2016).
In addition to facilitating transcription factor function, INO80 complex activity was also recently reported to be important in the propagation of heterochromatin. INO80 complex member LEC5 was shown to control the inheritance of pericentric heterochromatin in yeast (Shan et al., 2020). Other chromatin remodelers, such as BRG1, have been demonstrated to regulate meiotic progression and found to interact with the testis-specific PRC1 member SCML2 (Kim et al., 2012; Menon et al., 2019), which was shown to be necessary for H3K27me3 establishment at bivalent domains (Maezawa et al., 2018). Thus, there may be unique roles for different chromatin remodelers in regulating poised chromatin to lead to the same developmental fate.
PRC2 has been proposed to be a crucial regulator of spermatogenesis. Loss of the PRC2 subunit SUZ12 in murine spermatocytes led to defects in synapsis and homologous recombination (Mu et al., 2014), which is a similar phenotype to that observed in Ino80cKO spermatocytes that reach the pachytene stage (Serber et al., 2016). In Ino80cKO spermatocytes, derepression of many genes were observed due to reduced H3K27me3 levels in the absence of SUZ12. As a result, the upregulation of pathways related to somatic development occurred. Therefore, we conclude that de-repression of somatic genes due to reduced PRC2 activity because of INO80 depletion culminates in the cellular phenotypes and developmental arrest seen in Ino80cKO spermatocytes.
The specialized histone variant H2A.Z is a known target of INO80 action, and turnover of H2A.Z translates the effect of INO80 in different cells (Giaimo et al., 2019; Morrison and Shen, 2009). There have been several reports proposing a role for INO80 in the removal of H2A.Z/H2B from chromatin to facilitate the incorporation of H2A/H2B dimers that can be necessary for genome integrity and DNA repair (Alatwi and Downs, 2015; Brahma et al., 2017; Papamichos-Chronakis et al., 2011). In addition, incorporation of H2A.Z into chromatin was proposed to be facilitated by SWR1 complex in yeast (Mizuguchi et al., 2004) and by either SRCAP or the EP400 complex in mammalian cells (Gévry et al., 2007; Ruhl et al., 2006). However, several recent studies have demonstrated either a lack of change or a reduction of H2A.Z occupancy in cells in the absence of INO80 (Locations et al., 2015; Tramantano et al., 2016; Yang et al., 2020; Yu et al., 2021), suggesting a cell type-specific role of INO80 in regulation of H2A.Z incorporation. Alternatively, it is possible that either SRCAP or TIP60/EP400 complex may play a role in INO80-regulated H2A.Z incorporation in spermatocytes. However, mechanisms related to H2A.Z dynamics have not been well studied in spermatocytes and warrant further investigation.
Genome-wide H2A.Z occupancy also differs among different organisms and cell types. For example, in yeast, H2A.Z is associated with both active and repressed genes (Raisner et al., 2005). In ES cells, H2A.Z occupancy was shown to be remarkably similar to SUZ12 occupancy at the promoters of developmentally important genes (Creyghton et al., 2008). In spermatocytes, distribution of H2A.Z was observed in both poised and activated chromatin, both of which are regulated by INO80. H2A.Z was also reported to regulate PRC2-dependent H3K27me3 deposition in ES cells (Wang et al., 2018; Yu et al., 2021). In spermatocytes, high concurrence of loss of H2A.Z with the loss of H3K27me3 in spermatocytes suggests a possible role for H2A.Z in the establishment of H3K27me3 by allowing PRC2 activity.
We propose a mechanism for INO80-mediated regulation of transcription in meiotic spermatocytes, whereby the complex promotes transcription of meiotic genes while suppressing the transcription of somatic genes. INO80 facilitates SUZ12 binding at specific sites in chromatin to establish PRC2 complexes followed by incorporation of H3K27me3 modifications. INO80-mediated H2A.Z incorporation in spermatocytes may facilitate this process (Fig. 6) by providing a more accessible chromatin landscape.
Fig. 6.
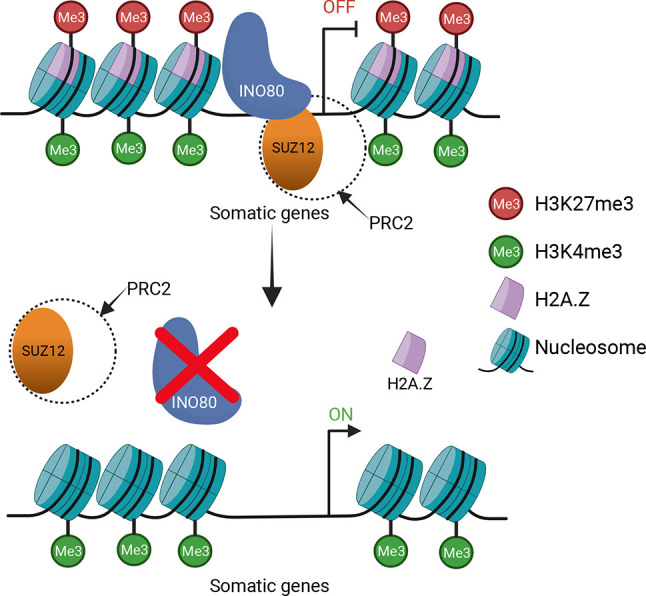
Schematic describing INO80-mediated regulation of poised chromatin in developing spermatocytes. INO80 regulates transcription during spermatogenesis by turning somatic genes to a poised state by facilitating binding of SUZ12, which activates PRC2 localization, leading to the introduction of repressive H3K27me3 modifications. INO80 deletion leads to de-repression of these genes, leading to a mis-regulated transcriptome. Created with BioRender.com.
MATERIALS AND METHODS
Animals and genotyping
Ino80 floxed (Serber et al., 2016) and Stra8-Cre (Sadate-Ngatchou et al., 2008) mice (Mus musculus) were maintained on an outbred CD1 background. Ino80f/f females were crossed with Stra8-CreTg/0 males to obtain Ino80f/+; Stra8CreTg/0 males. Ino80f/f females were crossed with Ino80f/+; Stra8CreTg/0 males to obtain male Ino80fl+ (Ino80WT), Ino80fl+; Stra8-CreTg/0 (Ino80Het) and Ino80Δ/f; Stra8CreTg/0 (Ino80cKO) littermates. Genotyping was performed using primers listed in Table S4. Mice were housed in a climate-controlled environment with 12 h light and 12 h dark cycles and fed ad libitum. All animal experiments were performed according to the protocol approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill.
Isolation of spermatogenic cells
Spermatogenic cells were isolated from freshly harvested testes according to a protocol described previously (Chang et al., 2011). All the experiments were performed twice or more with cells isolated from separate litters. Briefly, seminiferous tubules were digested with collagenase (1 mg/ml) in DMEM/F12 media or HBSS for 15 min followed by a second digestion with collagenase (1 mg/ml) and trypsin (0.1%) in DMEM/F12 media or HBSS for 15 min. Following the incubation, equal amounts of soybean trypsin inhibitor were added to inactivate trypsin and the digested product filtered through a 70 µm cell strainer and again through a 40 µm cell strainer. The flow through was centrifuged at 500 g for 10 mins to precipitate the cells, which were then washed twice with HBSS before resuspending in the appropriate buffer.
Immunofluorescence staining
Testes were embedded and frozen in Optimum Cutting Temperature (OCT) embedding medium. Cryosections (7 μm) were fixed in 10% freshly made paraformaldehyde solution in PBS for 10 min at 4°C. Spermatocyte single cell spreads were prepared following a previously described protocol (Wojtasz et al., 2009) with modifications (Biswas et al., 2018). Tissue sections or spreads were permeabilized, blocked and incubated overnight with primary antibody (listed in Table S5). The following day, samples were washed and incubated in Alexa Fluor-tagged secondary antibody before mounting. TUNEL staining was performed using Roche In Situ Cell Death Detection Kit, Fluorescein (MilliporeSigma) according to the manufacturer's instructions. Images were captured using a fluorescent microscope (Leica). Detailed protocols are available in the supplementary Materials and Methods.
Co-immunoprecipitation
Nuclear extract samples were prepared from spermatocytes (a detailed protocol is available in the supplementary Materials and Methods) isolated on P18 for co-immunoprecipitation. Samples were incubated with primary antibody (listed in Table S5) for 30 min, followed by the addition of protein A-conjugated Dynabeads and incubated overnight on a nutator at 4°C. The following day, Dynabeads were washed and eluted in Laemmli buffer and denatured. A detailed protocol is available in the supplementary Materials and Methods.
Western blotting
Polyacrylamide gel electrophoresis was performed to separate protein samples, followed by wet transfer to PVDF (polyvinylidene difluoride) membranes for fluorescence detection (Bio-Rad) overnight. Blots were blocked by Li-COR intercept blocking buffer and probed with primary antibody (listed in Table S5), followed by Li-COR secondary antibody.
RNA-seq
Total RNA samples were extracted from Ino80WT and Ino80cKO spermatocytes (n=5) on P18. Cell lysis was performed by TRIzol reagent (Invitrogen) and total RNA was purified using the Direct-zol RNA kit (Zymo). Sequencing libraries were made from 1 µg total RNA using the KAPA mRNA-Seq kit according to the manufacturer's protocol. Libraries were sequenced yielding 50 bp single end reads on an Illumina Hiseq 4000. Data analysis details are included in the supplementary Materials and Methods.
ChIP-seq
INO80 ChIP-seq was performed from Ino80WT spermatocytes isolated from P18 testes, with two biological replicates, as described previously (Runge et al., 2018). In short, 4×107 spermatocytes were isolated, followed by nuclear isolation and MNase digestion to obtain a mix of mono-, di- and tri-nucleosome fragments. Next, nuclei were lysed, and the extract incubated overnight with the primary antibody (listed in Table S5) attached to Dynabeads. The next day, samples were washed and eluted followed by de-crosslinking at 65°C overnight. On the following day, samples were treated with RNase A and proteinase K, and the DNA was purified and quantitated. Library preparation was carried out with the KAPA Hyper prep kit followed by sequencing on a Hiseq 2500 with 50 bp single end reads. H2A.Z ChIP-seq was performed in Ino80WT and Ino80cKO spermatocytes from P18 testes with the same protocol and sequenced on a Hiseq 4000 with 50 bp single end reads. H3K27me3 and H3K4me3 ChIP-seq data have been previously published (Mu et al., 2014) and are available under GEO accession number GSE61902. Data analysis details are included in the supplementary Materials and Methods.
ATAC-seq
ATAC-seq was performed on either Ino80WT or Ino80cKO spermatocytes, with three biological replicates, isolated on P18 following a method as described previously (Corces et al., 2017). Size selection was performed on the resulting libraries using 0.5× and 1.8× Kapa pure beads to generate a size range of ∼150 bp to ∼2 kb. ATAC-seq libraries were pooled and sequenced on a Novaseq 6000 platform yielding 50 bp paired-end reads. Data analysis details are included in the supplementary Materials and Methods.
CUT&RUN
CUT&RUN (cleavage under targets and release using nuclease) was performed with 250,000 cells for each sample, with three biological replicates from Ino80WT and Ino80cKO spermatocytes following the protocol previously described (Meers et al., 2019) with minor modifications. Briefly, spermatocytes were attached to concanavalin A-coated magnetic beads followed by permeabilization and incubation with either IgG or antigen-specific primary antibody (listed in Table S5) overnight. The next day, beads were washed, followed by binding to protein A/G-MNase fusion protein, chromatin digestion at 0°C for 30 min and release at 37°C for 30 min. The samples obtained were purified using DNA purification columns (Zymo ChIP DNA Clean & Concentrator). The eluate was quantitated and the library made with the KAPA Hyper prep kit following the manufacturer's protocol. Samples were sequenced using a Novaseq 6000 yielding paired end 50 bp reads. Data analysis details are included in the supplementary Materials and Methods.
Statistical analysis
Comparisons of signal from ChIP-seq, CUT&RUN and ATAC-seq experiments were analyzed by either the Wilcoxon rank sum test, in the case of unpaired observations, or the Wilcoxon signed rank test, in the case of paired observations. Multiple comparisons of gene expression levels from RNA-seq data were analyzed by one-way ANOVA, followed by Tukey's post-hoc test. All the tests performed were two-tailed.
Supplementary Material
Acknowledgements
We thank all the Magnuson lab members for their valuable input and comments in the study and with manuscript preparation. We thank Dr Jesse Raab for his input with ChIP-seq data analysis. We thank the Duke University School of Medicine for use of the Sequencing and Genomic Technologies Shared Resource, which provided the next-generation sequencing service.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: P.C., T.M.; Methodology: P.C.; Validation: P.C.; Formal analysis: P.C., T.M.; Investigation: P.C.; Data curation: P.C., T.M.; Writing - original draft: P.C.; Writing - review & editing: T.M.; Supervision: T.M.; Project administration: T.M.; Funding acquisition: T.M.
Data availability
The sequencing data from RNA-seq, ChIP-seq, ATAC-seq and CUT&RUN experiments have been deposited in GEO under accession number GSE179584.
Funding
This study was supported by the National Institutes of Health (R01GM101974 to T.M.) and by a Lalor Foundation Postdoctoral Fellowship (P.C.). Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.200089.
References
- Alajem, A., Biran, A., Harikumar, A., Sailaja, B. S., Aaronson, Y., Livyatan, I., Nissim-Rafinia, M., Sommer, A. G., Mostoslavsky, G., Gerbasi, V. R.et al. (2015). Differential association of chromatin proteins identifies BAF60a/SMARCD1 as a regulator of embryonic stem cell differentiation. Cell Rep. 10, 2019-2031. 10.1016/j.celrep.2015.02.064 [DOI] [PubMed] [Google Scholar]
- Alatwi, H. E. and Downs, J. A. (2015). Removal of H2A.Z by INO 80 promotes homologous recombination. EMBO Rep. 16, 986-994. 10.15252/embr.201540330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., Fry, B., Meissner, A., Wernig, M., Plath, K.et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315-326. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Biswas, U., Stevense, M. and Jessberger, R. (2018). SMC1α substitutes for many meiotic functions of SMC1β but cannot protect telomeres from damage. Curr. Biol. 28, 249-261.e4. 10.1016/j.cub.2017.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahma, S., Udugama, M. I., Kim, J., Hada, A., Bhardwaj, S. K., Hailu, S. G., Lee, T.-H. and Bartholomew, B. (2017). INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat. Commun. 8, 15616. 10.1038/ncomms15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska, U., Hisano, M., Erkek, S., Ramos, L., Oakeley, E. J., Roloff, T. C., Beisel, C., Schübeler, D., Stadler, M. B. and Peters, A. H. F. M. (2010). Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679-687. 10.1038/nsmb.1821 [DOI] [PubMed] [Google Scholar]
- Cao, L., Ding, J., Dong, L., Zhao, J., Su, J., Wang, L., Sui, Y., Zhao, T., Wang, F., Jin, J.et al. (2015). Negative regulation of p21Waf1/Cip1 by human INO80 chromatin remodeling complex is implicated in cell cycle phase G2/M arrest and abnormal chromosome stability. PLoS ONE 10, e0137411. 10.1371/journal.pone.0137411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y.-F., Lee-Chang, J. S., Panneerdoss, S., MacLean, J. A. and Rao, M. K. (2011). Isolation of Sertoli, Leydig, and spermatogenic cells from the mouse testis. BioTechniques 51, 341-344. 10.2144/000113764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Cai, Y., Jin, G., Florens, L., Swanson, S. K., Washburn, M. P., Conaway, J. W. and Conaway, R. C. (2011). Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J. Biol. Chem. 286, 11283-11289. 10.1074/jbc.M111.222505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier, C. R. and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273-304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Corces, M. R., Trevino, A. E., Hamilton, E. G., Greenside, P. G., Sinnott-Armstrong, N. A., Vesuna, S., Satpathy, A. T., Rubin, A. J., Montine, K. S., Wu, B.et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959-962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton, M. P., Markoulaki, S., Levine, S. S., Hanna, J., Lodato, M. A., Sha, K., Young, R. A., Jaenisch, R. and Boyer, L. A. (2008). H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135, 649-661. 10.1016/j.cell.2008.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle, J. A., Mehta, M., Kass, E. M., Vuong, B. Q., Inagaki, A., Egli, D., Jasin, M., Keeney, S., Flaus, A., Owen-Hughes, T.et al. (2013). Mouse BAZ1A (ACF1) is dispensable for double-strand break repair but is essential for averting improper gene expression during spermatogenesis. PLoS Genet. 9, e1003945. 10.1371/journal.pgen.1003945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gévry, N., Ho, M. C., Laflamme, L., Livingston, D. M. and Gaudreau, L. (2007). p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 21, 1869-1881. 10.1101/gad.1545707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaimo, B. D., Ferrante, F., Herchenröther, A., Hake, S. B. and Borggrefe, T. (2019). The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 12, 37. 10.1186/s13072-019-0274-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud, S. S., Nix, D. A., Zhang, H., Purwar, J., Carrell, D. T. and Cairns, B. R. (2009). Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473-478. 10.1038/nature08162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud, S. S., Low, D. H. P., Yi, C., Carrell, D. T., Guccione, E. and Cairns, B. R. (2014). Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15, 239-253. 10.1016/j.stem.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Handel, M. A. and Schimenti, J. C. (2010). Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11, 124-136. 10.1038/nrg2723 [DOI] [PubMed] [Google Scholar]
- Hasegawa, K., Sin, H.-S., Maezawa, S., Broering, T. J., Kartashov, A. V., Alavattam, K. G., Ichijima, Y., Zhang, F., Bacon, W. C., Greis, K. D.et al. (2015). SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev. Cell 32, 574-588. 10.1016/j.devcel.2015.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, L., Jothi, R., Ronan, J. L., Cui, K., Zhao, K. and Crabtree, G. R. (2009). An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U. S. A. 106, 5187-5191. 10.1073/pnas.0812888106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højfeldt, J. W., Laugesen, A., Willumsen, B. M., Damhofer, H., Hedehus, L., Tvardovskiy, A., Mohammad, F., Jensen, O. N. and Helin, K. (2018). Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol. 25, 225-232. 10.1038/s41594-018-0036-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, G., Cui, K., Northrup, D., Liu, C., Wang, C., Tang, Q., Ge, K., Levens, D., Crane-Robinson, C. and Zhao, K. (2013). H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12, 180-192. 10.1016/j.stem.2012.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, Y., Biot, M., Clément, J. A. J., Teragaki, M., Urbach, S., Robert, T., Baudat, F., Grey, C. and de Massy, B. (2020). Prdm9 activity depends on hells and promotes local 5-hydroxymethylcytosine enrichment. Elife 9, e57117. 10.7554/eLife.57117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Fedoriw, A. M. and Magnuson, T. (2012). An essential role for a mammalian SWI/SNF chromatin-remodeling complex during male meiosis. Development 139, 1133-1140. 10.1242/dev.073478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopf, E., Schmidt, H. A., Clauder-Münster, S., Steinmetz, L. M. and Schüller, C. (2017). INO80 represses osmostress induced gene expression by resetting promoter proximal nucleosomes. Nucleic Acids Res. 45, 3752-3766. 10.1093/nar/gkw1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic, I., González-Medina, A., Gaspa, L., Hidalgo, E. and Ayté, J. (2018). The INO80 complex activates the transcription of S-phase genes in a cell cycle-regulated manner. FEBS J. 285, 3870-3881. 10.1111/febs.14640 [DOI] [PubMed] [Google Scholar]
- Kota, S. K. and Feil, R. (2010). Epigenetic transitions in germ cell development and meiosis. Dev. Cell 19, 675-686. 10.1016/j.devcel.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Ku, M., Jaffe, J. D., Koche, R. P., Rheinbay, E., Endoh, M., Koseki, H., Carr, S. A. and Bernstein, B. E. (2012). H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 13, R85. 10.1186/gb-2012-13-10-r85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon, A., Taranum, S., Pietrocola, F., Dingli, F., Loew, D., Brahma, S., Bartholomew, B. and Papamichos-Chronakis, M. (2015). INO80 chromatin remodeler facilitates release of RNA polymerase II from chromatin for ubiquitin-mediated proteasomal degradation. Mol. Cell 60, 784-796. 10.1016/j.molcel.2015.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, J. S. and Herr, W. (1992). Ethidium bromide provides a simple tool for identifying genuine DNA- independent protein associations. Proc. Natl. Acad. Sci. U. S. A. 89, 6958-6962. 10.1073/pnas.89.15.6958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen, A., Højfeldt, J. W. and Helin, K. (2019). Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell 74, 8-18. 10.1016/j.molcel.2019.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, I., West, J., Yan, Z., Gao, X., Fang, P., Dennis, J. H., Gnatovskiy, L., Wang, W., Kingston, R. E. and Wang, Z. (2015). BAF250a protein regulates nucleosome occupancy and histone modifications in priming embryonic stem cell differentiation. J. Biol. Chem. 290, 19343-19352. 10.1074/jbc.M115.637389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch, B. J. and Page, D. C. (2014). Poised chromatin in the mammalian germ line. Development 141, 3619-3626. 10.1242/dev.113027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Wu, J., Kim, S. Y., Zhao, M., Hearn, S. A., Zhang, M. Q., Meistrich, M. L. and Mills, A. A. (2014). Chd5 orchestrates chromatin remodelling during sperm development. Nat. Commun. 5, 3812. 10.1038/ncomms4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locations, H. A. Z. I., Kaplan, C. D., Peterson, C. L., Watanabe, S., Kaplan, C. D., Peterson, C. L., Jeronimo, C., Watanabe, S., Kaplan, C. D., Peterson, C. L.et al. (2015). The histone chaperones FACT and Spt6 Restrict H2A.Z from intragenic locations. Mol. Cell 58, 1113-1123. 10.1016/j.molcel.2015.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa, S., Hasegawa, K., Yukawa, M., Kubo, N., Sakashita, A., Alavattam, K. G., Sin, H. S., Kartashov, A. V., Sasaki, H., Barski, A.et al. (2018). Polycomb protein SCML2 facilitates H3K27me3 to establish bivalent domains in the male germline. Proc. Natl. Acad. Sci. U. S. A. 115, 4957-4962. 10.1073/pnas.1804512115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, G., Khil, P. P., Kim, J., Bellani, M. A. and Camerini-Otero, R. D. (2014). Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics 15, 39. 10.1186/1471-2164-15-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers, M. P., Bryson, T. D., Henikoff, J. G. and Henikoff, S. (2019). Improved CUT&RUN chromatin profiling tools. Elife 8, e46314. 10.7554/eLife.46314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, D. U., Shibata, Y., Mu, W. and Magnuson, T. (2019). Mammalian SWI/SNF collaborates with a polycomb-associated protein to regulate male germline transcription in the mouse. Development 146, dev174094. 10.1242/dev.174094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, G., Shen, X., Landry, J., Wu, W. H., Sen, S. and Wu, C. (2004). ATP-Driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343-348. 10.1126/science.1090701 [DOI] [PubMed] [Google Scholar]
- Morrison, A. J. and Shen, X. (2009). Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 10, 373-384. 10.1038/nrm2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, W., Starmer, J., Fedoriw, A. M., Yee, D. and Magnuson, T. (2014). Repression of the soma-specific transcriptome by polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 28, 2056-2069. 10.1101/gad.246124.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman, S. D., Ihry, R. J., Gruetzmacher, K. M. and Bashirullah, A. (2014). INO80-dependent regression of ecdysone-induced transcriptional responses regulates developmental timing in Drosophila. Dev. Biol. 387, 229-239. 10.1016/j.ydbio.2014.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis, M., Watanabe, S., Rando, O. J. and Peterson, C. L. (2011). Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200-213. 10.1016/j.cell.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti, A. and Shilatifard, A. (2016). Epigenetic balance of gene expression by polycomb and compass families. Science 352, aad9780. 10.1126/science.aad9780 [DOI] [PubMed] [Google Scholar]
- Poli, J., Gerhold, C. B., Tosi, A., Hustedt, N., Seeber, A., Sack, R., Herzog, F., Pasero, P., Shimada, K., Hopfner, K. P.et al. (2016). Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 30, 337-354. 10.1101/gad.273813.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli, J., Gasser, S. M. and Papamichos-Chronakis, M. (2017). The INO80 remodeller in transcription, replication and repair. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160290. 10.1098/rstb.2016.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner, R. M., Hartley, P. D., Meneghini, M. D., Bao, M. Z., Liu, C. L., Schreiber, S. L., Rando, O. J. and Madhani, H. D. (2005). Histone variant H2A.Z Marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123, 233-248. 10.1016/j.cell.2005.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan, A., Mizuguchi, G., Fitzgerald, P. C., Wei, D., Wang, F., Huang, Y., Luk, E., Woodcock, C. L. and Wu, C. (2013). Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell 154, 1232-1245. 10.1016/j.cell.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, N., Salmon-Divon, M., Dvinge, H., Hynes-Allen, A., Balasooriya, G., Leaford, D., Behrens, A., Bertone, P. and Hendrich, B. (2012). NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 31, 593-605. 10.1038/emboj.2011.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl, D. D., Jin, J., Cai, Y., Swanson, S., Florens, L., Washburn, M. P., Conaway, R. C., Conaway, J. W. and Chrivia, J. C. (2006). Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 45, 5671-5677. 10.1021/bi060043d [DOI] [PubMed] [Google Scholar]
- Runge, J. S., Raab, J. R. and Magnuson, T. (2018). Identification of two distinct classes of the human INO80 complex genome-wide. G3 Genes, Genomes, Genet. 8, 1095-1102. 10.1534/g3.117.300504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadate-Ngatchou, P. I., Payne, C. J., Dearth, A. T. and Braun, R. E. (2008). Cre recombinase activity specific to post natal, premeiotic male germ cells in transgenic mice. Genesis 46, 738-742. 10.1002/dvg.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, H. and Matsui, Y. (2008). Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 9, 129-140. 10.1038/nrg2295 [DOI] [PubMed] [Google Scholar]
- Serber, D. W., Runge, J. S., Menon, D. U. and Magnuson, T. (2016). The mouse INO80 chromatin-remodeling complex is an essential meiotic factor for spermatogenesis. Biol. Reprod. 94, 1-9. 10.1095/biolreprod.115.135533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, C. M., Bao, K., Diedrich, J., Chen, X., Lu, C., Yates, J. R. and Jia, S. (2020). The INO80 Complex Regulates Epigenetic Inheritance of Heterochromatin. Cell Rep. 33, 108561. 10.1016/j.celrep.2020.108561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin, H. S., Kartashov, A. V., Hasegawa, K., Barski, A. and Namekawa, S. H. (2015). Poised chromatin and bivalent domains facilitate the mitosis-to-meiosis transition in the male germline. BMC Biol. 13, 1-15. 10.1186/s12915-014-0111-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruce, C., Dlamini, S., Ananda, G., Bronkema, N., Tian, H., Paigen, K., Carter, G. W. and Baker, C. L. (2020). HELLS and PRDM9 form a pioneer complex to open chromatin at meiotic recombination hot spots. Genes Dev. 34, 398-412. 10.1101/gad.333542.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana, M., Nozaki, M., Takeda, N. and Shinkai, Y. (2007). Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 26, 3346-3359. 10.1038/sj.emboj.7601767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, Y., Naruse, C., Costa, Y., Shirakawa, T., Tachibana, M., Sharif, J., Kezuka-Shiotani, F., Kakiuchi, D., Masumoto, H., Shinkai, Y.et al. (2011). HP1γ links histone methylation marks to meiotic synapsis in mice. Development 138, 4207-4217. 10.1242/dev.064444 [DOI] [PubMed] [Google Scholar]
- Tramantano, M., Sun, L., Au, C., Labuz, D., Liu, Z., Chou, M., Shen, C. and Luk, E. (2016). Constitutive turnover of histone H2A.Z at yeast promoters requires the preinitiation complex. Elife 5, 1-30. 10.7554/eLife.14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Gu, H., Lin, H. and Chi, T. (2012). Essential roles of the chromatin remodeling factor BRG1 in spermatogenesis in mice. Biol. Reprod. 86, 186. 10.1095/biolreprod.111.097097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Du, Y., Ward, J. M., Shimbo, T., Lackford, B., Zheng, X., Miao, Y.-L., Zhou, B., Han, L., Fargo, D. C.et al. (2014). INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14, 575-591. 10.1016/j.stem.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Long, H., Yu, J., Dong, L., Wassef, M., Zhuo, B., Li, X., Zhao, J., Wang, M., Liu, C.et al. (2018). Histone variants H2A.Z and H3.3 coordinately regulate PRC2-dependent H3K27me3 deposition and gene expression regulation in mES cells. BMC Biol. 16, 1-18. 10.1186/s12915-018-0568-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz, L., Daniel, K., Roig, I., Bolcun-Filas, E., Xu, H., Boonsanay, V., Eckmann, C. R., Cooke, H. J., Jasin, M., Keeney, S.et al. (2009). Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5, e1000702. 10.1371/journal.pgen.1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C., Yin, L., Xie, F., Ma, M., Huang, S., Zeng, Y., Shen, W. H., Dong, A. and Li, L. (2020). AtINO80 represses photomorphogenesis by modulating nucleosome density and H2A.Z incorporation in light-related genes. Proc. Natl. Acad. Sci. U. S. A. 117, 33679-33688. 10.1073/pnas.2001976117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, K., Vinayachandran, V. and Pugh, B. F. (2013). XSWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154, 1246-1256. 10.1016/j.cell.2013.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Wang, J., Lackford, B., Bennett, B., Li, J. and Hu, G. (2021). INO80 promotes H2A.Z occupancy to regulate cell fate transition in pluripotent stem cells. Nucleic Acids Res. 49, 6739-6755. 10.1093/nar/gkab476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B., Wang, L., Zhang, S., Bennett, B. D., He, F., Zhang, Y., Xiong, C., Han, L., Diao, L., Li, P.et al. (2016). INO80 governs superenhancer-mediated oncogenic transcription and tumor growth in melanoma. Genes Dev. 30, 1440-1453. 10.1101/gad.277178.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.