Abstract
Human genetic studies identified a strong association between loss of function mutations in RBFOX2 and hypoplastic left heart syndrome (HLHS). There are currently no Rbfox2 mouse models that recapitulate HLHS. Therefore, it is still unknown how RBFOX2 as an RNA binding protein contributes to heart development. To address this, we conditionally deleted Rbfox2 in embryonic mouse hearts and found profound defects in cardiac chamber and yolk sac vasculature formation. Importantly, our Rbfox2 conditional knockout mouse model recapitulated several molecular and phenotypic features of HLHS. To determine the molecular drivers of these cardiac defects, we performed RNA-sequencing in Rbfox2 mutant hearts and identified dysregulated alternative splicing (AS) networks that affect cell adhesion to extracellular matrix (ECM) mediated by Rho GTPases. We identified two Rho GTPase cycling genes as targets of RBFOX2. Modulating AS of these two genes using antisense oligos led to cell cycle and cell-ECM adhesion defects. Consistently, Rbfox2 mutant hearts displayed cell cycle defects and inability to undergo endocardial-mesenchymal transition, processes dependent on cell-ECM adhesion and that are seen in HLHS. Overall, our work not only revealed that loss of Rbfox2 leads to heart development defects resembling HLHS, but also identified RBFOX2-regulated AS networks that influence cell-ECM communication vital for heart development.
INTRODUCTION
The RNA binding protein RBFOX2 has an emerging role in heart diseases (1–6). Low RBFOX2 expression or activity are associated with heart failure (1) and cardiac complications of diabetes (2). Furthermore, RBFOX2 is also one of the high-risk genes for congenital heart defects (1–6). Heterozygous loss of function mutations in RBFOX2 are identified in patients with hypoplastic left heart syndrome (HLHS) and are linked to the left ventricle obstruction phenotype observed in these patients (3–5). HLHS is one of the most complex congenital heart diseases with a high mortality rate (6,7) and it is thought to be multigenic (8,9). HLHS patients typically exhibit circulation problems due to the hypoplasia within the left side of the heart, including the cardiac valves, left ventricle and aorta. Cross-talk between myocardial cells that pump the heart and the endocardial cells that line the interior of the heart is essential for normal cardiac development (10). Endocardial-mesenchymal transition (Endo-MT) and myocardial proliferation defects have been identified in HLHS patients and implicated in developmental defects of HLHS (11,12). Genome-wide gene expression and alternative splicing (AS) abnormalities have been identified in infants with HLHS (13). We have recently shown that RBFOX2 is a major contributor to transcriptome changes observed in HLHS patients (14). Despite the prominent genetic and molecular association of RBFOX2 to HLHS pathogenesis, the mechanistic underpinnings of RBFOX2 function in cardiovascular development and disease remain elusive.
RBFOX2 belongs to the RBFOX family of RNA-binding proteins that are involved in alternative splicing (AS) regulation (15). RBFOX2 controls AS in large macromolecular complexes via its interactions with several splicing regulators (16). RBFOX2 binding sites ((U)GCAUG) are enriched near alternative exons regulated during post-natal murine heart development (17). However, not much is known regarding RBFOX2 targets in the embryonic heart. A comprehensive identification of RBFOX2-regulated AS networks in the embryonic heart is necessary to fully understand RBFOX2′s role in heart development and disease.
RBFOX2 is critical for heart function. Knockdown of both Rbfox2 and its paralog Rbfox1 in zebrafish lowered heart rate and negatively affected myofibril formation (18). Conditional loss of Rbfox2 in committed embryonic cardiomyocytes using myosin light chain promoter driven Cre (mlc2v::Cre) resulted in dilated cardiomyopathy at postnatal stages partly due to defects in excitation-contraction coupling (19). However, embryonic origins of postnatal dilated cardiomyopathy were not reported (1,19,20). A recent study has shown that conditional deletion of Rbfox2 using neural crest-specific Cre drivers did not affect cardiovascular development (21). In sum, previous Rbfox2 conditional knockout studies in mice did not reveal cardiovascular development defects. Therefore, it is unclear how RBFOX2 contributes to congenital heart defects.
Based on our findings that RBFOX2 has a widespread and early expression pattern in the embryonic heart, we conditionally deleted Rbfox2 in multiple cell types at an earlier embryonic stage using Nkx2.5 Cre, different than the other studies. We found that Rbfox2 is required for embryonic survival and proper formation of the cardiac chambers, outflow tract (OFT) and yolk sac vasculature. To determine the mechanisms underlying these defects, we performed RNA-seq and analyzed global AS patterns. We identified AS changes in genes involved in cytoskeletal organization, cell-extracellular matrix (ECM) adhesion and Rho GTPase signaling/cycling in Rbfox2 mutant hearts. Modifying AS of just two of these RBFOX2 targets (guanine exchange factors for Rho GTPases) impaired cell-ECM adhesion and cell cycle progression. Consistently, Rbfox2 mutant embryos displayed defects in cell cycle progression and Endo-MT that are dependent on cell-ECM adhesion. Notably, our Rbfox2 conditional mouse model recapitulated some phenotypic and molecular features of HLHS. Our data also suggest that Rbfox2 deletion in multiple cell lineages collectively contribute to cardiovascular defects. In summary, our data demonstrate that Rbfox2 is required for heart development and the precise regulation of RBFOX2-dependent AS networks contributes to cell-ECM communication in the embryonic heart.
MATERIAL AND METHODS
Generation of Rbfox2 conditional knockout mice
All animal experiments were conducted in accordance with the NIH Guidelines for the care and the use of animals approved by the Institutional Animal Care and Use Committee of UTMB. E0.5 was estimated as the noon of the day the plugs. The timed-pregnant uterus was dissected and suspended in cold phosphate-buffered saline (PBS) till the embryos and/or embryonic heart were harvested. Tail or yolk sac genomic DNA was used for genotyping PCR. Rbfox2flox/flox mice were purchased from Jackson labs (Stock no: 014090). Nkx2.5Cre/+ mice were obtained from Dr Robert Schwartz's lab. These mice have reduced Nkx2-5 mRNAs due to the Cre knock in into the Nkx2-5 locus (22). To generate Rbfox2 mutant embryos, Rbfox2flox/flox mice were crossed with Nkx2.5Cre/+ mice and the Rbfox2-HetCKO (Rbfox2lox/+, Nkx2.5Cre/+) male animals from this mating were crossed back with Rbfox2flox/flox females to generate Rbfox2-CKO (Rbfox2flox/flox, Nkx2.5Cre/+) embryos.
Whole mount embryos processing and paraffin sectioning
A total number of six embryos for each genotype (control - Rbfox2flox/flox; Rbfox2-HetCKO -Rbfox2lox/+, Nkx2.5Cre/+ & Rbfox2-CKO - Rbfox2flox/flox, Nkx2.5Cre/+) at two developmental stage (E9.5 and E10.5) were fixed for 24 h in 4% freshly prepared from paraformaldehyde buffered with 0.1 M sodium phosphate (pH 7.2) at 4°C. Subsequently, embryos were washed with 1× PBS (#46-013-CM, Corning) followed by dehydration with increasing concentration of ethanol (#E7023, Sigma) from 70 to 100%. Embryos were cleared with Xylene (#534056, Sigma) before embedding in paraffin (#76258, Sigma) block with preferred orientation. 7 μm-thick sections of whole mount embryos were obtained using a microtome (Microtome, HM2035) and collected on positively charged glass slides.
Immunofluorescence of embryo sections
Immunofluorescence staining was performed on paraffin sections from each genotype using three sections per embryo, from three embryos, obtained from three different litters. Paraffin sections were incubated at 56°C for 12–14 h, followed by deparaffinization and dehydration in xylene for 20 min. Slides were washed in decreasing concentrations (100–50%) of ethanol for 5 min at each concentration. Antigens were then exposed by incubating the sections with sodium citrate buffer (10 mM, pH 6.0) for 20 min in a steam chamber. Blocking was performed in 3% BSA in PBST (0.2% Triton X-100) at RT for 1 hr. Sections were then incubated with the following primary antibodies: Rbfox2 (A300-864A, Bethyl laborites inc.); cardiac troponin I (ab47003, Abcam); CD31/PECAM1(AF3628, R&D system); alpha-smooth muscle actin (NB300-978, Novus biologicals); phospho-Histone H3 (#9701, Cell signaling technology); vinculin (V9131, Sigma) for 14–16 h at RT in a humidifying chamber. Slides were washed with PBS containing 0.1% Triton X-100 and then incubated with the appropriate fluorescently labeled appropriate secondary antibodies for 2 h at 37°C. Slides were then washed four times with PBS containing 0.1% Triton X-100, followed by incubation with 4′,6-diamino-2-phenylindole dihydrochloride (DAPI, #MP01306, Invitrogen) or TO-PRO-3 (#T3605, Life technology) stain for 30 min at room temperature in the dark. Excess stain was washed away with PBS before mounting the coverslips using Mowiol mounting media and then the slides were sealed using nail polish. Fluorescence images were obtained with a confocal laser-scanning microscope (LSM 880META, Carl Zeiss) at the University of Texas Medical Branch, Optical Microscopy Core facility. All the image quantification was performed using ImageJ software and significance were calculated by either t-test or one-way ANOVA using GraphPad PRISM software.
Hematoxylin and eosin (H&E) staining
Complete whole mount embryo (E9.5 and E10.5) sections were stained with hematoxylin and eosin (H&E; Hemotoxylin, Gill no 2, Sigma, GHS232; Eosin Y, Sigma, HT110332). 7 μm sections were dewaxed twice in xylene and rehydrated in decreasing concentrations of ethanol from 100% to 50% before proceeding with further staining. Sections were incubated in Hemotoxylin for 2 min, de-stained (1% hydrochloric acid in 70% ethanol) for 5 s, washed in 80% ethanol for 1 min, followed by a brief incubation (1 s) in eosin before dehydrating in ethanol (95% and 100%) and clearing in xylene. Stained slides were mounted using DPX mountant (Sigma, 06522). Images were obtained with a light microscope (Olympus, IX51) at the University of Texas Medical Branch, Optical Microscopy Core facility. All the image quantification was performed using ImageJ software and significance were calculated by either t-test or one-way ANOVA using GraphPad PRISM.
Alternative splicing validations
The semi-quantitative RT-PCR was performed to assess splicing patterns of genes using a previously described protocol (2,23). Primer sequences to validate AS events were designed to detect inclusion/exclusion of alternative exons of the following genes: ABI1 exon10, ECT2 exon 4, FN1 exon 25 and CTTN exon 11 (Supplementary Table S2). For all genes, PCR was carried out using 5μl cDNA and 100ng primer pair with Biolase Taq polymerase (Bioline BIO-21042) using following amplification condition 95°C 45 s; 59°C 45 s; 72°C 1 min for 25 cycles. Data from at least three independent experiments were used for quantification and statistical analysis, and significance was calculated using an unpaired t-test or one-way ANOVA with Bonferroni correction using GraphPad PRISM.
TUNEL staining
Apoptotic cells were identified using the DeadEnd Fluorometric TUNEL System (G3250, Promega) following the manufacturer's protocol. In brief, paraffin sections were incubated at 56°C for 12–14 h, followed by deparaffinization and dehydration in xylene for 20 min. Slides were washed in decreasing concentrations (100–50%) of ethanol for 5 min at each concentration. Tissue sections were then fixed in 4% formaldehyde at room temperature after washing with 0.85% sodium chloride for 5 min. Tissue was permeabilized by proteinase K (20 μg/ml) at room temp for 20 min then fixed with 4% formaldehyde and incubated with equilibration buffer prior to the rTdT reaction in dark, humidified chamber at 370°C for 90 min. The reaction was stopped with 2× SSC followed by incubation with 4′,6-diamino-2-phenylindole dihydrochloride (DAPI, MP01306, Invitrogen)) stain for 30 min at room temperature in the dark. Slides were processed and imaged using a confocal laser-scanning microscope (LSM 880META, Carl Zeiss) at the University of Texas Medical Branch, Optical Microscopy Core facility. TUNEL positive nuclei were counted using ImageJ software and quantified by one-way ANOVA using GraphPad PRISM 6.
Real time RT-qPCR
The cDNA was synthesized from 2.0 μg RNA using Biolase DNA polymerase (Bioline) with random OligodT primers (Invitrogen) as detailed previously (14). cDNA was diluted five times and used for real time qPCR reaction using LightCycler® 480 SYBR Green I Master mix (Roche) and Roche LightCycler 480 (Roche diagnostic). Specific primers for qPCR were designed to detect the total mRNA levels for Twist1 and Cdh2 genes (Supplementary Table S2). EEF1A1 was used as internal control to normalize the mRNA levels. Relative mRNA level in comparison to controls were determined using 2−ΔΔct method.
RNA-sequencing and genome wide alternative splicing analysis
RNA was extracted from E9.5 mice heart tissues from control (Rbfox2flox/flox), Rbfox2-HetCKO (Rbfox2lox/+, Nkx2.5Cre/+) and Rbfox2-CKO (Rbfox2flox/flox, Nkx2.5Cre/+; two hearts per group, from two different litters) using TRIzol (#15596018, Invitrogen) according to the manufacturer's protocol. RNA quality was assessed by Agilent Bioanalyzer and quantified using Qubit Fluorometer by the Next Generation Sequencing Core Facility at UTMB. Sequencing libraries were prepared, and 75-base paired-end sequencing was performed using an Illumina NextSeq 550 system. Data were analyzed for AS changes using mixture of isoform (MISO) algorithm (24). We compared Rbfox2-CKO to Rbfox2-HetCKO embryos to avoid AS changes that could result from low Nkx2-5 levels. Percent spliced in is defined as % exon or intron inclusion. ΔPSI is defined as the difference between Rbfox2-HetCKO PSI and Rbfox2-CKO PSI in embryonic hearts as tabulated in the excel file (diff). For RBFOX2 binding site overlay, the BLAT tool from UCSC Genome browser was used to map these sequences to the human genome (hg18) and to download RBFOX2 CLIP-density for each alternative exon and flanking 250nt long upstream and downstream intronic regions.
Cell-ECM adhesion assay
HUVECs were treated with either control siRNA (#4390844, Thermo fisher) or RBFOX2 specific siRNA (#4390816, Thermo fisher) and Control SSO (standard control, Gene Tools LLC, ‘seq- CCTCTTACCTCAGTTACAATTTATA’) or Abi1 + Ect2 SSO for 48 h. Cell viability was monitored using trypan blue. 2 × 104 cells were seeded in each well of 96-well plate coated with 40 μg/ml of Collagen-I (#C9791, Sigma). Cells were allowed to attach for 45 min before washing off the non-adherent cells. EGM-2 complete endothelial growth media was then added, and the plate was incubated at 37°C in a 5% CO2 humidified incubator for 4 h to recover. Live cells attached to the plates were quantified by MTT ((3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide)) assay (#30101K, ATCC) following the manufacturer's protocol. Data from at least three independent experiments were used for statistical analysis, and significance was calculated using an unpaired t-test using GraphPad PRISM software.
Focal adhesion and stress fiber analysis
HUVECs were treated with either control siRNA (#4390844, Thermo fisher) or RBFOX2 specific siRNA (#4390816, Thermo fisher) for 48 h in each chamber of a four-chambered cover glass slide. Cells were washed with PBS and fixed with 4% paraformaldehyde solution in PBS at RT for 15 min. Cell membranes were partially perforated by incubating with 0.5% Triton X-100 at RT. Cells were stained with an anti-vinculin antibody (#V9131, Sigma) to mark focal adhesions and phalloidin (#A12379, Invitrogen) to label stress fibers. Fluorescence images were obtained with a confocal laser-scanning microscope (LSM 880META, Carl Zeiss) at UTMB Optical Microscopy Core facility. All the image quantification was determined using Image J software and significance were calculated by t-test or one-way ANOVA with Bonferroni correction using GraphPad PRISM software.
Cell cycle analysis
HUVECs were treated with either control siRNA (#4390844, Thermo fisher) or RBFOX2 specific siRNA (#4390816, Thermo fisher) and Control SSO (10 μM) or Abi1 (6 μM) +Ect2 (10 μM) SSO for 48 h. Cells were collected and pelleted down and were washed once with 1× PBS. Pellets were resuspended in 500 μl fresh PBS. Then, 500 μl 100% ethanol was added dropwise while vortexing and cells were placed in −20°C overnight. The cells were then pelleted down and washed with 1× PBS followed by addition of 500 μl propidium iodide (#P4170, Sigma) + RNAse (#R4875, Sigma) solution at 50 μg/ml. Cells were analyzed with a Beckham Coulter CytoFlex Flow Cytometer for 50 000 cell counts. Data obtained from the flow cytometer was analyzed with FlowJo software.
HUVEC in vitro mesh network formation
HUVECs (2 × 104 cells) treated with either control (#4390844, Thermo fisher) or RBFOX2 specific (#4390816, Thermo fisher) siRNAs were seeded on matrigel I (#CB-40234C, Corning) coated in a 96-well plate and cultured in EGM-2 complete endothelial growth media (#CC-3162, Lonza) and incubated at 37°C in a 5% CO2 humidified incubator. Bright field images of each well were taken every 2 h to monitor network formation for 7 h. HUVECs were stained with 6 μM Calcein AM (#C1430, Invitrogen) dye for 15 min at 37°C in a 5% CO2 humidified incubator at the end of experiment and GFP fluorescence was imaged using an Olympus microscope fluorescent microscope. Images from three independent experiments were used for quantifying various vessel morphometric and spatial parameters using AngioTool software (version 0.5, 25 May 2011)(25) and significance was determined using an unpaired t-test using GraphPad PRISM software.
Flp-in HEK293 stable cells
Human FLAG-tagged RBFOX2WT and RBFOX2RRM mutant was cloned into pcDNA5/FRT/TO expression vectors. Flp-in T-REx 293 cells plated on six-well plates were co-transfected with 4 μg pOG44 recombinase and 0.4 μg RBFOX2WT or 0.4 μg RBFOX2RRM plasmid using Lipofectamine 2000 (#11668019, Invitrogen). Cells stably expressing the constructs were selected in 100 μg/ml hygromycin. To induce RBFOX2 expression, stable cells were treated with 1.0 μg/ml of doxycycline for 24 h.
Western blot
HEK293 protein lysates (30–50 μg/sample) were separated on 10% SDS-PAGE gels and were then transferred to a PVDF membrane. Membranes were blocked with 5% dry fat-free milk solution in PBST (PBS containing 0.1% Tween 20) and then were cut into two pieces below 50 kDa. Each membrane was incubated with different primary antibodies: RBFOX2 (#ab57154, Abcam) and FLAG (#F7425, Sigma) at 4°C overnight. Membranes were washed with PBST 4 times 15 min for each wash and then incubated with HRP-labeled secondary antibody for 2 h at room temperature. HRP activity was determined using Immobilon Western chemiluminescent (Millipore P90720) or SuperSignal West Femto Chemiluminescent (Pierce PI34095) HRP substrate followed by exposure to X-ray film or imaged using a Bio-Rad Chemidoc Touch Imaging system. All the bands were quantified using Image Lab software (Bio-Rad laboratories).
Statistical analysis
Prism 9 was used to perform all statistical analyses (GraphPad Software). Student's t-test was used to determine significance between two groups. Multiple groups were compared using one-way analyses of variance (ANOVAs) with Bonferroni's post hoc test. Details for statistical analysis for specific experiments were detailed for each experiment in Figure legends.
RESULTS
Conditional deletion of RBFOX2 results in embryonic lethality and profound cardiovascular developmental defects
To investigate the role of RBFOX2 in heart development, we first examined the developmental stage specific expression pattern of Rbfox2 in the developing mouse heart. We isolated hearts from wild type (WT) newborn and adult (6 months-old) mice as well as from embryos at embryonic day 8.5 (E8.5), 9.5 (E9.5) and 12.5 (E12.5) and assessed Rbfox2 mRNA levels. Rbfox2 mRNA was detectable in the heart at all developmental stages (E8.5, E9.5, E12.5, newborn and adult), but was downregulated in adult hearts, as previously described (Supplementary Figure S1A) (17,23). To determine the cell type specific RBFOX2 expression, we examined the localization of RBFOX2 protein in different cell types in WT E13.5 mouse hearts. RBFOX2 protein colocalized with endothelial/endocardial cell marker platelet endothelial cell adhesion molecule (PECAM1/CD31), with smooth muscle cell marker alpha-smooth muscle actin (α-SMA) and with cardiomyocte marker cardiac troponin I (cTNI) (Supplementary Figure S1B). As RBFOX2 was expressed in multiple cell types in the embryonic heart at E13.5, we examined expression at earlier embryonic stages. At E9.5 and E10.5 RBFOX2 protein was broadly detected throughout the embryo, with widespread localization in the cardiac region colocalizing with α-SMA that marks contractile cells at these embryonic stages (Supplementary Figure S1C and D).
To determine how Rbfox2 contributes to cardiovascular development, we conditionally deleted in the embryonic heart. Based on the timing and widespread expression of RBFOX2 in multiple cell types in the embryonic heart, we investigated the role of Rbfox2 in the embryonic heart by crossing mice containing a conditional allele of Rbfox2 flox/flox (Rbfox2flox/flox) (26) to Nkx2.5 Cre knock-in mice (22) to delete Rbfox2 (Rbfox2flox/flox; Nkx2.5Cre/+, i.e. in short Rbfox2-CKO) between E7.5 and E8.5. Rbfox2 heterozygous conditional cardiac knockout (Rbfox2flox/+; Nkx2.5Cre/+, i.e. in short Rbfox2-HetCKO) pups were born with the expected Mendelian ratios. However, no live pups were obtained with the Rbfox2-CKO genotype, indicating that conditional homozygous cardiac knockout of Rbfox2 is embryonic lethal. Rbfox2-CKO embryos were viable until embryonic day 11.5 (E11.5) but were severely growth impeded when compared to Rbfox2-HetCKO and control (Rbfox2flox/flox) littermate embryos (Figure 1A). Importantly, RBFOX2 protein was absent in the heart of Rbfox2-CKO embryos marked with α-SMA, confirming effective recombination of the conditional allele and deletion of Rbfox2 as by the Nkx2.5 Cre driver (Supplementary Figure S2).
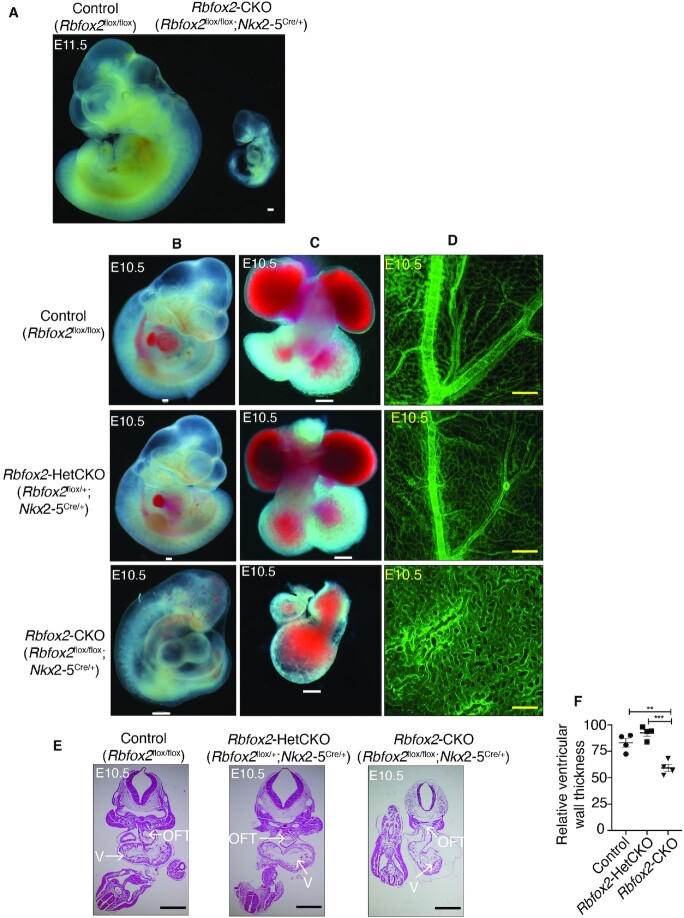
Figure 1.
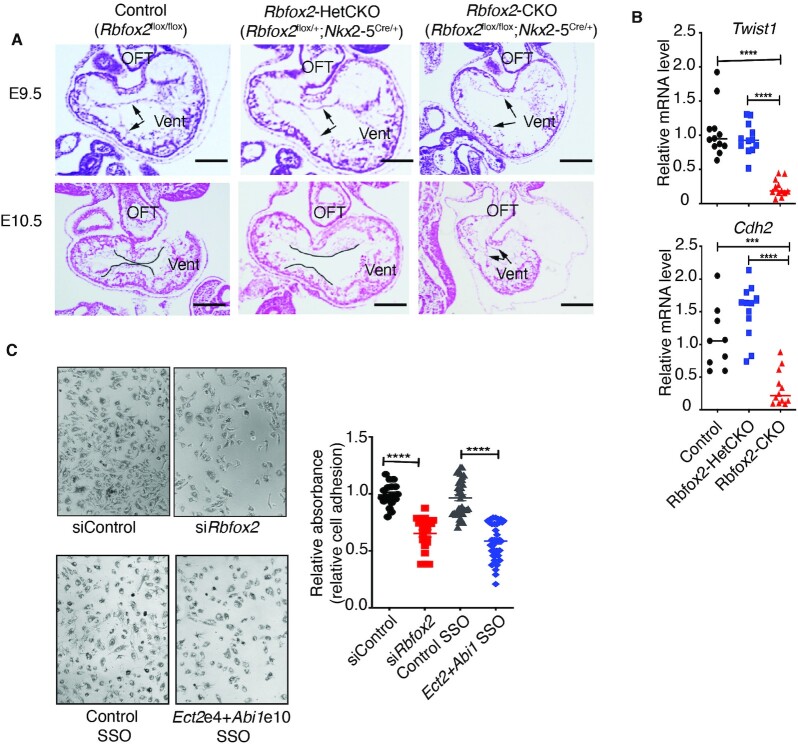
Severe cardiovascular developmental defects in Rbfox2-CKO embryos. (A) Gross images of E11.5 Control (Rbfox2flox/flox) or Rbfox2-CKO (Rbfox2flox/flox;Nkx2-5Cre/+) embryos. (B) Representative images of Control (Rbfox2flox/flox) (E10.5: n = 44), Rbfox2-HetCKO (Rbfox2flox/+;Nkx2-5Cre/+) (E10.5: n = 23) and Rbfox2-CKO (Rbfox2flox/flox;Nkx2-5Cre/+) (E10.5: n = 22) embryos isolated at E10.5. (C) Representative images of hearts isolated from Control, Rbfox2-HetCKO and Rbfox2-CKO embryos at E10.5. (D) PECAM1 immunostaining of yolk sac vasculature of Control, Rbfox2-HetCKO and Rbfox2-CKO embryos at E10.5. All scale bars = 100 μm. (E) Representative images of hematoxylin and eosin (H&E) stained embryonic sections at E10.5. Arrows mark specific cardiac regions. V: ventricular chamber, OFT: outflow tract. (F) Relative ventricular wall thickness in E10.5 embryos was quantified using ImageJ. Data are mean ± standard deviation. ** P = 0.0029, *** P = 0.0003; by analysis of variance (ANOVA) with Bonferroni's post hoc test.
Grossly, at E10.5 100% of Rbfox2-CKO embryos were noticeably smaller in size and displayed hemorrhage and pericardial edema, which are commonly associated with early congestive heart failure (Figure 1B, bottom left panel). The hearts of control and Rbfox2-HetCKO embryos featured the normal morphological arrangement of four cardiac chambers whereas Rbfox2-CKO embryos (100%, N = 22) failed to display four chambered heart and progress this far at E10.5 (Figure 1C, bottom middle panel). Rbfox2-CKO embryo demonstrated failure to thrive but with beating hearts at the time of harvest at E10.5 (Supplementary Movie S1). At this stage, we also noticed that the yolk sac vasculature appeared abnormal and anemic in Rbfox2-CKO embryos. PECAM1 staining of E10.5 yolk sacs confirmed the lack of remodeled vasculature and the persistence of primitive plexus in the yolk sacs of Rbfox2-CKO embryos, unlike littermate controls (Figure 1D, bottom right panel). Histological analysis of tissue sections provided evidence that Rbfox2-CKO embryos exhibited underdeveloped hearts with a thin and hypoplastic OFT (Figure 1E). In addition, ventricle walls were significantly thinner in E10.5 Rbfox2-CKO embryos compared to Rbfox2-HetCKO and controls (Figure 1F). We also examined Rbfox2-CKO embryos at E9.5. Rbfox2 mutant embryos (Supplementary Figure S3A) and their isolated hearts (Supplementary Figure S3B) from these embryos were indistinguishable from the control embryos at this stage grossly and histologically (Supplementary Figure S3C). PECAM1 staining of E9.5 yolk sacs (Supplementary Figure S3D) revealed no difference in yolk sac vasculature between controls and Rbfox2 mutants. Overall, these results show that Rbfox2 conditional ablation using Nkx2.5 Cre mice impairs proper heart development and causes embryonic lethality.
Genome wide AS changes impact genes involved in cytoskeleton organization, cell-ECM adhesion and Rho GTPase signaling/cycling in Rbfox2 mutant hearts
Not much is known regarding RBFOX2′s role in the embryonic heart and its target RNAs regulated via AS. To determine the molecular drivers underlying the developmental defects observed in Rbfox2-CKO mutants, we performed RNA-sequencing using polyadenylated RNA collected from E9.5 embryonic hearts prior to the manifestation of severe morphological defects observed at E10.5. We obtained 70–100 million paired-end reads per sample, which are needed for detection of AS variants, and mapped these reads to the mouse genome with high efficiency and reproducibility (Supplementary Table S1).
RBFOX2 has a role in AS, so we checked AS patterns using the mixture of isoforms (MISO) algorithm based on the criteria that percent spliced in (PSI) is ≥ 15% and significance (defined as Bayes factor: B ≥ 1). We identified 986 aberrant AS patterns that were altered specifically in Rbfox2-CKO but not in Rbfox2-HetCKO embryos (Supplementary Dataset S1). Violin plots show different types of AS events and the extent of AS changes in Rbfox2-CKO hearts (Figure 2A). The most commonly affected AS event was the cassette exon (CE) splicing (435 out of 986, 44%). 65% of cassette exons were more included and 35% were more excluded in Rbfox2 mutant hearts (Figure 2B), suggesting that RBFOX2 predominantly represses alternative exon inclusion in the embryonic heart.
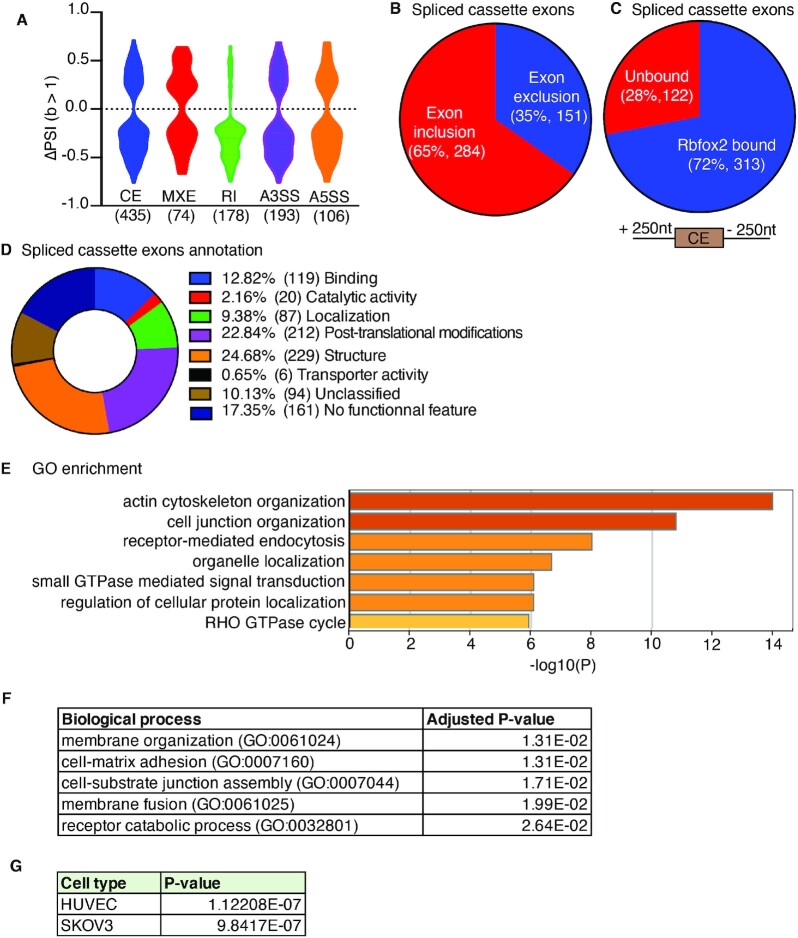
Figure 2.
Global AS changes in Rbfox2-CKO embryonic hearts impact cytoskeletal organization, Rho GTPase cycling and cell-ECM adhesion. (A) Violin plot representation of different types of AS events and the extent of AS changes in Rbfox2-CKO (Rbfox2flox/flox;Nkx2-5Cre/+) E9.5 hearts in comparison to Rbfox2-HetCKO (Rbfox2flox/+;Nkx2-5Cre/+) E9.5 hearts. CE: cassette exon (435 events; 387 genes), MXE: mutually exclusive exons (74 events; 71 genes), RI: retention of introns (170 events; 170 genes), A3SS: Alternative 3′ splice site (193 events; 189 genes), A5SS: Alternative 5′ splice site (106 events; 106 genes). PSI: Percent spliced in (Rbfox2-hetCKO – Rbfox2-CKO), significance determined by Bayes factor (B) >1. (B) Cassette exon inclusion and exclusion in embryonic hearts at E9.5. (C) RBFOX2 eCLIP comparisons to identify RBFOX2 targets that undergo AS changes in Rbfox2-CKO E9.5 embryo hearts. Alternative exons or 250nt intronic regions upstream or downstream of alternative exons were used for this analysis. Pie chart of RBFOX2-binding clusters in 435 transcripts that are mis-spliced in Rbfox2-CKO E9.5 embryo hearts. Cassette exon splicing events affected in Rbfox2-CKO hearts with at least one CLIP peak at a Bayes factor 1 were designated as ‘‘significant’’ and <1 as ‘not significant’. (D) Exon Ontology analysis of 435 RBFOX2-regulated cassette exons and the corresponding encoded protein properties of these cassette exons. (E) GO enrichment analysis of genes affected in Rbfox2-CKO E9.5 hearts via cassette exon splicing. (F) Biological processes affected in E9.5 Rbfox2-CKO hearts via cassette exon splicing. (G) Major cell types affected by mis-splicing of genes with cassette exon changes in Rbfox2-CKO embryos using Enrichr server.
To determine which of these genes with cassette exon changes are direct targets of RBFOX2, we used the publicly available RBFOX2 enhanced crosslinking immunoprecipitation (eCLIP) datasets from ENCODE project that identified RNAs bound to RBFOX2 and specific RBFOX2 binding sites within these pre-mRNAs in HEPG2 and K562 cells (27,28). RBFOX2 mainly binds to intronic regions in the pre-mRNAs flanking alternative exons to regulate AS, therefore, we examined RBFOX2 binding sites within 250nt upstream and downstream intronic regions flanking cassette alternative exons, which are mis-spliced in Rbfox2-CKO embryonic hearts (Figure 2C). Strikingly, 72% (313 out of 435) of these genes displayed RBFOX2 binding sites within 250nt flanking alternative exons (Figure 2C). These results suggest that 72% of genes that undergo cassette exon splicing changes in Rbfox2-CKO hearts are RBFOX2 targets.
Cassette exon splicing can directly influence protein coding region via excluding or including alternative coding exons. To determine how RBFOX2-regulated exons influence the functionality of the encoded protein, we used the Exon Ontology Interface (29). This analysis revealed distinct protein features encoded by RBFOX2-regulated alternative exons (Figure 2D). 24.7% of RBFOX2-regulated alternative exons contain known structural domains, 22.8% contain regions of proteins that are post-translationally modified, while 12.8% are within binding domains and 9.4% are predicted to affect subcellular localization (Figure 2D). These results indicate that RBFOX2 induced AS changes is predicted to affect regulatory regions of proteins via AS of cassette exons in the embryonic heart.
Most of the RBFOX2-regulated cassette exons contain regulatory and structural regions of the encoded proteins and these cassette exons displayed the most common AS change in Rbfox2 mutants. Thus, we searched which biological processes and signaling pathways are affected via AS of cassette exons in Rbfox2-CKO embryos that could explain the cardiac defects. GO enrichment analysis of genes with cassette exon changes revealed cytoskeleton organization and Rho GTPase cycle and small GTPase signaling to be the most affected (Figure 2E). The top biological processes affected were membrane organization, cell matrix adhesion and cell-substrate junction assembly (Figure 2F). The top cell types were endothelial (human umbilical endothelial cells:HUVECs) and SKOV3 (ovarian cancer cell with epithelial like morphology) cells (Figure 2G). Rho GTPases are known regulators of cytoskeletal organization and cell-ECM adhesion. These results suggest that RBFOX2-regulated AS networks impact Rho GTPase cycling/signaling that is important for cell-ECM adhesion and cytoskeletal organization in the embryonic heart.
RBFOX2 regulates AS of genes critical for Rho GTPase cycling, cell-ECM adhesion and proliferation
Among these RBFOX2 regulated AS targets, we further analyzed AS of 3 specific genes: Abi1 (Abl Interactor 1) and Ect2 (Epithelial Cell Transforming 2) and Fn1 (Fibronectin 1). These genes are important regulators of cell-ECM adhesion and cell cycle. In addition, they display extensive (top changers) cassette exon changes in Rbfox2-CKO embryos. Importantly, Abi1 and Ect2 act as guanine exchange factors (GEFs) of Rho GTPase family of proteins.
Ect2 encodes for ECT2 protein, a GEF of the Rho family of small GTPases, that controls cytokinesis during cell cycle, and cell polarity (30). Structural studies found that exon 4, which codes for part of the BRCT 0 domain of ECT2, contributes to GEF activation and interactions with proteins important for cell spreading (31,32) (Figure 3A). RNA-seq revealed that exon 4 of Ect2 is more excluded in Rbfox2-CKO hearts.
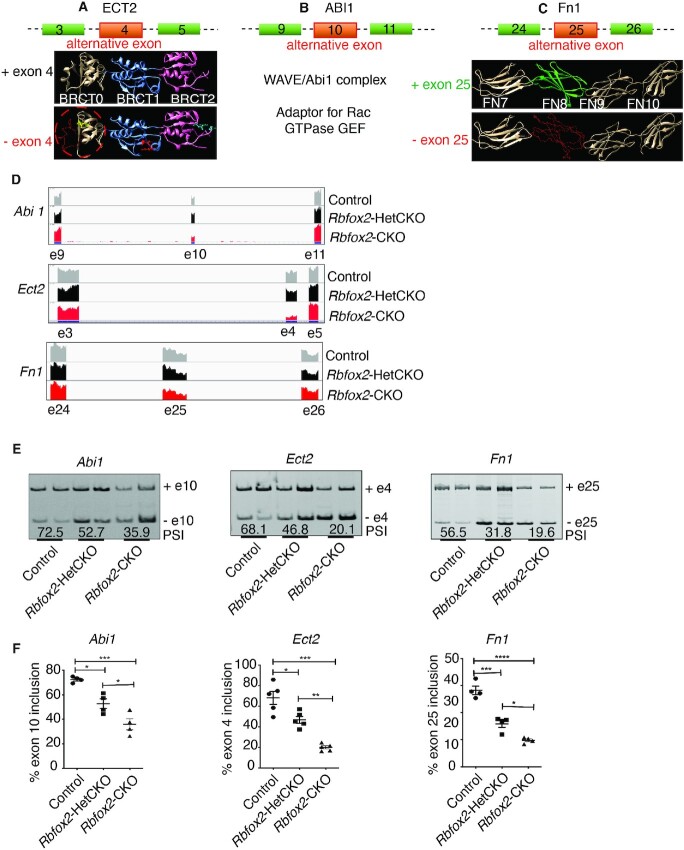
Figure 3.
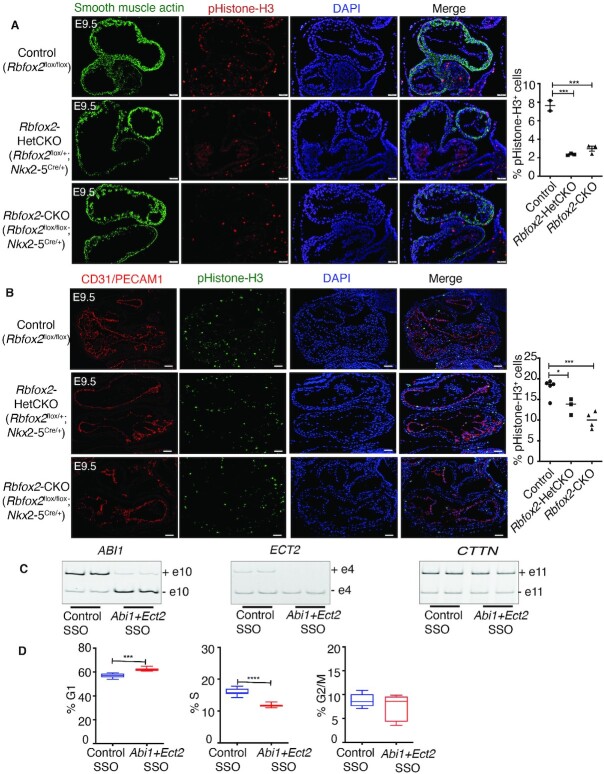
Validation of AS defects in Rbfox2-CKO embryonic hearts and predicted effects of specific AS changes on encoded proteins. (A) Graphical representation of alternative exon 4 of ECT2. BRCT protein domains are shown as ribbon and red backbone marked in BRCT0 is encoded by alternative exon 4. Predicted protein lacking exon 4 prediction model is derived from PDB id-4N40 (68). (B) Schematic representation of alternative exon 10 of ABI1 and its predicted effect on downstream cellular processes. Proposed model is adopted from (35). (C) Graphical representation of alternative exon 25 of FN1. Exon-25 encodes protein domain FN8 that plays important role in angiogenesis and cell adhesion. Predicted protein lacking exon 25 prediction model is derived based on PDB id-1FNF (69). (D) Representative Integrative Genomics Viewer (IGV) images of AS changes in Abi1, Ect2 and Fn1 genes identified in Control, Rbfox2-HetCKO and Rbfox2-CKO E9.5 hearts. (E) Representative gel images of AS changes in Abi1, Ect2 and Fn1 in Control, Rbfox2-HetCKO and Rbfox2-CKO E9.5 hearts. Mean PSI values were included below the gel images (n = 4 heart per genotype). (F) Quantification of AS changes in Abi1, Ect2 and Fn1 in Control, Rbfox2-HetCKO and Rbfox2-CKO E9.5 hearts. Data are mean ± standard deviation (n = 4 heart per genotype). *P = 0.0102; *P = 0.0263; ***P = 0.0001; *P = 0.0125; **P = 0.0024; ***P = 0.0002 by analysis of variance (ANOVA) with Bonferroni's post hoc test.
Abelson interactor 1 (Abi1) is an adaptor for the oncogene tyrosine kinase, ABL necessary for cell proliferation. ABI1 is also an adaptor for multiprotein complex that act as a GEF for Rac GTPase, critical for cell adhesion (33). ABI1 is also a component of the SCAR/WAVE complex that regulates cytoskeletal remodeling (34). Structural studies showed that exon 10 codes part of the proline-rich domain important for cell adhesion and proliferation (35–38) (Figure 3B). The spliced variant of Abi1 that lacks exon 10 is favored in Rbfox2 mutant hearts.
Fn1 encodes for Fibronectin 1, which is one of the main extracellular matrix component necessary for cell adhesion to ECM (39). AS change in exon 25 removes FN8 domain, part of the protein region (FN7-FN10 domains) that is important for cell adhesion (40,41) (Figure 3C).
Integrative Genomics Viewer (IGV) shows the cassette exon exclusion of Abi1 (53% AS change or PSI: 53), Ect2 (58% AS change or PSI: 58) and Fn1 (28% AS change or PSI: 28) in Rbfox2-CKO compared to Rbfox2-HetCKO and control embryos (Figure 3D). We successfully validated exon exclusions in these genes in E9.5 Rbfox2-CKO hearts (Figure 3E and F).
In addition, we depleted RBFOX2 in primary HUVECs and analyzed AS of ABI1, FN1 and ECT2. We used HUVECs for most of the in vitro analyses because our unbiased RNA-seq analysis revealed HUVECs as the top cell type affected via AS in Rbfox2-CKO mutants (Figure 2G). Similar to AS changes in Rbfox2-CKO mutant hearts (Figure 3E and F), alternative exons of ABI1, ECT2 and FN1 were more excluded in RBFOX2 depleted cells (Figure 4A). In agreement with these data, RBFOX2 binding clusters were identified within intronic/exonic regions near the alternative exons of these genes (ENCODE RBFOX2-eCLIP data, Figure 4B).
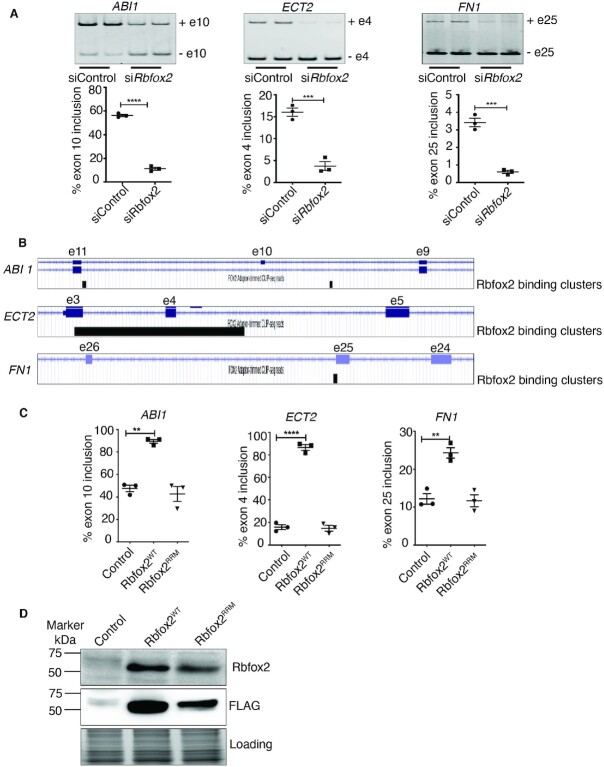
Figure 4.
RBFOX2 regulates AS of ABI1, ECT2 and FN1. (A) Verification of AS changes in ABI1 exon 10, ECT2 exon 4 and FN1 exon 25 in scrambled versus RBFOX2 siRNA treated HUVEC cells using RT-qPCR. Data are mean ± standard deviation (n = 6 from three independent experiments). ****P = 0.0001; ***P = 0.0009; ***P = 0.0004 by Student's t-test. (B) RBFOX2 eCLIP-clusters in ABI1, ECT2 and FN1 pre-mRNAs. RBFOX2 binding clusters within the exonic and intronic regions flanking alternative exons of Abi1, Ect2 and Fn1 pre-mRNAs were identified by RBFOX2 eCLIP-seq (ENCODE project). (e = exon). (C) Quantification of AS changes in ABI1, ECT2 and FN1 in HEK293 cells stably expressing RBFOX2WT or RNA binding mutant RBFOX2RRM. Data are mean ± standard deviation (n = 3 from three independent experiments). **P = 0.006; ****P = 0.0001; **P = 0.003 by analysis of variance (ANOVA) with Bonferroni's post hoc test. (D) Western blot analysis of RBFOX2 in control HEK293 cells or HEK293 cells inducibly expressing RBFOX2WT or RNA binding mutant RBFOX2RRM. No stain gel was used to monitor even protein loading.
To further delineate RBFOX2-mediated AS regulation of these genes, we used stable HEK293 cells engineered to express WT (RBFOX2WT), or a mutant form of RBFOX2 (RBFOX2RRM) that is unable to bind RNA (42–44). Expression of RBFOX2WT and RBFOX2RRM was induced by doxycycline treatment. RBFOX2WT induction increased exon inclusion of ABI1, ECT2 and FN1 genes in opposition to RBFOX2 depletion studies as expected (Figure 4A versus C). In contrast to RBFOX2WT, mutant RBFOX2RRM did not increase inclusion of these cassette exons (Figure 4C). RBFOX2WT and RBFOX2RRM proteins were successfully induced in HEK293 cells, which express very low levels of endogenous RBFOX2 (Figure 4D). These results demonstrate that RBFOX2 controls AS of these genes with RBFOX2 binding sites and that RBFOX2 RNA binding activity is necessary for AS regulation of these genes.
RBFOX2-mediated AS regulation of Abi1 and Ect2 is critical for cell cycle progression
Since Rbfox2 mutant hearts were hypoplastic and RBFOX2 targets Abi1 and Ect2 have prominent roles in cell cycle, we assessed whether cell cycle is affected in Rbfox2-CKO mutants at E9.5. Nkx2.5 knock in Cre mice recombines in myocardial and endothelial/endocardial cells (45–47). Therefore, we determined the percentage of myocardial and endothelial/endocardial cells in the cardiac region undergoing mitosis by marking mitotic cells using phospho-histone H3 antibody. The number of SMA+ contractile cells as well as PECAM1+ endocardial/endothelial cells were significantly decreased in both Rbfox2-HetCKO and Rbfox2-CKO embryos compared to the littermate controls at E9.5 (Figure 5A and B). To determine whether increased apoptosis contributed to hypoplastic hearts, we assessed apoptosis by TUNEL assay. At E9.5, there was no significant difference in the number of apoptotic cells between Rbfox2-CKO embryos and Rbfox2-HetCKO or control embryos (Supplementary Figure S4). These results indicate that conditional loss of Rbfox2 in the embryonic heart adversely affects cell cycle.
Figure 5.
AS regulation of Ect2 and Abi1 by Rbfox2 is important for cell cycle progression. (A) Immunofluorescence of phospho-histone H3 positive contractile cells in Control, Rbfox2-HetCKO and Rbfox2-CKO embryonic hearts at E9.5. Contractile cells were stained for α-smooth muscle actin (α-SMA) and nuclei with DAPI at E9.5. % proliferating contractile cells were quantified using ImageJ. Data are mean ± standard deviation. *** P = 0.0001; by analysis of variance (ANOVA) with Bonferroni's post hoc test. Scale bars = 50 μm. (B) Immunofluorescence of phospho-histone H3 positive endocardial/endothelial cells in Control, Rbfox2-HetCKO and Rbfox2-CKO embryonic hearts at E9.5. Endocardial/endothelial cells were stained for PECAM1 and nuclei with DAPI. Data are mean ± standard deviation. * P = 0.0452, *** P = 0.0009; by analysis of variance (ANOVA) with Bonferroni's post hoc test. Scale bars = 50 μm. (C) The effect of control or ECT2 and ABI1 targeting SSOs on alternative splicing of ABI1, ECT2 and CTTN determined by RT-PCR 48 h post-treatment. (n = 4). (D) Cell cycle was examined in Control-SSO or ECT2 and ABI1-SSO treated HUVECs by flow cytometry using propidium iodide DNA labeling and histogram was analyzed using FlowJo. Data are mean ± standard deviation (n = 6). ***P = 0.0003, ****P = 0.0001 by unpaired Student's t-test.
To determine whether differential splicing of Abi1, and Ect2 contribute to the cell cycle defects seen in Rbfox2 mutants, we used splice switching antisense oligonucleotides (SSOs) to modulate AS of these two genes to mimic their AS patterns observed in Rbfox2-CKO embryos (Figure 3E and F). Compared to control oligos, HUVECs treated with a mixture of ABI1 and ECT2 specific SSOs exhibited almost a complete switch to the exon skipped isoforms (Figure 5C), similar to AS changes seen in Rbfox2 mutants (Figure 3E and F) and RBFOX2 depleted cells (Figure 4A). Non-targeted alternative exon 11 of CTTN, which was used as a negative control, was unaffected (Figure 5C, right panel), demonstrating the specificity of our targeting oligos.
To determine if these alternatively spliced variants could potentially explain low mitotic cells observed in Rbfox2-CKO mutants, we examined cell cycle progression in HUVECs treated with these SSOs. Remarkably, altering AS of just ABI1, and ECT2 (out of 986 total AS) delayed cell cycle progression reflected as an increased number of cells in G1 phase and decreased number of cells in S phase (Figure 5D). These data suggest that dysregulation of Abi1 and Ect2 AS induces cell cycle defects and is likely one of the contributors to cell cycle defects seen in Rbfox2-CKO embryos.
RBFOX2-mediated AS regulation of Abi1 and Ect2 is important for cell adhesion to ECM
Analysis of AS changes in Rbfox2 mutants revealed endothelial cells and cell-ECM adhesion to be affected. In the embryonic heart, adherence of specialized endothelial (endocardial) cells to ECM is necessary for Endo-MT. Endo-MT is a process in which endocardial cells transdifferentiate into migratory mesenchymal cells that is critical for the septation of cardiac chambers, the formation of OFT and valves (10,48).
Rbfox2 mutants exhibited cardiac chamber and OFT defects. Moreover, the timing of the appearance of the heart defects in the Rbfox2-CKO embryos between E9.5 and E10.5 also coincided with Endo-MT process. Therefore, we examined the mesenchymal cells in the atrioventricular canal of embryos between E9.5 and E10.5; a known site for active Endo-MT during valve formation. We labeled the endocardial cells that are present in the atrioventricular canal before the mesenchymal transition with arrows (Figure 6A). We marked the boundaries of the endocardial cushions containing newly derived mesenchyme cells following EMT between E9.5 and E10.5 with lines (Figure 6A). E9.5 embryos showed normal endocardial cells in all embryos. E10.5 embryos revealed that control embryos displayed normal atrioventricular cushions populated by mesenchymal cells, indicative of normal Endo-MT, while Rbfox2-CKO embryos showed few, if any mesenchymal cells within this region at E10.5, suggestive of Endo-MT defects (Figure 6A, bottom right panel). Consistent with these phenotypic changes in Endo-MT, we found that transcription factor Twist1, which is critical for mesenchymal transition, was significantly downregulated in Rbfox2-CKO embryos (Figure 6B). In addition, the mesenchymal marker gene Cadherin-2 was also decreased in Rbfox2 mutants when compared to controls (Figure 6B). These results indicate that Endo-MT is defective in Rbfox2-CKO embryo hearts.
Figure 6.
RBFOX2-mediated AS regulation of Ect2 and Abi1 is critical for cell adhesion to ECM. (A) Representative images of H&E stained embryonic sections of Control, Rbfox2-HetCKO and Rbfox2-CKO at E9.5 and E10.5. Arrows indicate endocardial cells in cardiac cushions of the atrioventricular canal at E9.5. Lines represent the mesenchymal cells at E10.5. Scale bars = 100 μm. (B) RT-qPCR analysis of transcription factor Twist1, which promotes mesenchymal transition and mesenchymal cell marker Cadherin2 and in Control, Rbfox2-HetCKO and Rbfox2-CKO embryo hearts at E9.5. Data are mean ± standard deviation. *** P = 0.0004, **** P = 0.0001, by analysis of variance (ANOVA) with Bonferroni's post hoc test. (C) Left panel: Brightfield microscope images of MTT reagent treated control or RBFOX2 depleted (top) and Control-SSO or ABI1-ECT2 targeting SSO treated (bottom) HUVECs after adhesion to collagen I matrix within 45 min (n = 4). Right panel: Quantification of relative cell adherence (absorbance at 570) of scrambled or RBFOX2 siRNA treated HUVECs to collagen I determined by the MTT assay. Data are mean ± standard deviation (n = 16). ****P = 0.0001, ****P = 0.0001 using Student's t-test.
Rbfox2 mutants displayed Endo-MT defects coincident with defects in chamber and OFT formation, similar to endocardial cell defects observed in HLHS patients (11). For that reason, we tested whether RBFOX2 regulates the adherence of endothelial cells to ECM in vitro (Supplementary Figure S5). Briefly, we depleted RBFOX2 in cultured HUVECs and examined cell viability, which was found to be similar between control and RBFOX2 knocked down cells (98% versus 97%). We then plated HUVECs treated with control or RBFOX2 specific siRNAs and quantified their ability to attach to ECM component collagen I (Figure 6C, top panels). Cell-ECM adhesion was significantly reduced in RBFOX2 knocked down HUVECs compared to cells treated with scrambled siRNAs (Figure 6C).
To determine whether AS changes in Abi1 and Ect2 contributes to cell-ECM adhesion defects seen in Rbfox2 mutants, we designed SSOs to mimic exon exclusions in these genes observed in Rbfox2 mutants (Figure 3E and F). Altering AS of Abi1 and Ect2 was adequate to impair cell adhesion to ECM (Figure 6C, bottom panels), similar to that is observed in RBFOX2 depleted HUVECs (Figure 6C, top panels). These results suggest that RBFOX2 mediated AS of Abi1 and Ect2 is critical for cell adhesion to ECM and maybe one of the possible contributors to Endo-MT defects in Rbfox2 mutants.
RBFOX2 depletion modulates focal adhesions
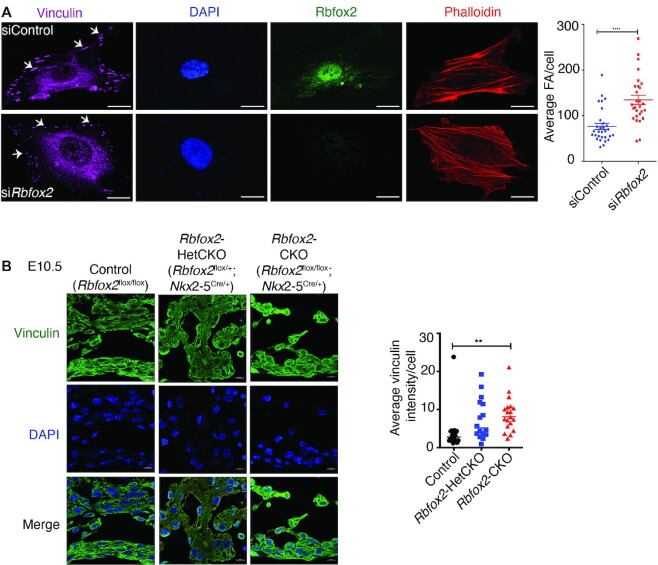
To determine the mechanism for defective cell-ECM adhesion in RBFOX2 depleted cells, we examined focal adhesions and actin stress fibers, both are essential structures for cell-ECM adhesion. We visualized the actin stress fiber assembly labeling F-actin with fluorescently conjugated phalloidin while focal adhesion formations were labeled by vinculin staining. The number of focal adhesions were increased in RBFOX2 knocked down HUVECs (Figure 7A). Noticeably, focal adhesion structures were different in RBFOX2 depleted cells such that there were more of nascent focal adhesions (dot type) than the mature adhesions (dash type), which are present in adherent cells (49) (Figure 7A, white arrows). These results demonstrate that RBFOX2 depletion can influence focal adhesion structures.
Figure 7.
RBFOX2 depletion alters focal adhesions. (A) Immunohistochemical staining of focal adhesions (FA) using vinculin antibody (magenta) and stress fibers using phalloidin (red) in control or RBFOX2 (green) knocked down HUVECs. Nuclei were stained with DAPI. The number of focal adhesions were quantified using control (n = 30) and RBFOX2 (n = 28) knocked down HUVECs with ImageJ. Data are mean ± standard, ****P = 0.0001 using Student's t-test. Scale bars = 10 μm. (B) Vinculin immunofluorescence in Control, Rbfox2-HetCKO and Rbfox2-CKO embryonic hearts at E10.5. Nuclei were stained with DAPI. Vinculin intensity per cells were quantified using Image J. Data are mean ± standard deviation. ** P = 0.0054; by analysis of variance (ANOVA) with Bonferroni's post hoc test. Scale bars = 50 μm.
Changes in focal adhesions can affect endothelial cell migration and tube formation, which can be tested in vitro using an endothelial network formation assay. We checked the network forming ability of HUVECs depleted of RBFOX2. RBFOX2 knockdown did not significantly alter the assembly of the networks determined by the total junctions formed, total vessel length, total vessel area and vessel length, although the total number of end points were significantly increased in RBFOX2-depleted HUVECs (Supplementary Figure S6).
To investigate whether focal adhesions are also affected in Rbfox2-CKO mutant embryos, we performed vinculin staining to mark focal adhesions in E10.5 embryonic heart tissues. Vinculin signal was increased in Rbfox2-CKO mutants at E10.5 (Figure 7B), consistent with increased vinculin positive focal adhesions in RBFOX2 depleted cells (Figure 7A). Loss of Rbfox2 alters focal adhesions that are crucial for cell-ECM adhesion.
DISCUSSION
The RNA binding protein Rbfox2 has an emerging role in cardiovascular diseases and congenital heart defects (1,3–6). However, it remains elusive how RBFOX2 impacts heart development. In this study, we addressed this clinically relevant question using a combination of genetics, genomics, and molecular and cellular biology tools. We discovered that Rbfox2 is essential for cardiac chamber, OFT and yolk sac vasculature formation. Rbfox2 mutants developed congestive heart failure as well as yolk sac vasculature defects at E10.5. However, it is unclear whether defects in yolk vasculature or cardiac output are the primary reasons for embryonic lethality and cardiovascular developmental defects. Our in vitro studies indicate that Rbfox2 is not necessary for endothelial cell mesh network formation (Supplementary Figure S6), which can be used to assess the tube formation in an in vitro setting (50,51). Rbfox2 mutant embryos displayed normal yolk sacs at E9.5 (Supplementary Figure S3). Our results indicate that both yolk sac vasculature and cardiac defects occur between E9.5 and E10.5. Additional studies are needed to determine the causal defects in these embryos.
RBFOX2 is a risk allele for HLHS and heterozygous loss of function mutations in patients are consistently linked to the HLHS phenotype (3–5). HLHS patients are born with circulation problems due to the hypoplasia of left part of the heart affecting the left ventricle, valves and the aorta (7). There is very little known regarding the mechanisms of these developmental defects in patients with HLHS. Cardiomyocyte proliferation defects are thought to be one of the contributors to the underdevelopment of the left heart in HLHS (8,12). In this study, we found phenotypic and molecular similarities between our conditional Rbfox2 knockout mice and HLHS patients. First, Rbfox2-HetCKO embryos exhibited profound changes in proliferation of contractile cells similar to that seen in HLHS. Loss of one allele of Rbfox2 in mice was adequate to impact mitosis of contractile and endocardial/endothelial cells. Second, Rbfox2 conditional ablation led to hypoplastic hearts and OFT, similar to HLHS in humans (6,52). Third, in our conditional knockout mouse model, mesenchymal cells in the developing cardiac valves were not detectable consistent with low expression of mesenchymal cell markers, suggestive of compromised Endo-MT that leads to valve formation defects, also observed in HLHS patients (8,12). Fourth, we identified AS changes in genes that regulate cell-ECM adhesion and found that RBFOX2 is important in influencing focal adhesions. Notably, genes with functions in focal adhesions and cell-ECM adhesion were also identified as the most affected processes in an HLHS mouse model (8), and cell-ECM adhesion and endocardial cell defects have also been identified in HLHS patients (11). Overall, our results indicate that Rbfox2 conditional loss in the embryonic heart partially recapitulates some of the features of HLHS. This could be attributed to the multigenic etiology of the disease. Even though Rbfox2-HetCKO embryos displayed cell proliferation and adhesion defects but these defects were not as severe as the Rbfox2-CKO embryos. Introducing HLHS patient specific mutations to mouse Rbfox2 locus may better phenocopy human HLHS.
HLHS patients can have neuronal developmental defects (3). RBFOX2 has an important role in regulating AS networks in the brain and is required for motor neuron function and the development of the cerebellum in mice (26). It is possible that loss of Rbfox2 contributes to both heart and brain problems in HLHS patients. Neural crest cell specific ablation of Rbfox2 did not impact cardiovascular development. It is possible that RBFOX1 and RBFOX3 might compensate for the loss of Rbfox2 in neural crest cells. Moreover, ablation of Rbfox2 in neural crests after mesenchymal transition (53) might have hindered its role on EMT of neural crest cells and on cardiovascular development. In our conditional knockout mouse model Rbfox2 is ablated in three cell lineages in the embryonic heart. These mutants displayed cardiovascular defects that resembled some features of HLHS. These results suggest that Rbfox2 deletion in multiple cell lineages collectively contribute to cardiovascular developmental defects.
RBFOX2 is a well-known regulator of AS and its binding sites were identified in genes that undergo dramatic AS transitions during post-natal heart development (17). RBFOX2 regulated AS networks in the embryonic heart are widely unknown. In this study, we identified RBFOX2 regulated AS networks that can direct specific cell behavior in the embryonic heart.
During cardiovascular development, the adherence of cells to the ECM via focal adhesions is critical for modulating cell behavior and is highly regulated by family of Rho GTPases (54,55) (56). Interaction of cells with ECM activates several GTPases and kinases to regulate cell polarity, migration, proliferation and differentiation (57). Although the importance of cell-ECM adhesion during development is well established, it is widely unknown whether post-transcriptional mechanisms are involved in regulation of cell-ECM adhesion during heart development. Our unbiased analyses identified AS networks that alter cytoskeletal organization and cell-ECM adhesion, processes mediated by Rho GTPases in Rbfox2 mutant hearts. Indeed, RBFOX2 depletion in cultured endothelial cells adversely affected cell adhesion to ECM associated with changes in focal adhesions. Examining the consequences of RBFOX2-regulated AS demonstrated that RBFOX2-regulated exons encode for regulatory and structural regions of proteins. Using splice switching oligos, we demonstrated that RBFOX2 regulated AS has physiological consequences such that modifying only two RBFOX2-regulated AS events (GEFs for Rho GTPases) was sufficient to impair cell-ECM adhesion and cell cycle progression similar defects seen in Rbfox2 mutant hearts. Importantly, different knockout mouse models of these GTPase regulators (ABI1, ECT2 and FN1) displayed embryonic lethality or cardiovascular development defects (37,58–61), supporting our findings regarding their AS dysregulation in Rbfox2 mutants.
Our results demonstrate that Rbfox2 has a previously unrecognized role in influencing cell-ECM adhesion properties. Genome wide analysis of RBFOX2 target RNAs using CLIP-seq in several different cell and tissues types identified cytoskeletal genes as RBFOX2 targets (26,62). However, the functional relationship between RBFOX2 and the cytoskeleton has been unresolved. Our new findings provide insights into the functional connection between RBFOX2 and cytoskeleton. Our in vitro studies unveiled that RBFOX2 is important for focal adhesions. The changes in focal adhesions may be one of the contributors to cell-ECM adhesion defects. Importantly, focal adhesions and cell-ECM adhesion were also identified as the most affected processes in an HLHS mouse model (8). Cell-ECM adhesion is critical for Endo-MT. In our mouse model, we identified Endo-MT defects associated with changes in transcription factor Twist 1 and mesenchymal cell marker Cdh2. Our finding is consistent with previous findings that RBFOX2 is implicated in EMT (63–65).
Our successful use of SSOs to modulate RBFOX2 regulated AS may lead to new ways to correct aberrant AS in HLHS patients that harbor RBFOX2 loss of mutations and suffer from long-term cardiac complications. FDA-approved antisense oligonucleotides have been used in clinics to treat children with spinal muscle dystrophy and adults with Duchenne's Muscular Dystrophy (66,67).
In summary, our results demonstrate that Rbfox2 is required for proper cardiac chamber, OFT and yolk sac vasculature formation. Our findings also indicate that establishment of RBFOX2 regulated AS networks in the embryonic heart is vital for cell-ECM adhesion, Endo-MT and cell proliferation.
DATA AVAILABILITY
RNA-sequencing data was deposited to GEO repository and can be accessed with GSE135152 accession number.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Mariano Garcia-Blanco for critically reading the manuscript. The authors acknowledge the UTMB Next Generation Sequencing Core and the Texas Advanced Computing Center at The University of Texas at Austin. We also thank the ENCODE project for RBFOX2-eCLIP. The contents of the manuscript are solely the responsibility of the authors and do not necessarily represent the official views of NHLBI of NIH.
Contributor Information
Sunil K Verma, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX 77555, USA.
Vaibhav Deshmukh, Department of Molecular Physiology and Biophysics, Baylor College of Medicine, Houston, TX 77030, USA.
Kaitlyn Thatcher, Department of Pediatrics, Medical College of Wisconsin, Division of Pediatric Cardiology, The Herma Heart Institute, Children's WI, Milwaukee, WI 53226, USA.
KarryAnne K Belanger, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX 77555, USA.
Alexander M Rhyner, Department of Molecular Physiology and Biophysics, Baylor College of Medicine, Houston, TX 77030, USA; Center for Organ Repair and Renewal, Baylor College of Medicine, Houston, TX 77030, USA.
Shu Meng, Houston Methodist Research Institute, Department of Cardiovascular Sciences, Houston, TX 77030, USA.
Richard Joshua Holcomb, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX 77555, USA.
Michael Bressan, Department of Cell Biology and Physiology, McAllister Heart Institute, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
James F Martin, Department of Molecular Physiology and Biophysics, Baylor College of Medicine, Houston, TX 77030, USA; Center for Organ Repair and Renewal, Baylor College of Medicine, Houston, TX 77030, USA; Cardiomyocyte Renewal Lab;Texas Heart Institute, Houston, TX, 77030, USA.
John P Cooke, Houston Methodist Research Institute, Department of Cardiovascular Sciences, Houston, TX 77030, USA.
Joshua D Wythe, Department of Molecular Physiology and Biophysics, Baylor College of Medicine, Houston, TX 77030, USA; Center for Organ Repair and Renewal, Baylor College of Medicine, Houston, TX 77030, USA; Cardiomyocyte Renewal Lab;Texas Heart Institute, Houston, TX, 77030, USA.
Steven G Widen, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX 77555, USA.
Joy Lincoln, Department of Pediatrics, Medical College of Wisconsin, Division of Pediatric Cardiology, The Herma Heart Institute, Children's WI, Milwaukee, WI 53226, USA.
Muge N Kuyumcu-Martinez, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, Galveston, TX 77555, USA; Department of Neuroscience, Cell Biology and Anatomy, Institute for Translational Sciences, University of Texas Medical Branch, 301 University Blvd. Galveston, TX 77555, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
AHA Grant [15GRNT22830010, 20TPA35490206, in part], Additional Ventures Single Ventricle Research Fund [4873, in part], John Sealy Memorial Endowment Fund [in part] and [NIH/NHLBI [1R01HL135031 to M.N.K.-M., 1R01HL132801 and R01HL142685 to J.L., R01HL146626 to M.B, 1R01HD099026-01, 1R01HL133254 to J.P.C.]; NIH [HL127717, HL130804, HL118761 to J.F.M.]; Vivian L. Smith Foundation, Fondation LeDucq Transatlantic Networks of Excellence in Cardiovascular Research [14CVD01] to J.F.M; Cancer Prevention Institute of Texas grant [RP200619, in part] to J.P.C; and Additional Ventures Single Ventricle Research Fund [4873, in part] to J.D.W. Funding for open access charge: John Sealy Memorial Endowment Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wei C., Qiu J., Zhou Y., Xue Y., Hu J., Ouyang K., Banerjee I., Zhang C., Chen B., Li H.et al.. Repression of the central splicing regulator RBFox2 is functionally linked to pressure overload-induced heart failure. Cell Rep. 2015; 10:1521–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nutter C.A., Jaworski E.A., Verma S.K., Deshmukh V., Wang Q., Botvinnik O.B., Lozano M.J., Abass I.J., Ijaz T., Brasier A.R.et al.. Dysregulation of RBFOX2 is an early event in cardiac pathogenesis of diabetes. Cell Rep. 2016; 15:2200–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J.et al.. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015; 350:1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Glessner J.T., Bick A.G., Ito K., Homsy J.G., Rodriguez-Murillo L., Fromer M., Mazaika E., Vardarajan B., Italia M., Leipzig J.et al.. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ. Res. 2014; 115:884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McKean D.M., Homsy J., Wakimoto H., Patel N., Gorham J., DePalma S.R., Ware J.S., Zaidi S., Ma W., Patel N.et al.. Loss of RNA expression and allele-specific expression associated with congenital heart disease. Nat. Commun. 2016; 7:12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morris S.A., Ethen M.K., Penny D.J., Canfield M.A., Minard C.G., Fixler D.E., Nembhard W.N.. Prenatal diagnosis, birth location, surgical center, and neonatal mortality in infants with hypoplastic left heart syndrome. Circulation. 2014; 129:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parker S.E., Mai C.T., Canfield M.A., Rickard R., Wang Y., Meyer R.E., Anderson P., Mason C.A., Collins J.S., Kirby R.S.et al.. Updated national birth prevalence estimates for selected birth defects in the united states, 2004-2006. Birth Defects Res. A Clin. Mol. Teratol. 2010; 88:1008–1016. [DOI] [PubMed] [Google Scholar]
- 8. Liu X., Yagi H., Saeed S., Bais A.S., Gabriel G.C., Chen Z., Peterson K.A., Li Y., Schwartz M.C., Reynolds W.T.et al.. The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 2017; 49:1152–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yagi H., Liu X., Gabriel G.C., Wu Y., Peterson K., Murray S.A., Aronow B.J., Martin L.J., Benson D.W., Lo C.W.. The genetic landscape of hypoplastic left heart syndrome. Pediatr. Cardiol. 2018; 39:1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang H., Lui K.O., Zhou B.. Endocardial cell plasticity in cardiac development, diseases and regeneration. Circ. Res. 2018; 122:774–789. [DOI] [PubMed] [Google Scholar]
- 11. Miao Y., Tian L., Martin M., Paige S.L., Galdos F.X., Li J., Klein A., Zhang H., Ma N., Wei Y.et al.. Intrinsic endocardial defects contribute to hypoplastic left heart syndrome. Cell Stem Cell. 2020; 27:574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bohlmeyer T.J., Helmke S., Ge S., Lynch J., Brodsky G., Sederberg J.H., Robertson A.D., Minobe W., Bristow M.R., Perryman M.B.. Hypoplastic left heart syndrome myocytes are differentiated but possess a unique phenotype. Cardiovasc. Pathol. 2003; 12:23–31. [DOI] [PubMed] [Google Scholar]
- 13. Ricci M., Xu Y., Hammond H.L., Willoughby D.A., Nathanson L., Rodriguez M.M., Vatta M., Lipshultz S.E., Lincoln J.. Myocardial alternative RNA splicing and gene expression profiling in early stage hypoplastic left heart syndrome. PLoS One. 2012; 7:e29784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Verma S.K., Deshmukh V., Nutter C.A., Jaworski E.A., Jin W., Wadhwa L., Abata J., Ricci M., Lincoln J., Martin J.F.et al.. Rbfox2 function in RNA metabolism is impaired in hypoplastic left heart syndrome patient hearts. Sci. Rep. 2016; 6:30896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Underwood J.G., Boutz P.L., Dougherty J.D., Stoilov P., Black D.L.. Homologues of the caenorhabditis elegans fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005; 25:10005–10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Damianov A., Ying Y., Lin C.H., Lee J.A., Tran D., Vashisht A.A., Bahrami-Samani E., Xing Y., Martin K.C., Wohlschlegel J.A.et al.. Rbfox proteins regulate splicing as part of a large multiprotein complex LASR. Cell. 2016; 165:606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A.. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Nat. Acad. Sci. U.S.A. 2008; 105:20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallagher T.L., Arribere J.A., Geurts P.A., Exner C.R., McDonald K.L., Dill K.K., Marr H.L., Adkar S.S., Garnett A.T., Amacher S.L.et al.. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol. 2011; 359:251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei C., Xiao R., Chen L., Cui H., Zhou Y., Xue Y., Hu J., Zhou B., Tsutsui T., Qiu J.et al.. RBFox2 binds nascent RNA to globally regulate polycomb complex 2 targeting in mammalian genomes. Mol. Cell. 2016; 62:875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu J., Gao C., Wei C., Xue Y., Shao C., Hao Y., Gou L.T., Zhou Y., Zhang J., Ren S.et al.. RBFox2-miR-34a-Jph2 axis contributes to cardiac decompensation during heart failure. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:6172–6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cibi D.M., Mia M.M., Guna Shekeran S., Yun L.S., Sandireddy R., Gupta P., Hota M., Sun L., Ghosh S., Singh M.K.. Neural crest-specific deletion of rbfox2 in mice leads to craniofacial abnormalities including cleft palate. Elife. 2019; 8:e45418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moses K.A., DeMayo F., Braun R.M., Reecy J.L., Schwartz R.J.. Embryonic expression of an nkx2-5/cre gene using ROSA26 reporter mice. Genesis. 2001; 31:176–180. [DOI] [PubMed] [Google Scholar]
- 23. Verma S.K., Deshmukh V., Liu P., Nutter C.A., Espejo R., Hung M.L., Wang G.S., Yeo G.W., Kuyumcu-Martinez M.N.. Reactivation of fetal splicing programs in diabetic hearts is mediated by protein kinase c signaling. J. Biol. Chem. 2013; 288:35372–35386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Katz Y., Wang E.T., Airoldi E.M., Burge C.B.. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010; 7:1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zudaire E., Gambardella L., Kurcz C., Vermeren S.. A computational tool for quantitative analysis of vascular networks. PLoS One. 2011; 6:e27385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gehman L.T., Meera P., Stoilov P., Shiue L., O’Brien J.E., Meisler M.H., Ares M. Jr, Otis T.S., Black D.L. The splicing regulator rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012; 26:445–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K.et al.. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tranchevent L.C., Aube F., Dulaurier L., Benoit-Pilven C., Rey A., Poret A., Chautard E., Mortada H., Desmet F.O., Chakrama F.Z.et al.. Identification of protein features encoded by alternative exons using exon ontology. Genome Res. 2017; 27:1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tatsumoto T., Xie X., Blumenthal R., Okamoto I., Miki T.. Human ECT2 is an exchange factor for rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J. Cell Biol. 1999; 147:921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schneid S., Wolff F., Buchner K., Bertram N., Baygun S., Barbosa P., Mangal S., Zanin E.. The BRCT domains of ECT2 have distinct functions during cytokinesis. Cell Rep. 2021; 34:108805. [DOI] [PubMed] [Google Scholar]
- 32. Chen M., Pan H., Sun L., Shi P., Zhang Y., Li L., Huang Y., Chen J., Jiang P., Fang X.et al.. Structure and regulation of human epithelial cell transforming 2 protein. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Innocenti M., Frittoli E., Ponzanelli I., Falck J.R., Brachmann S.M., Di Fiore P.P., Scita G.. Phosphoinositide 3-kinase activates rac by entering in a complex with eps8, abi1, and Sos-1. J. Cell Biol. 2003; 160:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L.B., Steffen A., Stradal T.E., Di Fiore P.P., Carlier M.F., Scita G.. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 2004; 6:319–327. [DOI] [PubMed] [Google Scholar]
- 35. Kotula L. Abi1, a critical molecule coordinating actin cytoskeleton reorganization with PI-3 kinase and growth signaling. FEBS Lett. 2012; 586:2790–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ryu J.R., Echarri A., Li R., Pendergast A.M.. Regulation of cell-cell adhesion by abi/diaphanous complexes. Mol. Cell. Biol. 2009; 29:1735–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dubielecka P.M., Ladwein K.I., Xiong X., Migeotte I., Chorzalska A., Anderson K.V., Sawicki J.A., Rottner K., Stradal T.E., Kotula L.. Essential role for abi1 in embryonic survival and WAVE2 complex integrity. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7022–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang R., Liao G., Wang Y., Tang D.D.. Distinctive roles of abi1 in regulating actin-associated proteins during human smooth muscle cell migration. Sci. Rep. 2020; 10:10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Astrof S., Crowley D., Hynes R.O.. Multiple cardiovascular defects caused by the absence of alternatively spliced segments of fibronectin. Dev. Biol. 2007; 311:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schiefner A., Gebauer M., Skerra A.. Extra-domain b in oncofetal fibronectin structurally promotes fibrillar head-to-tail dimerization of extracellular matrix protein. J. Biol. Chem. 2012; 287:17578–17588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benito-Jardon M., Klapproth S., Gimeno L.I., Petzold T., Bharadwaj M., Muller D.J., Zuchtriegel G., Reichel C.A., Costell M.. The fibronectin synergy site re-enforces cell adhesion and mediates a crosstalk between integrin classes. Elife. 2017; 6:e22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Auweter S.D., Fasan R., Reymond L., Underwood J.G., Black D.L., Pitsch S., Allain F.H.. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006; 25:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mayeda A., Munroe S.H., Caceres J.F., Krainer A.R.. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994; 13:5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun S., Zhang Z., Fregoso O., Krainer A.R.. Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA. 2012; 18:274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakano H., Liu X., Arshi A., Nakashima Y., van Handel B., Sasidharan R., Harmon A.W., Shin J.H., Schwartz R.J., Conway S.J.et al.. Haemogenic endocardium contributes to transient definitive haematopoiesis. Nat. Commun. 2013; 4:1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakashima Y., Yanez D.A., Touma M., Nakano H., Jaroszewicz A., Jordan M.C., Pellegrini M., Roos K.P., Nakano A.. Nkx2-5 suppresses the proliferation of atrial myocytes and conduction system. Circ. Res. 2014; 114:1103–1113. [DOI] [PubMed] [Google Scholar]
- 47. Harmon A.W., Nakano A.. Nkx2-5 lineage tracing visualizes the distribution of second heart field-derived aortic smooth muscle. Genesis. 2013; 51:862–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kovacic J.C., Mercader N., Torres M., Boehm M., Fuster V.. Epithelial-to-mesenchymal and endothelial-to-mesenchymal transition: from cardiovascular development to disease. Circulation. 2012; 125:1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuo J.C., Han X., Hsiao C.T., Yates J.R. 3rd, Waterman C.M.. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 2011; 13:383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Padget R.L., Mohite S.S., Hoog T.G., Justis B.S., Green B.E., Udan R.S.. Hemodynamic force is required for vascular smooth muscle cell recruitment to blood vessels during mouse embryonic development. Mech. Dev. 2019; 156:8–19. [DOI] [PubMed] [Google Scholar]
- 51. Garcia M.D., Larina I.V.. Vascular development and hemodynamic force in the mouse yolk sac. Front. Physiol. 2014; 5:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Feinstein J.A., Benson D.W., Dubin A.M., Cohen M.S., Maxey D.M., Mahle W.T., Pahl E., Villafane J., Bhatt A.B., Peng L.F.et al.. Hypoplastic left heart syndrome: current considerations and expectations. J. Am. Coll. Cardiol. 2012; 59(Suppl. 1):S1–S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Debbache J., Parfejevs V., Sommer L.. Cre-driver lines used for genetic fate mapping of neural crest cells in the mouse: an overview. Genesis. 2018; 56:e23105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fukata M., Kaibuchi K.. Rho-family GTPases in cadherin-mediated cell-cell adhesion. Nat. Rev. Mol. Cell Biol. 2001; 2:887–897. [DOI] [PubMed] [Google Scholar]
- 55. Etienne-Manneville S., Hall A.. Rho GTPases in cell biology. Nature. 2002; 420:629–635. [DOI] [PubMed] [Google Scholar]
- 56. Daley W.P., Peters S.B., Larsen M.. Extracellular matrix dynamics in development and regenerative medicine. J. Cell Sci. 2008; 121:255–264. [DOI] [PubMed] [Google Scholar]
- 57. Geiger B., Yamada K.M.. Molecular architecture and function of matrix adhesions. Cold Spring Harb. Perspect. Biol. 2011; 3:a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cook D.R., Solski P.A., Bultman S.J., Kauselmann G., Schoor M., Kuehn R., Friedman L.S., Cowley D.O., Van Dyke T., Yeh J.J.et al.. The ect2 rho guanine nucleotide exchange factor is essential for early mouse development and normal cell cytokinesis and migration. Genes Cancer. 2011; 2:932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Windmueller R., Leach J.P., Babu A., Zhou S., Morley M.P., Wakabayashi A., Petrenko N.B., Viatour P., Morrisey E.E.. Direct comparison of mononucleated and binucleated cardiomyocytes reveals molecular mechanisms underlying distinct proliferative competencies. Cell Rep. 2020; 30:3105–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. George E.L., Georges-Labouesse E.N., Patel-King R.S., Rayburn H., Hynes R.O.. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993; 119:1079–1091. [DOI] [PubMed] [Google Scholar]
- 61. Konstandin M.H., Volkers M., Collins B., Quijada P., Quintana M., De La Torre A., Ormachea L., Din S., Gude N., Toko H.et al.. Fibronectin contributes to pathological cardiac hypertrophy but not physiological growth. Basic Res. Cardiol. 2013; 108:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yeo G.W., Coufal N.G., Liang T.Y., Peng G.E., Fu X.D., Gage F.H.. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009; 16:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Baraniak A.P., Chen J.R., Garcia-Blanco M.A.. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol. Cell. Biol. 2006; 26:1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Venables J.P., Brosseau J.P., Gadea G., Klinck R., Prinos P., Beaulieu J.F., Lapointe E., Durand M., Thibault P., Tremblay K.et al.. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol. Cell. Biol. 2013; 33:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Braeutigam C., Rago L., Rolke A., Waldmeier L., Christofori G., Winter J.. The RNA-binding protein rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene. 2014; 33:1082–1092. [DOI] [PubMed] [Google Scholar]
- 66. Mendell J.R., Rodino-Klapac L.R., Sahenk Z., Roush K., Bird L., Lowes L.P., Alfano L., Gomez A.M., Lewis S., Kota J.et al.. Eteplirsen for the treatment of duchenne muscular dystrophy. Ann. Neurol. 2013; 74:637–647. [DOI] [PubMed] [Google Scholar]
- 67. Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R.. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011; 478:123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zou Y., Shao Z., Peng J., Li F., Gong D., Wang C., Zuo X., Zhang Z., Wu J., Shi Y.et al.. Crystal structure of triple-BRCT-domain of ECT2 and insights into the binding characteristics to CYK-4. FEBS Lett. 2014; 588:2911–2920. [DOI] [PubMed] [Google Scholar]
- 69. Leahy D.J., Aukhil I., Erickson H.P.. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996; 84:155–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data was deposited to GEO repository and can be accessed with GSE135152 accession number.