Abstract
Purpose
In this study, we analyzed the clinical distribution and drug resistance of carbapenem-resistant Klebsiella pneumoniae (CRKP) strains, the minimum inhibitory concentrations (MIC), MIC50 and MIC90, and geographical distribution in Hebei Province, China. We aimed to provide epidemiological research data, formulate appropriate combined treatment schemes, reasonably select antibiotics, and standardize nosocomial infection control schemes.
Patients and Methods
A total of 6328 strains of CRKP were collected from 2017 to 2019. The MIC was determined for the drug sensitivity test, and the experimental data were statistically analyzed using WHONET5.6.
Results
The detection rate of CRKP increased annually from 13.4% in 2017 to 14.5% in 2018, and 14.6% in 2019. The ratio of males to females was approximately 2:1; 53.6% were elderly, 39% were adults, 4.8% were minors, and 2.5% were newborns. The specimens collected were mainly sputum (70.9%). The resistance rate of CRKP to carbapenems and other β-lactam antibiotics was found to be increasing, with resistance rates generally greater than 90%. The resistance rate to aminoglycoside antibiotics decreased yearly to approximately 50%, and the resistance rate to quinolones remained unchanged at approximately 80%. From 2017 to 2019, the resistance rate of CRKP in Hebei Province to various antibiotics was high, and the resistance rate to β-lactam antibiotics increased each year.
Conclusion
The situation of CRKP resistance is severe in Hebei Province, China. The resistance rate to most antibiotics is very high and shows an upward trend. Among them, the resistance rate to polymyxin is low; however, few resistant strains do exist. MIC50 and MIC90 are higher than their MICs. It mainly causes lung infection in elderly men. This study is helpful to improve the diagnosis, treatment, and prevention of CRKP infection in our province.
Keywords: carbapenems, clinical distribution, drug resistance, minimum inhibitory concentration, geographic distribution, Klebsiella pneumoniae
Video abstract

Point your SmartPhone at the code above. If you have a QR code reader the video abstract will appear. Or use:
Introduction
Klebsiella pneumoniae is an important bacterium that is normally found in the flora of the human intestinal tract,1,2 and is a common microorganism in the environment. It may acquire the carbapenemase-coding, AmpC-producing, or extended-spectrum β-lactamase (ESBLs) with outer membrane porin deletion genes, which leads to carbapenem resistance.3 Carbapenem-coding genes are mostly located on transferable elements such as plasmids or transposons, resulting in the spread of drug-resistant strains.4 Bacterial drug resistance has become a major global challenge in the field of public health and is a serious threat to human health. Since the discovery of carbapenem-resistant K. pneumoniae (CRKP) in the United States in 1996, carbapenem-resistant Enterobacteriaceae (CRE) has spread rapidly worldwide.5 In 2019, the US Centers for Disease Control and Prevention listed CRE as an “emergency threat” pathogen threatening human health.
The mortality of patients infected with CRKP is higher than that of carbapenem-sensitive K. pneumoniae.6 According to the European Antimicrobial Resistance Surveillance Network (EARSN) database, the detection rate of K. pneumoniae is higher in Italy than in other European countries.7 Globally, the detection rate of CRKP is the highest in Greece (54.9%), followed by eastern Europe (22.5%), Argentina (14%), and the Philippines (11%).8 Data from the China Antimicrobial Resistance Surveillance System (CARSS) shows that the national average isolation rate of CRKP increases annually, from 6.4% in 2014 to 10.9% in 2019. The close communication between people all over the world has increased the risk of CRKP transmission.9–11
Tigecycline and polymyxin exhibit a high activity against CRKP.12 The emergence of new antibiotics such as ceftazidime-avibactam and cefiderocol has resulted in additional treatment options for CRKP infection.13–15 Zhang et al14 reported that the resistance rate of CRKP to ceftazidime-avibactam was only 3.7%. The aim of this study was to determine the distribution of specimens by statistically analyzing regional, sex, and age differences; antimicrobial resistance; minimum inhibitory concentration (MIC) 50/90; geographical distribution; and other aspects of CRKP. The study was conducted to provide epidemiological research data, formulate appropriate combined treatment schemes, reasonably select antibiotics, and standardize nosocomial infection control schemes.
Materials and Methods
Strain Selection and Characteristics
A total of 6328 Clinical CRKP strains were isolated from sputum, blood, secretions, and other specimens of patients between 2017 and 2019. These specimens were collected from 43 tertiary Hospitals in Hebei Province, China. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains. Duplicate strains were removed, and only the first tested sample of the patient was retained. The bacterial drug sensitivity test recommended by Clinical and Laboratory Standards Institute (CLSI) was used, and drug sensitivity was determined using the CLSI drug sensitivity break point of the 2019 edition.
Antimicrobial Susceptibility
Identification and determination of antimicrobial susceptibility of K. pneumoniae were performed using the Phoenix 100, FX-200 automatic blood culture instrument (Becton, Dickinson and Company, New Jersey, USA). Blood culture was performed using the BacT/ALERT 3D Microbial Identification System (BIO PARTICIPATIONS Co., LTD, Marcy-l’étoile, France). MacConkey agar plates (Guangzhou Dijing Microorganism Technology Co., Ltd.) were used to culture and screen the target bacteria.
Susceptibility testing was performed using the minimum inhibitory concentration (MIC) according to Clinical and Laboratory Standards Institute (CLSI, 2019). Isolates with an MIC ≥ 4 mg/L for meropenem and imipenem were defined as CRKP.16 The stock solution of drugs was prepared and diluted according to the guidelines formulated by Clinical and Laboratory Standards Institute (CLSI). The cultured bacteria to be tested were used to prepare a 0.5 McFarland bacterial suspension. Within 15 min of preparation of the bacterial suspension, 25 μL of the bacterial suspension and 45 μL of a colorific indicator were added into the broth and thoroughly mixed. The remaining bacterial suspension was poured into the biochemical well of a BD drug-sensitive plate (Becton, Dickinson and Company, New Jersey, USA), and the solution in the broth tube was poured into the drug-sensitive well in the drug-sensitive plate. The plate was covered, placed in the Phoenix 100 automated system, cultured for 24 h, and then observed.
Statistical Analysis
Antimicrobial susceptibility data were analyzed with the WHONET 5.6 software (WHO, Geneva, Switzerland). GraphPad prism 8 software (GraphPad Software, LaJolla, CA, USA) was used for drawing, and all counts were expressed as percentages (%).
Results
Specimen Distribution of CRKP
Among the 6328 CRKP isolates, sputum accounted for 4428 (70.9%) isolates, urine accounted for 669 (10.7%) isolates, and blood was the third most common (351 isolates; 5.6%). Secretions accounted for 193 (3.1%) isolates. Ascites accounted for 90 isolates (1.4%), as shown in Table 1.
Table 1.
Distribution Characteristics of CRKP Specimens from 2017 to 2019
| Specimen Type | 2017 (n = 1516) | 2018 (n = 2249) | 2019 (n = 2477) | Total | ||||
|---|---|---|---|---|---|---|---|---|
| N | P (%) | N | P (%) | N | P (%) | N | P (%) | |
| Sputum | 1044 | 68.87 | 1701 | 75.63 | 1738 | 70.17 | 4483 | 70.84 |
| Urine | 172 | 11.35 | 196 | 8.71 | 232 | 9.37 | 600 | 9.48 |
| Blood | 93 | 6.13 | 124 | 5.51 | 138 | 5.57 | 355 | 5.61 |
| Secretion | 38 | 2.51 | 60 | 2.67 | 90 | 3.63 | 188 | 2.97 |
| Ascites | 19 | 1.25 | 25 | 1.11 | 28 | 1.13 | 72 | 1.14 |
| Balf | 11 | 0.73 | 12 | 0.53 | 29 | 1.17 | 52 | 0.82 |
| Bile | 12 | 0.79 | 16 | 0.71 | 20 | 0.81 | 48 | 0.76 |
| Fine needle aspiration | 2 | 0.13 | 17 | 0.76 | 23 | 0.93 | 42 | 0.66 |
| Pus | 20 | 1.32 | 6 | 0.27 | 14 | 0.57 | 40 | 0.63 |
| Drainage | 3 | 0.20 | 14 | 0.62 | 14 | 0.57 | 31 | 0.49 |
| Excreta | 10 | 0.66 | 21 | 0.93 | 11 | 0.44 | 42 | 0.66 |
| Hydrothorax | 8 | 0.53 | 15 | 0.67 | 11 | 0.44 | 34 | 0.54 |
| Other | 84 | 5.54 | 128 | 5.48 | 129 | 5.21 | 341 | 5.39 |
| Total | 1516 | 100 | 2335 | 100 | 2477 | 100 | 6328 | 100.00 |
Abbreviations: N, number; P, proportion.
Geographical Distribution
The geographical distribution remained stable in Hebei Province, from a total of 10 cities counted, and the proportion varied widely. More than 40% of the isolates were from Shijiazhuang, and less than 2% of the isolates were from Langfang as shown in Table 2.
Table 2.
Regional Distribution of CRKP in Hebei Province from 2017 to 2019
| Territory Name | Year | Total | Proportion (%) | ||
|---|---|---|---|---|---|
| 2017(N) | 2018(N) | 2019(N) | |||
| Shijiazhuang | 891 | 991 | 893 | 2775 | 43.85 |
| Tangshan | 189 | 525 | 554 | 1268 | 20.04 |
| Cangzhou | 76 | 287 | 280 | 643 | 10.16 |
| Qinhuangdao | 89 | 167 | 180 | 436 | 6.89 |
| Xingtai | 94 | 82 | 128 | 304 | 4.80 |
| Handan | 46 | 66 | 120 | 232 | 3.67 |
| Zhangjiakou | 25 | 110 | 88 | 223 | 3.52 |
| Baoding | 63 | 49 | 87 | 199 | 3.14 |
| Chengteh | 22 | 41 | 99 | 162 | 2.56 |
| Langfang | 21 | 17 | 48 | 86 | 1.36 |
| Total | 1516 | 2335 | 2477 | 6328 | 100.00 |
Abbreviation: N, number.
Clinical Data
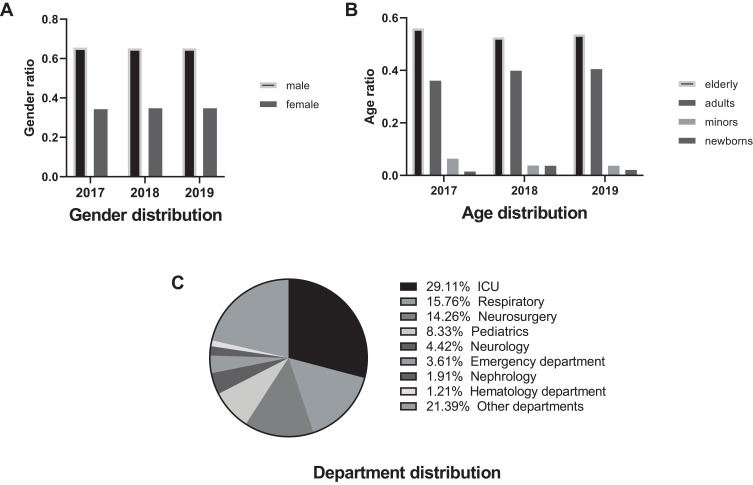
From 2017 to 2019, the proportion of CRKP, carbapenem-resistant P. aeruginosa (CRPae) and carbapenem-resistant E. coli (CRECO) in all infected patients and the population were 2.577%, 2.662%, 0.586% and 0.028%, 0.029%, and 0.006% respectively (Supplementary Table 1); CRKP accounted for 13.4% (1516/11,323), 14.5% (2335/16,100), and 14.6% (2477/16,737) of all isolates of K. pneumoniae, respectively. The ratio of males to females was almost unchanged at approximately 2:1, as shown in Figure 1A. The elderly (> 60 years old) accounted for 53.6%, adults (18–60 years old) accounted for 39%, minors (1–18 years old) accounted for 4.84%, and newborns (< 1 year old) accounted for 2.56%. The details are shown in Figure 1B. CRKP was mainly detected in the intensive care unit (ICU) (29.0%), respiratory (15.7%), neurosurgery (14.2%), pediatrics (6.4%), and emergency (3.6%) departments. The details are shown in Figure 1C.
Figure 1.
Overall distribution characteristics of carbapenem-resistant Klebsiella pneumoniae (CRKP). (A) Gender distribution characteristics of CRKP. (B) Age distribution characteristics of CRKP. (C) Distribution characteristics of CRKP departments.
Drug Susceptibility Analysis
Resistance trend of CRKP to common antibiotics: the resistance of CRKP to cephalosporin antibiotics showed an upward trend, except cefepime, for which the resistance first increased and then decreased. The resistance to penicillin antibiotics (piperacillin) showed an upward trend; the resistance to carbapenem antibiotics, including meropenem and imipenem, showed an upward trend; the resistance to single ring β-lactam antibiotics (aztreonam) increased initially and then decreased; the resistance to β-lactams/β-lactam inhibitors, including amoxicillin/clavulanate acis and piperacillin/tazobactam, showed an upward trend. The resistance to aminoglycoside antibiotics including tobramycin, amikacin, and gentamicin showed a downward trend, with the highest decrease observed in the case of tobramycin. The resistance to quinolones, including levofloxacin and ciprofloxacin, showed an upward trend. The resistance to sulfonamides showed a downward trend, with a large decline. In studies from 2017 to 2019, the observed polymyxin resistance rates were 0%, 1.5%, and 0%, respectively, showing a trend of increase at first and then a decrease.
The average resistance rate of CRKP to common antibiotics: the resistance rate to penicillin and β-lactamase/β-lactamase inhibitors was high: the resistance rate to piperacillin, amoxicillin/clavulanate acid, and piperacillin/tazobactam was 98.3%, 90.8%, and 92.5%, respectively. The resistance rate to cephalosporins was generally high. The resistance rate to First to fourth generation cephalosporins represented by cefazolin, cefuroxime, cefotaxime, ceftriaxone, ceftazidime, and cefepime was 95.8%, 97.2%, 94%, 97.6%, 94.1%, and 91.8%, respectively. The resistance rate to the fourth generation cephalosporins was lower than that to the second and third generation cephalosporins. The resistance rate to carbapenems was relatively high: 92.3% for imipenem and 93.2% for meropenem. The resistance rate to aztreonam was 95.7%; the resistance rate to quinolones was relatively low: 81.9% for ciprofloxacin and 79.3% for levofloxacin. The resistance rate to aminoglycoside antibiotics was relatively low: 59.6% to tobramycin, 47.7% to amikacin, and 64.3% to gentamicin. The resistance rate to cefoxitin was relatively low at 84.6%. The resistance rate to compound sulfamethoxazole antibiotics was low at 53.3%. The average resistance rate to polymyxin was very low, at 0.5% (Table 3).
Table 3.
Drug Sensitivity Data of CRKP to Various Antibacterial Drugs from 2017 to 2019
| Antibiotic Name | 2017 | 2018 | 2019 | Average Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | R(%) | S(%) | N | R(%) | S(%) | N | R(%) | S(%) | N | R (%) | S (%) | |
| Amoxicillin/clavulanic acid | 322 | 80.1 | 16.8 | 474 | 94.9 | 4.2 | 593 | 97.5 | 1.7 | 1389 | 90.8 | 7.6 |
| Cefotaxime | 551 | 90.4 | 9.1 | 809 | 96.2 | 3.8 | 561 | 95.5 | 4.1 | 1921 | 94.0 | 5.7 |
| Cefoxitin | 647 | 81.6 | 15.3 | 1033 | 85 | 10.4 | 1028 | 87.2 | 10.0 | 2708 | 84.6 | 11.9 |
| Piperacillin | 1063 | 97.7 | 0.9 | 1308 | 98.4 | 0.7 | 1317 | 98.8 | 0.5 | 3688 | 98.3 | 0.7 |
| Tobramycin | 833 | 71.2 | 22.1 | 1385 | 51.3 | 41.7 | 1405 | 56.2 | 33 | 3623 | 59.6 | 32.3 |
| Aztreonam | 1018 | 93.4 | 6.4 | 1513 | 97.4 | 2.4 | 1570 | 96.4 | 3.2 | 4101 | 95.7 | 4.0 |
| Cefuroxime | 1344 | 95.8 | 4.0 | 1823 | 97.9 | 1.8 | 1733 | 97.8 | 2.0 | 4900 | 97.2 | 2.6 |
| Ceftriaxone | 1262 | 95.2 | 4.7 | 1840 | 98.9 | 0.9 | 1972 | 98.7 | 1.1 | 5074 | 97.6 | 2.2 |
| Compound sulfamethoxazole | 1381 | 68.4 | 31.4 | 1875 | 47.8 | 52.1 | 2025 | 43.8 | 56.2 | 5281 | 53.3 | 46.6 |
| Cefazolin | 1427 | 91.5 | 8.5 | 2213 | 98.1 | 1.9 | 1960 | 97.8 | 2.2 | 5600 | 95.8 | 4.2 |
| Amikacin | 1507 | 56.6 | 42.1 | 2142 | 42.7 | 56.4 | 2452 | 43.9 | 55.5 | 6101 | 47.7 | 51.3 |
| Ciprofloxacin | 1439 | 81.2 | 13.6 | 2152 | 80.3 | 16.1 | 2183 | 84.2 | 13.7 | 5774 | 81.9 | 14.5 |
| Piperacillin/tazobactam | 1504 | 90.5 | 6.7 | 2169 | 92.9 | 4.6 | 2387 | 94.0 | 3.9 | 6060 | 92.5 | 5.1 |
| Meropenem | 1501 | 90.7 | 6.9 | 2260 | 94.5 | 4.4 | 2219 | 94.4 | 4.1 | 5980 | 93.2 | 5.1 |
| Levofloxacin | 1491 | 79.3 | 17.9 | 2290 | 76.8 | 20.1 | 2447 | 81.9 | 15.4 | 6228 | 79.3 | 17.8 |
| Ceftazidime | 1455 | 92.4 | 6.7 | 1981 | 95.8 | 3.0 | 2422 | 94.1 | 4.5 | 5858 | 94.1 | 4.7 |
| Gentamicin | 1452 | 71.4 | 27.1 | 2303 | 57.0 | 40.6 | 2169 | 64.5 | 33.2 | 5924 | 64.3 | 33.6 |
| Imipenem | 1510 | 90.4 | 7.0 | 2322 | 92.0 | 5.0 | 2456 | 94.5 | 2.9 | 6288 | 92.3 | 5.0 |
| Cefepime | 1507 | 91.8 | 5.6 | 2335 | 92.5 | 4.9 | 2462 | 91.1 | 4.9 | 6304 | 91.8 | 5.1 |
| Polymyxin | 132 | 0 | 100 | 69 | 1.5 | 98.5 | 99 | 0 | 100 | 99 | 0.5 | 99.5 |
Abbreviations: N, number of strains; R, drug resistance rate; S, sensitivity rate.
MIC50 and MIC90
In addition to amikacin, the MIC50 and MIC90 of CRKP was considerably higher than the MICs, and the MIC50 of meropenem, imipenem, and ertapenem was as high as 4 times the drug resistance break point. The MIC50 of cephalosporin antibiotics was 2–16 times the drug resistance break point, and the MIC50 of penicillin and its enzyme inhibitor was equal to its drug resistance break point, as shown in Table 4
Table 4.
CRKP MIC50 and MIC90 of CRKP
| Projects | Quantity | Break Point | MIC50 | MIC90 | MIC Range |
|---|---|---|---|---|---|
| Tigecycline | 962 | S≤0.03 R≥0.25 | 0.5 | 2 | 0.02–64 |
| Compound sulfamethoxazole | 4978 | S≤2 R≥4 | 4 | 16 | 0.25–128 |
| Ciprofloxacin | 5430 | S≤1 R≥4a | 4 | 8 | 0.01–64 |
| Etapenan | 724 | S≤0.5 R≥2 | 8 | 16 | 0.25–64 |
| Levofloxacin | 5646 | S≤2 R≥8 | 8 | 16 | 0.06–64 |
| Tobramycin | 3546 | S≤4 R≥16 | 16 | 16 | 0.5–32 |
| Meropenem | 5359 | S≤1 R≥4 | 16 | 32 | 0.06–128 |
| Gentamicin | 5370 | S≤4 R≥16 | 16 | 16 | 0.5–32 |
| Amikacin | 5479 | S≤16 R≥64 | 16 | 64 | 1–128 |
| Imipenem | 5658 | S≤1 R≥4 | 16 | 32 | 0.12–64 |
| Amoxicillin/clavulanic acid | 1027 | S≤8 R≥32 | 32 | 32 | 0.5–64 |
| Cefoxitin | 2218 | S≤8 R≥32 | 32 | 64 | 1–128 |
| Cefepime | 5694 | S≤2 R≥16 | 32 | 64 | 0.12–128 |
| Cefotaxime | 1327 | S≤1 R≥4 | 64 | 64 | 0.12–128 |
| Aztreonam | 3738 | S≤4 R≥16 | 64 | 128 | 0.25–128 |
| Cefuroxime | 4528 | S≤8 R≥32 | 64 | 128 | 1–128 |
| Ceftriaxone | 4624 | S≤1 R≥4 | 64 | 128 | 0.12–128 |
| Cefazolin | 5074 | S≤16 R≥32 | 64 | 128 | 1–128 |
| Ceftazidime | 5364 | S≤4 R≥16 | 64 | 64 | 0.06–128 |
| Piperacillin | 3414 | S≤16 R≥128 | 128 | 256 | 2–256 |
| Piperacillin/tazobactam | 5643 | S≤16 R≥128 | 128 | 256 | 0.5–256 |
Note: MIC50/MIC90, MIC50 and MIC90 can inhibit 50% or 90% of the tested bacteria.
Abbreviation: MIC, minimum inhibitory concentration.
Discussion
CRE often causes infection in immunocompromised patients, resulting in prolonged hospital stays and increased mortality.17 Although there are new antibiotics on the market for CRE infection, it is difficult to overcome antibiotic resistance. This is because there are only a few effective antibiotics and a lack of clinical data, especially data from large randomized controlled clinical research. Therefore, diagnosing, treating, and preventing CRE infection are challenging. It is worth noting that CRKP is listed as a critical-priority bacteria among CREs.18 The most important resistance mechanism of CRKP involves carbapenemases, and the common ones include K. pneumoniae carbapenemases (KPC), New Delhi metallo-β-lactamase (NDM), imipenemase metallo-β-lactamase (IMP), Verona integron-encoded metallo-β-lactamase (VIM), and oxacillinase-48-type carbapenemases (OXA-48).19 KPC-2 and KPC-3 are the most popular serine carbapenemases. KPC-2 is mainly detected in K. pneumoniae. In addition, gram-negative Enterobacteriaceae members carrying KPC-2 are resistant to all β-lactamase/β-lactamase inhibitors, except ceftazidime-avibactam.20 It further increases the limitations of antimicrobial selection and improves mortality. The metalloenzyme does not hydrolyze aztreonam, and the drug sensitivity results are sensitive to aztreonam, but cannot be inactivated or inhibited by avibactam, vaborbactam and relebactam. The strains producing metalloenzyme can be effectively screened using EDTA collaborative experiment. Metalloenzymes encoded by plasmid are a clinical threat because they are located in a mobile genetic structure and can spread and hide resistance genes to other antibiotics.21 OXA-48 enzymes are common in Klebsiella pneumoniae. They only hydrolyze penicillins and carbapenems, but do not hydrolyze extended spectrum cephalosporins. They often show low-level resistance to carbapenem antibiotics, whereas they are sensitive to the third and fourth generations cephalosporins. Their enzyme activity can be inhibited by avibactam, but cannot be inhibited by vaborbactam and relebactam.20 Therefore, the synergistic effects of different carbapenems may lead to a higher level of resistance to carbapenems and other antibiotics. A study of the characteristics of CRKP is particularly important for controlling pathogens, diagnosing and treating patients.
In recent years, there has been an increasing number of CRKP strains worldwide, but there are some differences in their distributions in different regions.8,22 This study showed that CRKP in Hebei Province mainly caused pneumonia, followed by urinary tract infection, bloodstream infection, wound infection, and abdominal infection (Table 1). A study in the United States showed that CRKP strains were mainly derived from urine.23 However, in Poland, CRKP strains were mainly derived from urine and feces.24 An Italian study showed that CRKP mainly caused urinary tract infections.7 A meta-analysis showed that CRKP mainly caused bloodstream infections,6 and a study in South Africa showed that 75% of CRKP strains in 2010–2012 were from bloodstream infections.25 CARSS showed that from 2017 to 2019, CRKP accounted for 13.4%, 14.5%, and 14.6%, respectively, showing an upward trend. In addition, data from the China Antimicrobial Surveillance Network (CHINET) indicate that the detection rate of CRKP in China increased from 0.7% in 2007 to 19.9% in 2018.26 However, a New York study showed that the CRKP infection rate in New York City showed a downward trend from 2006 to 2014, especially urinary tract infection.27
The geographical distribution of CRKP is different in Hebei, and its distribution in other cities in China is uneven (Table 2). The latest CARSS data showed that Henan, Shanghai, and Beijing were the top three cities in the national CRKP separation rate in 2019, and Qinghai and Tibet were the least. Hu et al28 calculated the separation rate of CRKP in various regions of China, and the separation rates in Inner Mongolia, Guangxi, Shanxi, Heilongjiang, Hainan, Gansu, Jilin, Ningxia, Qinghai, and Tibet were less than 5%, those in Jiangsu, Anhui, Jiangxi, Liaoning, Hebei, Hubei, Zhejiang, Yunnan, and Hunan were 10% to 20%, those in Beijing, Shanghai, and Henan were more than 20%. It was observed that CRKP distribution is related to the population distribution in China. The distribution of CRKP is greater in areas with larger populations and more developed economies, and less in areas with smaller populations and less developed economies. However, in contrast to the conclusion of this study, the detection rate of CRKP in underdeveloped countries such as Greece, Argentina, and the Philippines is much higher than that in developed countries all over the world.8 In this study, there were twice as many males as females with CRKP infection. A study in Taiwan29 compared the mortality rate of patients with urinary tract infections caused by CRKP, with a higher level of infection among males than females. Analysis from the age of the patient, we found that elderly patients in Hebei Province werethe most important CRKP group, which is common across China and the world.22,30 Patients with CRKP infection in our province are mainly in ICU, which agrees with previously reported findings in HeBei Province.16 In Ireland, CRKP infection is mainly concentrated in ICU and surgical ward.24 A study from Egypt showed that CRKP was mainly separated from the surgical ICU.31
The treatment of CRKP infection remains an intractable problem in clinics.7,32 The resistance rate of CRKP to penicillin and penicillin with β-lactamase inhibitors, especially to amoxicillin/clavulanic acid, is increasing yearly (Table 3). Amoxicillin/clavulanic acid is a commonly used antibiotic in pediatrics, and piperacillin/tazobactam is commonly used for the empirical treatment of nosocomial infection.33,34 Clavulanic acid and tazobactam mainly inhibit the activity of serine-β-lactamases, including CTX-M β-lactamases and the ESBL derivatives TEM-1, TEM-2, and SHV-1.20 The wide clinical use and limited drug action of these drugs lead to a high drug resistance rate. In our province, the resistance rate of CRKP to carbapenems is high and increasing yearly. CRKP is resistant to a variety of antibiotics, leading to a limited choice of therapeutic drugs. Treatment options for CRKP infection include the classic polymyxin, tigecycline, aminoglycosides, fosfomycin, a few quinolones or sulfonamides, and new drugs β-lactamase/β-lactamase inhibitors. Tigecycline and colistin are optional drugs for the treatment of CRKP; however, unfortunately, there are strains resistant to them.32 In 2004, a CRKP strain resistant to polymyxin was first reported in Greece.23 Since then, additional reports show that CRKP strains resistant to polymyxin have been isolated in a Hospital environment.35 From 2009 to 2018, the rate of resistance of CRKP to fosfomycin in China decreased from 81.8% to 50.8%, while that to tigecycline increased from 3.8% to 21.7%, and that to polymyxin B increased from 1.6% in 2013 to 18.3% in 2017.26 Fosfomycin inhibits the synthesis of bacterial cell walls and shows synergistic effects when combined with a variety of antibiotics. The drug is mainly excreted from the kidney in its original form and is highly concentrated in the urine. Oral administration of fosfomycin is mainly used to treat urinary tract infections. In Hebei Province, the resistance rate to polymyxin was 0.5%, and only a few drug-resistant strains emerged in 2018. The use of polymyxin is associated with several challenges, including high-dose nephrotoxicity and drug resistance. In the study of synergism between polymyxin and carbapenems, polymyxin exerted a synergistic effect on 50% of carbapenems.36 A meta-analysis showed37 that the mortality of patients on polymyxin/tigecycline and polymyxin/carbapenem combination therapy was lower than that of patients on polymyxin alone; polymyxin alone is not recommended in routine clinical practice. The polymyxin-resistant CRKP strain was less sensitive to amikacin and tigecycline, but more sensitive to gentamicin.23 Therefore, polymyxin can be combined with gentamicin and carbapenem for the treatment of CRKP infection. Polymyxin E methanesulfonate (CMS) is recommended for the treatment of CRE urinary tract infection because of its high colistin concentration in the urine. Tigecycline is a time-dependent drug with a long post-antibiotic effect. The drug level after intake is relatively low in the blood and cerebrospinal fluid,38 and therefore, it is not routinely recommended for CRKP-induced bloodstream infections and central nervous system infections. A systematic retrospective study found that increasing the dose of tigecycline can improve the clinical efficiency and microbial clearance in the treatment of hospital-acquired pneumonia (HAP). In addition, the mortality of tigecycline alone was higher than that of combined use.39 It is recommended to combine tigecycline with two or three drugs. The resistance rate of CRKP to cephalosporins in Hebei Province showed an increasing trend (Table 3). The resistance rates to ceftriaxone and cefuroxime were close to 100%. The resistance rate to cefepime was slightly lower than that of the second and third generation cephalosporins. El Badawy et al31 showed that clinically isolated CRKP was 100% resistant to cefazolin, cefradine, cefoperazone, and cefotaxime. In this study, it was also found that among the quinolones, levofloxacin was better than ciprofloxacin. Notably, the rate of resistance of CRKP to sulfamethoxazole in our province decreased (similar to the CHINET data26); similarly, the rate of resistance to amikacin also reduced. An et al40 showed that CRKP had the lowest rate of resistance to amikacin. The resistance rate to tobramycin and gentamicin was maintained at approximately 60%, which was lower than that of β-lactam antibiotics. A multivariate study showed that the treatment of CRKP infection with gentamicin (MIC ≤ 2 mg/L) was associated with a lower mortality of patients.41 Van Duin et al reported that amikacin was effective against 83% of CRKP42 cases. Aminoglycoside antibiotics exhibit more efficient microbial clearance in urinary tract infections caused by CRKP.43 In our province, aminoglycoside antibiotics is empirically applied to treat CRKP infection. In Hebei Province, the resistance rate of CRKP to sulfamethoxazole is low, which is consistent with the results of He et al and Lombardi.44,45 A recent study found that in vitro, compound sulfamethoxazole combined with polymyxin can quickly kill clinically isolated CRKP within 2–24 h, but this result needs further evidence-based medicine and clinical research certification.46 From 2017 to 2019, the MIC50/MIC90 of CRKP to imipenem and meropenem in Hebei Province was 16/32 µg/mL (Table 4). Studies have found that when the MIC of meropenem is ≤8 mg/L, adding a large dose of meropenem (2 g once every 8 h) to another active drug can reduce the mortality of patients with CRE infection.36 If the MIC of the strain to carbapenems is > 8 mg/L, the combination regimen containing carbapenems is no longer recommended. In China, from 2007 to 2018, the MIC90 of CRKP to imipenem fluctuated between 32 and 256, and the MIC90 for meropenem increased from 64 in 2009 to 256 in 2018.26 Studies have shown that the use of carbapenem is an independent risk factor for CRKP infection,47,48 and therefore, clinicians should use carbapenem antibiotics cautiously to reduce the risk of CRKP infection. Recent reports from Europe, Canada, the United States, Asia, and Africa show that CRKP is spreading worldwide.49 Based on the increasing trend of the CRKP detection rate worldwide, the prevalence of CRKP needs to be studied in a global context to understand its transmission rules and to provide data support for the development of prevention and control measures.
Conclusion
In summary, from 2017 to 2019, in Hebei Province, CRKP primarily caused lung infection; males accounted for about two-thirds, and older patients represented the majority of the patient population. Regional distribution varied greatly and was mainly concentrated in the economically developed and populous cities such as Shijiazhuang and Tangshan. The resistance rate of CRKP to β-lactam antibiotics was higher than 90%, and it is increasing annually. The resistance rate to sulfamethoxazole and aminoglycoside antibiotics was lower than 70%, showing a downward trend annually, and MIC50/MIC90 was higher for all kinds of antibacterial drugs. Antibiotics should be selected according to the results of drug sensitivity tests for the clinical treatment of CRKP infection, and positive and effective isolation measures should be taken to reduce the risk of nosocomial infection. This retrospective study was performed to analyze the clinical characteristics and the results of susceptibility testing of CRKP in Hebei Province, China, which has the advantage of broad coverage and large sample sizes. Hence, it can accurately reflect the current situation of CRKP infection in Hebei Province, provide epidemiological data for future clinical prevention and treatment, reduce the infection rate, and improve the success rate of the treatment of CRKP infection. The current study was performed using data of small number of samples, and the representativeness may be biased. In order to solve the above problem, we collected 6328 CRKP samples. A limitation of this study is that there is no genotyping of CRKP, and the link between the strains cannot be constructed. Therefore, advanced monitoring of the changes in CRKP resistance mechanism is required in the future. Considering that CRKP is highly resistant to commonly used antibiotics, attention should be paid to the training and assessment of clinicians on the rational use of antibiotics to reduce the infection rate of multidrug-resistant bacteria in the future.
Acknowledgments
We thank Clinical Laboratory, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Science and Peking Union Medical College and Clinical Laboratory, and Laboratory of Peking University People’s Hospital for administrative and technical support. Many thanks to graduate students Zhan Xuli, Yue Weiye, Si Liyan, Yang Zhicong, Li Jingqi and Wang Yuandi for their contributions to the preservation of strains. The author thanks Editage (www.editage.cn) for English language editing.
Funding Statement
This work was supported by National Key R & D Plan Research on Standardized Diagnosis, Investigation and Treatment Technology System of New and Major Foodborne Diseases (2017YFC1601502), Scientific Research Fund Project of Hebei Provincial Health Commission (20180843, 20190904, and 20210702) and Key R&D project of Zhangjiakou City (2121098D, 2121064D).
Data Sharing Statement
The data used and/or analyzed in this study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
The protocol has been reviewed by the Ethics Committee (IRB) of the First Affiliated Hospital of Hebei North University. As this was an observational study and strains cultured from residual samples were used in clinical diagnosis, the confidentiality of patient data is preserved and compliance with the Declaration of Helsinki is ensured. As the data did not affect patient care, the exemption criteria were met. After consulting the IRB of the First Affiliated Hospital of Hebei North University, a formal ethical review approval was obtained; written informed consent from the patient was not required (ethical approval no.: K2019147).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in relation to this study.
References
- 1.Zhu J, Wang T, Chen L, Du H. Virulence factors in hypervirulent Klebsiella pneumoniae. Front Microbiol. 2021;12:642484. doi: 10.3389/fmicb.2021.642484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin RM, Bachman MA. Colonization, Infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi: 10.3389/fcimb.2018.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Tella D, Tamburro M, Guerrizio G, Fanelli I, Sammarco ML, Ripabelli G. Molecular epidemiological insights into colistin-resistant and carbapenemases-producing clinical Klebsiella pneumoniae isolates. Infect Drug Resist. 2019;12:3783–3795. doi: 10.2147/IDR.S226416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guh AY, Limbago BM, Kallen AJ. Epidemiology and prevention of carbapenem-resistant Enterobacteriaceae in the United States. Expert Rev Anti Infect Ther. 2014;12(5):565–580. doi: 10.1586/14787210.2014.902306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18. doi: 10.1186/s12941-017-0191-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbadoro P, Dichiara A, Arsego D, et al. Spread of carbapenem-resistant Klebsiella pneumoniae in hub and spoke connected health-care networks: a case study from Italy. Microorganisms. 2019;8(1):37. doi: 10.3390/microorganisms8010037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diallo OO, Baron SA, Abat C, Colson P, Chaudet H, Rolain JM. Antibiotic resistance surveillance systems: a review. J Glob Antimicrob Resist. 2020;23:430–438. doi: 10.1016/j.jgar.2020.10.009 [DOI] [PubMed] [Google Scholar]
- 9.Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29(12):1099–1106. doi: 10.1086/592412 [DOI] [PubMed] [Google Scholar]
- 10.Grundmann H, Glasner C, Albiger B, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis. 2017;17(2):153–163. doi: 10.1016/S1473-3099(16)30257-2 [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Chan EW, Zhou H, Chen S. Prevalence and genetic characteristics of carbapenem-resistant Enterobacteriaceae strains in China. Lancet Infect Dis. 2017;17(3):256–257. doi: 10.1016/S1473-3099(17)30072-5 [DOI] [PubMed] [Google Scholar]
- 12.Yu L, Zhang J, Fu Y, et al. Synergetic effects of combined treatment of colistin with meropenem or amikacin on carbapenem-resistant Klebsiella pneumoniae in vitro. Front Cell Infect Microbiol. 2019;9:422. doi: 10.3389/fcimb.2019.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao W, Liu Y, Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist. 2020;23:174–180. doi: 10.1016/j.jgar.2020.09.004 [DOI] [PubMed] [Google Scholar]
- 14.Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microb Infect. 2020;26(1):124.e121–124.e124. doi: 10.1016/j.cmi.2019.08.020 [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto S, Singley CM, Hoover J, et al. Efficacy of cefiderocol against carbapenem-resistant gram-negative bacilli in immunocompetent-rat respiratory tract infection models recreating human plasma pharmacokinetics. Antimicrob Agents Chemother. 2017;61(9). doi: 10.1128/AAC.00700-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Li J, Hu T, et al. Five-year change of prevalence and risk factors for infection and mortality of carbapenem-resistant Klebsiella pneumoniae bloodstream infection in a tertiary hospital in North China. Antimicrob Resist Infect Control. 2020;9(1):79. doi: 10.1186/s13756-020-00728-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25(4):682–707. doi: 10.1128/CMR.05035-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 19.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bush K, Bradford PA. Interplay between β-lactamases and new β-lactamase inhibitors. Nat Rev Microbiol. 2019;17(5):295–306. doi: 10.1038/s41579-019-0159-8 [DOI] [PubMed] [Google Scholar]
- 21.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458, table of contents. doi: 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Liu C, Shen Z, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect. 2020;9(1):1771–1779. doi: 10.1080/22221751.2020.1799721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojas LJ, Salim M, Cober E, et al. Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin Infect Dis. 2017;64(6):711–718. doi: 10.1093/cid/ciw805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi: 10.1016/S1473-3099(13)70190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malinga NZZ, Shobo CO, Molechan C, Amoako DG, Zishiri OT, Bester LA. Molecular surveillance and dissemination of Klebsiella pneumoniae on frequently encountered surfaces in South African Public Hospitals. Microb Drug Resist. 2021. doi: 10.1089/mdr.2020.0546 [DOI] [PubMed] [Google Scholar]
- 26.Gao L, Lv Y, Li Y. Analysis of the drug resistance of carbapenem-resistant Klebsiella pneumoniae in the China antimicrobial resistance surveillance trial program, 2007–2018. Microb Drug Resist. 2020;26(8):944–950. doi: 10.1089/mdr.2019.0299 [DOI] [PubMed] [Google Scholar]
- 27.Park SO, Liu J, Furuya EY, Larson EL. Carbapenem-resistant Klebsiella pneumoniae infection in three New York City Hospitals Trended downwards from 2006 to 2014. Open Forum Infect Dis. 2016;3(4):ofw222. doi: 10.1093/ofid/ofw222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu F, Zhu D, Wang F, Wang M. Current status and trends of antibacterial resistance in China. Clin Infect Dis. 2018;67(suppl_2):S128–s134. doi: 10.1093/cid/ciy657 [DOI] [PubMed] [Google Scholar]
- 29.Chuang C, Su CF, Lin JC, et al. Does antimicrobial therapy affect mortality of patients with carbapenem-resistant Klebsiella pneumoniae bacteriuria? A nationwide multicenter study in Taiwan. Microorganisms. 2020;8(12):2035. doi: 10.3390/microorganisms8122035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Li Y, Song N, Chen Y. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection: a meta-analysis. J Glob Antimicrob Resist. 2020;21:306–313. doi: 10.1016/j.jgar.2019.09.006 [DOI] [PubMed] [Google Scholar]
- 31.El-Badawy MF, El-Far SW, Althobaiti SS, Abou-Elazm FI, Shohayeb MM. The First Egyptian report showing the co-existence of bla (NDM-25), bla (OXA-23), bla (OXA-181), and bla (GES-1) among carbapenem-resistant K. pneumoniae clinical isolates genotyped by BOX-PCR. Infect Drug Resist. 2020;13:1237–1250. doi: 10.2147/IDR.S244064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang R, Dong N, Huang Y, et al. Evolution of tigecycline- and colistin-resistant CRKP (carbapenem-resistant Klebsiella pneumoniae) in vivo and its persistence in the GI tract. Emerg Microbes Infect. 2018;7(1):127. doi: 10.1038/s41426-018-0129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horita N, Shibata Y, Watanabe H, Namkoong H, Kaneko T. Comparison of antipseudomonal β-lactams for febrile neutropenia empiric therapy: systematic review and network meta-analysis. Clin Microb Infect. 2017;23(10):723–729. doi: 10.1016/j.cmi.2017.03.024 [DOI] [PubMed] [Google Scholar]
- 34.Nimmich EB, Bookstaver PB, Kohn J, et al. Development of institutional guidelines for management of gram-negative bloodstream infections: incorporating local evidence. Hosp Pharm. 2017;52(10):691–697. doi: 10.1177/0018578717720506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchaim D, Chopra T, Pogue JM, et al. Outbreak of colistin-resistant, carbapenem-resistant Klebsiella pneumoniae in metropolitan Detroit, Michigan. Antimicrob Agents Chemother. 2011;55(2):593–599. doi: 10.1128/AAC.01020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev. 2018;31(2). doi: 10.1128/CMR.00079-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou SY, Wu D, Feng XH. Polymyxin monotherapy versus polymyxin-based combination therapy against carbapenem-resistant Klebsiella pneumoniae: a systematic review and meta-analysis. J Glob Antimicrob Resist. 2020;23:197–202. doi: 10.1016/j.jgar.2020.08.024 [DOI] [PubMed] [Google Scholar]
- 38.Peterson LR. A review of tigecycline–the first glycylcycline. Int J Antimicrob Agents. 2008;32(Suppl 4):S215–222. doi: 10.1016/S0924-8579(09)70005-6 [DOI] [PubMed] [Google Scholar]
- 39.Ni W, Han Y, Liu J, et al. Tigecycline treatment for carbapenem-resistant Enterobacteriaceae infections: a systematic review and meta-analysis. Medicine. 2016;95(11):e3126. doi: 10.1097/MD.0000000000003126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.An X, Xie W, Zheng X, Zhao L, Wu Y, Qu Y. [Detection and analysis of carbapenem-resistant Klebsiella pneumoniae resistance genes in five hospitals in Qingdao City]. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2020;32(2):145–149. Chinese. doi: 10.3760/cma.j.cn121430-20200110-00027 [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez-Padilla M, Torre-Cisneros J, Rivera-Espinar F, et al. Gentamicin therapy for sepsis due to carbapenem-resistant and colistin-resistant Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70(3):905–913. doi: 10.1093/jac/dku432 [DOI] [PubMed] [Google Scholar]
- 42.van Duin D, Cober E, Richter SS, et al. Impact of therapy and strain type on outcomes in urinary tract infections caused by carbapenem-resistant Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70(4):1203–1211. doi: 10.1093/jac/dku495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satlin MJ, Kubin CJ, Blumenthal JS, et al. Comparative effectiveness of aminoglycosides, polymyxin B, and tigecycline for clearance of carbapenem-resistant Klebsiella pneumoniae from urine. Antimicrob Agents Chemother. 2011;55(12):5893–5899. doi: 10.1128/AAC.00387-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He G, Huang J, Huang S, et al. Risk factors affecting clinical outcome in patients with carbapenem-resistant K. pneumoniae: a retrospective study. Med Sci Monitor. 2020;26:e925693. doi: 10.12659/MSM.925693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lombardi F, Gaia P, Valaperta R, et al. Emergence of carbapenem-resistant Klebsiella pneumoniae: progressive spread and four-year period of observation in a cardiac surgery division. Biomed Res Int. 2015;2015:871947. doi: 10.1155/2015/871947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su J, Li D, Guo Q, Guo Y, Zheng Y, Xu X. In vitro bactericidal activity of trimethoprim-sulfamethoxazole/colistin combination against carbapenem-resistant Klebsiella pneumoniae clinical isolates. Microb Drug Resist. 2019;25(2):152–156. doi: 10.1089/mdr.2018.0085 [DOI] [PubMed] [Google Scholar]
- 47.Gagliotti C, Giordani S, Ciccarese V, et al. Risk factors for colonization with carbapenemase-producing Klebsiella pneumoniae in hospital: a matched case-control study. Am J Infect Control. 2014;42(9):1006–1008. doi: 10.1016/j.ajic.2014.05.028 [DOI] [PubMed] [Google Scholar]
- 48.Lee SO, Kim NJ, Choi SH, et al. Risk factors for acquisition of imipenem-resistant Acinetobacter baumannii: a case-control study. Antimicrob Agents Chemother. 2004;48(1):224–228. doi: 10.1128/AAC.48.1.224-228.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi: 10.1093/femsre/fux013 [DOI] [PubMed] [Google Scholar]