Abstract
Background
A vast majority of the commercially available lateral flow immunoassay (LFIA) is used to detect SARS-CoV-2 antibodies qualitatively. Recently, a novel fluorescence-based lateral flow immunoassay (LFIA) test was developed for quantitative measurement of the total binding antibody units (BAUs) (BAU/mL) against SARS-CoV-2 spike protein receptor-binding domain (S-RBD).
Aim
This study aimed to evaluate the performance of the fluorescence LFIA FinecareTM 2019-nCoV S-RBD test along with its reader (Model No.: FS-113).
Methods
Plasma from 150 reverse trancriptase–PCR (RT-PCR)-confirmed positive individuals and 100 prepandemic samples were tested by FincareTM to access sensitivity and specificity. For qualitative and quantitative validation of the FinCareTM measurements, BAU/mL results of FinCareTM were compared with results of 2 reference assays: the surrogate virus-neutralizing test (sVNT, GenScript Biotech, USA) and the VIDAS®3 automated assay (BioMérieux, France).
Results
FinecareTM showed 92% sensitivity and 100% specificity compared with PCR. Cohen's Kappa statistic denoted moderate and excellent agreement with sVNT and VIDAS®3, with values being 0.557 (95% CI: 0.32–0.78) and 0.731 (95% CI: 0.51–0.95), respectively. A strong correlation was observed between FinecareTM/sVNT (r = 0.7, p < 0.0001) and FinecareTM/VIDAS®3 (r = 0.8, p < 0.0001).
Conclusion
FinecareTM is a reliable assay and can be used as a surrogate to assess binding and neutralizing antibody response after infection or vaccination, particularly in none or small laboratory settings.
Keywords: SARS-CoV-2, COVID-19, Serology, Lateral Flow Assay, Fluorescence immunoassay
1. Introduction
Soon after the emergence of SARS-CoV-2 in December 2019 and its declaration as a pandemic by the World Health Organization (WHO), the need for accurate, sensitive, and rapid detection of SARS-CoV-2 for the control and prevention of the disease was urgent (Theel et al., 2020). Several commercial COVID-19 test kits were developed in response to this urgent need for the detection of either nucleic acid or antibodies (Van Walle et al., 2021). Although reverse trancriptase–PCR (RT-PCR) is used as a gold standard test, it requires specialized conditions, expensive equipment, and qualified personnel for sampling and testing. These limitations pose a challenge in a pandemic setting, which requires rapid and reliable tests that can be used to screen populations (Dortet et al., 2021). Thus, serological testing, particularly point‐of‐care approaches such as lateral flow immunoassay (LFIA), provides a rapid, portable, and cost effective method that is complementary to RT-PCR. In addition, serological tests have important applications in public health; they can be used in epidemiological survey studies, detect past infections, and determine herd immunity in the community. Furthermore, serological tests have a vital role in convalescent plasma treatment and determining vaccine effectiveness (Nadoushan et al., 2020).
Conventional LFIA used in clinical settings for diagnosing SARS-CoV-2 infection are done manually within 15 minutes, and the results are interpreted qualitatively by the naked eye. As a result, errors due to manual operation and inadequate visual sensitivity interpretation can occur. Recently, Wondfo Biotech developed fluorescence-based lateral flow immunoassay (LFIA) to detect total anti-S-RBD binding antibodies (binding antibody unit [BAU]) against SARS-CoV-2. Finecare™ 2019-nCoV RBD Antibody Test is a fluorescence immunoassay that is carried out semiautomatically along with a small portable device. The combination of fluorescence and LFIA provides higher sensitivity, allows quantitative detection of antibodies, and is easily affordable and accessible in small clinical laboratories, research settings, or point-of-care testing after infection or vaccination.
To the best of our knowledge, there is only 1 report in the literature evaluating the rapid fluorescence‐based LFIA immunoassays, which aimed to validate the performance of rapid SARS‐CoV‐2 IgM and IgG test kits based on fluorescence immunochromatography (Kang et al., 2021). In this study, we aimed to evaluate the performance of a rapid semiautomated fluorescence-based LFIA. This assay is designated to provide qualitative and quantitative measurements of SARS‐CoV‐2 anti-S-RBD total antibodies. One of the biggest advantages of this assay over the commercially available LFIAs is that not only are the results quantitative but they are also converted to BAU/mL (instead of AU/mL) as recently recommended by WHO.
2. Materials and Methods
2.1. Study design and ethical approval
We evaluated the performance of FinecareTM 2019-nCOV Antibody test and its reader (Model No.: FS-113) (Guangzhou Wondfo Biotech Co., China) (from here on abbreviated as FinecareTM). The serum samples were collected from volunteer individuals between July 26 and September 9, 2020 as a nationwide survey substudy in Qatar (Al-Thani et al., 2020, Syed et al., 2021). The clinical symptoms of the samples varied between asymptomatic and symptomatic cases (S1 table). The project was approved by the institutional review board of Qatar University (QU-IRB 1492-E/21 and QU-IRB 1469-E/21). These samples were used in previous studies (Al-Jighefee et al., 2021, Yassine et al., 2021, Younes et al., 2021).
2.2. Sensitivity and specificity determination
The sensitivity of FinecareTM was determined using sera from 150 RT-PCR–confirmed individuals (7–>21 days). Qiagen extraction kit was used to extract RNA from nasopharyngeal swab specimens. Then, the SuperscriptIII One-Step RT-PCR kit was used to test for SARS-CoV-2 according to manufacturer's instruction (Cat No. 12594100, ThermoFisher, USA) (Yassine et al., 2021). Samples were tested using 2 sets of primers targeting the E gene and confirmed with 2 different sets of primers targeting the RdRp gene (Corman et al., 2020, Yassine et al., 2021). Complete descriptions for these participants are summarized in Table S1. The specificity was examined using 100 prepandemic plasma samples collected before 2019, which were used in previous studies (Nasrallah et al., 2018, Nasrallah et al., 2019, Smatti et al., 2020). The panel included plasma samples seropositive for (a) dengue virus (n = 26), (b) parvovirus B19 (n = 8), and (c) nonrespiratory viruses (n = 66).
2.3. Serological assays
2.3.1. Finecare™ 2019-nCoV RBD Antibody Test
Finecare™ 2019-nCoV RBD Antibody Test is a fluorescence immunoassay technology, specifically the sandwich immunodetection method. It uses fluorescently labeled SARS-CoV-2 S-RBD proteins to form immune complexes by binding to SARS-CoV-2 S-RBD antibodies present in the specimen. It is used along with the Finecare™ FIA Meters (Model No.: FS-113) for the quantitative detection of total antibodies against SARS-CoV-2 S-RBD (Co.). The test can be done using various specimens, including fingerstick whole blood, venipuncture blood, serum, or plasma. There are 2 test modes for Finecare™ FIA Meters: standard test mode and quick test mode. The difference is that samples are incubated inside the cartridge holder of Finecare™ FIA Meters in the standard mode, whereas the samples are incubated at room temperature in the quick mode. The test was done according to the manufacturer's instructions. In brief, 20 µL of plasma was added to the provided buffer tube and mixed for 45 seconds. Then, 75 µL was added to the test cartridge, incubated for 15 minutes at room temperature (RT), and then inserted in the test cartridge holder of Finecare™ FIA Meters holder for measurement on quick mode. Finecare™ FIA Meter displays the test result automatically on the screen. The results are given as relative fluorescence units (RFU, arbitrary units [AU]/mL). This test has a WHO international standardization factor to convert readings to from AU/mL to binding antibody units (BAU)/mL (1 AU/mL = 20 BAU/mL). Readings ≥1 AU/mL or ≥20 BAU/ml indicate positive results.
2.3.2. cPass GeneScript sVNT
Food and Drug Administration (FDA)-approved SARS-CoV-2 surrogate virus neutralization test (sVNT) developed by GeneScript is widly used in the literature as a reference assay to measure neutralizing antibodies (Cat. No. L00847-C, GenScript Biotech, USA) (Ismail et al., 2021, Meyer et al., 2020, Younes et al., 2021). This assay detects antibodies that inhibit the interaction between SARS-CoV-2 S-RBD–horseradish peroxidase (HRP) fusion protein and ACE2 that is coated in a 96-well plate (Meyer et al., 2020). The assay highly corelates with conventional pseudovirus neutralization tests (pVNTs, R2 = 0.84) and showed high specificity (99.9%) and sensitivity (100%) (Tan et al., 2020). The test was done according to the manufacturer's instructions. An inhibition value of ≥30% signal inhibition was considered positive and <30% signal inhibition was considered negative. The WHO international standardization factor for this assay was used to convert readings from percent inhibition to IU/mL by applying the recently published formula (Zhu et al., 2021).
2.3.3. BioMérieux VIDAS®3
VIDAS®3 SARS-CoV-2 IgG (REF 424114, BioMérieux, France) is a widely used automated CE-marked in vitro diagnostic (IVD) assay, which was granted the emergency use authorization (EUA) by FDA in early 2021 The assay principle is based on a 2-step enzyme immunoassay combined with an enzyme-linked fluorescent assay (ELFA) detection technique. The test uses a solid-phase receptacle (SPR) coated with SARS-CoV-2 S-RBD of the spike protein. The test was done according to the manufacturer's instructions. In brief, the test was calibrated with standard (S1), a positive control (C1), and a negative control (C2). Then, 100 µL of the sample was added to the test strip. The results are generated as relative fluorescence value (RFV) and automatically calculated by the instrument; according to the S1 standard and sample RFV, an index value (i) is obtained (where i = RFVsample/RFVS1). The test is interpreted as negative when i < 1.00 and positive when i ≥ 1.00 (Renard et al., 2021). All readings were standardized to BAU/mL by applying the WHO international standard (20.33 BAU/mL) for the VIDAS®3 SARS-CoV-2 IgG.
2.4. Statistical analysis
Using RT-PCR as the reference test, sensitivity, specificity, overall percent agreement, and Cohen's Kappa coefficient (κ) were calculated to assess the performance of FinecareTM. κ measures inter-rater reliability as well as the likelihood that an agreement will occur by chance (Ben-David, 2008, McHugh, 2012). κ ≤ 0 indicates no agreement, κ= 0.01–0.20 indicates poor agreement, κ=0.21–0.40 indicates fair agreement, κ= 0.41–0.60 indicates moderate agreement, κ= 0.61–0.80 indicates substantial agreement, and κ= 0.81–1.00 indicates almost perfect agreement (McHugh, 2012). Concordance analysis between FinecareTM and sVNT and VIDAS®3 were conducted. The concordance measures included overall, positive, and negative percent agreement and Cohen's Kappa statistic. The correlation between FinecareTM and each immunoassay was examined using Pearson's correlation coefficients (r) with 95% CI. A coefficient of 0–0.19 suggests very weak correlation, 0.2–0.39 suggests weak correlation, 0.40–0.59 suggests moderate correlation, 0.6–0.79 suggests strong correlation, and 0.8–1 suggests very strong correlation (Swinscow and Campbell, 2002). Receiving operating characteristic (ROC) curve and Youden's index (J) were used to assess the assay threshold (cutoff indexes), identify optimized ones, and measure the area under the curve (AUC). The relation between AUC and diagnostic accuracy is direct. The larger the AUC the more accurate a test can be considered in its overall performance. Statistically, an AUC of <0.5 suggests no discrimination, 0.7–0.8 is considered acceptable, 0.8–0.9 is considered excellent, and >0.9 is considered outstanding. To determine the optimal threshold for optimal sensitivity and specificity for FinecareTM, J was calculated using the following formula: J = max (sensitivity + specificity) – 1 (Fluss et al., 2005, Unal and medicine, 2017). All statistical analyses were performed using GraphPad Prism version 9.3.0.
3. Results
3.1. Diagnostic Performance of FinecareTM using RT-PCR as a reference test
The overall diagnostic performance of Finecare analyzer compared with RT-PCR is summarized in Table 1 . The sensitivity and specificity of FinecareTM were 92% (95% CI: 86.44%–95.80%) and 100% (95% CI: 96.38%–100.00%), respectively. The overall percent agreement with RT-PCR was 96.4% (95% CI: 93.2%–98.1%). Cohen's Kappa statistic with RT-PCR denoted an excellent agreement with κ coefficient of 0.881 (95% CI: 0.816–0.946).
Table 1.
Diagnostic assessment of Finecare for S-RBD total antibodies detection* RT-PCR was used as a reference test.
| Immunoassay | FinecareTM |
|---|---|
| Sensitivity, % (95% CI) | 92% (95% CI: 86.44%–95.80%) |
| Specificity, % (95% CI) | 100% (95% CI: 96.38%–100.00%) |
| Overall agreement, % (95% CI) | 96.4% (95% CI: 93.2%–98.1%) |
| Cohen's kappa coefficient, κ (95% CI) | 0.881 (95% CI: 0.816–0.946) |
| ROC curves optimized cutoff index | >10.70 |
| Sensitivity using optimized cutoff indexes, % (95% CI) | 94.67% (95%CI: 89.83%–97.27%) |
| Specificity using optimized cutoff indexes, % (95% CI) | 100% (95% CI: 96.34%–100%) |
Abbreviations: ROC, receiver operating characteristic; RT-PCR, reverse transcriptase PCR; S-RBD, SARS-CoV-2 spike protein receptor-binding domain.
3.2. Concordance assessment between FinecareTM test, sVNT, and VIDAS®3
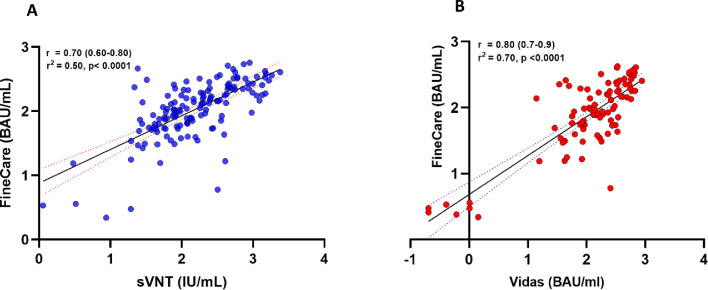
The tests’ concordance assessment is summarized in Table 2 . The overall percent agreement for sVNT and VIDAS®3 was 92% (95% CI: 86.5–95.4) and 94.4% (95% CI: 87.6–97.6), respectively. The positive percent agreement ranged from 95.1% (88.0–98.1) for Finecare versus VIDAS®3 to 97.7% (93.5–99.2) for FinecareTM versus sVNT. The negative percent agreement ranged from 50% (95% CI: 29–71.0) for FinecareTM versus sVNT to 88.9% (95% CI: 56.5–98.0) for FinecareTM versus VIDAS®3. Cohen's Kappa statistic denoted moderate to excellent agreement and ranged between 0.557 (95% CI: 0.32–0.78) for FinecareTM/sVNT test combination and 0.731 (95% CI: 0.51–0.95) for FinecareTM /VIDAS®3 test combination. Correlation analysis of the readings obtained from FinecareTM, sVNT, and VIDAS®3 is illustrated in Figure 1 . Both immunoassays showed strong correlation with FinecareTM. Pearson's correlation coefficients (r) ranged from 0.70 for FinecareTM/sVNT to 0.8 for FinecareTM/VIDAS®3.
Table 2.
Concordance assessment between the FinecareTM sVNT and VIDAS®3 tests
| Compared With | Overall Percent Agreement%(95% CI) | Positive Percent Agreement%(95% CI) | Negative Percent Agreement%(95% CI) | Cohen's Kappa Coefficientκ (95% CI) | |
|---|---|---|---|---|---|
| Finecare | sVNT | 92.0%(86.5–95.4) | 97.7%(93.5–99.2) | 50%(29–71.0) | 0.558(0.32–0.78) |
| VIDAS 3 | 94.4%(87.6–97.6) | 95.1%(88.0–98.1) | 88.9%(56.5–98.0) | 0.731(0.51–0.95) |
Abbreviations: sVNT, surrogate virus-neutralizing test.
Figure 1.
Correlation analysis of the assays standardized readings obtained by each immunoassay. (A) Correlation plot of FinecareTM with the sVNT. (B) Correlation plot of FinecareTM with VIDAS®3. Pearson's correlation coefficient (r) and p-value are indicated. Pearson's r of 0–0.19 is regarded as very weak, 0.2–0.39 as weak, 0.40–0.59 as moderate, 0.6–0.79 as strong, and 0.8–1 as very strong correlation, but these are rather arbitrary limits, and the context of the results should be considered. Data are presented for 150 RT-PCR–confirmed SARS-CoV-2–positive samples. BAU, binding antibody unit; RT-PCR, reverse transcriptase–PCR; sVNT, surrogate virus-neutralizing test.
3.3. Receiver Operating Characteristic (ROC) Curve Analysis
ROC curve analyses showed excellent performance for FinecareTM with an AUC of 0.9627 and p < 0.0001 (Figure 2 ). The optimized cutoff for detecting total antibodies against RBD was derived on the basis the ROC curves and the calculated J. The cutoff obtained was >10.70 compared with the manufacturer's cutoff ≥20. Applying this new cutoff showed improved sensitivity, from 92% to 94.67%, whereas the specificity remained 100% (Table 1).
Figure 2.
ROC curve for Finecare 2019-nCoV RBD Antibody Test. An AUC of 0.9–1.0 is considered excellent, 0.8–0.9 is considered very good, 0.7–0.8 is considered good, 0.6–0.7 is considered sufficient, 0.5–0.6 is considered bad, and <0.5 is considered not useful. AUC, area under the curve; ROC, receiver Operating Characteristic.
4. Discussion
This study validated the performance of Finecare™ 2019-nCoV RBD fluorescence immunoassay for detecting total antibodies against SARS-CoV2 S-RBD after infection. A panel of 150 samples collected from RT-PCR–confirmed individuals and 100 prepandemic sera were used to evaluate the assays’ performance. To our knowledge, this is the first study conducted to validate the fluorescence-LFIA-based Finecare 2019-nCoV RBD Antibody test, which marks the novelty of this research work.
Finecare test showed high sensitivity and specificity comparable to sVNT and VIDAS®3 and other immunoassays in the market. VIDAS®3 was reported to have a sensitivity and specificity of 88.3% and 98.4%, respectively (Younes et al., 2021). Similarly, sVNT reported a high specificity (99.9%) and sensitivity (95%–100%) (Tan et al., 2020). It is noteworthy that our chohort samples were collected from Qatar community volunteers; thus, the cases varied between symptomatic and asymptomatic. It was noticed that of the 12 negative samples by Finecare, 8 were asymptomatic. Earlier studies reported that severity of the case affects humoral response in patients (Wu et al., 2021, Zhao et al., 2020). Previously, we reported that the sensitivity of immunoassay was higher in symptomatic patients than in the asymptomatic patient group (Younes et al., 2021). In addition, the sensitivity of Fincare assay for estimating anti-S-RBD or neutralizing antibodies could be overestimated because the S-RBD antigen in this assay is designed to detect antibodies specific to the wild-type (Wuhan) but not to other SARS-CoV-2 variants. Some of these variants, such as Omicron, accumulate significant numbers of mutations in the S-RBD domain. These mutations might decrease the affinity of binding and neutralizing antibodies to S-RBD, which in turn affect the sensitivity of the assay.
We evaluated the degree of correlation between FinecareTM with sVNT and VIDAS®3. A strong correlation was obtained between FinecareTM and both assays (Figure 1); however, VIDAS®3 showed a slightly higher correlation. This is because both VIDAS®3 and FinecareTM detection methods are based on enzyme-linked fluorescent and target antibodies against the S-RBD domain. This is particularly important; the FinecareTM rapid antibody test could detect neutralizing antibodies and serve as a surrogate for the advanced automated immunoassays in clinical settings to measure the humoral immune response after vaccination or infection and research context. ROC curve analysis was also performed to determine the optimal cutoff indexes for Finecare (Figure 2). The new cutoff value showed improved sensitivity without affecting the specificity. However, the cutoff values could be adjusted depending on the clinical setting or research context. For instance, in high-prevalence settings, higher thresholds may be desirable for screening purposes, whereas lower cutoff may be helpful for diagnosis purposes (Ismail et al., 2021).
Our study had some limitations; most of our RT-PCR samples were collected from asymptomatic individuals (Table S1), which might have underestimated the assay's sensitivity. In addition, the control group did not include samples for other coronaviruses or influenza that might cross-react with SARS-CoV-2, which could have led to an overestimated specificity.
In conclusion, depending on characteristics of emerging variants, our data showed that Finecare 2019-RBD antibody test showed excellent performance in terms of sensitivity, specificity, and overall agreement, with RT-PCR as a reference test. In addition, in terms of correlation with the FDA-approved sVNT from GenScript and the automated analyzer VIDAS®3 from bioMérieux, FinecareTM immunoassay showed an outstanding performance in detecting total antibodies in serum samples against SARS-CoV-2. Thus, this assay could be reliable for the quantitative detection of antibodies in the vaccinated population and recovered patients.
Acknowledgments
Conflict of interest
GKN would like to declare that all test kits used in this study were provided as in-kind support for his laboratory to test seroprevalence of anti-SARS-CoV-2 and antibody response among vaccinated and infected individuals in Qatar.
Funding
This work was made possible by grant number UREP28-173-3-057 from the Qatar National Research Fund (a member of Qatar Foundation). The statements made herein are solely the responsibility of the authors.
Acknowledgment
We would like to thank Qatar National Research Fund (a member of Qatar Foundation) for funding this work.
Author contributions
Conceptualization: GKN, NL. Participant recruitment and demographic data collection: HQ. Laboratory testing: FMS, DWA, NY. Supervision: GKN. Data analysis: GKN, FMS, NY, NL. First draft writing: GKN, FMS, NL. Review and editing: LJA, GKN. Funding: GKN, LJA.
Ethical statement
The study was conducted according to the guidelines of the Declaration of Helsinki and was reviewed and approved by the institutional review board of Qatar University (QU-IRB 1492-E/21 and QU-IRB 1469-E/21). Informed consent was obtained from all subjects involved in the study before collecting samples.
Data availability
All data produced in this study are available upon reasonable request to the authors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.02.052.
Appendix. Supplementary materials
References
- Al-Jighefee HT, Yassine HM, Nasrallah GKJP. Evaluation of antibody response in symptomatic and asymptomatic COVID-19 patients and diagnostic assessment of new IgM/IgG ELISA kits. Pathogens. 2021;10(2):161. doi: 10.3390/pathogens10020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Thani MH, Farag E, Bertollini R, Al Romaihi HE, Abdeen S, Abdelkarim A, et al. Seroprevalence of SARS-CoV-2 infection in the craft and manual worker population of Qatar. medRxiv. 2020 doi: 10.1101/2020.11.24.20237719. [DOI] [Google Scholar]
- Ben-David AJESwA. Comparison of classification accuracy using Cohen's Weighted Kappa. Expert Systems with Applications. 2008;34(2):825–832. [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L, Ronat J-B, Vauloup-Fellous C, Langendorf C, Mendels D-A, Emeraud C, et al. Evaluating 10 commercially available SARS-CoV-2 rapid serological tests by use of the STARD (Standards for Reporting of Diagnostic Accuracy Studies) method. Journal of clinical microbiology. 2021;59(2):e02342. doi: 10.1128/JCM.02342-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluss R, Faraggi D, BJBJJoMMiB Reiser. Estimation of the Youden Index and its associated cutoff point. Journal of Mathematical Methods in Biosciences. 2005;47(4):458–472. doi: 10.1002/bimj.200410135. [DOI] [PubMed] [Google Scholar]
- Ismail A, Shurrab FM, HT Al-Jighefee, Al-Sadeq DW, Qotba H, Al-Shaar IA, et al. Can commercial automated immunoassays be utilized to predict neutralizing antibodies after SARS-CoV-2 infection? A comparative study between three different assays. Front Biosci. (Landmark Ed) 2021;26(7):198–206. doi: 10.52586/4934. Jul 30PMID: 34340267. [DOI] [PubMed] [Google Scholar]
- Kang K, Huang L, Ouyang C, Du J, Yang B, Chi Y, et al. Development, performance evaluation, and clinical application of a Rapid SARS-CoV-2 IgM and IgG Test Kit based on automated fluorescence immunoassay. Journal of Medical Virology. 2021;93(5):2838–2847. doi: 10.1002/jmv.26696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh MLJBm. Interrater reliability: the kappa statistic. Biochemia medica. 2012;22(3):276–282. [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Reimerink J, Torriani G, Brouwer F, Godeke G-J, Yerly S, et al. Validation and clinical evaluation of a SARS-CoV-2 surrogate virus neutralisation test (sVNT) Emerging microbes & infections. 2020;9(1):2394–2403. doi: 10.1080/22221751.2020.1835448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadoushan MJ, Ahmadi S, Nadoushan PJJIJoP. Serology testing for SARS-CoV-2: Benefits and challenges. Iranian Journal of Pathology. 2020;15(3):154. doi: 10.30699/IJP.2020.39841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah GK, Dargham SR, Mohammed LI, Abu-Raddad LJJJomv. Estimating seroprevalence of herpes simplex virus type 1 among different Middle East and North African male populations residing in Qatar. Journal of medical virology. 2018;90(1):184–190. doi: 10.1002/jmv.24916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah GK, Dargham SR, Sahara AS, Elsidiq MS. Abu-Raddad LJJJoCV. Performance of four diagnostic assays for detecting herpes simplex virus type 2 antibodies in the Middle East and North Africa. Journal of Clinical Virology. 2019;111:33–38. doi: 10.1016/j.jcv.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Renard N, Daniel S, Cayet N, Pecquet M, Raymond F, Pons S, et al. Performance Characteristics of the Vidas SARS-CoV-2 IgM and IgG Serological Assays. Journal of clinical microbiology. 2021;59(4):e02292. doi: 10.1128/JCM.02292-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smatti MK, Nasrallah GK, Al Thani AA, Yassine HMJV. Measuring influenza hemagglutinin (HA) stem-specific antibody-dependent cellular cytotoxicity (ADCC) in human sera using novel stabilized stem nanoparticle probes. Vaccine. 2020;38(4):815–821. doi: 10.1016/j.vaccine.2019.10.093. [DOI] [PubMed] [Google Scholar]
- Swinscow TDV, Campbell MJ. Statistics at square one: Bmj London, 2002.
- Syed MA, Al Nuaimi AS, Nasrallah GK, Althani AA, Yassine HM, Zainel AA, et al. Epidemiology of SARS-CoV2 in Qatar's primary care population aged 10 years and above. BMC infectious diseases. 2021;21(1):1–11. doi: 10.1186/s12879-021-06251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CW, Chia WN, Qin X, Liu P, Chen MI-C, Tiu C, et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nature biotechnology. 2020;38(9):1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Theel ES, Slev P, Wheeler S, Couturier MR, Wong SJ, KJJocm Kadkhoda. The role of antibody testing for SARS-CoV-2: is there one? Journal of clinical microbiology. 2020;58(8):e00797. doi: 10.1128/JCM.00797-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal IJC, medicine mmi. Defining an optimal cut-point value in ROC analysis: an alternative approach. Computational and mathematical methods in medicine. 2017. [DOI] [PMC free article] [PubMed]
- Van Walle I, Leitmeyer K, Broberg EKJE. Meta-analysis of the clinical performance of commercial SARS-CoV-2 nucleic acid and antibody tests up to 22 August 2020. MedRxiv. 2021;26(45) doi: 10.2807/1560-7917.ES.2021.26.45.2001675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liang B, Chen C, Wang H, Fang Y, Shen S, et al. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nature communications. 2021;12(1):1–9. doi: 10.1038/s41467-021-22034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HM, Al-Jighefee H, Al-Sadeq DW, Dargham SR, Younes SN, Shurrab F, et al. Performance evaluation of five ELISA kits for detecting anti-SARS-COV-2 IgG antibodies. International Journal of Infectious Diseases. 2021;102:181–187. doi: 10.1016/j.ijid.2020.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes S, Al-Jighefee H, Shurrab F, Al-Sadeq DW, Younes N, Dargham SR, et al. Diagnostic efficiency of three fully automated serology assays and their correlation with a novel surrogate virus neutralization test in symptomatic and asymptomatic SARS-COV-2 individuals. Microorganisms. 2021;9(2):245. doi: 10.3390/microorganisms9020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clinical infectious diseases. 2020;71(16):2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Althaus T, Tan CW, Costantini A, Chia WN, Chau NVV, et al. WHO international standard for SARS-CoV-2 antibodies to determine markers of protection. The Lancet Microbe. 2021 doi: 10.1016/S2666-5247(21)00307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data produced in this study are available upon reasonable request to the authors.