Abstract
Background:
Carbapenem-resistant Klebsiella pneumoniae (CRKP) are a global threat. We analyzed clinical outcomes of patients with CRKP in different countries and associated bacterial characteristics.
Methods:
The second Consortium on Resistance Against Carbapenems in Klebsiella and other Enterobacteriaceae (CRACKLE-2, ClinicalTrials.gov: NCT03646227) is a prospective, multicenter, cohort study. Patients were hospitalized in Argentina, Australia, Chile, China, Colombia, Lebanon, Singapore, and the US, with clinical cultures positive for CRKP in 2017–2018. Bacterial characteristics, all-cause mortality, and desirability of outcome ranking (DOOR) at 30 days were compared between patients from China, South America, and the US.
Findings:
Of 991 patients with CRKP, 502 (51%) were infected, and 489 (49%) colonized. CRKP within various regions were genetically similar. Infected patients from the US were more acutely ill as compared to patients from China or South America (median [IQR] Pitt bacteremia score 3 [2, 6] vs. 2 [0, 4], and 2 [0, 4]) and had more comorbid conditions (median [IQR] Charlson comorbidity index 3 [2, 5] vs. 1 [0, 3] and 1 [0, 2]). IPW-adjusted DOOR outcomes were similar in patients from China, South America, and the US. Unadjusted 30-day mortality after CRKP infection was 12% (29/246) in China, vs. 23% (30/130) in the US, vs. 28% (31/109) in South America. Adjusted mortality was similar between China and the US, but higher in South America (vs. China aOR 4·82, 95% CI 2·22–10·50, and vs. US aOR 3·34, 95% CI 1·50–7·47). Patients infected with strains carrying the O2v2 O locus were at lower risk for 30-day mortality vs. other O loci (aOR 0·34, 95% CI 0·15–0·78).
Interpretation:
The global CRKP epidemic has important regional differences in baseline characteristics of at-risk patients, bacterial characteristics, and clinical outcomes. Research findings from one region may not be generalizable to other regions.
Funding:
National Institutes of Health
Introduction
Antimicrobial resistance (AMR) is a global catastrophe that threatens progress in a variety of medical fields. Among multi-drug resistant organisms (MDRO), CRE are of specific concern given limited treatment options and potential for community spread. The World Health Organization (WHO) recognizes CRE among the highest priority pathogens.1 Within CRE, carbapenem-resistant K. pneumoniae (CRKP) are the most common bacterial species.2 In previous studies, the pooled mortality associated with CRKP infections has been estimated between 33% and 42%.3,4
In most regions of the world, Klebsiella pneumoniae carbapenemases (KPC) are the most common etiology of carbapenem resistance in CRKP.2,5,6 In China, carbapenem resistance in K. pneumoniae increased from 3% in 2005, to 21% in 2017; primarily mediated through KPC.6 In contrast, CRE cases in hospitalized patients in the US remained relatively stable from 2012 to 2017.3
From the second Consortium on Resistance against Carbapenems in Klebsiella and other Enterobacterales (CRACKLE-2), we recently reported on the molecular and clinical epidemiology of CRE in US hospitals between 2016 and mid-2017.2 Enrollment in CRACKLE-2 continued within and beyond the US. Here, we compared clinical characteristics and outcomes of this new international cohort of patients with CRKP. We also analyzed differences between bacterial isolates from eight countries around the world.
Methods
Patients
CRACKLE-2 was previously described.2 Briefly, CRACKLE-2 was a prospective, observational, international, multicenter study with consecutive enrollment of patients with CDC-defined CRE isolated in a clinical culture from any anatomic site during hospitalization. Here, from 71 hospitals in Argentina, Australia, Chile, China, Colombia, Lebanon, Singapore, and the United States, the first qualifying culture episode during the first admission for each unique patient enrolled during the study period (June 13, 2017 – November 30, 2018, details in the Supplementary Materials) with an available CRKP isolate was included. The study was approved by the Institutional Review Boards of all the health systems involved, with a waiver of consent.
Clinical data and outcomes
Clinical data were obtained from the health record. Standardized definitions for infection were applied centrally (see Supplementary Materials).2 Positive cultures that did not meet infection criteria were considered colonization.2 All included cultures were obtained as part of routine clinical care; cultures obtained for surveillance purposes only were excluded. At 90 days after discharge, data on post-hospitalization death and readmission were collected from the health record. The primary outcome was a desirability of outcome ranking (DOOR) analysis, as previously described.2 Briefly, this outcome assessed three deleterious events; lack of clinical response, prolonged hospitalization (hospitalization ≥ 30 days after first positive culture or readmission within 30 days), and adverse events (new renal failure and/or Clostridioides difficile infection), in addition to survival at 30 days after the index culture (see Supplementary Materials).2 The best outcome was defined as being alive without deleterious events. The worst outcome was death. Three levels in between these two extremes were: alive with 1, 2, and 3 deleterious events, respectively. As only 2 out of 502 patients with CRKP infection fell into the “alive with 3 events” level, that level was grouped post hoc with the “alive with 2 events” level for analysis, for 4 total levels of outcomes.
Microbiology
Determination of initial eligibility of CRKP isolates was performed in local microbiology laboratories (see Supplementary Materials). Meropenem and ertapenem susceptibility testing was later performed in the Antibacterial Resistance Leadership Group (ARLG) Laboratory Center laboratory using broth microdilution on all isolates that did not carry a carbapenemase gene. Carbapenemase genes were determined through whole genome sequencing of all included isolates. Patients were excluded from the study if their isolate did not harbor a carbapenemase gene and tested susceptible or intermediate to both carbapenems upon ARLG Laboratory Center testing.
Whole genome sequencing and genomic analysis
Sequencing was performed on all isolates as previously described at UTHealth (Illumina HiSeq 4000, NextSeq 2000, and MiSeq), Molecular Resource Facility, Rutgers (Rutgers; Illumina NextSeq500), University of El Bosque (Illumina MiSeq, HiSeq 4000 and NextSeq 2000), and Beijing Genomics Institute (Illumina, Hiseq X).2 Draft genomes were assembled using SPAdes v3.13.0.7 Klebsiella pneumoniae complex subspecies, MLST, wzi allele, capsule (K), O antigen (LPS) serotype, and acquired virulence loci were analyzed by Kleborate v2.0.1 and Kaptive v0.7.3.8–11 Resistance genes were called by AMRFinderPlus v3.9.8 and ARIBA v2.14.6.12,13 Core genome alignment was generated by snippy v4.6.0 (https://github.com/tseemann/snippy), and a maximum likelihood phylogenetic tree was constructed in RAxML v8.2.4.14 The genomes sequenced in this study were deposited in GenBank bioproject accession no. PRJNA658369. Further details are available in the Supplementary Materials.
Statistical analysis
Three regions were defined: South America (Argentina, Chile, and Colombia), the US, and China. Due to the limited sample size compared to other regions, patients from Australia, Lebanon, and Singapore were described but not included in comparative outcome analyses. Regions were compared using pairwise desirability of outcome ranking (DOOR) analyses.15 The following variables were used in inverse probability weighting (IPW): Origin (home vs. other), Charlson comorbidity index (CCI >3 vs. ≤3), age at culture, Pitt Bacteremia Score, and anatomical source (blood, respiratory, urine, other).16,17 The Pitt Bacteremia Score was previously validated for non-bacteremic infections.18 Bacterial risk factors (MLST type, yersiniabactin, colibactin, OmpK35/36, K locus, and O locus) for all-cause mortality were evaluated using multivariable logistic regression models. The same clinically relevant confounders used in the model to calculate IPW weights were included in all adjusted logistic regression models. To visualize all-cause mortality within 30 days of initial culture, Kaplan Meier curves with log-rank tests of unadjusted survival probability without censoring were created. Censoring was absent as, unless known to have died, patients were assumed to be alive at 30 days from initial cultureFurther details are available in the Supplementary Materials.
Role of the funding source
The sponsor of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
From 71 hospitals in eight countries, 991 unique patients were included (Table 1). Of these, 502 (51%, 95% CI 48–54) patients met criteria for CRKP infection, and the remaining 489 (49%, 95% CI 46–53) were colonized (Supplementary Table 1). A higher proportion of patients in South America (64%, 95% CI 57–71 [109/170]) were infected, as compared to China (51%, 95% CI 46–55 [246/485]) and the US (46%, 95% CI 40–52 [130/284]), p=0·0007. Patients in the US had more comorbidities (median CCI = 3, interquartile range [IQR] 1–5), as compared to China (median CCI = 1, IQR 0–2), and South America (median CCI = 1, IQR 0–3), p<0·0001. Notable specific comorbidities that were more common in the US included diabetes mellitus, chronic kidney disease, and dementia; absolute differences vs. China 24% (95% CI 18–31), 21% (95% CI 16–27), and 9% (95% CI 5–13), respectively, and absolute differences vs. South America 16% (95% CI 7–25), 15% (95% CI 8–22), and 8% (95% CI 4–13), respectively. In China, CRKP were most frequently isolated from respiratory cultures (62%, 95% CI 58–67 [302/485]), whereas urine was the most common single source in South America (44%, 95% CI 36–51 [74/170]) and the US (40%, 95% CI 35–46 [115/284]). Patients were hospitalized for a shorter duration prior to their first positive CRKP culture in the US (median 2 days from admission to culture; IQR 0–17) as compared to those in China and South America (median 8 days from admission to culture; IQR 2–18 in China; IQR 1–22 in South America). In 334/991 (34%, 95% CI 31–37) patients, another pathogen was isolated from the same source within 7 days of the index culture. Antibiotic treatment is summarized in Supplementary Table 2.
Table 1.
Patient Characteristics
| Characteristic | China | South America | U.S. | otherf | all infected patients | all | P g |
|---|---|---|---|---|---|---|---|
| n | 485 (49) | 170 (17) | 284 (29) | 52 (5) | 502 (51) | 991 | |
| age, median (IQR) | 60 (46, 69) | 63 (42, 73) | 63 (50, 73) | 67 (55, 76) | 62 (47, 71) | 62 (47, 72) | 0·029 |
| sex, male | 323 (67) | 102 (60) | 144 (51) | 29 (56) | 297 (59) | 598 (60) | <0·0001 |
| Charlson comorbidity index, median (IQR)a | 1 (0, 2) | 1 (0, 3) | 3 (1, 5) | 2 (1, 4) | 2 (0, 4) | 1 (0, 3) | <0·0001 |
| Pitt Bacteremia Score, median (IQR)b | 2 (0, 4) | 2 (0, 4) | 2 (1, 5) | 1 (0, 3) | 2 (0, 4) | 2 (0, 4) | 0·0002 |
| intensive care unitc | 263 (54) | 44 (26) | 101 (36) | 25 (48) | 202 (40) | 433 (44) | <0·0001 |
| time to positive culture, days, median (IQR)d | 8 (2, 18) | 8 (1, 22) | 2 (0, 17) | 15 (3, 34) | 8 (1, 22) | 7 (1, 19) | <0·0001 |
| admitted frome | <0·0001 | ||||||
| home | 171 (35) | 125 (74) | 127 (45) | 43 (83) | 250 (50) | 466 (47) | |
| hospital transfer | 310 (64) | 43 (25) | 50 (18) | 4 (8) | 194 (39) | 407 (41) | |
| long-term chronic care | 3 (1) | 0 | 80 (28) | 2 (4) | 44 (9) | 85 (9) | |
| long term acute care | 0 | 0 | 26 (9) | 2 (4) | 11 (2) | 28 (3) | |
| transferred from foreign country | 0 | 0 | 1 (0) | 1 (2) | 2 (0) | 2 (0) | |
| hospice | 1 (0) | 1 (0) | 0 | 0 | 1 (0) | 2 (0) | |
| culture | <0·0001 | ||||||
| blood: infection | 41 (8) | 34 (20) | 49 (17) | 6 (12) | 130 (26) | 130 (13) | |
| urine: infection | 30 (6) | 38 (22) | 41 (14) | 4 (8) | 113 (23) | 113 (11) | |
| urine: colonization | 32 (7) | 36 (21) | 74 (26) | 10 (19) | 152 (15) | ||
| respiratory: infection | 118 (24) | 5 (3) | 14 (5) | 1(2) | 138 (27) | 138 (14) | |
| respiratory: colonization | 184 (38) | 11 (6) | 50 (18) | 2 (4) | 247 (25) | ||
| wound: infection | 11 (2) | 13 (8) | 13 (5) | 0 | 37 (7) | 37 (4) | |
| wound: colonization | 5 (1) | 10 (6) | 23 (8) | 3 (6) | 41 (4) | ||
| intra-abdominal: infection | 46 (9) | 18 (11) | 13 (5) | 5 (10) | 82 (16) | 82 (8) | |
| other: infection | 0 | 1 (1) | 0 | 1 (2) | 2 (0) | 2 (0) | |
| other: colonization | 18 (4) | 4 (2) | 7 (2) | 20 (38) | 49 (5) |
All data is shown as n (%) unless otherwise specified.
Charlson comorbidity index. is a chronic comorbidity score with a range from 0 to 37, with higher scores indicating the presence of more comorbid conditions. A patient with a score of 3 could have three level 1 comorbid conditions (e.g. dementia, chronic pulmonary disease, and congestive heart failure), or one level 1 (e.g. dementia) and one level 2 comorbid condition (e.g. leukemia), or one level 3 condition (moderate or severe liver disease).29
Pitt bacteremia score is an acute severity of illness score. Higher scores indicate more severe illness. A patient with a score of 3 would have one level 1 marker (e.g. disoriented mental status) and one level 2 marker of acute illness (e.g. hypotension).18
Intensive care unit location on the day of first positive culture.
Time to first positive culture indicates the number of days from admission to the collection date of the index culture, with 0 indicating that the index culture was obtained on the day of admission.
For analysis purposes grouped as home/transferred from foreign country, long term acute care/hospital transfer, and long term chronic care/hospice. One person from South America had missing data for origin.
Australia, Lebanon, and Singapore.
p value comparing China, South America, and US, and distributions where applicable.
Based on whole genome sequence data, 97% (95% CI 96–98 [963/991]) of bacterial isolates were K. pneumoniae sensu stricto. Other K. pneumoniae complex species included K. variicola subsp. variicola (n=12), K. quasipneumoniae subsp. quasipneumoniae (n=8), and K. quasipneumoniae subsp. similipneumoniae (n=8). Carbapenemase genes were present in 90% (95% CI 88–92 [888/991]) of isolates (Table 2, and Supplementary Table 3); blaKPC was the most common (81%, 95% CI 79–84 [807/991]). In China, blaKPC-2 was the predominant carbapenemase gene (94%, 95% CI 91–96 [454/485]). In South America and the US, most CRKP carried blaKPC-2 (39%, 95% CI 32–46 [66/170] and 44%, 95% CI 38–49 [124/284]) or blaKPC-3 (30%, 95% CI 23–37 [51/170] and 37%, 95% CI 31–43 [105/284]). blaOXA-48-like genes were the most common family of carbapenemases in Lebanon (75%, 95% CI 56–94 [15/20]) and Singapore (94%, 95% CI 82–100 [15/16]). In Lebanon, six isolates carried both blaOXA-48 and blaNDM-5. ESBL genes were more common in isolates from China. blaCTX-M genes were found in 60% (95% CI 57–63 [598/991]) of isolates and were more common in China (81%, 95% CI 78–85 [395/485]) compared to South America (56%, 95% CI 48–63 [95/170]) or the US (27%, 95% CI 22–32 [77/284]). blaCTX-M-65 accounted for most blaCTX-M genes in China (Table 2); blaCTX-M-65 was only found in one isolate outside of China. In South America and the US, blaCTX-M-15 was the predominant blaCTX-M gene. ESBL blaSHV genes were also more common in China (45%, 95% CI 41–49 [218/485]) vs South America (11%, 95% CI 6–15 [18/170]) and the US (33%, 95% CI 27–38 [93/284]).
Table 2.
Bacterial Characteristics
| China (n=485) | South America (n=170) | U.S. (n=284) | Otherg (n=52) | all infected patients (n=502) | All (n=991) | ph | |
|---|---|---|---|---|---|---|---|
| Carbapenemases a | |||||||
| Carbapenemase(s) present | 473 (98) | 127 (75) | 249 (88) | 39 (75) | 443 (88) | 888 (90) | <0·0001 |
| bla KPC-2 | 454 (94) | 66 (39) | 124 (44) | 2 (4) | 324 (65) | 646 (65) | <0·0001 |
| bla KPC-3 | 0 | 51 (30) | 105 (37) | 0 | 78 (16) | 156 (16) | <0·0001 |
| other blaKPCb | 2 (0) | 0 | 3 (1) | 0 | 3 (1) | 5 (1) | 0·285 |
| bla NDM-1 | 8 (2) | 14 (8) | 6 (2) | 3 (6) | 16 (3) | 31 (3) | <0·0001 |
| other blaNDMc | 4 (1) | 0 | 0 | 9 (17) | 7 (1) | 13 (1) | 0·153 |
| bla OXA-48 | 0 | 0 | 7 (2) | 25 (48) | 10 (2) | 32 (3) | 0·0003 |
| other blaOXA-48-liked | 3 (1) | 1 (1) | 7 (2) | 5 (10) | 8 (2) | 16 (2) | 0·053 |
| othere | 4 (1) | 3 (2) | 0 | 1 (2) | 4 (1) | 8 (1) | 0·102 |
| No carbapenemase detected | 12 (2) | 43 (25) | 35 (12) | 13 (25) | 59 (12) | 103 (10) | |
| Extended spectrum β-lactamase | |||||||
| bla CTX-M | 395 (81) | 95 (56) | 77 (27) | 31 (60) | 302 (60) | 598 (60) | <0·0001 |
| bla CTX-M-15 | 81 (17) | 77 (45) | 75 (26) | 26 (50) | 121 (24) | 259 (26) | <0·0001 |
| bla CTX-M-65 | 300 (62) | 0 | 1 (0) | 0 | 151 (30) | 301 (30) | <0·0001 |
| bla SHV f | 218 (45) | 18 (11) | 93 (33) | 4 (8) | 155 (31) | 333 (34) | <0·0001 |
| bla TEM f | 0 | 3 (2) | 0 | 0 | 1 (0) | 3 (0) | 0·001 |
| bla AmpC | 51 (11) | 2 (1) | 5 (2) | 6 (12) | 31 (6) | 64 (6) | <0·0001 |
| Multi-locus sequence type | <0·0001 | ||||||
| ST11 | 379 (78) | 76 (45) | 16 (6) | 2 (4) | 250 (50) | 473 (48) | |
| ST258 | 0 | 13 (8) | 163 (57) | 1 (2) | 78 (16) | 177 (18) | |
| ST15 | 78 (16) | 1 (1) | 15 (5) | 2 (4) | 44 (9) | 96 (10) | |
| ST147 | 3 (1) | 2 (1) | 5 (2) | 15 (29) | 14 (3) | 25 (3) | |
| other | 25 (5) | 78 (46) | 85 (30) | 32 (62) | 116 (23) | 220 (22) | |
| K locus | <0·0001 | ||||||
| KL64 | 298 (61) | 3 (2) | 6 (2) | 1 (2) | 165 (33) | 308 (31) | |
| KL107 | 0 | 12 (7) | 96 (34) | 0 | 46 (9) | 108 (11) | |
| KL19 | 69 (14) | 0 | 2 (1) | 0 | 31 (6) | 71 (7) | |
| KL106 | 0 | 1 (1) | 57 (20) | 2 (4) | 29 (6) | 60 (6) | |
| KL47 | 58 (12) | 0 | 1 (0) | 0 | 29 (6) | 59 (6) | |
| KL105 | 2 (0) | 45 (26) | 3 (1) | 1 (2) | 34 (7) | 51 (5) | |
| other | 58 (12) | 109 (64) | 119 (42) | 48 (92) | 168 (33) | 334 (34) | |
| O locus | <0·0001 | ||||||
| O2v1 | 307 (63) | 5 (3) | 14 (5) | 7 (13) | 175 (35) | 333 (34) | |
| O2v2 | 6 (1) | 66 (39) | 171 (60) | 6 (12) | 119 (24) | 249 (25) | |
| other | 172 (35) | 99 (58) | 99 (35) | 39 (75) | 208 (41) | 409 (41) | |
| porin genes | |||||||
| OmpK35 mutation | 441 (91) | 72 (42) | 200 (70) | 13 (25) | 358 (71) | 726 (73) | <0·0001 |
| OmpK36 mutation | 433 (89) | 80 (47) | 89 (31) | 19 (37) | 316 (63) | 621 (63) | <0·0001 |
| Putative virulence genes | |||||||
| aerobactin | 299 (62) | 0 | 4 (1) | 8 (15) | 156 (31) | 311 (31) | <0·0001 |
| colibactin | 3 (1) | 13 (8) | 61 (21) | 3 (6) | 33 (7) | 80 (8) | <0·0001 |
| rmpA2 | 284 (59) | 0 | 1 (0) | 6 (12) | 148 (29%) | 291 (29) | <0·0001 |
| rmpADC | 185 (38) | 0 | 1 (0) | 2 (4) | 97 (19) | 188 (19) | <0·0001 |
| yersiniabactin | 460 (95) | 104 (61) | 119 (42) | 23 (44) | 357 (71) | 706 (71) | <0·0001 |
All data is shown as n (%), unless otherwise noted. WT wild-type; MT mutant.
Totals exceed 100%, as 17 isolates carried more than one carbapenemase gene.
Other blaKPC included blaKPC-12 (2), blaKPC-28 (1), blaKPC-31 (1), blaKPC-34 (1).
Other blaNDM included blaNDM-4 (2), blaNDM-5 (10), blaNDM-7 (1).
Other blaoxa-48-like included blaoxa-163 (1), blaoxa-181 (5), blaoxa-232 (10).
Other carbapenemases included blaVIM-2 (1), blaVIM-24 (2), blaIMP-1 (1), blaIMP-4 (4).
Limited to blaSHV and blaTEM genes that are considered extended spectrum β-lactamase genes, including blaSHV-12 (279), blaSHV-2A (32), blaSHV-110 (8), blaSHV-27 (3), blaSHV-2 (3), blaSHV-30 (2), blaSHV-32 (2), blaSHV-5 (2), blaSHV-100 (1), blaSHV-42 (1), and blaTEM-26 (3).
Australia, Lebanon, and Singapore.
Comparisons between China, South America, and the US.
Limited intra-country genetic variation was observed (Figure 1, and Interactive Figure available at http://arlg.med.unc.edu/crackle/). Multi-locus sequence types (MLST) were strongly associated with region. Strain type ST11 was predominantly found in in China (78%, 95% CI 75–82 [379/485]) and South America (45%, 95% CI 37–52 [76/170]). The ST11 strains in China mainly harbor K locus type (KL) 64 and 47, while South American ST11 isolates carry KL105 and KL39 (Table 2). South American ST11 isolates located at different phylogenetic clades as the Chinese ST11 KL64 and KL47 strains (Figure 1). ST11 KL64 and KL47 strains from China differ by an average of 22 core SNPs (range 1–53), while they differ with South American KL105 and KL39 strains by average of 53 (31–81) and 67 (54–93) core SNPs, respectively. In the US, 57% of CRKP isolates were ST258 strain types, harboring primarily KL107 and KL106. ST11 isolates from China were associated with four specific plasmid replicons (Figure 1).
Figure 1. Bacterial population structure.
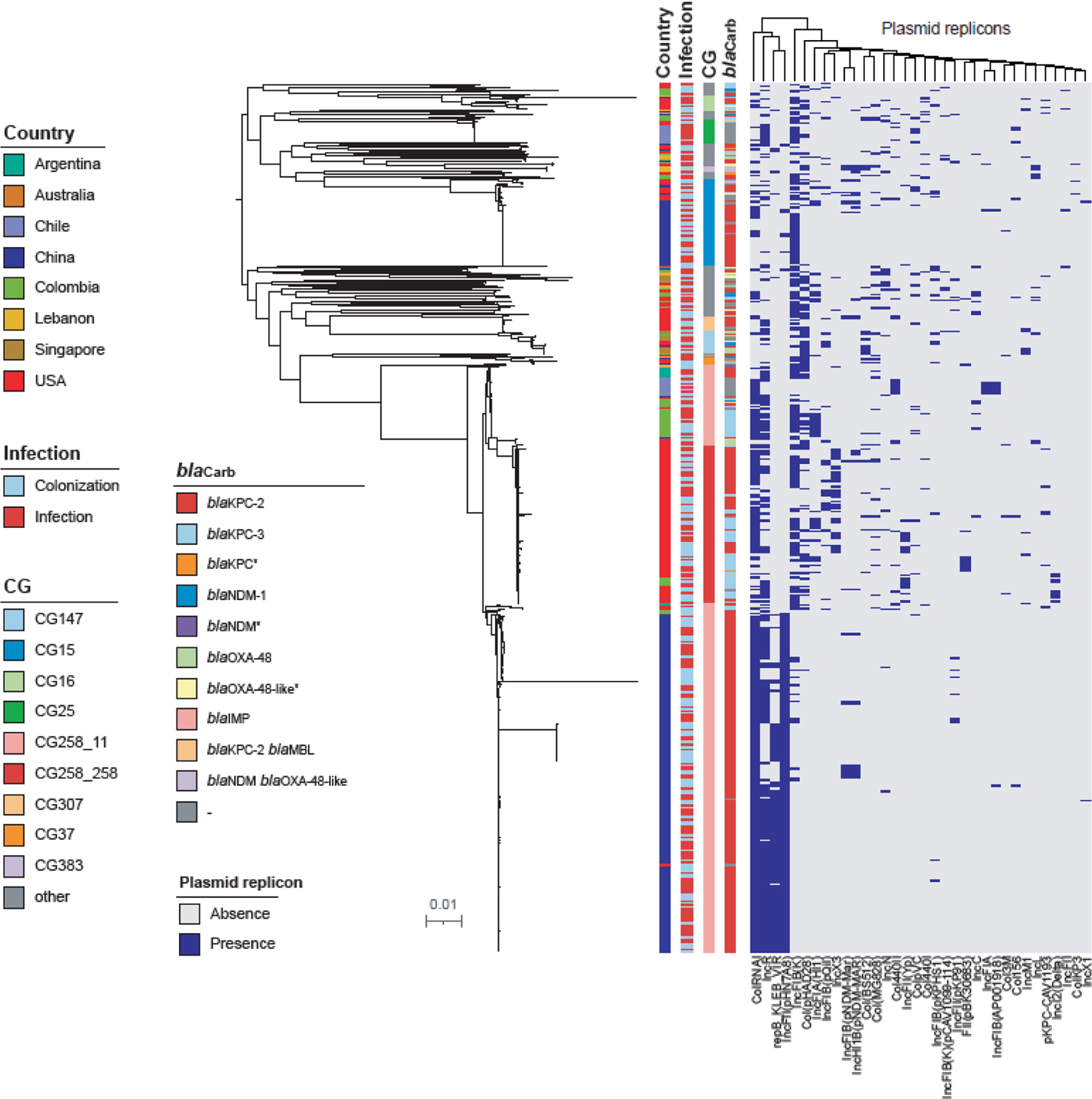
Maximum likelihood phylogenetic tree limited to K. pneumoniae sensu stricto is shown with corresponding metadata indicating country, clonal group, carbapenemase genes, and plasmid replicons present in each strain. An interactive figure for CRACKLE-2 data can be found here: http://arlg.med.unc.edu/crackle/.
The distribution of selected putative virulence genes is summarized in Table 2. Specific putative virulence genes more common in Chinese isolates included rmpA2 (59%, 95% CI 54–63 [284/485] vs. 0% [0/170] in South America vs. 0%, 95% CI 0–1 [1/284] in the US), rmpADC (38%, 95% CI 34–43 [185/485] vs. 0% [0/170] in South America vs. 0%, 95% CI 0–1 [1/284] in the US), yersiniabactin (95%, 95% CI 93–97 [460/485] vs. 61%, 95% CI 54–69 [104/170] in South America vs. 42%, 95% CI 36–48 [119/284] in the US), and aerobactin (62%, 95% CI 57–66 [299/485] vs. 0% [0/170] in South America vs. 1%, 95% CI 0–3 [4/284] in the US). In contrast, colibactin was less common in isolates from China (1%, 95% CI 0–1 [3/485] vs. 8%, 95% CI 4–12 [13/170] in South America vs. 21%, 95% CI 17–26 [61/284] in the US).
The distribution of unadjusted DOOR outcomes is shown in Figure 2. IPW-adjusted DOOR outcomes were similar in infected patients from China (n=246), South America (n=109), and the US (n=130). The IPW-adjusted DOOR probability estimates were: China vs. South America 53% (95% CI 42–65), US vs. China 50% (95% CI 41–61), and US vs. South America 53% (95% CI 41–66). The proportion of patients with infections who were alive at 30 days after first positive culture without deleterious events was lowest in China (31%, 95% CI 26–37 [77/246]), compared to 45% (95% CI 36–54 [49/109]) in South America, and 41% (95% CI 32–49 [53/130]) in the US. Among patients with infections who were alive at 30 days, lack of clinical response was observed in 62% (95% CI 55–68 [135/217]) of patients from China, compared to 28% (95% CI 18–38 [22/78]) in South America, and 24% (95% CI 16–32 [24/100]) in the US. For infected patients, length of stay was shorter in the US (median 19 days; IQR 8–46 days) compared to China (median 28 days; IQR 17–47 days), and South America (median 25 days, IQR 14–49 days), p=0·0055. Readmissions within 90 days in patients with infection, who were discharged alive were more common in the US (50%, 95% CI 40–60 [50/100]) vs. China (7%, 95% CI 3–10 [14/213]) and vs. South America (23%, 95% CI 13–33[17/74]), p<0·0001.
Figure 2. Distribution of desirability of outcome ranking (DOOR) outcomes in 485 patients with CRKP infections in China (n=246), South America (n=109), and the US (n=130).
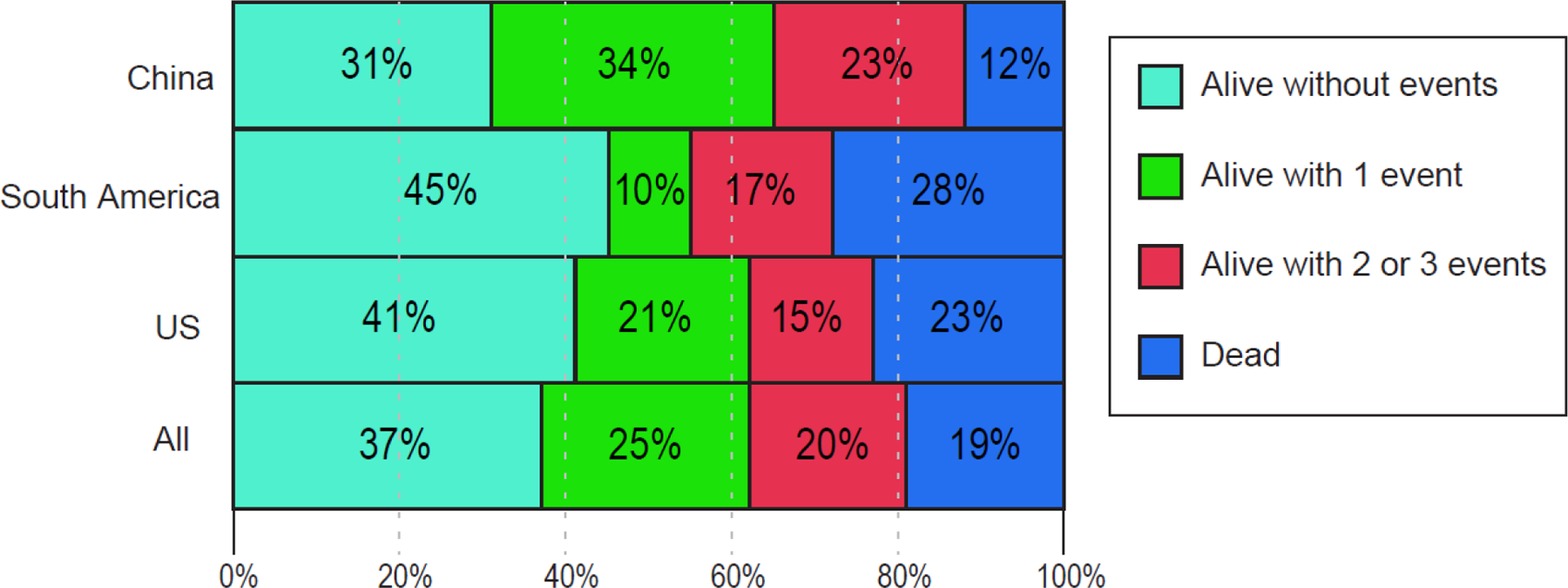
Outcome at 30 days after index culture is shown. The best outcome was defined as being alive without deleterious events. The worst outcome was death (red). Two levels in between are: alive with 1 (yellow), or with 2 or 3 events (orange), respectively. Events included were: lack of clinical response, prolonged hospitalization, and adverse events (defined in Supplementary Materials).
The all-cause 30- and 90-day mortality in patients with CRKP infections were 19% (95% CI 15–22 [93/502]), and 22% (95% CI 19–26 [111/501]), respectively (Figure 3.A.). In patients with CRKP infections (see Supplementary Table 4), CCI > 3 (aOR 2·93, 95% CI 1·53–5·61), and Pitt bacteremia score (aOR per point increase 1·45, 95% CI 1·31–1·60) were independently associated with increased 30-day mortality, whereas urinary infection was associated with lower 30-day mortality (vs bacteremia aOR 0·13, 95% CI 0·05–0·34, and vs respiratory infection aOR 0·26, 95% CI 0·09–0·78). In all patients with CRKP bacteremia, 30-day mortality was 34% (95% CI 26–42 [44/130]) overall, 24% (95% CI 11–38 [10/41]) in China, 56% (95% CI 39–73 [19/34]) in South America, and 31% (95% CI 18–44 [15/49]) in the US.
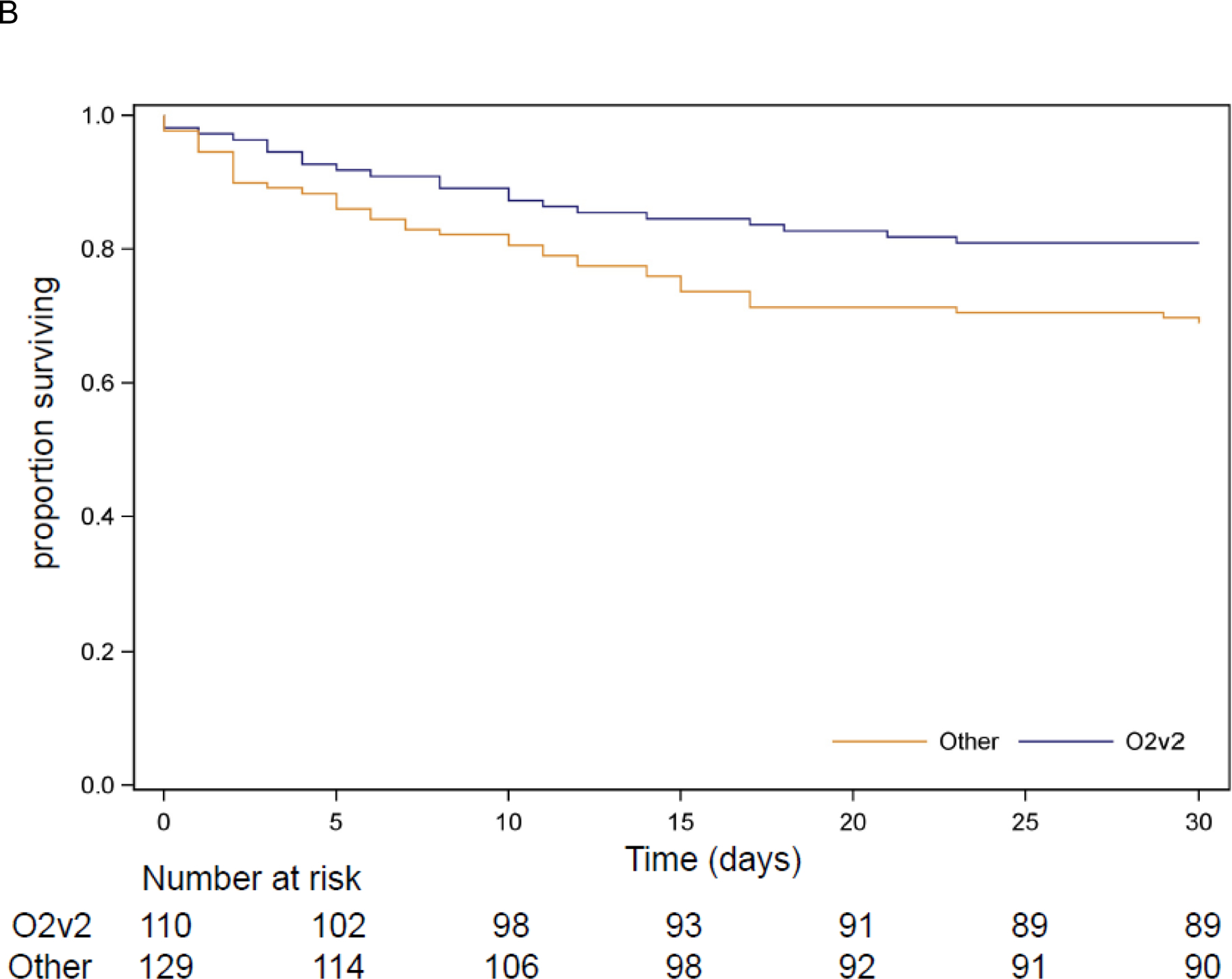
Figure 3. Kaplan-Meier Curves for all-cause mortality.
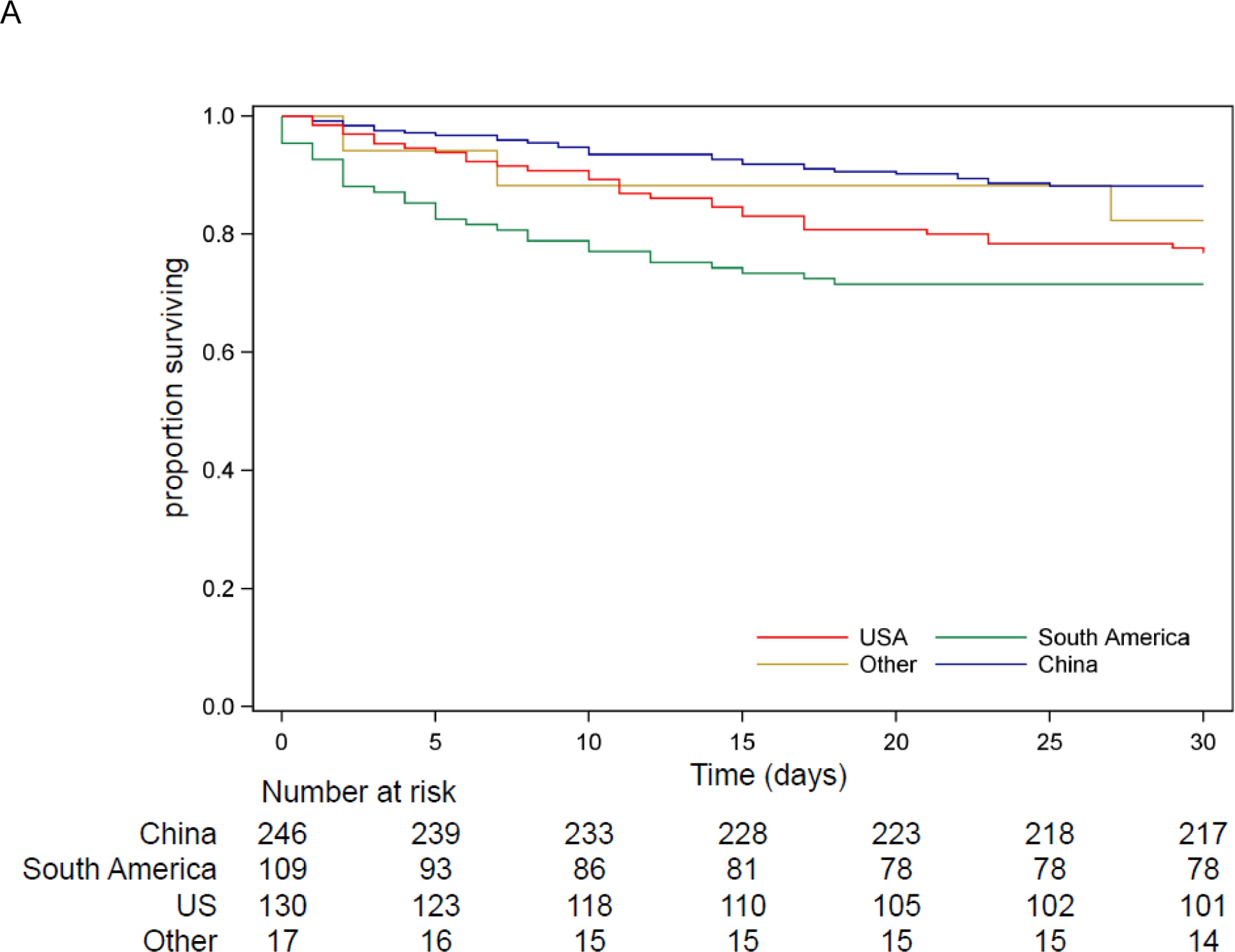
A. Survival for 502 patients with CRKP infection by region
B. Survival for 239 patients from South America and the US with CRKP infection by O-locus
In patients with CRKP infections, unadjusted all-cause 30- and 90-day mortality rates were lower in China (12%, 95% CI 8–16 [29/246] and 13%, 95% CI 9–17 [32/246]) compared to South America (28%, 95% CI 20–37 [31/109] and 35%, 95% CI 26–44 [38/109]) or the US (23%, 95% CI 16–30 [30/130] and 28%, 95% CI 20–36 [36/129]), p=0·0003 and p<0·0001, respectively (Figure 3.A.). After adjusting for age, origin, CCI, Pitt bacteremia score, and culture source, mortality was higher in South America (vs. China aOR 4·82, 95% CI 2·22–10·50, and vs. US aOR 3·34, 95% CI 1·50–7·47), with the mortality difference between the US and China no longer being significant (aOR 1·44, 95% CI 0·70–2·96).
No independent association was found between 30-day mortality and ST, capsule type, yersinibactin, colibactin, or OmpK35/36 porin genes (data not shown). There was insufficient diversity in the distribution of aerobactin, rmpA2, and rmpADC to allow for inclusion in adjusted models. The association between O locus and 30-day mortality was evaluated in US and South American patients, excluding patients from China where only 5 isolates had the O2v2 locus, and all five associated patients survived to 30 days. In patients from South America and the US, the O2v2 O locus was associated with lower 30-day mortality (Figure 3.B.) when compared to other O loci (aOR 0·34, 95% CI 0·15–0·78).
Discussion
In this large, multinational, contemporaneous, prospective cohort study, mortality rates were lower in China as compared to South America and the US. High levels of chronic comorbidities and severity of illness in US patients accounted for the observed mortality difference between China and the US. In patients with CRKP infections from South America, the odds of dying within 30 days was three to four-fold higher as compared to China and the US after adjusting for chronic and acute illness. The increased mortality rates in South America when compared to the US may be related to the limited availability of novel anti-CRE antibiotics such as ceftazidime-avibactam during the study period.19,20 In addition, factors that we did not evaluate in this study, such as healthcare system characteristics and resulting differences in healthcare-seeking behaviors may play a role in the observed increased mortality in South America. For example, inequality in access to care has been shown to be an important factor in mortality associated with COVID-19.21
Another possible explanation for variance in mortality rates is bacterial virulence. The genetically homogeneous CRKP from China may represent bacteria that are less likely to cause severe disease and/or detrimental host responses. However, most putative virulence genes we evaluated were more common in isolates from China. Among other bacterial factors that we evaluated, only the O2v2 O locus – uncommon in China – was associated with survival among patients in South America and the US. Genes encoded in the O locus are involved in the composition of bacterial lipopolysaccharide (LPS). LPS interacts with innate immune receptors including Toll-like receptor 4 to drive the host response to Gram-negative bacterial infection.22 Therefore, the observed association between the O locus and mortality has biologic plausibility. This should be considered hypothesis generating and requires confirmation in independent cohorts and animal studies.
The 30-day all-cause mortality in patients with CRKP infections was 19%. Previous estimates of mortality after CRKP infections are mostly based on retrospective studies. The prospective European cohort study EURECA has recently finished enrollment, with no data yet available.23 A meta-analysis of 62 studies published between 1999–2015 estimated pooled mortality after CRKP infections at 42%.4 The pooled mortality was 33% in KPC-producing CRKP infections in a more recent meta-analysis of 21 studies published from the US, Greece, Italy, Brazil, China, Spain and Israel during 2007–2018.3 The decreased mortality rate in our study may reflect the type of infections included, as well as advances in treatment of CRKP infections over time.
Overall, the 30-day mortality in patients with CRKP bacteremia in our cohort was 34%, lower than reported previously. In the INCREMENT study, the 30-day mortality rate was 43% in a retrospective cohort of patients with CPE bacteremia predominantly from hospitals in Europe in 2004–2013.24 Similarly, the 30-day mortality rate was 45% in patients with KPC-producing CRKP bacteremia in two Italian ICUs during 2015–2018.25 In South-Africa in 2015–2018, in-hospital mortality associated with CRE bacteremia was 38%.26
In DOOR analyses, no differences in the overall likelihood of a better outcome between infected patients in China, South America, and the US were seen. DOOR estimates are equally impacted by shifts between any of the ordinal outcomes. While mortality was lower in China, the proportion of patients without a clinical response was higher. This illustrates the complementary value of the DOOR outcome approach to determination of all-cause mortality. The underlying reasons for the observed discrepancy between clinical response rates and overall mortality remain to be determined.
While CRKP are a global threat, characteristics of the CRKP epidemic vary by region. For instance, in China, most CRKP were recovered from respiratory cultures. This likely reflects the microbiologic testing pattern in China. In the China Antimicrobial Surveillance Network (ChiNet), 40% of over 200,000 bacterial isolates were cultured from the respiratory tract, as compared to 19% from the urine and 15% from blood.27 Whole genome sequencing data revealed that the CRKP epidemic is also genetically different in different parts of the world. In China, a genetically homogeneous set of isolates was responsible for most CRKP infections. These isolates were characterized by ST11 strain type, KL64 capsule type, and carriage of blaKPC-2, blaCTX-M-65, and four common plasmid replicons uncommon in other regions. The emergence of a ST11-KL64-blaKPC-2 strain around 2016 was reported in a Chinese single-center retrospective study.28 Of note, that strain was not reported to carry blaCTX-M-65.28 These markers may be used to monitor clonal spread of other CRKP strains into China, or conversely of ST11 strains out of China. The predilection of specific CRKP strain types for certain regions was also observed in Europe in the EuSCAPE study.5 In this study, CRKP spread in Europe was determined to be primarily nosocomial in 2013–2014.5 These regional differences may have implications on whether studies evaluating diagnostics, treatment, and prognosis can be extrapolated from one region of the world to another.
Limitations
This study has several important limitations. First, the contribution of patients from Lebanon, Singapore and Australia was relatively small, which prohibited inclusion of these countries in comparative analyses. Similarly, while this study had a broad geographic reach, important areas of the world were not represented. Data from Europe will be forthcoming through the EURECA study.23 Several other regions with known high AMR incidence were similarly not included. Second, within each country, patients were predominantly enrolled from larger hospitals. The epidemiology of patients admitted to smaller community-based hospitals may be different. Furthermore, a relatively small number of hospitals participated per country, and for South America, only a limited number of countries participated. Our results should not be interpreted as being representative of the epidemiology of all types of hospitalized patients with CRKP in participating countries. Nonetheless, a strength of our approach is that we used a standardized, contemporaneous approach to include hospitalized patients with CRKP with a broad geographic area, combined with detailed clinical and bacterial genetic analyses. Third, the use of a waiver of consent results in consecutive enrollment without selection bias. A limitation of this approach is that only data that is collected as part of routine clinical practice can be recorded. These data may vary between regions. Fourth, we compared a large number of variables across three regions, which may raise issues with multiple comparisons. However, only two outcome variables were evaluated; the DOOR outcome, which was adjusted through IPW, and all-cause mortality for which we used multivariable logistic regression.
Conclusions
In summary, this evaluation of the CRKP epidemic in different parts of the world revealed more differences than similarities. Strain types, carbapenemase genes, and plasmid replicons were strongly associated with regions. Hospitalized patients with CRKP in China had lower unadjusted mortality rates, and lower levels of comorbidities and severity of illness than other regions. CRKP in China were also genetically homogeneous. In South America, mortality rates associated with CRKP infections were highest, even after adjusting for other contributing factors. These findings raise questions about the external generalizability of clinical studies on CRKP performed in any specific global region.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed and Google Scholar on February 1, 2021, using the terms “carbapenem resistant Klebsiella pneumoniae”, “carbapenemase”, “multi-locus sequence type” and “mortality”, without restrictions on language or dates. The results of these searches included primarily observational studies on epidemiology, risk factors, and outcomes associated with carbapenem-resistant Klebsiella pneumoniae (CRKP). Multi-locus sequence types (MLST) belonging to clonal group 258 are most common globally distributed CRKP. In two meta-analyses, the pooled mortality associated CRKP infections was estimated between 33% and 42%. Reported risk factors for mortality included host factors such as comorbid conditions, as well as treatment-related variables such as delayed time to effective antibiotics, and the use of polymyxin-based treatments as compared to treatment with novel β-lactam/β-lactamase antibiotics. Four randomized trials evaluated the use of novel antibiotics with activity against CRKP in pathogen-directed trials, which enrolled patients with a range of infections caused by various carbapenem-resistant Gram-negative bacteria. In three of these four trials, a numerical mortality benefit was associated with novel agents as compared to best available therapy.
Added value of this study
In this study, we used a prospective, standardized, contemporaneous approach to evaluate an all-inclusive cohort of hospitalized patients with CRKP in eight countries around the world. We showed that the genetic epidemiology of CRKP was unique within each specific region. All-cause 30-day mortality rates in patients with CRKP infections were lower in China (12%, 95% CI 8–16 [29/246]), as compared to the US (23%, 95% CI 16–30 [30/130]) and South America (28%, 95% CI 20–37 [31/109]).
After adjustment for source, Pitt bacteremia score, Charlson comorbidity index, origin, and age, mortality was higher in South America 4·82, 95% CI 2·22–10·50, and vs. US aOR 3·34, 95% CI 1·50–7·47), and the mortality difference between the US and China was no longer significant (aOR 1·44, 95% CI 0·70–2·96).
Implications of all the available evidence
Together with previous evidence, these results support the notion that the characteristics of the CRKP epidemic in various parts of the world are different. Strain types, plasmid replicons, and carbapenemase genes are strongly associated with regions. Clinical outcomes in patients with CRKP infections are driven by acute and chronic level of illness and vary according to region. These findings raise questions about the external generalizability of clinical studies on CRKP performed in any specific global region.
Acknowledgments:
The investigators would like to thank all the patients and their families, and also all contributing clinical microbiology laboratory personnel. The investigators would like to thank Dr. Sara Cosgrove and Dr. Antony Harris for their detailed review of an earlier version of this paper.
David van Duin, Michelle Earley, Lauren Komarow, Carol Hill, and Keri Baum had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Role of Funder:
This study is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIAID) under Award Number UM1AI104681. NIAID had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript nor the decision to submit the manuscript for publication, or to veto publication, or to control which journal the paper was submitted to.
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1AI104681. VGF was supported by a Mid-Career Mentoring Award 2K24-AI093969 from the National Institutes of Health. In addition, research reported in this publication was supported in part by the National Institutes of Health under Award Numbers R01AI143910 (DvD), R01AI090155 (BNK), R21AI135250 (BNK), R21AI117338 (LC), R01AI100560 (RAB), R01AI063517 (RAB), R01AI072219 (RAB), K24AI121296 (CAA), R01AI134637 (CAA), R01- AI148342–01 (CAA), P01AI152999 (CAA), T32GM086330 (CL), R01AI104895 (YD), R21AI123747 (YD), and R21AI135522 (YD). This study was supported in part by funds and/or facilities provided by the National Natural Science Foundation of China under Award numbers 81773785 (MW), 81991531 (MW), the Department of Veterans Affairs, Award Numbers 1I01BX001974 (RAB), VISN 10 (RAB), 5I01 BX003741 (BCF), UTHealth Searle Award (BH), UTHealth Presidential Collaborative Award (CAA), FONDECYT 1171805 *5I01 BX00374 (JMM), and the Government of Chile ANID Millennium Science Initiative, MICROB-R, NCN17_081 (JMM).
Conflict of Interest.
M.W., M.E., L.C., Y.Y., L.Z., S.S., E.C., L.L., S.S.K., H.G., K.M., L.K., S.R., S.H.M., C.M., J.R., M.V., C.H., R.A., K.B., B.F., B.N.K, R.A.B., report funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study. B.M.H. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study, and from NIH/NIAID K01AI148593–01 grant outside the submitted work. J.M. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; grants from Pfizer, grants from MSD, grants from bioMerieux, outside the submitted work. K.O. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; payments for educational events and presentations from Pfizer, MSD, AstraZeneca, and Farma de Colombia; and meeting support from Pfizerm MSD, and Gilead outside the submitted work. G.W. reports NIH/NIAID, and Allergan funding support. M.J.S. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; reports contracts payments, to his institution, with Merck, Allergan, BioFire Diagnostics, Affinity Biosensors; personal consulting fees from Achaogen and Shionogi; board participation with Spero Therapeutics outside the submitted work. S.V. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; personal fees from Merck Sharp and Dhome, and Biotoscana outside the submitted work. M.E.S. reports grants from National Institutes of Health (NIH), during the conduct of the study; speaker fees from Pfizer (Argentina), advisory board participation for Wockhardt, consultant for Basilea, outside the submitted work. C.L. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; salary support from the National Institute of General Medical Sciences of the National Institutes of Health outside the submitted work. D.P. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; grants/contracts with MERCK, Pfizer, Shionogi; consulting fees from Merck, Shionogi, and Qpex; payments and financial support from Sumitomo, Merck, Pfizer, bioMerieux, Shionogi; board participation for Symvivo outside the submitted work. S.E. reports grants from NIAID/NIH and Degruter (Editor in Chief: Statistical Communications in Infectious Diseases); Royalties from Taylor & Francis; Consulting fees from Genentech, AstraZeneca, Cardinal Health, Microbiotix, Stryker, Atricure, Roivant, Neovasc, Nobel Pharma, Horizon, International Drug Development Institute, SVB Leerink; payments from Analgesic, Anesthetic, and Addiction Clinical Trial Translations, Innovations, Opportunities, and Networks (ACTTION), Osaka University, and National Cerebral and Cardiovascular Center of Japan; Meeting support from FDA, Deming Conference on Applied Statistics, Clinical Trial Transformation Initiative, Council for International Organizations of Medical Sciences, Antimicrobial Resistance and Stewardship Conference; and Board member for NIH, Breast International Group, University of Pennsylvania, Duke University, Roche, Pfizer, Takeda, Novartis, Amgen, Teva, Vir, Shire, Alexion, Gilead, Tracon, Rakuten, Abbvie, Nuvelution, Clover, FHI Clinical, Lung Biotech, SAB Biopharm, Advantagene, American Statistical Association, Society for Clinical Trials, Frontier Science Foundation outside the submitted work. Y.D. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; grants and personal fees from Janssen, grants from Pfizer, grants and personal fees from MSD, grants from Shionogi, Astellas, Kanto Chemical, personal fees from Entasis, Janssen, VenatoRx, AstraZeneca, Gilead, FUJIFILM Toyama Chemical, bioMerieux, Meiji Seika Pharma, outside the submitted work. R.P. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; reports grants from Merck, ContraFect, TenNor Therapeutics Limited and Shionogi; is a consultant to Curetis, Specific Technologies, Next Gen Diagnostics, PathoQuest, Selux Diagnostics, 1928 Diagnostics, PhAST, and Qvella were monies are paid to Mayo Clinic; consultant to Netflix; has a patent on Bordetella pertussis/parapertussis PCR issued, a patent on a device/method for sonication with royalties paid by Samsung to Mayo Clinic, and a patent on an anti-biofilm substance issued; receives an editor’s stipend from IDSA, and honoraria from the NBME, Up-to-Date and the Infectious Diseases Board Review Course outside the submitted work. H.F.C. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; personal fees from Merck; stock ownership Moderna, outside the submitted work. V.G.F. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; personal fees from Novartis, Novadigm, Durata, Debiopharm, Genentech, Achaogen, Affinium, Medicines Co., Cerexa, Tetraphase, Trius, MedImmune, Bayer, Theravance, Basilea, Affinergy, Janssen, xBiotech, Contrafect, Regeneron, Basilea, Destiny, Amphliphi Biosciences. Integrated Biotherapeutics; C3J , grants from NIH, MedImmune, Cerexa/Forest/Actavis/Allergan, Pfizer, Advanced Liquid Logics, Theravance, Novartis, Cubist/Merck; Medical Biosurfaces; Locus; Affinergy; Contrafect; Karius; Genentech, Regeneron, Basilea, Janssen, from Green Cross, Cubist, Cerexa, Durata, Theravance; Debiopharm, other from UpToDate outside the submitted work; a patent for sepsis diagnostics pending. C.A.A reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; other financial support from University of Texas Health Science Center at Houston, grants from Merck, MeMed Diagnostics, Entasis Therapeutics, personal fees from UptoDate, American Society for Microbiology, Harrison Principles of Internal Medicine, Mandell Principles and Practice of Infectious Diseases - Chapters, personal fees from NIH/NIAID - Study section member, Grant reviewer, other from Infectious Diseases Society of America, IDSA, and American Society for Microbiology outside the submitted work. D.v.D. reports funding support from NIH/NIAID ARLG UM1AI104681 during the conduct of this study; is a consultant for Actavis, Tetraphase, Sanofi-Pasteur, MedImmune, Astellas, Merck, Allergan, T2Biosystems, Roche, Achaogen, Neumedicine, Shionogi, Pfizer, Entasis, QPex, Wellspring, Karius, and Utility. D.v.D. receives an editor’s stipend from BSAC. D.v.D. reports grants from NIH, outside the submitted work.
MDRO Investigators (listed by center alphabetically):
American University of Beirut Medical Center, Beirut, Lebanon: Souha S. Kanj; Case Western Reserve University School of Medicine, Cleveland, Ohio, United States of America: Robert A. Bonomo, Steven H. Marshall, Susan D. Rudin; Case Western Reserve University, Cleveland, Ohio, United States of America: Robert A. Salata; Centro de Educación Médica e Investigaciones Clínicas, Buenos Aires, Argentina: Martin Stryjewski, Valentina Di Castelnuovo; Centro Medico Imbanaco, Cali, Colombia: Jose Millan Oñate Gutierrez; Cleveland Clinic, Cleveland, Ohio, United States of America: Eric Cober; Duke Clinical Research Institute, Duke University, Durham, North Carolina, United States of America: Vance G. Fowler, Jr., Heather R. Cross, Carol Hill, Rebekka Arias, Keri Baum, Beth Evans; Duke University, Durham, North Carolina, United States of America: Deverick J. Anderson; E.S.E Hospital Universitario, San Jorge de Pereira, Pereira, Colombia: Karen Ordoñez; Emory University, Atlanta, Georgia, United States of America: Jesse T. Jacob; Hackensack Meridian Health, Nutley, New Jersey, United States of America: Barry N. Kreiswirth, Claudia Manca, Liang Chen, Samit Desai; Henry Ford Hospital, Detroit, Michigan, United States of America: Erica Herc; Hospital San Ignacio, Bogotá, Colombia: Sandra Valderrama; Huashan Hospital, Fudan University, Shanghai, China: Minggui Wang, Jianping Jiang, Yang, Jiachun Su; Instituto de Ciencias e Innovación en Medicina, Clínica Alemana, Universidad del Desarrollo, Santiago, Chile: Jose Munita, Maria Spencer; Mayo Clinic, Rochester, Minnesota, United States of America: Robin Patel, Kerryl Greenwood-Quaintance, Suzannah Schmidt-Malan; MedStar Washington Hospital Center, Washington, District of Columbia, United States of America: Glenn Wortmann; MetroHealth Medical Center, Cleveland, Ohio, United States of America: Robert C. Kalayjian; Montefiore Medical Center, Albert Einstein College of Medicine, Bronx, New York, United States of America: Greg Weston; North Shore University Hospital, Manhasset, New York, United States of America: Angela Kim; Ochsner Clinic Foundation, New Orleans, Louisiana, United States of America: Julia Garcia-Diaz; Organizacion Clinica General del Norte, Barranquilla, Atlántico, Colombia: Soraya Salcedo; Peking Union Medical College Hospital, Beijing, China: Fujie Zhang, Zhengyin Liu; Royal Brisbane and Women’s Hospital, Brisbane, Queensland, Australia: David L. Paterson; Shulan Hangzhou Hospital, Shulan Health, Hangzhou, Zhejiang Province, China: Hainv Gao; Sir Run Run Shaw Hospital, Zhejiang University-School of Medicine, Hangzhou, Zhejiang, China: Yunsong Yu; St. Vincent’s Hospital, Melbourne, Victoria, Australia: Mary Waters; Stony Brook University, Stony Brook, New York, United States of America: Bettina C. Fries; SUNY Downstate Medical Center, New York, New York, United States of America: Brandon Eilertson; Tan Tock Seng Hospital, Singapore: Kalisvar Marimuthu, Paul Ananth Tambyah, Nares Smitasin, Kean Lee Chew, Oon Tek Ng, Partha Pratim De; The Alfred Hospital, Melbourne, Victoria, Australia: Anton Peleg; The First Affiliated Hospital of Medical School of Zhejiang University, Hangzhou, Zhejiang, China: Lanjuan Li; The George Washington University, Washington, District of Columbia, United States of America: Scott R. Evans. Michelle Earley, Lauren Komarow; Universidad El Bosque, Bogotá, Colombia: Jinnethe Reyes, Maroa Virginia Villegas Botero, Lorena Diaz; University of Alabama at Birmingham, Birmingham, Alabama, United States of America: Todd McCarty; University of California San Francisco, San Francisco, California, United States of America: Henry F. Chambers; University of California, Los Angeles, California, United States of America: Omai B. Garner; University of Miami Miller School of Medicine and Jackson Health System, Miami, Florida, United States of America: Lilian M. Abbo; University of Michigan, Ann Arbor, Michigan, United States of America: Keith S. Kaye; University of North Carolina, Chapel Hill, North Carolina, United States of America: David van Duin, Courtney Lauterbach; University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, United States of America: Yohei Doi; University of Southern California, Los Angeles, California, United States of America: Darren Wong; University of Texas Health Science Center at Houston, Houston, Texas, United States of America: Cesar A. Arias, Blake Hanson, An Q. Dinh; Wayne State University, Detroit, Michigan, United States of America: Sorabh Dhar; Weill Cornell Medicine, New York-Presbyterian Hospital, New York, New York, United States of America: Michael J. Satlin.
| MDRO Investigators (listed by last name alphabetically) | ||
|---|---|---|
| First Name | Middle Initial | Last Name |
| Lilian | M. | Abbo |
| Deverick | J. | Anderson |
| Rebekka | Arias | |
| Cesar A. | A. | Arias |
| Keri | Baum | |
| Robert | A. | Bonomo |
| Henry | F. | Chambers |
| Liang | Chen | |
| Kean Lee | Chew | |
| Eric | Cober | |
| Heather | R. | Cross |
| Partha Pratim | De | |
| Samit | Desai | |
| Sorabh | Dhar | |
| Valentina | Di Castelnuovo | |
| Lorena | Diaz | |
| AN | Q. | Dinh |
| Yohei | Doi | |
| Michelle | Earley | |
| Brandon | Eilertson | |
| Beth | Evans | |
| Scott | Evans | |
| Vance | G. | Fowler, Jr. |
| Bettina | C. | Fries |
| Hainv | Gao | |
| Julia | Garcia-Diaz | |
| Omai | B. | Garner |
| Kerryl | Greenwood-Quaintance | |
| Blake | Hanson | |
| Erica | Herc | |
| Carol | Hill | |
| Jesse | T. | Jacob |
| Jianping | Jiang | |
| Robert | C. | Kalayjian |
| Souha | S. | Kanj |
| Keith | S. | Kaye |
| Angela | Kim | |
| Lauren | Komarow | |
| Barry | N. | Kreiswirth |
| Courtney | Lauterbach | |
| Lanjuan | Li | |
| Zhengyin | Liu | |
| Claudia | Manca | |
| Kalisvar | Marimuthu | |
| Steven | H. | Marshall |
| Todd | McCarty | |
| Jose | Munita | |
| Oon Tek | Ng | |
| Jose Millan | Oñate Gutierrez | |
| Karen | Ordoñez | |
| Robin | Patel | |
| David | L. | Paterson |
| Anton | Peleg | |
| Jinnethe | Reyes | |
| Susan | D. | Rudin |
| Robert | A. | Salata |
| Soraya | Salcedo | |
| Michael | J. | Satlin |
| Suzannah | Schmidt-Malan | |
| Nares | Smitasin | |
| Maria | Spencer | |
| Martin | Stryjewski | |
| Jiachun | Su | |
| Paul Ananth | Tambyah | |
| Sandra | Valderrama | |
| David | van Duin | |
| Maria Virginia | Villegas Botero | |
| Minggui | Wang | |
| Mary | Waters | |
| Greg | Weston | |
| Darren | Wong | |
| Glenn | Wortmann | |
| Yang | Yang | |
| Yunsong | Yu | |
| Fujie | Zhang | |
Footnotes
Publisher's Disclaimer: Disclaimer. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs.
References:
- 1.Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018; 18(3): 318–27. [DOI] [PubMed] [Google Scholar]
- 2.van Duin D, Arias CA, Komarow L, et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis 2020; 20(6): 731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agyeman AA, Bergen PJ, Rao GG, Nation RL, Landersdorfer CB. A systematic review and meta-analysis of treatment outcomes following antibiotic therapy among patients with carbapenem-resistant Klebsiella pneumoniae infections. Int J Antimicrob Agents 2020; 55(1): 105833. [DOI] [PubMed] [Google Scholar]
- 4.Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob 2017; 16(1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 2019; 4(11): 1919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu F, Zhu D, Wang F, Wang M. Current Status and Trends of Antibacterial Resistance in China. Clin Infect Dis 2018; 67(S2): S128–34. [DOI] [PubMed] [Google Scholar]
- 7.Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of computational biology: a journal of computational molecular cell biology 2012; 19(5): 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bialek-Davenet S, Criscuolo A, Ailloud F, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 2014; 20(11): 1812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. Genomic surveillance framework and global population structure for Klebsiella pneumoniae. BioRxiv 2021; 10.1101/2020.12.14.422303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyres KL, Wick RR, Gorrie C, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2016; 2(12): e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wick RR, Heinz E, Holt KE, Wyres KL. Kaptive Web: User-Friendly Capsule and Lipopolysaccharide Serotype Prediction for Klebsiella Genomes. J Clin Microbiol 2018; 56(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldgarden M, Brover V, Haft DH, et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob Agents Chemother 2019; 63(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt M, Mather AE, Sanchez-Buso L, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 2017; 3(10): e000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamatakis A RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30(9): 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans SR, Rubin D, Follmann D, et al. Desirability of Outcome Ranking (DOOR) and Response Adjusted for Duration of Antibiotic Risk (RADAR). Clin Infect Dis 2015; 61(5): 800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robins JM, Hernan MA, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology 2000; 11(5): 550–60. [DOI] [PubMed] [Google Scholar]
- 17.Hernan MA, Brumback B, Robins JM. Marginal structural models to estimate the causal effect of zidovudine on the survival of HIV-positive men. Epidemiology 2000; 11(5): 561–70. [DOI] [PubMed] [Google Scholar]
- 18.Henderson H, Luterbach CL, Cober E, et al. The Pitt Bacteremia Score Predicts Mortality in Non-Bacteremic Infections. Clin Infect Dis 2020; 70(9): 1826–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Duin D, Lok JJ, Earley M, et al. Colistin Versus Ceftazidime-Avibactam in the Treatment of Infections Due to Carbapenem-Resistant Enterobacteriaceae. Clin Infect Dis 2018; 66(2): 163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shields RK, Nguyen MH, Chen L, et al. Ceftazidime-Avibactam Is Superior to Other Treatment Regimens against Carbapenem-Resistant Klebsiella pneumoniae Bacteremia. Antimicrob Agents Chemother 2017; 61(8): e00883–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mena GE, Martinez PP, Mahmud AS, Marquet PA, Buckee CO, Santillana M. Socioeconomic status determines COVID-19 incidence and related mortality in Santiago, Chile. Science 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosadini CV, Kagan JC. Early innate immune responses to bacterial LPS. Curr Opin Immunol 2017; 44: 14–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez-Gutierrez B, Sojo-Dorado J, Bravo-Ferrer J, et al. EUropean prospective cohort study on Enterobacteriaceae showing REsistance to CArbapenems (EURECA): a protocol of a European multicentre observational study. BMJ open 2017; 7(4): e015365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutierrez-Gutierrez B, Salamanca E, de Cueto M, et al. Effect of appropriate combination therapy on mortality of patients with bloodstream infections due to carbapenemaseproducing Enterobacteriaceae (INCREMENT): a retrospective cohort study. Lancet Infect Dis 2017; 17(7): 726–34. [DOI] [PubMed] [Google Scholar]
- 25.Falcone M, Bassetti M, Tiseo G, et al. Time to appropriate antibiotic therapy is a predictor of outcome in patients with bloodstream infection caused by KPC-producing Klebsiella pneumoniae. Crit Care 2020; 24(1): 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perovic O, Ismail H, Quan V, et al. Carbapenem-resistant Enterobacteriaceae in patients with bacteraemia at tertiary hospitals in South Africa, 2015 to 2018. Eur J Clin Microbiol Infect Dis 2020; 39(7): 1287–94. [DOI] [PubMed] [Google Scholar]
- 27.Hu F, Guo Y, Yang Y, et al. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur J Clin Microbiol Infect Dis 2019; 38(12): 2275–81. [DOI] [PubMed] [Google Scholar]
- 28.Zhou K, Xiao T, David S, et al. Novel Subclone of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 11 with Enhanced Virulence and Transmissibility, China. Emerg Infect Dis 2020; 26(2): 289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40(5): 373–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


