Abstract
The fate-mapping mouse has become an essential tool in the immunologist’s toolbox. Although traditionally used by developmental biologists to trace the origins of cells, immunologists are turning to fate-mapping to better understand the development and function of immune cells. Thus, an expansion in the variety of fate-mapping mouse models has occurred to answer fundamental questions about the immune system. These models are also being combined with new genetic tools to study cancer, infection, and autoimmunity. In this review, we summarize different types of fate-mapping mice and describe emerging technologies that may allow immunologists to leverage this valuable tool and expand our functional knowledge of the immune system.
Keywords: lineage tracing, mouse models, fate mapping, Cre-loxP, immune development
The origins of fate-mapping mice in immunology
Immunologists have long desired the ability to track individual immune cells within an animal to better understand their role in the immune system. Historically, adoptive cell transfers [1] or bone marrow chimeras [2-4] were relied upon to track the fates of donor cells in congenically marked recipients. Other methods, such as deuterated water [5,6] or glucose, were used to label certain populations of immune cells to understand how these are produced and maintained in vivo. However, a major limitation of these experimental approaches is that they do not allow the study of distinct populations of immune cells in a physiological manner. As a result, immunologists recently turned their attention to fate-mapping mice to genetically mark and track specific cells of interest in situ.
Fate-mapping mice originated in the developmental biology field, used to understand how cells of different developmental origins contributed to the assembly of organs and tissues. In general, the approach involved using Cre recombinase to drive a fluorescent protein that was continually expressed in a specific lineage of cells. Later, geneticists developed inducible fate mappers so that the expression of a marker in a certain lineage of cells could be temporally controlled by the researcher. Cell marking using inducible fate-mapping mice was a significant advance in the field because it was more precise, highly reproducible, and long lasting. Thus, it was not long before immunologists took advantage of this robust system to study immune cell lineages. Today, fate-mapping mice are used to study a wide range of functions in many immune cell types.
In this review, we focus on the application of murine models of fate-mapping in immunological studies. Unless otherwise noted, all highlighted studies were conducted in mice. We start with a brief primer on the technical aspects of genetic fate-mapping; specifically, models using inducible systems. We then synthesize recent discoveries in specific cell types and immune functions, as well as in the context of disease, and note opportunities for new frontiers. Lastly, we consider how to streamline workflows when incorporating emerging technologies, and finally highlight the strengths of lineage tracing systems and important caveats when designing experiments.
Lineage tracing in different immune populations
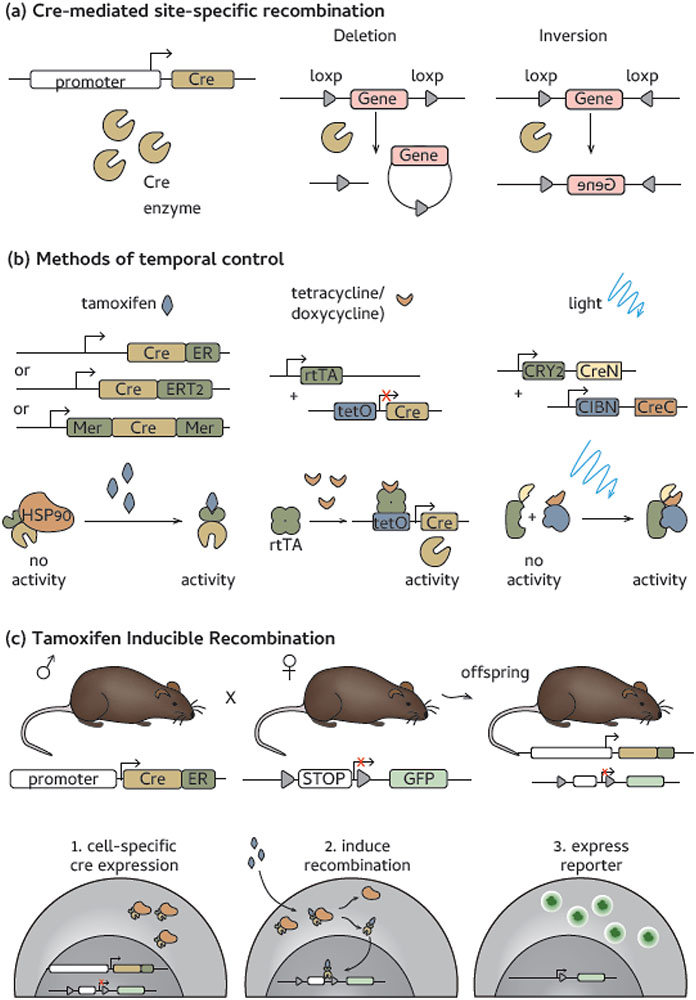
Lineage tracing is the ability to track a cell from the point of its creation. The primary method for lineage tracing is to use a temporally controlled Cre recombinase (see Box 1) under a relevant promoter combined with a floxed-stop reporter expressed in a ubiquitously expressed permissive locus, such as Rosa26 (Figure 1) [7], Temporal control can be achieved, for instance, through the expression of a Cre-estrogen receptor (ER)-ligand binding domain fusion protein which is sequestered in the cytoplasm by HSP90 [8,9]. When synthetic estrogen (tamoxifen) is administered, it is bound by the ER fusion protein, resulting in the release of the Cre-ER fusion protein from HSP90. The Cre-ER fusion protein then translocates to the nucleus, where it can mediate DNA recombination (Figure 1) [9]. Currently, most mouse models use one of two improved fusion proteins, CreERT2 [10] or MerCreMer [11], that have beenengineered to increase sensitivity to tamoxifen and reduce cytotoxic effects.
Box 1. Marking a moment in space and time.
Successful fate-mapping requires tracking the trajectory of a cell from its origins. Most methods of lineage tracing rely on temporally controlled site-specific recombination (SSR) systems in conjunction with appropriate reporter constructs.
Site-specific recombination (SSR) systems.
SSR systems integrate DNA recombinases and their target DNA sequences into the genome of the species of interest (e.g., mice) for the purpose of editing genetic information. The most used recombinase-target pair is Cre-loxP, which is derived from the P1 bacteriophage. The placement and orientation of the flanking target sequences dictate the type of gene editing that occurs: if a region of interest is flanked by two targeting sequences in the same orientation, that region of interest is excised; if the two targeting sequences are in opposite orientation, the region will be inverted. In the case of Cre-loxP, this flanking process is commonly referred to as floxing. By linking the expression of the recombinase to a cell-specific promoter, you can restrict deletion (or expression) of genes to a specific population of interest [81].
Temporal control of expression.
The selection of the promoter is crucial to effective lineage tracing. Restricting reporter expression to a specific cell type is often not sufficient for experimental success; one must also control when the reporter is turned on and off. Temporal expression can be achieved through various mechanisms, but a popular mechanism is drug induction. The most common in fate-mapping studies is the tamoxifen-estrogen receptor (ER) system, which relies on the Cre-ER fusion protein being sequestered in the cytoplasm unless tamoxifen is administered [117]. Other mechanisms that can provide temporal control are doxycycline-reverse tetracycline transactivator (rtTA) systems or light-induced split-Cre systems, where Cre is split into N- and C-terminal halves that can only make functional Cre when expressing cells are exposed to light (e.g. CRY2) [118].
Choosing a reporter.
Probably the most frequently used type of reporter in SSR systems is a simple floxed-stop-fluorescent protein. Another popular option is an enzyme like β-galactosidase or luciferase. Through bicistronic expression (via 2A sequence or similar), reporters can have multiple components to simultaneously report multiple colors or report on function. Other reporters switch colors upon recombination [13]. Reporters that allow for stochastic marking, either by randomly marking with several colors [14] or by generating a unique genetic barcode [15,16], are useful for tracking multiple clonal populations simultaneously.
Figure 1. How fate-mapping mice are made.
(a) Cre cassettes can be placed under the control of cell specific promoters. Cre is a site-specific recombinase that recognizes loxP sites as targets for DNA cleavage. Recognition of loxP sites in the same direction result in gene cassette deletion, while gene inversion occurs when loxP sites are placed in opposing directions. (b) Temporal expression is key for most current fate-mapping models. Tamoxifen mediated induction is a common method of inductions and is accomplished through one of three drivers: Cre-estrogen receptor (ER), Cre-ERT2 [10] or MerCreMer [11]. Other methods of temporal control include tetracycline induction [124] or split-Cre constructs that can be activated by a protein conformational change induced by light exposure [118]. (c) A fully functional fate-mapping model is often made by crossing driver and reporter lines to make a F1 generation with each transgene. In offspring, (1) Cre expression is cell specific, upon addition of tamoxifen, (2) the Cre-ER fusion protein translocates to the nucleus and mediates the recombination event and this (3) allows the reporter protein(s) to be expressed in a cell specific manner [10].
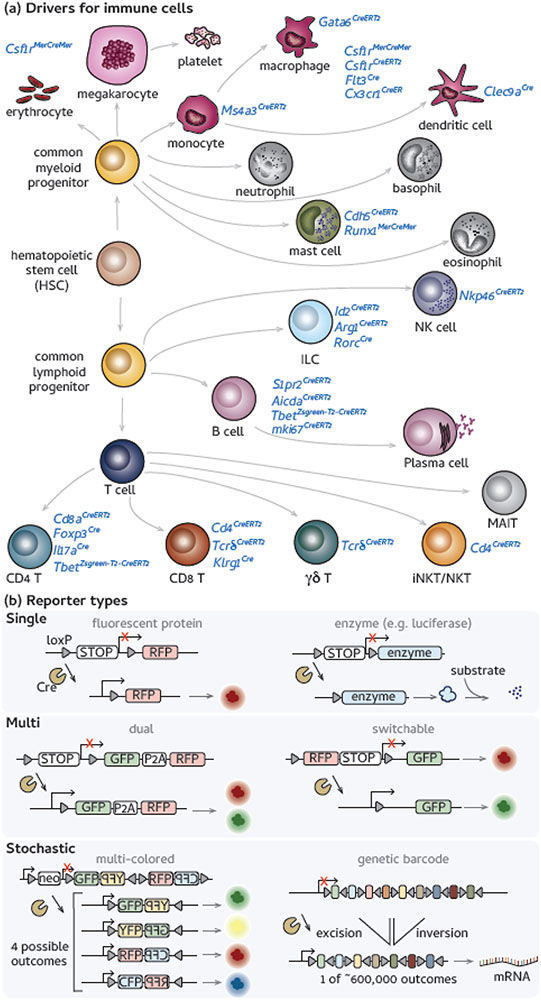
To study any immune cell type, a relevant promoter must first be identified, allowing specific cell types or subsets to be followed (Figure 2, Key Figure). Frequently, the reporter is a fluorescent protein, such as the green fluorescent protein (GFP) or red fluorescent protein (RFP), used as tracers for an entire cell population. However, as fate-mapping techniques have matured, reporters have become more specialized. Multiple markers can be produced simultaneously by the same genetic insertion through bicistronic expression [12]. Markers can also be made “switchable”, depending on the placement of LoxP target sites [13]. For example, confetti mice have gene cassettes that allow for the random expression of one to four fluorescent proteins upon gene recombination [14]. The polylox reporter has been developed to make genetic barcodes to identify individual cells using a combination of 10 equally spaced loxP sites in alternating orientations, such that excisions and inversion recombination events can generate ~600,000 unique genetic identifiers [15,16]. The newer reporters (e.g. confetti [17], polylox) offer the opportunity to study population dynamics within a particular lineage of cells.
Figure 2 (Key Figure). Examples of current immune cell-specific drivers and reporter options.
(a) Simplified diagram of hematopoiesis and cell lineages of the mouse immune system. In blue are the drivers that have been used to perform fate-mapping on specific cell types with corresponding references: megakaryocytes [125]; macrophages [2,18-37,63,126]; monocytes [96]; dendritic cells [69]; mast cells [39,86]; natural killer (NK) cells [52,53,95]; ILCs [57,58]; B cells [46-51]; CD4+T cells [41,43-45,77,127]; CD8+ T cells [40,42,99]; γδ T cells [54,55,89,128]; invariant natural killer T (iNKT) cells [59]. (b) Reporter constructs are usually inserted into a permissive locus. Reporters can be classified into three major types: single reporters, such as those that express a single fluorescent protein or an enzyme; multi reporters, such as those that bicistronically express two proteins [12] or are switchable [13]; and stochastic reporters in which recombination leads to random expression of a fluorescent marker (confetti) [14] or a genetic barcode (polylox) [15,16].
All immune cells derive from hematopoietic stem cells (HSCs) in the developmental process of hematopoiesis, giving rise to common myeloid and lymphoid progenitors (Figure 2). Myeloid lineages include erythrocytes, platelets, monocytes, macrophages, and granulocytes. Immune cells of the myeloid lineage are associated with innate immune function. The lymphoid lineage is composed of cells associated with adaptive immune responses, such as B cells and T cells as well as innate lymphoid cells (ILCs). Below is a summary of fate-mapping models currently developed for different lineages of immune cell populations (Figure 2).
Myeloid lineages
Macrophages and monocytes dominate the fate-mapping literature due to their well-known promoters, including Cx3cr1 [18-36] (non-classical/patrolling), Csfr1 [19,22,23,33,37] (yolk sacderived) and Flt3 (fetal-derived) [22,37,38]. Fate-mapping mice have been particularly useful in mapping diverse macrophage subsets in various tissues. For example, Cx3cr1CreERT2 mice helped establish that resident synovial and cardiac macrophages were self-renewing [23,24], and that kidney macrophages were derived from yolk sac macrophages [22]. Fate-mapping mice have also been used to define new subsets and functions of monocyte-derived subsets in the brain [25,28,30,35]. As for other myeloid lineage cells, Runx1MerCreMer (embryonic/fetal origin) and Csf1rjcre (early erythromyeloid progenitors) mice have revealed tissue-specific genes in mast cells [39]. Granulocytes such as basophils, neutrophils, and eosinophils, have not yet been studied, though promoters have been identified for ancestors of these cells. It is likely only a matter of time before fate-mapping mice are available to study the roles of granulocytes in health and disease.
Lymphoid lineages
Fate-mapping mice have also been used to study CD4+ and CD8+ T cells. Cd4creERT2 mice [40], for example, can be used to track the fates of CD8+ T cells leaving the thymus, since they only express CD4 briefly during thymic development. Similarly, Cd8creERT2 [41] mice have been recently developed to map the fates of naïve CD4+ T cells, since CD8 expression is temporally restricted to CD4+ T cells undergoing thymic development. Fate-mapping mice can also be used to label CD8+ and CD4+ T cells during the effector stage of the response to infection. For example, Klrg1cre was used to identify a novel subset of effector CD8+ T cells protecting the host against Listeria monocytogenes and cancer in mice [42]. The Foxp3CreERT2 driver has been useful in examining how T regulatory cells (Tregs) prevent rejection of heart transplants by reducing the numbers of pro-inflammatory cells [43]. Using a TbetZsGreen-T2A-CreERT2 driver, a subset of T follicular helper cells (Tfh) were revealed to express the pro-inflammatory cytokine IFN-γ in germinal centers (GC) post vaccination with a peptide, AS15 [44]. A report using the IL 17cre driver to study Th17 CD4+ T cells demonstrated context-dependent cellular behavior whereby the experimental autoimmune encephalitis (EAE) disease model of multiple sclerosis resulted in Th17 cells producing non-Th17 cytokines, while fungal Candida albicans infection triggered classical Th17 behavior [45]. B cell subsets have also been analyzed. B cell specific drivers revealing the dynamics of B cells in GCs, as well as the ability of B cells to undergo clonal selection and affinity maturation following vaccination, include AicdaCreERT2 (memory B cells and plasma cells) [46-48] S1pr2CreERT2 (gut associated GCs) [48,49] and Mki67CreERT2 (GC B cells) [50]. The TbetZsGreen-T2A-CreERT2 driver was also linked to a role of age-associated B cells in driving lupus in mice [51].
Fate mappers have informed our understanding of the development and function of ILCs. NKp46CreERT2 has allowed the tracking of natural killer (NK) cells to interrogate NK cell memory during toxoplasmosis [52] and murine cytomegalovirus infections [53]. Cx3cr1CreERT2 (which labels dendritic epidermal T cells) has been used to examine the γδ T cells role in epidermal wound healing [54] and TCRδCreER mice, for assessing the role of γδ T cells in causing psoriasis [55]. Moreover, fate-mapping is especially useful when studying rare immune populations, such as ILCs, which are difficult to detect using flow cytometry [56]. Id2creERT2 (important for the developmental origin of all ILCs) [57], Rorccre (needed for ILC3) [57] and Arg1RFPCreERT2 (needed for ILC2) [58] have been used to trace tissue seeding of various ILC populations. Similarly, our understanding of invariant natural killer T (iNKT) cell development has been enhanced in studies of Cd4CreERt2xVα14istopFxTraj18KO (knockout) mice, in which tamoxifen administration marked waves of iNKT cells emerging from the thymus [59]. Although many lymphoid lineages are well represented in the fate-mapping literature, there is still ample room for defining other immune cell populations. For example, mucosal-associated invariant T (MAIT) cells, which express Tcrδ or Cd4 during development, might be studied using TcrδCreERT2 or Cd4CreERT2 mice [60]. As MAIT cells are abundant in humans [60], fate-mapping experiments could potentially help unravel their unknown roles in disease.
Increasing the Breadth and Specificity of Fate-mapping
Currently, most fate-mapping models are designed to track a single population, but appropriately designed model systems can be multiplexed for orthogonal analysis of multiple factors and/or cell types. For example, the Tbiluc mouse displays constitutive T cell-specific green luciferase expression to report location, and NFAT-driven red luciferase to report cell activity [61]. Questions concerning population heterogeneity are well suited to concurrent fate-mapping strategies, as demonstrated by a pancreatic development study, using both Cre-loxP and Dre-roxP recombination to simultaneously track two lineages of β-cells establishing population heterogeneity and linking it to the transcription factor Pft1a [62]. A key to tracking multiple lineages is unique recombinase-target pairs for each population. Therefore, immune cell-specific mouse models using other DNA recombination enzyme systems need to be developed. Such systems include Flp-frt [41], Dre-roxP [63], the more recently developed sCre-sloxP and vCre-vloxP systems [64], as well as CRISPR-Cas9 based systems of gene editing [65]. Moreover, using complimentary induction agents such as tetracycline [66] or photoactivation [67], can also further refine experimental models.
Immunologists have harnessed temporally-controlled Cre recombination to develop numerous fate-mapping mouse models. As a result, lineage tracing can be performed on most immune cell types. The continued development of mouse models that can track multiple cell types simultaneously can thus provide new insights into cellular diversity and function.
Fate-mapping, immune function, and new frontiers
Early immunological applications of fate-mapping mice primarily focused on addressing the stem cell origins of immune cell types [68]. Embryonic and non-embryonic population sources were identified and observed until maturity. A natural extension of these studies was to ask whether functional consequences arose from different developmental origins. Determining the basis of differential behavior is key to understanding how immune cells contribute to homeostatic and disease processes.
Host defense
One role of the immune system is to protect the body from infectious disease. The immune response is comprised of multiple immune cell types with unique functions, but even within cell types, diverse behavior is observed. Fate-mapping has provided insights into varying infection responses. By following different fluorescently-labeled populations of dendritic cells (DC) with the Clec9aConfetti fate-mapping model, influenza A virus was found to not drive local clonal expansion of conventional dendritic cells (cDCs) [69]. Instead, bone marrow-derived circulating pre-cDCs were rapidly recruited to the lung and were the primary source of cDCs during infection [69]. In another study, the Cd4creERT2 driver was used to follow CD8+ T cells of different developmental origins. CD8+ T cells generated early in life were biased towards a short-lived effector fate following L. monocytogenes infection, showing that the age at which cells were produced influenced their long-term fate [40]. This CD8+ T cell study highlighted how fate-mapping models might be useful for evaluating age-related differences in immune responses across lifespans. Indeed, currently, little is known about the developmental origins of aging immune cells and their contribution to a variety of diseases.
Immunization serves to activate protective immune responses without actual infection, thereby ideally preventing severe disease should infection occur. A mediator of protection is that of antibody production by memory B cells once neutralizing antibody titers decline. Leveraging a GC B cell-specific promoter -- identified by scRNA-seq, S1pr2creERT2 reporter mice were given tamoxifen to fluorescently label B cells; consequently, cellular behavior could be monitored post-immunization [49]. T cell responses to immunization, particularly tissue-specific responses, are also important for containing infections [70,71], and further insights may be gained about these responses using existing fate-mapping models. Immune priming of HSCs may also be better understood by fate-mapping immune cell populations such as monocytes [72].
Inflammation and tissue repair
Recent work defined the developmental origins of immune cells that mediate inflammation and tissue repair. In particular, fate-mapping mice studies have provided new insights into the roles that tissue-specific macrophages play during responses to, and resolution of injury. Studies showing that cavity macrophages directly contribute to tissue repair have typically relied on genetic deletion of Gata6-expressing cells [73,74]. However, a recently developed fate-mapping strategy demonstrated a more nuanced picture of cavity macrophage behavior. The model used a CD45-driven Dre-rox system to restrict Gata6iCreERT2 to CD45+ cells, specifically reporting fluorescence in peritoneal cavity macrophages and allowing to track the cavity macrophage response to cryo- and heat-induced injury in the liver; results showed that while cavity macrophages were recruited to the liver parenchyma, they did not penetrate deep into the tissue, suggesting that they did not directly mediate repair in this model [63]. Tissue resident macrophages can be tracked using a Cx3cr1CreER mouse to drive the expression of a fluorescent reporter protein [24]. In a study examining cardiac infarction, Cx3cr1CreER x R26Td/DTR mice were bred to selectively delete tissue-resident cardiac macrophages, demonstrating that tissue-resident cardiac macrophages prevented pathological remodeling after cardiac infarction [24]. Like macrophages, many immune cell types contribute to tissue repair and inflammation regulation, including Tregs [75], and their roles may be further deciphered using fate-mapping models.
Cancer
Immune cells can promote or suppress cancer, depending on context; therefore, balancing their responses is crucial for achieving effective cancer therapies. Fate-mapping mice have been used to understand cancer-driving and protective immune populations. In one example, using a combination of Cx3cr1CreERT2, Lyz2Cre and Csf1rCreERT2 mice, the embryonic origin of a cancer-driving macrophage population was determined by fate-mapping embryonic macrophages, which where genetically ablated in a model of metastatic murine epithelial ovarian cancer [19]. Their ablation resulted in reduced tumor growth [19] – an outcome that might eventually be further investigated for creating targeted therapeutics. Similarly, a FoxP3YFPcre x Id2EmGFP mouse that both reports and over-expresses Id2, revealed a genetic basis for the cancer-exacerbating potential of Tregs [76]. In this system, Id2 was only over-expressed when doxycycline was administered; mice over-expressing Id2 exhibited reduced mouse melanoma B16.F10 tumor growth relative to the control mouse with normal Id2 protein expression, thus identifying another potential candidate drug target in this setting [76]. Moving forward. fate-mapping mice can contribute to further unraveling the heterogeneity of responses to immunomodulators and checkpoint inhibitors during immunotherapy, and might provide valuable insight into variable clinical outcomes.
Autoimmunity and allergy
When immune cells aberrantly react to self or innocuous foreign proteins, they can cause disease. Fate-mapping mice can track specific subpopulations of cells that contribute to allergic and/or autoimmune conditions. For example, Cx3cr1creERT2 and Cx3cr1cre mice were used to characterize synovial macrophages using microscopy and single cell sequencing [23]. These synovial lining macrophages were shown to be important in preventing collagen-induced arthritis as evidenced by the increased clinical score reported when the macrophages were ablated in a deleter mouse model restricting diphtheria toxin receptor to Cx3cr1 expressing cells [23]. In an EAE study, IL-17Cre mice were bred to mTORC1 knockout reporter mice; deleting mTORC1 led to increased resistance to EAE, as indicated by decreased clinical score and reduced spinal cord immunopathology when compared with mice expressing endogenous mTORC1 This suggested that disease resistance may have been due to metabolic changes in the fate-mapped CD4+ Th17 cells, although this remains to be further studied [77]. While B cells [78], CD4+ Th2 cells [79] and mast cells [80] are also important cellular mediators of allergy, fate-mapping mice remain to be used to further explore the different nuances and contributions of these cell types to allergic disease.
Taken together, recent applications of fate-mapping mice have yielded fundamental information about cellular origins and the roles of cell subsets in diverse aspects of immune function; however, there are still new frontiers that remain to be explored using these valuable tools.
Working with fate-mapping mouse models
Experimental design considerations
Inducible Cre-driven models of fate-mapping can reduce the likelihood of issues associated with unexpected Cre expression because Cre activity is restricted in the absence of an induction agent (i.e tamoxifen). However, promoters can display wider or more transient gene expression patterns than anticipated. For instance, even minimal Cre expression can edit at target loxP sites and this so-called “leakiness” results in unintended reporter expression, which causes complications in data interpretation due to the loss of fidelity between reporter expression and specific induction of Cre [81]. Moreover, Cre expression, even in the absence of recombination, can have deleterious effects, including DNA damage and chromosomal abnormalities [82]. Additionally, ERT2-inducible Cre mouse models may show reduced proliferation in rapidly dividing cells, as has been shown for the clonal expansion of T cells [83]. Moreover, the effects of concurrent drug treatments should be considered. For example, ivermectin, an anti-parasitic medication widely used in routine animal husbandry as a treatment for pinworms, can activate ERT2-mediated Cre recombination, thus resulting in fluorescence reporter expression in the absence of tamoxifen administration, an outcome that can confound data interpretation [84].
Furthermore, the induction agent tamoxifen, a synthetic estrogen, can also have unintended side effects. Specifically, in pregnant dams, tamoxifen administration can result in embryonic lethality, difficulty in labor, and reduced live birth rates [85]. Therefore, the co-administration of progesterone can reduce some of these side-effects [86]. Also, in males, tamoxifen administration has demonstrated ill effects on reproductive fitness [87]. Generally, long-term exposure to tamoxifen can be quite toxic. Therefore, it is desirable to administer tamoxifen at the lowest effective dose for short time intervals [87], and oral administration can reduce the occurrence of drug delivery-related inflammation in mice [88].
Lastly, fate-mapping models frequently use breeding schemes in which one parent bears the promoter-driven Cre while the other bears the floxed-stop reporter to generate offspring with single copies of each fate-mapping element (Figure 1) [81]. While a single copy of these transgenic elements is usually sufficient for their expression in most cells, cellular marking can be incomplete. For example, green fluorescence driven by TCRδcreERT2 in such transgenic mice varies from 60-90% in splenic γδ T cells, depending on the genotype of the offspring [89], suggesting that gene copy number may impact reporter expression.
Combining fate-mapping with other experimental tools
There isn't a singular way of experimentally using fate-mapping mice, and taking advantage of current and emerging techniques (Box 2) has many benefits. The specific research question will dictate which techniques are most useful, and oftentimes using a combination of techniques yields additional insights (Figure 3).
Box 2. A fate mapper’s toolbox.
For maximal discovery, fate-mapping models should take advantage of cutting-edge scientific technologies and methods:
Sequencing technologies.
RNA sequencing comprehensively describes the transcriptome of a cellular population, while ATAC sequencing is a method to identify areas of open chromatin. Both of these technologies are now routinely being applied to single cells, most frequently using the 10x genomics platform [119]. Additionally, for those interested in clonal populations of B and T cells, receptor identification and transcriptional profiling can be assessed simultaneously at the individual cell level. The genomics field is continually developing new computational algorithms and machine learning techniques to identify important signaling networks and underlying regulatory mechanisms.
High parameter flow cytometry:
Cytometry is a workhorse of immunology research. Optical advances, including full spectrum cytometry, now allow for the analysis of upwards of twenty-five parameters in any given sample for the identification of cell types and subpopulations with unique functions [120]. While conventional one- and two-dimensional analysis can result in useful findings, additional interpretation (and visual representation) of high-dimensional data utilizes dimensional reduction algorithms, such as FlowSOM, as well as t-SNE and UMAP [101].
Mathematical modeling:
Experiments designed to track turnover of cellular populations over time yield data that can be used to construct mathematical models describing kinetics of expansion, contraction, differentiation, and survival. These models can, in turn, be used to form hypotheses about cellular behavior that can then be experimentally tested.
Imaging Technologies:
Many of the experimental tools used by immunologists rely on the removal of cells from their native environment. Use of fate-mapping models with newer imaging modalities can provide essential information about how populations of interest act within tissues. Multiphoton techniques take advantage of high powered near-infrared lasers and 2+ photon absorption, allowing for deep imaging within tissues [121]. Alternatively, whole tissues can be imaged at high resolution by newer methods of tissue clearance (e,g.3DISCO) [122], in conjunction with confocal and light sheet microscopy. Non-invasive techniques such as a positron emission tomography (PET) scan can be used to track marked populations over time within the same animal [123].
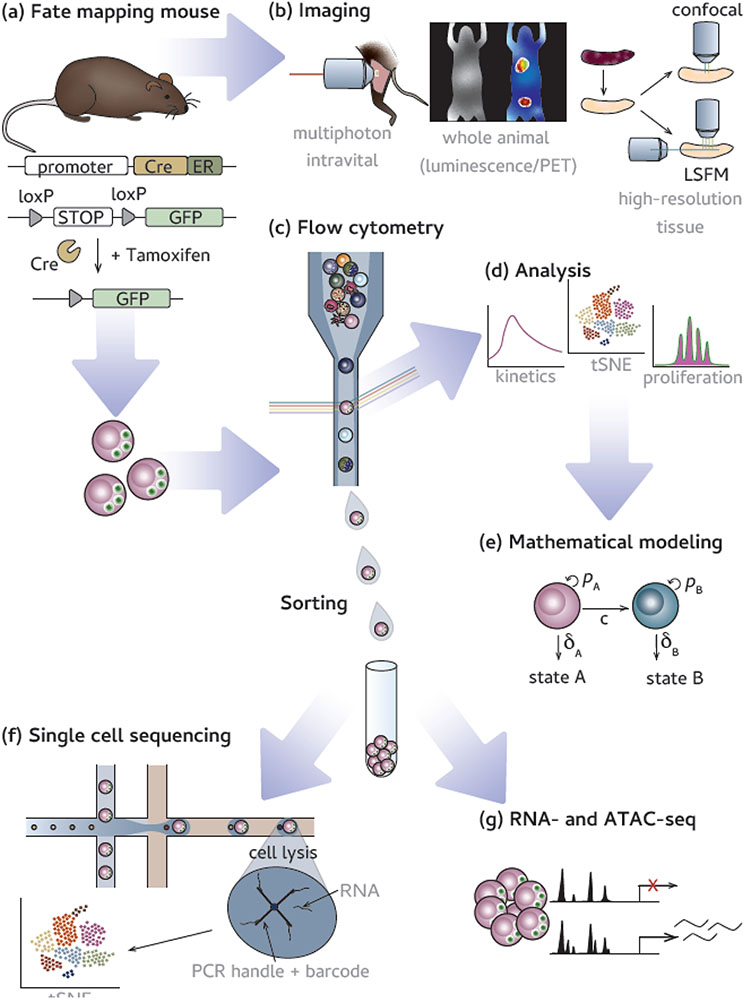
Figure 3. Examples of experimental techniques that are compatible with fate-mapping mice.
(a) Fate-mapping begins with the temporal expression of a reporter allows for fate-mapping. Following fate-mapping, researchers can use many tools to understand how cellular origin can dictate phenotype and function. (b) Imaging technologies are a powerful collection of tools to assess in situ localization and interactions with other cell types. (left) Multiphoton imaging is used to perform intravital imaging deep into animal tissues of interest [121] (an example is live imaging of cellular interactions and dynamics in lymph nodes); (middle) whole animal imaging, either by luminescence or positron emission tomography (PET) modalities, allows for non-invasive longitudinal study; (right) advances in tissue clearance (e.g. 3DISCO) [122] improve high resolution microscopy of tissues with techniques such as confocal and light sheet fluorescent microscopy (LSFM). (c) Flow cytometry uses laser excitation to measure fluorescence of labeled markers on cells. (d) Traditional and high parameter analyses can be performed via flow cytometry data. (e) Mathematical modeling uses accumulated data to model cellular behavior. Flow cytometry can also sort cells, based on specific marker expression, for a variety of downstream technologies such as (f) single cell sequencing analysis in which an individual cell’s transcripts are associated with a unique barcode and (e) bulk RNA and ATAC sequencing in which cellular populations are sequences for transcriptional (RNA) or chromatin accessibility (ATAC) information [94,129].
High throughput and single-cell sequencing technologies are rapidly-evolving and improved technologies which assess the transcriptional and epigenetic state of cells. Many important questions have been addressed with population level RNA-seq and ATAC-seq, including defining lineage-specific transcription factors relevant to CD4+ T cells in the gut [90]. To correlate gene expression patterns in cells of interest to expression patterns of defined subsets [40], the data collected through these techniques can be compared to curated data sets, such as in the ImmGen databasei [91-93]. Recent advances using sequencing data to infer cell-cell interactions can be valuable in yielding future insights into the behavior of fate-mapped cells [94]. Single-cell RNA sequencing (scRNA-seq) was used in conjunction with a model that fluorescently-barcoded NK cells to identify two distinct differentiation paths for ILC1-like and NK cells [95]. Additionally, multiple mouse models were developed based on scRNA-sequencing that identified Ms4a3 as a promoter that could efficiently fate-map monocyte lineages [96]. scRNA-seq is currently an expensive endeavor, which can result in having to balance the number of individual cells sequenced and sequencing depth [97]. This trade-off can sometimes make it hard to identify populations based on known markers whose genes are expressed at low amounts. CITE-seq [98] was developed in part, to overcome this limitation by using oligonucleotide-conjugated antibodies so that protein expression of markers of cellular identity could be correlated with gene expression during sequencing.
Immunologists have relied on flow cytometry for decades because it is an invaluable tool for looking at protein expression at the cellular level. Flow cytometry tracks the phenotype and population dynamics of fate-mapped cells [99]. As cytometric technologies continue to be refined, the number of biomarkers that can be tracked on an individual cell has increased dramatically. The synthesis of high dimensional data has necessitated the adoption of machine learning techniques, such as FlowSOM, which can help to understand, describe, and predict cellular behavior [100,101]. For example, FlowSOM was used to identify and track the fates of GM-CSF-producing cells in the Frog mouse system [100].
While much has been learned by studying fate-mapped cells in isolation, either in vitro or ex vivo, the ability to see how cellular lineages interact with their natural environment is essential for deepening our understanding of cellular fate and function. Confocal images have helped identify yolk sac-derived mast cells in a CD5creERT2 driven fate-mapping mice [54], while clonal dynamics of B cell selection in gut GCs has been visualized using, for instance, the B cell-specific confetti reporter AciadacreERT2 , coupled with multiphoton imaging [48]. Light sheet fluorescence microscopy (LSFM) is also currently being used for lineage tracing by developmental biologists [102,103] and is of great advantage for further answering immunological questions. In addition, the continued development of reporter mice, such as the Tri-modal reporter mouse, which concurrently reports bioluminescence, fluorescence, and positron emission tomography can allow for imaging at multiple scales within the same mouse model [104].
The studies highlighted above demonstrate how careful experimental design along with rapid technological advances in high throughput sequencing, cytometry, and imaging are enhancing the power of fate-mapping mouse models to answer questions about immune cell origins, phenotypes, and functions. We anticipate that technological advances will continue to improve the utility of fate-mapping mouse models in future studies.
Concluding remarks
Genetic fate-mapping of immune cells has led to discoveries about the origins of cell lineages, but recently, has been used to further inform on the specific roles of these lineages in health and disease. Additional discoveries in this realm can contribute to our understanding of why and how cellular origins impact immunologic outcomes, but also inform future therapeutic targets in cancer and infection, aid in the design of effective vaccines, and shed light on unexpected treatment outcomes (Box 3. Clinician’s Corner). Opportunities exist for developing new tools to better examine the cellular fates of specific responding populations. Engineering viral and bacterial pathogens that express Cre to mark infected cells could also generate insight into host-pathogen interactions, for example for pathogens with immune tropism [105].
Box 3. Clinician’s Corner.
Fate-mapping mice track defined populations of cells with specificity that is not achievable in humans. The singular method of fate-mapping within humans involves administering deuterated water, which only allows proliferation to be tracked broadly.
By marking specific populations, fate-mapping mice present an ideal model for studying heterogeneity in immune cell responses with high clinical relevance. For example, fate-mapping mice may help uncover why some patients respond to immunomodulators or checkpoint inhibitors, while others do not. This approach is especially powerful when combined with emerging techniques such as barcoding individual cells.
Findings from fate-mapping mice can be verified in humans. Tools, such as sequencing and imaging, may reveal markers and tissue specific properties that might be correlated to human samples.
Results from fate-mapping mice studies can also have translational benefit in designing new therapeutics. Once the dynamics of disease-causing populations are defined, drugs may then be designed and tested.
In addition, leveraging such highlighted technologies can provide new insights into immune homeostasis and function. As single cell technologies continue to evolve and become more affordable, we posit that they might become the gold standard for understanding the genomics of fate-mapped populations. Technologies that combine aspects of imaging, cytometry, and sequencing, including spatial transcriptomics [106], scope-seq [107] and chip cytometry [108], are providing new ways to interrogate fate-mapping systems. New analysis techniques, such as pseudotime analysis [109], can also push the field toward a deeper understanding of cell function and drive new hypotheses (outstanding questions).
Finally, new fate-mapping mouse models can also exploit emerging genetic tools, such as CRISPR-Cas9 gene editing [110-112], polycistronic expression systems [113] and intersectional approaches [114] . For instance, improved reporters that can reveal cellular function and identity [115] may help deepen our understanding of biological processes in cell lineages. Implementing genetic barcoding [111,116] in cells of interest can also help uncover lineage diversity during homeostasis or disease. We eagerly anticipate the rapid development of additional fate-mapping tools combined with emerging technologies to enhance our fundamental knowledge of how cell origin can influence immune cell fates and functions.
Outstanding Questions.
How can we integrate multiple fate-mapping modalities to mark more specialized populations in mice? By combining several types of drivers and reporters, multiple populations might be tracked simultaneously.
Many immune cell types have been described using fate-mapping mice, but we still lack mouse models for many rare cell types. What new mice might be designed to lineage-trace other immune cell types/subtypes? The field currently lacks fate-mapping mice for certain populations of granulocytes, ILCs, and MAIT cell.
How can we leverage imaging techniques in conjunction with fate-mapping mice, such as multiphoton imaging, to better understand the developmental origins of tissue distribution during homeostasis and disease?
How can viruses or bacteria expressing Cre be utilized to mark and track infected cells in mice?
Can we apply previously created fate-mapping mice to answer new questions related to infection and cancer, particularly to understand the developmental origins of the heterogeneous response to treatments and vaccines?
Highlights Box.
Fate-mapping mice have revealed the developmental origins of multiple types of immune cells.
When combined with technologies such as scRNAseq, multiphoton imaging, and multiparameter flow cytometry, fate-mapping mice can define novel cell populations.
When utilized in the context of infection and cancer, fate-mapping mice can both aid in understanding immune cell responses and help uncover new putative therapeutic targets that are unique to cells of specific developmental origins.
Emerging fate-mapping models take advantage of newer genetic tools, such as cellular barcoding and stochastic multi-color reporters, thus allowing further resolution of the dynamics of immune cell populations in mice.
Acknowledgements
This work was supported by multiple National Institute of Health awards: U01 AI131348, R01 AI110613, R01 AI105265, R01 HD07798 (B.D.R.) and R21 AI138025 (N.L.S.). S.E.L. was supported by training fellowship 5T32 OD011000. The authors would also like to thank Kate Kallal and Zachary Hilt for helpful discussion and feedback during the writing of this review.
Glossary
- Affinity maturation
biochemical process B cells undergo to produce higher affinity antibodies
- ATAC-seq
high throughput sequencing technique assessing chromatin accessibility
- Bicistronic
translation of two genes from one mRNA transcript
- Bone marrow chimera
mouse model in which the immune compartment is reconstituted with donor mouse stem cells
- Checkpoint inhibitor
immunotherapeutic drug targeting immune regulators (checkpoints)
- Chip cytometry
immobilized cell arrays for single cell analysis
- Confetti mouse
reporter mouse stochastically expressing different fluorescent proteins
- Cre
Bacteriophage P1-derived site-specfic DNA recombinase
- Epigenetic
regulation of gene expression without changing DNA sequence
- ERT2
triple mutant of estrogen receptor with high specificity for tamoxifen
- Fate-mapping
method used to study how the origin of cells influences their trajectory
- Flow cytometry
technique that rapidly analyzes cells for parameters such as size, granularity, and protein expression
- FlowSOM
self-organized map algorithm used on flow cytometry data
- Floxed
flanked by two loxP sequences
- Genetic barcoding
marking unique identifiers with short DNA sequences
- Germinal center
location within a lymphoid organ follicle where high affinity antibodies are produced by B cells
- HSP90
molecular chaperone aiding in protein folding
- Invariant natural killer T cell (iNKT cell)
NKT cell lymphoid population recognizing specific lipids
- Light sheet fluorescent microscopy (LSFM)
excitation occurs in a plane perpendicular to the observational direction
- LoxP
the DNA sequence is a target for Cre
- Machine Learning
computer systems that learn and adapt by using algorithms to analyze data patterns
- MerCreMer
fusion protein of Cre flanked by two modified ER binding domains
- MAIT cell
innate like T cell bearing an invariant TCR
- Multi-photon microscopy
using near-infrared light to excite fluorescent molecules with 2+ photons to image deep into animal tissue while minimizing damage
- Permissive locus
gene region allowing genetic insertion without transgene silencing or dysregulation of neighboring genes. In this review, different from transcriptionally permissive
- Polycistronic expression
translation of multiple genes from a single mRNA transcript
- Polylox reporter
system whereby Cre drives the generation of unique genetic barcodes
- Positron emission tomography (PET)
functional imaging technique using radioactive substances
- Pseudotime analysis
dynamic analysis ordering cells along a lineage based on gene expression profiles
- RNA-seq
high throughput sequencing analyzing transcriptional profiles
- Rosa26
permissive gene locus for ubiquitous expression
- SCOPE-seq
sequencing method merging scRNAseq and live cell imaging
- Sequencing depth
average number of times nucleotides are read in high throughput sequencing
- scRNA-seq
high throughput sequencing analyzing transcriptional profiles of individual cells
- tSNE
statistical method for visualizing high-dimensional data
- T follicular helper cells
CD4+ T cells that aid B cell maturation
- Tregs
immunosuppressive CD4+ T cells
- Th17 cells
CD4+ T cells producing IL-17
- UMAP
dimension reduction algorithm for high parameter data
- 2A
self-cleaving peptide sequences placed between genes of interest; used to generate individual, instead of fusion proteins
- 3DISCO
tissue-clearing method for 3D imaging of entire organs without sectioning
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Komatsu N et al. (2014) Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat Med 20, 62–68. 10.1038/nm.3432 [DOI] [PubMed] [Google Scholar]
- 2.Bain CC et al. (2014) Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 15, 929–937. 10.1038/ni.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogan T et al. (2015) Temporal fate mapping reveals age-linked heterogeneity in naive T lymphocytes in mice. Proc Natl Acad Sci U S A 112, E6917–6926. 10.1073/pnas.1517246112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogan T et al. (2017) Generation of Busulfan Chimeric Mice for the Analysis of T Cell Population Dynamics. Bio Protoc 7, e2650. 10.21769/BioProtoc.2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonhoeffer S et al. (2000) Quantification of cell turnover kinetics using 5-bromo-2'-deoxyuridine. J Immunol 164, 5049–5054. 10.4049/jimmunol.164.10.5049 [DOI] [PubMed] [Google Scholar]
- 6.Westera L et al. (2013) Closing the gap between T-cell life span estimates from stable isotope-labeling studies in mice and humans. Blood 122, 2205–2212. 10.1182/blood-2013-03-488411 [DOI] [PubMed] [Google Scholar]
- 7.Zambrowicz BP et al. (1997) Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci U S A 94, 3789–3794. 10.1073/pnas.94.8.3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Indra AK et al. (1999) Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res 27, 4324–4327. 10.1093/nar/27.22.4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzger D et al. (1995) Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci U S A 92, 6991–6995. 10.1073/pnas.92.15.6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seibler J et al. (2003) Rapid generation of inducible mouse mutants. Nucleic Acids Res 31, e12. 10.1093/nar/gng012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y et al. (1996) Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res 24, 543–548. 10.1093/nar/24.4.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YT et al. (2012) R26R-GR: a Cre-activable dual fluorescent protein reporter mouse. PLoS One 7, e46171. 10.1371/journal.pone.0046171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muzumdar MD et al. (2007) A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- 14.Snippert HJ et al. (2010) Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144. 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- 15.Pei W et al. (2017) Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460. 10.1038/nature23653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei W et al. (2019) Using Cre-recombinase-driven Polylox barcoding for in vivo fate mapping in mice. Nat Protoc 14, 1820–1840. 10.1038/s41596-019-0163-5 [DOI] [PubMed] [Google Scholar]
- 17.Livet J et al. (2007) Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62. 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- 18.Gallerand A et al. (2021) Brown adipose tissue monocytes support tissue expansion. Nat Commun 12, 5255. 10.1038/s41467-021-25616-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang N et al. (2021) LYVE1+ macrophages of murine peritoneal mesothelium promote omentum-independent ovarian tumor growth. J Exp Med 218. 10.1084/jem.20210924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bain CC et al. (2020) Rate of replenishment and microenvironment contribute to the sexually dimorphic phenotype and function of peritoneal macrophages. Sci Immunol 5. 10.1126/sciimmunol.abc4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chappell-Maor L et al. (2020) Comparative analysis of CreER transgenic mice for the study of brain macrophages: A case study. Eur J Immunol 50, 353–362. 10.1002/eji.201948342 [DOI] [PubMed] [Google Scholar]
- 22.Ide S et al. (2020) Yolk-sac-derived macrophages progressively expand in the mouse kidney with age. Elife 9. 10.7554/eLife.51756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Culemann S et al. (2019) Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature 572, 670–675. 10.1038/s41586-019-1471-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dick SA et al. (2019) Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol 20, 29–39. 10.1038/s41590-018-0272-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordao MJC et al. (2019) Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363. 10.1126/science.aat7554 [DOI] [PubMed] [Google Scholar]
- 26.Kolter J et al. (2019) A Subset of Skin Macrophages Contributes to the Surveillance and Regeneration of Local Nerves. Immunity 50, 1482–1497.e1487. 10.1016/j.immuni.2019.05.009 [DOI] [PubMed] [Google Scholar]
- 27.Lin JD et al. (2019) Single-cell analysis of fate-mapped macrophages reveals heterogeneity, including stem-like properties, during atherosclerosis progression and regression. JCI Insight 4. 10.1172/jci.insight.124574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes NJ et al. (2019) Fate Mapping In Vivo to Distinguish Bona Fide Microglia Versus Recruited Monocyte-Derived Macrophages in Retinal Disease. Methods Mol Biol 1834, 153–164. 10.1007/978-1-4939-8669-9_11 [DOI] [PubMed] [Google Scholar]
- 29.Soysa R et al. (2019) Fetal origin confers radioresistance on liver macrophages via p21(cip1/WAF1). J Hepatol 71, 553–562. 10.1016/j.jhep.2019.04.015 [DOI] [PubMed] [Google Scholar]
- 30.Van Hove H et al. (2019) A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci 22, 1021–1035. 10.1038/s41593-019-0393-4 [DOI] [PubMed] [Google Scholar]
- 31.De Schepper S et al. (2018) Self-Maintaining Gut Macrophages Are Essential for Intestinal Homeostasis. Cell 175, 400–415.e413. 10.1016/j.cell.2018.07.048 [DOI] [PubMed] [Google Scholar]
- 32.Puranik AS et al. (2018) Kidney-resident macrophages promote a proangiogenic environment in the normal and chronically ischemic mouse kidney. Sci Rep 8, 13948. 10.1038/s41598-018-31887-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stremmel C et al. (2018) Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat Commun 9, 75. 10.1038/s41467-017-02492-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gundra UM et al. (2017) Vitamin A mediates conversion of monocyte-derived macrophages into tissue-resident macrophages during alternative activation. Nat Immunol 18, 642–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tay TL et al. (2017) A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat Neurosci 20, 793–803. 10.1038/nn.4547 [DOI] [PubMed] [Google Scholar]
- 36.Meghraoui-Kheddar A et al. (2020) Revising CX3CR1 Expression on Murine Classical and Non-classical Monocytes. Front Immunol 11, 1117. 10.3389/fimmu.2020.01117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M et al. (2021) Two populations of self-maintaining monocyte-independent macrophages exist in adult epididymis and testis. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2013686117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clemente-Casares X et al. (2017) A CD103(+) Conventional Dendritic Cell Surveillance System Prevents Development of Overt Heart Failure during Subclinical Viral Myocarditis. Immunity 47, 974–989.e978. 10.1016/j.immuni.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 39.Li Z et al. (2018) Adult Connective Tissue-Resident Mast Cells Originate from Late Erythro-Myeloid Progenitors. Immunity 49, 640–653.e645. 10.1016/j.immuni.2018.09.023 [DOI] [PubMed] [Google Scholar]
- 40.Smith NL et al. (2018) Developmental Origin Governs CD8(+) T Cell Fate Decisions during Infection. Cell 174, 117–130.e114. 10.1016/j.cell.2018.05.029 [DOI] [PubMed] [Google Scholar]
- 41.Andrews LP et al. (2021) A Cre-driven allele-conditioning line to interrogate CD4(+) conventional T cells. Immunity 54, 2209–2217.e2206. 10.1016/j.immuni.2021.08.029 [DOI] [PubMed] [Google Scholar]
- 42.Herndler-Brandstetter D et al. (2018) KLRG1(+) Effector CD8(+) T Cells Lose KLRG1, Differentiate into All Memory T Cell Lineages, and Convey Enhanced Protective Immunity. Immunity 48, 716–729.e718. 10.1016/j.immuni.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheen JH et al. (2017) TLR-Induced Murine Dendritic Cell (DC) Activation Requires DC-Intrinsic Complement. J Immunol 199, 278–291. 10.4049/jimmunol.1700339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang D et al. (2018) Transient T-bet expression functionally specifies a distinct T follicular helper subset. J Exp Med 215, 2705–2714. 10.1084/jem.20180927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirota K et al. (2011) Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol 12, 255–263. 10.1038/ni.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Gallou S et al. (2017) The AID-Cre-ERT2 Model: A Tool for Monitoring B Cell Immune Responses and Generating Selective Hybridomas. Methods Mol Biol 1623, 243–251. 10.1007/978-1-4939-7095-7_19 [DOI] [PubMed] [Google Scholar]
- 47.Le Gallou S et al. (2018) A splenic IgM memory subset with antibacterial specificities is sustained from persistent mucosal responses. JExp Med 215, 2035–2053. 10.1084/jem.20180977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nowosad CR et al. (2020) Tunable dynamics of B cell selection in gut germinal centres. Nature 588, 321–326. 10.1038/s41586-020-2865-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glaros V et al. (2021) Limited access to antigen drives generation of early B cell memory while restraining the plasmablast response. Immunity 54, 2005–2023.e2010. 10.1016/j.immuni.2021.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verheijen M et al. (2020) Fate Mapping Quantifies the Dynamics of B Cell Development and Activation throughout Life. Cell Rep 33, 108376. 10.1016/j.celrep.2020.108376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ricker E et al. (2021) Altered function and differentiation of age-associated B cells contribute to the female bias in lupus mice. Nat Commun 12, 4813. 10.1038/s41467-021-25102-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivanova DL et al. (2019) The IL-12- and IL-23-Dependent NK Cell Response Is Essential for Protective Immunity against Secondary Toxoplasma gondii Infection. J Immunol 203, 2944–2958. 10.4049/jimmunol.1801525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nabekura T and Lanier LL (2016) Tracking the fate of antigen-specific versus cytokine-activated natural killer cells after cytomegalovirus infection. J Exp Med 213, 2745–2758. 10.1084/jem.20160726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gentek R et al. (2018) Epidermal gammadelta T cells originate from yolk sac hematopoiesis and clonally self-renew in the adult. J Exp Med 215, 2994–3005. 10.1084/jem.20181206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandrock I et al. (2018) Genetic models reveal origin, persistence and non-redundant functions of IL-17-producing gammadelta T cells. J Exp Med 215, 3006–3018. 10.1084/jem.20181439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tait Wojno ED and Beamer CA (2018) Isolation and Identification of Innate Lymphoid Cells (ILCs) for Immunotoxicity Testing. Methods Mol Biol 1803, 353–370. 10.1007/978-1-4939-8549-4_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones R et al. (2018) Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur J Immunol 48, 1481–1491. 10.1002/eji.201847511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider C et al. (2019) Tissue-Resident Group 2 Innate Lymphoid Cells Differentiate by Layered Ontogeny and In Situ Perinatal Priming. Immunity 50, 1425–1438.e1425. 10.1016/j.immuni.2019.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bortoluzzi S et al. (2021) Brief homogeneous TCR signals instruct common iNKT progenitors whose effector diversification is characterized by subsequent cytokine signaling. Immunity 54, 2497–2513.e2499. 10.1016/j.immuni.2021.09.003 [DOI] [PubMed] [Google Scholar]
- 60.Legoux F et al. (2020) MAIT Cell Development and Functions: the Microbial Connection. Immunity 53, 710–723. 10.1016/j.immuni.2020.09.009 [DOI] [PubMed] [Google Scholar]
- 61.Kleinovink JW et al. (2019) A Dual-Color Bioluminescence Reporter Mouse for Simultaneous in vivo Imaging of T Cell Localization and Function. Frontiers in Immunology 9, 3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C et al. (2019) Evidence of a developmental origin for β-cell heterogeneity using a dual lineage-tracing technology. Development 146. 10.1242/dev.164913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin H et al. (2021) Genetic fate-mapping reveals surface accumulation but not deep organ invasion of pleural and peritoneal cavity macrophages following injury. Nat Commun 12, 2863. 10.1038/s41467-021-23197-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshimura Y et al. (2018) Novel reporter and deleter mouse strains generated using VCre/VloxP and SCre/SloxP systems, and their system specificity in mice. Transgenic Res 27, 193–201. 10.1007/s11248-018-0067-0 [DOI] [PubMed] [Google Scholar]
- 65.Yang H et al. (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Connolly KA et al. (2021) A reservoir of stem-like CD8(+) T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Sci Immunol 6, eabg7836. 10.1126/sciimmunol.abg7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Medaglia C et al. (2017) Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science 358, 1622–1626. 10.1126/science.aao4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Höfer T et al. (2016) Fate Mapping and Quantitation of Hematopoiesis In Vivo. Annu Rev Immunol 34, 449–478. 10.1146/annurev-immunol-032414-112019 [DOI] [PubMed] [Google Scholar]
- 69.Cabeza-Cabrerizo M et al. (2019) Tissue clonality of dendritic cell subsets and emergency DCpoiesis revealed by multicolor fate mapping of DC progenitors. Sci Immunol 4. 10.1126/sciimmunol.aaw1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swain SL et al. (2012) Expanding roles for CD4+ T cells in immunity to viruses. Nat Rev Immunol 12, 136–148. 10.1038/nri3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin MD and Badovinac VP (2018) Defining Memory CD8 T Cell. Front Immunol 9, 2692. 10.3389/fimmu.2018.02692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen L and Ozato K (2021) Innate Immune Memory in Hematopoietic Stem/Progenitor Cells: Myeloid-Biased Differentiation and the Role of Interferon. Front Immunol 12, 621333. 10.3389/fimmu.2021.621333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deniset JF et al. (2019) Gata6(+) Pericardial Cavity Macrophages Relocate to the Injured Heart and Prevent Cardiac Fibrosis. Immunity 51, 131–140.e135. 10.1016/j.immuni.2019.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J and Kubes P (2016) A Reservoir of Mature Cavity Macrophages that Can Rapidly Invade Visceral Organs to Affect Tissue Repair. Cell 165, 668–678. 10.1016/j.cell.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 75.Panduro M et al. (2016) Tissue Tregs. Annu Rev Immunol 34, 609–633. 10.1146/annurev-immunol-032712-095948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hwang SM et al. (2018) Inflammation-induced Id2 promotes plasticity in regulatory T cells. Nat Commun 9, 4736. 10.1038/s41467-018-07254-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karmaus PWF et al. (2019) Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 565, 101–105. 10.1038/s41586-018-0806-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saunders SP et al. (2019) Non-classical B Cell Memory of Allergic IgE Responses. Front Immunol 10, 715. 10.3389/fimmu.2019.00715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.León B and Ballesteros-Tato A (2021) Modulating Th2 Cell Immunity for the Treatment of Asthma. Frontiers in Immunology 12, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Méndez-Enríquez E and Hallgren J (2019) Mast Cells and Their Progenitors in Allergic Asthma. Frontiers in Immunology 10, 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song AJ and Palmiter RD (2018) Detecting and Avoiding Problems When Using the Cre-lox System. Trends Genet 34, 333–340. 10.1016/j.tig.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Janbandhu VC et al. (2014) Cre recombinase induces DNA damage and tetraploidy in the absence of loxP sites. Cell Cycle 13, 462–470. 10.4161/cc.27271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurachi M et al. (2019) Hidden Caveat of Inducible Cre Recombinase. Immunity 51, 591–592. 10.1016/j.immuni.2019.09.010 [DOI] [PubMed] [Google Scholar]
- 84.Corbo-Rodgers E et al. (2012) Oral ivermectin as an unexpected initiator of CreT2-mediated deletion in T cells. Nat Immunol 13, 197–198. 10.1038/ni.2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ved N et al. (2019) Tamoxifen administration in pregnant mice can be deleterious to both mother and embryo. Lab Anim 53, 630–633. 10.1177/0023677219856918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gentek R et al. (2018) Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity 48, 1160–1171.e1165. 10.1016/j.immuni.2018.04.025 [DOI] [PubMed] [Google Scholar]
- 87.Patel SH et al. (2017) Low-dose tamoxifen treatment in juvenile males has long-term adverse effects on the reproductive system: implications for inducible transgenics. Sci Rep 7, 8991. 10.1038/s41598-017-09016-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Donocoff RS et al. (2020) Optimization of tamoxifen-induced Cre activity and its effect on immune cell populations. Sci Rep 10, 15244. 10.1038/s41598-020-72179-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang B et al. (2015) Differential Requirements of TCR Signaling in Homeostatic Maintenance and Function of Dendritic Epidermal T Cells. J Immunol 195, 4282–4291. 10.4049/jimmunol.1501220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.London M et al. (2021) Stepwise chromatin and transcriptional acquisition of an intraepithelial lymphocyte program. Nature Immunology 22, 449–459. 10.1038/s41590-021-00883-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heng TS et al. (2008) The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094. 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- 92.Maslova A et al. (2020) Deep learning of immune cell differentiation. Proc Natl Acad Sci U S A 117, 25655–25666. 10.1073/pnas.2011795117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Project IG (2020) ImmGen at 15. Nat Immunol 21, 700–703. 10.1038/s41590-020-0687-4 [DOI] [PubMed] [Google Scholar]
- 94.Armingol E et al. (2021) Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet 22, 71–88. 10.1038/s41576-020-00292-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flommersfeld S et al. (2021) Fate mapping of single NK cells identifies a type 1 innate lymphoid-like lineage that bridges innate and adaptive recognition of viral infection. Immunity 54, 2288–2304.e2287. 10.1016/j.immuni.2021.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Z et al. (2019) Fate Mapping via Ms4a3-Expression History Traces Monocyte-Derived Cells. Cell 178, 1509–1525.e1519. 10.1016/j.cell.2019.08.009 [DOI] [PubMed] [Google Scholar]
- 97.Zhang MJ et al. (2020) Determining sequencing depth in a single-cell RNA-seq experiment. Nat Commun 11, 774. 10.1038/s41467-020-14482-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stoeckius M et al. (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14, 865–868. 10.1038/nmeth.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reynaldi A et al. (2019) Fate mapping reveals the age structure of the peripheral T cell compartment. Proc Natl Acad Sci U S A 116, 3974–3981. 10.1073/pnas.1811634116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Komuczki J et al. (2019) Fate-Mapping of GM-CSF Expression Identifies a Discrete Subset of Inflammation-Driving T Helper Cells Regulated by Cytokines IL-23 and IL-1beta. Immunity 50, 1289–1304.e1286. 10.1016/j.immuni.2019.04.006 [DOI] [PubMed] [Google Scholar]
- 101.Van Gassen S et al. (2015) FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645. 10.1002/cyto.a.22625 [DOI] [PubMed] [Google Scholar]
- 102.Wolff C et al. (2018) Multi-view light-sheet imaging and tracking with the MaMuT software reveals the cell lineage of a direct developing arthropod limb. Elife 7. 10.7554/eLife.34410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McDole K et al. (2018) In Toto Imaging and Reconstruction of Post-Implantation Mouse Development at the Single-Cell Level. Cell 175, 859–876.e833. 10.1016/j.cell.2018.09.031 [DOI] [PubMed] [Google Scholar]
- 104.Yan X et al. (2013) A transgenic tri-modality reporter mouse. PLoS One 8, e73580. 10.1371/journal.pone.0073580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dutia BM et al. (2009) A novel Cre recombinase imaging system for tracking lymphotropic virus infection in vivo. PLoS One 4, e6492. 10.1371/journal.pone.0006492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stahl PL et al. (2016) Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82. 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- 107.Yuan J et al. (2018) SCOPE-Seq: a scalable technology for linking live cell imaging and single-cell RNA sequencing. Genome Biol 19, 227. 10.1186/s13059-018-1607-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nitta N et al. (2018) Intelligent Image-Activated Cell Sorting. Cell 175, 266–276.e213. 10.1016/j.cell.2018.08.028 [DOI] [PubMed] [Google Scholar]
- 109.Campbell KR and Yau C (2018) Uncovering pseudotemporal trajectories with covariates from single cell and bulk expression data. Nat Commun 9, 2442. 10.1038/s41467-018-04696-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bowling S et al. (2020) An Engineered CRISPR-Cas9 Mouse Line for Simultaneous Readout of Lineage Histories and Gene Expression Profiles in Single Cells. Cell 181, 1693–1694. 10.1016/j.cell.2020.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kalhor R et al. (2018) Developmental barcoding of whole mouse via homing CRISPR. Science 361. 10.1126/science.aat9804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Spanjaard B et al. (2018) Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol 36, 469–473. 10.1038/nbt.4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Z et al. (2017) Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci Rep 7, 2193. 10.1038/s41598-017-02460-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ahmadzadeh E et al. (2020) A collection of genetic mouse lines and related tools for inducible and reversible intersectional mis-expression. Development 147. 10.1242/dev.186650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Daigle TL et al. (2018) A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell 174, 465–480.e422. 10.1016/j.cell.2018.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Leeper K et al. (2021) Lineage barcoding in mice with homing CRISPR. Nat Protoc 16, 2088–2108. 10.1038/s41596-020-00485-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fliss AE et al. (2000) Control of estrogen receptor ligand binding by Hsp90. J Steroid Biochem Mol Biol 72, 223–230. 10.1016/s0960-0760(00)00037-6 [DOI] [PubMed] [Google Scholar]
- 118.Meador K et al. (2019) Achieving tight control of a photoactivatable Cre recombinase gene switch: new design strategies and functional characterization in mammalian cells and rodent. Nucleic Acids Res 47, e97. 10.1093/nar/gkz585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wagner DE and Klein AM (2020) Lineage tracing meets single-cell omics: opportunities and challenges. Nature Reviews Genetics 21, 410–427. 10.1038/s41576-020-0223-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Futamura K et al. (2015) Novel full-spectral flow cytometry with multiple spectrally-adjacent fluorescent proteins and fluorochromes and visualization of in vivo cellular movement. Cytometry A 87, 830–842. 10.1002/cyto.a.22725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hor JL and Germain RN (2021) Intravital and high-content multiplex imaging of the immune system. Trends Cell Biol. 10.1016/j.tcb.2021.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Molbay M et al. (2021) A guidebook for DISCO tissue clearing. Mol Syst Biol 17, e9807. 10.15252/msb.20209807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thunemann M et al. (2017) Cre/lox-assisted non-invasive in vivo tracking of specific cell populations by positron emission tomography. Nat Commun 8, 444. 10.1038/s41467-017-00482-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schönig K and Bujard H (2003) Generating conditional mouse mutants via tetracycline-controlled gene expression. Methods Mol Biol 209, 69–104. 10.1385/1-59259-340-2:69 [DOI] [PubMed] [Google Scholar]
- 125.Iturri L et al. (2021) Megakaryocyte production is sustained by direct differentiation from erythromyeloid progenitors in the yolk sac until midgestation. Immunity 54, 1433–1446.e1435. 10.1016/j.immuni.2021.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ginhoux F et al. (2010) Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shalaby KH et al. (2018) Pathogenic TH17 inflammation is sustained in the lungs by conventional dendritic cells and Toll-like receptor 4 signaling. J Allergy Clin Immunol 142, 1229–1242.e1226. 10.1016/j.jaci.2017.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yao Y et al. (2018) Embryonic Fate Mapping Uncovers the Critical Role of microRNAs in the Development of Epidermal gammadelta T Cells. J Invest Dermatol 138, 236–239. 10.1016/j.jid.2017.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gomes T et al. (2019) Immunology Driven by Large-Scale Single-Cell Sequencing. Trends Immunol 40, 1011–1021. 10.1016/j.it.2019.09.004 [DOI] [PubMed] [Google Scholar]