Abstract
Objective:
To propose EV-derived mRNA as a potential diagnostic biomarker detecting the presence of clear cell renal cell carcinoma (ccRCC). There is currently no kidney cancer specific screening or diagnostic technology. Therefore, one-third of kidney cancer diagnoses occur after the cancer has metastasized and is past curative measures
Materials and Methods:
Urine, plasma, normal tumor adjacent tissue, and tumor tissue was collected from a limited population of ccRCC patients. Extracellular vesicle (EV) isolation was performed on each sample, followed by mRNA extraction from isolated EVs. NanoString® nCounter technology was utilized to count the mRNA transcripts present in matched plasma, urine, tumor tissue, and normal tumor adjacent tissue samples.
Results:
770 mRNA transcripts related to gene’s affecting cancer’s progression and metastasis processes were evaluated. Four EV derived mRNA transcripts (ALOX5, RBL2, VEGFA, TLK2) were found specific to urine and cancer tissue samples.
Conclusion:
Four candidate RCC-specific urine EV biomarkers were identified. However, due to the lack of a true negative control and urine collection techniques, further re-examination is necessary for validation. This study demonstrates the promise of defining disease-specific EV biomarkers in liquid biopsy patient samples.
Keywords: renal cell carcinoma, clear cell renal cell carcinoma, extracellular vesicles, liquid biopsy, biomarker
Introduction
Kidney cancer was globally responsible for the deaths of 179,368 men and women in 2020 [1, 2], contributing to 2% of cancer diagnoses and cancer-related deaths. [1] There are several types and subtypes of kidney cancer that can affect one or both kidneys. Renal cell carcinoma (RCC) makes up 90% of all kidney cancer diagnoses. [3] The most common subtype of RCC, making up 70% of all kidney cancer diagnoses, is clear cell RCC (ccRCC). [3, 4] Due to the asymptomatic and non-kidney specific symptomatic nature of the disease, one-third of kidney cancers are diagnosed after the disease has progressed to locally advanced disease or metastasized. [4–6] Metastatic RCC has a five-year survival rate of 12% [7], highlighting the urgency of sensitive and specific diagnostic biomarkers for earlier disease diagnosis.
There are currently no noninvasive diagnostic tests for kidney cancer. ccRCC is typically diagnosed following routine imaging tests and a core needle biopsy. [8] According to a recent study on the effectiveness of core biopsies for kidney cancers, 36.7% of individuals (n=3113) with a negative biopsy had malignant disease on surgical pathology. [8] Imaging as a screening technique is used in developed countries, such as the United States; however, the absence of a kidney cancer-specific biomarker does not allow for image-based diagnoses. [9]
Liquid biopsy is a promising diagnostic tool for RCC. Other liquid biopsy biomarkers have been explored for RCC, including circulating tumor cells (CTCs) and cell-free tumor (ctDNA). [6, 10] In this study, we specifically interrogate extracellular vesicles (EVs) as a possible source for diagnostic liquid biopsy biomarkers for kidney cancer. EVs are lipid bilayer membrane enclosed particles that are released by cells and have been identified in all bodily fluids in abundance. [11–13] EVs contain heterogeneous cargo, including varying amounts of nucleic acids, proteins, lipids, and other small molecules, dependent on the cell or tissue of origin. [11] Coding mRNA has been identified as EV cargo, including from RCC cells, by many groups. [4, 14–17] The specificity of mRNA detection assays and the relative low input required makes EV-derived mRNA a strong candidate as a potential EV liquid biopsy diagnostic biomarker prior to detection through conventional diagnostic techniques.
In this study, we evaluate mRNA cargo from EVs isolated from ccRCC patient biofluids, urine and plasma, as well as, EVs isolated directly from RCC tumor tissue and tumor adjacent tissue. Applying this strategy to a pilot study group, we aimed to optimize the identification of candidate RCC EV biomarkers for diagnostic validation through comparison to tumor tissue derived EVs.
Methods & Materials
Sample Collection
Blood, urine, and tissue samples were collected from eleven patients diagnosed with ccRCC prior to radical nephrectomy. (Table 1) Two patients (not included in the majority of the analyses) received neoadjuvant immunotherapy prior to collection. (Supplementary Table 1) Median age of patients was 59 (range 47-86). Six of the patients were male and three were female. ISUP scores spanned 2-4. Size and pT stages were reported as stated in Table 1. Samples were collected following written informed consent in accordance with the Declaration of Helsinki and approval by the Johns Hopkins Office of Human Subjects Research Institutional Review Board. Patient selection was based on clinical tumor size ≥4 cm to ensure sufficient tumor tissue was available for clinical pathology. Blood samples were collected in standard EDTA collection tubes. Urine was collected via catheter during the patient’s surgical procedure. Tumor tissue and tumor adjacent tissue were collected immediately following the procedure, prior to formalin fixation.
Table 1: Clinical information of ccRCC patient population.
Clinical information for patients consented for study collections. Patients were diagnosed with ccRCC and underwent no treatment prior to collection. pTNM and ISUP scores were provided from pathology reports following surgery. Tumor size refers to the maximum diameter of tumor listed on pathology reports
| Patient ID | Age | Sex | Ethnicity | Race | Size (cm) | pTNM Score | ISUP |
|---|---|---|---|---|---|---|---|
| 1 | 61 | F | Not Hispanic | White or Caucasian | 4.3 | pT3aNx | 2 |
| 2 | 59 | M | Not Hispanic | White or Caucasian | 5.4 | pT1bNxM1 | 3 |
| 3 | 52 | F | Not Hispanic | White or Caucasian | 15 | pT4Nx | 4 |
| 4 | 52 | M | Not Hispanic | White or Caucasian | 4.1 | pT1bNx | 3 |
| 5 | 86 | M | Not Hispanic | White or Caucasian | 6 | pT3aNx | 2 |
| 6 | 74 | M | Not Hispanic | White or Caucasian | 5.6 | pT3aNx | 3 |
| 7 | 57 | M | Not Hispanic | White or Caucasian | 4 | pT3aNx | 2 |
| 8 | 47 | F | Not Hispanic | White or Caucasian | 6.6 | pT3aNx | 2 |
| 9 | 60 | M | Not Hispanic | Asian | 9 | pT2aN0 | 2 |
Sample inclusion was based on availability of tissue; sufficient non-atrophic tumor and tumor adjacent tissue had to be present in the surgical specimen to warrant the clinical pathological procedures.
Tissue-conditioned media for EV isolation
Tumor tissue and tumor adjacent tissue from each patient were processed as previously reported. [12] Following removal of fat and capsule, at least 225 mg of tissue was cut into 2 mm pieces and PBS-washed. Washed tissue was incubated in 15 mL of serum-free media with added collagenase D and DNase I. After 30 minutes incubation, the tissue was filtered from the media using a 70 μm cell strainer. Differential centrifugation at 4 °C was used to pellet and remove live cells, dead cells, and debris. The sequential centrifuge settings were 500 × g for 5 minutes, 2,000 × g for 20 minutes, and 10,000 × g for 20 minutes. The tissue-conditioned media (TCM) containing EVs was then filtered twice using 0.8 μm and 0.45 μm hydrophilic polyether sulfone syringe filters (PALL). Filtered TCM was stored at minus 80 °C. Prior to EV isolation, thawed media was centrifuged at 1,000 x g to clear salt precipitates.
EV Isolation from patient samples
EV isolation was performed as previously described. [12] EV isolation techniques for each sample type (plasma, urine, tissue) are included in the supplemental material.
mRNA isolation from sample derived EVs
Total RNA was isolated from sample derived EVs. 1 mL plasma, 17 mL urine, and 225 mg of tissue derived EVs were input for RNA isolation. Isolated EVs were thawed on ice prior to RNA extraction. For isolation, the miRNeasy micro kit (Qiagen) was used according to manufacturer’s protocol, including one-column treatment with DNase I (Qiagen). RNA was eluted in 20 μL RNase-free water and stored at minus 80 °C.
mRNA expression profiling using NanoString®
mRNA was assayed by the nCounter PanCancer Progression Panel (NanoString Technologies). NanoString hybridization is optimized for cellular quantities of bioparticles, and, therefore, an amplification step prior to hybridization is necessary for the determination of EV derived mRNA. The targeted genes were amplified using the nCounter Low RNA Input Kit (NanoString Technologies) following manufacturer’s instructions with no modification, as previously validated. [18] Amplified products were used as input for hybridization to the nCounter PanCancer Progression Panel probes. Hybridized samples were loaded into an nCounter Sprint cartridge and run on a nCounter SPRINT (NanoString Technologies).
Analysis of EV derived mRNA transcript counts
Data generated by the nCounter PanCancer Progression Panel were processed by nSolver Analysis Software v 4.0 (NanoString Technologies) as previously described. [18] All samples were normalized to the total counts of the nCounter-defined positive controls to reduce lane-to-lane variation from cartridge loading and normalize binding affinity across all samples surveyed. mRNA transcript reads of less than 40 were considered undetected. EVs isolated from samples of untreated patients were analyzed together. To be considered a positive “hit” in a particular sample type, an mRNA had to be detected in at least three of nine patients per sample type. For the two previously treated patients, an mRNA had to be detected in one patient per sample type.
Results
mRNA EV cargo from ccRCC patient tumor, tumor adjacent tissue, plasma, and urine
The NanoString PanCancer Progression panel targets 770 mRNA transcripts related to genes affecting cancer’s progression and metastasis processes. nCounter was used to enumerate mRNA transcripts present as EV cargo. Samples were normalized to standard nCounter defined loading controls and mRNA counts of greater than 40 were considered present and mRNA counts of 40 or fewer were defined as not detected. (Supplemental Figure 1)
The number of mRNAs detected in the cargo of EV varied by sample and by patient. (Table 2) 37-81 transcripts per patient (average 61.625, median 61.625) were detected in urine derived EVs. The number of mRNA transcripts detected in the cargo of tumor derived EV ranged from 1-170 transcripts per patient (average 50.667, median 23). The number of mRNA transcripts present in tumor adjacent tissue EV samples ranged from 3-144 transcripts per patient (average 61.556, median 50). The fewest number of mRNAs were detected in plasma derived EVs, with 1-5 transcripts per patient detected (average 1.778, median 1). (Table 2)
Table 2: Number of mRNA transcripts present per sample.
Enumeration of mRNA transcripts exceeding 40 counts detected per sample per patient type.
| Patient ID | mRNA transcripts present at 40+ counts per sample type per patient | |||
|---|---|---|---|---|
| Plasma | Urine | Tumor | Tumor Adjacent | |
| 1 | 1 | 71 | 6 | 9 |
| 2 | 1 | N/A | 16* | 118* |
| 3 | 1 | 81 | 24* | 144 |
| 4 | 1 | 71 | 75* | 71* |
| 5 | 1 | 56 | 23 | 50 |
| 6 | 3 | 55 | 1 | 3 |
| 7 | 5 | 67 | 4 | 10 |
| 8 | 1 | 55 | 170 | 18 |
| 9 | 2 | 37 | 137 | 131 |
Please note there was no urine collected from patient 2. Asterisks denote samples with EV outputs below normalization. These samples had lowered EV input for mRNA isolation compared to the remaining samples.
Defining an RCC-specific EV mRNA signature
To identify candidate RCC-specific EV mRNA cargo, mRNA transcripts found in EVs derived from tumor tissue, tumor adjacent tissue, urine, and plasma were directly compared. (Figure 1) To be scored as present in a particular sample type, we determined the mRNA had to be detected in at least 3 patients. Using these criteria, 49 mRNAs were detected in tumor derived EVs, 67 in tumor adjacent tissue derived EVs, and 79 in urine derived EVs. Only one mRNA transcript, STAT1, was identified in EVs isolated from plasma. STAT1 was found in abundance across all four sample types, and, therefore, plasma was excluded from further analysis.
Figure 1. Comparison of mRNA transcripts present per sample type.
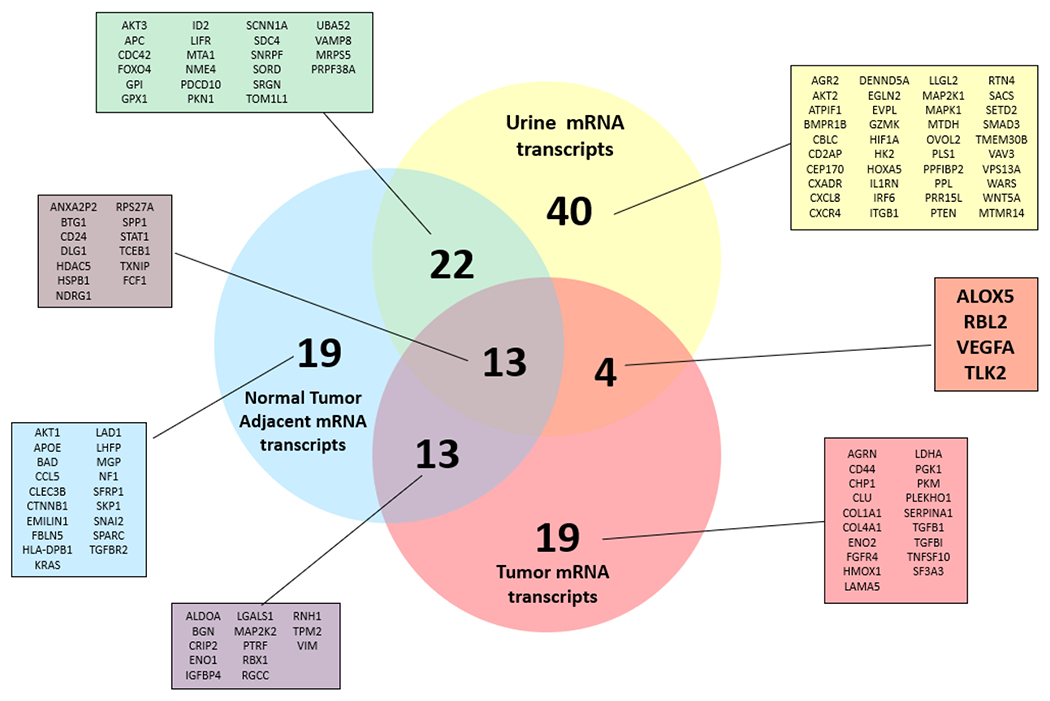
Venn diagram depicting the relationships between mRNA transcripts detected per sample type. To be considered present, mRNA transcript counts must exceed 40 in at least 3 patients’ samples per sample type. Four mRNA transcripts were found unique to urine and tumor derived EVs: ALOX5, RBL2, VEGFA, TLK2.
mRNA cargo was compared among the remaining three sample types to determine any mRNA transcripts that were unique to a particular sample type or shared between multiple sample types. (Figure 1) Each sample type had unique mRNA EV cargo. 40 mRNA transcripts were found to be unique to urine derived EVs, 19 mRNA transcripts were unique to tumor adjacent tissue derived EVs, and 19 mRNAs were only detected in tumor tissue derived EVs. 13 mRNA transcripts were found to be present in all three samples.
The ideal mRNA EV biomarker would be present in tumor derived EV samples and urine derived EV samples, but absent from tumor adjacent tissue derived EV cargo. 13 mRNA transcripts were detected in both tumor tissue derived EVs and tumor adjacent tissue derived EVs, likely representing EV characteristics that are shared among all renal cells. The greatest overlap in shared mRNA transcripts was observed between tumor adjacent tissue derived EV samples and urine derived EV samples with 22 shared genes. In contrast, only four mRNAs were common in both tumor tissue derived and urine derived EV samples: ALOX5, RBL2, VEGFA, and TLK2. (Figure 1)
While at least one of these four mRNAs was detected in tumor derived or urine derived EVs from all patients, no single patient had all four of these mRNA transcripts detected in both tumor derived EVs and urine derived EVs. (Table 3) There was not a clear pattern of mRNA detection between tumor derived EV and urine derived EV within patients, with no concordance between the mRNA detected or absent in both sample types.
Table 3: Distribution of Urine, Tumor unique mRNA transcripts across patient samples.
Distribution of the four mRNA transcripts unique to urine and tumor samples across patients. Please note, patient 2 did not have urine collected.
| mRNA Transcript | Urine | Tumor | Tumor Adjacent | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| ALOX5 | + | N/A | − | + | + | − | − | − | − | − | − | − | + | − | − | − | + | + | − | − | − | − | − | − | − | − | − |
| RBL2 | + | N/A | + | + | + | − | + | + | − | + | − | − | − | + | − | + | − | − | − | + | + | − | − | − | − | − | − |
| VEGFA | + | N/A | + | − | − | − | − | − | + | − | − | − | + | + | − | − | + | + | − | − | − | − | − | − | − | − | + |
| TLK2 | + | N/A | + | + | + | + | + | + | − | + | − | − | − | + | − | − | + | − | − | − | − | − | − | − | − | + | − |
EV derived mRNA transcripts in patients receiving immunotherapy prior to collection
In addition to the treatment-naïve RCC patient samples, EVs were also isolated from tumor, tumor adjacent tissue, plasma, and urine from two patients with neoadjuvant therapy. The number of mRNAs detected in EVs differed by sample type: 49 and 67 transcripts per patient were identified from tumor derived EVs; 10 and 41 mRNAs from tumor adjacent tissue derived EVs, 167 and 170 transcripts per patient detected in urine derived EVs, and only 1 and 2 transcripts identified in the cargo of plasma derived EVs. (Supplemental Table 2) As in the treatment-naïve patients, there were mRNAs unique to a particular sample type and those that were shared among multiple sample types. (Supplemental Figure 2, Supplemental Table 3) Two of the four identified mRNAs from non-treated patients that were present in the tumor and urine derived EVs and absent from the tumor adjacent tissue derived EVs — ALOX5 and VEGFA — were also present in the same EV population in treated patients. An additional nine mRNAs met these criteria: CAMK2D, DLG1, EN02, PPP3R1, ROCK2, SORD, TNFSF10, FCF1, and SF3A3.
Discussion
There is an unmet clinical need for a noninvasive diagnostic test with high sensitivity and specificity for RCC. Liquid biopsy work in the RCC field has brought attention to potential prognostic biomarkers detailing overall survival and disease outcome in patients. [6, 15, 19–22] The study of EVs within RCC, while limited, has followed a similar route. It is hypothesized that tumor derived EVs may promote disease progression and metastasis, indicating poor prognosis. [4, 15] We have previously described EV isolation from kidney and RCC tissue in culture, demonstrating that isolation of excreted EVs directly from tissue is a robust and feasible method. [23]
This is the first study to query the mRNA cargo of RCC-specific liquid biopsy derived EVs comparative to tissue biopsy EVs. EVs are present at high amounts in the plasma and urine, ideal biofluids for liquid biopsy. Those biofluids, however, will include EVs from normal cells and the tumor itself. To assess the identity of RCC derived EVs directly, we harvested EVs from media conditioned from fresh viable tissue. The PanCancer Progression panel was utilized to assess a targeted set of 770 mRNAs implicated in the development and progression of cancer. This strategy enables two important analyses: determining the mRNA signature of RCC patient’s urine and plasma EVs and identification of biofluid EV cargo specific to the positive control RCC tumor tissue.
This study highlights the heterogeneity of mRNA cargo in EVs, both within sample types of a single patient and among patients. While EV mRNA cargo was detected in at least one sample from every patient, there was no correlation of mRNA cargo detected in EVs from different sample types of the same patient. For example, Patient 9 had the second highest number of mRNA transcripts present in both tumor tissue (137) and tumor adjacent tissue (131), but lowest number of mRNA transcripts in urine (37). There was also no correlation of mRNAs detected in the same sample types from different patients, as seen with the range of tumor tissue derived EV mRNA (1-170 transcripts). Overall, unique urine derived EVs provided the highest number of mRNA transcripts detected with more than twice as many unique mRNAs detected as from tumor adjacent tissue and tumor tissue derived EVs. [12] Notably, plasma derived EVs had very low levels of mRNA cargo detected, with only a single mRNA, STAT1, reaching the required threshold. STAT1 has been linked to poor prognosis and, specifically within RCC, lowered overall survival rates in patients with solid tumors. [22]
The ideal RCC biomarker would be present in biofluid EV samples and tumor derived EVs, but absent in tumor adjacent tissue derived EVs. As such, any mRNA transcripts that were detected in the tumor adjacent EV samples were excluded as potential diagnostic biomarkers for ccRCC. 13 mRNA transcripts including AKT1, APOE, BAD, CCL5, CLEC3B, CTNNB1, EMILIN1, FBLN5, HLA-DPB1, KRAS, LAD1, LHFP, MGP, NF1, SFRP1, SKP1, SNAI2, SPARC, and TGFBR2 were detected in EV cargo from both tumor derived EVs and tumor adjacent tissue derived EVs. (Figure 1) As both sample types arise from the kidney, this mRNA signature may represent kidney-specific EV cargo. Additionally, the presence of tumor may alter the composition of the tumor adjacent tissue, making it not a true negative control.
To identify candidate liquid biopsy biomarkers, mRNAs from each sample type were compared to identify urine and RCC tumor tissue unique transcripts not detected tumor adjacent tissue EVs. Four mRNA transcripts met this criterion: ALOX5, RBL2, TLK2, and VEGFA. These transcripts are also present in EV samples from treated patients, however, not all were urine and tumor tissue specific. RBL2 and TLK2 were found to be unique to treated urine samples and were not present in either tissue sample. ALOX5 and VEGFA were also found unique to tumor and urine derived EVs from treated patients.
Wierzbicki et al report high expression of VEGFA in ccRCC cancer tissue correlated to worsened survival and progression-free survival rates in ccRCC patients. [21] Elevated expression of ALOX5 predicts reduced survival rates in ccRCC patients and was consistently found upregulated in tumor samples comparative to healthy tissue samples. [24] RBL2 and TLK2 are regulators of cell division processes. RBL2 regulates the cell’s entrance into cell division and plays a role in determining the cell’s fate by cell cycle arrest or apoptosis. [20, 25] TLK2 regulates chromatin assembly in S phase and regulates histone chaperone H3/H4v. [26]
In a previous study, we defined the mRNA signature of EVs released from common RCC cell lines using the same strategy as we did here. [23] None of the mRNA cargo identified from ccRCC tumor derived EVs were detected in the RCC cell line derived EVs. [23] This important finding highlights the limitation of in vitro modeling to understand RCC biology, EV biology, and in informing biomarker research.
This study was the first characterization of mRNA cargo of EVs isolated directly from RCC tumor tissue, providing a unique opportunity to identify tumor-specific EV biomarkers present in the urine. While this study is an advancement in identifying sensitive and specific RCC diagnostic biomarkers, it has limitations that should be resolved in future studies. First, the urine for this study was collected via the catheter at time of nephrectomy, possibly affecting EV yield and cargo. Catheter urine may contain a greater number of contaminating cells and EVs than a fresh catch sample due to catheter placement. For clinical utility, a liquid biopsy must be readily available at a routine clinic visit, such as fresh catch urine. Fresh catch urine may produce fewer EVs than catheter urine, however, we are not dissuaded by this limitation affecting the validity of tumor specific EVs detected in urine. Any contaminating cell derived EVs isolated from catheter urine should be non-tumor specific when compared to the positive control tumor tissue. Using tumor adjacent tissue EVs as an exclusionary factor also eliminated any overlap in contaminating cell EV analysis. The robust exclusion criteria support the likelihood of ALOX5, RBL2, TLK2, and VEGFA as RCC-specific EV cargo, future work should confirm the presence of these in fresh urine samples of newly diagnosed RCC patients. Second, this study was designed to identify candidate EV biomarkers specific to ccRCC, but does not include any non-ccRCC or non-cancer controls. Using tumor adjacent tissue as a negative control was a stringent way to assess biomarkers. However, this approach is limited as the EV milieu of tumor adjacent tissue may be influenced by the tumor (vs a non-affected tissue sample). Lastly, the limited population size is too small to conclusively validate the candidate RCC-specific EV biomarkers; however, it is encouraging that the determination of four candidate biomarkers was still possible from such a limited sample pool. Forty transcripts were identified in urine derived EVs that were not detected in tissue derived EV samples. These may represent possible disease-specific biomarkers that were below the limit of detection in our studies. Notably, this work found that plasma is unlikely to be a good source for EV derived biomarkers in RCC. These data, while limited, suggest that EV biomarkers may be promising as prognostic biomarkers. We believe the technology in this study has the potential to produce a sensitive and specific biomarker for the diagnosis of ccRCC. The application of such methods to a larger sample size and controls will be necessary for validation, but this pilot study holds great implications for the RCC liquid biopsy field.
Conclusion
This study demonstrates the promise of defining disease-specific EV biomarkers in a liquid biopsy sample. While this study focused on RCC-specific biomarkers, a similar strategy could be utilized for other cancer or disease types. On the basis of this work, four candidate RCC-specific urine EV biomarkers were identified. While no single gene was found in the urine and tumor derived EVs from all patients, every patient had at least one of these four mRNA transcripts detected in urine derived EV. With validation, such strategies could be utilized in the clinical setting for diagnostic purposes in RCC and other cancers.
Supplementary Material
Supplemental Figure 1 Distribution of mRNA transcripts present in patients’ samples Visual representation of the number of mRNA transcripts exceeding 40 counts detected per sample type per patient. There is no distinct pattern within patient samples or sample types.
Supplemental Figure 2 Comparison of mRNA transcripts present per sample type in patients receiving treatment Venn diagram depicting the relationships between mRNA transcripts detected per treated sample type in 2 patients receiving neoadjuvant treatment. To be considered present, mRNA transcript counts must exceed 40 in at least 1 patient per sample type. Eleven mRNA transcripts were found unique to urine and tumor derived EVs: ALOX5, CAMK2D, DLG1, ENO2, PPP3R1, ROCK2, SORD, TNFSF10, VEGFA, FCF1, and SF3A3.
Acknowledgements:
The authors thank Tina Wlajnitz and Dr. Phil Pierorazio from the Johns Hopkins University School of Medicine for their contributions in sample collection. We would like to acknowledge the Kiernan Family Fund, who support Dr. Pierorazio’s team and RCC projects for the department. The authors thank NanoString Technologies for their expertise and technical support. RC Zieren is supported by Stichting Cure for Cancer foundation, Amsterdam, The Netherlands. This work was funded by US Department of Defense CDMRP/PCRP (W81XWH-20-10353), the Patrick C. Walsh Prostate Cancer Research Fund and the Prostate Cancer Foundation to SR Amend; and NCI grants U54CA143803, CA163124, CA093900, and CA143055, and the Prostate Cancer Foundation to KJ Pienta.
Conflicts of Interest:
KJ Pienta is a consultant for CUE Biopharma, Inc., is a founder and holds equity interest in Keystone Biopharma, Inc., and receives research support from Progenies, Inc. SR Amend also holds equity interest in Keystone Biopharma, Inc. The other authors declare no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 2018. 68(6): p. 394–424. [DOI] [PubMed] [Google Scholar]
- 2.Global cancer observatory: cancer today, I.A.f.R.o. Cancer, Editor. 2020, World Health Organization: Lyon, France. [Google Scholar]
- 3.Feng X, Zhang L, Tu W, et al. , Frequency, incidence and survival outcomes of clear cell renal cell carcinoma in the United States from 1973 to 2014: A SEER-based analysis. Medicine, 2019. 98(31): p. e16684–e16684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin Z, Xu Q, Hu H, et al. , Extracellular Vesicles in Renal Cell Carcinoma: Multifaceted Roles and Potential Applications Identified by Experimental and Computational Methods. Frontiers in oncology, 2020. 10: p. 724–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns P, Renal cell carcinoma. Cancer biomarkers : section A of Disease markers, 2010. 9(1-6): p. 461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakshminarayanan H, Rutishauser D, Schraml P, et al. , Liquid Biopsies in Renal Cell Carcinoma—Recent Advances and Promising New Technologies for the Early Detection of Metastatic Disease. Frontiers in Oncology, 2020. 10: p. 2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Padala SA, Barsouk A, Thandra KC, et al. , Epidemiology of Renal Cell Carcinoma. World journal of oncology, 2020. 11(3): p. 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel HD, Johnson MH, Pierorazio PM, et al. , Diagnostic Accuracy and Risks of Biopsy in the Diagnosis of a Renal Mass Suspicious for Localized Renal Cell Carcinoma: Systematic Review of the Literature. (1527–3792 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeCastro GJ and McKiernan JM, Epidemiology, Clinical Staging, and Presentation of Renal Cell Carcinoma. Urologic Clinics of North America, 2008. 35(4): p. 581–592. [DOI] [PubMed] [Google Scholar]
- 10.Broncy L and Paterlini-Bréchot P, Circulating Tumor Cells for the Management of Renal Cell Carcinoma. Diagnostics (Basel, Switzerland), 2018. 8(3): p. 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong L, Zieren RC, Wang Y, et al. , Recent advances in extracellular vesicle research for urological cancers: From technology to application. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer, 2019. 1871(2): p. 342–360. [DOI] [PubMed] [Google Scholar]
- 12.Dong L, Zieren RC, Horie K, et al. , Comprehensive evaluation of methods for small extracellular vesicles separation from human plasma, urine and cell culture medium. Journal of extracellular vesicles, 2020. 10(2): p. e12044–e12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crescitelli R, Lässer C, and Lötvall J, Isolation and characterization of extracellular vesicle subpopulations from tissues. Nature Protocols, 2021. 16(3): p. 1548–1580. [DOI] [PubMed] [Google Scholar]
- 14.Kim KM, Abdelmohsen K, Mustapic M, et al. , RNA in extracellular vesicles. Wiley interdisciplinary reviews. RNA, 2017. 8(4): p. 10.1002/wrna.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grange C, Brossa A, and Bussolati B, Extracellular Vesicles and Carried miRNAs in the Progression of Renal Cell Carcinoma. International journal of molecular sciences, 2019. 20(8): p. 1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen G, Zhang Y, and Wu X, 786-0 Renal cancer cell line-derived exosomes promote 786-0 cell migration and invasion in vitro. Oncology letters, 2014. 7(5): p. 1576–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gai C, Pomatto MAC, Grange C, et al. , Extracellular vesicles in onconephrology. Experimental & Molecular Medicine, 2019. 51(3): p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong LA-O, Huang CY, Johnson EJ, et al. , High-Throughput Simultaneous mRNA Profiling Using nCounter Technology Demonstrates That Extracellular Vesicles Contain Different mRNA Transcripts Than Their Parental Prostate Cancer Cells. (1520–6882 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J-A, Tan Y, Wang X, et al. , Comprehensive functional analysis of the tousled-like kinase 2 frequently amplified in aggressive luminal breast cancers. Nature Communications, 2016. 7(1): p. 12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knudsen ES and Knudsen KE, Tailoring to RB: tumour suppressor status and therapeutic response. Nature reviews. Cancer, 2008. 8(9): p. 714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wierzbicki PM, Klacz J, Kotulak-Chrzaszcz A, et al. , Prognostic significance of VHL, HIF1A, HIF2A, VEGFA and p53 expression in patients with clear-cell renal cell carcinoma treated with sunitinib as first-line treatment. (1791–2423 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Wang F, Liu F, et al. , Predicting STAT1 as a prognostic marker in patients with solid cancer. (1758–8340 (Print)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zieren RC, Dong L, Pierorazio PM, et al. , Extracellular vesicle isolation from human renal cancer tissue. Medical Oncology, 2020. 37(4): p. 28. [DOI] [PubMed] [Google Scholar]
- 24.Cui H, Shan H, Miao MZ, et al. , Identification of the key genes and pathways involved in the tumorigenesis and prognosis of kidney renal clear cell carcinoma. (2045–2322 (Electronic)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pentimalli F, Forte IM, Esposito L, et al. , RBL2/p130 is a direct AKT target and is required to induce apoptosis upon AKT inhibition in lung cancer and mesothelioma cell lines. Oncogene, 2018. 37(27): p. 3657–3671. [DOI] [PubMed] [Google Scholar]
- 26.Carrera P, Moshkin YM, Gronke S, et al. , Tousled-like kinase functions with the chromatin assembly pathway regulating nuclear divisions. Genes & development, 2003. 17(20): p. 2578–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Distribution of mRNA transcripts present in patients’ samples Visual representation of the number of mRNA transcripts exceeding 40 counts detected per sample type per patient. There is no distinct pattern within patient samples or sample types.
Supplemental Figure 2 Comparison of mRNA transcripts present per sample type in patients receiving treatment Venn diagram depicting the relationships between mRNA transcripts detected per treated sample type in 2 patients receiving neoadjuvant treatment. To be considered present, mRNA transcript counts must exceed 40 in at least 1 patient per sample type. Eleven mRNA transcripts were found unique to urine and tumor derived EVs: ALOX5, CAMK2D, DLG1, ENO2, PPP3R1, ROCK2, SORD, TNFSF10, VEGFA, FCF1, and SF3A3.


