Abstract
The use of non-Saccharomyces yeast strains in winemaking is becoming a common trend. In fact, consumers are demanding new and healthier styles of wine. On the other hand, these strains are a challenge for the starting process due to winery-resident strains, especially with regard to industrial-scale fermentations. Current assay focuses on the scale-up of the laboratorial inoculum inside the winery environment to ferment 15,000 and 25,000 L of Vitis labrusca Bordô must, using a Hanseniaspora uvarum β-glucosidase-producer strain as starter culture. This scale-up could confirm the viability of using non-Saccharomyces yeast, as it presented promising results on a laboratory scale. The non-Saccharomyces strain was selected in a previous study since it proved to increase resveratrol concentration in lab scale winemaking. The yeast diversity was followed by the plate culturing method. Species identification and strain typing were determined by ITS-RFLP and PCR-fingerprinting, respectively. Physical and chemical analyses and resveratrol quantification were performed in the elaborated wines.
Keywords: Laboratory scale, Non-Saccharomyces yeast, Resveratrol, Table wine, Winemaking, Wine yeast
Introduction
Wine is the result of the alcoholic fermentation of grape must where, essentially, sugars are used to produce ethanol and carbon dioxide. "Yeasts play an important role in the production of numerous fermented beverages, including wines, due to their ability to produce high levels of ethanol, carbon dioxide and highly desirable aromatic compounds, as they follow a seemingly regulated pattern of fermentation (Ashaolu 2019; Ashaolu and Reale 2020)". Saccharomyces cerevisiae is the yeast species associated with the process. Fortunately, yeasts are related with hundreds of chemical reactions that contribute towards wine quality and complexity, especially aroma compounds. Non-Saccharomyces species are present in the first stages of fermentation, greatly contributing to wine aroma and complexity as a result of the production of various metabolites and the activity of several enzymes (Aranda et al. 2011; Pretorius 2000).
β-glucosidases are enzymes secreted by Saccharomyces and non-Saccharomyces yeast and are responsible for hydrolyzing aromatic glycosylated precursors into free volatile compounds to improve wine flavor (Bonciani and Vero 2018; Polizzotto et al. 2016). Several studies have shown that Saccharomyces cerevisiae present low β-glucosidase activity and may be inhibited under certain winemaking conditions (Spagna et al. 2002; Plessis et al. 2017). In this context, species of non-Saccharomyces yeasts seem more promising. A study by Manzanares et al. (2011) has indicated Pichia and Hanseniaspora strains as important β-glucosidase producing strains because they exhibit satisfactory activity on pH of wine and in the presence of ethanol.
β-glucosidases are also involved in the increase of trans-resveratrol, the most biological active form of this stilbene (Todaro et al. 2008). Resveratrol is a natural polyphenolic compound with an important antioxidant activity associated with beneficial effects in neuroprotection and against cardiovascular diseases (Chen et al. 2017; Ling et al. 2017). In a previous study by our research group, it had been demonstrated that four autochthonous strains of the species Hanseniaspora uvarum showed adequate oenological characteristics and hydrolyzed resveratrol-glucosides during the alcoholic fermentation in laboratory-scale winemaking (Gaensly et al. 2015).
Fermentation studies employing autochthonous non-Saccharomyces strains as starter culture, either on a pilot or on a semi-industrial scale, are rather common. Lleixà et al. (2016) used 100 L of Macabeo must fermented by a strain of Hanseniaspora vineae as starter; Padilla et al. (2017) employed four species of non-Saccharoymces as starter cultures in a 1050 L of Grenache and Carignan must; and Belda et al. (2016) conducted winemaking with 700 kg of Tempranillo grapes using a Metschnikowia pulcherrima strain. However, in all the above-mentioned studies, the inoculum has been multiplied only within a laboratory environmental, since the volume of must to be inoculated allowed this strategy.
Since several consumers are willing to pay a higher price for a bottle of wine with increased resveratrol concentration (Barreiro-Hurlé et al. 2008), current study employs one of the previous selected β-glucosidase-producer strains of Hanseniaspora uvarum 187 as starter culture for industrial-scale winemaking, in order to obtain greater complexity, as well as levels of resveratrol increased in wines.
Materials and methods
Chemicals and reagents
Glucose, peptone, yeast extract and agar were obtained from Merck (São Paulo SP Brazil). Glycerol was acquired from Merck (São Paulo SP Brazil). Maurivin B yeast was obtained from Amazon Group® and dNTP, ITS1 and ITS4 primers were acquired from Sigma (USA). Taq polymerase, endonucleases CfoI, HaeIII and HinfI were obtained from Promega (USA). Trans-resveratrol was purchased from Sigma-Aldrich (São Paulo SP Brazil). Formic acid, methanol and hydrochloric acid were acquired from LAS do Brasil (Aparecida de Goiânia GO Brazil).
Strains and biomass production
A β-glucosidase-producer strain of the species Hanseniaspora uvarum 187, isolated from Vitis labrusca grapes in the 2015 vintage, was used as the non-Saccharomyces inoculum. Microorganisms were deposited at the Yeast Culture Collection (WDCM 1056) of the Centro Nacional de Pesquisa de Uva e Vinho (CNPUV), Bento Gonçalves RS Brazil, belonging to the Brazilian Agricultural Research Corporation (EMBRAPA). The strain´s long-term preservation was performed at -20 °C with glycerol as protective agent. For biomass production, cells were primarily grown in YPD medium (glucose 20 g/L, peptone 20 g/L, yeast extract 10 g/L), pH 5.5, at 28 °C, during 18 h using an orbital shaker at 150 rpm. Cells were inoculated on a 5 L bioreactor (New Brunswick Scientific Co) containing 3.6 L of Vitis labrusca grapes var. Bordô (Ives). Growth conditions comprised initial pH 3.8 and constant temperature 28 °C. Aeration and agitation settings were adjusted to 2 vvm (gas volume flow, per unit of liquid volume, per minute) and 300 rpm. After 18 h, the cells were centrifuged, washed with sterile distilled water, and maintained under refrigeration. These steps were repeated to obtain 800 g of cells to start the industrial inoculum.
A commercial strain of Saccharomyces cerevisiae (Maurivin B) was purchased as active dry yeast and rehydrated for control fermentation, following the manufacturer´s instructions.
Industrial-scale fermentation
Biomass produced in the laboratory was transported to the winery and used as starter in a 20 L fermenter. After 24 h at 28 °C the inoculum was added to 120 L of must (Vitis labrusca grapes var. Bordô/Ives) in a 250 L stainless steel fermenter for 48 h at 28 °C, under aerobic conditions.
Industrial winemaking was carried out in 15,000 L and 25,000 L stainless steel vessels, during 2017 and 2018 vintage, respectively. The must had a density of 1050 and 1060 g/L, respectively. The alcoholic fermentation occurred between 18 and 22 °C, during 144 h.
Sampling and yeast isolation
Samples were collected at different times to monitor microbial dynamics, during 2018 vintage: (a) laboratory inoculum, just before delivery to the winery; (b) industrial inoculum, just before its use in the winemaking process; (c) must immediately after inoculation; (d) 24 h after inoculation; (e) 139 h after inoculation; (f) 188 h after inoculation.
After serial dilutions with distilled water, samples were spread on YPD agar plates (pH 5.5). Plates were incubated at 28 °C during 48 h. After the growth, yeasts were isolated by randomly transferring the colonies to individual tubes containing the same culture medium.
Yeast identification
Yeast was identified by Restriction Fragment Length Polymorphism Analysis of PCR-Amplified Fragments (PCR–RFLP) of the ribosomal region, spanning ITS1 (internal transcribed spacer), 5.8S rRNA gene and ITS2. DNA extractions were carried out by the freeze-thawing process described by Silva et al. (2012). The primer pairs were ITS1 and ITS4, described in White et al. (1990). PCR was performed in 25 µL reaction volume containing 100 µM of each dNTP, 1 × PCR buffer, 1.5 mM MgCl2, 0.8 µM of each primer, 1.5 U Taq polymerase and 1 µL DNA template. Amplifications were carried out in a ProFlex thermocycler (Applied Biosystems, USA) with the following PCR program: 94 °C for 5 min, followed by 25 cycles at 94 °C for 30 s; 60 °C for 45 s; 72 °C for 30 s; and a final step at 72 °C for 5 min. The endonucleases used were CfoI, HaeIII and HinfI (Promega, USA). Digestion reactions were performed according to manufacturer’s instruction and temperature and incubation time followed the manufacturer´s recommendations for each enzyme. PCR products were solved in 1% agar gel electrophoresis, while restriction fragments were solved in 3% agar gel electrophoresis. Gels were stained with ethidium bromide and the stained DNA was visualized under UV light on the Gel Doc XR + Documentation System (Bio-Rad, USA). The fragments’ sizes were estimated by 100-bp DNA ladder.
PCR fingerprinting
Isolates from Saccharomyces cerevisiae and Hanseniaspora uvarum were genetically characterized with primer (GTG)5. PCR was performed in 25 µL reaction volume containing 200 µM of each dNTP, 1 × PCR buffer, 1.5 mM MgCl2, 1 µM of each primer, 1.5 U Taq polymerase and 1 µL DNA template. Amplifications were carried out in a ProFlex thermocycler (Applied Biosystems, USA) with the following PCR program: 94 °C for 5 min followed by 40 cycles of 94 °C for 15s; 35 °C for 45 s; 72 °C for 90s, and a final step at 72 °C for 4 min. PCR products were solved in 1.8% agar gel electrophoresis and stained as above.
Chemical analysis of wines
Total sugar content, pH, ethanol, total acidity, volatile acidity and fixed acidity, sulfate, total sulfur dioxide, total and reduced dry extract were analyzed. All parameters were determined according to methodology described by the Brazilian Ministry of Agriculture, Livestock and Food Supply (Brasil 2005).
SPE cartridges (Strata-X-C 200 mg/3 mL, Phenomenex) were used for sample cleanup to determine resveratrol. Cartridges were conditioned with 1 mL methanol and 1 mL ultrapure water. After the conditioning step, 1 mL of the wine sample was run through the cartridge and the analyte was eluted with 1 mL HCl 0.1 N into a polypropylene tube.
Determinations of resveratrol was carried out by HPLC–DAD, following Silva et al. (2017). A Shimadzu system (Kyoto, Japan) equipped with DAD detector (SPD-M10AVP), two dual piston solvent delivery pumps (LC-10AD), a controller module (UFLC CBM-20A) and CLASS VP 6.12 were used. The column selected was a C8 (vertical) 150 × 4.6 mm, 5 µm particle size, carbon load 9%, surface area 450 m2/g and pore size 10 nm, protected by a guard column of C8 material (Phenomenex). Detection was conducted at 306 nm, according to kmax of each analyte. Injection volume was 20 µL and flow rate was 1 mL/min. Whereas mobile phase A consisted of formic acid and water (1:1000, V/V), mobile phase B comprised methanol, formic acid and water (900:1:100, V/V/V). Linear gradient ranged from 30 to 45% of B in 7 min; 45% of B for 7 min; from 45 to 55% of B in 1 min; from 55 to 65% of B in 9 min; from 65 to 90% of B in 1 min; 90% of B for 2 min; from 90 to 30% of B in 0.5 min; 30% of B for 2.5 min. The total time of analysis was 30 min.
Analyte was quantified by external calibration curve constructed by six concentration levels (1.0, 5.0, 10.0, 20.0, 30.0 and 40.0 µg/mL). Ethanol 70% (V/V in water) was used for dilution. Linear regression equations were calculated by the least squares method and linearity was evaluated by ANOVA.
Results and discussion
Yeast population dynamics
Microbial dynamics were followed by yeast colonies grown on YPD plates at the end of the laboratory and industrial inoculum and at the middle and end of fermentation. Taxonomic identification was based on ITS-RFLP, whilst colonies isolated from laboratory inoculum were all identified as Hanseniaspora uvarum, showing a 770 bp ITS-amplicon and a restriction profile, following Agustini et al. (2014).
After a 48-h inoculum multiplication in the winery, 35 colonies were isolated and identified. All of them resulted in an ITS-amplicon size of 880 pb with a restriction profile indicating Saccharomyces cerevisiae, as described in Table 1. The use of an autochthonous starter culture is a challenge when made in non-sterile conditions as in a winery environment since the winery/cellar environment is acknowledged as an important source of microbial material (Varela et al. 2017).
Table 1.
Identification of species with ITS-RFLP profile for isolated yeasts
| Species identification | Time | Amplicon | Endonuclease CfoI | Endonuclease HaeIII | Endonuclease HinfI | Endonuclease DdeI |
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | Industrial inoculum | 880 pb | 360,320,130 | 320,240,180,130 | – | – |
| Saccharomyces cerevisiae | Time zero | 880 pb | 360,320,130 | 320,240,180,130 | – | – |
| Hanseniaspora uvarum | Time zero | 750 pb | – | – | 340, 190, 160 | 300, 180, 90, 80 |
| Hanseniaspora opuntiae | Time zero | 750 pb | 320, 310, 105 | – | 340, 190, 160 | 380, 180, 90, 80 |
| Candida californica | Time zero | 450 pb | 220, 105, 80, 70 | 290, 80, 60 | 240, 210 | – |
| Hanseniaspora uvarum | 24 h | 750 pb | – | – | 340, 190, 160 | 300, 180, 90, 80 |
| Hanseniaspora opuntiae | 24 h | 750 pb | – | – | – | 380, 170, 90, 80 |
| Saccharomyces cerevisiae | 24 h | 880 pb | – | 320,240,180,130 | – | – |
| Saccharomyces cerevisiae | 139 h | 880 pb | – | 320,240,180,130 | – | – |
| Saccharomyces cerevisiae | 188 h | 880 pb | – | 320,240,180,130 | – | – |
When employing a plate culture method to monitor population dynamics, the experiment is susceptible to lose some information on the whole biodiversity present (Lleixà et al. 2016). Since enzymatic mechanism is related to increase in resveratrol yield and although no Hanseniaspora uvarum was detected in the industrial inoculum, the winemaking process was concluded. It has been debated that S. cerevisiae is not acknowledged as a good producer of extracellular enzymes and non-Saccharomyces wine yeasts have been described as potential sources of glycosidase. Mateo and Maicas (2016) have shown that, depending on the strain, β-glucosidase activity could be related to cell wall bound, intra or extra-cellular activity.
Immediately after inoculation, a sample consisting of grape microbiota added to the industrial inoculum was collected from the industrial vessel. Eighteen colonies were isolated at this stage and four different species were detected. The species Saccharomyces cerevisiae amounted to 50% of the strains, followed by Hanseniaspora uvarum, Hanseniaspora opuntiae and Candida californica, respectively with 28, 17 and 6% of isolates. Table 1 shows ITS-RFLP profiles of the strains, compatible with the literature (Agustini et al. 2014, 2018; Wang and Liu 2013).
Twenty-four hours after inoculation, the yeast population dynamics were Saccharomyces cerevisiae (61%), Hanseniaspora uvarum (33%) and Hanseniaspora opuntiae (6%). Finally, the last samples collected (139 and 188 h) provided all isolates as strains from the species Saccharomyces cerevisiae.
Yeast typing
Although no strain of Hanseniaspora uvarum was detected at the end of the industrial inoculum by the plate-culture method, the species appeared during the subsequent sampling. Hence, to test the presence of the inoculated H. uvarum during the fermentation, isolates of the referred species were typified at strain level using PCR-fingerprinting. None of the strains isolated in times 0 (lines 1 to 4) and 24 h (lines 5 to 10) of fermentation has presented an amplification profile similar to the strain grown in the lab scale inoculum (line 11), employing primer (GTG)5 (Fig. 1). The different strains found in the early stages of fermentation probably demonstrate the grape microbiota biodiversity.
Fig. 1.
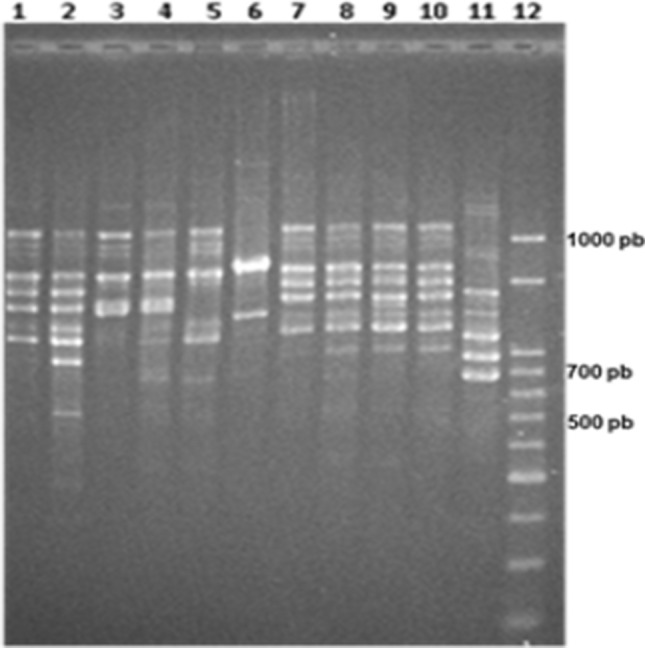
PCR-RAPD profile employing initiator (GTG)5 to compare Hanseniaspora uvarum strains. Lines 1–4 represent yeasts isolated at time 0; lines 5 to 10 show yeasts after 24 h inoculation; line 11 exhibits the laboratory inoculum strain
Considering resident yeasts in the winery environment, the origin of the Saccharomyces cerevisiae strain encountered at the end of the industrial inoculum was investigated. Hence, the strains of Saccharomyces cerevisiae isolated from the industrial inoculum and from the end of fermentation were compared to the commercial strain frequently used in the winery. Figure 2 shows that the commercial strain´s profile is the same as that of the two colonies isolated at the end of industrial inoculum. In fact, it reinforces previous studies which point out that commercial Saccharomyces cerevisiae remained in the cellar and participated in non-inoculated winemaking (Martiniuk et al. 2016; Blanco, 2020). Figure 2 also shows that, at the end of fermentation (lines 4 and 5), a different strain of Saccharomyces cerevisiae had contributed to the process, probably originated from the vineyard. This occurrence corroborated results by Martiniuk et al. (2016) and revealed that the winemaking process is not conducted by a single strain of Saccharomyces cerevisiae. In fact, it demonstrated the complexity of the process based on yeast population composition during the fermentation process.
Fig. 2.
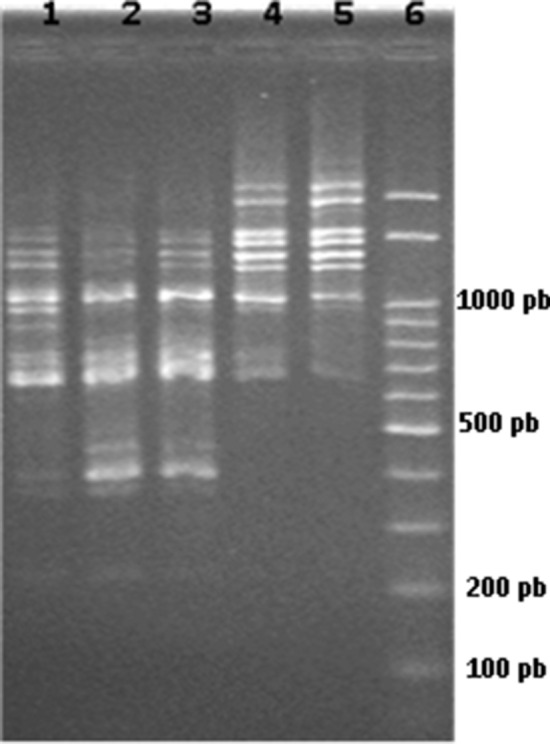
PCR-RAPD profiles employing initiator (GTG)5 with Saccharomyces cerevisiae strains. Line 1 corresponds to commercial strain; lines 2 and 3 represent yeasts from industrial inoculum; lines 4 and 5 exhibit yeasts at 139 and 188 h after inoculation, respectively
Industry-scale fermentation studies using autochthonous non-Saccharomyces yeasts, in which the scale-up of the laboratory inoculum was conducted inside the winery environmental, are rare. In their study, Tristezza et al. (2016) used a mixed culture of autochthonous Hanseniaspora uvarum and Saccharomyces cerevisiae to ferment seven tons of Negroamaro must. The laboratory inoculum was scaled-up in cellar environmental by adding the laboratorial volume (30 L) to 300 kg of Negroamaro grapes during six hours.
The short time used to scale-up the laboratory inoculum into industrial inoculum has permitted the prevalence of the inoculated strains during the process. Contrastingly, in current study, the industrial inoculum grew during 48 h inside the winery environment. Probably this fact caused a resident Saccharomyces cerevisiae strain as the prevalent species in the industrial inoculum.
Lombardi et al. (2018) conducted 9 hL winemaking employing Hanseniaspora guilliermondii as an autochthonous starter culture and 3 days after a commercial strain of Saccharomyces cerevisiae was inoculated. The study, however, did not specify whether the non-Saccharomyces inoculum was made in laboratory or cellar conditions. The dominance of the inoculated strains was confirmed by molecular analysis (PCR-RAPD) in the above-mentioned studies.
Chemical analyses and industrial fermentation
Table 2 shows the oenological parameters of wines elaborated with Hanseniaspora uvarum 187 as starter culture in 2017 and 2018 vintages. The two wines contained less than 4 g/L of residual sugars, whilst alcohol level lay between 8.60 and 14% v/v, following the Brazilian regulation for wine (Brasil 2014, 2018). The amount of sulfur dioxide was lower than the maximum acceptable limit (300 mg/L) established by The International Organization of Vine and Wine (2011) for both vintages.
Table 2.
Physico-chemical characterization of 2017 and 2018 wines employing H. uvarum 187 as starter culture
| Parameters | 2017 | 2018 | Brazilian legislation boundaries |
|---|---|---|---|
| Sugar content (g/L) | 1.60 ± 0.40 | 2.90 ± 0.36 | ≤ 4 * |
| Alcohol content (% vol; 20 ºC) | 10.57 ± 0.35 | 11.27 ± 0.38 | between 8.60 and 14 ** |
| Total dry extract (g/L) | 26.00 ± 0.17 | 25.90 ± 0.17 | – |
| Reduced dry extract (g/L) | 25.40 ± 0.55 | 24.00 ± 0.53 | 21 ** |
| Total acidity (meq/L) | 115.71 ± 1.04 | 103.20 ± 1.379 | between 40 and 130 ** |
| Volatile acidity (meq/L) | 11.01 ± 0.51 | 13.86 ± 0.37 | 20 ** |
| Fixed acidity (meq/L) | 104.70 ± 1.21 | 89.34 ± 1.40 | – |
| pH values | 3.39 ± 0.01 | 3.29 ± 0.01 | – |
| Sulfates (g/L in potassium sulfate) | Less than 0.70 | Less than 0.70 | 1.20 ** |
| Total sulfur dioxide (mg/L) | 52.70 ± 1.00 | 53.80 ± 3.00 | 300 *** |
As previous mentioned, the selected strain of H. uvarum 187 is capable of hydrolyzing the commercial resveratrol-glucoside (piceid) in synthetic media and in lab scale with the natural piceid present in Vitis labrusca Bordô must. This strain exhibited a β-glucosidase activity under winemaking conditions, increasing the resveratrol concentration from 3.18 ± 0.01 mg/L in the must to 6.50 ± 0.40 mg/L in the final wine (Gaensly et al. 2015).
In the case of resveratrol concentrations in current assay for the 2017 vintage, the elaborated wine employing only a Saccharomyces cerevisiae commercial strain produced 3.18 ± 0.04 mg/L of trans-resveratrol and the wine elaborated with Hanseniaspora uvarum 187 achieved 3.38 ± 0.29 mg/L of trans-resveratrol. These rates are not statistically different (p > 0.05). The wine, elaborated in the 2018 vintage employing Hanseniaspora uvarum 187 as non-Saccharomyces inoculum, achieved 3.58 ± 0.12 mg/L of trans-resveratrol, which is not statistically different from the wines produced in the 2017 vintage. Once the β-glucosidase-producer strain of Hanseniaspora uvarum was not able to neither dominate the industrial inoculum nor be found during the fermentation process, the resveratrol concentration between the obtained wines failed to diverge statistically.
Conclusion
Consumer demands for typical and healthier wines have triggered alternative practices in winemaking. The employment of β-glucosidase-producer non-Saccharomyces strains in winemaking is associated with aroma enhancing and increase in resveratrol levels. However, the use of autochthonous non-Saccharomyces yeasts is a challenge with regard to cellar-resident microorganisms. The industrial inoculum multiplication time has to be short to ensure the prevalence of the inoculated strain. The scale-up confirmed the viability of the non-Saccharomyces yeast and it could become a potential strain for producing functional wines.
Acknowledgements
The authors would like to thank the technicians of the Embrapa Grape and Wine for their technical support in resveratrol and molecular analyses.
Authors contributions
CS carried out the experiments and wrote the MS; TMBB was responsible for conceiving the idea, leader of the study; BCA and DB edited the manuscript and supervised the experiments; FD assisted in the execution of the experiments; MGAS edited the manuscript; GAS supervised the work and edited the manuscript.
Funding
Current study was funded by the Brazilian Coordination for the Upgrading of Higher Education Personnel (CAPES).
Availability of data and material
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cintia Rompkovksi, Email: cintia.s.rk@gmail.com.
Bruna Carla Agustini, Email: brunagustini@gmail.com.
Flavia Deffert, Email: flaviadeffert@gmail.com.
Maria Goreti Amboni Stadtlober, Email: goreti08@gmail.com.
Debora Brand, Email: dbrandufpr@gmail.com.
Gildo Almeida da Silva, Email: gildo.almeida@embrapa.br.
Tania Maria Bordin Bonfim, Email: tbonfim@gmail.com.
References
- Agustini BC, da Silva GA, Bonfim TMB. MALDI-TOF MS Supplementary database for species identification employing the yeast diversity encountered on southern Brazil grapes. Folia Microbiol (praha) 2018;63(6):685–693. doi: 10.1007/s12223-018-0607-2. [DOI] [PubMed] [Google Scholar]
- Agustini BC, Silva LP, Bloch C, et al. Evaluation of MALDI-TOF mass spectrometry for identification of environmental yeasts and development of supplementary database. Appl Microbiol Biotechnol. 2014;98:5645–5654. doi: 10.1007/s00253-014-5686-7. [DOI] [PubMed] [Google Scholar]
- Aranda A, Matallana E, del Marcell lı´ O. Molecular Wine Microbiology. 1. Spain: Elsevier; 2011. Saccharomyces Yeasts I: Primary Fermentation; pp. 1–31. [Google Scholar]
- Ashalou TJ. A review on selection of fermentative microorganisms for functional foods and beverages: the production and future perspectives. Food Sci Technol. 2019;54:2511–2519. doi: 10.1111/ijfs.14181. [DOI] [Google Scholar]
- Ashalou TJ. A holistic review on Euro-Asian lactic acid bacteria fermented cereals and vegetables. MDPI. 2020;8:1176. doi: 10.3390/microorganisms8081176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro-Hurlé J, Colombo S, Cantos-villar E. Is there a market for functional wines? Consumer preferences and willingness to pay for resveratrol-enriched red wine. Food Qual Prefer. 2008;19:360–371. doi: 10.1016/j.foodqual.2007.11.004. [DOI] [Google Scholar]
- Belda I, Conchillo LB, Ruiz J, et al. Selection and use of pectinolytic yeasts for improving clarification and phenolic extraction in winemaking. Int J Food Microbiol. 2016;223:1–8. doi: 10.1016/j.ijfoodmicro.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Blanco P, et al. Saccharomyces cerevisiae strain diversity associated with spontaneous fermentations in organic wineries from Galicia (NW Spain) Fermentation. 2020;6:89. doi: 10.3390/fermentation6030089. [DOI] [Google Scholar]
- Bonciani T, De VL. Qualitative and quantitative screening of the b-glucosidase activity in Saccharomyces cerevisiae and Saccharomyces uvarum strains isolated from refrigerated must. Lett Appl Microbiol. 2018;67(1):72–78. doi: 10.1111/lam.12891. [DOI] [PubMed] [Google Scholar]
- Brasil (2005) Instrução normativa nº 24, de 08 de setembro de 2005. Aprova o Manual Operacional de Bebidas e Vinagres do Ministério da Agricultura, Pecuária e Abastecimento. Caderno 04. Fermentados Alcoólicos. Diário Oficial da União. Brasília, DF, 20 set.2005
- Brasil (2014) Decreto nº 8.198 de 20 de fevereiro de 2014. Regulamenta a Lei nº 7.678, de 8 de novembro de 1988, que dispõe sobre a produção, circulação e comercialização do vinho e derivados da uva e do vinho. Diário Oficial da União. Brasília, DF, 21 fev.2014
- Brasil (2018) Instrução normativa nº 14 de 08 de fevereiro de 2018. Estabelece a complementação dos Padrões de Identidade e Qualidade do Vinho e Derivados da Uva e do Vinho. Diário Oficial da União. Brasília, DF, 09 mar. 2018
- Chen AC, Shyu LY, Hsin YL, et al. Resveratrol relieves Angiostrongylus cantonensis—Induced meningoencephalitis by activating sirtuin-1. Acta Trop. 2017;173:76–84. doi: 10.1016/j.actatropica.2017.05.023. [DOI] [PubMed] [Google Scholar]
- Gaensly F, Agustini BC, da Silva GA, et al. Autochthonous yeasts with β-glucosidase activity increase resveratrol concentration during the alcoholic fermentation of Vitis labrusca grape must. J Funct Foods. 2015;19:288–295. doi: 10.1016/j.jff.2015.09.041. [DOI] [Google Scholar]
- International Organization of Vine and Wine (2011) Maximum acceptable limits of various substances contained in wine. Commendium of international methods of analysis.
- Ling L, Gu S, Cheng Y. Resveratrol activates endogenous cardiac stem cells and improves myocardial regeneration following acute myocardial infarction. Mol Med Rep. 2017;15:1188–1194. doi: 10.3892/mmr.2017.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lleixà J, Martín V, del Portillo MC, et al. Comparison of fermentation and wines produced by inoculation of Hanseniaspora vineae and Saccharomyces cerevisiae. Front Microbiol. 2016;7:1–12. doi: 10.3389/fmicb.2016.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi SJ, Pannella G, Iorizzo M, et al. Sequential inoculum of Hanseniaspora guilliermondii and Saccharomyces cerevisiae for winemaking Campanino on an industrial scale. World J Microbiol Biotechnol. 2018;34:161. doi: 10.1007/s11274-018-2540-6. [DOI] [PubMed] [Google Scholar]
- Manzanares P, Vallés S, Viana F. Carrascosa A V, Muñoz R, Gonzales R Molecular Wine Microbiology. 1. UK: Elsevier; 2011. Non-Saccharomyces Yeasts in the Winemaking Process; pp. 85–111. [Google Scholar]
- Martiniuk JT, Pacheco B, Russell G, et al. Impact of commercial strain use on Saccharomyces cerevisiae population structure and dynamics in pinot noir vineyards and spontaneous fermentations of a canadian Winery. PLoS ONE. 2016;11:1–19. doi: 10.1371/journal.pone.0160259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo JJ, Maicas S. Application of Non-Saccharomyces Yeasts to Wine-Making Process. Fermentation. 2016;2:14. doi: 10.3390/fermentation2030014. [DOI] [Google Scholar]
- Padilla B, Zulian L, Ferreres À, et al. Sequential inoculation of native non-saccharomyces and saccharomyces cerevisiae strains for wine making. Front Microbiol. 2017;8:1–12. doi: 10.3389/fmicb.2017.01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis HW, Toit M, Hoff JW, et al. Characterisation of non-saccharomyces yeasts using different methodologies and evaluation of their compatibility with malolactic fermentation. S Afr J Enol Vitic. 2017;38:46–63. [Google Scholar]
- Polizzotto G, Barone E, Ponticello G, et al. Isolation, identification and oenological characterization of non-Saccharomyces yeasts in a Mediterranean island. Lett Appl Microbiol. 2016;63(2):131–138. doi: 10.1111/lam.12599. [DOI] [PubMed] [Google Scholar]
- Pretorius IS. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- da Silva LF, Guerra CC, Klein D, Bergold AM. Solid cation exchange phase to remove interfering anthocyanins in the analysis of other bioactive phenols in red wine. Food Chem. 2017;227:158–165. doi: 10.1016/j.foodchem.2017.01.087. [DOI] [PubMed] [Google Scholar]
- Silva GA, Bernardi TL, Schaker PDC, et al. Rapid yeast DNA extraction by boiling and freeze-thawing without using chemical reagents and dna purification. Brazilian Arch Biol Technol. 2012;55:319–327. doi: 10.1590/S1516-89132012000200020. [DOI] [Google Scholar]
- Spagna G, Barbagallo RN, Palmeri R, et al. Properties of endogenous β-glucosidase of a Pichia anomala strain isolated from Sicilian musts and wines. Enzyme Microb Technol. 2002;31:1036–1041. doi: 10.1016/S0141-0229(02)00239-9. [DOI] [Google Scholar]
- Todaro A, Palmeri R, Barbagallo RN, et al. Food Chemistry Increase of trans-resveratrol in typical Sicilian wine using β-Glucosidase from various sources. Food Chem. 2008;107:1570–1575. doi: 10.1016/j.foodchem.2007.09.075. [DOI] [Google Scholar]
- Tristezza M, Tufariello M, Capozzi V, et al. The oenological potential of Hanseniaspora uvarum in simultaneous and sequential co-fermentation with Saccharomyces cerevisiae for industrial wine production. Front Microbiol. 2016;7:1–14. doi: 10.3389/fmicb.2016.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela C, Barker A, Tran T, et al. Sensory profile and volatile aroma composition of reduced alcohol Merlot wines fermented with Metschnikowia pulcherrima and Saccharomyces uvarum. Int J Food Microbiol. 2017;252:1–9. doi: 10.1016/j.ijfoodmicro.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu Y. Dynamic study of yeast species and Saccharomyces cerevisiae strains during the spontaneous fermentations of Muscat blanc in Jingyang, China. Food Microbiol. 2013;33:172–177. doi: 10.1016/j.fm.2012.09.014. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee E, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


