Abstract
Hemodialysis patients are at high risk for bloodstream infections associated with highest morbidity and mortality rates. Bacterial species not commonly related to such infections has been hardly identified by traditional methods. Pseudocitrobacter is a novel genus of the order Enterobacterales that is associated with carbapenemase genes and nosocomial infection. In this context, we have investigated nine cases of bloodstream infections by carbapenem-resistant Gram-negative bacilli in patients assisted at a hemodialysis unit in Brazil. The infections were caused by a metallo-β-lactamase (IMP-1)-producing clone (> 90% XbaI-PFGE similarity) of Pseudocitrobacter vendiensis, displaying a multidrug-resistant profile to broad-spectrum cephalosporins, carbapenems, chloramphenicol, and trimethoprim-sulfamethoxazole. S1-PFGE and Southern blot hybridization revealed that blaIMP-1 was carried by a 200-kb IncC/ST3 plasmid. Patients were successfully treated with amikacin, and strict disinfection procedures and hand washing protocols were reinforced. We report the emergence of P. vendiensis, a recently described species of the genus, in bloodstream infections of patients undergoing hemodialysis. Considering the epidemic potential of carbapenemase-producing Enterobacterales in hospital settings, surveillance of this emerging pathogen is of utmost importance.
Keywords : Nosocomial infection, Metallo-β-lactamase, Carbapenemase, Enterobacterales, Outbreak
Patients undergoing hemodialysis are at high risk for bloodstream infections (BSI), with both morbidity and mortality being highest in this population [1]. Because of the dialysis process and conditions, they are vulnerable to acquiring such infections by water-borne bacteria not commonly related to them, and the consequent identification of isolates by standard methods is not trustworthy [2, 3]. Pseudocitrobacter is a novel genus of the order Enterobacterales with recent taxonomic revaluation, which has differentiated two species, Pseudocitrobacter faecalis (with Pseudocitrobacter anthropi being a posterior heterotypic synonym) isolated from feces of patients attending military hospitals in Pakistan, and Pseudocitrobacter vendiensis, recently isolated from a hospitalized patient transferred from Spain to Denmark [4, 5]. Worryingly, P. faecalis and P. vendiensis have been associated with production of NDM-1 and KPC-2 carbapenemases, respectively [4, 5]. We hereby report the emergence of IMP-1 metallo-β-lactamase (MβL)-producing P. vendiensis causing bloodstream infection in hemodialysis patients in Brazil.
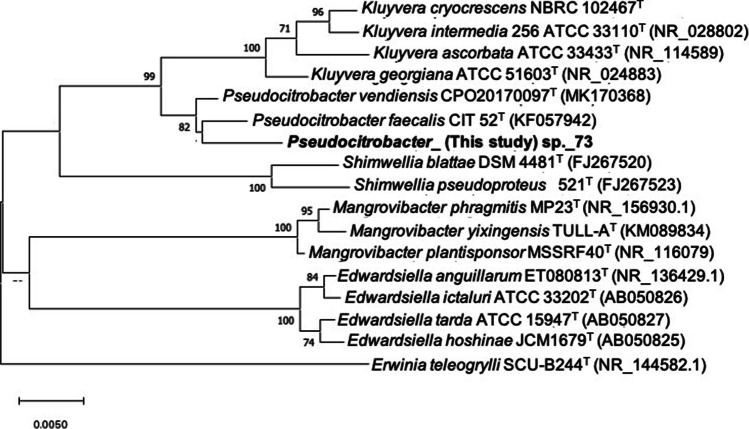
In January 2018, a 56-year-old male patient undergoing hemodialysis in a tertiary-care hospital presented symptoms of bacteremia (i.e., fever, chills and shivering). In this regard, a Gram-negative bacilli (GNB) labeled as HMD01 was isolated from blood culture. Over the following 9 months, eight novel cases of bloodstream infections by GNB (strains HMD02 to HMD09) were confirmed in patients assisted at the same hemodialysis unit. Patients were successfully treated with amikacin 500 mg IV every 48 h for 10 days, whereas strict disinfection procedures and hand washing protocols were reinforced. Initially strains were identified and tested for antimicrobial susceptibility using the VITEK 2 Compact system (bioMérieux). The automated system identified five isolates (HMD03, HMD04, HMD05, HMD06, and HMD09) as Pantoea sp., and four (HMD01, HMD02, HMD07, and HMD08) as Enterobacter cloacae complex. All nine isolates presented identical resistance profile to ampicillin, cefepime, ceftazidime, ceftriaxone, ertapenem (MIC > 32 µg/mL), imipenem (MIC > 32 µg/mL), meropenem (MIC > 32 µg/mL), chloramphenicol, and trimethoprim-sulfamethoxazole according to the CLSI guidelines [6]. Molecular typing was performed by XbaI-PFGE, and carbapenemase genes were screened by PCR and DNA sequencing [7]. Localization of carbapenemase encoding gene was determined by S1-PFGE and Southern blot hybridization [8]. Molecular typing by XbaI-PFGE revealed clonal relatedness (> 93%) among carbapenem-resistant isolates, which carried the blaIMP-1 MβL gene onto a 200-kb IncC/ST3 plasmid. Total DNA of a representative clone (strain HMD06) was extracted and sequenced using the MiSeq platform (Illumina Inc., San Diego, CA), with paired-end reads (250 bp). Reads were submitted to de novo assembly using Unicycler v.0.4.0, and then subjected to automatic annotation using NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v.3.2 (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/). Prediction of bacterial species, resistome, and plasmidome were analyzed using online tools (http://www.genomicepidemiology.org/). Separation of contigs into different networks concerning plasmids and the chromosome was obtained using the PLACNETw tool (https://castillo.dicom.unican.es/upload/). Since 16S ribosomal DNA sequence analysis (SpeciesFinder-2.0) and fast K-mer algorithm (KmerFinder-3.1) failed to resolve the species identification, the GTDB Toolkit (GTDB-Tk) [9] was used to achieve a taxonomic assignment for the HMD006 genome. This tool allocated the strain within the genus Pseudocitrobacter, but it did not match perfectly with the only validly accepted species of this genus until that moment, P. faecalis. Therefore, a phylogenetic analysis of the 16S rRNA gene was performed with type strains of P. faecalis and the recently validly published P. vendiensis, as well as with other type strains of related Enterobacterales species [4, 5]. 16S rRNA gene sequences were obtained from public databases. Multiple alignments were constructed based upon the CLUSTAL W (http://www.clustal.org/clustal2/). The phylogenetic tree was constructed using the neighbor-joining method by using the MEGA X software (https://www.megasoftware.net/). Distance matrices were calculated by the Kimura 2-parameter method and bootstrap analysis was performed based on 1000 re-samplings. Type strains of the genus Pseudocitrobacter formed a monophyletic cluster (Fig. 1), and the strain HMD006 did not cluster with any of the Pseudocitrobacter spp., although it shared 16S rRNA gene similarities of 99.0% and 99.2% with the strain P. faecalis CIT 52 T and P. vendiensis CP020170097T, respectively. Therefore, average nucleotide identity (ANI) and Tetra analyses were performed by using online tools (http://jspecies.ribohost.com/jspeciesws/; https://www.ezbiocloud.net/), in order to confirm the taxonomic position of this strain. The ANI between HMD006 and P. faecalis CIT 52 T strains ranged from 90.8 to 92.2%, and between HMD006 and P. vendiensis CP020170097T ranged from 97.5 to 98.2%; therefore, considering a threshold ≥ 95% for new species definition [10, 11], the strain HMD006 was confirmed as belonging to the species P. vendiensis.
Fig. 1.
Neighbor-joining phylogenetic tree of the 16S rRNA gene (1357 bp) of Enterobacterales species showing the position of Pseudocitrobacter sp. strain 73 (HMD006) within the genus Pseudocitrobacter and other close genera. Bootstrap values (> 70%) based on 1000 replications are shown at the nodes of the tree. Bar, 5 substitutions per 1000 nt
Resistome analysis of P. vendiensis HMD06 revealed the presence of genes conferring resistance to β-lactams [blaIMP-1, blaOXA-1, blaCTX-M-15], aminoglycosides [aac(6’)-31, aac(6’)-Ib-cr, ant(3″)-Ia, aph(3’’)-Ib, and aph(6)-Id], fluoroquinolones [aac(6’)-Ib-cr, qnrB1], tetracycline [tet(A)], phenicols [catB3 and floR], sulphonamides [sul1, sul2], trimethoprim [dfrA14], and quaternary ammonium compounds [qacE]. On the other hand, plasmidome analysis confirmed only the presence of the IncC/ST3 plasmid. In order to better characterize the plasmid involved in the dissemination of the blaIMP-1 gene, the distinct FASTA files containing the separated contigs were additionally submitted to the ResFinder 4.1 tool, and we obtained the localization of each antimicrobial resistance gene. The plasmid IncC/ST3 was determined as presenting about 206.3 kb and containing blaIMP-1, blaOXA-1, tet(A), catB3, floR, qacE, sul1, sul2, aac(6')-31, aph(3'')-Ib, aph(6)-Id, ant(3’’)-Ia, aac(6')-Ib-cr, and qnrB1. A second plasmid of about 77.5 kb, also identified on S1-PFGE, able for mobilization and transferring since it possesses MOB genes, was not identified because it did not present any known replicon or antimicrobial resistance genes (according to the analysis performed on PLACNETw). The blaCTX-M-15 (preceded by an ISEcp1 and succeeded by a Tn3 truncated by an IS26) and dfrA14 genes were identified inserted into the chromosome of P. vendiensis HMD06.
Genomic approaches in microbiology are elucidating genetic backgrounds and taxonomic changes of clinically relevant pathogens [12]. Clinical isolates of Pseudocitrobacter spp. and other species of the order Enterobacterales are commonly incorrectly identified due to the use of traditional methods in clinical laboratories. Phenotypic tests are only partially reliable to distinguish species of bacteria not recognized as usually pathogens to humans, resulting in misidentification, such as the observed in the present study and in others [13, 14]. Genome sequencing is an important taxonomic tool to resolve these issues, and correct identification of potentially pathogenic bacteria is important to establish the appropriated treatment. In this study, we report the emergence of IMP-1-producing P. vendiensis causing an outbreak of invasive infections in hemodialysis patients, in Brazil. Considering the epidemic potential of carbapenemase-producing Enterobacterales in hospital settings, and unfavorable prognosis of critical patients with carbapenem-resistant infections [15], surveillance of this emerging pathogen is of utmost importance.
Acknowledgements
The authors are grateful to CEFAP-GENIAL (Centro de Facilidades para a Pesquisa – Genome Investigation and Analysis Laboratory) facility for Illumina sequencing and Dr Peter Kämpfer from Institut für Angewandte Mikrobiologie (Universität Giessen, Germany), for the valuable assistance with taxonomy.
Author contribution
Letícia Kellen de Andrade, Arturo Levican, Louise Cerdeira, Tiago Casella, Quézia Moura, Bruna Fuga, and Nilton Lincopan performed the investigation process. Andressa Batista Zequini de Morais, Melissa Maia Braz, and Evelin Rodrigues Martins performed the investigation and data curation about the laboratory characterization of P. vendiensis. Nilton Lincopan and Mara Corrêa Lelles Nogueira conducted the project administration and supervision. All authors contributed to the analysis and the writing of the final manuscript.
Funding
This study was funded by the Conselho Nacional de Desenvolvimento Científico e Tecnológico under Grant (CNPq 433128/2018–6, 443819/2018–1 and 312249/2017–9). N.L. is a research grant fellow of CNPq. L.K.A and E.R.M receive scholarships from CAPES.
Data availability
The draft genome sequence of strain HMD006 was deposited at DDBJ/ENA/GenBank, as Pseudocitrobacter sp. 73, under accession number VTZO00000000.1 and BioProject PRJNA563339.
Code availability
Not applicable.
Declarations
Ethics approval
The study was approved by the Research Ethics Committee of the Faculdade de Medicina de São José do Rio Preto under the approval #E: 3.300.442. This study does not require a consent form from the participants.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nishiwaki H, Sasaki S, Hasegawa T, Sasai F, Kawarazaki H, Minatoguchi S, Uchida D, Koitabashi K, Ozeki T, Koiwa F. External validation of the quick sequential organ failure assessment score for mortality and bacteraemia risk evaluation in Japanese patients undergoing haemodialysis: a retrospective multicentre cohort study. BMJ Open. 2019;9(7):e028856. doi: 10.1136/bmjopen-2018-028856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsang C-C, Xiong L, Poon RWS, Chen JHK, Leung K-W, Lam JYW, Wu AKL, Chan JFW, Lau SKP, Woo PCY. Gordoniahongkongensis sp. nov., isolated from blood culture and peritoneal dialysis effluent of patients in Hong Kong. Int J Syst Evol Microbiol. 2016;66(10):3942–3950. doi: 10.1099/ijsem.0.001292. [DOI] [PubMed] [Google Scholar]
- 3.Shahraki AH, Trovato A, Droz S, Haidarieh P, Borroni E, Mirsaeidi M, Mannino R, Hashemzadeh M, Mariottini A, Cirillo DM, Tortoli E. Mycobacteriumaquaticum sp. nov., a rapidly growing species isolated from haemodialysis water. Int J Syst Evol Microbiol. 2017;67(9):3279–3282. doi: 10.1099/ijsem.0.002103. [DOI] [PubMed] [Google Scholar]
- 4.Kämpfer P, Glaeser SP, Raza MW, Abbasi SA, Perry JD. Pseudocitrobacter gen. nov., a novel genus of the Enterobacteriaceae with two new species Pseudocitrobacterfaecalis sp. nov., and Pseudocitrobacteranthropi sp. nov, isolated from fecal samples from hospitalized patients in Pakistan. Syst Appl Microbiol. 2014;37(1):17–22. doi: 10.1016/j.syapm.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Kämpfer P, Fuglsang-Damgaard D, Overballe-Petersen S, Hasman H, Hammerum AM, Fuursted K, Blom J, Glaeser SP, Hansen F. Taxonomic reassessment of the genus Pseudocitrobacter using whole genome sequencing: Pseudocitrobacteranthropi is a later heterotypic synonym of Pseudocitrobacterfaecalis and description of Pseudocitrobactervendiensis sp. nov. Int J Syst Evol Microbiol. 2020;70(2):1315–1320. doi: 10.1099/ijsem.0.003918. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute . M100-S29. Performance standards for antimicrobial susceptibility testing: 29th informational supplement. Wayne: CLSI; 2019. [Google Scholar]
- 7.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JDD. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50(12):3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villa L, Carattoli A. Plasmid typing and classification. Methods Mol Biol. 2020;2075:309–321. doi: 10.1007/978-1-4939-9877-7_22. [DOI] [PubMed] [Google Scholar]
- 9.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019;36(6):1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 11.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102(7):2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillonetto M, Arend L, Gomes SMT, Oliveira MAA, Timm LN, Martins AF, Barth AL, Mazzetti A, Hersemann L, Smits THM, Mira MT, Rezzonico F. Molecular investigation of isolates from a multistate polymicrobial outbreak associated with contaminated total parenteral nutrition in Brazil. BMC Infect Dis. 2018;18:397. doi: 10.1186/s12879-018-3287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng A, Liu C-Y, Tsai H-Y, Hsu M-S, Yang C-J, Huang Y-T, Liao C-H, Hsueh P-R. Bacteremia caused by Pantoea agglomerans at a medical center in Taiwan, 2000–2010. J Microbiol Immunol Infect. 2013;46(3):187–194. doi: 10.1016/j.jmii.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Jean SS, Lee WS, Lam C, Hsu CW, Chen RJ, Hsueh P. Carbapenemase producing Gram-negative bacteria: current epidemics, antimicrobial susceptibility and treatment options. Future Microbiol. 2015;10(3):407–425. doi: 10.2217/fmb.14.135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The draft genome sequence of strain HMD006 was deposited at DDBJ/ENA/GenBank, as Pseudocitrobacter sp. 73, under accession number VTZO00000000.1 and BioProject PRJNA563339.
Not applicable.