Abstract
Klebsiella variicola is generally known as endophyte as well as lignocellulose-degrading strain. However, their roles in goat omasum along with lignocellulolytic genetic repertoire are not yet explored. In this study, five different pectin-degrading bacteria were isolated from a healthy goat omasum. Among them, a new Klebsiella variicola strain HSTU-AAM51 was identified to degrade lignocellulose. The genome of the HSTU-AAM51 strain comprised 5,564,045 bp with a GC content of 57.2% and 5312 coding sequences. The comparison of housekeeping genes (16S rRNA, TonB, gyrase B, RecA) and whole-genome sequence (ANI, pangenome, synteny, DNA-DNA hybridization) revealed that the strain HSTU-AAM51 was clustered with Klebsiella variicola strains, but the HSTU-AAM51 strain was markedly deviated. It consisted of seventeen cellulases (GH1, GH3, GH4, GH5, GH13), fourteen beta-glucosidase (2GH3, 7GH4, 4GH1), two glucosidase, and one pullulanase genes. The strain secreted cellulase, pectinase, and xylanase, lignin peroxidase approximately 76–78 U/mL and 57–60 U/mL, respectively, when it was cultured on banana pseudostem for 96 h. The catalytically important residues of extracellular cellulase, xylanase, mannanase, pectinase, chitinase, and tannase proteins (validated 3D model) were bound to their specific ligands. Besides, genes involved in the benzoate and phenylacetate catabolic pathways as well as laccase and DiP-type peroxidase were annotated, which indicated the strain lignin-degrading potentiality. This study revealed a new K. variicola bacterium from goat omasum which harbored lignin and cellulolytic enzymes that could be utilized for the production of bioethanol from lignocelluloses.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-021-00660-7.
Keywords: Goat omasum strain, Genome comparison, Klebsiella variicola, Lignocellulose, Molecular docking
Introduction
Lignocellulolytic bacteria consist of a variety of hydrolase enzymes that are responsible for the biodegradation of lignocellulosic materials. These enzymes play a key role in the production of biofuel [1]. Biofuel such as bioethanol production is an alternative strategy to mitigate the global fossil fuel crisis. Like other countries, Bangladesh still depends on fossil fuel for daily transport over vehicles on the road and aviation, which contributes to a significant effect on carbon dioxide emission and global warming. Meanwhile, global warming has had a tremendous impact on agriculture and living conditions in Bangladesh. Only in 2019–2020, more than 1100 people died by electricity produced from the firing blaze of the sky during farming practice on lands. To enlighten the darkness of global warming, the utilization of clean energy or bioenergy is increasing locally. However, biofuel production from lignocellulose is under consideration and may be a promising route to mitigate global warming. The sources of lignocellulolytic enzyme such as cellulase, xylanase, and pectinase have not been declared locally yet. Therefore, aside from using pulp, paper, and textile industries, these enzymes’ production is of great importance to developing sustainable saccharification and fermentation technology [2].
The conversion of biomass to biofuel by microorganisms has gained significant momentum over the last several years [3]. Lignin-degrading strains are capable of reducing the lignin content and enhance the digestibility of cellulose [4, 5]. For efficient saccharification, cellulose hydrogen bonds are required to be disrupted to make it easily accessible for cellulase. However, multiple types of pretreatment strategies were applied to lignocellulose, together with the massive amount of commercial enzymes needed for the efficient degradation of celluloses and hemicelluloses. Therefore, the effective enzyme cocktail required to generate a more economic degradation process renders a cheaper bioethanol production [6]. Energy crops like maize and banana are massively cultivated all over the country especially in the northern part of Bangladesh. These plants are only wasted and discarded after harvesting the fruits, which can be promising lignocellulosic sources or secondary substrates of microorganisms for enzyme production. Therefore, the wasted stem and stalk of maize and banana may produce a large amount of monosaccharides.
Several bacterial strains like Bacillus sp., Brevibacillus sp., Cellulomonas sp., Streptomyces sp., and Pseudomonas sp. can decompose lignocellulose by secreting cellulase, hemicellulase, ligninase, swollenin, and pectinase enzymes [7–12]. These enzymes are extendedly categorized into glycosyl hydrolases (GH), carbohydrate esterases (CE), polysaccharide lyases (PL), and auxiliary activities (CBM) [11]. Lignocellulolytic proteins from thermophiles such as Anoxybacillus sp. and Geobacillus sp. have received considerable attention because of their enzyme stability [13, 14]. Moreover, the ligninolytic activity has been found in bacterial genera Pseudomonas, Cellulomonas, Burkholderia, Sphingomonas, Cupriavidus Aquitalea, Streptomyces, Rhodococcus, Gordonia, Clostridiales, and Bacillus isolated from forest soils [15].
The lignin-degrading enzymes such as peroxidases, laccases, monooxygenases, dioxidases, and phenol oxidases were found in bacterial strains such as Escherichia coli K-12, Thermobifida fusca YX, Rhodococcus jostii RHA1, Streptomyces viridosporus strain T7A, Streptomyces coelicolor A3(2), Amycolatopsis sp. 75iv2, Pseudomonas sp. strain YS-1p, Bacillus sp. HSTU-2, Citrobacter sp. HSTU-AAJ4, Acinetobacter sp. HSTU-6, and Enterobacter sp. HSTU-AAH8 [2, 10]. Bacteria affiliated with Klebsiella variicola are ethanol-producing prokaryotes [16]. To date, limited studies reported on the lignocellulose degradation of this group of bacteria. K. variicola–affiliated strains retrieved from untreated wheat straw consortia showed endoglucanase/xylanase activities [17]. Several other members of bacteria, such as Klebsiella oxytoca, Klebsiella Pneumonia, and Klebsiella variicola isolated from the intestine of Diatraea saccharalis larvae, were able to produce cellulolytic enzymes [6]. Moreover, K. variicola has not only lignocellulolytic activities but also has the ability to utilize diverse types of carbohydrates [18]. Klebsiella pneumonia is a common species in the omasum content, feces, and alleyways, but the Klebsiella oxytoca and Klebsiella variicola were the most frequent among isolates from soil and feed crops. It was reported in a similar story that heterogeneity of Klebsiella sp. in rumen content and feces, with a median of 80% [19].
The study was aimed to investigate the pectin-degrading strain biochemical characterization, lignocellulose-degrading enzyme production capacities, and genomic features. The genome investigation of goat intestinal K. variicola’s, CAZyme, ligninolytic enzyme activities, and molecular docking results were underexplored. Here, we described the genomic investigation of a K. variicola bacterium, isolated from fresh goat omasum, which secreted lignocellulose-degrading enzymes. The analyses of whole-genome and housekeeping genes of the K. variicola strain HSTU-AAM51 suggested that the strain has markedly deviated from homologs K. variicola strains. In addition, the presence of CAZYyme gene details in the K. variicola HSTU-AAM51 genome leads us to hypothesize that the strain might secrete CAZyme and ligninolytic enzymes to degrade lignocellulose in goat omasum as well as facilitates grass or other feed crops for nitrogen fixation in the environment.
Materials and methods
Omasum contents sampling and isolation
The goat omasum from a freshly slaughtered goat was collected from the town meat market at Bahadurbazar (25.6263° N; 88.6333° E), Dinajpur. The sample was transported immediately to the laboratory of Biochemistry and Molecular Biology at Hajee Mohammad Danesh Science and Technology University, Basherhat nearby Dinajpur, under sterile conditions. Appropriate dilutions (10−4) and (10−6) of goat omasum contents were spread on agar plates containing 1% yeast extract, 1.5% Tryptic Soy Broth (TSB), 1.5% agar, and pH 7.0 and the plates were incubated at 37 °C for 24 h. Bacterial colonies were picked from each plate and streaked on agar plates for further purification. The purified colonies were subjected to Congo red agar media (composition: 0.1% (NH4)2SO4, 0.05%KH2PO4, 0.05%K2HPO4, 0.02%MgSO4.7H2O, 0.01% CaCl2, 0.01% NaCl, 0.1% yeast extract, 1.5% agar, and Congo red 0.5%) for the rapid and sensitive screening test of cellulase producers by observing clear zones [20, 21]. The grown colonies that showed discoloration of Congo red plate were considered cellulose-degrading bacteria and stored for further experiments. Several strains were isolated as cellulose-degrading bacteria. Among these isolates, a strain that showed potentialities was selected for the rest of this study. The strain was identified and named Klebsiella variicola HSTU-AAM51.
Biochemical characterization
The metabolic pattern of the strain was analyzed using different biochemical tests, including MR-VP, KOH string, catalase, oxidase, triple sugar iron (TSI), citrate utilization (CIU), motility indole urease (MIU), urease, and individual sugar fermentation/carbohydrates (dextrose, lactose, maltose, and sucrose) test [21]. The detection of the extracellular hydrolytic enzyme activity of the isolate was done by the agar diffusion method. The strain was grown on different enzyme activity indicator medium to detect cellulase, xylanase, amylase, lignin-degrading, and protease. The bacterial growth indicator medium was prepared in distilled water with carboxymethyl cellulose, xylan, and starch; Casein powder was supplemented as the carbon source. Then, the medium was sterilized at 121 °C with 15 psi for 15 min. The purified colonies were subjected to the specific substrate agar plate and incubated at 37 °C for 24 h [20–22].
DNA extraction, genome sequencing, assembly, and annotation
The genomic DNA of the HTU-AAM51 strain was extracted using the Genomic DNA Extraction Kit (Promega, Madison-Wisconsin, USA) as the manufacturer’s instructions. The purity and concentration of the isolated genomic DNA were measured using a DNA spectrophotometer (Promega, Madison-Wisconsin, USA). The whole-genome shotgun sequencing of strain HSTU-AAM51 was performed using pair-end sequencing in an Illumina Miniseq sequencing platform (Illumina, CA, USA). The genomic library was prepared from purified DNA fragments using Nextera XT Library Kit following the manufacturer’s instructions. Raw sequencing reads were processed by FASTQC v. 10.1. FASTQ ToolKit used the manipulation of FASTQ files including adapter trimming, quality trimming, and length filtering [23]. The appropriate paired reads of length ≥ 30 bp were chosen from the pool of corrected reads and the remaining singleton reads were considered single-end reads according to Li et al. [24]. Next, the paired-end and single-end corrected reads were analyzed in k-mer-based de novo assembly using the SOAPDenovo, version 2.04 [25]. The set of scaffolds with largest N50 was identified by evaluating k-mers in the range 29–99 [25]. Furthermore, the scaffolds were subjected to gap closing by utilizing the corrected paired-end reads. Finally, the resulted scaffolds of length ≥ 300 bp were chosen for assembly [24]. The quality-filtered data (248.95 Mbp data size) was de novo assembled using SPAdes version 3.9.0 into contigs and then scaffolds. The respectively assembled reads were finally merged with Progressive Mauve v.2.4.0 (http://darlinglab.org/mauve/user-guide/reordering.html). The resulting assembled genome was annotated and analyzed using NCBI Prokaryotic Genome Annotation Pipeline (PGAP) version 4.5, Prokka. The prediction of multilocus sequence typing (MLST) and pMLST was conducted by using a bacterial analysis pipeline v 1.0.4 Illumina server (www.illumina.com). The functional prediction of protein-coding genes was accomplished by searching against Clusters of Orthologous Groups (COG) via the rapid annotations using subsystems technology (RAST) server (https://rast.nmpdr.org/).
Gene bank accession number
The 16S rRNA gene sequence of the strains HSTU-AAM51 has been deposited in the national center for biotechnology information (NCBI) and the obtained accession number is MW674660. The whole-genome datasets generated in this manuscript can be found on NCBI under the Klebsiella variicola HSTU-AAM51 complete genome BioProject number PRJNA594144 and BioSample number SAMN13505812 and accession number WSET00000000, respectively.
Comparative genomic analyses
Phylogenetic tree and average nucleotide identity (ANI)
The sequences of housekeeping genes were acquired from genome data, aligned separately, and concatenated in the following order: 16S rRNA, rpoB, recA, tonB, gyrB. The phylogenetic trees of the 16S rRNA gene and concatenated housekeeping genes were constructed by the neighbor-joining method using Molecular Evolutionary Genetics Analysis (MEGA X). The phylogenetic tree of the HSTU-AAM51 strain along with the whole-genome sequence of the nearest strains was built using REALPHY1.12 online server (http://www.realphy.unibas.ch/realphy/). The JSpeciesWS server (http://jspecies.ribhost.com/jspeciesws) was used to determine the ANI values based on BLAST (ANIb) between strain HSTU-AAM51 and its closely related taxa.
Genome comparison
To reveal the genome features, the HSTU-AAM5 genome was compared with the nearest recently reported genome sequences of Klebsiella species. The circular and linear genomic maps of each genome were generated using Interactive Microbial Genomic Visualization with cGView (http://www.cgview.) and Ring Image Generator (BRIG, version 0.95). Each circular and linear genomic map was generated with BLAST+, with strand parameters (70% lower and 90% upper cutoff for identity and E value of 10), using Klebsiella variicola HSTU-AAM51 as the alignment reference genome. Moreover, the synteny block/genome colinearity of the strain HSTU-AAM51 was analyzed with the nearest homologous genome of Klebsiella variicola strain 13,450, Klebsiella sp. strain P1CD1, Klebsiella variicola strain ACCHC, and Klebsiella variicola strain FDA ARGOS627 using Progressive Mauve server (http://darlinglab.org/mauve/mauve.html). The in silico DNA-DNA hybridization of the Klebsiella variicola strain HSTU-AAM51 with the top fifteen nearest strains was performed in the GGDC server (http://ggdc.dsmz.de).
Carbohydrate-active enzyme (CAZy) gene annotation
Essential genes for active carbohydrate enzymes (CAZymes) encoded in the genome of strain HSTU-AAM51 were classified by dbCAN (using HMMER), CAZy (using DIAMOND), and PPR (using Hotpep) databases, respectively, in the integrated dbCAN2 meta server using default settings. The gene functions were investigated by the following different databases: the carbohydrate-active enzymes (CAZy) database (http://www.cazy.org/), the KEGG database (http://www.genome.jp.kegg/pathway.html), the non-redundant protein database (http://www.ncbi.nlm.nih.gov/COG/), the gene database (http://www.geneontology.org/), and the clusters of orthologous groups (COG) database (http://www.ncbi.nlm.nih.gov/COG/).
Lignin-degrading pathways
The lignin degradation pathways such as benzoate, phenylacetate, and β-ketoadipate pathway genes of the HSTU-AAM51 strain were screened from the prokaryotic genome annotation pipeline (PGAP) annotation file provided by NCBI. The genes involved in lignin degradation were screened according to previously reported Kumar et al. [26]. The plant growth–promoting genes were screened in a PGAP annotated file according to Guo et al. [27].
Enzyme production on banana fiber
The isolate HSTU-AAM51 was inoculated into the production medium contained 1% (w/v) of banana fiber with yeast extracts and mineral salts [21, 22]. The banana fiber was extracted from the culture after 6 days and dried overnight in an oven at 55 °C. The culture solution was centrifuged at 6000 g for 10 min to separate the debris and cells. The supernatant was used as an enzyme source. The secreted protein concentration was determined using the Bradford method. The crude enzyme solution’s cellulase activity was assayed by measuring the level of reducing sugars liberated from the avicel substrate using the DNS method [28]. The reaction mixture was composed of 0.4 mL of 0.05 M sodium citrate (pH 5.2) buffer with avicel (1%) and 0.1 mL of culture supernatant. The mixture was incubated at 55 °C for 10 min, and then the reaction was stopped by adding 0.5 mL DNS reagent and kept in a boiling water bath for 10 min. For control, the culture supernatant was added after mixing DNS reagents. Then, the solutions were cooled at room temperature, and the absorbance was measured at 540 nm. One unit of cellulase was considered the amount of enzyme required to release 1 µmol of reducing sugars per minute under the mentioned assay conditions. The xylanase activity was determined by measuring the amount of reducing sugars released from 1% (w/v) oat-spelt xylan as a substrate-like cellulase assay. The pectinase activity was determined by measuring the release of D-galacturonic acid from commercial 1% (w/v) pectin as substrates under the same conditions mentioned for cellulase and xylanase. The lignin peroxidase assay method adapted from Magalaes et al. [29]. The assay mixture contained 3.0 mL: 2.2 mL of supernatant, 0.1 mL of 1.2 mM methylene blue dye, and 0.6 mL of 0.5 M sodium tartrate buffer (pH 4.0). The reaction was started after adding 0.1 mL of 2.7 mM H2O2. The gradually declined absorbance of methylene blue was recorded at 664 nm and the unit of lignin peroxidase was calculated as per the change of absorbance per minute.
Structural analysis and molecular docking of CAZy enzymes
To sort the CAZy enzyme for molecular docking, the signal peptide of the CAZy enzyme was identified using the SignalP5.0 server (www.cbs.dtu.dk). Afterward, the 3D structure of the noncytoplasmic domain–containing enzymes of HSTU-AAM51 strain was built according to the homology modeling in I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The 3D structure of CAZy enzymes was evaluated before and after the energy minimization step to assess the quality of the constructed models using several tools provided in the UCLA-DOE server (http://servicesn.mbi.ucla.edu/). The stereochemical quality of each enzyme model was validated by PROCHECK (https://servicesn.mbi. ucla.edu/PROCHECK/) and Varify3D (https://servicesn.mbi.ucla.edu/Verify3D/) to check the atomic model and its amino acid quality. The docking interaction of the signal peptide–containing lignocellulose-degrading twelve enzymes with interested substrates was conducted to validate the binding affinity. Firstly, blind docking was performed using the EDock online server (https://zhanglab.ccmb.med.umich.edu/EDock/). Secondly, the molecular docking of the proteins with ligand was confirmed using Auto dock vina, and PyRx. The standard docking was performed by centralizing the ligand in the 3D structure utilizing the default box dimension. The molecular visualization of the docked complex was performed by Pymol software (Schrodinger, LLC, 2010). The protein–ligand interaction was analyzed by Ligplot plus and protein–ligand interaction profiler.
Results
Isolation and characterization of goat omasum strains
The cellulose-degrading bacterial strains were isolated based on the discoloration of the Congo red agar plate. A simple and conclusive idea about a strain can be inferred from biochemical tests. The biochemical tests were conducted immediately after the strain’s isolation. A total of five different pectin-degrading strains were selected and characterized (Fig. S1). The strains Pec-22 and Pec-24 are gram-negative, while the other three strains are gram-positive. All five strains showed catalase and oxidase activities but with varying degrees (Table 1). Importantly, Pec-23 and Pec-24 have degraded methyl red but all others did not. In contrast, none of them was capable of showing positive results in the VP test. Strains Pec-17 and Pec-23 were positive in motility, indole, urease (MIU), whereas strains Pec-22 and Pec-24 were negative in MIU tests. Strain Pec-16 showed positive in the urease test. All strains showed positive activity towards carbohydrates like dextrose, maltose, sucrose, and lactose utilization tests. In addition, carboxymethyl cellulose, oat-spelt xylan, and lignin-related dye utilization were also confirmed by the strain Pec-24/HSTU-AAM51 (Table 1). Based on the biochemical tests, the pectin hydrolytic strains were varied among each other. Importantly, only the Pec-24/HSTU-AAM51 strain was grown on diazinon-enriched media, which suggested that the strain is capable of scavenging insecticides related to toxic chemicals in goat omasum.
Table 1.
Biochemical characterization of pectin-degrading bacterial isolates from goat omasum
| Test | Pec-16 | Pec-17 | Pec-22 | Pec-23 | Pec-24/HSTU-AAM51 |
|---|---|---|---|---|---|
| Gram stain | + Ve | + Ve | -Ve | + Ve | -Ve |
| Catalase | + + | + + + | + + + | + + | + + + |
| Oxidase | + | + | + + + | + + + | + + + |
| Methyl red | - | - | - | + | + |
| VogesProskauer | - | - | - | - | - |
| Triple sugar iron | + | + | + | + | + |
| Citrate utilization test | + | + + + | + + + | + + + | - |
| Motility, indole, urease | -,-, + | + , + , + | -,-,- | + , + , + | -,-,- |
| Maltose | + | + | + | + | + |
| Sucrose | + | + | + | + | + |
| Dextrose | + | + | + | + | + |
| Lactose | - | + | + | + | + |
| Cellulose hydrolysis | + | + | + | + | + |
| Oat-spelt xylan hydrolysis | + | + | + | + | + |
| Lignin hydrolysis | + | + | + | + | + |
| Diazinon | - | - | - | - | + + + |
The lignin hydrolysis was confirmed by the presence of the strain’s growth in lignin-related dye (Congo red, methylene blue, thymol blue, commercial brilliant blue) agar plate and in broth solution
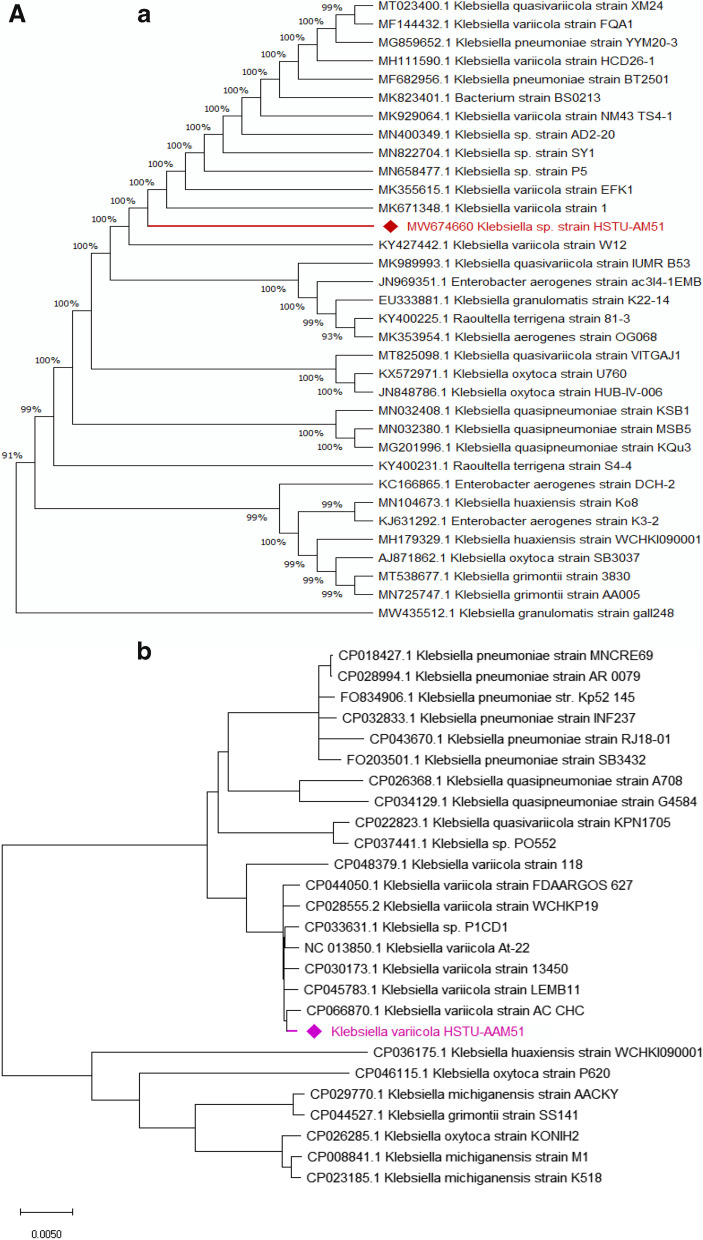
The fastQC analysis of the raw sequence file reported 57.2% that indicated sequence quality was good for further processing of sequence assembly. The MLST analysis confirmed that the strain belonged to the genus Klebsiella variicola. The alignment of housekeeping genes, namely 16S rRNA, tonB, gyrB, and rpoB sequences of the strain HSTU-AAM51, is presented in Fig. 1. Figure 1Aa shows that the gene 16S rRNA has good coverage with K. variicola strain1 (MK671348.1) and K. variicola strain W12 (KY427442.1), but was placed in a separate taxon. Figure 1Ab shows that the whole-genome sequence has also high similarities with K. variicola AC CHC (CP066870.1) and K. variicola LEMB11 (CP045783.1). The phylogenetic tree of whole-genome sequences of HSTU-AAM51 showed that the strain clustered with both K. variicola P1CD1 (CP033631.1) and K. variicola 13,450 (CP030173.1) (Fig. 1Ab). Again, the strain HSTU-AAM51 branches near to other closely related genera, namely, K. grimontii strain SS141 (CP044527.1), K. michiganensis strain AACKY (CP029770.1), K. oxytoca strain P620 (CP046115.1), K. huaxiensis strain WCHKI090001 (CP036175.1), and much far from K. pneumonia (F0203501.1).
Fig. 1.
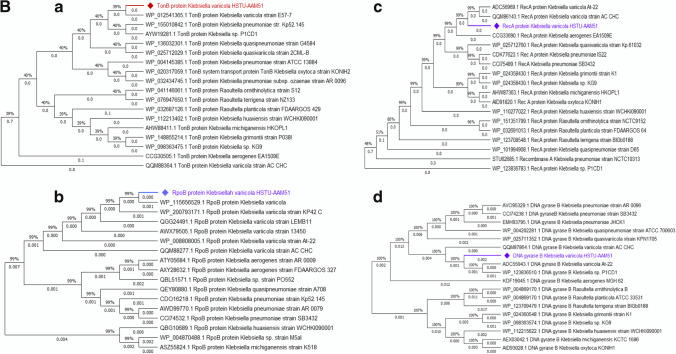
A (a) Phylogenetic tree based on 16S rRNA gene sequence, and (b) a midpoint rooted phylogenetic tree of K. variicola HSTU-AAM51 strain with closely related strains based on whole-genome sequences. All sequences obtained in this study are currently available in the gene bank database. B phylogenetic tree of housekeeping genes: (a) TonB protein, (b) rpoB protein, (c) RecA, (d) DNA gyrase B protein of K. variicola HSTU-AAM51 strain with closely related strains. All the trees were made in MegaX.0 software
The tonB gene has 95% similarities with K. variicola strain E57-7. Moreover, the strain HSTU-AAM51 demonstrated a different cluster with K. variicola At-22 at 0.001 distances. Similarly, the rpoB possessed a separate cluster with K. variicola strain LEMB11, whereas it was placed from the 2nd node of its nearest neighbor’s belongings to K. variicola, and K. variicola KP42C, respectively (Fig. 1Bb). In addition, the recA gene of strain HSTU-AAM51 showed 99% similarities with K. variicola At-22 in a separate cluster (Fig. 1Bc). Similar things were observed for DNA gyrase B since gyrase B of K. variicola HSTU-AAM51 placed from the 2nd node of gyrase B of neighbor strains K. variicola At-22 and K. variicola P1CD1, respectively.
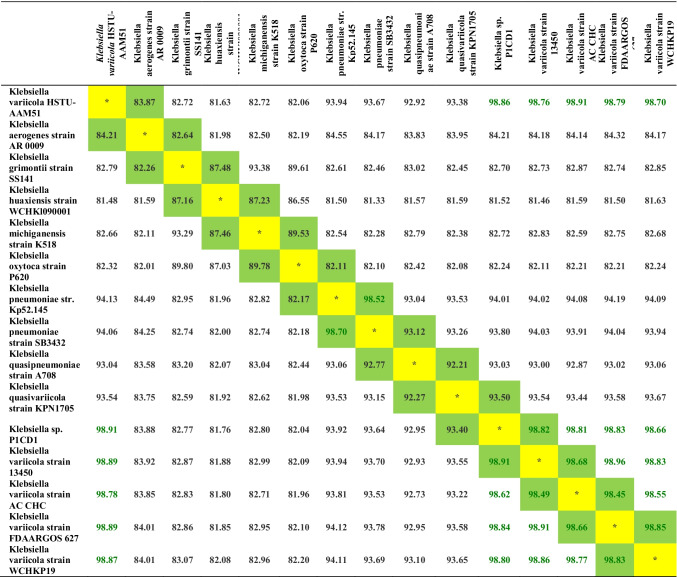
The pairwise ANI blast values between HSTU-AAM51 and K. variicola strain ACCHC were 98.91% (Table 2). K. variicola strain FDA ARGOS 327 and K. variicola strain WCHKP19 showed 98.79% and 98.70% ANIb, whereas other Klebsiella species showed below 90% ANIb, indicating that the strain HSTU-AAM51 might be a new member of K. variicola. The digital DNA-DNA hybridization of the HSTU-AAM51 strain with its nearest homologs showed that except for K. variicola strain 118 (74.5%), all other K. variicola strains showed DDH 92–94% (Table 3), while K. pneumonia related and other Klebsiella species showed 25.7–58.4% DDH.
Table 2.
Average nucleotide identity by Blast (ANIb) analyses of the Klebsiella variicola strain HSTU-AAM51
Table 3.
Digital DNA-DNA hybridization (DDH) of Klebsiella variicola strain HSTU-AAM51 with its nearest homologs
| Reference strains genome compared | Formula: 1 (HSP length/total length) DDH | Formula: 2 (identities/HSP length) DDH recommended | Formula: 3 (identities/total length) DDH | Difference in % G + C (interpretation:distinct species) |
|---|---|---|---|---|
| Klebsiella aerogenes strain AR0009 | 46.80% | 29.90% | 41.80% | 2.26 |
| Klebsiella grimontii strain SS141 | 48.70% | 27.00% | 42.00% | 1.30 |
| Klebsiella huaxiensisstrain WCHKl | 41.30% | 25.70% | 36.20% | 1.27 |
| Klebsiella oxytoca strain P620 | 46.30% | 26.30% | 39.90% | 3.11 |
| Klebsiella pneumonia str. Kp52.145 | 77.80% | 58.40% | 76.40% | 0.17 |
| Klebsiella pneumonia strain SB3432 | 71.20% | 57.30% | 70.50% | 0.02 |
| Klebsiella quasipneumoniae strain A708 | 79.70% | 52.50% | 76.20% | 0.66 |
| Klebsiella quasivariicola strain KPN1705 | 75.60% | 55.90% | 73.90% | 0.29 |
| Klebsiella variicola P1CD1 | 90.50% | 93.30% | 93.40% | 0.12 |
| Klebsiella variicola At-22 | 91.00% | 93.60% | 93.80% | 0.35 |
| Klebsiella variicola strain 15WZ-82 | 92.40% | 94.00% | 94.90% | 0.18 |
| Klebsiella variicola strain 118 | 88.20% | 74.50% | 88.70% | 0.00 |
| Klebsiella variicola strain 13,450 | 90.90% | 92.90% | 93.60% | 0.20 |
| Klebsiella variicola strain AC CHC | 84.30% | 94.10% | 88.70% | 0.11 |
| Klebsiella variicola strain FDAARGOS_627 | 91.70% | 92.50% | 94.10% | 0.40 |
| Klebsiella variicola strain LEMB11 | 92.90% | 93.40% | 95.10% | 0.30 |
| Klebsiella variicola strain WCHKP19 | 89.90% | 92.70% | 92.90% | 0.32 |
| Klebsiella variicola strain X39 | 88.80% | 93.60% | 92.10% | 0.12 |
Logistic regression is used for reporting the probabilities that DDH is > = 70% and > = 79%. Percent G + C content cannot differ by > 1 within a single species but by < = 1 between distinct species
Genome metrics of strain HSTU-AAM51
The assembly of the genome sequence generated a total of 290 contigs (with PEGs) and the largest contig size obtained 657,932 bp. The total genome size of the strain HSTU-AAM51 was determined to be 5,564,045 bp with a GC content of 57.2%, N50 value 207,699, L50 value 9, 5312 coding sequences, and 102 RNA genes according to PGAP annotation of NCBI. In particular, the strain HSTU-AAM51 has 5150 protein-coding genes. The genome size of HSTU-AAM51 was slightly greater than the strain of K. variicola 13,450 (5,511,705 bp). The GC content of strain HSTU-AAM51 was similarly related to K. variicola strain 13,450 (57.4%), K. variicola strain WCHKP19 (57.6%), K. quasivariicola strain KPN1705 (56.9%), and K. pneumonia subsp. rhinoscleromatis strain SB3432 (57.2), respectively. The RAST analysis of the strain HSTU-AAM51 genome was conducted and described in the subsections below (Fig. 2A). The genomic feature in the subsystem was about 59% genome coverage. A total of 3038 proteins are involved in the subsystem, including 858 proteins were predicted to be involved in carbohydrate metabolism and for about 594 proteins are involved in amino acid metabolism, whereas the nearest relative, K. variicola strain 13,450, has a total of 438 and 432 proteins for carbohydrate and amino acid metabolism, respectively. The CGview analysis shows all the proteins of a bacterium in a proper genomic map. We hereby transcribed the K. variicola HSTU-AAM51 bacterial protein into a genomic map where 5187 protein-coding sequence (CDS), and 88 tRNAs were shown in 5.5 mbp length sequences. In Fig. 2B, the GC-skew positive indicated that the CDS was present in downstream and the GC-skew negative indicated that the CDS was present in upstream.
Fig. 2.
General features of the genomic organization of Klebsiella variicola HSTU-AAM51 strain. A Features in the subsystem. B Circular map of the complete genome of K. variicola HSTU-AAM51 strain
Comparative genomic analyses
Pangenome analyses
The pangenome graph clearly showed that some parts of the genome sequence of HSTU-AAM51 were unmatched compared to six other strains studied. Specifically, it was focused on the toxin genes in the pangenome study and represented in Fig. 3Aa. The genome sequence of HSTU-AAM51 after 5500 kbp was almost abolished; however, a little irregular homology of genome sequence appeared. The human pathogenicity related genes available in K. pneumonia such as clbJ, clbL, rtlK, gt, gcl, icl, htpB, clpP, ybtA, ybtX, ybtQ, clpX, ybtP, and bet (from 6100 to 7100 kbp) were absent in the K. variicola strain HSTU-AAM51 genome. Interestingly, the top five nearest homologs strain genome comparison (Fig. 3Ab) also showed varied genome assembly.
Fig. 3.
Genomic comparison of K. variicola HSTU-AAM51 strain. A Pangenome analysis of K. variicola HSTU-AAM51 strain with closely related strains. B Synteny analysis of K. variicola HSTU-AAM51 strain with other close related strains. The same color module represents the colinear region. Good colinearity of the strain HSTU-AAM51 and other Klebsiella variicola are shown
Mauve alignment
The block outlines of the K. variicola HSTU-AAM51 genome encompassed a sort of sequence that was homologous to part of other genomes compared. It was assumed that the homologous LCBs are internally free from genomic rearrangement of genomes compared. The LCB in the genome of HSTU-AAM51 was connected by lines to similarly colored LCBs in the genomes of P1CD1, 13,450, and FDA ARGOS strains, respectively. The boundaries of LCBs of the HSTU-AAM51 and other strains that have taken comparison are considered break-points of genome rearrangements. As seen in Fig. 3B, the LCBs of the HSTU-AAM51 genome were not exactly matched with LCBs of genomes taken for comparison. A part of the light blue–colored LCB of the HSTU-AAM51 genome was deleted that appeared in other genomes. Moreover, the yellow-colored LCB of the HSTU-AAM51 genome was intact, whereas sorts of sequences are lost from the same colored LCBs of other genomes studied. In addition, the reshuffling or rearrangements of sorts of sequences were found in various LCBs compared to the LCBs of other nearest strain genomes. Interestingly, some LCBs of the K. variicola FDA ARGOS genome were located below the line. These results indicated that this region was inverted compared to the HSTU-AAM51 and other genome sequences studied.
Lignocellulose-degrading enzymes of HSTU-AAM51 strain
A total of 36 CAZy enzymes involved in lignocellulose degradation were observed in the genome of the HSTU-AAM51 strain (Table 4). In particular, a total of seventeen enzymes were categorized as cellulase included GH1, GH3, GH4, GH5, and GH13 families. One cellulase and endoglucanase were found that have non cytoplasmic peptides and used Sec/SPI path for their secretions. Moreover, thirteen different beta-glucosidases (2GH3, 7GH4, and 4GH1) were annotated in the HSTU-AAM51 strain genome. In addition, 6-phospho-alpha-glucosidase, maltodextrin glucosidase, pullulanase type alpha-1,6-glucosidase associated with CBM41 were observed in the genome of the HSTU-AAM51 strain. The pullulanase has shown the signal peptide predicted to use Sec/SPII path. The beta-1,4-mannanase has also contained signal peptide (Sec/SpI), fibronectin type III (FNIII), and carbohydrate-binding module (CBM35) domain. Fifteen different xylanases and mannanases were observed as shown in Table 4. Among them, the hemicellulase, galactosidase, and polysaccharide deacetylase enzymes used Sec/SpI path, whereas glycosyl hydrolase (GH10) used lipoprotein Sec/SpII path of the cell membrane for their secretion (Table 4). Moreover, GH43 family protein xylan 1,4 beta-xylosidase, rhamnogalactouronyl hydrolase, and rhamnosidase were also observed in the genome of HSTU-AAM51. In addition, some β-1,4-mannanase, α-galactosidase, β-galactosidase, pectinase, and chitinase belong to the GH18 family protein that holds chitin-binding domain and glycoside hydrolase domain were also annotated in the genome of HSTU-AAM51 strain (Table 4).
Table 4.
List of potential lignocellulose-degrading CAZYzyme found in the genome of Klebsiella variicola strain HSTU-AAM51
| Category | CAZY family | Activities in the family | E.C. number | Locus tag/scaffold | Secretion pathwaysº |
|---|---|---|---|---|---|
| Cellulase | GH3 | Cellulase | 3.2.1.4 | GO309_06680 | Sec/SPI |
| GH5 | Endoglucanase | - | GO309_06640 | Sec/SPI | |
| GH3 | β-glucosidaseBglX | 3.2.1.21 | GO309_13125 | Sec/SPI | |
| GH4 | 6-phospho-alpha-glucosidase | - | GO309_08325 | Others | |
| GH4 | 6-phospho-Beta-glucosidase | 3.2.1.86 | GO309_09810 | Others | |
| GH1 | Beta-glucosidase | 3.2.1.23 | GO309_00745 | Others | |
| GH13 | Maltodextrin glucosidase | 3.2.1.20 | GO309_03805 | Others | |
| GH3 | β-glucosidase | - | GO309_09355 | Others | |
| GH4 | 6-phospho-Beta-glucosidase | 3.2.1.86 | GO309_13085 | Others | |
| GH4 | 6-phospho-Beta-glucosidase | 3.2.1.86 | GO309_18650 | Others | |
| GH4 | 6-phospho-Beta-glucosidase | - | GO309_19150 | Others | |
| GH4 | 6-phospho-Beta-glucosidase | - | GO309_22465 | Sec/SPI | |
| GH4 | 6-phospho-alpha-glucosidase | - | GO309_08325 | Others | |
| GH13/CBM41 | Pullulanase type alpha-1,6-glucosidase | - | GO309_16300 | Sec/SPII | |
| GH13 | Glucohydrolase | - | GO309_24225 | Others | |
| GH1 family | β-glucosidase | - | GO309_20960 | Others | |
| GH1 | Glycoside hydrolase | - | GO309_11750 | Others | |
| GH1 | β-glucosidase | - | GO309_09895 | Others | |
| Xylanase | GH31 | Alpha xylosidase | 3.2.1.177 | GO309_25310 | Others |
| GH4 family | Alpha galactosidase | GO309_14666 | Others | ||
| GH4 family | Alpha galactosidase | - | GO309_21195 | Others | |
| GH53 | Galactosidase | - | GO309_04245 | Sec/SPI | |
| GH42 family | Beta-galactosidase | 3.2.1.23 | GO309_07895 | Others | |
| GH42 family | Beta-galactosidase | - | GO309_04250 | Others | |
| CE4 | Polysaccharide deacetylase | - | GO309_16175 | Sec/SPI | |
| CE4 | Polysaccharide deacetylase | - | GO309_20045 | Others | |
| GH43 family | Beta-xylosidase | - | GO309_26190 | Others | |
| GH43 family | Glycosyl hydrolase | - | GO309_16050 | Others | |
| GH43 family | Xylan 1,4 beta-xylosidase | - | GO309_13940 | Others | |
| GH10 family | Glycosyl hydrolase | - | GO309_08230 | Lipoprotein Sec/SPII | |
| GH105 family | Rhamnogalactouronyl hydrolase | - | GO309_03125 | Others | |
| GH78 family | Rhamnosidase | - | GO309_00360 | Others | |
| Mannanase | GH26/CBM35 | Beta 1,4-Mannanase | - | GO309_09900 | Sec/SPI |
| Chitinase | GH18 | Chitinase | - | GO309_21965 | Sec/SPI |
| Pectin esterase | CE8 | Pectin esterase | Sec/SPII | ||
| Feruloyl esterase | Tannase family | Tannase/Feruloyl esterase | - | GO309_22885 | Sec/SPI |
Enzyme secretion
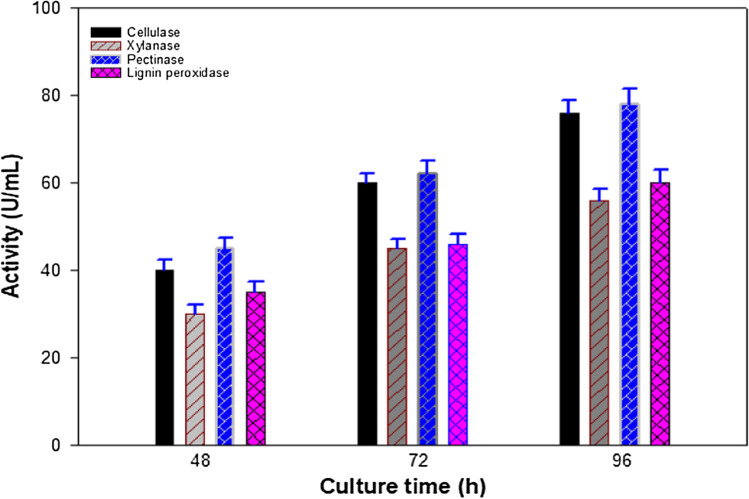
The secretion of lignocellulolytic enzymes such as cellulase, xylanase, pectinase, and lignin peroxidase of HSTU-AAM51 strain was monitored at various incubation periods (Fig. 4). The cellulase and pectinase enzymes had shown higher activities compared to the xylanase and lignin peroxidase at all incubation periods. All tested enzymes showed higher unit activities following increased incubation time as seen (Fig. 4). In particular, the activities of cellulase, xylanase, pectinase, and lignin peroxidase were increased by 1.9-, 1.86-, 1.73-, and 1.7-fold, respectively, after 96 h of incubation in banana pseudostem–enriched media.
Fig. 4.
Lignocellulose-degrading enzyme production by the K. variicola HSTU-AAM51 strain on banana pseudostem. The enzyme production was continued in growth media where banana fiber was used as the sole carbon
source in a shaking incubator at 37° with 140 rpm
Molecular docking of CAZy enzymes with substrate
Homology modeling and assessment
The quality of the protein models obtained from K. variicola HSTU-AAM51 was assessed beforehand. Various factors were taken into consideration including template modeling score (TM), RMSD value, sequence identity, model coverage, structural orientation quality (α-helix, β-sheet, ɳ-coil, unstructured %), overall quality score, compatibility analysis (3D-1D score in %), and Ramachandran plot for amino acid positional quality score analysis (most favorable regions, additionally allowed regions, allowed regions and disallowed regions) (Table 5). Sequence identity and model coverage of the 12 proteins are within an acceptable range (Table 5). All the protein models scored TM values exceeding 0.5 proving their viability (Table 5). The average RMSD of the twelve modeled proteins is 0.87 (less than 1) while 66% of them were below 1.
Table 5.
Model quality of extracellular CAZYzyme of Klebsiella variicola strain HSTU-AAM51
| Model protein | Best PDB hit | TM score; RMSD, Iden.,Cov | α -helix, β-strand, η-coil, π, unstructured (%) | ERRAT (quality score) | VERIFY (3D-1D score)% | Ramachandran plot (core, allow, gener., disallow) % |
|---|---|---|---|---|---|---|
| Cellulase GH3 | 4q2b | 0.906; 0.35; 0.534; 0.908 | 42, 12.4, 0.8, 0, 44.7 | 94.98 | 95.9 | 89.4, 9, 0.3, 1.3 |
| Beta-glucosidase | 6r5i | 0.954; 0.68; 0.626; 0.658 | 26.7, 16.7, 4.2, 0.8, 51.6 | 89.17 | 94.3 | 87.6, 10.3, 1.1, 1.1 |
| Galactosidase | 2gft | 0.889; 0.96; 0.407; 0.902 | 26.5, 11.7, 4.7, 0, 57.0 | 82.9 | 92.5 | 80, 17.1, 0.3, 2.6 |
| Endoglucanase | 5czl | 0.944; 0.28; 0.832; 0.946 | 42.6, 6.9, 2.7, 0, 47.7 | 96.9 | 88.6 | 87.7, 9.9, 0.7, 1.7 |
| 6-phospho β-D-glucosidase | 1up7 | 0.913; 1.36; 0.266; 0.939 | 48.1, 15.6, 2.7, 0, 13.4 | 93.04 | 96.4 | 85.9, 12.5, 1.3, 0.3 |
| Glycosyl hydrolase GH10 | 5oq2 | 0.824; 0.52; 0.438; 0.828 | 33.0, 11.8, 5.5, 1.1, 59.5 | 86.73 | 80.0 | 79.1, 16.1, 1.3, 3.5 |
| Mannase | 5awo | 0.792; 1.15; 0.109; 0.803 | 15.2, 7.5, 1.9, 0, 75.3 | 62.5 | 80.0 | 70.3, 21.6, 3.5, 4.6 |
| Pectinase | 3grh | 0.929; 0.21; 0.761; 0.93 | 7.7, 24.8, 2.1, 0, 65.3 | 74.8 | 90.4 | 81.6, 15.2, 1.4, 1.9 |
| Polysaccharide deacetylase | 6go1 | 0.714; 1.48; 0.235; 0.736 | 20.1, 9.3, 1.4, 0, 69.1 | 93.04 | 96.4 | 85.9, 12.5, 1.3, 0.3 |
| Tannase | 6fat | 0.788; 2.36; 0.255; 0.828 | 21.5, 12.8, 4.4, 0, 61.2 | 76.99 | 76.0 | 73.1, 19.3, 3.4, 4.2 |
| Pullulanase | 2fhf | 0.953; 0.41; 0985; 0.955 | 21, 20.6, 5.2, 0, 53.1 | 88.85 | 80.0 | 88.1, 9.6, 1.3, 0.9 |
| Chitinase | 3qok | 0.924; 0.78; 0.95; 0.93 | 27.8, 17.7, 5.2, 0, 49.1 | 97.55 | 90.89 | 85.2, 11.2, 1.7, 1.4 |
The protein models were fine-tuned using loop refining and energy minimization. The loop-refined and energy-minimized cellulase model protein showed an overall quality factor of 94.9818% and Verify 3D score of 95.93%. The model quality for the other proteins also falls at a satisfactory level. All the protein models except tannase (75.77%) showed above 80.0–96.923% verify 3D score > = 0.2 (Table 5). Protein models for mannase (62.46%), pectinase (74.87%), and tannase (76.98%) demonstrated comparatively lower ERRAT scores while the nine other modeled proteins showed an excellent level (83–97%) of quality for ERRAT in the SAVES server (Table 5).
The Ramachandran plot analysis for cellulase showed 89.4% amino acid residues to be in the most favorable regions, 9% in the additional allowed regions, 0.3% in the generously allowed region, and 1.3% in the disallowed regions. The corresponding values for the best hit cellulase protein (PDB ID: 4q2b) showed 92.2%, 7.4%, and 0.3%, respectively. In addition, beta-glucosidase, endoglucanase, 6-phospho β- D-glucosidase, polysaccharide deacetylase, pullulanase, and chitinase showed above 85% amino acids in the most favorable regions in the Ramachandran plot. Galactosidase and pectinase showed about 80.0% amino acids in the most favorable regions. Moreover, glycosyl hydrolase GH10, mannase, and tannase showed amino acids about 70–79% in the most favorable regions in the Ramachandran plot.
The structural orientation quality (percentage of α-helix, β-sheet, ɳ-coil, the unstructured portion in the protein model, respectively) has also been exhibited (Table 5) which also falls into direct resemblance with the data found in previous articles (mention viable reference). The modeled cellulase for example has a globular structure containing 42%, 12.4%, 0.8%, and 44.7% of α -helix, β-strand, η-coil, and unstructured regions. After considering all possible factors, the modeled proteins were subjected to the next phase which is molecular docking.
Docking analyses
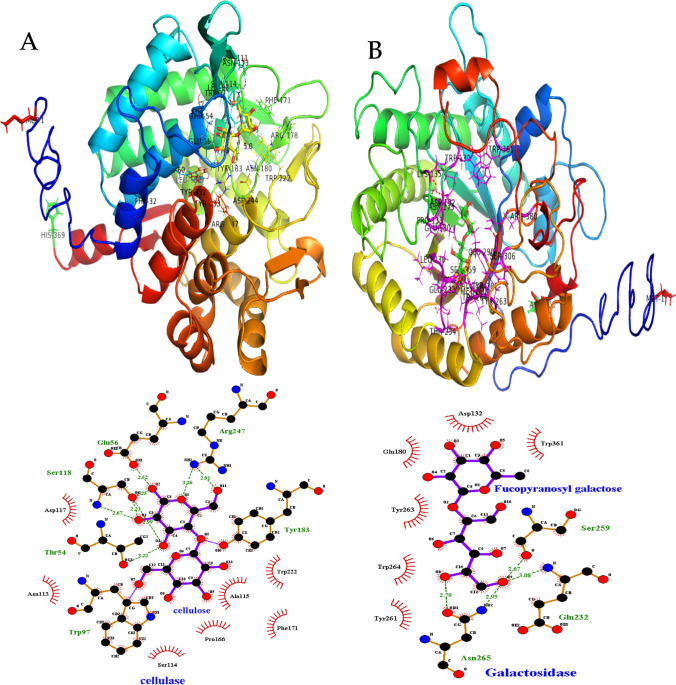
The best docking complex based on docking score was used to analyze the interaction of enzyme and substrate (Table S1). As seen in Fig. 5A, the cellobiose molecule successfully interacted with the cellulase enzyme. The important catalytic residues of cellulase enzyme include Ser118, Glu56, Arg247, Tyr183, Trp97, and Thr54. These residues have formed H-bonds with cellobiose with an average bond length of 2.23–3.26A°. Moreover, hydrophobic interactions were made by the residues Asp117, Asn113, Ser114, Pro166, Ala115, Phe171, and Trp222 with a maximum bond length of 3.9A°. The β-glucosidase BglX (GH3) residues Arg169, Lys208, Asp287, and Glu518 were interacted with cellobiose through H-bonds, but its residues His288, Met319, Phe81, Met252, trp155, and Trp450 were facilitated hydrophobic interactions with cellobiose ligands (Fig. S2). The other cellulase-type enzyme endoglucanase (GH5), 6-phospho-Beta-glucosidase (GH4), and pullulanase-type alpha-1,6-glucosidase (GH13/CBM41) have shown considerable H-bonds, non-bonded hydrophobic, and salt bridge interactions (Fig. S2). In Fig. 5B, xylanase family enzyme galactosidase has formed H-bonds via residues Ser259, Glu232, and Asn265 with fucopyranosyl galactose. All these important catalytic residues were within 3A° of the ligand. Non-bonded hydrophobic interactions were also made by Asp132, Glu180, Tyr161, Tyr263, Trp264, and Trp361 residues of galactosidase.
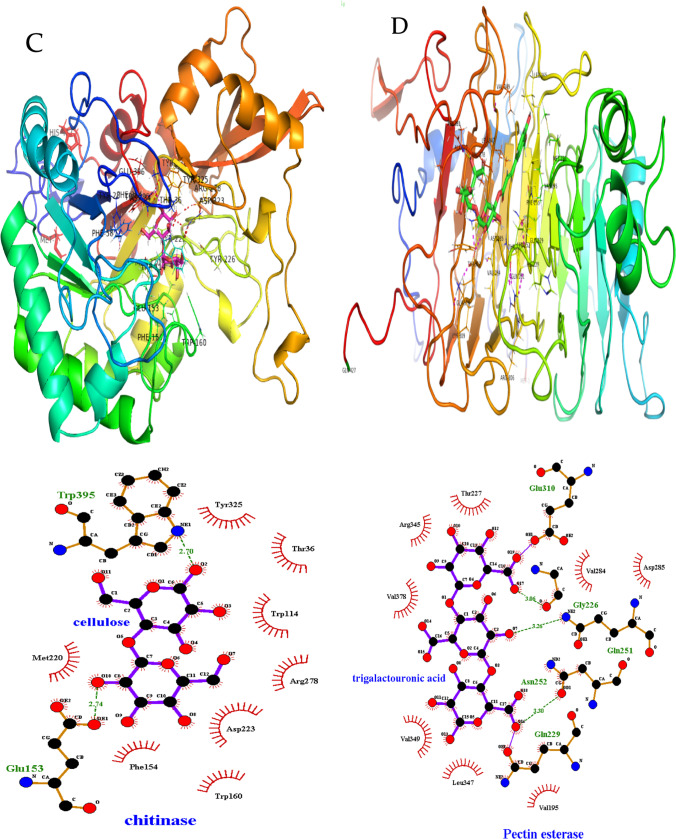
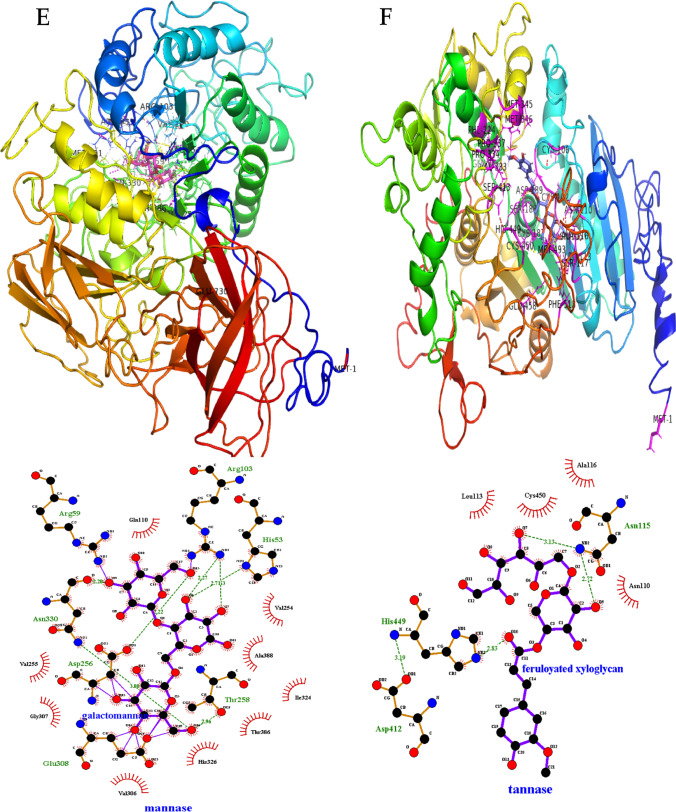
Fig. 5.
Molecular docking and catalytic interactions of extracellular signal peptide–containing lignocellulose-degrading enzymes. A Cellulase. B Galactosidase. C Chitinase. D Pectin esterase. E Mannase. F Tannase/feruloyl esterase
The 3D model of pectinase had a folded globular structure consisting of five α-helixes, twenty-five β-strands, and three coils. In particular, cellulose showed the existence of 7.26% α-helixes, 22.7% β-strands, 2.1% coils, and 62.34% non-structured region (Fig. 5D). Structural analysis showed that all catalytically important residues were within 5 Å of the ligands. The binding affinity of feruloyated xyloglucan (FX) was slightly higher than polygalacturonic acid (PGA) as indicated by the docking score (Supplementary Table S1). As seen in Fig. 5F, the Gln229, Gln251, Asn252, Asp285, Glu310, and Gly379 contributed to H-bonds and Arg345 contributed a salt bridge interaction with TGA. In contrast, FX made hydrophobical (Val378), hydrogen bonds (Gln251, Asn381), and salt bridge (Arg345) interactions. The residues Asp285 and Glu310 were found to have bond lengths of 1.86 Å and 3.09 Å, whereas Ser288 was located about 5 Å away from the PGA.
The 3D surface models of tannase (Fig. 5F) consist of 20.82% α-helixes, 12.63% β-strands, 3.75% randon coils, and 64% unstructured regions. The corresponding substrates of the 3D model of tannases were methyl ferulate, feruloyated xyloglucan, and conferyl alcohol. Among these three substrates, the highest docking score and binding affinity were found for feruloyated xyloglucan (Supplementary Table S1).
The endoglucanase showed hydrogen bond interaction (Glu53, Asp104, Ser109, Gly111, Pro158, Tyr175, Trp233, and ASP240) with cellulose (Fig. S2C). The best binding energy was − 6.5 kcal/mole and − 7.0 kcal/mole which was found for cellulase and endoglucanase with cellulose as the substrate. The binding energy indicated superior alignment between the active site residues of cellulase and its corresponding substrates at individual sites shown in Fig. 5A, B, C, D. The docking score for the other cellulase enzymes such as beta-glucosidase, galactosidase, 6-phospho β-D-glucosidase, pullulanase was − 6.6 to − 6.8, − 6.7 to − 6.8, − 5.9 to − 6.5, and − 7.0 to − 8.0 kcal/mole with substrates, namely methyl β-D-galactopyranoside, cellulose, 4-Nitrophenyl β-D-glucopyranose, cellobiose, maltose, and maltrose, respectively (Supplementary Table S1).
Subsequently, hemicellulase enzyme (glycosyl hydrolase, polysaccharide deacetylase, and mannase) interacted with substrate, namely cellulose, 4-nitrophenyl β-D-glucopyranose, galactomannan, feruloyated arabinose, feruloyated xyloglucan, and dextrin with free-binding energies ranging from − 7.6 to − 8.3, − 6.5 to − 7.4, and − 7.0 to − 7.1 kcal/mole (Supplementary Table S1). Residues involving possible hydrogen bond interactions are represented in Supplementary Table S1. In addition, chitinase has shown chitin and cellulose-binding interaction with docking scores ranging from − 6.3 to − 6.5 kcal/mole (Fig. 5C, Supplementary Table S1).
Ligninolytic genetic repertoire
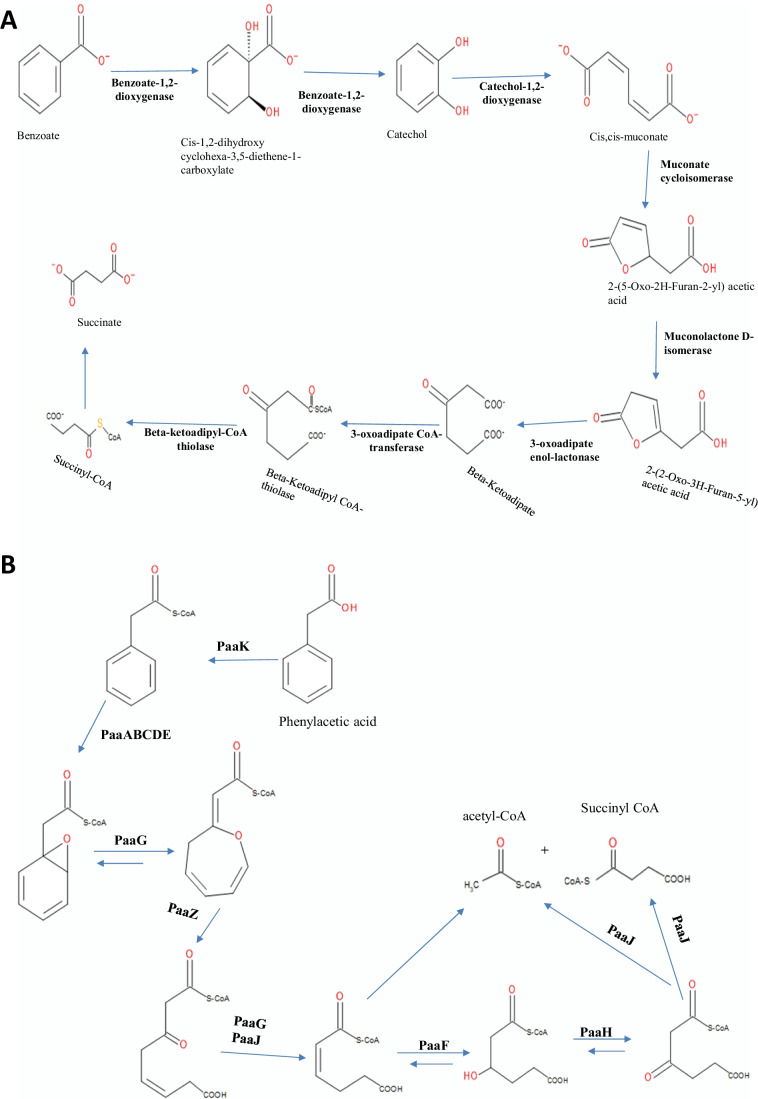
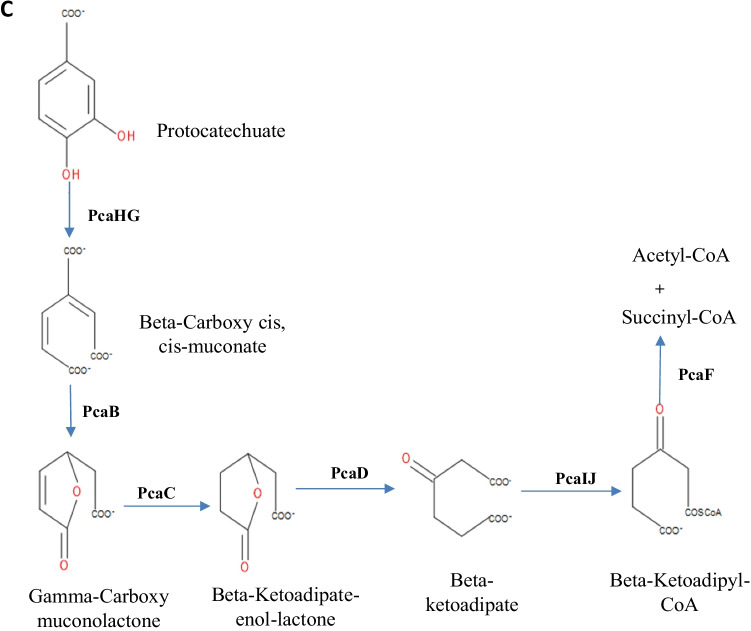
The genes involved in the β-keto adipate pathway are shown in Table 6. In particular, the catechol branch, run by cat gene products, was involved in the conversion of catechol (generated from benzoate), benzene, and some lignin monomers into β-ketoadipate. The pob gene products catalyzed the conversion of 4-hydroxy benzoate to protocatechuate.
Table 6.
Genes encoding the β-ketoadipate pathway and peripheral reactions
| Branch name | Product name | Gene | Locus tag | Ref accession no: | E.C. number |
|---|---|---|---|---|---|
|
Catechol (cat gene) |
Catechol 1,2-dioxygenase | catA | GO309_00275 | YP_004594166.1 | 1.13.11.1 |
| Muconolactone δ-isomerase | catC | GO309_00280 | YP_004594167.1 | 5.3.3.4 | |
| Muconatecycloisomerase | catB1 | GO309_00285 | YP_004594168.1 | - | |
| Extracellular solute binding receptor protein | catX | GO309_20910 | WP_004124644.1 | - | |
| 3-carboxyethylcatechol 2,3-dioxygenase | mphB | GO309_01715 | WP_016947516.1 | 1.13.11.16 | |
|
Benzoate (ben gene) |
Benzoate 1,2-dioxygenase small subunit | benB | GO309_00265 | YP_004594164.1 | 1.14.12.10 |
| Benzoate/H( +) symporterBenE family transporter | benE | GO309_00615 | WP_004898576.1 | - | |
| 2,3-dihydro-2,3-dihydroxy benzoate dehydrogenase | dhbA | GO309_5240 | YP_004592926.1 | 1.3.1.28 | |
|
4-hydroxy benzoate (pob) gene |
4-hydroxy benzoate 3-monooxygenase | pobA | GO309_17060 | YP_004592293.1 | 1.14.13.2 |
| 4-hydroxybenzoate octaprenyltransferase | - | GO309_22995 | YP_004591886.1 | 2.5.1.39 | |
| DNA-binding response regulator | pobR | GO309_17070 | WP_008807539.1 | - | |
|
Protocatechuate (pca) genes |
LysR family transcriptional regulator | pcaQ | GO309_00455 | WP_019705703.1 | - |
| 4-carboxy muconolactone decarboxylase | pcaC | GO309_17485 | WP_004143067.1 | 4.1.1.44 | |
| 3-oxoadipate enol-lactonase | pcaD | GO309_17490 | WP_012968330.1 | 3.1.1.24 | |
| 3-carboxy-cis,cis-muconate cycloisomerase | pcaB | GO309_17495 | YP_004593823.1 | 5.5.1.2 | |
| 3-oxoadipyl-CoA thiolase | pcaF | GO309_17500 | YP_004593822.1 | 2.3.1.174 | |
| 3-oxoacid CoA-transferase subunit B | - | GO309_17505 | YP_004593821.1 | - | |
| 3-oxoacid CoA-transferase subunit A | - | GO309_17510 | YP_004593820.1 | - | |
| Protocatechuate 3,4-dioxygenase β-subunit | pcaH | GO309_00675 | YP_004594243.1 | 1.13.11.3 | |
| Protocatechuate 3,4-dioxygenase α-subunit | pcaG | GO309_00680 | YP_004594244.1 | 1.13.11.3 | |
| 3-oxoadipyl-CoA thiolase | pcaF | GO309_17860 | YP_17900111.1 | 2.3.1.174 | |
| 2,3-dehydroadipyl-CoA hydratase | GO309_17880 | YP_004594284.1 | 4.2.1.17 | ||
| 4-carboxy-4-hydroxy-2-oxoadipate aldolase | ligK | GO309_25155 | YP_019725153.1 | - |
Benzoate pathway
The strain K. variicola sp. HSTU-AAM51 genome showed gene products of catechol, benzoate, 4-hydroxy benzoate, and protocatechuate branches (Table 7). The pcaQ gene was found in the vicinity of the genes pcaC, pcaD, pcaB, pcaF, pcaH, and pcaG in the genome sequence of the AAM51 strain, which suggested the transcriptional regulation was maintained by the pcaQ gene product in the protocatechuate branch. In this study, two proteins identified 3-oxoacid CoA-transferase subunit B and 3-oxoacid CoA-transferase subunit A, which was located in the pca genes cluster.
Table 7.
Genes encoding catabolic pathways for benzene, toluene, (methyl) phenols, 3-hydroxy phenyl-propionate, and 3-hydroxy anthranilate
| Products name | Genes | Locus tag | Ref accession no | E.C. number |
|---|---|---|---|---|
| Replication initiation negative regulator | seqA | GO309_05745 | YP_004593003.1 | - |
| Putative membrane protein | ymiA | GO309_19345 | WP_002901776.1 | - |
| Transcriptional regulator ChbR | chbR | GO309_19145 | YP_004594490.1 | - |
| Reductase subunit | paaK | GO309_17885 | YP_004594286.1 | - |
| Hydroxylase subunit HpxD | hpxD | GO309_08190 | WP_002904779.1 | - |
| Bacterioferritin-associated ferredoxin | bfd | GO309_25925 | YP_004591201.1 | - |
| Polyphenol oxidase | pgeF | GO309_09130 | YP_004590421.1 | 1.10.3.- |
| Phenolic acid decarboxylase | kpdC | GO309_06850 | YP_004591347.1 | - |
| 3-phenyl-propionate MFS transporter | GO309_18435 | YP_004590381.1 | - | |
| Ferredoxin–NADP( +) reductase | GO309_06005 | YP_004591496.1 | 1.18.1.2 | |
| 2Fe-2S ferredoxin-like protein | GO309_12680 | YP_004595022.1 | - | |
| Pyruvate:ferredoxin (flavodoxin) oxidoreductase | nifJ | GO309_18025 | WP_009486361.1 | - |
| 2-octaprenyl-6-methoxyphenyl hydroxylase | ubiH | GO309_20470 | YP_004590795.1 | 1.14.13.- |
| FAD-dependent 2-octaprenylphenol hydroxylase | ubiI | GO309_20475 | YP_004590794.1 | - |
| 2-octaprenyl-6-hydroxy phenol methylase | GO309_12695 | WP_015959798.1 | - | |
| Catechol 1,2-dioxygenase | catA | GO309_00275 | YP_004594166.1 | 1.13.11.1 |
| 3-(3-hydroxy-phenyl)propionate/3-hydroxycinnamic acid hydroxylase" | GO309_01720 | WP_004891230.1 | 1.14.13.127 | |
| 5-carboxymethyl-2-hydroxymuconate semialdehyde dehydrogenase | hpaE | GO309_1689 | YP_004592265.1 | - |
| 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase | GO309_16900 | YP_004592266.1 | - | |
| 4-oxalocrotonate tautomerase | GO309_01255 | WP_004175772.1 | - | |
| 4-hydroxy-2-oxovalerate aldolase | dmpG | GO309_01695 | WP_019705824.1 | 4.1.3.39 |
| Acetaldehyde dehydrogenase (acetylating) | dmpF | GO309_01700 | WP_014907417.1 | 1.2.1.10 |
| 4-oxalocrotonate tautomerase | GO309_01255 | WP_004175772.1 | - | |
| LysR family transcriptional regulator | GO309_19585 | WP_008805220.1 | - | |
| FAD-dependent 2-octaprenylphenol hydroxylase | ubiI | GO309_20475 | YP_004590794.1 | - |
| acrEF/envCD operon transcriptional regulator | envR | GO309_25365 | WP_008806701.1 | - |
| 3-(3-hydroxy-phenyl)propionate transporter MhpT | mhpT | GO309_01690 | WP_014907419.1 | - |
| 2-keto-4-pentenoate hydratase | mhpD | GO309_01705 | WP_016947515.1 | 4.2.1.80 |
| 2-oxo-hepta-3-ene-1,7-dioic acid hydratase | hpaH | GO309_16880 | YP_004592262.1 | - |
| 3-carboxyethyl catechol 2,3-dioxygenase | mhpB | GO309_01715 | WP_016947516.1 | 1.13.11.16 |
| Transcriptional activator MhpR | mhpR | GO309_01725 | WP_004221425.1 | - |
Phenylacetate pathways
The phenylacetate-degrading (Paa) pathway is involved in the series degradation of phenylacetate. Above, eleven different Paa genes are observed after searching in the K. variicola HSTU-AAM51 genome, which encodes for phenylacetic acid degradation (Table 8). Three different transcriptional units such as PaaZ, PaaABCDEFGHIJK, and PaaXY appeared in K. variicola HSTU-AAM51. All these gene clusters were aligned in a row. The existence of the three gene clusters indicates that HSTU-AAM51 can degrade phenylacetate into TCA cycle ingredients acetyl CoA and succinyl CoA.
Table 8.
Genes encoding pathways for phenylacetate, 4-hydroxy phenylacetate, 3-hydroxyl phenyl-propionate, and benzoate proceeding through aryl-CoA intermediates
| Products name | Gene | Locus tag | Ref accession no | E.C. number |
|---|---|---|---|---|
| Phenylacetic acid degradation protein PaaY | paaY | GO309_17845 | WP_008805114.1 | - |
| Phenylacetic acid degradation operon negative regulatory protein | paaX | GO309_17850 | WP_016947252.1 | - |
| Phenylacetate–CoA ligase | paaF | GO309_17855 | YP_004594279.1 | 6.2.1.30 |
| 3-oxoadipyl-CoA thiolase | pcaF | GO309_17860 | YP_004594280.1 | 2.3.1.174 |
| Hydroxyphenylacetyl-CoA thioesterasePaaI | paaI | GO309_17865 | YP_004594281.1 | - |
| 3-hydroxyacyl-CoA dehydrogenase | paaH | GO309_17870 | YP_004594282.1 | 1.1.1.35 |
| 2-(1,2-epoxy-1,2 dihydrophenyl) acetyl-CoA isomerase | paaG | GO309_17875 | YP_004594283.1 | |
| 2,3-dehydroadipyl-CoA hydratase | paaD | GO309_17880 | YP_004594284.1 | 4.2.1.17 |
| Phenylacetate-CoA oxygenase/reductase subunit | paaK | GO309_17885 | YP_004594286.1 | - |
| Phenylacetate-CoA oxygenase subunit PaaJ | paaJ | GO309_17890 | YP_004594287.1 | - |
| Phenylacetate-CoA oxygenase subunit PaaC | paaC | GO309_17895 | YP_004594288.1 | - |
| 1,2-phenylacetyl-CoA epoxidase subunit B | paaB | GO309_17900 | YP_004594289.1 | 1.14.13.149 |
| 1,2-phenylacetyl-CoA epoxidase subunit A | paaA | GO309_17905 | WP_020803990.1 | 1.14.13.149 |
| Phenylacetic acid degradation bifunctional protein | paaZ | GO309_17910 | YP_004594291.1 | - |
| 4-hydroxy phenylacetate 3-monooxygenase reductase subunit | hpaC | GO309_16855 | YP_004592257.1 | - |
| 4-hydroxy phenylacetate 3-monooxygenase | hpaB | GO309_16860 | WP_000801462.1 | 1.14.14.9 |
| 4-hydroxyphenylacetate catabolism regulatory protein HpaA | hpaA | GO309_16865 | YP_004592259.1 | - |
| 4-hydroxyphenylacetate permease | hpaX | GO309_16870 | NP_460080.1 | - |
| 4-hydroxy-2-oxoheptanedioate aldolase | hpaI | GO309_16875 | WP_016809283.1 | 4.1.2.52 |
| 2-oxo-hepta-3-ene-1,7-dioic acid hydratase | hpaH | GO309_16880 | YP_004592262.1 | - |
| 5-carboxymethyl-2-hydroxymuconate Delta-isomerase | hpaF | GO309_16885 | YP_004592263.1 | 5.3.3.10 |
| 3,4-dihydroxyphenylacetate 2,3-dioxygenase | hpaD | GO309_16890 | YP_004592264.1 | 1.13.11.15 |
| 5-carboxymethyl-2-hydroxymuconate semialdehyde dehydrogenase | hpaE | GO309_16895 | YP_004592265.1 | - |
| 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase | hpaG | GO309_16900 | YP_004592266.1 | - |
| 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase | hpaG | GO309_16905 | YP_004592267.1 | - |
| Homoprotocatechuate degradation operon regulator HpaR | hpaR | GO309_16910 | YP_004592268.1 | - |
| O-succinyl benzoate synthase | menC | GO309_12605 | YP_004595037.1 | 4.2.1.113 |
| O-succinylbenzoate–CoA ligase | menE | GO309_12610 | WP_014599221.1 | 6.2.1.26 |
| 3-(3-hydroxy-phenyl)propionate transporter MhpT | mhpT | GO309_01690 | WP_014907419.1 | - |
| 4-hydroxy-2-oxovalerate aldolase | mhpE | GO309_01695 | WP_019705824.1 | 4.1.3.39 |
| Acetaldehyde dehydrogenase (acetylating) | mhpF | GO309_01700 | WP_014907417.1 | 1.2.1.10 |
| 2-keto-4-pentenoate hydratase | mhpD | GO309_01705 | WP_016947515.1 | 4.2.1.80 |
| Alpha/beta fold hydrolase | mhpC | GO309_01710 | NP_308431.3 | - |
| 3-carboxyethylcatechol 2,3-dioxygenase | mhpB | GO309_01715 | WP_016947516.1 | 1.13.11.16 |
| (3-hydroxy-phenyl)propionate/3-hydroxycinnamic acid hydroxylase | mhpA | GO309_01720 | WP_004891230.1 | 1.14.13.127 |
| Transcriptional activator MhpR | mhpR | GO309_01725 | WP_004221425.1 | - |
In the HSTU-AAM51 genome, the operons of the 4-hydroxy phenylacetic acid degradation pathway were observed (Table 8). The pathway was composed of hpaBC transcriptional unit (4-HPA hydroxylase), hpaGEDFHI (meta-cleavage), and regulatory genes hpaR and hpaA, respectively. The operons database indicated that HSTU-AAM51 could degrade 4-hydroxy phenylacetic acid into TCA cycle precursors like pyruvate and succinate, while 3-hydroxyl phenyl-propionate–degrading operon genes have existed in the genome of the HSTU-AAM51 strain. From the above analysis, it was therefore assumed that the strain HSTU-AAM51 genome reveals putative genes that encode enzymes leading to ring oxidation and cleavage of lignin and related aromatic compounds via both ortho- and meta-cleavage pathways. Besides, proteins peroxiredoxin, cytochrome oxidase, oxidoreductase, ferredoxin, amino deoxychorismate-lyase, dehydrogenase, acetyl-CoA C-acetyl transferase, enoyl CoA hydratase, glyoxalase, glutaredoxin, alkyl hydroperoxidase, alcohol dehydrogenase, formate dehydrogenase, alkene reductase, GNAT family N-acetyl transferase, glycine betaine/L-proline ABC transporter, etc. are observed in the genome of K. variicola HSTU-AAM51. To further confirm, we have searched additional gene products that are potentially involved in lignin degradation and enlisted them in Supplementary Table S2–5.
Likewise, glycine betaine ABC transporter was also identified in the genome of HSTU-AAM51 (Supplementary Table S2). As seen in Supplementary Table S3, a laccase domain–containing protein was observed. Besides, a total of 23 different types of oxidoreductases including GMC family genes were annotated (Supplementary Table S3). Furthermore, 13 different NADH-Quinone oxidoreductase subunits were observed in an operon (Supplementary Table S3), while Dyp-type peroxidases were the fungal counterpart of peroxidases (LiP or MnP) found in the genome (Supplementary Table S3) of the HSTU-AAM51 strain. In particular, glutathione-S-transferase, glutathione reductase, glutathione peroxidase, glutathione hydroxylase, glutathione dehydrogenase, S-formyl glutathione hydroxylase, catalase, superoxide dismutase, thioredoxin, peroxiredoxin, glyoxalase, glutaredoxin, alkyl hydroperoxide reductase, thiol peroxidase, and deferrochelatase encoding genes are identified (Supplementary Table S4) and four separate genes that encode formate dehydrogenase were apparent in an operon (Supplementary Table S5).
Plant growth–promoting traits and abiotic stress tolerance
The nitrogen-fixing genes (nifJKEHABMSUNTKV, iscU, and ntrB) added with norR and norV were annotated in the genome of K. variicola HSTU-AAM51. Moreover, ACC deaminase producing vital genes such as dcyD and rimM were found in the genome of the HSTU-AAM51strain. Besides, IAA hormone–producing genes (trpCF, trpS, trpB, trpD) and siderophore-producing genes (fes, entDFS, fhuABCD, tonB, febBG, exbB) existed in the K. variicola HSTU-AAM51 genome. Interestingly, genes involved in phosphate metabolism, e.g., pitA, pstABCS, phoABHRU, tomB, and pntAB, and biofilm formation genes such as luxS, efp, and hfq are common in the HSTU-AAM51 strain genome. Moreover, flagellar motility–related genes flgABCGHIJKLM and motAB were predicted in the K. variicola HSTU-AAM51 genome (Table 9). The ranges of phosphate uptake, inorganic phosphate transporter, and phosphate starvation genes, e.g., pitA, pstABCU, ugpBE, phoABEHQR, and pntAB, were observed too. Importantly, abiotic stress tolerance genes such as cold shock, heat shock, homeostasis of heavy metals, e.g., arsenic, chromium, and drought resistance genes were annotated in the K. variicola strain HSTU-AAM51 genome (Supplementary Table S5). Interestingly, several kinds of pesticide and herbicide-degrading genes (ampD, glpABQ, pepABD, phnFDGHKLMP, phosphodiesterase), and a number of esterases were annotated in the genome of the strain (Supplementary Table S6).
Table 9.
Genes involved in plant growth–promoting activity
| PGP activities description | Gene name | Locus tag | Product | CDS | E.C. number |
|---|---|---|---|---|---|
| Nitrogen fixation | iscU | GO309_18480 | Fe-S cluster assembly scaffold IscU | 49,393..49779 | - |
| fnifJ | GO309_18025 | Pyruvate:ferredoxin (flavodoxin) oxidoreductase | 122,184..125711 | - | |
| glnK | GO309_04155 | P-II family nitrogen regulator | 198,581..198919 | - | |
| ntrB | GO309_02285 | Nitrate ABC transporter permease | 472,449..473333 | - | |
| nifE | GO309_24775 | Nitrogenase iron-molybdenum cofactor biosynthesis protein NifE | 38,288..39661 | - | |
| nifH | GO309_24750 | Nitrogenase iron protein | 32,547..33428 | 1.18.6.1 | |
| nifA | GO309_24830 | Nif-specific transcriptional activator NifA | 48,777..50351 | - | |
| nifB | GO309_24835 | Nitrogenase cofactor biosynthesis protein NifB | 50,516..51922 | - | |
| nifM | GO309_24815 | Nitrogen fixation protein NifM | 45,623..46423 | - | |
| nifS | GO309_24795 | Cysteine desulfuraseNifS | 42,563..43765 | 2.8.1.7 | |
| nifU | GO309_24790 | Fe-S cluster assembly protein NifU | 41,708..42547 | - | |
| nifN | GO309_24780 | Nitrogenase iron-molybdenum cofactor biosynthesis protein NifN | 39,671..41056 | - | |
| nifT | GO309_24765 | Putative nitrogen fixation protein NifT | 36,551..36769 | - | |
| nifK | GO309_24760 | Nitrogenase molybdenum-iron protein subunit beta | 34,949..36511 | 1.18.6.1 | |
| nifV | GO309_24800 | Homocitrate synthase | 43,781..44923 | 2.3.3.14 | |
| Nitrosative stress | norR | GO309_09770 | Nitric oxide reductase transcriptional regulator NorR | 125,015..126571 | - |
| norV | GO309_09775 | Anaerobic nitric oxide reductaseflavorubredoxin | 126,759..128207 | - | |
| Ammonia assimilation | gltB | GO309_14435 | Glutamate synthase large subunit | 231,718..236178 | 1.4.1.13 |
| ACC deaminase | dcyD | GO309_19925 | D-cysteine desulfhydrase | 34,486..35472 | 4.4.1.15 |
| rimM | GO309_09210 | Ribosome maturation factor RimM | 16,698..17246 | - | |
| Siderophore | |||||
| Siderophoreenterobactin | fes | GO309_05185 | Enterochelin esterase | 419,528..420736 | 3.1.1.- |
| entF | GO309_05195 | Enterobactin non-ribosomal peptide synthetaseEntF | 420,978..424859 | 6.3.2.14 | |
| entS | GO309_05215 | Enterobactin transporter EntS | 427,824..429065 | - | |
| fepA | |||||
| entD | GO309_05175 | Enterobactin synthase subunit EntD | 416,344..416973 | 6.3.2.14 | |
| fhuA | GO309_16320 | FerrichromeporinFhuA | 165,154..167361 | - | |
| fhuB | GO309_16335 | Fe(3 +)-hydroxamate ABC transporter permeaseFhuB | 169,094..171076 | - | |
| fhuC | GO309_16325 | Fe3 + -hydroxamate ABC transporter ATP-bindingproteinFhuC | 167,407..168204 | - | |
| fhuD | GO309_16330 | Fe(3 +)-hydroxamate ABC transporter substrate-binding protein FhuD | 168,204..169097 | - | |
| tonB | GO309_02050 | TonB system transport protein TonB | 417,432..418169 | - | |
| fepB | GO309_05220 | Fe2 + -enterobactin ABC transporter substrate-binding protein | 429,422..430381 | - | |
| fepG | GO309_05205 | Iron-enterobactin ABC transporter permease | 425,715..426704 | - | |
| exbB | GO309_13485 | Tol-pal system-associated acyl-CoA thioesterase | 42,198..42929 | - | |
| Plant hormones | |||||
| IAA production | trpCF | GO309_19275 | Bifunctional indole-3-glycerol-phosphate synthase TrpC/phosphoribosylanthranilateisomeraseTrpF | 56,470..57828 | 4.1.1.48/5.3.1.24 |
| trpS | GO309_07325 | Tryptophan–tRNA ligase | 311,227..312231 | 6.1.1.2 | |
| trpB | GO309_19270 | Tryptophan synthase subunit beta | 55,267..56460 | 4.2.1.20 | |
| trpD | GO309_19280 | Bifunctionalanthranilate synthase glutamate amidotransferase component TrpG/anthranilatephosphoribosyltransferaseTrpD | 57,832..59427 | 2.4.2.18/4.1.3.27 | |
| Phosphate metabolism | pitA | GO309_06800 | Inorganic phosphate transporter PitA | 196,804..198300 | - |
| pstS | GO309_22625 | Phosphate ABC transporter permeasePstA | 54,729..55619 | - | |
| pstC | GO309_22630 | Phosphate ABC transporter permeasePstC | 55,619..56578 | - | |
| pstA | GO309_22625 | Phosphate ABC transporter permeasePstA | 54,729..55619 | - | |
| pstB | GO309_22620 | Phosphate ABC transporter ATP-binding protein PstB | 53,908..54681 | - | |
| phoU | GO309_22615 | Phosphate signaling complex protein PhoU | 53,132..53857 | 3.5.2.6 | |
| ugpB | GO309_07025 | Sn-glycerol-3-phosphate ABC transporter substrate-binding protein UgpB | 241,896..243212 | - | |
| ugpE | GO309_07035 | Sn-glycerol-3-phosphate ABC transporter permeaseUgpE | 244,203..245048 | - | |
| phoA | GO309_03715 | Alkaline phosphatase | 107,973..109388 | 3.1.3.1 | |
| phoE | GO309_03485 | PhosphoporinPhoE | 60,858..61910 | - | |
| phoB | GO309_03780 | Phosphate response regulator transcription factor PhoB | 121,081..121770 | - | |
| phoR | GO309_03785 | Phosphate regulon sensor histidine kinase PhoR | 121,792..123087 | 2.7.13.3 | |
| phoH | GO309_10720 | Phosphate starvation-inducible protein PhoH | 676..1734 | - | |
| pntA | GO309_17575 | Re/Si-specific NAD(P)( +) transhydrogenase subunit alpha | 23,311..24840 | 1.6.1.2 | |
| pntB | GO309_17570 | Re/Si-specific NAD(P)( +) transhydrogenase subunit beta | 21,912..23300 | - | |
| phoQ | GO309_25065 | Two-component system sensor histidine kinase PhoQ | 35,712..37178 | 2.7.13.3 | |
| Biofilm formation | tomB | GO309_04305 | Hha toxicity modulator TomB | 224,892..225266 | - |
| luxS | GO309_09640 | S-ribosylhomocysteinelyase | 104,383..104898 | 4.4.1.21 | |
| efp | GO309_14975 | Elongation factor P | 101,008..101574 | - | |
| hfq | GO309_15100 | RNA chaperone Hfq | 124,744..125052 | - | |
| Sulfur assimilation | cysZ | GO309_18970 | Sulfate transporter CysZ | 156,243..157004 | - |
| cysK | GO309_18965 | Cysteine synthase A | 155,095..156066 | 2.5.1.47 | |
| cysM | GO309_18925 | Cysteine synthase CysM | 148,124..149035 | 2.5.1.47 | |
| cysA | GO309_18920 | Sulfate/thiosulfate ABC transporter ATP-binding protein CysA | 146,911..148005 | - | |
| cysW | GO309_18915 | Sulfate/thiosulfate ABC transporter permeaseCysW | 146,046..146921 | - | |
| cysC | GO309_10115 | Adenylyl-sulfate kinase | 194,395..195000 | 2.7.1.25 | |
| cysN | GO309_10120 | Sulfateadenylyltransferase subunit CysN | 195,000..196427 | 2.7.7.4 | |
| cysD | GO309_10125 | Sulfateadenylyltransferase subunit CysD | 196,437..197345 | 2.7.7.4 | |
| cysH | GO309_10140 | Phosphoadenosinephosphosulfatereductase | 200,127..200861 | 1.8.4.8 | |
| cysI | GO309_10145 | Assimilatory sulfitereductase (NADPH) hemoproteinsubunit | 200,960..202672 | 1.8.1.2 | |
| cysJ | GO309_10150 | NADPH-dependent assimilatory sulfitereductaseflavoprotein subunit | 202,672..204471 | 1.8.1.2 | |
| cysT | GO309_18910 | Sulfate/thiosulfate ABC transporter permeaseCysT | 145,213..146046 | - | |
| Sulfur metabolism | cysC | GO309_10115 | Adenylyl-sulfate kinase | 194,395..195000 | 2.7.1.25 |
| cysN | GO309_10120 | Sulfateadenylyltransferase subunit CysN | 195,000..196427 | 2.7.7.4 | |
| cysD | GO309_10125 | Sulfateadenylyltransferase subunit CysD | 196,437..197345 | 2.7.7.4 | |
| cysH | GO309_10140 | Phosphoadenosinephosphosulfatereductase | 200,127..200861 | 1.8.4.8 | |
| cysI | GO309_10145 | Assimilatory sulfitereductase (NADPH) hemoprotein subunit | 200,960..202672 | 1.8.1.2 | |
| cysJ | GO309_10150 | NADPH-dependent assimilatory sulfitereductaseflavoprotein subunit | 202,672..204471 | 1.8.1.2 | |
| cysE | GO309_06280 | Serine O-acetyltransferase | 70,686..71507 | 2.3.1.30 | |
| cysQ | GO309_15285 | 3′(2′),5′-bisphosphate nucleotidaseCysQ | 158,377..159120 | 3.1.3.7 | |
| cysK | GO309_18965 | Cysteine synthase A | 155,095..156066 | 2.5.1.47 | |
| cysS | GO309_04505 | Cysteine–tRNA ligase | 269,952..271337 | 6.1.1.16 | |
| cysZ | GO309_18970 | Sulfate transporter CysZ | 156,243..157004 | - | |
| cysM | GO309_18925 | Cysteine synthase CysM | 148,124..149035 | 2.5.1.47 | |
| cysA | GO309_18920 | Sulfate/thiosulfate ABC transporter ATP-binding protein CysA | 146,911..148005 | - | |
| cysW | GO309_18915 | Sulfate/thiosulfate ABC transporter permeaseCysW | 146,046..146921 | - | |
| fdxH | GO309_21120 | Formate dehydrogenase subunit beta | 24,484..25386 | - | |
| Antimicrobial peptide | pagP | GO309_05455 | Lipid IV(A) palmitoyltransferasePagP | 482,608..483177 | 2.3.1.251 |
| sapB | GO309_19460 | Peptide ABC transporter permeaseSapB | 95,370..96335 | - | |
| Synthesis of resistance inducers | |||||
| 2,3-butanediol | ilvB | GO309_22405 | Acetolactate synthase large subunit | 9103..10791 | - |
| 2,3-butanediol | ilvA | GO309_23220 | Threonine ammonia-lyase, biosynthetic | 8479..10023 | 4.3.1.19 |
| ilvC | GO309_23230 | Ketol-acid reductoisomerase | 11,065..12540 | 1.1.1.86 | |
| ilvY | GO309_23225 | HTH-type transcriptional activator IlvY | 10,020..10910 | - | |
| ilvD | GO309_23215 | Dihydroxy-acid dehydratase | 6626..8476 | - | |
| ilvM | GO309_23205 | Acetolactate synthase 2 small subunit | 5359..5616 | 2.2.1.6 | |
| Methanethiol isoprene | metH | GO309_23130 | Methionine synthase | 75,514..79197 | 2.1.1.13 |
| gcpE/ispG | GO309_18555 | Flavodoxin-dependent (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase | 68,399..69520 | 1.17.7.1 | |
| ispE | GO309_02360 | 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase | 485,846..486697 | 2.7.1.148 | |
| Hydrolase | ribA | GO309_19355 | GTP cyclohydrolase II | 76,756..77358 | 3.5.4.25 |
| folE | GO309_13000 | GTP cyclohydrolase I FolE | 226,451..227119 | 3.5.4.16 | |
| bglF | GO309_13080 | PTS beta-glucoside transporter subunit IIABC | 242,386..244248 | - | |
| bglX | GO309_13125 | Beta-glucosidaseBglX | 253,401..255698 | .2.1.21 | |
| malZ | GO309_03805 | Maltodextrin glucosidase | 126,431..128248 | 3.2.1.20 | |
| amyA | GO309_19935 | Alpha-amylase | 36,569..38056 | 3.2.1.1 | |
| Symbiosis-related | gcvT | GO309_20480 | Glycine cleavage system aminomethyltransferaseGcvT | 16,096..17190 | 2.1.2.10 |
| phnC | GO309_03520 | Phosphonate ABC transporter ATP-binding protein | 67,851..68693 | - | |
| tatA | GO309_23525 | Sec-independent protein translocase subunit TatA | 73,386..73637 | - | |
| pyrC | GO309_23855 | Dihydroorotase | 54,965..56011 | 3.5.2.3 | |
| zur | GO309_22965 | Zinc uptake transcriptional repressor Zur | 42,733..43248 | - | |
| Root colonization | |||||
| Chemotaxis | malE | GO309_23020 | Maltose/maltodextrin ABC transporter substrate-binding protein MalE | 54,343..55533 | - |
| rbsB | GO309_22750 | Ribose ABC transporter substrate-binding protein RbsB | 82,965..83855 | - | |
| Adhesive structure | hofC | GO309_16020 | Protein transport protein HofC | 97,395..98600 | - |
| Adhesin production | pgaA | GO309_14720 | Poly-beta-1,6 N-acetyl-D-glucosamine export porinPgaA | 47,429..49897 | - |
| pgaB | GO309_14715 | Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylasePgaB | 45,405..47420 | 3.5.1.- | |
| pgaC | GO309_14710 | Poly-beta-1,6 N-acetyl-D-glucosamine synthase | 44,084..45412 | - | |
| pgaD | GO309_14705 | Poly-beta-1,6-N-acetyl-D-glucosamine biosynthesis protein PgaD | 43,635..44084 | - | |
| Flagellar protein | murJ | GO309_23825 | Murein biosynthesis integral membrane protein MurJ | 48,906..50441 | - |
| Superoxide dismutase | sodA | GO309_21265 | Superoxide dismutase [Mn] | 54,764..55384 | 1.15.1.1 |
| sodB | GO309_00910 | Superoxide dismutase [Fe] | 184,360..184941 | 1.15.1.1 | |
| sodC | GO309_00860 | Superoxide dismutase [Cu–Zn] SodC2 | 176,890..177411 | 1.15.1.1 | |
| Trehalose metabolism | treB | GO309_15495 | PTS trehalose transporter subunit IIBC | 203,366..204784 | 2.7.1.201 |
| treC | GO309_15490 | Alpha,alpha-phosphotrehalase | 201,659..203314 | 3.2.1.93 | |
| treR | GO309_15500 | HTH-type transcriptional regulator TreR | 204,915..205862 | - | |
| otsA | GO309_03140 | Alpha,alpha-trehalose-phosphate synthase | 646,115..647539 | 2.4.1.15 | |
| otsB | GO309_03145 | Trehalose-phosphatase | 647,514..648302 | 3.1.3.12 | |
| lamB | GO309_23010 | MaltoporinLamB | 51,489..52778 | - |
Discussion
A few Klebsiella spp. are known for the production of lignocellulose-degrading enzymes from different lignocellulose materials [6, 20, 30]. In this study, we demonstrated a new pectinolytic bacteria namely Klebsiella variicola strain HSTU-AAM51. The biochemical tests of the strain were conducted and found that the pectin hydrolytic strains were varied among each other. In fact, they are different from the strains that showed inverse results in MR and VP, respectively [2]. Importantly, Pec-24/HSTU-AAM51 strain’s diazinon mineralization capability concluded that the strain is related to insecticide or toxic chemical degradation role in goat omasum strains. The assembly of extracellular cellulase, xylanase, and pectinase is required to degrade straw and grass materials in the omasum of goats [31]. These results suggested that strain HSTU-AAM51 may be reasoned with potential lignocellulose degradation. Therefore, we selected the HSTU-AAM51 strain for genomic analysis to reveal its lignocellulolytic potentialities.
The quality of the genome sequence was predicted according to GC% of strains DNA [32]. It is accepted that the optimum GC% should be around 40–70% that is covered by the HSTU-AAM51 strain. Therefore, it was ensured that the sequence quality was good for further processing of sequence assembly. MLST analysis, housekeeping genes alignment, phylogenetic tree of whole-genome sequences, digital DNA-DNA hybridization (DDH), ANIb values confirmed that the strain has an intimate intragenus relationship with K. variicola species. Only the environmental strain K. variicola P1CD1 with ligninolytic genetic repertoire is closer (93.3%) according to DDH analysis. But all types of gene and genome alignments lead us to conclude that the K. variicola HSTU-AAM51 strain is much far away from K. pneumonia, etc. The GC content of strain HSTU-AAM51 with three nearest K. variicola strains, namely 13,450 (NZ_CP026013.1), WCHKP19 (NZ_CP020847.1), P1CD1 (CP033631), had shown total CDS 5274, 6066, 5454 gene coding 5163, 5809, 5313, and RNA 124, 128, 118, respectively. However, the strain K. variicola HSTU-AAM51 had shown total CDS 5212, gene coding 5150, and RNA 102 which is markedly deviated from that nearest K. variicola strain sourced from sick humans [33]. The presence of CDS in downstream and upstream was meaningful according to relevant studies. The GC skew is observed in local genomic regions primarily introduced by RNA synthesis, but the analysis of GC skew was first utilized for the computational prediction of ori and ter positions by examining available genome sequences [34].
The bacterial pangenome analysis revealed a framework to answer the genomic diversity within a species. More specifically, bacterial pangenome analysis can reveal individual strain’s single-nucleotide polymorphisms, insertion/deletion variants. and structural variants, at the level of absence or presence of whole genes, annotated functions, and mobile genetic elements such as integrons or prophages [35]. The pangenome graph clearly showed that some part of the genome sequence of HSTU-AAM51 was unmatched compared to the six other strains studied (Fig. 3). Especially, the potential pathogenic genes did not appear in the pangenome analysis. Therefore, it is suggested that the HSTU-AAM51 strain genome has an evolutionary character with no potential pathogenic inducers genes. Besides, the locally collinear blocks (LCB) of the genomes of four nearest strains, namely Klebsiella sp. P1CD1 (CP033631.1), K. variicola 13,450 (CP030173.1), K. variicola FDA ARGOS (CP044050.1) with K. variicola strain HSTU-AAM51, were inspected using Progressive Mauve. The addition, deletion, reshuffling or rearrangements, and inverted of sort of K. variicola HSTU-AAM51 genome sequences were found in various LCBs compared to the LCBs of other nearest strains (CP033631.1, CP030173.1, CP044050.1) genome. Therefore, it is suggested that the K. variicola HSTU-AAM51 strain might be a new member of the K. variicola species and it is a newly evolved strain in Bangladesh.
The depolymerization of plant lignocellulose into monosaccharides requires the combined action of a wide range of lignocellulolytic enzymes like glycosyl hydrolase (GH), polysaccharide deacetylase (CE), polysaccharide lyase (PL), and associated domain of carbohydrate-binding modules (CBM) [36]. Interestingly, cellulase, endoglucanase, 6-phospho-alpha-glucosidase, maltodextrin glucosidase, pullulanase type alpha-1,6-glucosidase, fibronectin type III (FNIII), chitinase, pectinase, feruloyl esterase, pullulanase, beta-1,4-mannanase, hemicellulase, galactosidase, and polysaccharide deacetylase were observed in the genome of the HSTU-AAM51 strain. The pectinase attacked pectin-lignin-cellulose ester bonds to degum the lignocellulose, while feruloyl esterase synergistically acts with xylanase to release arabinose and ferulic acid from lignocellulose [36, 37]. As a consequence, the lignocellulolytic enzymes such as cellulase, xylanase, pectinase, and lignin peroxidase activities of strain HSTU-AAM51 were significantly observed in banana pseudostem–enriched media, while Lam et al. found that the activities of lignocellulolytic enzymes (endoglucanase, exoglucanase, β-1,4-xylanase, β-mannanase) in oil palm empty fruit bunch–enriched media were diminished after 72 h [36]. Interestingly, the Klebsiella sp. MD21 showed maximum endoglucanase, exoglucanase, and β-1,4-xylanase activity on sawdust substrate at 96 h of incubation [38], which is consistent with HSTU-AAM51 strain. The enzyme activities variance in this study may come out due to variation in the lignocellulose source. The lignocellulose components such as cellulose, xylose, galactose, and pectin can be degraded by the carbohydrate-active enzymes [39]. The banana pseudostem contains approximately 63–64% cellulose, 10–14% hemicellulose, and 8–10% lignin [40]; therefore, strain K. variicola HSTU-AAM51 might use these lignocellulosic compositions as their sole source of carbon. This is likely that the strain HSTU-AAM51 has shown enormous lignin-degrading enzymes and extracellular CAZy zyme as well as feruloyl esterase and glycosyl hydrolase domain-containing chitinase that might participate in the degradation of banana fibers [34, 36]. The banana plants usually discard into nature without further use due to a lack of the recycling process in Bangladesh. Thus, recycling banana fibers into valuable lignocellulolytic enzymes of the K. variicola HSTU-AAM51 strain may contribute to the production of bioethanol [2, 40].
However, the knowledge regarding catalytic interaction of cellulase, beta-glucosidase, and endoglucanase of K. variicola with cellulose as the substrate is limited. Since the lignocellulose-degrading capacity of the strain K. variicola HSTU-AAM51 accounts for its comprising CAZy enzymes [39], a molecular docking study was performed for the CAZy enzymes having signal peptide with lignocellulose substrates to verify their efficacy. The root mean square distance of a protein model indicated its acceptance when compared against a standard protein model (protein modeling using X-ray crystallography or NMR). The lower RMSD is always preferable as it indicated fewer errors. The average RMSD of the 12 modeled proteins is below one indicating the acceptance of the analyses. It is reported that the RMSDs of the model proteins ranged between 0.35 and 2.36 Å with their nearest homologs is satisfactory [41]. In the ERRAT server, a model is evaluated based on non-bonded interactions between different types of an atom to assess error rates with standard optimized models while the 3D to 1D comparisons are made based on the surrounding environment and locations of the α-helix, β-sheets, loops, etc. The protein models were fine-tuned using loop refining and energy minimization. Protein models for mannase (62.46%), pectinase (74.87%), and tannase (76.98%) demonstrated comparatively lower ERRAT scores while the nine other modeled proteins showed an excellent level (83–97%) of quality for ERRAT in the SAVES server. In addition, a similar analysis for the modeled cellulase of Acinetobacter sp. demonstrated 81.1% of residues in the most favored regions, 14.5% in the allowed regions, and 2.5% in the generously allowed regions [38]. The corresponding values for the best hit cellulase protein (PDB ID: 4q2b) showed 92.2%, 7.4%, and 0.3%, respectively. In consequence, the best hit beta-glucosidase (PDB ID: 6r5i), galactosidase (PDB ID: 2gft), endoglucanase (PDB ID: 5czl), 6-phospho β- D-glucosidase (PDB ID: 1up7), glycosyl hydrolase (PDB ID: 5oq2), Mannase (PDB ID: 5awo), pectinase (PDB ID: 3grh), polysaccharide deacetylase (PDB ID: 6go1), tannase (PDB ID: 6fat), pullulanase (PDB ID: 2fhf), chitinase (PDB ID: 3qok) were shown 91.3%, 88%, 88.4%, 93.5%, 85.4%, 90.6%, 86.9%, 86.7%, 87.8%, 87.4%, 88.2% amino acids in the most favorable regions in the Ramachandran plot, which is slightly higher to the corresponding values of the modeled CAZy zyme proteins as observed for the HSTU-AAM51 strain (Table 5). These results conclude that the model proteins’ structural quality was fine enough for molecular docking and catalytic interactions analyses. But the mannase and tannase proteins’ average amino acid residues appeared below 80% in the most favorable regions, which might be due to their novel gene sequences.
The best docking complex based on docking score was used to analyze the interaction of enzyme and substrate. The cellobiose molecule successfully interacted with the cellulase group enzymes. The cellulase residues Trp223, His216, Ala157, and Leu156 were bounded to cellobiose via hydrogen bonds [39], and represented docking score shown − 6.15 kJ/mole. A similar docking score (− 5.9 to 6.5 kJ/mole) with different H-bonds interactions was observed for the cellulase (GH3) and cellobiose complex in the present study. On the other hand, AtBgl3 and AtBgl3.5 beta-glucosidase residues Arg106, His140, Tyr186, Asp218 and Ser36, Asp96 were reported to establish H-bonds interactions with docking score − 5.25 to − 8.89 kJ/mole [40], which is corroborated with β-glucosidase BglX (GH3) and cellobiose complex (Supplementary Figure S2). Interestingly, the catalytic important residues of beta-glucosidase with cellobiose complex were identified as Asp106, Asp287, Tyr255, Arg170, and Glu514 [41], which is greatly similar to the β-glucosidase BglX (GH3) residues Arg169, Lys208, Asp287, and Glu518 interactions with cellobiose (Supplementary Figure S2). These results suggested that the model β-glucosidase BglX (GH3) protein has interacted with cellobiose and corroborated with the relevant previous reports. Besides, pectin esterase had shown multiple H-bonds interactions with substrates PGA. The presence of H-bonds interaction was observed within the pectinase active site and polygalactouronic acid [42] with binding energies − 5.73 to − 7.27 kJ/mole. Especially, the pectinase from Erwinia carotovora had shown an interaction to ligands by the residues ASN84, CYS99, ALA101, GLN155, ASN157, and ASN200, but pectinase from Pseudoalteromonas haloplanktis played a similar role by LYS167, ARG192, TRP242, ASN265, ARG297, and SER329 residues [42], which is corroborated with the pectin esterase of K. variicola HSTU-AAM51 strain. The galactosidase interacted with fucopyranosyl galactose via H-bonds and non-bonded hydrophobic interactions as well, which might be capable of degrading xylan since galactose and fucopyranose are essential components of hemicellulose [37]. Besides, mannase and tannase are key enzymes for the degumming of hemicellulose from lignin [43]. The presence of multiple H-bonds, hydrophobic, and salt bridge interactions between the active sites of tannase/feruloyl esterase (Asn115, Cys450, His449, Asp412, Gly101, and Asn110) and ligands proves the efficacy of the enzymes with their subsequent substrates [44, 45]. Since the existence of multiple H-bonds and non-bonded interactions of mannase and tannase residues (Fig. 5E, F) was observed with their respective ligands, we concluded that this enzyme could play a significant role in the biodegradation of banana fibers. The docking score of both cellulase enzymes with substrates CMC and cellulose varied from − 4.6 to − 9.1 kcal/mole which elucidates the highest and lowest docking score [7]. These results suggested that the extracellular signal peptide–containing enzymes of K. variicola HSTU-AAM51 were capable of degrading banana fibers and utilize the degraded products as their sole nutrients for growth and reproduction [2].
The lignin degradation pathways were observed in Rhodococcus sp. SYK-6, Streptomyces viridosporus T7A, Sphingobium sp. SYK-6, Nocardia pseudomonas, Bacillus, and sulfate-reducing bacteria [46–49]. The pathways of lignin degradation are different among bacteria [50]. To date, lignin-degrading reports by Klebsiella have not been revealed yet. Some bacteria modify lignin by using hydroxylation, demethylation or using oxidative enzyme cyt-P450 monooxygenase, dye decolorizing peroxide (DyP), laccase, and superoxide dismutase [51–54]. However, some bacteria use the β-keto adipate pathway to degrade lignin. The β-ketoadipate pathway plays a central role in lignin and other aromatic compound degradation. In particular, the catechol branch, run by cat gene products, was involved in the conversion of catechol (generated from benzoate), benzene, and some lignin monomers into β-ketoadipate. The ben gene products assisted the conversion of benzoate into catechol. The pob gene products catalyzed the conversion of 4-hydroxy benzoate to protocatechuate, while in protocatechuate branch, gene (pca) products convert the protocatechuate (derived from 4-hydroxy benzoate) and lignin monomers into β-ketoadipate [55]. In this study, two proteins have low sequence similarity with β-ketoadipate succinyl-CoA transferase. We assumed that it is played like pcaIJ to accomplish the degradation of β-ketoadipate into succinyl CoA and acetyl CoA (Fig. 6A). Two additional steps lead by these gene sequences are highly conserved in diverse organisms but some soil bacteria had shown great diversity in terms of gene organization and regulations involved in the pathway [56].
Fig. 6.
Ligninolytic pathways of Klebsiella variicola strain HSTU-AAM51. A Benzoate catabolic pathway. B Beta-ketoadipyl catabolic pathway, C Phenylacetate catabolic pathway
The catA gene starts the catechol branch of β-KAP has clustered together with catC, catB1, and benB gene, which are essential for funneling benzoate into this pathway. The enzyme muconate cycloisomerase and enol lactone hydrolase were the product of the catC gene, which converts muconate to β-ketoadipate. The gene catD in the cat operon is absent in the genome of the K. variicola HSTU-AAM51 strain, which is consistent with the previously reported strain Pseudomonas [57]. The pcaQ gene product is the transcriptional regulator for pcaBDCFGH genes. The pcaQ gene is found in the vicinity of the genes pcaC, pcaD, pcaB, pcaF, pcaH, and pcaG in the genome sequence of the AAM51 strain, which suggested the transcriptional regulation is maintained by the pcaQ gene product in the protocatechuate branch. However, the pca genes are organized as previously established for Agrobacterium tumefaciens and Sinorhizobium meliloti except for pcaIJ orthologues [57]. MacLean [57] identified and characterized two similar proteins with low-sequence similarity that functions like β-ketoadipate succinyl-coenzyme A (CoA) transferase in S. meliloti. It is assumed that though these proteins have low-sequence similarity with β-ketoadipate succinyl-CoA transferase, it is played like pcaIJ to accomplish the degradation of β-ketoadipate into succinyl CoA and acetyl CoA (Fig. 6B,C). A plausible explanation of the absence of these genes in protocatechuate branches is due to the divergence of the pathways which was observed in the case of A. tumefaciens and S. meliloti [57] and some soil bacteria [56].
The phenylacetate is a vital intermediate in the catabolism of numerous aromatic compounds including ethylbenzene, styrene, tropate, and phenylethylamine [58]. The phenylacetate-degrading (Paa) pathway is involved in the series degradation of phenylacetate. It is important to note that in E. coli, three different transcriptional units such as PaaZ, PaaABCEDEFGHIJK, and PaaXY were identified [59], which is consistent with the current study. However, C. necator JMP134 had shown 19 different Paa genes [55], which might vary due to far genetic distances of the HSTU-AAM51 strain with C. necator JMP134. The existence of the three gene clusters (as PaaZ, PaaABCEDEFGHIJK, and PaaXY) indicated that HSTU-AAM51 can degrade phenylacetate into TCA cycle ingredients acetyl CoA and succinyl CoA (Fig. 6B). However, in the vicinity of the hpaA gene, the transmembrane facilitator encodes hpaX that is observed which is cotranscribed with hpaA [55]. Importantly, two hpaG genes encode 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase that was located in a similar genetic context. A similar transcriptional unit was identified for E. coli as previously described [55]. The transcriptional unit’s mhpABCDFET and mhpRT are observed, suggesting that the strain HSTU-AAM51 was able to degrade 3-hydroxyl phenyl-propionate and utilized the degraded products as their carbon source [59–61]. From the above analysis, it is therefore assumed that the strain HSTU-AAM51 genome reveals putative genes that encode enzymes leading to ring oxidation and cleavage of lignin and related aromatic compounds via both ortho- and meta-cleavage pathways. Besides, the presence of oxidase, reductase, dehydrogenase, transferase, hydratase, glyoxalase, and glycine betaine/L-proline ABC transporter was reported to be involved in the degradation of lignin compounds [26].
Lignin degradation is mainly accomplished by four different enzymes named laccases, lignin peroxidase, manganese-dependent peroxidase, lignin peroxidases, and versatile peroxidases, which are secreted by white rot fungi [62]. However, these enzymes are not available in bacteria. Therefore, bacteria used different pathways (already mentioned above) for lignin degradation and mineralization. It was reported that laccases cleave the ether linkage (aryl β-O-4) and β-1 bond on lignin using various mediators [26], while Dyp-type peroxidases are the fungal counterpart of peroxidases (LiP or MnP) found in the genome (Supplementary Table S3) of the HSTU-AAM51 strain, which participates in lignin degradation. The Dyp-type peroxidases are involved in the depolymerization, dimer formation, and degradation of aryl ether bonds in lignin and lignin-containing compounds [26, 63, 64]. The Dyp-type peroxidases are equivalent to fungal oxidases that contributed to lignin degradation, which was found in the ligninolytic genetic repertoire of K. variicola P1CD1 [30]. Therefore, the presence of laccase and Dyp-type peroxidases in the HSTU-AAM51 strain genome indicated its capability to degrade lignin through Fenton reactions. Moreover, the presence of tannase indicates the strain’s capability to degrade the covalent bond between lignin and hemicellulose.
The existence of formate dehydrogenase in the HSTU-AAM51 genome indicated the involvement of induction of manganese-peroxidase in the absence of H2O2 (Supplementary Table S5). In addition, glycolate oxidase, oxidase, aldehyde oxidase, and GMC family oxidoreductase were acting as accessory enzymes for lignin degradation [65–68]. The NADH-Quinone oxidoreductases were involved in lignin depolymerization and degradation by Fenton reactions in Pandoraea A514 [26], Phanerochaete chrysosporium [69], and Trametes versicolor [70]. Therefore, the HSTU-AAM51 strain is capable of degrading lignin through Fenton reactions. Moreover, the presence of feruloyl esterase/tannase in the HSTU-AAM51 genome might indicate its capability to degrade the covalent bond between lignin and hemicellulose. It is noteworthy to mention that due to having several lignin degradation pathways, a huge amount of oxidative stress were generated inside the bacterial cells, and to defend against several types of antioxidant and stress response, proteins are identified [26]. In consequence, the presence of huge numbers of oxidoreductase, dismutase, hydroxylase, peroxidase, dehydrogenase, etc., enzymes in the HSTU-AAM51 genome conclude that the strain HSTU-AAM51 is capable of scavenging oxidative stress for lignin degradation [26].
The Klebsiella variicola strain HSTU-AAM51 had shown a range of plant growth–promoting genes. Moreover, the phosphate uptake, inorganic phosphate transporter, and phosphate starvation genes were observed too. These genes were also found in the genome of the sugarcane growth–promoting Enterobacter roggenkampii ED5 strain [27]. Besides, sulfur-metabolizing genes existed in the genome of the HSTU-AAM51 strain, which might conclude its involvement in the oxidation of sulfur and sulfur conjugates metabolites [71]. The oxidation of sulfur affects soil pH, which enhanced N, P, K, Mg, and Zn’s solubility to the spreading of the micronutrients to the host plants [72]. Importantly, the presence of abiotic stress tolerance genes in the K. variicola strain HSTU-AAM51 genome suggested its involvement in tolerance against abiotic stress of the plant. These results indicated that the strain was evolved from some endophytic Klebsiella sp. and later adopted to survive in goat omasum’s hard gastric environment. The ampD, glp, pep, and phn gene products from various bacteria are involved in degrading the organophosphorus insecticides and weed killer [8], and the presence of these genes indicated that the strain is capable of degrading pesticides and herbicides (Supplementary Table S7). These results indicated that the strain HSTU-AAM51 might be involved in degrading lignocellulose in the omasum and detoxifying ingested insecticides to maintain the sound intestine.
Conclusion
The K. variicola HSTU-AAM51 strain is a novel strain, isolated from goat’s omasum. This strain is evidenced by the degradation of lignocellulose by secreting lignocellulose-degrading extracellular enzymes. To the best of our knowledge, this study firstly reports a new Klebsiella variicola strain from goat omasum (intestine), harboring lignocellulose-degrading genetic repertoire. As a consequence, the banana fiber pectin coat and the lignin–hemicellulose network are degraded by its pectinases and tannases enzyme, while the hemicellulose is degraded by its mannases and hemicellulases. Finally, the cellulose network is degraded by the cellulase, beta-glucosidase enzyme, glycosyl hydrolase, and chitinase. The strain K. variicola HSTU-AAM51 is a potential candidate for lignocellulose degradation and bioethanol production upon considering their lignocellulolytic secretion profile, CAZy analysis, and molecular docking results. The nitrogen fixation capacity of the K. variicola HSTU-AAM51 is needed to be checked in the laboratory to utilize it as a biofertilizer for sustainable agriculture.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Md. Azizul Haque would like to express gratitude to the TWAS staff for their cordial and sincere cooperation. The authors are also grateful to Invent Technologies, Banani, Dhaka, Bangladesh, for their support of next-generation sequencing.
Author contribution
Md. Abdullah-Al-Mamun & Md. Shohorab Hossain: experiment, genome analysis, partial writing.
Gautam Chandra Debnath, Md. Ashikujjaman Ashik, Zoherul Hasan: bioinformatics, analysis and data curation.
Sharmin Sultana, Snygdha Rani Das, Shefali Aktar: strain isolation, biochemical analysis.
Aminur Rahman: bioinformatics, data analysis, critical reviewing and editing.
Md. Yeasin Prodhan, Kye Man Cho: resources and proof reading.
Md. Azizul Haque: fund acquisition, conceptualization, genome sequencing, experiments, analysis, manuscript writing, edition, supervision.
Funding
The research was supported by The World Academy of Sciences (TWAS), Trieste, Italy (Research Grants: 17–475 RG/BIO/AS_I, January, 2018-June, 2020).
Declarations
Ethical approval
This research does not contain any human or animal experiments. Therefore, ethical approval was not required.
Consent to publication
The authors agreed to submit the manuscript.
Conflict of interest
The manuscript is not under consideration by another journal as the same time as “Brazilian Journal of Microbiology” Journal. All authors have approved the manuscript for the submission to “Brazilian Journal of Microbiology” Journal.
Footnotes
Responsible Editor: Solange I. Mussatto
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abdullah-Al-Mamun and Shohorab Hossain contributed equally
References
- 1.Chukwuma OB, Rafatullah M, Tajarudin HA, Ismail N. Lignocellulolytic enzymes in biotechnological and industrial processes: a review. Sustainability. 2020;12:7282. doi: 10.3390/su12187282. [DOI] [Google Scholar]
- 2.Haque MA, Ashik MA, Akter MS, Halilu A, Haque MA, Islam MR, Abdullah-Al-Mamun M, Nahar MNN, Das SR, Das KC, Ahmed I, Manir MS, Islam MK, Shahadat MRB. Rapid deconstruction of cotton, coir, areca, and banana fibers recalcitrant structure using a bacterial consortium with enhanced saccharifcation. Waste Biomass Valorization. 2020 doi: 10.1007/s12649-020-01294-w. [DOI] [Google Scholar]
- 3.Kumar R, Kumar P. Future microbial applications for bioenergy production: a perspective. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva JP, Ticona ARP, Hamann PRV, Quirino BF, Noronha EF. Deconstruction of lignin: from enzymes to microorganisms. Molecules. 2021;26:2299. doi: 10.3390/molecules26082299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su Y, Yu X, Sun Y, Wang G, Chen H, Chen G. Evaluation of screened lignin-degrading fungi for the biological pretreatment of corn stover. Sci Rep. 2018;8:5385. doi: 10.1038/s41598-018-23626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dantur KI, Enrique R, Welin B, Castagnaro AP. Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express. 2015;5:15. doi: 10.1186/s13568-015-0101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juturu V, Wu JC. Microbial cellulases: engineering, production and applications. Renew Sust Energ Rev. 2014;33:188–203. doi: 10.1016/j.rser.2014.01.077. [DOI] [Google Scholar]
- 8.Juturu V, Wu JC. Microbial xylanases: engineering, production and industrial applications. Biotechnol Adv. 2012;30:1219–1227. doi: 10.1016/j.biotechadv.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Malgas S, van Dyk JS, Pletschke BI. A review of the enzymatic hydrolysis of mannans and synergistic interactions between β-mannanase, β-mannosidase and α-galactosidase. World J Microbiol Biotechnol. 2015;31:1167–1175. doi: 10.1007/s11274-015-1878-2. [DOI] [PubMed] [Google Scholar]
- 10.de Gonzalo G, Colpa DI, Habib MH, Fraaije MW. Bacterial enzymes involved in lignin degradation. J Biotechnol. 2016;236:110–119. doi: 10.1016/j.jbiotec.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Kamsani N, Salleh MM, Yahya A, Chong CS. Production of lignocellulolytic enzymes by microorganisms isolated from Bulbitermes sp. termite gut in solid-state fermentation. Waste Biomass Valorization. 2016;7:357–371. doi: 10.1007/s12649-015-9453-5. [DOI] [Google Scholar]
- 12.Sharma HK, Xu C, Qin W. Biological pretreatment of lignocellulosic biomass for biofuels and bioproducts: an overview. Waste Biomass Valori. 2019;10:235–251. doi: 10.1007/s12649-017-0059-y. [DOI] [Google Scholar]
- 13.Goh KM, Kahar UM, Chai YY, Chong CS, Chai KP, Ranjani V, Illias RM, Chan KG. Recent discoveries and applications of Anoxybacillus. Appl Microbiol Biotechnol. 2013;97:1475–1488. doi: 10.1007/s00253-012-4663-2. [DOI] [PubMed] [Google Scholar]
- 14.Potprommanee L, Wang XQ, Han YJ, Nyobe D, Peng YP, Huang Q, Liu JY, Liao YL, Chang KL. Characterization of a thermophilic cellulase from Geobacillus sp. HTA426, an efficient cellulase-producer on alkali pretreated of lignocellulosic biomass. PLoS One. 2017;12:e0175004. doi: 10.1371/journal.pone.0175004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seta K, Suzuki T, Kiyoshi K, Shigeno T, Kambe TN. Potential use of methane fermentation digested slurry as a low-cost, environmentally-friendly nutrient for bioethanol production from crude glycerol by Klebsiella variicola TB-83D. New Biotechnol. 2018;44:1–5. doi: 10.1016/j.nbt.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Woo HL, Hazen TC, Simmons BA, DeAngelis KM. Enzyme activities of aerobic lignocellulolytic bacteria isolated from wet tropical forest soils. Syst Appl Microbiol. 2014;37:60–67. doi: 10.1016/j.syapm.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Jiménez DJ, Dini-Andreote F, van Elsas JD. Metataxonomic profiling and prediction of functional behaviour of wheat straw degrading microbial consortia. Biotechnol Biofuels. 2014;7:92. doi: 10.1186/1754-6834-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Islam MA, Karim A, Woon CW, Ethiraj B, Cheng CK, Yousuf A, Khan MMR. Augmentation of air cathode microbial fuel cell performance using wild type Klebsiella variicola. RSC Adv. 2017;7:4798. doi: 10.1039/C6RA24835G. [DOI] [Google Scholar]
- 19.Zadoks RN, Griffiths HM, Munoz MA, Ahlstrom C, Bennett GJ, Thomas E, Schukken YH. Sources of Klebsiella and Raoultella species on dairy farms: be careful where you walk. J Dairy Sci. 2011;94:1045–1051. doi: 10.3168/jds.2010-3603. [DOI] [PubMed] [Google Scholar]
- 20.Haque MA, Cho KM, Barman DN, Kim MK, Yun HD. A potential cellulose microfibril swelling enzyme isolated from Bacillus sp. AY8 enhances cellulose hydrolysis. Process Biochem. 2015;50:807–815. doi: 10.1016/j.procbio.2015.02.003. [DOI] [Google Scholar]
- 21.Haque MA, Lee JH, Cho KM. Endophytic bacterial diversity in Korean kimchi made of Chinese cabbage leaves and their antimicrobial activity against pathogens. Food Control. 2015;56:24–33. doi: 10.1016/j.foodcont.2015.03.006. [DOI] [Google Scholar]
- 22.Kelley DR, Schatz MC, Salzberg SL. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 2010;11(11):R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman A, Nahar N, Nawani NN, Jass J, Ghosh S, Olsson B, Mandal A. Comparative genome analysis of Lysinibacillus B1-CDA, a bacterium that accumulates arsenics. Genomics. 2015;106:384–392. doi: 10.1016/j.ygeno.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar M, Verma S, Gazara RK, Kumar M, Pandey A, Verma PK, Thakur IS. Genomic and proteomic analysis of lignin degrading and polyhydroxyalkanoate accumulating β-proteobacterium Pandoraea sp. ISTKB Biotechnol Biofuels. 2018;11:154. doi: 10.1186/s13068-018-1148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo DJ, Singh RK, Singh P, Li DP, Sharma A, Xing YX, Song XP, Yang LT, Li YR. Complete genome sequences of Enterobacter roggenkampii ED5, a nitrogen fixing plant growth promoting endophytic bacterium with biocontrol and stress tolerance properties, isolated from sugarcane root. Front Microbial. 2020;11:580081. doi: 10.3389/fmicb.2020.580081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 28.Magalães DB, de Carvalho MEA, Bon E, Neto JSA, Kling SH. Colorimetric assay for lignin peroxidase activity determination using methylene blue as substrate. Biotechnol Tech. 1996;10:273–276. doi: 10.1007/BF00184028. [DOI] [Google Scholar]
- 29.dos Santos Melo-Nascimento AO, Mota Moitinho SantÁnna B, Goncalves CC, Santos G, Noronha E, Parachin N, de Aberue Roque MR, Bruce T. Complete genome reveals genetic repertoire and potential metabolic strategies involved in lignin degradation by environmental ligninolytic Klebsiella variicola P1CD1. PLoS ONE. 2020;15:e0243739. doi: 10.1371/journal.pone.0243739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castillo-González AR, Burrola-Barraza ME, Domínguez-Viveros J, Chávez-Martínez A. Rumen microorganisms and fermentation. Arch Med Vet. 2014;46:349–361. doi: 10.4067/S0301-732X2014000300003. [DOI] [Google Scholar]
- 31.Chen YC, Liu T, Yu CH, Chiang TY, Hwang CC. Effects of GC bias in next-generation-sequencing data on De Novo genome assembly. PLoS ONE. 2013;8(4):e62856. doi: 10.1371/journal.pone.0062856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Medina N, Martínez-Romero E, la Cruz MAD, Ares MA, Valdovinos-Torres H, Silva-Sanchez J, Lozano-Aguirre L, Martínez-Barnetche J, Andrade V, Garza-Ramos U. A Klebsiella variicola plasmid confers hypermucoviscosity-like phenotype and alters capsule production and virulence. Front Microbiol. 2020 doi: 10.3389/fmicb.2020.579612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arakawa K, Tomita M. The GC skew index: a measure of genomic compositional asymmetry and the degree of replicational selection. Evol Bioinform Online. 2007;3:159–168. [PMC free article] [PubMed] [Google Scholar]
- 34.Marschall T, Marz M, Abeel T, Dijkstra L, Dutilh BE, Ghaffaari A, et al. Computational pan-genomics: status, promises and challenges. Brief Bioinform. 2018;19:118–135. doi: 10.1093/bib/bbw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam MQ, Oates NC, Thevarajoo S, Tokiman L, Goh KM, McQueen-Mason SJ, Bruce NC, Chong CS. Genomic analysis of a lignocellulose degrading strain from the underexplored genus Meridianimaribacter. Genomics. 2020;112:952–960. doi: 10.1016/j.ygeno.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Huang L, Zheng D, Xu Z, Li Y, Shao S, Zhang Y, Ge X, Lu F. Biochemical characterization of a novel GH43 family β-xylosidase from Bacillus pumilus. Food Chem. 2019;295:653–661. doi: 10.1016/j.foodchem.2019.05.163. [DOI] [PubMed] [Google Scholar]
- 37.Dar MA, Shaikh AA, Pawar KD, Pandit RS. Exploring the gut of Helicoverpa armigera for cellulose degrading bacteria and evaluation of a potential strain for lignocellulosic biomass deconstruction. Process Biochem. 2018;73:142–153. doi: 10.1016/j.procbio.2018.08.001. [DOI] [Google Scholar]
- 38.Botha J, Mizrachi E, Myburg AA, Cowan DA. Carbohydrate active enzyme domains from extreme thermophiles: components of a modular toolbox for lignocellulose degradation. Extremophiles. 2018;22:1–12. doi: 10.1007/s00792-017-0974-7. [DOI] [PubMed] [Google Scholar]
- 39.Pereira ALS, Nascimento DMD, Souza MDSM, Cassales AR, Morais JPS, Paula RCMD, Rosa MDF, Feitosa JPA (2014) Banana (Musa sp. Cv. Pacovan) pseudostem fibers are composed of varying lignocellulosic composition throughout the diameter. BioResources 9: 7749–7763. 10.15376/biores.9.4.7749-7763
- 40.Selvam K, Senbagam D, Selvankumar T, Sudhakar C, Kamala-Kannan S, Senthilkumar B, Govarthanan M. Cellulase enzyme: homology modeling, binding site identification and molecular docking. J Mol Struct. 2017;1150:61–67. doi: 10.1016/j.molstruc.2017.08.067. [DOI] [Google Scholar]
- 41.Dadheech T, Jakhesara S, Chauhan PS, Pandit R, Hinsu A, Kunjadiya A, Rank D, Joshi C. Draft genome analysis of lignocellulolytic enzymes producing Aspergillus terreus with structural insight of β-glucosidases through molecular docking approach. Biomac. 2018 doi: 10.1016/j.ijbiomac.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 42.Shah MP, Reddy GV, Banerjee R, Babu PR, Kothari IL. Microbial degradation of banana waste under solid state bioprocessing using two lignocellulolytic fungi (Phylosticta spp. MPS-001 and Aspergillus spp. MPS-002) Process Biochem. 2005;40(1):445–451. doi: 10.1016/j.procbio.2004.01.020. [DOI] [Google Scholar]
- 43.Suzuki K, Hori A, Kawamoto K, Thangudu RR, Ishida T, Igarashi K, Samejima M, Yamada C, Arakawa T, Wakagi T, Koseki T, Fushinobu S. Crystal structure of a feruloyl esterase belonging to the tannase family: a disulfide bond near a catalytic triad. Proteins. 2014;82:2857–2867. doi: 10.1002/prot.24649. [DOI] [PubMed] [Google Scholar]
- 44.Lee HY, Cho DY, Ahmad I, Patel HM, Kim MJ, Jung JG, Jeong EH, Haque MA, Cho KM. Mining of a novel esterase (est3S) gene from a cow rumen metagenomic library with organosphosphorus insecticides degrading capability: catalytic insights by site directed mutations, docking, and molecular dynamic simulations. Int J Biol Macromol. 2021;190:441–455. doi: 10.1016/j.ijbiomac.2021.08.224. [DOI] [PubMed] [Google Scholar]
- 45.Seto M, Kimbara K, Shimura M, Hatta T, Fukuda M, Yano K. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl Environ Microbiol. 1995;61:3353–3358. doi: 10.1128/AEM.61.9.3353-3358.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis JR, Goodwin L, Teshima H, Detter C, Tapia R, Han C, Huntemann M, Wei CL, Han J, Chen A, Kyrpides N, Mavrommatis K, Szeto E, Markowitz V, Ivanova N, Mikhailova N, Ovchinnikova G, Pagani I, Pati A, Woyke T, Pitluck S, Peters L, Nolan M, Land M, Sello JK. Genome sequence of Streptomyces viridosporus strain T7A ATCC39115, a lignin-degrading actinomycete. Genome Announc. 2013;1(4):e00416–e513. doi: 10.1128/genomeA.00416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meux E, Prosper P, Masai E, Mulliert G, Dumarçay S, Morel M, Didierjean C, Gelhaye E, Favier F. Sphingobium sp. SYK-6 LigG involved in lignin degradation is structurally and biochemically related to the glutathione transferase omega class. FEBS Lett. 2012;586:3944–3950. doi: 10.1016/j.febslet.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 48.Janusz G, Pawlik A, Sulej J, Świderska-Burek U, Jarosz-Wilkołazka A, Paszczyński A. Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol Rev. 2017;6:941–962. doi: 10.1093/femsre/fux049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bugg TDH, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol. 2011;22:394–400. doi: 10.1016/j.copbio.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 50.Couto SR, Herrera JLT. Laccase production at reactor scale by filamentous fungi. Biotechnol Adv. 2007;25:558–569. doi: 10.1016/j.biotechadv.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Galhaup C, Wagner H, Hinterstoisser B, Haltrich D. Increased production of laccase by the wood-degrading basidiomycete Trametes pubescens. Enzyme Microb Technol. 2002;30:529–536. doi: 10.1016/S0141-0229(01)00522-1. [DOI] [Google Scholar]
- 52.Koroleva OV, Gavrilova VP, Stepanova EV, Lebedeva VI, Sverdlova NI, Landesman EO, Yavmetdinov IS, Yaropolov AI. Production of lignin modifying enzymes by cocultivated white-rot fungi Cerena maxima and Coriolus hirsutus and characterization of laccase from Cerrena maxima. Enzyme Microb Technol. 2002;30:573–580. doi: 10.1016/S0141-0229(02)00021-2. [DOI] [Google Scholar]
- 53.Andlar M, Rezi T, Marđetko N, Kracher D, Ludwig R, Santek B. Lignocellulose degradation: an overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng Life Sci. 2018;18:768–778. doi: 10.1002/elsc.201800039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Pantoja D, la Iglesia RD, H. Pieper D, Gonzalez B, Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cuprividus necator JMP1134. FEMS Microbiol Rev. 2008;32:736–794. doi: 10.1111/j.1574-6976.2008.00122.x. [DOI] [PubMed] [Google Scholar]
- 55.Harwood CS, Parales RE. The beta-keto adipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 56.MacLean AM, MacPherson G, Aneja P, M. Finan TM, Characterization of the β-Ketoadipate pathway in Sinorhizobium meliloti. Appl Environ Microbiol. 2006;72:5403–5413. doi: 10.1128/AEM.00580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luengo JM, García JL, Olivera ER. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol Microbiol. 2001;39:1434–1442. doi: 10.1046/j.1365-2958.2001.02344.x. [DOI] [PubMed] [Google Scholar]
- 58.Ferrandez A, Minambres B, Garcia B, Olivera ER, Luengo JM, Garcia JL, Diaz E. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J Biol Chem. 1998;273:25974–25986. doi: 10.1074/jbc.273.40.25974. [DOI] [PubMed] [Google Scholar]
- 59.Ferrandez A, Prieto MA, Garcia GL, Diaz E. Molecular characterization of PadA, a phenylcetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 1997;406:23–27. doi: 10.1016/s0014-5793(97)00228-7. [DOI] [PubMed] [Google Scholar]
- 60.Manso I, Torres B, Andreu JM, Menéndez M, Rivas G, Alfonso C, Díaz E, García JL, Galán, B. (2009). 3-Hydroxyphenylpropionate and phenylpropionate are synergistic activators of the MhpR transcriptional regulator from Escherichia coli. J Biol Chem 284(32), 21218–21228. 10. 1074/jbc.M109.008243 [DOI] [PMC free article] [PubMed]
- 61.Kracher D, Ludwig R. cellobiose dehydrogenase: an essential enzyme for lignocellulose degradation in nature-a review. J Land Manag Food Enviro. 2016;67:145–163. doi: 10.1515/boku-2016-0013. [DOI] [Google Scholar]
- 62.Rahmanpour R, Rea D, Jamshidi S, Fülöp V, Bugg TD. Structure of Thermobifda fusca DyP-type peroxidase and activity towards kraft lignin and lignin model compounds. Arch Biochem Biophys. 2016;594:54–60. doi: 10.1016/j.abb.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 63.Singh R, Grigg JC, Qin W, Kadla JF, Murphy ME, Eltis LD. Improved manganese-oxidizing activity of DypB, a peroxidase from a lignolytic bacterium. ACS Chem Biol. 2013;8:700–706. doi: 10.1021/cb300608x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma J, Zhang K, Liao H, Hector SB, Shi X, Li J, Liu B, Xu T, Tong C, Liu X. Genomic and secretomic insight into lignocellulolytic system of an endophytic bacterium Pantoea ananatis Sd-1. Biotechnol Biofuels. 2016;9(1):25. doi: 10.1186/s13068-016-0439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garrido-Pertierra A, Cooper RA. Identifcation and purification of distinct isomerase and decarboxylase enzymes involved in the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli. FEBS. 1981;J117:581–584. doi: 10.1111/j.1432-1033.1981.tb06377.x. [DOI] [PubMed] [Google Scholar]
- 66.Rather LJ, Knapp B, Haehnel W, Fuchs G. Coenzyme A-dependent aerobic metabolism of benzoate via epoxide formation. J Biol Chem. 2010;285:20615–20624. doi: 10.1074/jbc.M110.124156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc Natl Acad Sci. 2010;107(32):14390. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Constam D, Muheim A, Zimmermann W, Fiechter A. Purifcation and partial characterization of an intracellular NADH: quinone oxidoreductase from Phanerochaete chrysosporium. Microbiology. 1991;137:2209–2214. doi: 10.1099/00221287-137-9-2209. [DOI] [Google Scholar]
- 69.Lee S, Moon D, Choi HT, Song H. Purifcation and characterization of an intracellular NADH: quinone reductase from Trametes versicolor. J Microbiol. 2007;45(4):333. [PubMed] [Google Scholar]
- 70.Kwak MJ, Jeong H, Madhaiyan M, Lee Y, Sa TM, Oh TK, Kim JF. Genome information of Methylobacterium oryzae, a plant-probiotic methylotroph in the phyllosphere. PLoS ONE. 2014;9:e106704. doi: 10.1371/journal.pone.0106704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vidyalakshmi R, Paranthaman R, Bhakyaraj R. Sulfur oxidizing bacteria and pulse nutrition, a review world. J Agric Sci. 2009;5:270–278. [Google Scholar]
- 72.Singh BK, Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiol Rev. 2006;30:428–471. doi: 10.1111/j.1574-6976.2006.00018.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.