Abstract
Crude aqueous extracts from Arabidopsis leaves were subjected to chromatographic separations, after which the different fractions were monitored for antimicrobial activity using the fungus Neurospora crassa as a test organism. Two major fractions were obtained that appeared to have the same abundance in leaves from untreated plants versus leaves from plants challenge inoculated with the fungus Alternaria brassicicola. One of both major antimicrobial fractions was purified to homogeneity and identified by 1H nuclear magnetic resonance, gas chromatography/electron impact mass spectrometry, and gas chromatography/chemical ionization mass spectrometry as 4-methylsulphinylbutyl isothiocyanate (ITC). This compound has previously been described as a product of myrosinase-mediated breakdown of glucoraphanin, the predominant glucosinolate in Arabidopsis leaves. 4-Methylsulphinylbutyl ITC was found to be inhibitory to a wide range of fungi and bacteria, producing 50% growth inhibition in vitro at concentrations of 28 μm for the most sensitive organism tested (Pseudomonas syringae). A previously identified glucosinolate biosynthesis mutant, gsm1-1, was found to be largely deficient in either of the two major antimicrobial compounds, including 4-methylsulphinylbutyl ITC. The resistance of gsm1-1 was compared with that of wild-type plants after challenge with the fungi A. brassicicola, Plectosphaerella cucumerina, Botrytis cinerea, Fusarium oxysporum, or Peronospora parasitica, or the bacteria Erwinia carotovora or P. syringae. Of the tested pathogens, only F. oxysporum was found to be significantly more aggressive on gsm1-1 than on wild-type plants. Taken together, our data suggest that glucosinolate-derived antimicrobial ITCs can play a role in the protection of Arabidopsis against particular pathogens.
Living plants are under constant assault by microbial pathogens trying to gain access to the nutrients sequestered in plant tissues. To deter such potential pathogens, plants have evolved a complex set of defense mechanisms, of which some are preformed and others inducible. The former mechanisms are in place irrespective of whether or not the plant tissue is challenged by microorganisms, whereas the latter are activated in response to microbial attack.
Arabidopsis has emerged since the early 1990s as the leading model for the study of plant defense responses at the molecular level (Buell, 1998). In this plant three different genetic programs have been identified that are activated upon pathogen recognition leading to the production of inducible antimicrobial compounds. A first program controls the synthesis in cells surrounding the infection site of an antimicrobial sulfur-containing indole derivate, the phytoalexin camalexin (Tsuji et al., 1992). A mutation in the gene PAD3, likely to encode a camalexin biosynthesis enzyme, abolishes camalexin production following pathogen attack (Glazebrook and Ausubel, 1994; Zhou et al., 1999a) and causes enhanced susceptibility to the fungal pathogen Alternaria brassicicola (Thomma et al., 1999b), but not to the bacterial pathogen Pseudomonas syringae (Glazebrook and Ausubel, 1994). A second program leads to systemic activation of genes encoding antimicrobial proteins such as PR-1, PR-2 (a β-1,3-glucanase), and PR-5 (a thaumatin-like protein). Activation of this program requires amplification of the initial pathogen recognition event through an endogenous signaling circuit based on the production of salicylic acid. It has been shown that Arabidopsis plants with defects in either production or perception of salicylic acid are more susceptible than wild-type plants to the pathogens P. syringae and Peronospora parasitica (Cao et al., 1994; Delaney et al., 1994). A third program controls systemic activationof a set of genes encoding antimicrobial proteins such as PDF1.2 (a plant defensin), PR-3 (a chitinase), and PR-4 (a hevein-like protein). Full activation of this program depends on a signal amplification circuit involving production of the plant hormones ethylene and jasmonic acid (Penninckx et al., 1998). Arabidopsis mutants with defects in either ethylene or jasmonic acid perception show reduced resistance to the fungal pathogen Botrytis cinerea but not to P. parasitica (Thomma et al., 1998; Thomma et al., 1999a). Taken together, these studies reveal that Arabidopsis uses several pathogen-inducible defense programs, each contributing to resistance against particular pathogens.
In contrast to the wealth of information accumulated over the past 10 years on pathogen-inducible defense systems in Arabidopsis, very little is known in this plant about preformed antimicrobial compounds, generally called phytoanticipins (for review, see Osbourn, 1996). The only known potential source of constitutive antimicrobial components from Arabidopsis is a group of sulfur-containing glucosides termed glucosinolates (Hogge et al., 1988). Upon tissue damage, glucosinolates are converted by an endogenous thioglucosidase into breakdown products, some of which are known to inhibit microorganisms in vitro (Mithen et al., 1986; Kirkegaard et al., 1996; Manici et al., 1997). Nevertheless, except for a study on the relationship between glucosinolates and the development of clubroot disease in Arabidopsis (Ludwig-Müller et al., 1999), very little attention has been drawn to the possible role of glucosinolates in the resistance of this model plant against microbial pathogens. In this study we show that the glucosinolate breakdown product 4-methylsulphinylbutyl isothiocyanate (ITC) is one of the two main water-soluble phytoanticipins in Arabidopsis leaves. In addition, we demonstrate that a previously identified Arabidopsis mutant, gsm1-1, which lacks or has reduced amounts of many aliphatic glucosinolates found in its wild-type parental ecotype (Col-0; Haughn et al., 1991), is deficient in this component. To test the possible role of 4-methylsulphinylbutyl ITC as a preformed defense component, it was of interest to compare the in planta susceptibility of mutant gsm1-1 and wild-type Col-0 plants to different fungi and bacteria.
RESULTS
Separation of Preformed Antimicrobial Compounds from Arabidopsis Leaves
Four different solvents (water, 70% [v/v] methanol, 100% [v/v] ethanol, and 100% [v/v] acetone) were used to prepare extracts of leaves from untreated Arabidopsis plants. Following evaporation of the solvents, extract residues were tested for the presence of antimicrobial compounds. Highest levels of antimicrobial activity were found in the water extracts (Table I).
Table I.
In vitro antimicrobial activity (units mL−1) of four crude Arabidopsis leaf extracts (1 g fresh wt mL−1 extraction solvent) against the bacteria Escherichia coli and P. syringae and the fungi A. brassicicola, Fusarium culmorum, and B. cinerea
| Extraction Solvent | E. coli | P. syringae | A. brassicicola | F. culmorum | B. cinerea |
|---|---|---|---|---|---|
| Water | 3.5 | 8.3 | <0.9 | 7 | <0.9 |
| 70% (v/v) Methanol | 0.9 | 3.1 | 0.9 | 1.5 | <0.9 |
| Ethanol | <0.9 | <3.6 | <0.9 | 1.8 | <0.9 |
| Acetone | <0.9 | <4.2 | <0.9 | <0.9 | <0.9 |
The activity was calculated as described in “Materials and Methods.”
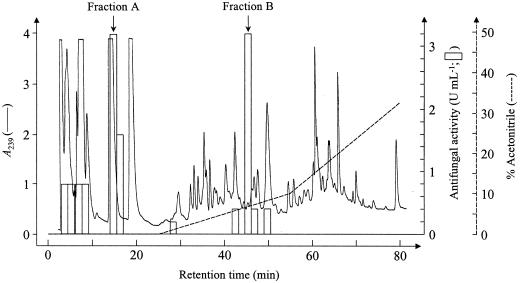
A crude aqueous extract from untreated Arabidopsis leaves was passed over several chromatographic matrices including an anion exchanger (Q-Sepharose), a cation exchanger (S-Sepharose), and a hydrophobic matrix (C8-silica). The major part of the antimicrobial activity, as monitored using an assay for growth inhibition of the fungus Neurospora crassa, was recovered in the unbound fraction of either of both ion-exchange columns but bound partially to a C8-silica column (Table II). Based on this information, a separation scheme was devised consisting of passage of the crude extract over a Q-Sepharose column equilibrated at pH 9 and passage of the unbound fraction over a C18-silica HPLC column. This HPLC column was eluted for 15 min with 0.1% (v/v) trifluoroacetic acid (TFA) followed by a biphasic linear gradient of acetonitrile in 0.1% (v/v) TFA. The first part of the gradient ranged from 0% to 10% (v/v) acetonitrile in 30 min and the second part from 10% to 45% (v/v) acetonitrile in 35 min. When the unbound Q-Sepharose fraction was separated following this elution scheme, the antimicrobial activity was mainly recovered in two fractions. Fraction A flowed through the column and eluted at 0.1% (v/v) TFA with slight retardation, whereas fraction B eluted at 8% (v/v) acetonitrile and 0.1% (v/v) TFA (Fig. 1).
Table II.
Percentage of recovered antimicrobial activity present in the crude aqueous extract from Arabidopsis leaves after passing the extract over different chromatographic matrices as described in “Materials and Methods”
| Chromatographic Matrix | Recovery of Antimicrobial Activity in Unbound Fraction |
|---|---|
| Q-Sepharose (pH 9) | >90% |
| S-Sepharose (pH 5) | >90% |
| C8-silica | <10% |
Figure 1.
HPLC profile obtained after loading Arabidopsis leaf extract from 4-g wild-type (Col-0) plants on a C18 reversed phase (RP)-HPLC column equilibrated in 0.1% (v/v) TFA. The column was eluted at a flow rate of 1 mL min−1, 15 min with 0.1% (w/v) TFA, followed by a linear gradient of acetonitrile in 0.1% (v/v) TFA from 0% to 10% (v/v) acetonitrile in 30 min and from 10% to 45% (v/v) acetonitrile in the following 35 min. The eluate was monitored by online measurement of the A239 (A239; ——) and the acetonitrile gradient (−−−−−−) was monitored with an online conductivity sensor. Fractions (1.5 mL) were evaporated and dissolved in 60 μL of distilled water, of which 20 μL was assayed for antifungal activity (indicated as bars) against N. crassa as described in “Materials and Methods.” The indicated fractions A and B were used for further purification.
Similar relative antimicrobial activities in fractions A and B were obtained when extracts from untreated leaves and leaves infected with the fungus A. brassicicola were processed as described above (Table III).
Table III.
Antimicrobial activity (units mL−1) in noninduced, A. brassicicola-infected, young and old rosette leaves from Arabidopsis
| Biological Material | Antimicrobial Activity (U)
|
|
|---|---|---|
| Fraction A | Fraction B | |
| Total leaves noninfected plants | 1.20 | 0.90 |
| Total leaves A. brassicicola-infected plants | 0.95 | 0.90 |
| Young noninfected rosette leaves | N.D.a | 1.45 |
| Old noninfected rosette leaves | N.D. | 0.07 |
For each condition, 4 g of leaves was extracted and separated by HPLC as described in the legend of Figure 1. Fraction A was collected as the fraction eluting between 12 and 18 min upon loading, and Fraction B was collected as the fraction eluting between 7% and 9% (v/v) acetonitrile. Antimicrobial activity was tested against N. crassa as described in “Materials and Methods.”
N.D., Not determined.
Very distinct chromatograms were obtained when extracts were prepared from young rosette leaves (seventh through 10th leaves of 4-week-old plants) and old rosette leaves (third through sixth leaves of 4-week-old plants). The antimicrobial compound B was at least 20-fold more abundant in young than in old leaves (Table III).
Characterization of the Antimicrobial Compounds
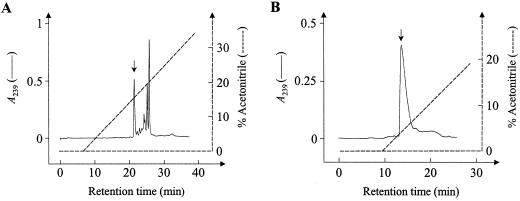
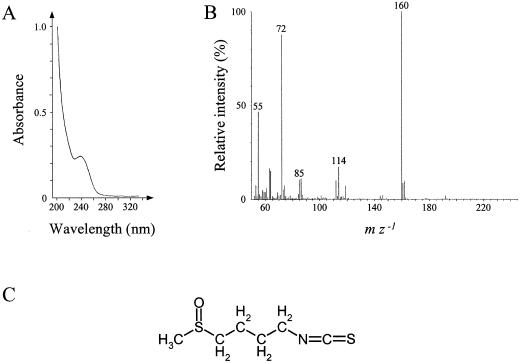
The antimicrobial compound in fraction B was further purified in two further chromatographic steps. First, fraction B was passed a second time over the C18-silica HPLC column where all of the antimicrobial activity was found in a single peak (Fig. 2A). This active fraction was purified to homogeneity on a phenyl-silica HPLC column resulting in a well-resolved peak that eluted at 2% (v/v) acetonitrile (Fig. 2B). The UV-absorption spectrum of this compound shows a characteristic peak at 239 nm (Fig. 3A). The molecular mass of the component was found to be 177 D as detected by GC/chemical ionization MS indicating an [M+H]+ ion at m/z 178. This is supported by the obtained EI mass spectrum showing key ions at m/z 160 (C6H10NS2, as determined by high-resolution MS), 114, 85, 72, and 55 (Fig. 3B) that correspond to the previously identified spectrum of 4-methylsulphinylbutyl ITC (Spencer and Daxenbichler, 1980). 1H NMR and 2D NMR measurements (gradient heteronuclear multiple bond correlation spectroscopy, gradient single quantum coherence spectroscopy, and gradient correlation spectrosopy; results not shown) also confirmed this structure.
Figure 2.
A, HPLC profile obtained after loading the combined antifungal fractions of Fraction B from Figure 1 on a C18 RP-HPLC column equilibrated in 0.1% (v/v) TFA. The column was eluted for 40 min with a linear gradient of acetonitrile in 0.1% TFA (v/v) from 0% to 40% (v/v) acetonitrile in 40 min at a flow rate of 1 mL min−1. B, HPLC profile obtained after loading the indicated (arrow) peak fraction from Figure 2A, containing all antimicrobial activity, on a phenyl RP-HPLC column equilibrated in 0.1% (v/v) TFA. The column was eluted for 30 min with a linear gradient of acetonitrile in 0.1% (v/v) TFA from 0% to 30% (v/v) acetonitrile in 30 min at a flow rate of 1 mL min−1. The indicated peak fraction (arrow) contained all antimicrobial activity. For both profiles, the eluate was monitored by online measurement of the A239 (A239; ——) and at the same time the acetonitrile gradient (−−−−−−) was monitored with an online conductivity sensor. Fractions (1.5 mL) were evaporated and dissolved in 60 μL distilled water, of which 20 μL was assayed for antifungal activity against N. crassa as described in “Materials and Methods.”
Figure 3.
A, Light absorption spectrum of 4- methylsulphinylbutyl isothiocyanate. B, Mass spectrum of 4-methylsulphinylbutyl isothiocyanate obtained by gas chromatography (GC)/electron impact (EI) mass spectrometry (MS). C, Chemical structure of 4-methylsulphinylbutyl isothiocyanate.
The antimicrobial activity of this compound was assessed against nine different fungi and four bacteria (Table IV). 4-Methylsulphinylbutyl ITC was found to have broad spectrum antimicrobial activity: It was active at concentrations below 1.13 mm against seven out of nine fungi tested as well as against all of four bacteria tested. The most sensitive microorganism was P. syringae, which displayed an inhibitory concentration (IC50) value of 28 μm.
Table IV.
Concentrations of 4-methylsulphinylbutyl isothiocyanate from Arabidopsis leaves required for 50% growth inhibition in vitro (IC50 value) on different microorganisms
| Microorganism | IC50 value |
|---|---|
| μm | |
| A. brassicicola | >1,130 |
| B. cinerea | >1,130 |
| F. culmorum | 124 |
| Fusarium oxysporum | 325 |
| N. crassa | 271 |
| Nectria hematococca | 260 |
| Plectosphaerella cucumerina | 147 |
| Penicillium expansum | 294 |
| Verticillium dahliae | 215 |
| E. coli | 282 |
| P. syringae pv tomato | 28 |
| Sarcina lutea | 294 |
| Xanthomonas campestris | 136 |
The growth inhibition assays were performed as described in “Materials and Methods.”
Antimicrobial Compounds in Leaves of the Arabidopsis Mutant gsm1-1
4-Methylsulphinylbutyl ITC (also known as sulforaphane) has previously been described as an enzymatic breakdown product of 4-methylsulphinylbutyl glucosinolate (also known as glucoraphanin; Benn, 1977; for review, see Fenwick et al., 1983) by the plant's thioglucosidase, an enzyme commonly referred to as myrosinase (for review, see Rask et al., 2000). In Arabidopsis leaves, up to 45% of the total amount of glucosinolates is represented by glucoraphanin: Therefore, it is by far the most abundant member of this class of secondary metabolites (Haughn et al., 1991). An Arabidopsis mutant with reduced aliphatic glucosinolate content, called gsm1-1, has been previously identified in which the presence of 4-methylsulphinylbutyl glucosinolate is reduced about 60-fold (Haughn et al., 1991). The GSM1 gene has so far not been cloned, but is believed to encode an enzyme involved in the biosynthesis of glucosinolates with aliphatic side chains containing four, five, or six carbon atoms (Haughn et al., 1991).
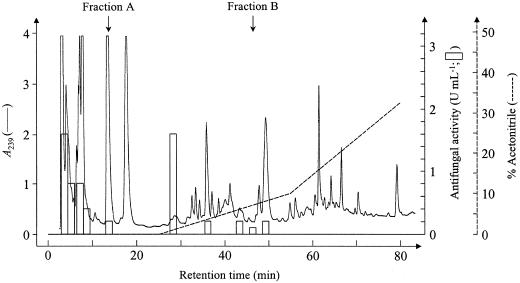
An aqueous extract was prepared from leaves of the gsm1-1 mutant and subjected to the purification scheme as described above for the crude extract from wild-type Arabidopsis leaves. The antimicrobial activity corresponding to fractions A and B (4-methylsulphinylbutyl ITC) was reduced 24- and 36-fold, respectively, in the extract of gsm1-1 mutants (Fig. 4) relative to an extract from wild-type (Col-0) plants (Fig. 1). It has to be noted that the fractions eluting around 2% (v/v) acetonitrile in the chromatograms had a higher antifungal activity for the gsm1-1 extract compared with the Col-0 extract (Figs. 4 and 1, respectively).
Figure 4.
HPLC profile obtained after loading Arabidopsis leaf extract from 4 g noninduced gsm1-1 leaf material on a C18 RP-HPLC column equilibrated in 0.1% (v/v) TFA. Elution of the column, fractionation, and determination of the antifungal activity as in Figure 1.
Disease Susceptibility of Mutant gsm1-1
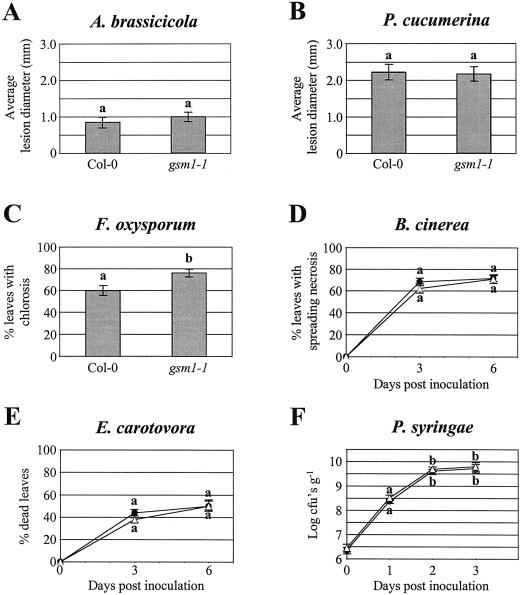
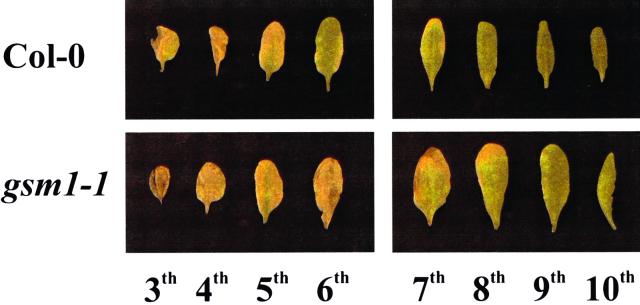
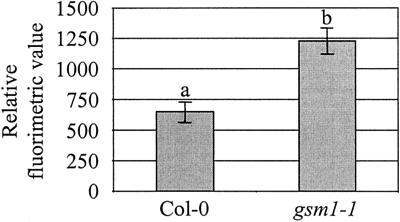
The above-described results indicate that: (a) The main water-soluble antimicrobial compounds in Arabidopsis leaves are glucosinolate breakdown products, including an ITC; and (b) the mutant gsm1-1 is deficient in these components. To test whether these antimicrobial glucosinolate breakdown products play a role as preformed defense compounds, we have compared the susceptibility of gsm1-1 and its wild-type parental ecotype (Col-0) with various fungal and bacterial leaf pathogens. The pathogens used were the necrotrophic bacterium Erwinia carotovora subsp. carotovora, the virulent biotrophic bacterium P. syringae pv tomato DC3000, the necrotrophic fungi A. brassicicola, B. cinerea, P. cucumerina, and F. oxysporum f. sp. matthiolae, and the biotrophic fungus P. parasitica. Disease ratings of gsm1-1 mutants did not significantly differ from those of wild-type Col-0 plants for A. brassicicola, P. cucumerina, B. cinerea, E. carotovora, or P. syringae (Fig. 5, A, B, and D–F). Glucosinolate-deficient gsm1-1 mutants and Col-0 plants were also equally resistant to P. parasitica strain Wela. Unlike what is seen after infection of susceptible Arabidopsis genotypes such as Weiningen, neither sporulation nor mycelium growth could be detected in Col-0 nor gsm1-1 leaves inoculated with this pathogen (data not shown). Therefore, the deficiency of gsm1-1 plants in 4-methylsulphinylbutyl ITC appears not to be important for resistance to these pathogens. In contrast, 8 d following inoculation, infection of gsm1-1 plants with the fungal pathogen F. oxysporum resulted in significantly more leaves showing chlorotic symptoms relative to Col-0 plants (76.3% and 60%, respectively; Fig. 5C). This test was repeated three times and each time data were significantly different at 95% confidence (results not shown). Mock-inoculated Col-0 or gsm1-1 plants showed no chlorotic symptoms and no visible differences were observed between both genotypes up to 8 d after mock inoculation (results not shown). F. oxysporum caused symptoms preferentially on the older leaves in both gsm1-1 and Col-0 plants, but relatively more of the younger leaves showed chlorosis in gsm1-1 compared with Col-0 (Fig. 6). Microscopic analysis of F. oxysporum-infected gsm1-1 or Col-0 leaves stained with lactophenol trypan blue revealed no obvious phenotypic differences in the growth of the fungus in infected mutant or wild-type plants (results not shown). The growth of F. oxysporum in infected plants was measured by quantification of fungal biomass using quantitative PCR. Reproducible results could not be obtained when older chlorotic leaves were assayed, probably due to inhibitory substances released from macerated tissue. Therefore, sampling was restricted to the younger leaves (seventh through 10th leaves) of the infected plants. As shown in Figure 7, the biomass of F. oxysporum at 8 d after inoculation was 1.9 times higher in gsm1-1 versus Col-0 plants. This experiment was repeated with similar results (not shown). During similar quantifications of mock-inoculated Col-0 and gsm1-1 plants, no F. oxysporum biomass could be detected by quantitative PCR at 8 d after inoculation (results not shown).
Figure 5.
Comparative disease rating of glucosinolate-deficient gsm1-1 mutants and wild-type Col-0 plants inoculated with different pathogens. Throughout the different graphs, different letter labels indicate that the corresponding data are significantly different (P > 0.95) according to Tukey's studentized range test (Neter et al., 1996). A, Average diameter of necrotic lesions formed on leaves of 4-week-old Arabidopsis plants 6 d after drop inoculation with a spore suspension of A. brassicicola. Bars represent averages ± se of measurements from 40 lesions on five different plants. B, Average diameter of necrotic lesions formed on leaves of 4-week-old Arabidopsis plants 6 d after drop inoculation with a spore suspension of P. cucumerina. Bars represent averages ± se of measurements from 40 lesions on five different plants. C, Percentage from a total of eight fully expanded and inoculated leaves per plant showing chlorotic symptoms 8 d after inoculation with a spore suspension of F. oxysporum f. sp. matthiolae. Bars represent averages ± se from 20 4-week-old Arabidopsis plants. Analysis was done on leaves three through 10, with leaf numbering starting at one for the first true leaf and reflecting the order of appearance after germination. D, Percentage of inoculated leaves showing spreading necrosis symptoms 3 and 6 d after inoculation of Col-0 (●) and gsm1-1 (▵) plants with a spore suspension of B. cinerea. Data represent averages ± se of inoculations on all expanded leaves of 20 4-week-old Arabidopsis plants per genotype. E, Percentage of dead inoculated leaves 3 and 6 d after inoculation of Col-0 (●) and gsm1-1 (▵) plants with a bacterial suspension of E. carotovora. Data points represent averages ± se of inoculations on five expanded leaves of 20 4-week-old Arabidopsis plants. F, Growth of P. syringae pv tomato DC3000 in Col-0 (●) and gsm1-1 (▵) plants dip inoculated with a bacterial suspension of 107 colony forming units (cfu) mL−1. Data points represent averages ± se of log-transformed data from three experiments of the average number cfu per gram fresh leaf material.
Figure 6.
Chlorosis symptoms of 4-week-old Arabidopsis Col-0 and gsm1-1 plants 8 d following inoculation on the leaves with F. oxysporum spores. Leaves are arranged from left to right in order of decreasing age. For each genotype, a series of eight leaves (third through 10th, in accordance with the scored leaves for the quantification of chlorosis symptoms as represented in Fig. 5C) from a representative plant is shown.
Figure 7.
Quantification of F. oxysporum biomass in pooled (seventh through 10th) Col-0 and gsm1-1 infected leaves. Bars represent average relative fluorimetric values ± se of six samples, taken as described in “Materials and Methods,” 8 d following spray inoculation. Relative fluorimetric values were obtained by quantitative PCR, as described in “Materials and Methods.” Values are based on the quantification of a standard dilution series of DNA extracted from in vitro-grown F. oxysporum. Different letter labels indicate that the corresponding data are significantly different (P > 0.95) according to Tukey's studentized range test (Neter et al., 1996). This test was repeated with similar results.
DISCUSSION
Crude aqueous extracts of noninfected Arabidopsis leaves contain two antimicrobial components, called A and B. Component B was identified as the glucosinolate breakdown product 4-methylsulphinylbutyl ITC by means of MS and NMR. The identity of this component was further corroborated by its strongly depressed level in leaf extract prepared from gsm1-1, an Arabidopsis mutant known to be largely deficient in aliphatic glucosinolates having a core butyl, pentyl, or hexyl group (Haughn et al., 1991). It was also observed that gsm1-1 leaf extracts contained a fraction in which the antifungal activity was about 6-fold higher compared with wild-type leaf extracts. This fraction might represent a breakdown product of a precursor of 4-methylsulphinylbutyl glucosinolate. Haughn et al. (1991) have previously reported that gsm1-1 accumulates about 3-fold more 3-methylsulphinylpropyl glucosinolate than wild-type plants. We did not succeed in purifying component A to homogeneity, precluding its characterization. The fact that component A was also almost completely absent in gsm1-1 leaf extracts suggests that this compound is also a breakdown product of an aliphatic glucosinolate with a core butyl, pentyl, or hexyl group. In aqueous extracts from Arabidopsis leaves, components A and B were the predominant antimicrobial factors, at least when antimicrobial activity was assessed against N. crassa. The amount of 4-methylsulphinylbutyl ITC that could be recovered from leaf extracts of Arabidopsis wild-type plants is about 65 μg g−1 fresh weight.
Production of glucosinolate breakdown products initially requires the hydrolytic action on glucosinolates by thioglucosidase enzymes, also known by the trivial name myrosinases (EC 3.2.3.1). In Brassicaceae, myrosinases are present as a family of isoenzymes. They are stored predominantly inside myrosin cells, protein-accumulating cells with deviant morphology that are scattered throughout root, stem, leaf, and seed tissues (Rask et al., 2000). Myrosinases are believed to be physically separated from their substrates, the glucosinolates, which accumulate in vacuoles of non-myrosin cells (Rask et al., 2000). Hence, glucosinolate breakdown products can only be generated after disruption of cellular compartments. This can occur during extract preparation, but also under “natural” conditions such as wounding, chewing, or maceration by herbivores or microbial pathogens.
Several authors have shown that glucosinolate breakdown products exhibit antimicrobial activity in vitro, whereas glucosinolates themselves are nontoxic (Mithen et al., 1986; Manici et al., 1997). In a comparison of the relative antifungal activity of several ITC breakdown products of different glucosinolates, it was found that aromatic ITCs are more toxic than aliphatic ITCs and that the fungal toxicity of aliphatic ITCs decreases with increasing length of the side chain (Mithen et al., 1986; Peterka and Schlosser 1989; Manici et al., 1997; Sarwar et al., 1998). 4-Methylsulphinylbutyl ITC has been described previously as an antimicrobial agent (Dornberger et al., 1975), but IC50 values had not yet been determined.
A number of research groups have addressed the question of whether glucosinolates or their breakdown products play a role in defense against pathogens by studying correlations between glucosinolate contents of different plant genotypes and disease resistance levels. Glucosinolate levels in different Brassica spp. did not correlate at all with resistance to the fungal pathogen Leptosphaeria maculans (Mithen and Magrath, 1992; Sexton et al., 1999), whereas there was even a negative correlation with resistance to Alternaria spp. (Doughty et al., 1991; Giamoustaris and Mithen, 1997). On the other hand, a general correlation trend was observed between the extent of pathogen-induced production of indole glucosinolates on Brassica napus and resistance to Sclerotinia sclerotiorum (Li et al., 1999a). Hence, the contribution of glucosinolates to microbial disease resistance appears to be strongly pathogen dependent. The main drawback of these studies, however, is that they have not been performed on isogenic lines, so that other genetic factors may have confused the analyses and the conclusions drawn from them.
The Arabidopsis gsm1-1 mutant, found to be largely devoid of the antimicrobial glucosinolate breakdown products A and B (4-methylsulphinylbutyl ITC) in leaf extracts, provides together with its wild-type parent (ecotype Col-0) an ideal set of isogenic plants allowing the study of the role of glucosinolates in disease resistance. When compared with Col-0 wild-type plants, the gsm1-1 mutant did not show significant differences in susceptibility to a range of different pathogens including A. brassicicola, P. cucumerina, B. cinerea, P. parasitica, E. carotovora, or P. syringae. These pathogens apparently have devised strategies to cope with the presence of glucosinolates in their hosts. Fungi such as B. cinerea and A. brassicicola are not inhibited in vitro by at least 4-methylsulphinylbutyl ITC (Table IV), although it is not excluded that they are inhibited by other types of glucosinolate breakdown products. The basis of such apparent tolerance is currently unclear. One possible mechanism may rely on metabolic conversion. For instance, it has been shown that the Brassica spp. pathogen L. maculans produces an enzyme that can convert nitrile compounds, such as some glucosinolate breakdown products, into less toxic compounds (Sexton and Howlett, 2000). It should be kept in mind, however, that susceptibility in vitro does not necessarily translate into susceptibility in planta because tolerance mechanisms may become activated specifically during pathogenesis. Moreover, it is clear that some of these pathogens, notably the biotrophs P. syringae and P. parasitica, cause little or no tissue damage during pathogenesis and thus avoid the release of glucosinolates from host cell vacuoles as well as their myrosinase-dependent conversion to antimicrobial breakdown products. Other pathogens in our test panel, namely B. cinerea, P. cucumerina, and E. carotovora, cause tissue maceration and necrosis and therefore it is likely that glucosinolate breakdown products will accumulate in or around the infection site during attack by these pathogens.
The only pathogen for which a relatively slight but significant difference in susceptibility between gsm1-1 and Col-0 was observed is F. oxysporum f. sp. matthiolae. The oldest leaves of gsm1-1 were equally susceptible as Col-0 leaves, whereas relatively more of the younger leaves of gsm1-1 showed more severe chlorosis symptoms compared with their counterparts in Col-0 (Fig. 6). The younger leaves of gsm1-1 were also shown to contain about twice as much F. oxysporum biomass relative to Col-0 leaves of the same age at 8 d after inoculation (Fig. 7). This indicates that the glucosinolate content in younger leaves may contribute to their protection against some pathogens such as F. oxysporum. It is worthwhile to note that the content of the glucosinolate breakdown product B (4-methylsulphinylbutyl ITC) was found to be more than one order of magnitude higher in young versus old leaves (Table III). This is consistent with previous studies demonstrating declining glucosinolate levels in aging leaves (Porter et al., 1991; Wallsgrove et al., 1993; Li et al., 1999b). Although we have only found one example of a microorganism whose interaction with Arabidopsis is affected by the glucosinolate content of the host plant, there may be numerous others, especially among the opportunistic microbial flora that can only cause infection under favorable conditions.
Ludwig-Müller et al. (1999) have previously found that gsm1-1 plants are equally susceptible as wild-type plants to the clubroot pathogen Plasmodiaphora brassicae, which is in line with our findings for most of the pathogens tested in our study. However, another glucosinolate mutant, namely TU3, was less susceptible to clubroot than either gsm1-1 or wild type. Mutant TU3 makes all glucosinolates that are lacking in gsm1-1 (i.e. aliphatic glucosinolates with butyl, pentyl, or hexyl core groups) but is deficient in aliphatic glucosinolates with heptyl and octyl core groups (Haughn et al., 1991). This suggests that P. brassicae, an obligate biotrophic pathogen on Brassicaceae, requires long-chain glucosinolates or long-chain glucosinolate breakdown products for full pathogenicity. Perhaps these molecules are recognized by P. brassicae and used to activate pathogenicity genes. Hence, glucosinolates can either be factors that contribute to protection against some opportunistic pathogens, such as F. oxysporum, or factors that are utilized or recognized to the advantage of specialist pathogens, such as P. brassicae.
Glucosinolates have also previously been proposed to play both positive and negative roles in interactions with insects. Glucosinolates can act as feeding stimulants and oviposition stimulants on a range of insect species (Louda and Mole, 1991; van Loon et al., 1992). For example, caterpillars of cabbage white butterflies (Pieris spp.) started feeding on non-Brassica plant tissues when painted with glucosinolates (Verschaffelt, 1910). On the other hand, glucosinolates and their breakdown products cause growth inhibition and even mortality when fed to other types of insects (Blau et al., 1978). In field trials, Giamoustaris and Mithen (1995) found that glucosinolate contents of different Brassica genotypes correlated with decreased damage by generalist insect herbivores but increased susceptibility to other insects specialized on Brassicaceae hosts. In conclusion, glucosinolates appear to act as chemical signals that are percieved differentially by different members of the ecological community and therefore are important in driving coevolution between plants and their pest and disease organisms.
MATERIALS AND METHODS
Biological Material
The mutant gsm1-1 (Haughn et al., 1991) was obtained from the Nottingham Arabidopsis Stock Centre online catalogue (http://nasc.nott.ac.uk/home.html). This mutant was isolated in a screen of ethyl methane sulfonate mutagenized Col-0 plants and backcrossed several times before handing over to the Arabidopsis Stock Centre (Nottingham, UK; G.W. Haughn, personal communication). Arabidopsis plants were essentially grown as described previously (Penninckx et al., 1996).
Growth and spore harvesting of the fungi Alternaria brassicicola (strain MUCL20297, Mycothèque Université Catholique de Louvain, Louvain-la-Neuve, Belgium), Botrytis cinerea (strain MUCL30158, Mycothèque Université Catholique de Louvain), Fusarium culmorum (strain MUCL30162, Mycothèque Université Catholique de Louvain), Fusarium oxysporum f. sp. matthiolae (strain 247.61, Centraalbureau voor Schimmelcultures, Baarn-Delft, The Netherlands), Nectria hematococca (strain 160-2-2, University of Basel), Neurospora crassa (strain FGSC2489, Fungal Genetics Stock Center, Kansas City, KS), Penicillium expansum (field isolate provided by David Sugar, Oregon State University, Corrallis), Plectosphaerella cucumerina (provided by Dr. B. Mauch-Mani, Université de Fribourg, Switzerland), and Verticillium dahliae (strain MUCL19210, Mycothèque Université Catholique de Louvain) were done as described previously (Broekaert et al., 1990). Peronospora parasitica strain Wela (Delaney et al., 1994) was maintained on living Arabidopsis plants of the Weiningen ecotype.
Erwinia carotovora (strain LMG6663, Laboratorium voor Microbiologie, Universiteit Gent, Belgium) was grown overnight at 28°C in Luria (L) broth (10 g L−1 peptone, 10 g L−1 NaCl, and 5 g L−1 yeast extract). Escherichia coli (strain DH5alpha; Hanahan, 1983) was grown overnight at 37°C in L broth. Pseudomonas syringae pv tomato DC3000 (provided by Jane Glazebrook, University of Maryland, College Park), was grown overnight at 28°C in King's B medium (King et al., 1954) supplemented with 25 μg mL−1 rifampicin. Sarcina lutea (strain ATCC9341, American Type Culture Collection, Manassas, VA) was grown overnight at 30°C in 2% (w/v) peptone. Xanthomonas campestris pv pelargonii (strain 10342, provided by Elisabeth Chevreau, Institut National de la Recherche Agronomique, Paris) was grown overnight at 30°C in L broth.
Antifungal and Antibacterial Activity Assays
Antifungal activity was monitored microscopically and measured by microspectophotometry as previously described (Broekaert et al., 1990). In the wells of a 96-well microplate, 20-μL samples (usually as 2-fold dilution series of test fractions) routinely were mixed with 80 μL of potato dextrose broth (12 g L−1, Difco, Franklin Lakes, NJ) containing 2 × 104 fungal spores mL−1, with addition of extra salts to a final concentration of 1 mm CaCl2 and 50 mm KCl. Plates were incubated at 25°C in the dark. The IC50 value (i.e. the concentration of the antifungal component that is required to inhibit 50% of the fungal growth) was calculated as described in Cammue et al. (1992).
Unless otherwise stated, the fungal test organism was N. crassa (strain FGSC2489). The antifungal activity of a fraction in units per mL is defined as the total volume of the assay mixture divided by the volume of the fraction in the assay mixture that gives 50% growth inhibition (=dilution factor for 50% growth inhibition). Percent growth inhibition was calculated as described in Cammue et al. (1995).
Antibacterial activity was measured microspectrophotometrically as follows. Bacteria were precultured overnight in a rotary shaker as described above. A soft agarose medium (1% [w/v] peptone and 0.5% [w/v] low melting point agarose) was inoculated with the bacteria to a cell density of 105 cfu mL−1. For P. syringae, the soft agarose medium consisted of King's B medium supplemented with 0.5% (w/v) low melting point agarose. Aliquots (80 μL) of the bacterial suspension in soft agarose were added to filter-sterilized samples (20 μL) in flat bottom 96-well microplates and allowed to solidify. The A595 of the culture was measured with the aid of a microplate reader after 30 min and 48 h of incubation at appropriate temperatures for each bacterium as described above. Percent growth inhibition was calculated as described by Cammue et al. (1995).
Extraction and Purification of Antimicrobial Compounds in Leaves of Arabidopsis
Small-scale tests for determining suitable chromatography matrices were performed as follows. Ten grams of leaves from five-week-old Arabidopsis plants were lyophilized, ground in a mortar, and extracted with 30 mL of boiling distilled water. The extract was stirred for 30 min at room temperature and heated for 5 min in a water bath at 100°C. Following centrifugation, the pH of the supernatant was adjusted to 9 or 5 by the addition of 500 mm NH4Ac at the appropriate pH (50-mm final concentration). Precipitated compounds were centrifuged and the supernatant was passed over a self-packed column containing either Q- or S-Sepharose Fast Flow (3 × 3 cm, Pharmacia, Peapack, NJ) equilibrated in 50 mm NH4Ac at pH 9 or pH 5, respectively. The unbound components were collected and this flow-through fraction was lyophilized and resuspended in distilled water. Passing of the crude leaf extract over the reversed phase C8 column (2 × 1.75 cm, International Sorbent Technology, Tucson, AZ) equilibrated in 0.1% (v/v) TFA was performed after addition of 0.1% (v/v) TFA and centrifugation. Unbound components were collected, evaporated under reduced pressure, and resuspended in distilled water. These three different flow-through fractions were analyzed for loss of antifungal activity compared with an equally concentrated crude leaf extract.
For the large-scale purification of the antimicrobial compounds in the crude leaf extract, 0.1% (v/v) TFA was added to the resuspended Q-Sepharose flow-through fraction and it was passed over a C18 RP-HPLC column (30 × 0.39 cm, Phenomenex, Torrance, CA) equilibrated in 0.1% (v/v) TFA. The column was eluted as described in the legend of Figure 1. Fractions with high antifungal activity were pooled for each peak, evaporated under reduced pressure, resuspended in 1 mL 0.1% (v/v) TFA, and passed a second time over the C18 RP-HPLC. The column was equilibrated in 0.1% (v/v) TFA and eluted as described in the legend of Figure 2A. Following passage of fraction B from Figure 1 over the C18 RP-HPLC column, the active fractions, eluting in one major peak (Fig. 2A), were pooled and passed over a phenyl RP-HPLC column (25 × 0.4 cm, Vydac, Hesperia, CA). This column was equilibrated in 0.1% (v/v) TFA and eluted as described in the legend of Figure 2B.
Characterization of 4-Methylsulphinylbutyl ITC
The GC/EIMS and GC/chemical ionization MS measurements were performed with a Voyager GC-MS-System (ThermoQuest, San Jose, CA) using the following conditions: 70 eV EI, source temperature 200°C and column DB-5MS (15 m × 0.25 mm, 0.25-μm film thickness, J&W, Folsom, CA), injection temperature 250°C, interface temperature 300°C, carrier gas He, flow rate 1.3 mL min−1, splitless injection, column temperature program (60°C for 1 min, then raised to 110°C at a rate of 25°C min−1, held for 1 min, and then raised to 300°C at a rate of 10°C min−1). The retention time of 4-methylsulphinylbutyl ITC was 10 min. The high-resolution MS of m/z 160.0251 (calculated for C6H10NS2, 160.0255) was carried out on a double-focusing mass spectrometer (AMD-402, AMD Intectra GmbH, Harpstedt, Germany). The 1H NMR measurements were obtained from a Varian UNITY 500 (499.82 MHz, solvent CD3OD).
Plant Inoculations
Inoculations of 4-week-old soil-grown Arabidopsis plants with A. brassicicola, B. cinerea, or P. parasitica were performed as described previously (Thomma et al., 1998) except that for A. brassicicola and B. cinerea, respectively, one and two 5-μL droplets of spore suspension were used per leaf. Inoculations with P. cucumerina were identical as described for A. brassicicola. For the inoculation of Arabidopsis plants with F. oxysporum, 4-week-old soil-grown plants were sprayed evenly with a spore suspension of 5 × 105 spores mL−1 in potato dextrose broth (12 g L−1, Difco). For inoculations with E. carotovora, two 5-μL droplets of a bacterial suspension in 10 mm MgSO4 (OD600 = 0.5) were placed on the leaves of 4-week-old soil-grown Arabidopsis plants. For inoculation of 4-week-old soil-grown Arabidopsis plants with P. syringae a protocol was followed based on the one previously described by Cao et al. (1994). In brief, plants were placed at 100% relative humidity 1 d before inoculation. At the day of inoculation, a bacterial suspension of approximately 107 cfu mL−1 (OD600 = 0.05) supplemented with 0.01% (v/v) Silwet l-77 was prepared. For each set of two plants, 35 mL of this suspension was used to dip infiltrate the leaves for 30 s. Plants were kept at 100% relative humidity for the remainder of the experiment.
Quantification of P. syringae pv tomato DC3000 and F. oxysporum in Inoculated Plants
For the quantification of P. syringae growth in inoculated plants, a protocol was used that is largely based on the one previously described by Cao et al. (1994). Sixteen inoculated leaves were excised at 0-, 1-, 2-, and 3-d time points. Four samples on d 0 and eight samples on days 1, 2, and 3 after inoculation of 20 ± 1 mg were taken from these leaves for measurement of bacterial growth. The bacteria were extracted from the leaf tissue in 0.5 mL 10 mm MgSO4 by maceration with a plastic pestle followed by vigorous vortexing. Serial dilutions were made from the resulting extracts, and 50 μL of each dilution was spread onto King's B medium agar plates containing 25 μg mL−1 rifampicin. The plates were incubated for 2 d at 25°C and the number of colonies for each sample was then recorded. Statistical analyses of the differences between two means of log-transformed data from three independent experiments were performed according to Tukey's studentized range test (P > 0.95).
For the determination of the fungal biomass in F. oxysporum-infected leaves, two plant sets were inoculated as described above under plant inoculations: one with a spore suspension in potato dextrose broth and the other with potato dextrose broth. The presence of F. oxyporum in inoculated plants was detected by microscopic observation of leaves stained with lactophenol trypan blue as described by Mauch-Mani and Slusarenko (1996). From the spore- and mock-inoculated plants, samples were collected at 0 and 8 d following inoculation. For Col-0 and gsm1-1, samples containing 12 leaf discs (0.36 cm2, cut with a cork borer) were taken at each time point from the seventh, eighth, ninth, and 10th leaf from three plants. This was done in six replicates for each sample. Fungal biomass was determined by quantitative PCR using specific primers for Fusarium spp. DNA (5′-AGTATTCTGGCGGGCATGCCTGT and 5′-ACAAATTACAACTCGGGCCCGAGA; Hue et al., 1999). DNA extraction was performed with a revised protocol of Ward (1990). A mix of 0.2-g glass beads (2 mm and 200–300 μm diameter; Sigma, St. Louis) was added to the samples together with 300 μL breaking buffer (2.5 m LiCl, 50 mm Tris-HCl [pH 8], 4% [v/v] Triton X-100, and 62.5 mm Na2EDTA) and 300 μL phenol:chloroform:iso-amylalcohol (25:24:1, v/v). Samples were shaken in a Phastprep (Savant Instruments, Inc., Farmingdale, NY; BIO 101, Inc., Vista, CA) high-speed shaker for 30 s at maximum speed, incubated on ice for 2 min, and centrifuged for 5 min at 11,000 rpm. Two volumes ethanol were added to the supernatant of the samples, incubated for 15 min at −20°C, and centrifuged for 5 min at 10,000 rpm. Pellets were washed with 500 μL 70% (v/v) ethanol, centrifuged for 5 min at 10,000 rpm, dried, and resuspended in 50 μL Tris-EDTA (pH 8). Quantitative PCR was performed on undiluted DNA extract from the plant samples with a LightCycler real-time quantitative PCR apparatus (Roche, Basel). A serial dilution of in vitro-grown F. oxysporum f. sp. matthiolae DNA was used as a standard. For each PCR reaction, samples (20 μL) contained 2 μL DNA extract, 2 μL LightCycler-FastStart DNA Master SYBR Green I (Roche), both primers (0.5-μm final concentration), and MgCl2 (3-mm final concentration). Forty cycles of amplification (95°C denaturation for 15 s, 68°C annealing for 5 s, and 72°C polymerization for 15 s) were carried out in sealed LightCycler Capillaries (Roche).
Footnotes
This work was partially supported by the Vlaams Instituut voor Bevordering van het Wetenschappelijk-Technologisch Onderzoek in de Industrie (grant no. G961052). K.T.F.M.-J. is the recipient of a predoctoral fellowship of this fund. B.P.H.J.T. is a research assistant of the fund Fonds voor Wetenschappelijk Onderzoek-Vlaanderen.
LITERATURE CITED
- Benn M. Glucosinolates. Pure Appl Chem. 1977;49:197–210. [Google Scholar]
- Blau PA, Feeney P, Contardo L, Tobson DS. Allylglucosinolate and herbivorous caterpillars: a contrast in toxicity and tolerance. Science. 1978;200:1296–1298. doi: 10.1126/science.200.4347.1296. [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Terras FRG, Cammue BPA, Vanderleyden J. An automated quantitative assay for fungal growth. FEMS Microbiol Lett. 1990;69:55–60. [Google Scholar]
- Buell CR. Arabidopsis: a weed leading the field of plant-pathogen interactions. Plant Physiol Biochem. 1998;36:177–186. [Google Scholar]
- Cammue BPA, De Bolle MFC, Terras FRG, Proost P, Van Damme J, Rees SB, Vanderleyden J, Broekaert WF. Isolation and characterization of a novel class of plant antimicrobial peptides from Mirabilis jalapa L. seeds. J Biol Chem. 1992;267:2228–2233. [PubMed] [Google Scholar]
- Cammue BPA, Thevissen K, Hendriks M, Eggermont K, Goderis IJ, Proost P, Van Damme JC, Broekaert WF. A potent antimicrobial protein from onion (Allium cepa L.) seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 1995;109:445–455. doi: 10.1104/pp.109.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon S, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Dornberger K, Böckel V, Heyer J, Schönfeld C, Tonew M, Tonew E. Untersuchungen über die isothiocyanate erysolin und sulforaphan aus Candaria draba L. Pharmazie. 1975;30:792–796. [PubMed] [Google Scholar]
- Doughty KJ, Porter AJR, Morton AM, Kiddle G, Bock CH, Wallsgrove R. Variation in the glucosinolate content of oilseed rape (Brassica napus L.) leaves: II. Response to infection by Alternaria brassicae (Berk.) Sacc. Ann Appl Biol. 1991;118:469–477. [Google Scholar]
- Fenwick GR, Heaney RK, Mullin WJ. Glucosinolates and their breakdown products in food and food-plants. CRC Crit Rev Food Sci Nutr. 1983;18:123–201. doi: 10.1080/10408398209527361. [DOI] [PubMed] [Google Scholar]
- Giamoustaris A, Mithen R. The effect of modifying the glucosinolate content of leaves on oilseed rape (Brassica napus spp. oleifera) on its interaction with specialist and generalist pests. Ann Appl Biol. 1995;126:347–363. [Google Scholar]
- Giamoustaris A, Mithen R. Glucosinolates and disease resistance in oilseed rape (Brassica napus spp oleifera) Plant Pathol. 1997;46:271–275. [Google Scholar]
- Glazebrook J, Ausubel FM. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Davin L, Giblin M, Underhill EW. Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana. Plant Physiol. 1991;97:217–226. doi: 10.1104/pp.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogge LR, Reed DW, Underhill EW, Haughn GW. HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thaliana and their identification using thermospray liquid chromatography-mass spectrometry. J Chromatogr Sci. 1988;26:551–556. [Google Scholar]
- Hue F-X, Huerre M, Rouffault MA, De Bievre Claude. Specific detection of Fusarium species in blood and tissues by a PCR technique. J Clin Microbiol. 1999;37:2434–2438. doi: 10.1128/jcm.37.8.2434-2438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE. Two simple media for demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- Kirkegaard JA, Wong PTW, Desmarchelier JM. In vitro suppression of fungal root pathogens of cereals by Brassica tissues. Plant Pathol. 1996;45:593–603. [Google Scholar]
- Li Y, Kiddle G, Bennett RN, Wallsgrove RM. Local and systemic changes in glucosinolates in Chinese and European cultivars of oilseed rape (Brassica napus L.) after inoculation with Sclerotinia sclerotiorum (stem rot) Ann Appl Biol. 1999a;134:45–58. [Google Scholar]
- Li YC, Kiddle G, Bennett R, Doughty K, Wallsgrove R. Variation in the glucosinolate content of vegetative tissues of Chinese lines of Brassica napus L. Ann Appl Biol. 1999b;134:131–136. [Google Scholar]
- Louda S, Mole S. Glucosinolates, chemistry and ecology. In: Rosenthal GA, Berenbaum RM, editors. Herbivores: Their Interactions with Secondary Plant Metabolites. Ed 2. Vol. 1. San Diego: Academic Press; 1991. pp. 123–164. [Google Scholar]
- Ludwig-Müller J, Pieper K, Ruppel M, Cohen JD, Epstein E, Kiddle G, Bennett R. Indole glucosinolate and auxin biosynthesis in Arabidopsis thaliana (L.) Heynh. glucosinolate mutants and the development of clubroot disease. Planta. 1999;208:409–419. doi: 10.1007/s004250050576. [DOI] [PubMed] [Google Scholar]
- Manici LM, Lazzeri L, Palmieri S. In vitro fungitoxic activity of some glucosinolates and their enzyme-derived products toward plant pathogenic fungi. Agric Food Chem. 1997;45:2768–2773. [Google Scholar]
- Mauch-Mani B, Slusarenko AJ. Production of salicylic acid precursors in a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell. 1996;8:203–212. doi: 10.1105/tpc.8.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithen RF, Lewis BG, Fenwick GR. In vitro activity of glucosinolates and their products against Leptosphaeria maculans. Trans Br Mycol Soc. 1986;87:433–440. [Google Scholar]
- Mithen RF, Magrath R. Glucosinolates and resistance to Leptosphaeria maculans in wild and cultivated Brassica species. Plant Breed. 1992;108:60–68. [Google Scholar]
- Neter J, Kutner MH, Nachtsheim CJ, Wasserman W. Applied Linear Statistical Models. Ed 4. Boston: WCB/McGraw-Hill; 1996. [Google Scholar]
- Osbourn AE. Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell. 1996;8:1821–1831. doi: 10.1105/tpc.8.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblanx GW, Buchala A, Métraux J-P, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Métraux J-P, Broekaert WF. Cooperative activation of jasmonate and ethylene response pathways in parallel is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2114. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterka S, Schlosser E. In vitro activity of glucosinolates against Leptosphaeria maculans in comparison to the glucosinolate content and susceptibility of seedlings of different Brassica spp. Med Fac Landbouww Rijksuniv Gent. 1989;54:439–446. [Google Scholar]
- Porter AJR, Morton AM, Kiddle G, Doughty KJ, Wallsgrove RM. Variation in the glucosinolate content of oilseed rape (Brassica napus L.): I. Effects of leaf age and position. Ann Appl Biol. 1991;118:461–467. [Google Scholar]
- Rask L, Andréasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000;42:93–113. [PubMed] [Google Scholar]
- Sarwar M, Kirkegaard JA, Wong PTW, Desmarchelier JM. Biofumigation potential of brassicas: III. In vitro toxicity of isothiocyanates to soil-borne fungal pathogens. Plant Soil. 1998;201:103–112. [Google Scholar]
- Sexton AC, Howlett BJ. Characterization of a cyanide hydratase gene in the phytopathogenic fungus Leptosphaeria maculans. Mol Gen Genet. 2000;263:463–470. doi: 10.1007/s004380051190. [DOI] [PubMed] [Google Scholar]
- Sexton AC, Kirkegaard JA, Howlett BJ. Glucosinolates in Brassica juncea and resistance to Australian isolates of Leptosphaeria maculans, the blackleg fungus. Australas Plant Pathol. 1999;28:95–102. [Google Scholar]
- Spencer GF, Daxenbichler Gas chromatography-mass spectrometry of nitriles, isothiocyanates and oxazolidinethiones derived from cruciferous glucosinolates. J Sci Food Agric. 1980;31:359–367. [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Tierens KFM-J, Broekaert WF. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 1999a;121:1093–1101. doi: 10.1104/pp.121.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Nelissen I, Eggermont K, Broekaert WF. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 1999b;19:163–171. doi: 10.1046/j.1365-313x.1999.00513.x. [DOI] [PubMed] [Google Scholar]
- Tsuji J, Jackson EP, Gage DA, Hammerschmidt R, Somerville SC. Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv syringae. Plant Physiol. 1992;98:1304–1309. doi: 10.1104/pp.98.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon JJ, Blaakmeer A, Griepink FC, van Beek TA, Schoonhoven LM, De Groot A. Leaf surface compound from Brassica oleracea (Cruciferae) induces oviposition by Pieris brassicae (Lepidoptera: Pieridae) Chemoecology. 1992;3:39–44. [Google Scholar]
- Verschaffelt E. The cause determining the selection of food in some herbivorous insects. Proc Royal Acad Amsterdam. 1910;13:536–542. [Google Scholar]
- Wallsgrove RM, Bennett R, Donald A, Kiddle G, Porter A, Doughty K. The biochemical basis for the differential response of oilseed rape varieties to infection and stress. Asp Appl Biol. 1993;34:155–161. [Google Scholar]
- Ward AC. Single-step purification of shuttle vectors from yeast for high frequency back-transformation into E. coli. Nucleic Acids Res. 1990;18:5319–5320. doi: 10.1093/nar/18.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J. Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell. 1999;11:2419–2428. doi: 10.1105/tpc.11.12.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]