Graphical abstract
Keywords: ras GTPase-activating protein-binding protein 1, SARS-CoV-2 nucleocapsid protein, stress granule, NTF2-like domain, pathogen-host interaction
Abstract
Stress granule (SG) formation mediated by Ras GTPase-activating protein-binding protein 1 (G3BP1) constitutes a key obstacle for viral replication, which makes G3BP1 a frequent target for viruses. For instance, the SARS-CoV-2 nucleocapsid (N) protein interacts with G3BP1 directly to suppress SG assembly and promote viral production. However, the molecular basis for the SARS-CoV-2 N − G3BP1 interaction remains elusive. Here we report biochemical and structural analyses of the SARS-CoV-2 N − G3BP1 interaction, revealing differential contributions of various regions of SARS-CoV-2 N to G3BP1 binding. The crystal structure of the NTF2-like domain of G3BP1 (G3BP1NTF2) in complex with a peptide derived from SARS-CoV-2 N (residues 1–25, N1–25) reveals that SARS-CoV-2 N1–25 occupies a conserved surface groove of G3BP1NTF2 via surface complementarity. We show that a φ-x-F (φ, hydrophobic residue) motif constitutes the primary determinant for G3BP1NTF2-targeting proteins, while the flanking sequence underpins diverse secondary interactions. We demonstrate that mutation of key interaction residues of the SARS-CoV-2 N1–25 − G3BP1NTF2 complex leads to disruption of the SARS-CoV-2 N − G3BP1 interaction in vitro. Together, these results provide a molecular basis of the strain-specific interaction between SARS-CoV-2 N and G3BP1, which has important implications for the development of novel therapeutic strategies against SARS-CoV-2 infection.
Introduction
The Covid-19 pandemic caused by the novel Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) is posing a grave threat to global public health, resulting in ∼28 million confirmed cases and ∼5 million deaths reported to date. SARS-CoV-2 is a beta-coronavirus containing a ∼30 kb, positive-sense single-stranded RNA genome, which makes it one of the largest genomes of all known RNA viruses. The SARS-CoV-2 genome is organized into 14 open reading frames (ORFs) encoding 27 proteins that are functionally divided into non-structural proteins (NSPs), structural proteins and accessory proteins. The structural proteins include Spike protein (S), Envelope protein (E), Membrane protein (M) and Nucleocapsid protein (N).1 Among these, the N protein is essential for virion assembly and viral RNA synthesis.2, 3 In addition, the N protein modulates a variety of host cellular activities, such as actin reorganization, cell cycle progression, apoptosis, and immune response4. The multifunctionality of the N protein makes it an attractive target for drug intervention against Covid-19.5
Recent studies have further demonstrated that SARS-CoV-2 N plays a role in disassembling host stress granule (SG) through an interaction with Ras GTPase-activating protein-binding protein 1 (G3BP1) and 2 (G3BP2).6, 7 SGs are dynamic ribonucleoprotein (RNP) assemblies formed in eukaryotic cells in response to various stresses, such as oxidative stress and viral infection,8, 9 with G3BP1/2 serving as key SG-nucleating agents.10, 11, 12, 13, 14, 15 Through restriction of protein synthesis and insulating viral mRNAs, SG serves as a key defense mechanism for the host to counter viral attack.16 G3BP1/2 contains an N-terminal nuclear transport factor 2-like (referred to as NTF2 herein) domain responsible for protein interaction and a C-terminal RRM domain responsible for RNA binding (Figure 1 (A)). The NTF2 domain is followed by the first intrinsically disordered region (IDR1) harboring an acidic segment, while the RRM domain is followed by an Arg-Gly-rich (RGG) region-containing IDR (IDR2) (Figure 1(A)).17 The G3BP1- and G3BP2-mediated SG assembly and disassembly is dynamically regulated via a network of competing protein and RNA interactions involving the NTF2 domain, the RRM domain and the IDR2, as described by the network theory proposed recently.10, 18, 19 It has been demonstrated that the SG-associated protein Caprin1, containing G3BP1/2 NTF2-binding, oligomerization and RNA-interaction modules, provides multiple intermolecular contacts (valences) to expand the protein-RNA interaction network within the SG, thereby promoting SG condensation.19, 20 In contrast, another SG-associated protein USP10, which possesses a G3BP1/2 NTF2-binding module but lacks an oligomerization or RNA-binding module, serves as a valence cap to inhibit the SG formation.19, 20 Given its important role in the formation of SG, the G3BP1/2 protein has become a recurrent target for viral proteins to suppress the SG assembly.21, 22, 23, 24 For instance, the non-structural protein 3 (nsP3) from the Old World alphavirus recruits G3BP1 to viral replication complex via an interaction with the NTF2 domain, leading to disruption of SG in virus-infected cells.21, 25, 26 Likewise, SARS-CoV-2 N was shown to partition into stress granules and interacts with the NTF2 domains of G3BP1 and G3BP2, resulting in inhibition of SG assembly to promote viral infection in complementation-based assay.7, 27 However, the underlying mechanism of SARS-CoV-2 N-mediated SG disassembly is unclear.
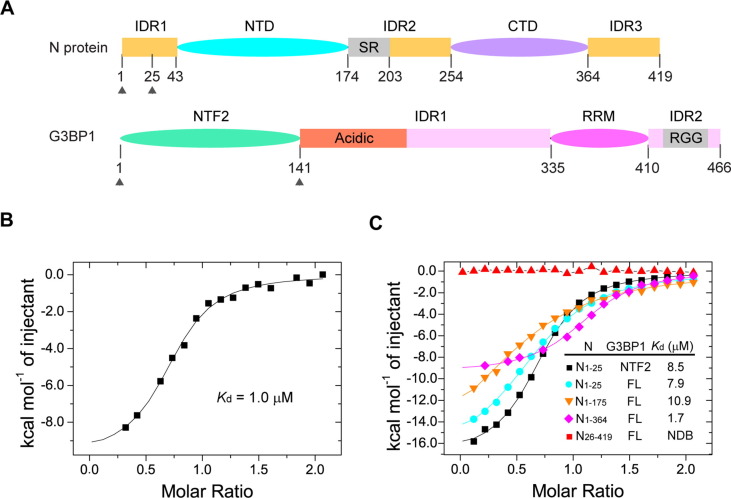
Figure 1.
Biochemical characterization of the interaction between SARS-CoV-2 N and G3BP1. (A) Domain architecture of SARS-CoV-2 N and G3BP1, with individual domains color coded. The SR-rich region within the IDR2 of SRS-CoV-2 N, the acidic region within the IDR1 of G3BP1 and the RG-rich region (RGG) within the IDR2 of G3BP1 are labeled. The protein fragments (residues 1–25 of SARS-CoV-2 N protein and 1–139 of G3BP1) used for crystallographic study are delimited by arrows. (B) ITC binding assays for full-length SARS-CoV-2 N and G3BP1. (C) ITC binding assays for the truncated fragments of SARS-CoV-2 N and G3BP1. FL, full length; NDB, no detectable binding.
The crystal structure of the G3BP1 NTF2 domain (G3BP1NTF2) in complex with a peptide derived from nucleoporin, Semliki Forest virus (SFV) nsP3 or Caprin1 has been reported.26, 28, 29 Common to these structures is an insertion of a phenylalanine residue from the G3BP1NTF2-interacting peptide into a hydrophobic pocket formed by the G3BP1NTF2 domain. On the other hand, these structures show large divergence for the Phe-flanking regions of the peptide sequence, suggesting high diversity for the G3BP1NTF2-mediated protein interactions.
SARS-CoV-2 N consists of an N-terminal domain (NTD) responsible for RNA binding and a C-terminal domain (CTD) that mediates both RNA binding and dimerization (Figure 1(A)).30, 31, 32 Both the NTD and CTD domains are flanked by IDRs to reinforce their RNA-binding activities,2, 33 with a serine-arginine (SR) rich segment located within the second IDR (IDR2) subjected to posttranslational modification for modulation of RNA binding.34, 35 Recent biochemical and cellular evidence indicated that the interaction between SARS-CoV-2 N protein and G3BP1NTF2 critically depends on the N-terminal IDR (IDR1) of SARS-CoV-2 N protein 7, 27, 36, 37. Deletion of the first 50 residues of SARS-CoV-2 N led to disruption of SG inhibition and reduced viral production,7 suggesting an important role of the IDR1 of SARS-CoV-2 N in viral infection. Consistently, introduction of an I15A/T16A/F17A/G18A quadruple mutation to the IDR1 of SARS-CoV-2 N led to abolished G3BP1 binding in an in vitro binding assay, indicating an ITFG motif-dependent interaction.37 However, due to the lack of structural information, the molecular basis for the interaction between SARS-CoV-2 N and G3BP1 remains elusive.
To determine the molecular basis of the SARS-CoV-2 N − G3BP1 interaction, we solved the crystal structure of the complex between the G3BP1NTF2 domain and a peptide derived from the IDR1 of SARS-CoV-2 N (comprising residues 1–25, N1–25) at a resolution of 2.35 Å. The structure revealed surface complementarity and hydrophobic groove-insertion mechanisms dominating the SARS-CoV-2 N1–25 − G3BP1NTF2 interaction. Comparative structural analyses of SARS-CoV-2 N − G3BP1NTF2 and other G3BP1NTF2 complexes further revealed a consensus φxF motif as a primary G3BP1NTF2-binding determinant, which insert into a surface groove of G3BP1NTF2 that is conserved among the NTF2 domains. On the other hand, the φxF-flanking sequences underpin diverse secondary interactions among various G3BP1NTF2 complexes, raising a possibility for the development of specific inhibitors toward the SARS-CoV-2 N − G3BP1 interaction. The φxF motif is conserved in SARS-CoV and SARS-CoV-2 but not in many other coronavirus strains, suggesting a strain-specific G3BP1NTF2 interaction. This study provides a framework for molecular understanding of the targeting of G3BP1 by SARS-CoV-2 N, with important implications in development of therapeutic interventions against SARS-CoV-2 infection.
Results
Biochemical characterization of the SARS-CoV-2 N − G3BP1/2 interaction
To identify the interaction elements between SARS-CoV-2 N and G3BP1, we performed isothermal titration calorimetry (ITC) on full-length or truncated fragments of SARS-CoV-2 N and G3BP1. Full-length SARS-CoV-2 N and G3BP1 interact strongly, with a dissociation constant (K d) of 1.0 µM (Figure 1(B) and Figure S1(A)), and enthalpy and entropy changes of ΔH = −9.9 kcal/mol and ΔS = −5.9 cal/mol/deg, respectively (Table S1). Similar binding thermograms were observed when full-length G3BP1 was titrated with the C-terminal IDR3-truncated SARS-CoV-2 N (residues 1–364, N1–364), which gave a K d of 1.7 µM (Figure 1(C) and Figure S1(B)), and enthalpy and entropy changes of ΔH = −9.4 kcal/mol and ΔS = −5.4 cal/mol/deg, respectively (Table S1). Next, we titrated full-length G3BP1 with the SARS-CoV-2 N fragment encompassing the IDR1 and NTD but not the CTD and IDR2 (residues 1–175, N1–175). We observed a K d of 10.9 µM (Figure 1(C) and Figure S1(C)), which is ∼11- and ∼6-fold weaker than those of full-length N or N1–364, respectively. Interestingly, comparison of the titration parameters for full-length SARS-CoV-2 N, N1–364 and N1–175 reveals that removal of the IDR2 and CTD led to increased reductions for both enthalpy (ΔH = −18.2 kcal/mol) and entropy (ΔS = −38.2 cal/mol/deg) (Table S1), suggesting that, in the context of full-length N or N1–364, the CTD and/or IDR2 contribute to the G3BP1 binding by reducing the entropic cost of the complex formation. Furthermore, we titrated full-length G3BP1, the G3BP1NTF2 domain and the G3BP2NTF2 domain with the peptide derived from the first 25 residues of the IDR1 of SARS-CoV-2 N protein (N1–25), which gave K ds of 7.9 µM, 8.5 µM and 10.9 µM, respectively (Figure 1(C) and Figure S1(D–F)), consistent with the previous observations that the IDR1 of SARS-CoV-2 N protein is primarily responsible for its interaction with G3BP1/2.7, 27, 36, 37 Notably, the enthalpy and entropy changes associated with the titrations by the N1–25 peptide are equivalent to those of SARS-CoV-2 N1–175 within experimental error (Table S1), suggesting a minimal effect of the NTD of SARS-CoV-2 N in G3BP1 binding. Finally, titration of full-length G3BP1 with SARS-CoV-2 N26–419 showed no appreciable binding (Figure 1(C) and Figure S1(G)), indicating that the N1–25 is indispensable for the SARS-CoV-2 N − G3BP1 interaction.
Together, these observations identified that the SARS-CoV-2 N1–25 and the G3BP1NTF2 domain as the major elements for the SARS-CoV-2 N protein − G3BP1 interaction, whereas the CTD and/or IDR2 of SARS-CoV-2 N protein contribute to the interaction entropically.
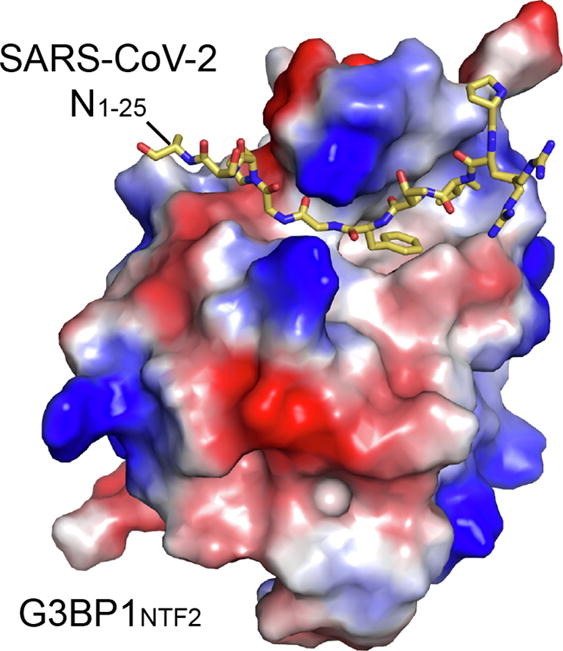
Crystal structure of the SARS-CoV-2 N1–25 − G3BP1NTF2 complex
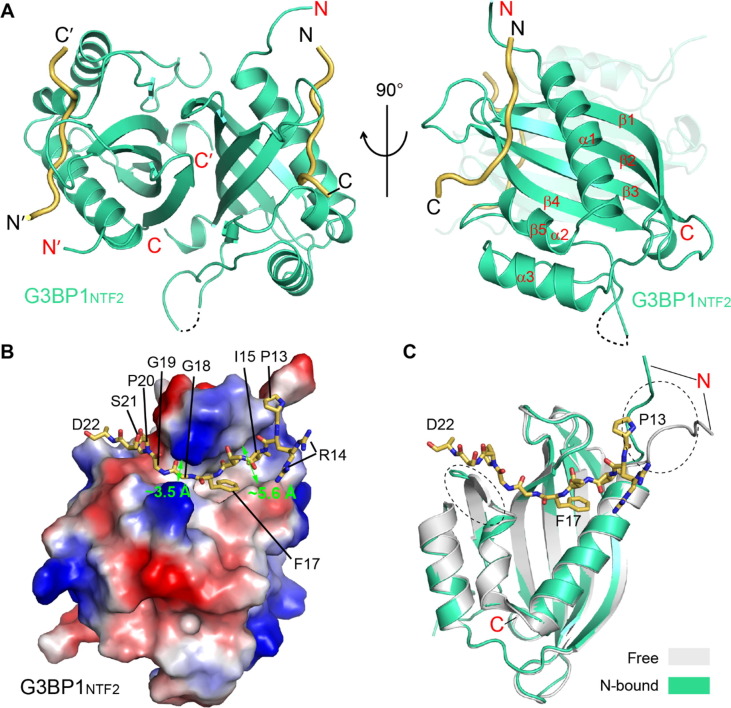
We then solved the crystal structure of the G3BP1NTF2 domain bound to the SARS-CoV-2 N1–25 peptide at 2.35 Å resolution (Figure 2 (A) and Table S2). The SARS-CoV-2 N1–25 − G3BP1NTF2 complex belongs to the P212121 space group, with each asymmetric unit containing a G3BP1NTF2 homodimer bound to the N1–25 peptide in a G3BP1NTF2:N1–25 ratio of 2:2. We were able to trace nearly the entire G3BP1NTF2 domain, spanning residues V2-F138 (except for S47) (Figure 2(A)), as well as residues P13-D22 of the SARS-CoV-2 N1–25 peptide (Figure S2(A)). Note that this segment of SARS-CoV-2 N is strictly conserved in SARS-CoV N, but not in the counterparts of related Middle East Respiratory Syndrome Coronavirus (MERS-CoV) or other coronaviruses (Figure S2(B)).
Figure 2.
Structural overview of the SARS-CoV-2 N1–25 − G3BP1NTF2 complex. (A) Orthogonal views of the SARS-CoV-2 N1–25 − G3BP1NTF2 complex, with SARS-CoV-2 N1–25 colored in yellow orange and G3BP1NTF2 colored in green. The traceable N- and C-termini of SARS-CoV-2 N1–25 or G3BP1NTF2 are labeled with “N” and “C”, respectively. The region (residue S47) with untraceable electron density is shown in dashed line. (B) Electrostatic surface view of the G3BP1NTF2 domain bound to the SARS-CoV-2 N1–25 peptide (stick representation). The widths of two distinct groove regions of G3BP1NTF2 are marked. For clarity, only one monomer of the G3BP1NTF2 homodimer is shown. (C) Structural overlay of the G3BP1NTF2 domain, free (grey) and in complex with SARS-CoV-2 N1–25 (green). The two structurally divergent regions are circled with dotted lines.
As previously characterized,26, 29, 38 the G3BP1NTF2 domain is dominated by a five-stranded antiparallel β-sheet, preceded by three α-helices packed on one side of the β-sheet (Figure 2(A)). Two of the G3BP1NTF2 domains further undergo a face-to-face β-sheet stacking with each other to form a homodimer. On the outer face of the β-sheet of each G3BP1NTF2 monomer, the α1- and α2-helices join with the loop connecting β3 and β4 (lβ3β4) to form a long surface groove, cradling the extended SARS-CoV-2 N1–25 peptide (Figure 2(B)). The association of SARS-CoV-2 N1–25 peptide with the G3BP1NTF2 is underpinned by their strong surface complementarity (Figure 2(B)). Notably, the bulky side chains of N1–25 I15 and F17 are accommodated by a ∼5.6 Å-wide groove, whereas the downstream residues G18 and G19 snug into a ∼3.5 Å-wide groove (Figure 2(B)). In fact, our structural modeling analysis indicated that replacement of N1–25 G18 with bigger-sized threonine would lead to a steric clash with G3BP1NTF2 K123 (Figure S2(C)). Together, the association of SARS-CoV-2 N1–25 with G3BP1NTF2 results in a buried surface area of ∼571 Å2. Structural superposition of the SARS-CoV-2 N1–25-bound G3BP1NTF2 with the previously reported apo form38 gives a root-mean-square deviation (RMSD) of 0.43 Å over 225 aligned Cα atoms (Figure 2(C)), indicative of high similarity. The most divergent regions include the N-terminal tail and the loop connecting α2 and β2, both of which are involved in the interaction with the N1–25 peptide (Figure 2(C)). These observations highlight that the G3BP1NTF2 domain adopts a preconfigured conformation for the interaction with N1–25 peptide.
Structural details of the SARS-CoV-2 N1–25 − G3BP1NTF2 interaction
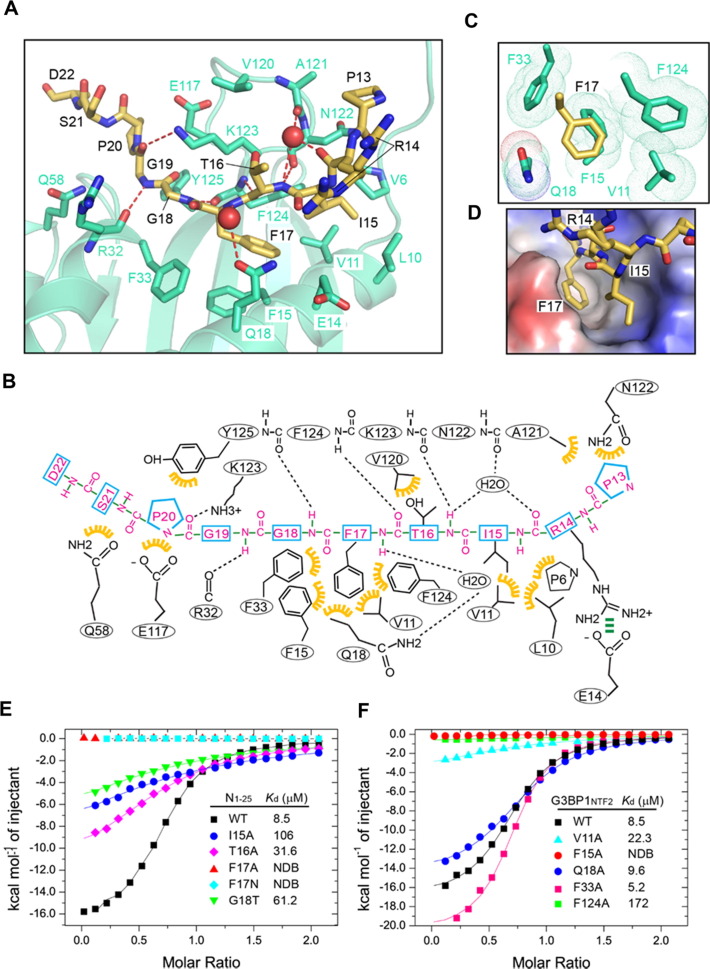
The assembly of G3BP1NTF2 with SARS-CoV-2 N1–25 is mediated by a network of hydrogen-bonding and van der Waals contacts (Figure 3 (A, B)). Of note, the aromatic ring of SARS-CoV-2 N1–25 F17 inserts into the hydrophobic cavity formed by the side chains of G3BP1NTF2 V11, F15, Q18, F33 and F124 (Figure 3(A–D)). The side chain of N1–25 I15 is positioned in parallel with that of N1–25 F17, engaging in nonpolar contacts with the side chains of G3BP1NTF2 P6, L10 and V11 (Figure 3(B, D)). The side chains of N1–25 P13, R14 and T16 point away from G3BP1NTF2, with N1–25 P13 surrounded by the side chains of G3BP1NTF2 A121 and N122, the guanidinium of N1–25 R14 positioned within a distance for an electrostatic contact with the sidechain carboxylate of G3BP1NTF2 E14, and N1–25 T16 in proximity with the side chain of G3BP1NTF2 V120 (Figure 3(A, B)). Along one side of the groove, the N1–25 I15-G18 segment pairs in parallel with the C-terminal end of G3BP1NTF2 β5 (residues A121-Y125), involving both direct and water-mediated main chain hydrogen bonds (Figure 3(A, B)). On the other side of the groove, G3BP1NTF2 Q18, R32 and K123 form water-mediated or direct hydrogen bonds with the backbone of N1–25 F17, G18 and G19, respectively (Figure 3(A, B)). Additional intermolecular interactions involve the van der Waals contacts between the backbone of N1–25 G18-S21 and the side chains of G3BP1NTF2 F33, Q58, E117 and Y125 (Figure 3(A, B)).
Figure 3.
Structural and biochemical characterizations of the SARS-CoV-2 N1–25 − G3BP1NTF2 interaction. (A) Close-up view of the SARS-CoV-2 N1–25 − G3BP1NTF2 interaction. The interacting residues are shown in stick representation. The hydrogen bonds are shown as dashed lines. The water molecules are shown as red spheres. (B) Schematic view of the SARS-CoV-2 N1–25 − G3BP1NTF2 interaction. Hydrogen bonds and electrostatic interactions are indicated by black and green dashed lines, respectively, and van der Waals contacts are indicated by yellow gears. (C) Close-up view of the hydrophobic pocket harboring residue F17 of SARS-CoV-2 N1–25. The van der Waals radii of the G3BP1NTF2 residues are shown in dots. (D) Close-up view of the electrostatic surface of G3BP1NTF2 harboring the side chains of I15 and F17 of SARS-CoV-2 N1–25. (E) ITC binding assays for wild-type G3BP1NTF2 titrated with wild-type or mutant SARS-CoV-2 N1–25 peptide. (F) ITC binding assays for wild-type or mutant G3BP1NTF2 titrated with the wild-type SARS-CoV-2 N1–25 peptide. NDB, no detectable binding.
To test the structural observation, we selected several key SARS-CoV-2 N1–25 − G3BP1NTF2 interacting residues for mutagenesis and performed ITC binding assays. Mutation of N1–25 I15 or T16 each to alanine reduced the binding by > 10-fold and ∼3-fold, respectively (Figure 3(E) and Figure S3(A, B)). Mutation of N1–25 F17 to alanine or asparagine abolished the interaction between SARS-CoV-2 N1–25 and G3BP1NTF2 (Figure 3(E) and Figure S3(C, D)), supporting the role of these residues in the G3BP1NTF2 interaction. Furthermore, we observed that introducing the N1–25 G18T mutation led to ∼6-fold reduction of the binding affinity (Figure S3(E)), supporting the notion that surface complementarity underpins the specific interaction between SARS-CoV-2 N1–25 and G3BP1NTF2. Conversely, we also observed impairment of the SARS-CoV-2 N1–25 − G3BP1NTF2 interaction by several G3BP1NTF2 mutations: Introducing G3BP1NTF2 V11A and F124A mutations reduced the binding by ∼10- and ∼20-fold, respectively, while introducing the G3BP1NTF2 F15A mutation abolished the binding.
It is worth noting that introduction of the G3BP1NTF2 Q18A or F33A mutations led to no appreciable change in binding (for Q18A) or even slightly enhanced the binding affinity (for F33A). The G3BP1NTF2 Q18A mutation modestly reduced both the enthalpy (ΔH = −15.1 kcal/mol for Q18A vs −17.6 kcal/mol for wild-type) and entropy (ΔS = −27.8 cal/mol/degree for Q18A vs −35.0 cal/mol/degree for wild-type) effects, whereas the G3BP1NTF2 F33A mutation led to an increase of both enthalpy (ΔH = −21.3 kcal/mol for Q18A vs −17.6 kcal/mol for wild-type) and entropy (ΔS = −51.6 cal/mol/degree for F33A vs −35.0 cal/mol/degree for wild-type) effects. These enthalpy–entropy compensation effects, the origin of which is currently unclear, suggest a certain extent of structural plasticity of the N1–25 F17-binding pocket of the G3BP1NTF2 domain.
Distinct intermolecular interaction mechanisms among G3BP1 complexes
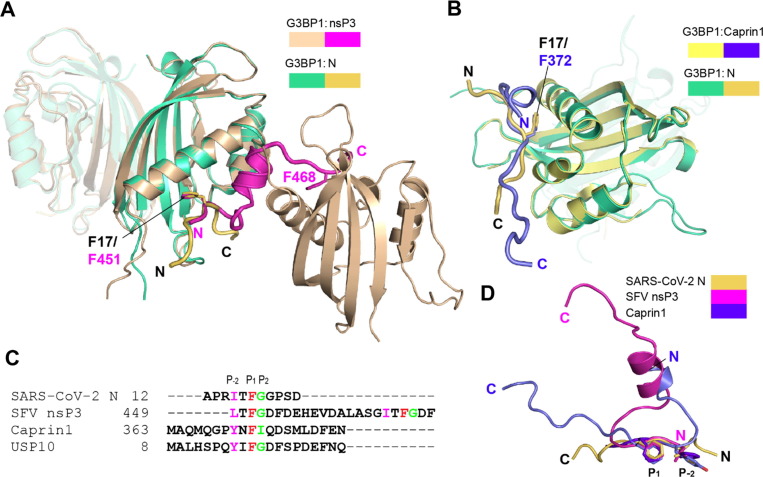
Structural comparison of the G3BP1NTF2 domain bound to SARS-CoV-2 N1–25 with that bound to a peptide derived from the SFV nsP3 protein (residues 449–471, nsP3449–471) or a peptide derived from Caprin1 (residues 363–382, Caprin1363–382) reveals different binding stoichiometry: In the SFV nsP3449–471 − G3BP1NTF2 complex, two FGDF motifs of SFV nsP3 each bind to one G3BP1NTF2 homodimer, thereby tethering the G3BP1 molecules into a poly-complex26; in contrast, in the SARS-CoV-2 N1–25 − G3BP1NTF2 and Caprin1363–382 − G3BP1NTF2 complexes, the G3BP1NTF2 domain binds to the N1–25 or Caprin1363–382 peptide in a 2:2 binding mode (Figure 4 (A, B)). Nevertheless, residues I15-G18 of N1–25, L449-G452 of the nsP3449–471 peptide and Y370-I373 of the Caprin1363–382 peptide are anchored to the surface groove of the G3BP1NTF2 domain in a similar fashion (Figure 4(A–C) and Figure S4(A)), with the aromatic rings of N1–25 F17, nsP13449–471 F451 and Caprin1363–382 F372 embraced by the same aromatic cage of the G3BP1NTF2 domain (Figure 4(A, B) and Figure S4(A, B)). In addition, nsP3449–471 L449 and Caprin363–382 Y370 insert their side chains into the groove similarly as the corresponding N1–25 I15 (Figure 4(C, D)). Beyond this region, nsP3449–471 and Caprin1363–382 interact with the G3BP1NTF2 domain in a distinct mode than that of N1–25 (Figure 4(A, B)). Unlike the N1–25 peptide that occupies the entire groove, the nsP3449-471 peptide exits from the surface groove at the D453 site to engage in a secondary contact with the α1-helix of G3BP1NTF2 via helical packing (Figure 4(A) vs Figure 2(B)). Subsequently, the C-terminal segment of nsP3449-471 extends into another G3BP1NTF2 homodimer and presents residue F468 for a groove-insertion interaction like that of F452 (Figure 4(A)). Similar to nsP3449–471, Caprin363–382 forms a helical turn at its N-terminal end to interact with the α1-helix of the G3BP1NTF2 domain (Figure 4(B)), albeit in an opposite direction (Figure 4(D)).
Figure 4.
Structural comparison of SARS-CoV-2 N1–25 − G3BP1NTF2 with other G3BP1NTF2 complexes. (A) Structural overlay between the SARS-CoV-2 N1–25 (yellow orange) − G3BP1NTF2 (green) and the SFV nsP3449–471 (magenta) − G3BP1NTF2 (wheat) complex. The phenylalanine residues inserting into the hydrophobic pocket of G3BP1NTF2 are labeled and shown in stick representation. (B) Structural overlay between the SARS-CoV-2 N1–25 (yellow orange) − G3BP1NTF2 (green) and the Caprin1363–382 (slate) − G3BP1NTF2 (yellow) complex. The phenylalanine residues inserting into the hydrophobic pocket of G3BP1NTF2 are labeled and shown in stick representation. (C) Sequence alignment of the G3BP1NTF2-interacting peptides, with the P-2, P1 and P2 sites colored in magenta, red and green, respectively. (D) Structural alignment of the G3BP1NTF2-interacting peptides, with the P–2 and P1 sites, as well as the N- and C-termini, labeled.
Together, our structural and sequence analyses of the G3BP1NTF2-binding peptides reveal a φxF (φ denotes a hydrophobic amino acid) consensus motif, in which an invariable phenylalanine (designated as P1 site) and a bulky hydrophobic amino acid at the P–2 site dominate the G3BP1NTF2 binding via a groove-insertion mechanism (Figure 4(C, D)). In addition, the P2 site is populated with a small amino acid (e.g. glycine), except for that in the Caprin1363–382 peptide, which contains an isoleucine (I373), coinciding with its structural divergence from the SARS-CoV-2 N1–25 and nsP3449–471 peptides (Figure 4(B, D)). The diverse sequence composition at the φxF-flanking regions introduces secondary protein interactions underpinning various binding outcomes of the G3BP1NTF2-interaction partners. Along the line, a recent study reported that residues K36, K50, K59 and K64 of G3BP1 are subjected to ubiquitination in response to heat shock in cells, leading to disassembly and autophagy-independent degradation of SG.39 Structural inspection of the SARS-CoV-2 N1–25 − G3BP1NTF2, SFV nsP3449-471 − G3BP1NTF2 and Caprin1363–382 − G3BP1NTF2 complexes indicate that the potential ubiquitination sites of G3BP1 are largely exposed in these complexes (Figure S4(C–E)). One exception lies in G3BP1 K59 in the Caprin1363–382 complex, which becomes partially shielded from solvent by the Caprin1363–382 binding (Figure S4(E)). These observations imply that the Caprin1 binding may affect the ubiquitylation of G3BP1 differently than the SARS-CoV-2 N1–25 and SFV nsP3449–471 bindings.
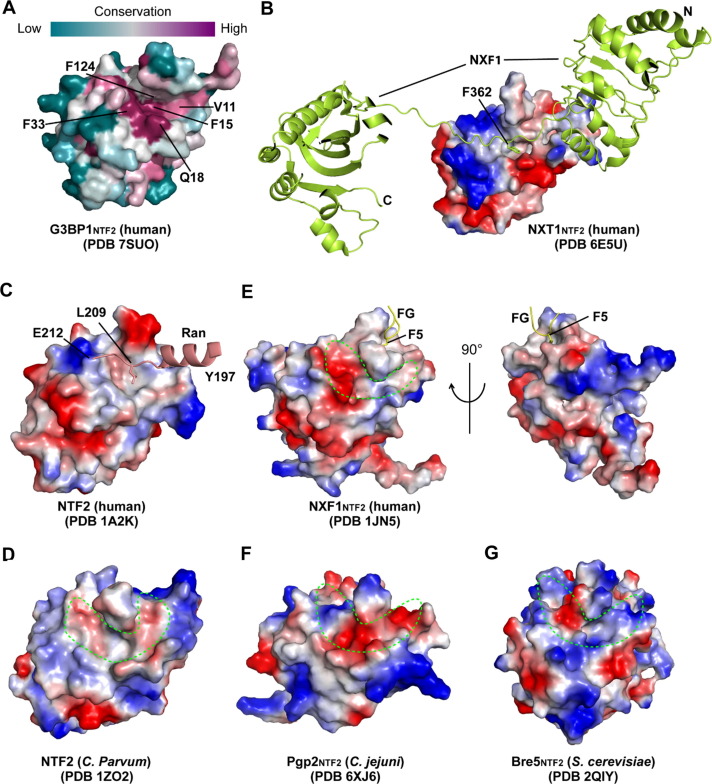
The SARS-CoV-2 N1–25-binding groove is recurrently present in NTF2 domains
The NTF2 domain is present in a wide array of proteins from diverse species.40 Sequence analysis of the G3BP1NTF2 domain from selected model species (Figure S5(A)) or using the ConSurf server41 reveals that the SARS-CoV-2 N1–25-binding groove evolves more slowly than the surrounding area, with the N1–25 F17-binding site falling into the most conserved region (Figure 5 (A)). Furthermore, structural survey of the 19 NTF2-containing proteins in the protein data bank reveals that the surface groove is evolutionarily preserved in the NTF2 domain of many other proteins. For instance, the NTF2 domain of nuclear transport factor 2-related export protein 1 (NXT1NTF2) forms a surface groove in the corresponding region to harbor the linker sequence connecting the leucine-rich repeat (LRR) and NTF2 domains of nuclear RNA export factor 1 (NXF1), resulting in formation of a heterodimer for mRNA export (Figure 5(B)).42 Likewise, human NTF2 protein and its counterpart from C. parvum, the founding members of the NTF2 family, also form a surface groove in the corresponding region (Figure 5(C, D)).43, 44 As with the G3BP1NTF2 domain, the NXT1 and NTF2 proteins form a hydrophobic cavity at the center of the groove, harboring NXF1 F362 and Ran L209, respectively (Figure 5(B, C)). These observations suggest groove insertion as a conserved interaction mechanism for the NTF2 domains.
Figure 5.
Structural evolution of the NTF2 domains. (A) Color-coded sequence conservation of the G3BP1NTF2 domain, analyzed using the ConSurf server (https://consurf.tau.ac.il). (B) Electrostatic surface of the NXT1NTF2 domain bound to the LRR (NXF1LRR)-NTF2 (NXF1NTF2) domain linker of the NXF1 protein (limon), with the side chain of NXF1 F362 shown in stick representation. (C) Electrostatic surface of human NTF2 domain bound to residues 197–212 of one Ran molecule (light pink), with the side chain of Ran L209 shown in stick representation. For clarity, the rest of Ran structure is not shown. (D-G) Electrostatic surface of C. parvum NTF2 (D), human NXF1NTF2 (E), C. jejuni Pgp2NTF2 (F) and S. cerevisiae Bre5NTF2 (G), with the individual surface grooves circled by dotting lines. Note that human NXF1NTF2 binds to the FG peptide through a hydrophobic pocket positioned separately from the groove.
A similar surface groove is formed in the NTF2 domain of some other proteins, such as human NXF1, C. jejuni PgP2 and S. cerevisiae Bre5 (Figure 5(E–G)).28, 45, 46 However, the electrostatic and steric properties of these grooves diverge substantially from that of the G3BP1NTF2 domain (Figure 5(E–G)), implying different interaction mechanisms associated with the groove. Indeed, it was shown that the NXF1NTF2 domain binds to the phenylalanine (F5) of an FG peptide derived from the nucleoporins (NUPs) repeat via a hydrophobic pocket distant to the surface groove corresponding to the N1–25-binding site of the G3BP1NTF2 domain (Figure 5(E)).28
Taken together, these observations suggest that the SARS-CoV-2 N1–25-binding groove serves as a recurrent structural feature of the NTF2 domain, which is subjected to electrostatic and steric fine tuning for diverse protein interaction behaviors.
Discussion
SARS-CoV-2 N becomes an increasingly attractive drug target not only because it plays a multifaceted role in packing of viral genome, virion assembly and viral transcription4, 47, but also due to the fact that it helps break host defense through inhibition of the G3BP1/2-mediated SG formation in cells.7 Development of an effective therapeutic strategy against this viral factor necessitates a detailed understanding of its molecular mechanism. Through combined structural and biochemical analyses, this study establishes the structural basis for the interaction between SARS-CoV-2 N and the G3BP1NTF2 domain, providing a framework for the development of novel therapeutic strategies against COVID-19 and related viral infection.
Molecular basis for the inhibition of G3BP1-mediated SG formation by SARS-CoV-2 N protein
The G3BP1-mediated SG formation is underpinned by a heterologous network of interactions involving the protein contacts mediated by the G3BP1NTF2 domain and the RNA bindings mediated by the G3BP1RRM domain.10, 11, 12, 13, 14, 15 As proposed in a network-based model,19 the interaction of G3BP1NTF2 with the “bridge” protein Caprin1, which possesses both protein and RNA binding-modules, creates multiple valences for the interaction network, thereby promoting the SG assembly.19 In contrast, the interaction with USP10, which is classified as “valence cap” due to lack of multivalent interaction module, serves to halt the propagation of the interaction network inside the SG.20 Surprisingly, the interaction of SARS-CoV-2 N with G3BP1 leads to inhibition of G3BP1-mediated liquid–liquid phase separation in vitro and in cells,7, 37 even though the former contains both protein and RNA interaction modules.
Through fragment-based ITC binding assays, this study identified that the interaction between SARS-CoV-2 N and G3BP1 is primarily mediated by N1–25 and the G3BP1NTF2 domain. In addition, the C-terminal regions of SARS-CoV-2 N, including IDR2 and/or CTD, contribute to the interaction by reducing the entropic cost of the complex formation. Note that both G3BP1 and SARS-CoV-2 N assume a dimeric form in solution, which presumably contributes to the fact that full-length SARS-CoV-2 N protein shows a higher G3BP1-binding affinity than N1–25.The observation that both the N- and C-terminal regions of SARS-CoV-2 N contribute to the G3BP1 binding further suggests a multivalent engagement between SARS-CoV-2 N and G3BP1, by which the C-terminal region of SARS-CoV-2 N-mediated secondary interaction may interfere with the SG-nucleating activity of G3BP1. In support of this notion, a recent study showed that the SARS-CoV-2 N1–175 fragment exhibits a reduced SG inhibition activity than full-length SARS-CoV-2 N.7 In addition, given the fact that the SARS-CoV-2 N and Caprin1 interact with G3BP1 in a mutually exclusive manner, it is conceivable that the interaction between SARS-CoV-2 N1–25 and G3BP1NTF2 also serves to inhibit the association of G3BP1 with Caprin1, providing another mechanism in attenuating the SG formation. A detailed mechanism by which SARS-CoV-2 N protein inhibits G3BP1-mediated SG formation remains to be investigated.
A groove-insertion mechanism dictating the SARS-CoV-2 N1–25 − G3BP1NTF2 interaction
The structure of the SARS-CoV-2 N1–25 − G3BP1NTF2 complex reveals strong surface complementarity between residues 13–22 of SARS-CoV-2 N1–25 and the surface groove of G3BP1NTF2. Most notably, the P1-F17 of SARS-CoV-2 N1–25 inserts its aromatic ring into a hydrophobic pocket at the center of the surface groove, which is flanked by a parallel sidechain insertion of P–2-I15. This dual groove-insertion mode is reminiscent of what was previously observed for the complexes of G3BP1NTF2 with SFV nsP3 and Caprin1,26, 29 attesting the φxF motif as a primary determinant for the G3BP1NTF2-mediated protein interaction. In addition, the embedding of N1–25 P2-G18 in a narrow region of the surface groove permits a parallel pairing between N1–25 I15-G18 and G3BP1NTF2 β5-strand, in line with the previous observation that a P2-glycine is favored by the G3BP1NTF2 domain.21, 26 Note that these G3BP1-binding sites of N1–25 is highly conserved among different variants of SARS-CoV-2 reported to date,48 suggesting that the SARS-CoV-2 N1–25 − G3BP1NTF2 interaction might constitute a critical factor for SARS-CoV-2 infection.
Unlike the Caprin1 and SFV nsP3 peptides that diverge their φxF-flanking regions out of the surface groove to engage helical packing with the α1-helix of G3BP1NTF2, the φxF-flanking regions of SARS-CoV-2 N1–25 peptide remain bound to the groove. Through surface complementarity, residues P13-R14 and G19-S21 of N1–25 are anchored to the two ends of the groove, engaging van der Waals and/or hydrogen-bonding interactions with G3BP1NTF2, respectively. These secondary binding sites in the SARS-CoV-2 N1–25 − G3BP1NTF2 complex may serve as attractive targets for the development of therapeutic agents that selectively inhibit the SARS-CoV-2 N − G3BP1 association.
NTF2 domain, a platform for diverse protein–protein interactions
The NTF2 domain represents an evolutionarily divergent protein interaction module involved in various cellular activities, such as SG formation7, 37, 10, 11, 12, 13, 14, 15 and mRNA transport.42 This study, through structural survey of the NTF2 domains from diverse proteins, uncovers that the surface groove of G3BP1NTF2 represents a recurrent feature of the NTF2 domains. Common to many of these NTF2 domains is the formation of a hydrophobic cavity at the center of the groove, which provides a primary docking site for a bulky, hydrophobic side chain from host or viral factors. On the other hand, the NTF2-interacting proteins, such as SARS-CoV-2 N, Caprin1 and NXF1, often extend beyond the hydrophobic pocket for secondary contacts, which presumably contributes to their distinct binding affinity and specificity. In this context, the structure of the SARS-CoV-2 N1–25 − G3BP1NTF2 complex not only provides a framework for understanding how SARS-CoV N and SARS-CoV-2 N have evolved to target the G3BP1/2, but also provides a basis for identification of other host factors that can potentially be targeted by the SARS-CoV-2 N.
Experimental procedures
Cloning, expression and purification of proteins
The DNA fragment encoding SARS-CoV-2 N, codon optimized for bacterial expression, was synthesized from Thermo Fisher Scientific. The cDNAs for full-length human G3BP1 and G3BP2 were purchased from DNASU plasmid repository (https://dnasu.org). The SARS-CoV-2 N1–25 peptide was cloned in a modified pRSF vector, in which N1–25 was preceded by an N-terminal His6-SUMO tag and ULP1 (ubiquitin-like protease) cleavage site. Full-length SARS-CoV-2 N, full-length G3BP1, G3BP1NTF2 (residues 1–139) and G3BP2NTF2 (residues 1–139) were cloned into an in-house His6-MBP vector, preceded by a TEV cleavage site. The plasmids were transformed into BL21 RIL (DE3) cell strain (Agilent technologies) for protein expression. The transformed cells were first grown at 37 °C until cells attained an A 600 of 0.8. The temperature was then lowered to 16 °C, after which the cells were induced by addition of 0.1 mM isopropyl β-D-galactoside and continued to grow overnight. For His6-SUMO-tagged N1–25, the fusion protein was purified using a Ni-NTA affinity column, followed by removal of the His6-SUMO tag with ULP1 protease and further purification by size-exclusion chromatography on a HiLoad 16/600 Superdex 75 pg column (GE Healthcare) pre equilibrated with 25 mM Hepes (pH 7.5), 0.3 M NaCl and 2 mM DTT. His6-MBP-tagged full-length G3BP1, G3BP1NTF2 and G3BP2NTF2 proteins were purified sequentially through Ni-NTA chromatography, Q HP column (GE Healthcare) followed by TEV protease treatment. Samples were then subjected to a second round of Ni-NTA chromatography for tag removal. Finally, the proteins were purified by size-exclusion chromatography on a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) pre equilibrated with 25 mM Hepes (pH 7.5), 300 mM NaCl, and 2 mM DTT. The purified proteins were confirmed by SDS-PAGE, concentrated to ∼20 mg/mL and stored at −80 °C for further use.
Crystallization, X-ray data collection and structure determination
For crystallization, ∼6 mg/mL G3BP1NTF2 protein dissolved in 25 mM Hepes (pH7.5), 300 mM NaCl and 2 mM DTT was mixed with the N1–25 peptide in a 1:2 molar ratio. The crystallization condition for the SARS-CoV-2 N1–25 − G3BP1NTF2 complex was initially identified through sparse-matrix screening (Hampton Research Inc.) at 4 °C. The crystals were then reproduced by hanging-drop vapor diffusion method under the same temperature, from drops mixed from 1 μL of SARS-CoV-2 N1–25 − G3BP1NTF2 complex and 1 μL of precipitant solution containing 20% propan-2-ol, 0.1 M MES monohydrate (pH 6.0), 20% PEG MME 20,000. Crystals were soaked for one minute in a cryoprotectant solution, comprised of crystallization buffer and 30% glycerol, before flash frozen in liquid nitrogen. The X-ray diffraction data for the SARS-CoV-2 N1–25 − G3BP1NTF2 complex were collected on the BL 5.0.3 beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory. The diffraction data were indexed, integrated and scaled using the HKL3000 program49. The structure was solved using the molecular replacement method in PHASER50 with the structure of human G3BP1NTF2 in complex with SFV nsP3449–471 (PDB ID: 5FW5) as search model. The structure was improved by iterative model building and refinement with Coot51 and PHENIX52 software packages. The same R-free test set was used throughout the refinement. The statistics for data collection and structural refinement of the SARS-CoV-2 N1–25 − G3BP1NTF2 complex is summarized in Table S2.
Isothermal titration calorimetry
A MicroCal iTC200 system (GE Healthcare) was used to conduct the ITC measurements. All proteins and peptides were dialyzed against buffer containing 25 mM Hepes (pH 7.5) and 300 mM NaCl before titration. For titration of G3BP1NTF2 or G3BP2NTF2 with SARS-CoV-2 N1–25, 0.1 mM human G3BP1NTF2 or G3BP2NTF2 and 1 mM SARS-CoV-2 N1–25 were used. For the rest of titrations, 0.03 mM full-length G3BP1 and 0.3 mM SARS-CoV-2 N, full-length or fragments, were used. A total of 20 injections with a spacing of 180 s and a reference power of 5 μcal/s were performed at 25 °C. The ITC curves were processed with ORIGIN (MicroCal) software by using a one-site fitting model.
CRediT authorship contribution statement
Mahamaya Biswal: Conceptualization, Investigation, Visualization, Data curation. Jiuwei Lu: Investigation. Jikui Song: Conceptualization, Supervision, Investigation, Visualization, Writing – review & editing.
Acknowledgments
Acknowledgments
We thank staff members at the Advanced Light Source (DE-AC02-05CH11231), Lawrence Berkeley National Laboratory for access to X-ray beamlines. We thank Jianbin Chen for assistance in protein purification.
Funding
This work was supported by NIH grants 1R21AI147057, R35GM119721, and R01AI153419.
Conflict of Interest
The authors declare no competing interest.
Edited by Michael Summers
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2022.167516.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
Coordinates and structure factors for the SARS-CoV-2 N1–25−G3BP1NTF2 complex have been deposited in the Protein Data Bank under accession code 7SUO.
References
- 1.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein–forms and functions. Antiviral Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., et al. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X., Verma A., Garcia G., Jr., Ramage H., Lucas A., Myers R.L., Michaelson J.J., Coryell W., et al. Targeting the coronavirus nucleocapsid protein through GSK-3 inhibition. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2113401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo L., Li Z., Zhao T., Ju X., Ma P., Jin B., Zhou Y., He S., et al. SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production. Science bulletin. 2021;66:1194–1204. doi: 10.1016/j.scib.2021.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchan J.R., Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White J.P., Lloyd R.E. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillen-Boixet J., Kopach A., Holehouse A.S., Wittmann S., Jahnel M., Schlussler R., Kim K., Trussina I., et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.049. 346–361 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bley N., Lederer M., Pfalz B., Reinke C., Fuchs T., Glass M., Moller B., Huttelmaier S. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuki H., Takahashi M., Higuchi M., Makokha G.N., Oie M., Fujii M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells: Devoted Mol. Cell. Mech. 2013;18:135–146. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- 14.Yang P., Mathieu C., Kolaitis R.M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.046. 325–345 e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tourriere H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.McCormick C., Khaperskyy D.A. Translation inhibition and stress granules in the antiviral immune response. Nature Rev. Immunol. 2017;17:647–660. doi: 10.1038/nri.2017.63. [DOI] [PubMed] [Google Scholar]
- 17.Parker F., Maurier F., Delumeau I., Duchesne M., Faucher D., Debussche L., Dugue A., Schweighoffer F., et al. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol. Cell. Biol. 1996;16:2561–2569. doi: 10.1128/mcb.16.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann S., Kedersha N., Anderson P., Ivanov P. Molecular mechanisms of stress granule assembly and disassembly. Biochim. Biophys. Acta, Mol. Cell. Res. 2021;1868 doi: 10.1016/j.bbamcr.2020.118876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders D.W., Kedersha N., Lee D.S.W., Strom A.R., Drake V., Riback J.A., Bracha D., Eeftens J.M., et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.050. 306–324 e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panas M.D., Varjak M., Lulla A., Eng K.E., Merits A., Karlsson Hedestam G.B., McInerney G.M. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol. Biol. Cell. 2012;23:4701–4712. doi: 10.1091/mbc.E12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Reineke L.C., Lloyd R.E. Diversion of stress granules and P-bodies during viral infection. Virology. 2013;436:255–267. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pager C.T., Schutz S., Abraham T.M., Luo G., Sarnow P. Modulation of hepatitis C virus RNA abundance and virus release by dispersion of processing bodies and enrichment of stress granules. Virology. 2013;435:472–484. doi: 10.1016/j.virol.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fros J.J., Domeradzka N.E., Baggen J., Geertsema C., Flipse J., Vlak J.M., Pijlman G.P. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J. Virol. 2012;86:10873–10879. doi: 10.1128/JVI.01506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulte T., Liu L., Panas M.D., Thaa B., Dickson N., Gotte B., Achour A., McInerney G.M. Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open Biol. 2016;6 doi: 10.1098/rsob.160078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savastano A., Ibanez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nature Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fribourg S., Braun I.C., Izaurralde E., Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- 29.Schulte T., Panas M.D., Williams L., Kedersha N., Fleck J.S., Tan T.J.C., Olsson A., Morro A.M., et al. Caprin-1 binding to the critical stress granule protein G3BP1 is regulated by pH. bioRxiv. 2021 doi: 10.1098/rsob.220369. 2021.2002.2005.429362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou R., Zeng R., von Brunn A., Lei J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid protein. Mol. Biomed. 2020;1:2. doi: 10.1186/s43556-020-00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng Y., Du N., Lei Y., Dorje S., Qi J., Luo T., Gao G.F., Song H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39 doi: 10.15252/embj.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R., Villa E., Cleveland D.W., Corbett K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nature Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng T.Y., Lee K.R., Tarn W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruse T., Benz C., Garvanska D.H., Lindqvist R., Mihalic F., Coscia F., Inturi R.T., Sayadi A., et al. Large scale discovery of coronavirus-host factor protein interaction motifs reveals SARS-CoV-2 specific mechanisms and vulnerabilities. bioRxiv. 2021 doi: 10.1038/s41467-021-26498-z. 2021.2004.2019.440086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W., Ju X., Tian M., Li X., Yu Y., Sun Q., Ding Q., Jia D. Molecular determinants for regulation of G3BP1/2 phase separation by the SARS-CoV-2 nucleocapsid protein. Cell Discovery. 2021;7:69. doi: 10.1038/s41421-021-00306-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vognsen T., Moller I.R., Kristensen O. Crystal structures of the human G3BP1 NTF2-like domain visualize FxFG Nup repeat specificity. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0080947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gwon Y., Maxwell B.A., Kolaitis R.M., Zhang P., Kim H.J., Taylor J.P. Ubiquitination of G3BP1 mediates stress granule disassembly in a context-specific manner. Science. 2021;372:eabf6548. doi: 10.1126/science.abf6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eberhardt R.Y., Chang Y., Bateman A., Murzin A.G., Axelrod H.L., Hwang W.C., Aravind L. Filling out the structural map of the NTF2-like superfamily. BMC Bioinf. 2013;14:327. doi: 10.1186/1471-2105-14-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., Ben-Tal N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K., Xie Y., Muñoz-Moreno R., Wang J., Zhang L., Esparza M., García-Sastre A., Fontoura B.M.A., et al. Structural basis for influenza virus NS1 protein block of mRNA nuclear export. Nature Microbiol. 2019;4:1671–1679. doi: 10.1038/s41564-019-0482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart M., Kent H.M., McCoy A.J. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J. Mol. Biol. 1998;277:635–646. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- 44.Vedadi M., Lew J., Artz J., Amani M., Zhao Y., Dong A., Wasney G.A., Gao M., et al. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Li K., Ossareh-Nazari B., Liu X., Dargemont C., Marmorstein R. Molecular basis for bre5 cofactor recognition by the ubp3 deubiquitylating enzyme. J. Mol. Biol. 2007;372:194–204. doi: 10.1016/j.jmb.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin C.S., Chan A.C.K., Vermeulen J., Brockerman J., Soni A.S., Tanner M.E., Gaynor E.C., McIntosh L.P., et al. Peptidoglycan binding by a pocket on the accessory NTF2-domain of Pgp2 directs helical cell shape of Campylobacter jejuni. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai Z., Cao Y., Liu W., Li J. The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses. 2021;13 doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohammad T., Choudhury A., Habib I., Asrani P., Mathur Y., Umair M., Anjum F., Shafie A., et al. Genomic Variations in the Structural Proteins of SARS-CoV-2 and Their Deleterious Impact on Pathogenesis: A Comparative Genomics Approach. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.765039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 50.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the SARS-CoV-2 N1–25−G3BP1NTF2 complex have been deposited in the Protein Data Bank under accession code 7SUO.