SUMMARY
Nuclear receptor binding SET domain protein 3 (NSD3), a gene located within the 8p11-p12 amplicon frequently detected in human cancers, encodes a chromatin modulator and an attractive onco-target. However, agents that effectively suppress NSD3-mediated oncogenic actions are currently lacking. We report the NSD3-targeting proteolysis targeting chimera (PROTAC), MS9715, which achieves effective and specific targeting of NSD3 and associated cMyc node in tumor cells. MS9715 is designed by linking BI-9321, an NSD3 antagonist, which binds NSD3’s PWWP1 domain, with an E3 ligase VHL ligand. Importantly, MS9715, but not BI-9321, effectively suppresses growth of NSD3-dependent hematological cancer cells. Transcriptome profiling demonstrates that MS9715, but not BI-9321, effectively suppresses NSD3- and cMyc-associated gene-expression programs, resembling effects of the CRISPR/Cas9-mediated knockout of NSD3. Collectively, these results suggest that pharmacological degradation of NSD3 as an attractive therapeutic strategy, which co-suppresses NSD3- and cMyc-related oncogenic nodes, is superior to blocking the PWWP1 domain of NSD3.
Keywords: Chromatin, histone, NSD3, cMyc, epigenetics, PROTAC, degrader
eTOC blurb
Xu et al. report the discovery and characterization of the NSD3 PROTAC, MS9715, which effectively and selectively depletes NSD3 and the associated cMyc oncogenic node. We provide evidence that pharmacological degradation of NSD3 is a superior therapeutic strategy to pharmacological inhibition of NSD3 for treating NSD3-dependent cancers.
Graphical Abstract

INTRODUCTION
Aberrations of chromatin modulators represent a central oncogenic pathway and targeting these aberrations may provide an attractive antitumor strategy (Chi et al., 2010; Dawson et al., 2012; Zhao et al., 2021). Among various chromatin-regulatory machineries, Nuclear receptor binding SET Domain protein 3 (NSD3; also known as KMT3F or WHSC1L1) is frequently altered in a range of human tumors, indicative of a high value cancer target (Bennett et al., 2017; Li et al., 2019; Li et al., 2017; Shen et al., 2015). The NSD3 gene, localized within an amplicon locus of 8p11-p12 in breast and squamous lung cancers, encodes two splicing variants, NSD3-short (NSD3S) (Shen et al., 2015) and NSD3-long (NSD3L) isoforms (Figure S1A). While NSD3L is a methyltransferase that catalyzes mono- and di-methylation of histone H3 Lysine 36 (H3K36), NSD3S lacks the C-terminal methyltransferase domain but retains the N-terminal domains including an acidic transactivation domain, an H3K36me3/2-binding domain (PWWP1) and the complex-scaffolding region (Bennett et al., 2017; Li et al., 2017; Shen et al., 2015). While the gained hyper-activity of NSD3L was previously reported to be responsible for oncogenesis in a subset of human acute myeloid leukemias (AML) that carry the NUP98-NSD3 fusion (Rosati et al., 2002; Wang et al., 2007) and to induce mammary tumors in NSD3 transgenic mice (Turner-Ivey et al., 2017), increasing evidence shows that NSD3S also exerts oncogenic effects in a wide spectrum of tumors. Approximately 6% of midline carcinoma is characterized by aberrant fusion between the NUTM1 gene and NSD3S, generally displaying poor prognosis (Chau et al., 2020). Additionally, a large-scale protein–protein interaction (PPI)-based interactome study points to the importance of the NSD3S-cMyc interaction for malignant growth of lung cancer (Li et al., 2017). Importantly, in AML with rearrangement of Mixed Lineage Leukemia (MLL-r AML), NSD3S was shown to form a complex with BRD4 (Shen et al., 2015), a chromatin modulator known to be essential for sustaining expression of cMyc in the same tumor type (Dawson et al., 2011; Zuber et al., 2011). Thus, cellular interactions between NSD3S and cMyc are wired through both PPI- and transcription-based regulations and agents that target the NSD3S and cMyc oncogenic nodes could be effective anticancer therapeutics.
To date, selective inhibitors of the SET domain of NSD3L have not been reported, despite recent advances in discovering enzymatic inhibitors of NSD1 (Huang et al., 2020) and NSD2 (Shen et al., 2019). Recently, BI-9321, a selective antagonist that blocks the NSD3 PWWP1 domain (a motif retained in both NSD3L and NSD3S; Figure S1A), was developed (Bottcher et al., 2019). However, BI-9321 does not target other functions of NSD3 and its inhibitory effect on the chromatin-reading function of the NSD3 PWWP1 domain does not lead to effective killing of cancer cells (Bottcher et al., 2019). Thus, a therapeutic strategy that simultaneously targets multivalent oncogenic functions of NSD3 in tumor is desirable.
Proteolysis targeting chimeras (PROTACs) have recently emerged as a promising class of therapeutic modalities (Dale et al., 2021; Khan et al., 2020; Lai and Crews, 2017; Nalawansha and Crews, 2020; Schapira et al., 2019; Sun et al., 2019). PROTACs are heterobifunctional small molecules that simultaneously bind the protein of interest (POI) and an E3 ligase such as von Hippel-Lindau (VHL) or cereblon (CRBN) and hijack the cellular ubiquitination-proteasome system, leading to selective polyubiquitination and subsequent degradation of the POI at the proteasome. In contrast to small-molecule inhibitors that rely on receptor occupancy pharmacology and do not typically target multiple functions of the POI, PROTACs pharmacologically deplete the POI, thus temporally eliminating all functions of the POI.
Here, we report the discovery and characterization of an NSD3 PROTAC degrader, MS9715, and present pharmacological degradation of NSD3 and concurrent suppression of the associated cMyc node as an attractive therapeutic strategy for treating NSD3-dependent cancers. Our biochemical, cellular, proteomic, transcriptomic results show that the NSD3 PROTAC degrader MS9715, which simultaneously targets NSD3- and cMyc-related oncogenic programs, is superior to BI-9321, which antagonizes the chromatin-reading function of the NSD3 PWWP1 domain.
RESULTS
Design and biophysical characterization of the NSD3 PROTAC, MS9715.
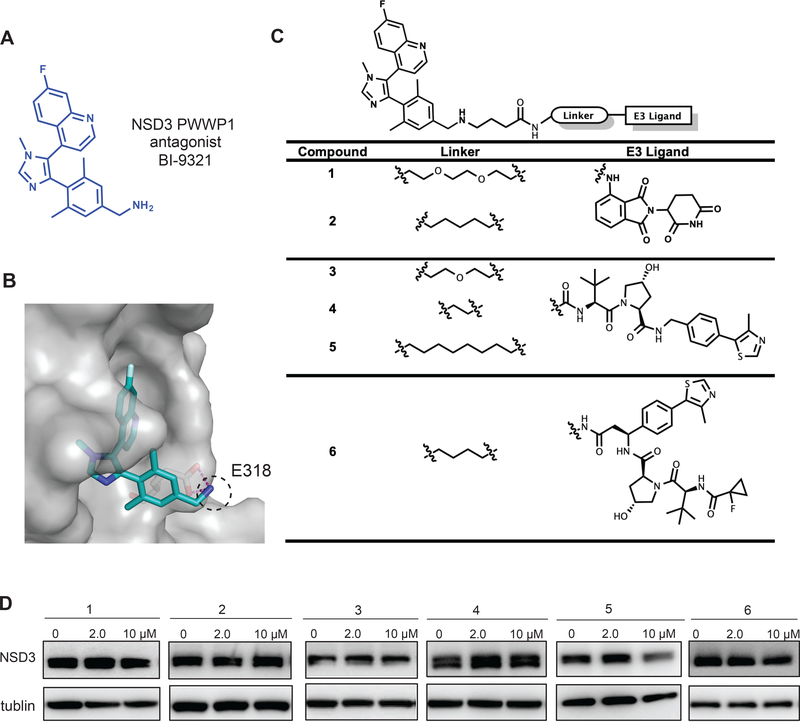
To discover an effective NSD3-targeting PROTAC degrader, we utilized BI-9321 (Figure 1A) as the NSD3 binder. The co-crystal structure of the NSD3-PWWP1 domain in complex with BI-9321 (Bottcher et al., 2019) revealed that the primary amino group of BI-9321 reaches out the binding pocket while making hydrogen bond interactions with E318 of NSD3 (Figure 1B). We hypothesized that converting the primary amino group into a secondary amino group would maintain the hydrogen bond interactions between the amino group and E318 of NSD3 and other key interactions, resulting in no significant loss in binding affinity, while the newly introduced butyric acid group could serve as a handle to attach a linker for connecting with an E3 ligase ligand (Figure 1C). Based on this hypothesis, we designed and synthesized a set of putative NSD3 degraders 1–6, by exploring various alkyl linkers and several ligands of E3 ligases such as CRBN and VHL, and evaluated their effect on degrading NSD3 (Figure 1C–D). Compounds 1 and 2, which link the CRBN ligand pomalidomide to BI-9321 through poly(ethylene glycol) (PEG) and five-carbon alkyl linkers, respectively, failed to degrade NSD3 (Figure 1D). On the other hand, among compounds 3, 4 and 5, which link BI-9321 to the E3 ligand VHL-1 via PEG, short alkyl and long alkyl linkers, respectively, compound 5 clearly induced NSD3 degradation at 10–40 μM (Figure 1D and S1B). We also explored another VHL binding ligand (Han et al., 2019) (shown in compound 6), but found that this ligand did not lead to effective degradation of NSD3 (Figure 1D).
Figure 1. Design and structure–activity relationship results of putative NSD3 PROTACs.
(A) Chemical structure of BI-9321, a selective binder of the NSD3 PWWP1 domain.
(B) Co-crystal structure (PDB code: 6G2O) of NSD3 (gray) in complex with BI-9321 (blue). The primary amino group, highlighted by the dashed black circle, reaches out the binding pocket while making hydrogen bond interactions with E318 of NSD3.
(C) Structures of the putative NSD3 degraders 1–6.
(D) Immunoblotting for NSD3S and GAPDH in 293FT cells after a 48-hour treatment with the indicated concentration of compounds 1–6. Results are representative of at least two independent experiments. See also Figure S1.
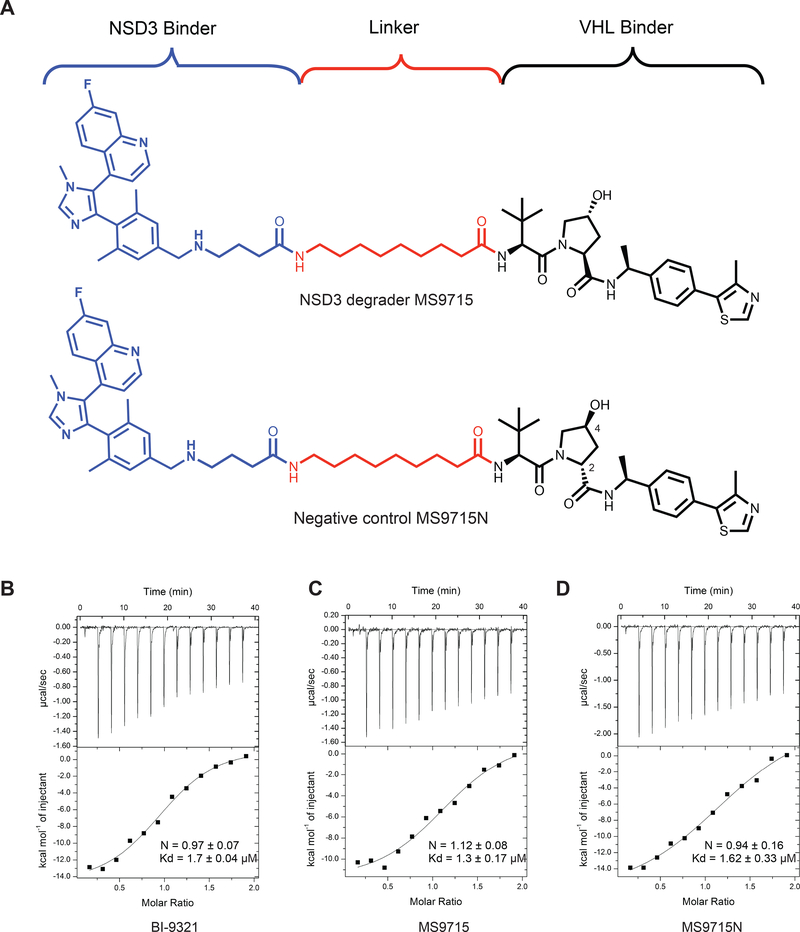
Since compound 5 showed promising NSD3 degradation effect, we designed and synthesized MS9715, by utilizing the VHL binder (S,R,S)-AHPC-Me (VHL-2) (Figure 2A top; Data S1), which was previously reported to result in more effective degraders than the ones derived from VHL-1 (Su et al., 2019; Wei et al., 2019). Of note, MS9715 and compound 5 contain the same NSD3 binding moiety and linker. To facilitate assessment and characterization of MS9715, we also designed and synthesized a close analog of MS9715, termed MS9715N (Figure 2A, bottom; Data S1), as a negative control compound. MS9715N was designed to be incapable of binding the VHL E3 ligase while maintaining a similar binding affinity to NSD3 by only changing the VHL binding portion of MS9715 to its diastereomer in MS9715N and keeping the linker and NSD3 binding portion of MS9715N identical to that of MS9715. Specifically, the stereochemistry of the 4-hydroxy group at the pyrrolidine ring is changed from (R) in MS9715 to (S) in MS9715N and the stereochemistry of the 2-substituent at the pyrrolidine ring is changed from (S) in MS9715 to (R) in MS9715N, which are known to abolish the binding to VHL (Cheng et al., 2020; Raina et al., 2016; Shen et al., 2020; Wei et al., 2019). Using isothermal titration calorimetry (ITC), we assessed binding affinities of MS9715 and MS9715N to NSD3. Comparing to BI-9321 (Kd = 1.7 ± 0.04 μM, Figure 2B), MS9715 (Kd = 1.3 ± 0.17 μM, Figure 2C) and MS9715N (Kd = 1.62 ± 0.33 μM, Figure 2D) maintained similar binding affinities to recombinant protein of the NSD3 PWWP1 domain, validating our design hypothesis.
Figure 2. Design and biophysical characterization of the NSD3 PROTAC MS9715.
(A) Chemical structures of the NSD3 PROTAC degrader MS9715 (top) and its analog control, MS9715N (bottom). See also Data S1.
(B-D) ITC titrations of BI-9321 (B), MS9715 (C) and MS9715N (D) into recombinant protein of the NSD3 PWWP1 domain. Binding affinity of BI-9321 (B), MS9715 (C) and MS9715N (D) to NSD3 was measured by ITC. The calculated values represent the mean ± SD from two independent experiments. First injection was removed from the fitting.
MS9715 effectively degrades NSD3 in hematologic cancer cells.
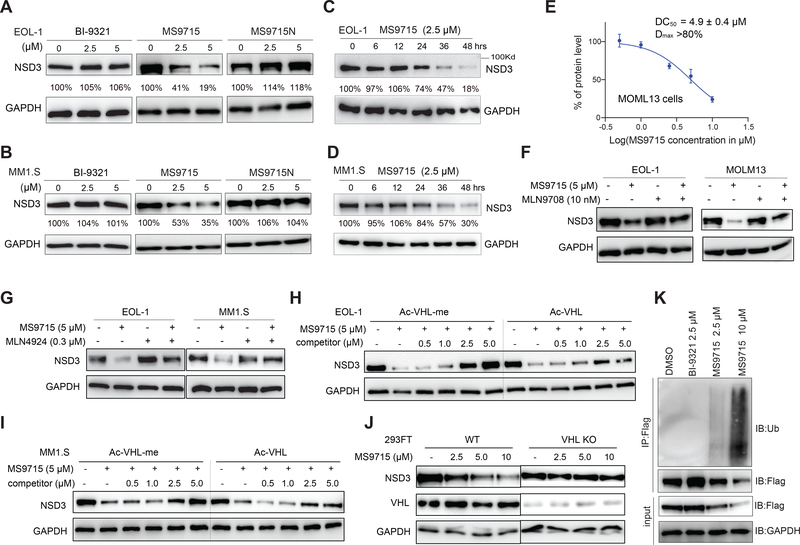
We next assessed the effect of MS9715 on degrading NSD3 in cancer cells. Using models of MLL-r AML (EOL-1 cells) and multiple myeloma (MM1.S cells), we first found that treatment of EOL-1 and MM1.S cells with MS9715, but not BI-9321 or MS9715N, led to depletion of both NSD3S and NSD3L isoforms (Figure S1C–D). Based on previous reports that NSD3S has essential and much broader oncogenic functions in AML (Shen et al., 2015), which is consistent with its higher level of abundance relative to NSD3L (Figure S1C–D), we mainly focused on NSD3S in subsequent studies. We observed a concentration-dependent depletion of cellular NSD3S by MS9715, but not BI-9321 or MS9715N, in EOL-1 (Figure 3A), MM1.S (Figure 3B) and MOLM13 cells (another MLL-r AML cell line, Figure S1E). In addition, the MS9715-induced NSD3S degradation was also found to be time-dependent in EOL-1 (Figure 3C) and MM1.S cells (Figure 3D), with significant degradation occurring at as early as 24 hours and with more profound degradation at 48 hours. In MOLM13 cells, MS9715 exhibited the half-maximal degradation concentration (DC50) value of 4.9 ± 0.4 μM and maximum degradation (Dmax) value of greater than 80% with a 48-hour treatment (Figure 3E and S1E). Together, these results show that MS9715 is an effective NSD3 PROTAC that consistently degrades NSD3 in multiple cell models of hematological cancer.
Figure 3. MS9715 degrades NSD3 in cells.
(A-B) Immunoblotting for NSD3S and GAPDH in EOL-1 (A) and MM1.S (B) cells after a 48-hour treatment with the indicated concentration of MS9715, BI-9321 or MS9715N.
(C-D) Immunoblotting for NSD3S in EOL-1 (C) and MM1.S (D) cells treated with 2.5 μM of MS9715 for the indicated duration.
(E) DC50 and Dmax values of MS9715 in MOLM13 cells, shown as the mean ± SD from three independent experiments. MOLM13 cells were treated with MS9715 for 48 hours. See also Figure S1.
(F-G) Immunoblots for NSD3S using the indicated cells pre-treated with DMSO, 10 nM of MLN9708 (F) or 0.3 μM of MLN4924 (G) for two hours, followed by a 48-hour treatment with 5 μM of MS9715.
(H-I) Immunoblots for NSD3S and GAPDH using EOL-1 (H) and MM1.S (I) cells pre-treated with the indicated concentration of Ac-VHL or Ac-VHL-Me relative to DMSO for two hours, followed by a 48-hour treatment with 5 μM of MS9715.
(J) Immunoblots for NSD3S, VHL and GAPDH after a 48-hour-treatment of 293FT cells, either wildtype (WT; left) or with the CRISPR/Cas9-mediated KO of VHL (right), with the indicated concentration of MS9715, relative to mock.
(K) Immunoblots (IB) for ubiquitin (Ub) after anti-Flag immunoprecipitation (IP) of extracts prepared from 293FT cells expressing Flag-tagged NSD3S, after a 48-hour treatment with the indicated concentration of DMSO, BI-9321 or MS9715.
We next determined the mechanism of action (MOA) of the MS9715-induced NSD3 degradation. First, MS9715-mediated degradation of NSD3 can be almost completely blocked by pretreatment of MLN9708 (Figure 3F), a proteasome inhibitor (Kupperman et al., 2010), or MLN4924 (Figure 3G), a NEDD8-activating enzyme inhibitor (Soucy et al., 2009). Additionally, pretreatment with a VHL ligand, either Ac-VHL or the higher affinity Ac-VHL-Me (Han et al., 2019), concentration-dependently blocked the MS9715-induced NSD3 degradation, with Ac-VHL-Me showing a stronger effect than Ac-VHL, in both EOL-1 (Figure 3H) and MM1.S cells (Figure 3I). Furthermore, the CRISPR/Cas9-mediated knockout (KO) of VHL in 293FT cells abrogated the MS9715-mediated NSD3 degradation, compared to the WT control (Figure 3J). Lastly, we found that the treatment of 293FT cells with MS9715, but not BI-9321, induced ubiquitination of cellular NSD3 in a concentration-dependent manner (Figure 3K). Taken together, these results demonstrate that MS9715 induces NSD3 degradation in the VHL- and ubiquitin-proteasome system-dependent manner.
MS9715 is a highly selective NSD3 degrader and effectively suppresses the NSD3-related gene-expression program.
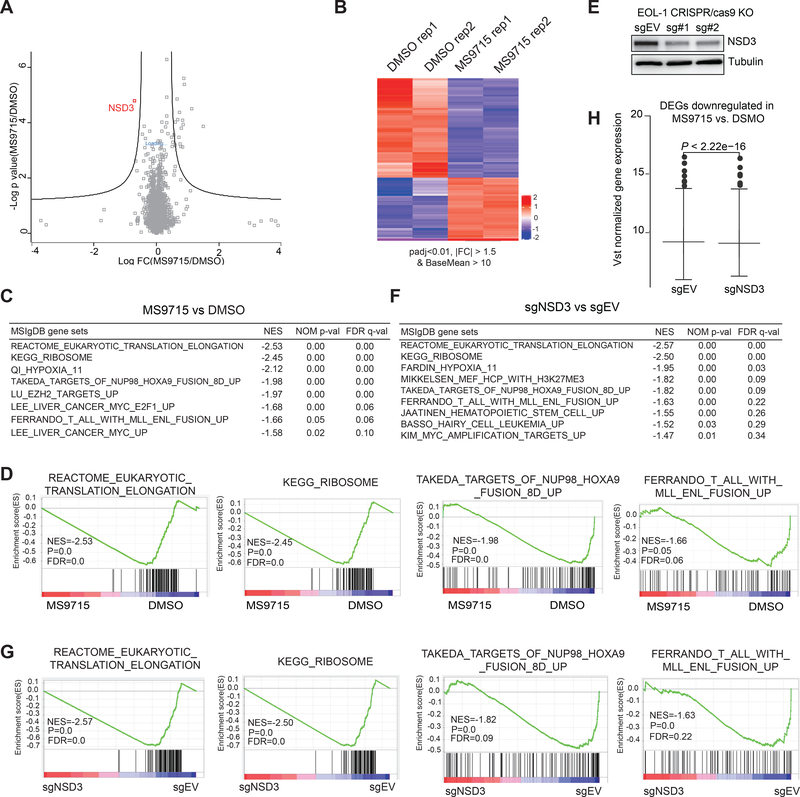
To assess selectivity of MS9715, we employed a mass spectrometry-based global proteomic profiling approach. We chose to treat EOL-1 cells with 2.5 μM of MS9715 for 30 hours, a relatively early time point, in order to capture early events rather than secondary effects. We found that, out of more than 5,000 proteins detected, NSD3 was the only protein showing significant downregulation in EOL-1 cells (Figure 4A and Table S1). Thus, MS9715 is a highly selective NSD3 PROTAC degrader.
Figure 4. Omics studies show that MS9715 selectively degrades NSD3 in cells, leading to suppression of the NSD3-related gene-expression program.
(A) Quantitative proteomics results showing relative abundance of proteins in EOL-1 cells treated with DMSO or 2.5 μM of MS9715 for 30 hours. Each dot in the plot represents one of a total of 5,452 proteins detected among all samples (n=3 biologically independent samples per group measured in a single 10-plex tandem mass tag [TMT] experiment), with x-axis and y-axis showing log transformed value of fold-change in expression and P value, respectively. Black lines were calculated using a false discovery rate of 5% and a hyperbolic curve threshold of S0 value of 0.15 using the Perseus software. See also Table S1. The method for calculating P value is described in STAR Methods section.
(B) Heatmap showing the relative expression levels of differentially expressed genes (DEGs) after a 4-day treatment with 2.5 μM of MS9715, relative to DMSO, in EOL-1 cells (n=2 biologically independent replicates [Rep] per group, i.e., Rep 1 and 2). Threshold of DEG is set at the adjusted DESeq P value (padj) less than 0.01 and fold-change (FC) over 1.5 for transcripts with mean tag counts of at least 10. The Wald test is used for calculating DESeq2 P value and Benjamin-Hochberg method for calculating padj value, See also Table S2.
(C) Summary of GSEA results showing correlation between the indicated gene signatures and MS9715 treatment in EOL-1 cells. The method for calculating P and FDR values in GSEA results is described in STAR Methods section.
(D) GSEA revealing that, relative to DMSO, MS9715 treatment is positively correlated with downregulation of the indicated gene sets related to translation elongation and ribosome activity or those activated in hematopoietic cancer due to the NUP98-HOXA9 or MLL-ENL oncogene.
(E) Immunoblotting for NSD3 after a four-day induction of Cas9 in the EOL-1 cells stably transduced with either empty vector (sgEV) or an NSD3-targeting sgRNA (sg#1 or sg#2).
(F) Summary of GSEA results showing correlation between the indicated gene signatures and NSD3 KO in EOL-1 cells. See also Figure S2 and Table S3.
(G) GSEA revealing that, relative to control (sgEV), the CRISPR/Cas9-mediated NSD3 KO is positively correlated with downregulation of the indicated gene sets related to translation elongation and ribosome activity or those activated in hematopoietic cancer due to the NUP98-HOXA9 or MLL-ENL oncogene.
(H) Box plot showing overall expression of the MS9715-downregulated DEGs (as defined in panel B) in EOL-1 cells after a four-day induction of Cas9 for NSD3 depletion (sg#1 as sgNSD3; right) or mock treatment (sgEV; left). A paired t-test was applied to compare the mean of normalized expression between the two treatment groups.
Since NSD3 is known to be a chromatin modulator involved in gene expression regulation, we next evaluated the gene-regulatory effect of MS9715 by carrying out RNA sequencing (RNA-seq)-based transcriptome profiling of EOL-1 cells treated with DMSO or 2.5 μM MS9715 for a longer duration (four days). The subsequent analysis of the RNA-seq data revealed the differentially expressed genes (DEGs) caused by the MS9715 treatment (Figure 4B and Table S2; with a cutoff of an absolute value of fold-change more than 1.50 and adjusted P value less than 0.01). Gene Set Enrichment Analysis (GSEA) revealed that, relative to DMSO, treatment of MS9715 is associated with the reduced expression of genes responsible for protein translation or ribosomal activities and with downregulation of transcripts activated by the AML oncoproteins such as NUP98-HOXA9 and MLL-r (Figure 4C–D and Figure S2A). To determine NSD3-regulated transcripts, we employed a genetic approach and used an inducible CRISPR-Cas9 system to knock out (KO) NSD3 in EOL-1 cells (Figure 4E). Subsequent RNA-seq (Figure S2B and Table S3) and GSEA analyses identified same correlations between NSD3 KO and downregulation of all of the above-mentioned gene signatures associated with protein translation and oncoproteins (Figure 4F–G and S2C, compared to Figure 4C–D and S2A). In agreement, NSD3 KO is also associated with upregulation of transcripts related to cell differentiation (Figure S2D). Additionally, overall expression levels of those MS9715-downregulated DEGs were also significantly reduced in NSD3 KO cells (Figure 4H).
Taken together, these results indicate that: (1) MS9715 is highly selective for NSD3; and (2) the transcriptomic alterations (such as significant downregulation of genes critical for oncogenesis, protein translation and cell proliferation) caused by MS9715 resemble those by KO of NSD3.
MS9715 has superior effects to the NSD3 PWWP1 antagonist on repressing oncogenic gene-expression programs in cancer cells.
Next, we queried whether MS9715 is superior to the NSD3 PWWP1 antagonist BI-9321 in suppressing NSD3’s functions. To this end, we performed additional RNA-seq experiments to compare gene-modulating effects of BI-9321, MS9715N and MS9715 treatment (Table S2, S4–S6). First, a survey of DEGs due to compound treatment versus mock showed that MS9715 exhibited a more profound effect on altering the cancer cell transcriptome, compared to BI-9321 and MS9715N (Figure 5A). Next, we further correlated the compound-induced changes with those by NSD3 KO and found that the NSD3-activated transcripts exhibited the significantly more downregulation post-treatment of MS9715, compared to BI-9321 or MS9715N (Figure 5B and Figure S3A–D). We also identified genes showing significant expression change in EOL-1 cells post-treatment with MS9715, compared to BI-9321 (Table S7), thus solidifying the stronger effect by MS9715 on transcriptomic modulation. By using both RT-qPCR (Figure 5C) and western blots (Figure 5D), we verified the superior effect of MS9715 to BI-9321 and MS9715N on suppressing expression of CBLB, IFITM1, SELL and MAP7, a set of genes showing downregulation upon NSD3 KO in same cells (Table S3). These results collectively support that MS9715 has much more profound effects than BI-9321 on repressing oncogenic transcripts, which prompted us to explore antitumor utilities of this NSD3 degrader.
Figure 5. RNA-seq-based profiling demonstrates that MS9715 has a superior effect to BI-9321 on inhibition of target gene transcription.
(A) Venn diagram showing overlap of the DEGs showing downregulation after treatment of EOL-1 cells with 2.5 μM of MS9715, BI-9321 or MS9715N, compared to DMSO, for four days. See also Tables S2, S4 and S6–7.
(B) Box plots showing the Log2 ratios for gene expression changes of DEGs downregulated post-KO of NSD3 relative to mock in EOL-1 cells across the indicated compound treatment conditions, i.e., BI-9321 vs. DMSO, MS9715 vs. DMSO, MS9715N vs. DMSO, and MS9715 vs. MS9715N, with P value for each comparison shown on the top of panels. A paired t-test was applied to compare the mean of normalized expression between treatment groups. See also Figure S3.
(C-D) RT-qPCR (C) and immunoblotting (D) for the indicated NSD3-upregulated gene targets post-treatment of EOL-1 (C, left panel and D) or MM1.S (C, right) cells with 2.5 μM of MS9715, BI-9321 or MS9715N, compared to DMSO, for 48 hours. Y-axis in panel C shows averaged fold-change ± SD of three independent experiments after normalization to beta-actin and to DMSO-treated.
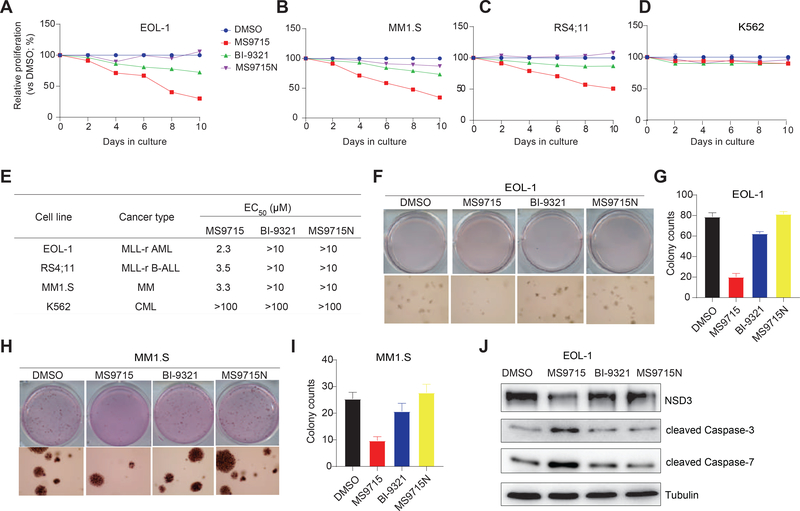
MS9715, but not the NSD3 PWWP1 antagonist, effectively suppresses the growth of hematological cancer cells.
Next, we evaluated anti-proliferative effects of MS9715 in MLL-r acute leukemia and MM cells. Using cell models that cover MLL-r AML (EOL-1; Figure 6A), MM (MM1.S; Figure 6B) and MLL-r B-ALL (RS4;11; Figure 6C), we found that MS9715, but not BI-9321 and MS9715N, effectively inhibited the growth of these blood cancer cells. Additionally, while MS9715 phenocopies NSD3 KO in EOL-1 cells (Figure S4A), it exhibited little growth inhibitory effect in K562 cells (Figure 6D), suggesting that MS9715 is not a non-selective cytotoxic agent. We also determined the half maximal effective concentration (EC50) values of MS9715, which range from 2 to 4 μM in EOL-1, RS4;11 and MM1.S cells, while BI-9321 and MS9715N were largely ineffective in these cells (EC50 > 10 μM) (Figure 6E and S4B). Furthermore, in contrast to little or no effect seen with BI-9321 and MS9715N, MS9715 treatment led to drastically decreased colony-forming capabilities in EOL-1 (Figure 6F–G) and MM1.S (Figure 6H–I) cells. Consistent with these results, MS9715, but not BI-9321 and MS9715N, also induced a prominent increase of apoptosis in these cells (Figure 6J and S4C–D). Collectively, MS9715, but not BI-9321, is an effective antitumor agent.
Figure 6. MS9715, but not BI-9321, effectively inhibits cancer cell proliferation and colony formation.
(A-D) The growth inhibitory activity of MS9715 (2.5 μM) was assessed in EOL-1 (A), MM1.S (B), RS4;11 (C) and K562 (D) cells, compared to DMSO, BI-9321 (2.5 μM) and MS9715N (2.5 μM). Cells were treated for ten days. Y-axis shows average ± SD of three independent experiments after normalization to DMSO-treated.
(E) Summary of EC50 values of MS9715, MS9715N and BI-9321 in the indicated cell lines after an eight-day treatment. EC50 values are the means of three independent experiments. CML, chronic myelogenous leukemia. See also Figure S4.
(F-I) Representative cell plate scan images (F and H) and quantifications of colony formation (G and I) using EOL-1 (F-G) or MM1.S cells (H-I) treated with DMSO or 2.5 μM of MS9715, BI-9321 or MS9715N. Colony numbers are presented in average ± SD of two independent experiments.
(J) Immunoblotting for the indicated apoptotic markers after a four-day treatment of EOL-1 cells with DMSO or 2.5 μM of MS9715, BI-9321 or MS9715N.
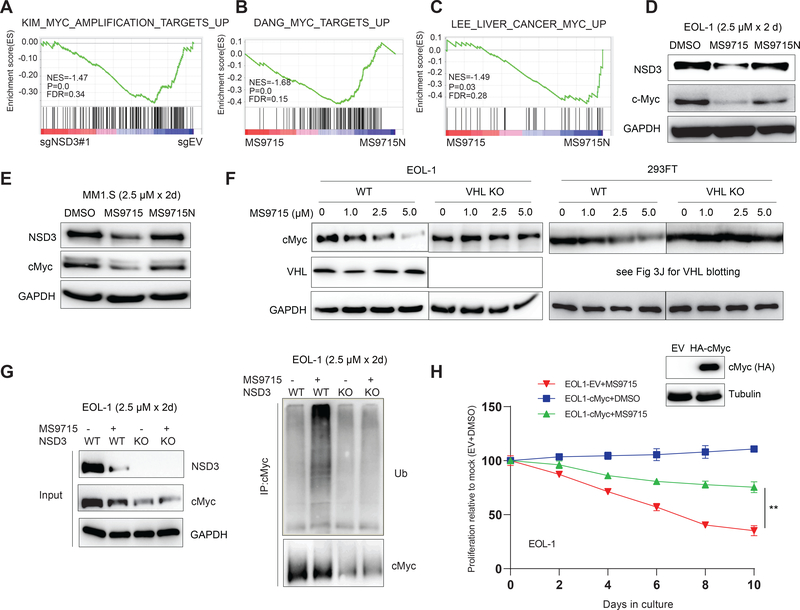
cMyc is simultaneously degraded by MS9715, leading to repression of the cMyc node in cancer cells.
NSD3S serves as an adaptor, which is associated with cMyc (Li et al., 2017) and BRD4 (Shen et al., 2015), a known upstream activator of cMyc. Cellular interactions between NSD3S and cMyc are wired through both PPI- and transcription-based regulations. We hypothesized that NSD3 degradation could result in a significant decrease in cMyc signaling in cells. Indeed, GSEA of the RNA-seq profiles in EOL-1 cells showed that, similar to NSD3 KO (Figure 7A), treatment with MS9715 is positively correlated with suppression of cMyc-related gene sets, relative to DMSO, BI-9321 or MS9715N (Figure 7B–C and S5). Furthermore, treatment with MS9715, but not MS9715N, led to a significant decrease in the protein levels of cMyc in both EOL-1 (Figure 7D) and MM1.S (Figure 7E) cells. Additionally, the MS9715-induced cMyc depletion was almost completely diminished in the VHL-deficient EOL-1 (Figure 7F, left) and 293FT (Figure 7F, right) cells, relative to WT control cells. We also found that the ubiquitination of cellular cMyc was significantly enhanced after treatment of EOL-1 cells with MS9715 (Figure 7G; right panel, lane 2 versus 1) and that such an effect was abolished in NSD3 KO cells (Figure 7G; right panel, lanes 3–4 versus 2), thereby further demonstrating the involvement of NSD3 in the MS9715-induced cMyc ubiquitination. It should be noted that NSD3 KO resulted in a notable loss of cMyc (Figure 7G; left panel, lane 3 versus 1), consistent with a previous report that NSD3 potentiates the transcriptional upregulation of cMyc (Shen et al., 2015). It is currently unclear that the observed cMyc loss is primarily due to the direct degradation of NSD3 and cMyc induced by MS9715 via the ubiquitin-proteasome system, or NSD3 loss-mediated downregulation of cMyc gene expression, or both, which warrants further investigation. Lastly, ectopic overexpression of cMyc in EOL-1 cells significantly rescued the suppressed growth caused by MS9715 (Figure 7H). Overall, our data show that, in addition to degrading NSD3, the NSD3 PROTAC MS9715 also represses the cMyc node, thereby co-suppressing both oncogenic circuitries in tumor cells.
Figure 7. MS9715 represses the cMyc oncogenic node in cancer cells.
(A) GSEA revealing the positive correlation between repression of cMyc-related gene signatures and KO of NSD3 (sg#1), relative to control (sgEV), in EOL-1 cells. The method for calculating P and FDR values in GSEA results is described in STAR Methods section.
(B-C) GSEA revealing the positive correlation between repression of cMyc-related gene signatures and treatment of EOL-1 cells with MS9715, relative to control. See also Figure S5.
(D-E) Immunoblotting for NSD3, cMyc and GAPDH using total cell lysates of EOL-1 (D) and MM1.S (E) cells treated with DMSO or 2.5 μM of MS9715 or MS9715N for 48 hours.
(F) Immunoblot for cMyc and VHL in EOL-1 (left) and 293FT cells (right), either wildtype (WT) or with KO of VHL, after a 48-hour treatment with the indicated concentration of MS9715.
(G) Right: Ubiquitin (Ub) immunoblots (IB) after immunoprecipitation (IP) with cMyc antibodies using total cell lysate of EOL-1 cells, either WT or with KO of NSD3, after a 48-hour treatment with DMSO or 2.5 μM of MS9715. Left: blots for NSD3, cMyc and GAPDH using the input samples of the indicated cells.
(H) Growth of EOL-1 cells, stably expressing either control vector (EV) or HA-tagged cMyc (insert: anti-HA immunoblot), after treatment with DMSO or 2.5 μM of MS9715 for the indicated duration. Y-axis shows the average ± SD of three independent experiments after normalization to DMSO-treated controls. **, P <0.01 by two-sided Student’s t-test.
DISCUSSION
In this study, we discovered the NSD3 degrader MS9715, which is a VHL-recruiting PROTAC based on a previously reported NSD3 antagonist BI-9321 that binds the NSD3 PWWP1 domain. We also developed a close analog of MS9715, MS9715N, which binds NSD3 but not VHL, as a negative control of MS9715. We show that MS9715, but not MS9715N and BI-9321, concentration- and time-dependently degrades NSD3 in hematological cancer models covering AML, ALL and MM. The MS9715-induced NSD3 degradation is dependent on the E3 ligase VHL and the ubiquitin-proteasome system. Results from our global proteomics studies support that MS9715 is a highly selective NSD3 degrader. We also show that MS9715 is superior to BI-9321 in repressing the NSD3-related gene-expression program in cancer cells. Consistent with these results, MS9715, but not BI-9321 or MS9715N, effectively inhibits the growth and induces apoptosis in hematological cancer cells. Moreover, we show that MS9715, but not MS9715N, also suppresses the cMyc-associated oncogenic node. In addition to the reported role for NSD3 in transcriptional potentiation of the cMyc gene (Shen et al., 2015), a part of the observed cMyc-suppressing effect by MS9715 could be ascribed to the MS9715-induced degradation of cMyc protein (most likely in form of a cMyc-NSD3S complex) because the induced cMyc ubiquitination by MS9715 is dependent on both MS9715-bound VHL and NSD3.
Our results that the NSD3 PWWP1 antagonist BI-9321 was largely ineffective in suppressing the growth of NSD3-dependent cancer cells are consistent with the previously reported data (Bottcher et al., 2019). This suggests that pharmacological inhibition of a binding function alone harbored within the NSD3 PWWP1 domain is unlikely an effective approach for treating NSD3-dependent cancers. In contrast, the NSD3 PROTAC degrader MS9715 simultaneously suppresses both NSD3- and cMyc-associated gene-expression programs, phenocopying NSD3 KO. The much stronger pharmacological effects observed here for NSD3 degradation versus inhibition, together with previously reported examples (Cromm et al., 2018; Ma et al., 2020) such as FAK degradation versus inhibition (Cromm et al., 2018), have demonstrated potential benefits of the degrader technology. While a future drug discovery effort that focuses on optimizing the NSD3 binding, linker and E3 ligase binding moieties of MS9715 will likely result in more potent NSD3 PROTAC degraders, the promising results from our current proof-of-concept study support that pharmacological degradation of NSD3 as a therapeutic strategy is superior to pharmacological blocking of the NSD3 reading function for treating NSD3-dependent cancers.
SIGNIFICANCE
NSD3 is frequently amplified and/or overexpressed in tumors such as hematological malignancy and lung cancer. Recently, BI-9321, a selective antagonist of the NSD3-PWWP1 domain, was reported. However, BI-9321 is largely ineffective in treating NSD3-dependent cancers, likely due to that it does not target all of the NSD3’s multivalent oncogenic functions. To address this issue, we discovered MS9715, a NSD3 small-molecule degrader based on BI-9321 using the PROTAC technology. Importantly, MS9715, which depletes cellular NSD3 and associated onco-partners (notably cMyc), effectively inhibits the proliferation in several cellular models that cover multiple hematological malignancy subtypes while BI-9321 does not. Results from global proteomic and transcriptomic profiling studies show that MS9715 is highly selective for NSD3 and that effects of MS9715 resemble those of NSD3 KO. Overall, we present pharmacological degradation of NSD3 as an attractive therapeutic strategy and provide evidence that this strategy is superior to pharmacological blocking of the NSD3 PWWP1 domain for treating NSD3-dependent cancers.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding authors, Greg Wang at greg_wang@med.unc.edu
Materials availability
All of the reagents reported in this study are available from the lead or correspondence contact with Materials Transfer Agreement as long as stocks remain available.
Data and code availability
All transcriptomic profiling datasets have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under accession number GSE158296. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Perez-Riverol et al., 2019) with the dataset identifier PXD021901. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Human hematological cancer cell lines used in the study include EOL-1 (Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ], ACC-386), RS4;11 (ATCC, CRL-1873), MOLM-13 (DSMZ, ACC-554), K562 (ATCC, CRL-243) and MM1.S (ATCC, CRL-2974). These lines were cultured in the RPMI 1640 base medium supplemented with 10% of FBS and 1% of penicillin plus streptomycin. 293FT cells (Thermo Fisher Scientific, R70007) were cultured in DMEM base medium supplemented with 10% of FBS and 1% of penicillin plus streptomycin. Authentication of cell line identities was ensured by the Tissue Culture Facility (TCF) of UNC Lineberger Comprehensive Cancer Center with the genetic signature profiling and fingerprinting analysis. Every month, a routine examination for potential mycoplasma contamination was carried out by using the commercially available detection kits from Lonza.
Bacterial strains
DH5a and Stabl3 competent cells were purchased from Thermo Fisher Scientific and used for plasmid transformation and propagation based on manufacturer’s instructions.
METHOD DETAILS
General chemistry
All chemical reagents were purchased from commercial vendors and used without further purification. The flash column chromatography was conducted using a Teledyne ISCO CombiFlash Rf+ instrument. This instrument was also equipped with a variable-wavelength UV detector and a fraction collector. RediSep Rf Gold C18 columns were used for purification. High-performance liquid chromatography (HPLC) spectra for compounds were acquired using an Agilent 1200 Series system with a DAD detector. Chromatography was performed on a 2.1 × 150 mm Zorbax 300SB-C18 5 μm column with water containing 0.1% formic acid as solvent A and acetonitrile containing 0.1% formic acid as solvent B at a flow rate of 0.4 mL/min. The gradient program was as follows: 1% B (0−1 min), 1−99% B (1−4 min), and 99% B (4−8 min). Ultra performance liquid chromatography (UPLC) spectra for compounds were acquired using a Waters Acquity I-Class UPLC system with a PDA detector. Chromatography was performed on a 2.1 × 30 mm ACQUITY UPLC BEH C18 1.7 μm column with water containing 3% acetonitrile, 0.1% formic acid as solvent A and acetonitrile containing 0.1% formic acid as solvent B at a flow rate of 0.8 mL/min. The gradient program was as follows: 1−99% B (1−1.5 min), and 99−1% B (1.5−2.5 min). High-resolution mass spectra (HRMS) data were acquired in the positive ion mode using Agilent G1969A API-TOF with an electrospray ionization (ESI) source. Nuclear magnetic resonance (NMR) spectra were acquired on a Bruker DXI 800 MHz spectrometer with 800 MHz for proton (1H NMR) or a Bruker DRX-500 spectrometer with 500 MHz for proton (1H NMR) or a Bruker DRX-600 spectrometer with 600 MHz for proton (1H NMR) or 151 MHz for carbon (13C NMR); Chemical shifts are reported in ppm (δ). Preparative HPLC was performed using an Agilent Prep 1200 series with UV detector set to 220 nm. Samples were injected into a Phenomenex Luna 75 × 30 mm, 5 μm, C18 column at room temperature. The flow rate was 40 mL/min. A linear gradient was used with 10% of Acetonitrile (A) in H2O (with 0.1% TFA) (B) to 100% (or 60%) of Acetonitrile (A). All final compounds had > 95% purity using the UPLC and HPLC methods described above. MLN4924 (cat # S7109) were purchased from Selleck Chemical. BI-9321, Ac-VHL and Ac-VHL-me were synthesized according to the published procedures (Bottcher et al., 2019; Han et al., 2019; Hu et al., 2019). Synthesis and characterization of compounds 1–6, MS9715, MS9715N and related intermediates are detailed below. 1H NMR and 13C NMR spectra of MS9715 and MS9715N are provided (Data S1).
Synthetic scheme for compounds 1–17, MS9715
Reaction Conditions: (a) NaBH(OAc)3, DCM, rt; (b) NEt3, DCM, (Boc)2O, rt; (c) DIPEA, Pd(amphos)Cl2, B2(OH)4, THF/MeOH, 75 °C, 3 h; (d) Pd(PPh3)4, K2CO3, Dioxane, H2O, Microwave 125 °C, 2 h; (e) DCM/TFA, rt; (f) HOAT, EDCI, NMM, DMSO, rt.
Synthesis and characterization of compounds 1–17, MS9715 and MS9715N
Compounds 1-6 were synthesized according to the synthetic procedure of MS9715.
N-(2-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethyl)-4-((4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylbenzyl)amino) butanamide (1).
1H NMR (600 MHz, Methanol-d4) δ 9.36 (s, 1H), 8.94 (d, J = 4.5 Hz, 1H), 7.92 (dd, J = 9.2, 5.7 Hz, 1H), 7.85 – 7.80 (m, 1H), 7.61 – 7.52 (m, 2H), 7.49 (d, J = 4.5 Hz, 1H), 7.26 (s, 1H), 7.14 (s, 1H), 7.10 (d, J = 8.5 Hz, 1H), 7.06 (d, J = 7.0 Hz, 1H), 5.07 (dd, J = 12.6, 5.5 Hz, 1H), 4.08 (s, 2H), 3.77 (s, 3H), 3.74 (t, J = 5.2 Hz, 2H), 3.70 – 3.66 (m, 2H), 3.66 – 3.62 (m, 2H), 3.56 (t, J = 5.5 Hz, 2H), 3.51 (t, J = 5.2 Hz, 2H), 3.40 – 3.33 (m, 2H), 3.03 (t, J = 7.2 Hz, 2H), 2.91 – 2.82 (m, 1H), 2.79 – 2.66 (m, 2H), 2.34 (t, J = 6.7 Hz, 2H), 2.30 (s, 3H), 2.15 – 2.08 (m, 1H), 2.08 (s, 3H), 1.95 – 1.85 (m, 2H). HPLC > 95%, tR = 3.58 min, HRMS (ESI-TOF) m/z: [M + H]+ calcd for C45H50FN8O7+, 833.3781; found, 833.3768.
N-(5-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)pentyl)-4-((4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylbenzyl)amino)butanamide (2).
1H NMR (600 MHz, Methanol-d4) δ 9.21 (s, 1H), 8.92 (d, J = 4.5 Hz, 1H), 7.90 (dd, J = 9.2, 5.7 Hz, 1H), 7.81 (dd, J = 9.7, 2.6 Hz, 1H), 7.59 – 7.50 (m, 2H), 7.46 (d, J = 4.4 Hz, 1H), 7.24 (s, 1H), 7.12 (s, 1H), 7.06 – 7.00 (m, 2H), 5.07 (dd, J = 12.8, 5.5 Hz, 1H), 4.06 (s, 2H), 3.75 (s, 3H), 3.38 – 3.32 (m, 2H), 3.20 (t, J = 6.9 Hz, 2H), 3.04 (t, J = 7.1 Hz, 2H), 2.87 (ddd, J = 17.5, 14.0, 5.3 Hz, 1H), 2.79 – 2.73 (m, 1H), 2.76 – 2.66 (m, 1H), 2.37 (t, J = 6.6 Hz, 2H), 2.27 (s, 3H), 2.16 – 2.09 (m, 1H), 2.07 (d, J = 1.2 Hz, 3H), 1.94 – 1.86 (m, 2H), 1.70 (p, J = 7.0 Hz, 2H), 1.57 (p, J = 7.1 Hz, 2H), 1.46 (tt, J = 9.1, 5.8 Hz, 2H). HPLC > 95%, tR = 3.72 min, HRMS (ESI-TOF) m/z: [M + H]+ calcd for C44H48FN8O5+,787.3726; found, 787.3728.
(2S,4R)-1-((S)-15-(tert-butyl)-1-(4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylphenyl)-6,13-dioxo-10-oxa-2,7,14-triazahexadecan-16-oyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (3).
1H NMR (500 MHz, Methanol-d4) δ 9.30 (s, 1H), 8.94 (s, 1H), 8.93 (s, 1H), 7.91 (dd, J = 9.2, 5.9 Hz, 1H), 7.82 (dd, J = 9.7, 2.6 Hz, 1H), 7.58 (td, J = 8.6, 2.6 Hz, 1H), 7.51 – 7.46 (m, 3H), 7.43 (d, J = 8.3 Hz, 2H), 7.26 (s, 1H), 7.15 (s, 1H), 4.69 – 4.65 (m, 1H), 4.62 – 4.55 (m, 1H), 4.54 – 4.46 (m, 2H), 4.40 (d, J = 15.3 Hz, 1H), 4.08 (s, 2H), 3.91 (d, J = 11.0 Hz, 1H), 3.83 (dd, J = 11.1, 4.0 Hz, 1H), 3.76 (s, 3H), 3.75 – 3.67 (m, 2H), 3.53 (t, J = 5.4 Hz, 2H), 3.37 (t, J = 5.4 Hz, 2H), 3.05 (t, J = 7.2 Hz, 2H), 2.59 – 2.39 (m, 2H), 2.49 (s, 3H), 2.38 (t, J = 6.7 Hz, 2H), 2.31 (s, 3H), 2.28 – 2.22 (m, 1H), 2.15 – 2.10 (m, 1H), 2.08 (s, 3H), 1.95 – 1.87 (m, 2H), 1.05 (d, J = 2.1 Hz, 9H). HPLC > 95%, tR = 3.68 min, HRMS (ESI-TOF) m/z: [M + H]+ calcd for C53H65FN9O6S+, 974.4757; found, 974.4738.
(2S,4R)-1-((S)-2-(3-(4-((4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylbenzyl)amino)butanamido)propanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (4).
1H NMR (500 MHz, Methanol-d4) δ 9.32 (s, 1H), 9.02 – 8.80 (m, 2H), 7.92 (dd, J = 9.2, 5.7 Hz, 1H), 7.85 – 7.74 (m, 1H), 7.58 (ddd, J = 9.2, 8.1, 2.6 Hz, 1H), 7.51 – 7.44 (m, 3H), 7.42 (dd, J = 8.3, 2.1 Hz, 2H), 7.27 (s, 1H), 7.16 (s, 1H), 4.63 (s, 1H), 4.59 – 4.55 (m, 1H), 4.53 – 4.45 (m, 2H), 4.42 – 4.34 (m, 1H), 4.09 (s, 2H), 3.95 (d, J = 11.0 Hz, 1H), 3.82 (dd, J = 11.1, 3.9 Hz, 1H), 3.76 (s, 3H), 3.52 – 3.38 (m, 2H), 3.05 (t, J = 7.2 Hz, 2H), 2.53 – 2.46 (m, 5H), 2.34 (t, J = 6.8 Hz, 2H), 2.31 (s, 3H), 2.28 – 2.21 (m, 1H), 2.14 – 2.10 (m, 1H), 2.09 (s, 3H), 1.95 – 1.87 (m, 2H), 1.04 (d, J = 2.5 Hz, 9H). HPLC > 95%, tR = 3.66 min, HRMS (ESI-TOF) m/z: [M + H]+ calcd for C51H61FN9O5S+, 930.4495; found, 930.4487.
(2S,4R)-1-((S)-2-(9-(4-((4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylbenzyl)amino)butanamido)nonanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (5).
1H NMR (500 MHz, Methanol-d4) δ 9.32 (s, 1H), 9.04 – 8.84 (m, 2H), 7.92 (dd, J = 9.3, 5.7 Hz, 1H), 7.82 (dd, J = 9.7, 2.6 Hz, 1H), 7.67 – 7.54 (m, 1H), 7.50 – 7.46 (m, 3H), 7.44 (d, J = 8.3 Hz, 2H), 7.27 (s, 1H), 7.16 (s, 1H), 4.66 (s, 1H), 4.61 – 4.56 (m, 1H), 4.54 – 4.49 (m, 2H), 4.38 (d, J = 15.5 Hz, 1H), 4.09 (s, 2H), 3.92 (d, J = 10.9 Hz, 1H), 3.83 (dd, J = 11.0, 3.9 Hz, 1H), 3.77 (s, 3H), 3.15 (t, J = 7.2 Hz, 2H), 3.05 (t, J = 7.2 Hz, 2H), 2.50 (s, 3H), 2.36 (t, J = 6.7 Hz, 2H), 2.33 – 2.29 (m, 4H), 2.29 – 2.24 (m, 1H), 2.24 – 2.21 (m, 1H), 2.12 (dd, J = 9.0, 4.5 Hz, 1H), 2.09 (s, 3H), 1.98 – 1.83 (m, 2H), 1.68 – 1.55 (m, 2H), 1.55 – 1.47 (m, 2H), 1.42 – 1.25 (m, 8H), 1.05 (s, 9H). HPLC > 95%, tR = 3.82 min, HRMS (ESI-TOF) m/z: [M + H]+ calcd for C57H73FN9O5S+, 1014.5434; found, 1014.5419.
(2S,4R)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-N-((S)-3-((4-(4-((4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylbenzyl)amino)butanamido)butyl)amino)-1-(4-(4-methylthiazol-5-yl)phenyl)-3-oxopropyl)-4-hydroxypyrrolidine-2-carboxamide (6).
1H NMR (800 MHz, Methanol-d4) δ 9.22 (s, 1H), 8.94 – 8.89 (m, 2H), 7.93 – 7.89 (m, 1H), 7.81 (d, J = 9.6 Hz, 1H), 7.59 – 7.56 (m, 1H), 7.47 (d, J = 11.7 Hz, 5H), 7.26 (s, 1H), 7.15 (s, 1H), 5.35 – 5.31 (m, 1H), 4.75 (s, 1H), 4.62 – 4.54 (m, 1H), 4.48 – 4.46 (m, 1H), 4.08 (s, 2H), 3.86 (d, J = 11.2 Hz, 1H), 3.81 – 3.77 (m, 1H), 3.76 (s, 3H), 3.16 – 3.01 (m, 8H), 2.86 (dd, J = 14.3, 6.3 Hz, 1H), 2.76 (dd, J = 14.2, 8.2 Hz, 1H), 2.49 (s, 3H), 2.32 (t, J = 6.7 Hz, 2H), 2.31 (s, 3H), 2.25 – 2.20 (m, 1H), 2.08 (s, 3H), 2.00 – 1.94 (m, 1H), 1.92 – 1.88 (m, 2H), 1.43 – 1.27 (m, 6H), 1.08 (s, 9H). HPLC > 95%, tR = 3.57 min, HRMS (ESI-TOF) m/z: [M + H]+ calcd for C58H71F2N10O6S+, 1073.5241; found, 1073.5204.
tert-butyl 4-((4-bromo-3,5-dimethylbenzyl)(tert-butoxycarbonyl)amino)butanoate (7).
To a solution of 4-bromo-3,5-dimethylbenzaldehyde (2.1 g, 10 mmol) and tert-butyl 4-aminobutanoate hydrochloride (2.0 g, 10 mmol) in DCM (30 mL) was added NaBH(OAc)3 (4.2 g, 20 mmol). The mixture was stirred at room temperature for 5 h. The reaction was monitored by UPLC. Upon completion, 100 mL water was added to quench the reaction. The mixture was then extracted with DCM (30 mL x 3). The combined organic phase was dried over Na2SO4 and then filtered.
The solvent was evaporated to give crude product without further purification. The crude product was dissolved in DCM (30 mL) and Boc2O (2.18 g, 10 mmol) and NEt3 (1.0 g, 10 mmol) were added to the solution. After being stirred at room temperature for 3 h, the mixture was purified by reverse-phase column chromatography (10−100% MeCN/0.1% TFA in H2O) to afford compound 7 (3.2 g, 71% yield) as a brown oil. 1H NMR (600 MHz, DMSO-d6) δ 7.03 (s, 2H), 4.29 (s, 2H), 3.18 – 3.10 (m, 2H), 2.35 (s, 6H), 2.16 (t, J = 7.2 Hz, 2H), 1.73 – 1.66 (m, 2H), 1.56 – 1.30 (m, 18H). 13C NMR (151 MHz, DMSO) δ 172.2, 155.5/155.1, 138.0, 127.9, 127.7, 125.6, 79.9, 79.3, 49.5/48.9, 46.0, 32.5, 28.5, 28.1, 23.9, 23.4. UPLC > 95%, tR = 1.61 min. MS (ESI) [M – Boc + 2H]+ = 356.1761.
(4-(((4-(tert-butoxy)-4-oxobutyl)(tert-butoxycarbonyl)amino)methyl)-2,6-dimethylphenyl)boronic acid (8).
To a solution of compound 7 (2.5 g, 5.6 mmol), B2(OH)4 (1.0 g, 11.2 mmol) and Pd(amphos)Cl2 (39.6 mg, 0.056 mmol) in MeOH (10 mL) and THF (10 mL) was added DIPEA (722 mg, 5.6 mmol). The mixture was stirred at 75 °C for 3 h. The reaction was monitored by UPLC. Upon completion, the mixture was purified by reverse-phase column chromatography (10−100% MeCN/0.1% TFA in H2O) to afford compound 8 (1.89 g, 80% yield) as a yellow oil. 1H NMR (600 MHz, Methanol-d4) δ 6.84 (s, 2H), 4.33 (s, 2H), 3.24 – 3.13 (m, 2H), 2.22 (s, 6H), 2.19 – 2.14 (m, 2H), 1.73 (p, J = 7.2 Hz, 2H), 1.55 – 1.33 (m, 18H). 13C NMR (151 MHz, DMSO) δ 172.3, 155.5/155.2, 139.1, 138.4, 137.8, 125.2, 80.0, 79.2, 49.9/49.4, 45.8/45.4, 32.5, 28.5, 28.2, 23.7/23.3, 22.5. UPLC > 95%, tR = 1.54 min. MS (ESI) [M + Na]+ = 444.2929.
4-(4-bromo-1-methyl-1H-imidazol-5-yl)-7-fluoroquinoline (9).
Compound 9 was synthesized based on the published procedures (Bottcher et al., 2019) and the details are provided below.
To a solution of 4-chloro-7-fluoro-quinoline (500 mg, 2.7 mmol) in NMP (20 ml) under argon atmosphere was added potassium acetate (814 mg; 8.3 mmol), 1-methyl-1H-imidazole (452 mg; 5.5 mmol) and palladium(II) acetate (60.8 mg; 0.27 mmol). The resulting mixture was stirred at 120 °C for 16 h. The progress of the reaction was monitored by UPLC and upon completion, the mixture was filtered through celite and concentrated under reduced pressure. Water was then added, and the mixture was extracted with DCM. The combined organic phases were dried over MgSO4 and concentrated under reduced pressure. The mixture was purified by normal phase chromatography (DCM/MeOH = 10/1) to afford the product as a yellow oil (380 mg, 60%), which was resuspended in MeCN (15 ml). NBS (288 mg, 1.62 mmol) was added to the above solution at 0 °C. The mixture is then slowly warmed to room temperature and stirred for 3 h. The progression of the reaction was monitored by UPLC and upon completion, 1N aqueous NaOH was added, and the mixture was extracted with DCM. The organic phases were collected, dried over MgSO4 and concentrated under reduced pressure. The mixture was purified by normal phase chromatography (DCM/MeOH = 20/1) to afford compound 9 as a yellow solid (254 mg, 51%) as previously reported. 1H NMR (600 MHz, DMSO-d6) δ 9.08 (d, J = 4.4 Hz, 1H), 8.00 (s, 1H), 7.90 (dd, J = 10.2, 2.7 Hz, 1H), 7.74 – 7.67 (m, 1H), 7.64 – 7.55 (m, 2H), 3.49 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 162.92 (d, J = 249.0 Hz), 151.97, 149.58 (d, J = 12.9 Hz), 140.23, 134.84, 128.96 (d, J = 9.9 Hz), 125.84, 123.85, 123.79 (d, J = 2.3 Hz), 118.29 (d, J = 25.3 Hz), 116.17, 113.50 (d, J = 20.0 Hz), 33.45. UPLC > 95%, tR = 1.18 min. MS (ESI) [M + H]+ = 306.0494.
tert-butyl-4-((tert-butoxycarbonyl)(4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylbenzyl)amino)butanoate (10).
To a solution of compound 8 (421 mg, 1.0 mmol), compound 9 (306 mg, 1.0 mmol) and K2CO3 (276 mg, 2.0 mmol) in dioxane (6 mL) and H2O (3 mL) was added Pd(PPh3)4 (115 mg, 0.1 mmol). The mixture was stirred in a microwave at 125 °C for 2 h. Upon completion, the reaction mixture was purified by reverse-phase column chromatography (10−100% MeCN/0.1% TFA in H2O) to afford compound 10 (427 mg, 71% yield) as a yellow oil. 1H NMR (600 MHz, DMSO-d6) δ 9.48 (s, 1H), 8.99 (d, J = 4.4 Hz, 1H), 7.96 – 7.90 (m, 1H), 7.87 (dd, J = 10.0, 2.7 Hz, 1H), 7.64 – 7.57 (m, 1H), 7.48 (s, 1H), 6.90 (s, 1H), 6.81 (s, 1H), 4.30 – 4.18 (m, 2H), 3.63 (s, 3H), 3.18 – 3.01 (m, 2H), 2.18 – 2.09 (m, 5H), 1.95 (s, 3H), 1.63 (p, J = 7.3 Hz, 2H), 1.48 – 1.02 (m, 18H). UPLC > 95%, tR = 1.44 min. MS (ESI) [M + H]+ = 603.4466.
4-((tert-butoxycarbonyl)(4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylbenzyl)amino)butanoic acid (11).
Compound 10 (427 mg, 0.71 mmol) was dissolved in DCM (4 mL) and TFA (4 mL). The resulting mixture was stirred for 30 min. Then it was concentrated to give the crude product without further purification. This crude product was dissolved in DCM. Boc2O (186 mg, 0.85 mmol) and NEt3 (72 mg, 0.71 mmol) were added to the solution. After being stirred at room temperature for 3 h, the mixture was purified by reverse-phase column chromatography (10−100% methanol/0.1% TFA in H2O) to afford compound 11 (330 mg, 85% yield) as a yellow oil. 1H NMR (600 MHz, DMSO-d6) δ 9.53 (s, 1H), 9.00 (d, J = 4.4 Hz, 1H), 7.98 – 7.91 (m, 1H), 7.88 (dd, J = 9.9, 2.7 Hz, 1H), 7.65 – 7.58 (m, 1H), 7.48 (s, 1H), 6.92 (s, 1H), 6.82 (s, 1H), 4.22 (d, J = 20.4 Hz, 2H), 3.63 (s, 3H), 3.20 – 3.06 (m, 2H), 2.21 – 2.11 (m, 5H), 1.99 – 1.92 (m, 3H), 1.65 – 1.62 (m, 2H), 1.50 – 1.05 (m, 9H). UPLC > 95%, tR = 1.21 min. MS (ESI) [M + H]+ = 547.2515.
(S)-3-((2S,4R)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxamido)-3-(4-(4-methylthiazol-5-yl)phenyl)propanoic acid (12).
Compound 12 was synthesized based on the published procedures (Han et al., 2019) as detailed below.
A solution of (S)-3-(4-bromophenyl)-3-((tert-butoxycarbonyl)amino)propanoic acid (200 mg, 0.58 mmol), 4-methylthiazole (115 mg, 1.16 mmol), KOAc (110 mg, 1.16 mmol) and Pd(OAc)2 (2.6 mg, 0.0116 mmol) in DMF/TEA (1.5 mL/ 1.5 mL) was stirred at 80 °C for 4 h. After the reaction completed, TEA was removed under reduced pressure then water was added into the mixture, the mixture was extracted by EA. The organic phases were collected together, and dried with NaSO4 and evaporated under reduced pressure to give (S)-3-((tert-butoxycarbonyl)amino)-3-(4-(4-methylthiazol-5-yl)phenyl)propanoic acid (149 mg, 71%). UPLC > 95%, tR = 1.38 min. MS (ESI) [M + H]+ = 363.1841. To a solution of (S)-3-((tert-butoxycarbonyl)amino)-3-(4-(4-methylthiazol-5-yl)phenyl)propanoic acid (362 mg, 1.0 mmol) in MeOH (10 mL) was added 0.5 mL HCl (conc.). The resulting mixture was stirred at 80 °C overnight. The solvent was evaporated under reduced pressure to give methyl (S)-3-amino-3-(4-(4-methylthiazol-5-yl)phenyl)propanoate as a yellow oil, which could be used without further purification. UPLC > 95%, tR = 1.04 min. MS (ESI) [M + H]+ = 277.0591.
A mixture of (S)-2-((tert-butoxycarbonyl)amino)-3,3-dimethylbutanoic acid (231 mg, 1.0 mmol), methyl (2S,4R)-4-hydroxypyrrolidine-2-carboxylate hydrogen chloride salt (182 mg, 1.0 mmol), HOAT (272 mg, 2.0 mmol), EDCI (382 mg, 2.0 mmol) and NMM (0.4 mL) in DMSO (5 mL) was stirred at room temperature overnight. Then the mixture was purified by reverse-phase column chromatography (10−100% MeCN/0.1% TFA in H2O) to afford methyl (2S,4R)-1-((S)-2-((tert-butoxycarbonyl)amino)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxylate (233 mg, 65%). UPLC > 95%, tR = 1.40 min. MS (ESI) [M + H]+ = 359.0351. (2S,4R)-1-((S)-2-((tert-butoxycarbonyl)amino)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxylate (358 mg, 1.0 mmol) was then dissolved in DCM/TFA (6 mL/3 mL). The resulting mixture was stirred at room temperature for 1 h. Then the solvent was evaporated under reduced pressure to give methyl (2S,4R)-1-((S)-2-amino-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxylate as a yellow oil, which could be used without further purification (1.0 mmol) and mixed with 1-fluorocyclopropane-1-carboxylic acid (104 mg, 1.0 mmol), HOAT (272 mg, 2.0 mmol), EDCI (382 mg, 2.0 mmol) and NMM (0.4 mL) in DMSO (5 mL) and the mixture allowed to stirr at room temperature overnight. Then the mixture was purified by reverse-phase column chromatography (10−100% MeCN/0.1% TFA in H2O) to afford methyl (2S,4R)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxylate (203 mg, 59%). UPLC > 95%, tR = 1.50 min. MS (ESI) [M + H]+ = 345.3384. A mixture of methyl (2S,4R)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxylate (172 mg, 0.5 mmol) and LiOH (60 mg, 2.5 mmol) in THF/H2O (2 mL/2 mL) was stirred at room temperature overnight. Then the mixture was purified by reverse-phase column chromatography (10−100% Methanol/0.1% TFA in H2O) to afford methyl (2S,4R)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxylic acid (155 mg, 94%). UPLC > 95%, tR = 1.17 min. MS (ESI) [M + H]+ = 331.1922.
A mixture of (S)-3-amino-3-(4-(4-methylthiazol-5-yl)phenyl)propanoate (138 mg, 0.5 mmol), HOAT (136 mg, 1.0 mmol), EDCI (192 mg, 1.0 mmol), (2S,4R)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxylic acid (166 mg, 0.5 mmol) and NMM (0.2 mL) in DMSO (5 mL) was stirred at room temperature overnight. Then the mixture was purified by reverse-phase column chromatography (10−100% MeCN/0.1% TFA in H2O) to afford methyl (S)-3-((2S,4R)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxamido)-3-(4-(4-methylthiazol-5-yl)phenyl)propanoate (159 mg, 54%). UPLC > 95%, tR = 1.80 min. MS (ESI) [M + H]+ = 589.0985. A mixture of LiOH (30 mg, 1.25 mmol) and (S)-3-((2S,4R)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxamido)-3-(4-(4-methylthiazol-5-yl)phenyl)propanoate (147 mg, 0.25 mmol) in THF/H2O (2 mL/2 mL) was stirred at room temperature overnight. Then the mixture was purified by reverse-phase column chromatography (10−100% Methanol/0.1% TFA in H2O) to afford the title compound (S)-3-((2S,4R)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxamido)-3-(4-(4-methylthiazol-5-yl)phenyl)propanoic acid (12) (130 mg, 91%). The NMR spectra of the product were in agreement with the reported spectra. UPLC > 95%, tR = 1.13 min. MS (ESI) [M + H]+ = 575.3002.
(2S,4R)-N-((S)-3-((4-aminobutyl)amino)-1-(4-(4-methylthiazol-5-yl)phenyl)-3-oxopropyl)-1-((S)-2-(1-fluorocyclopropane-1-carboxamido)-3,3-dimethylbutanoyl)-4-hydroxypyrrolidine-2-carboxamide (13).
To a solution of compound 12 (57 mg, 0.1 mmol) and tert-butyl (4-aminobutyl)carbamate (19 mg, 0.1 mmol) in DMSO (1 mL) were added HOAt (28 mg, 0.2 mmol), EDCI (38 mg, 0.2 mmol), and 4-methylmorpholine (50 mg, 0.5 mmol). The reaction mixture was stirred at room temperature overnight. The reaction was monitored by UPLC. Upon completion, the reaction mixture was purified by preparative HPLC to give the desired intermediate as a white solid. This intermediate then was dissolved in DCM (2 mL) and trifluoroacetic acid (2 mL). The reaction mixture was stirred at room temperature for 1 h. The reaction was monitored by UPLC. Upon completion, the solvent was evaporated, and the residue was purified by preparative HPLC to afford compound 13 (45.1 mg, 70% yield for two steps) as a white solid. 1H NMR (600 MHz, Methanol-d4) δ 8.98 (s, 1H), 7.50 – 7.47 (m, 4H), 5.38 – 5.32 (m, 1H), 4.78 – 4.73 (m, 1H), 4.62 – 4.56 (m, 1H), 4.48 – 4.43 (m, 1H), 3.88 – 3.83 (m, 1H), 3.78 (dd, J = 11.1, 3.8 Hz, 1H), 3.23 – 3.09 (m, 2H), 2.96 – 2.87 (m, 2H), 2.86 (dd, J = 14.3, 6.8 Hz, 1H), 2.76 (dd, J = 14.3, 7.9 Hz, 1H), 2.51 (s, 3H), 2.26 – 2.19 (m, 1H), 1.96 (ddd, J = 13.5, 9.3, 4.4 Hz, 1H), 1.64 – 1.54 (m, 2H), 1.56 – 1.48 (m, 2H), 1.44 – 1.27 (m, 4H), 1.08 (s, 9H). UPLC > 95%, tR = 1.04 min. MS (ESI) [M + H]+ = 645.4623.
2S,4R)-1-((S)-2-amino-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (14).
Compound 14 was synthesized based on the published procedures (Hu et al., 2019) as detailed below.
A mixture of (S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethan-1-amine (218 mg, 1.0 mmol), HOAT (272 mg, 2.0 mmol), EDCI (380 mg, 2.0 mmol), (2S,4R)-1-(tert-butoxycarbonyl)-4-hydroxypyrrolidine-2-carboxylic acid (231 mg, 1.0 mmol) and NMM (0.4 mL) in DMSO (5 mL) was stirred at room temperature for 24 h. Then the mixture was purified by reverse-phase column chromatography (10−100% methanol/0.1% TFA in H2O) to afford tert-butyl (2S,4R)-4-hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidine-1-carboxylate as a white solid (300 mg, 70%), which was dissolved in DCM/TFA (6 mL/3 mL). The resulting mixture was stirred at room temperature for 1 h. The mixture was then concentrated, and the residue was dried under vacuum to afford crude product, which was used in next step without further purification and mixed with HOAT (190 mg, 1.4 mmol), EDCI (268 mg, 1.4 mmol), (S)-2-((tert-butoxycarbonyl)amino)-3,3-dimethylbutanoic acid (162 mg, 0.7 mmol) and NMM (0.3 mL) in DMSO (5 mL). The resulting mixture was stirred at room temperature for 24 h. Then the mixture was purified by reverse-phase column chromatography (10−100% methanol/0.1% TFA in H2O) to afford tert-butyl ((S)-1-((2S,4R)-4-hydroxy-2-(((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)carbamate (335 mg, 88%), which was resuspended in DCM/TFA (6 mL/3 mL) and was stirred at room temperature for 1 h. The mixture was then concentrated, and the residue was dried under vacuum to afford compound 14, which could be used without further purification for the next steps.
(2S,4R)-1-((S)-2-(9-aminononanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (15).
To a solution of compound 14 (44 mg, 0.1 mmol) and 9-((tert-butoxycarbonyl)amino)nonanoic acid (27 mg, 0.1 mmol) in DMSO (1 mL) were added HOAt (28 mg, 0.2 mmol), EDCI (38 mg, 0.2 mmol), and 4-methylmorpholine (50 mg, 0.5 mmol). The reaction mixture was stirred at room temperature overnight. The reaction was monitored by UPLC. Upon completion, the reaction mixture was purified by preparative HPLC to give the desired intermediate (58.2 mg, 83% yield) as a white solid. This intermediate then was dissolved in DCM (2 mL) and trifluoroacetic acid (2 mL). The reaction mixture was stirred at room temperature for 1 h. The reaction was monitored by UPLC. Upon completion, the solvent was evaporated, and the residue was purified by preparative HPLC to obtain compound 15 (47.3 mg, 79% yield for two steps) as a white solid. 1H NMR (600 MHz, Methanol-d4) δ 8.95 (s, 1H), 7.36 (d, J = 8.5 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 4.91 (q, J = 6.9 Hz, 1H), 4.53 (s, 1H), 4.50 – 4.44 (m, 1H), 4.36 – 4.31 (m, 1H), 3.80 – 3.75 (m, 1H), 3.65 (dd, J = 11.0, 4.0 Hz, 1H), 2.81 (t, J = 7.7 Hz, 2H), 2.40 (s, 3H), 2.25 – 2.06 (m, 3H), 1.86 (ddd, J = 13.3, 9.0, 4.5 Hz, 1H), 1.60 – 1.46 (m, 4H), 1.41 (d, J = 7.0 Hz, 3H), 1.27 (tt, J = 14.6, 8.4 Hz, 8H), 0.94 (s, 9H). 13C NMR (151 MHz, Methanol-d4) δ 176.8, 174.1, 173.2, 154.3, 148.8, 146.9, 131.8, 131.4, 128.6, 128.4, 71.8, 61.4, 59.9, 58.8, 51.0, 41.6, 39.7, 37.5, 37.4, 31.0, 31.0, 30.8, 29.4, 28.2, 27.9, 27.8, 23.2, 16.2. UPLC > 95%, tR = 1.04 min. MS (ESI) [M + H]+ = 600.3870.
(2S,4R)-1-((S)-2-(9-(4-((4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylbenzyl)amino)butanamido)nonanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (MS9715)
To a solution of compound 11 (10.0 mg, 0.018 mmol) and compound 15 (10.0 mg, 0.019 mmol) in DMSO (1.0 mL) were added HOAt (4.9 mg, 0.036 mmol), EDCI (7.0 mg, 0.036 mmol) and NMM (9.1 mg, 0.09 mmol). The reaction mixture was stirred at room temperature overnight. The reaction was monitored by UPLC. Upon completion, the mixture was purified by reverse phase HPLC to give product. This product was dissolved in DCM (1 mL) and TFA (1 mL). The resulting mixture was stirred for 30 min. Then, it was concentrated and purified by preparative HPLC to give compound MS9715 (11.1 mg, 60% yield) as a white solid. 1H NMR (600 MHz, Methanol-d4) δ 9.38 (s, 1H), 9.04 – 8.99 (m, 1H), 8.95 (d, J = 4.5 Hz, 1H), 7.93 (dd, J = 9.2, 5.7 Hz, 1H), 7.82 (dd, J = 9.7, 2.6 Hz, 1H), 7.62 – 7.56 (m, 1H), 7.50 (d, J = 4.4 Hz, 1H), 7.51 – 7.42 (m, 4H), 7.28 (s, 1H), 7.16 (s, 1H), 5.02 (q, J = 6.9 Hz, 1H), 4.67 – 4.63 (m, 1H), 4.62 – 4.56 (m, 1H), 4.48 – 4.43 (m, 1H), 4.11 – 4.07 (m, 2H), 3.90 (d, J = 11.1 Hz, 1H), 3.78 (s, 3H), 3.80 – 3.74 (m, 1H), 3.16 (t, J = 7.1 Hz, 2H), 3.06 (t, J = 7.3 Hz, 2H), 2.53 – 2.48 (m, 3H), 2.35 (t, J = 6.8 Hz, 2H), 2.31 (s, 3H), 2.35 – 2.18 (m, 3H), 2.09 (s, 3H), 2.03 – 1.88 (m, 3H), 1.64 – 1.57 (m, 2H), 1.55 – 1.46 (m, 5H), 1.39 – 1.29 (m, 8H), 1.07 – 1.02 (m, 9H). 13C NMR (151 MHz, Methanol-d4) δ 174.6, 173.1, 171.9, 170.9, 163.5 (d, J = 252.7 Hz), 151.9, 151.0, 149.1 (d, J = 12.4 Hz), 146.9, 144.5, 140.3, 139.7, 137.7, 133.9, 132.5, 130.3, 129.7, 129.1, 128.8 (d, J = 3.6 Hz), 128.3, 127.3 (d, J = 10.0 Hz), 126.3, 126.2 (d, J = 16.1 Hz), 123.5 (d, J = 2.5 Hz), 123.4, 118.7 (d, J = 25.7 Hz), 112.8 (d, J = 20.9 Hz), 69.6, 59.2, 57.6, 56.6, 50.1, 48.8, 39.1, 37.4, 35.2, 35.1, 34.0, 32.5, 28.9, 28.9, 28.8, 28.8, 26.5, 25.6, 25.6, 21.5, 21.0, 19.2, 19.1, 14.1. HPLC > 99%, tR = 3.93 min, HRMS (ESI-TOF) m/z: [M + H]+ calcd for C58H75FN9O5S+, 1028.5590; found, 1028.5584.
(2R,4S)-1-((S)-2-amino-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (16).
Compound 16 was synthesized based on the published procedures (Hu et al., 2019) using (2R,4S)-1-(tert-butoxycarbonyl)-4-hydroxypyrrolidine-2-carboxylic acid following the same producers provided above for compound 14.
(2R,4S)-1-((S)-2-(9-aminononanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (17).
Using the above procedure for the synthesis of compound 15, compound 17 was obtained as a white solid (43.1 mg, 72% yield for two steps). 1H NMR (600 MHz, Methanol-d4) δ 9.07 – 9.05 (m, 1H), 7.61 – 7.52 (m, 2H), 7.52 – 7.44 (m, 2H), 5.03 (q, J = 7.0 Hz, 1H), 4.60 – 4.55 (m, 1H), 4.54 (s, 1H), 4.50 – 4.45 (m, 1H), 3.94 (dd, J = 10.8, 4.9 Hz, 1H), 3.71 (ddd, J = 10.9, 3.5, 1.2 Hz, 1H), 2.91 (q, J = 7.7, 6.3 Hz, 2H), 2.53 (d, J = 1.2 Hz, 3H), 2.34 – 2.26 (m, 1H), 2.26 – 2.17 (m, 2H), 2.15 – 2.08 (m, 1H), 1.67 – 1.57 (m, 4H), 1.48 (d, J = 6.9 Hz, 3H), 1.41 – 1.27 (m, 8H), 1.07 (s, 9H). 13C NMR (151 MHz, Methanol-d4) δ 174.9, 172.2, 170.7, 151.9, 146.7, 144.4, 132.6, 129.0, 126.9, 126.5, 69.2, 59.3, 57.9, 55.5, 48.5, 39.3, 37.6, 35.2, 34.4, 28.8, 28.7, 28.6, 27.1, 25.9, 25.6, 25.4, 21.2, 14.1. UPLC > 95%, tR = 1.20 min. MS (ESI) [M + H]+ = 600.1534.
(2R,4S)-1-((S)-2-(9-(4-((4-(5-(7-fluoroquinolin-4-yl)-1-methyl-1H-imidazol-4-yl)-3,5-dimethylbenzyl)amino)butanamido)nonanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide (MS9715N).
Using the above procedure for the synthesis of compound MS9715, compound MS9715N was obtained as a white solid (8.4 mg, 45% yield). 1H NMR (600 MHz, Methanol-d4) δ 9.40 (s, 1H), 9.09 (s, 1H), 8.97 – 8.93 (m, 1H), 7.96 – 7.88 (m, 1H), 7.86 – 7.80 (m, 1H), 7.62 – 7.56 (m, 1H), 7.57 – 7.52 (m, 2H), 7.53 – 7.44 (m, 3H), 7.28 (s, 1H), 7.16 (s, 1H), 5.04 (q, J = 6.9 Hz, 1H), 4.57 (dd, J = 8.2, 6.5 Hz, 1H), 4.54 – 4.49 (m, 1H), 4.47 (p, J = 8.4, 4.1 Hz, 1H), 4.11 – 4.07 (m, 2H), 3.95 (dd, J = 10.7, 4.9 Hz, 1H), 3.78 (d, J = 2.1 Hz, 3H), 3.71 (dd, J = 10.7, 3.5 Hz, 1H), 3.18 – 3.10 (m, 2H), 3.08 – 3.02 (m, 2H), 2.53 (s, 2H), 2.51 (d, J = 10.6 Hz, 1H), 2.38 – 2.26 (m, 6H), 2.26 – 2.17 (m, 2H), 2.16 – 2.08 (m, 1H), 2.11 – 2.07 (m, 3H), 1.96 – 1.87 (m, 2H), 1.62 – 1.52 (m, 1H), 1.49 – 1.43 (m, 4H), 1.35 – 1.25 (m, 10H), 1.07 (s, 9H). 13C NMR (151 MHz, Methanol-d4) δ 175.0, 173.1, 172.1, 170.8, 163.5 (d, J = 252.5 Hz), 152.1, 150.9, 149.0 (d, J = 13.0 Hz), 146.4, 144.5, 140.3, 139.7, 137.6, 133.9, 132.5, 130.2, 129.3, 129.0, 128.8 (d, J = 3.8 Hz), 128.3, 127.3 (d, J = 10.0 Hz), 126.5, 126.1, 123.5, 123.4, 118.7 (d, J = 25.8 Hz), 112.8 (d, J = 21.2 Hz), 69.1, 59.3, 58.1, 55.5, 50.1, 48.5, 39.1, 37.6, 35.2, 34.3, 34.1, 32.5, 28.9, 28.9, 28.8, 26.5, 25.6, 25.5, 21.5, 21.2, 19.2, 19.1, 13.9. HPLC > 99%, tR = 3.97 min, HRMS (ESI-TOF) m/z: [M + H]+ calcd for C58H75FN9O5S+, 1028.5590; found, 1028.5579.
Inducible CRISPR/Cas9-mediated gene knockout (KO)
The oligos of sgRNAs (with sequence information listed in Table S8) were designed based on the CRISPR sgRNA Database on GenScript website (https://www.genscript.com/gRNA-database.html) and cloned into a pLenti LRG-2.1_Neo vector (Addgene,125593). For human NSD3 sgRNA sequences, we used as designed before (Bottcher et al., 2019). A doxycycline (dox)-inducible lentiviral expression vector that contains SpCas9, pCW-Cas9, was obtained from Addgene (cat# 50661) and used for preparation of viral particles after co-transfection into 293FT cells with the packaging plasmids psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259). Viral infection was carried out as described before (Cai et al., 2018), followed by cell selection with 1 μg/mL of puromycin for 4 days. The final concentration of 2 μg/mL dox was added in cell culture for Cas9 induction.
Antibodies
Antibodies used for western blot included rabbit anti-NSD3 (WHSC1L1; Cell Signaling Technology, 92056), rabbit anti-GAPDH (Cell Signaling Technology, 5174), rabbit anti-CBLB (Cell Signaling Technology, 9498), rabbit anti-IFITM1 (Cell Signaling Technology, 13126), mouse anti-VHL (Santa Cruz, SC135657), rabbit anti-cMyc (Cell Signaling, 9402), rabbit anti-HA tag (Cell Signaling, 3724), mouse anti-Flag tag (Sigma, F1804), mouse anti-Ub (Santa Cruz, SC8017), rabbit anti-cleaved caspase-3 (Cell Signaling, 9661), rabbit anti-cleaved caspase-7 (Cell Signaling, 8438) and the HRP-linked secondary anti-mouse IgG antibody (7076) and anti-rabbit IgG antibody (7074) from Cell Signaling Technology. Anti-FLAG M2 magnetic bead was obtained from Sigma (cat# M8823).
Colony formation assay
20,000 cells per well were plated in triplicate into 6-well plates. Every four days, 1 mL of fresh media containing either compound or vehicle was used to replace the original culture medium. After 4 weeks’ incubation, plates were stained with 100 μg/mL of iodonitrotetrazoliuim chloride solution (Sigma) and numbers of cell colonies counted after incubation overnight.
Cell growth inhibition assay
Cell growth inhibition assay was performed as described previously (Xu et al., 2015). In brief, 0.5 million of cells per well were seeded in triplicate into 24-well plates, subjected to treatment with various final concentrations of compound. Fresh medium containing compound was changed every two days. All flow-growing cells were periodically diluted to keep the cell density less than 1×106/mL. Cell numbers were counted by an automated TC10 cell counter (BioRad) every two days. Effective control to 50% growth inhibition (EC50) values were calculated via a nonlinear regression analysis by using data from at least three experiments and presented as the mean ± SD.
Immunoprecipitation (IP) and ubiquitination assay
The cell pellets were lysed in the EBC buffer (50 mM Tris pH 8.0, 120 mM NaCl, 0.5% NP40, 0.1 mM EDTA and 10% glycerol; freshly supplemented with the protease inhibitor cocktail before use) on ice for 30 min. After brief sonication, cells were spun at 12,000 g for 5 min at 4°C to remove debris. 1 mg of proteins from the whole cell lysate was incubated with antibodies against protein-of-interest overnight at 4°C, followed by addition of 10 μL of protein A/G magnetic beads (BioRad) and rotation for an additional three hours at 4°C. For Flag-tagged protein IP, the Flag M2 magnetic beads (Sigma, #M8823) were incubated with the cell lysate overnight at 4°C. For the ubiquitination assay, cell pellets were lysed in 100 μL of EBC buffer containing 1% of SDS. Cell extracts were heat-denatured for 5 min at 95°C and diluted with 900 μL of EBC buffer. After sonication and centrifugation, cell lysates were subjected to IP with antibodies, followed by immunoblotting with anti-ubiquitin antibody.
Reverse transcription followed by quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted using the RNeasy Mini Kit (Qiagen #74104) according to manufacturer’s manual. 1 μg of total RNA was subjected to reverse transcription using cDNA Reverse Transcription kit according to the manufacturer’s protocols (Invitrogen). Then, real-time PCR using Power SYBR Green Master Mix (Thermo Fisher Scientific) was performed on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). The relative abundance of gene expression was calculated as previously described (Cai et al., 2018; Lu et al., 2016; Ren et al., 2019). In brief, the relative gene expression was calculated by using the 2−ΔΔCT method and normalized to the mean of an internal control (the housekeeping gene beta-actin). Primers used for RT-qPCR were listed in Table S8.
Isothermal titration calorimetry (ITC) assay
Construct of the NSD3-PWWP1 domain (residues 247–402) in pNICBio2 was expressed in E. coli and purified as described in a previous publication (Bottcher et al., 2019). Binding of BI9321, MS9715 and MS9715N was analyzed at 15 °C using a MicroCal iTC200 (Malvern) in a solution of 50 mM Tris-HCl pH 6.8, 150 mM NaCl, 1% DMSO and 3% NMP. After an initial 0.4 μL injection, 13 injections from the syringe solution (200 μM of compounds) were titrated into 300 μL of the protein solution (20 μM of NSD3) in the cell, which was stirred at 750 rpm. The data were fitted by single binding site model using Microcal Origin 7.0 (Malvern). The reported values represent the mean ± SD from two independent measurements.
RNA sequencing (RNA-seq)
RNA-seq was performed as described before (Cai et al., 2018; Fan et al., 2020; Ren et al., 2019). Briefly, EOL-1 cells, either treated with DMSO or 2.5 μM of compound (either BI-9321, MS9715 or MS9715N) for four days, were collected for isolation of total RNA using the RNeasy Mini Kit (Qiagen #74104). Also, cells, which were stably transduced with an sgRNA-expressing vector and a dox-inducible Cas9, were treated with 2μg/ml of dox for four days to induce Cas9 expression, followed by total RNA preparation. RNA was processed using NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, #E7490) and NEBNext Ultra II RNA library Prep kit (NEB, #E7770) following manufacturer’s instructions. The final multiplexed RNA-seq libraries were assessed for quality and quantity with Qubit and TapeStation (Aglient) and subjected to deep sequencing with an Illumina Sequencing platform as described before (Cai et al., 2018; Fan et al., 2020; Ren et al., 2019).
Proteomics profiling using a tandem mass tag (TMT) isobaric labeling method and an Orbitrap Eclipse mass spectrometer
EOL-1 cells were seeded at 3 × 106 cells/10mL in the 10 cm plates and treated with 2.5 μM of DMSO or degrader for 30 hours. Cells were harvested and washed three times with 1 x PBS. Then the cells were centrifuged at 300 g for 5 minutes to collect cell pellets in a refrigerated microfuge and frozen at −80 °C until further analysis. Proteins were reduced, alkylated, and purified by chloroform/methanol extraction prior to digestion with sequencing grade modified porcine trypsin (Promega). Tryptic peptides were labeled using tandem mass tag isobaric labeling reagents (Thermo) following the manufacturer’s instructions and combined into one 10-plex sample group. The labeled peptide multiplex was separated into 36 fractions on a 100 × 1.0 mm Acquity BEH C18 column (Waters) using an UltiMate 3000 UHPLC system (Thermo) with a 40 min gradient from 99:1 to 60:40 buffer A:B ratio under basic pH conditions (Buffer A = 0.1% formic acid, 0.5% acetonitrile; Buffer B = 0.1% formic acid, 99.9% acetonitrile; and both buffers adjusted to pH 10 with ammonium hydroxide for offline separation), and then consolidated into 18 super-fractions. Each super-fraction was then further separated by reverse phase XSelect CSH C18 2.5 um resin (Waters) on an in-line 150 × 0.075 mm column using an UltiMate 3000 RSLCnano system (Thermo). Peptides were eluted using a 60 min gradient from 97:3 to 60:40 buffer A:B ratio. Eluted peptides were ionized by electrospray (2.2 kV) followed by mass spectrometric analysis on an Orbitrap Eclipse Tribrid mass spectrometer (Thermo) using multi-notch MS3 parameters. MS data were acquired using the FTMS analyzer in top-speed profile mode at a resolution of 120,000 over a range of 375 to 1500 m/z. Following CID activation with normalized collision energy of 35.0, MS/MS data were acquired using the ion trap analyzer in centroid mode and normal mass range. Using synchronous precursor selection, up to 10 MS/MS precursors were selected for HCD activation with normalized collision energy of 65.0, followed by acquisition of MS3 reporter ion data using the FTMS analyzer in profile mode at a resolution of 50,000 over a range of 100–500 m/z.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using the GraphPad Prism version 8 software. The unpaired two-tailed Student’s t-test was used for experiments comparing two sets of data. Data are presented as mean ± SD from at least three independent experiments. *, **, and *** denote the P value of <0.05, 0.01 and 0.001, respectively. NS denotes not significant. As for mass spectrometry and RNA-seq datasets, the methods used for statistical analysis are described in the above sections.
For MS/MS data analysis, the obtained MS/MS data was searched against the most recent Uniprot human database containing both the Swiss Prot and the TREMBL entries using MaxQuant. Protein TMT MS3 reporter ion intensity values are assessed for quality using our in-house ProteiNorm app, a user-friendly tool for a systematic evaluation of normalization methods, imputation of missing values and comparisons of different differential abundance methods (proteiNorm; https://github.com/ByrumLab/proteiNorm). Popular normalization methods were evaluated including log2 normalization (Log2), median normalization (Median), mean normalization (Mean), variance stabilizing normalization (VSN) (Huber et al., 2002), quantile normalization (Quantile) (preprocessCore version 1.46.0, https://github.com/bmbolstad/preprocessCore), cyclic loess normalization (Cyclic Loess) (Ritchie et al., 2015), global robust linear regression normalization (RLR), and global intensity normalization (Global Intensity) (Chawade et al., 2014). The individual performance of each method was evaluated by comparing of the following metrices: total intensity, Pooled intragroup Coefficient of Variation (PCV), Pooled intragroup Median Absolute Deviation (PMAD), Pooled intragroup estimate of variance (PEV), intragroup correlation, sample correlation heatmap (Pearson), and log2-ratio distributions. The cyclic loess normalization performed the best on the data and was used to perform statistical analysis using Linear Models for Microarray Data (limma) with empirical Bayes (eBayes) smoothing to the standard errors (Ritchie et al., 2015). Proteins with an |Log10Fold-change| more than 0.5 and P value less than 0.01 were considered to be significant. Data processing and statistical analysis were performed on Perseus (version 1.6.2.2).
For RNA-seq data analysis, RNA-seq reads were first mapped to the reference genome followed by differential gene expression (DEG) analysis as we described before (Cai et al., 2018; Fan et al., 2020; Li et al., 2021; Ren et al., 2019). In brief, sequencing reads were mapped using MapSplice (Wang et al., 2010) and quantified using RSEM (Li and Dewey, 2011). Read counts were then upper-quantile normalized and log2 transformed. Raw read counts were used for DEG analysis by DESeq (Anders and Huber, 2010). GSEA was carried out by using the GSEA program (Subramanian et al., 2005) as described before (Fan et al., 2020; Li et al., 2021; Ren et al., 2019). For RNA-seq DESeq2 results, P value is calculated by using the Wald test and adjusted P (padj) value calculated using the Benjamini-Hochberg method for multiple test correction. The P value in GSEA results is calculated by an empirical phenotype-based permutation test and the respective false discovery rate (FDR) value is further adjusted for gene set size and multiple hypotheses testing while the P value is not.
Supplementary Material
Supplemental Table S2. RNAseq identifies differentially expressed genes (DEGs) showing significant up-regulation in the EOL-1 cells treated with 2.5 uM of MS9715, in comparison to DMSO, for 4 days. Related to Figure 4.
Supplemental Table S3. RNAseq identifies differentially expressed genes (DEGs) showing significant up-regulation in the EOL-1 cells with NSD3 knockout (KO) (sgNSD3#1), in comparison to empty vector control (sgEV). Related to Figure 4.
Supplemental Table S4. RNAseq identifies differentially expressed genes (DEGs) showing significant down-regulation in the EOL-1 cells treated with 2.5 μM of BI-9321, in comparison to control (DMSO), for 4 days. Related to Figure 5.
Supplemental Table S5. RNAseq identifies differentially expressed genes (DEGs) showing significant up-regulation in the EOL-1 cells treated with 2.5 μM of MS9715N, in comparison to control (DMSO), for 4 days. Related to Figure 5.
Supplemental Table S6. RNAseq identifies differentially expressed genes (DEGs) showing significant down-regulation in the EOL-1 cells treated with 2.5 μM of MS9715, in comparison to a negative control (MS9715N lacking VHL binding activity), for 4 days. Related to Figure 5.
Supplemental Table S7. RNAseq identifies differentially expressed genes (DEGs) showing significant down-regulation in the EOL-1 cells treated with MS9715, in comparison to BI-9321, for 4 days. Related to Figure 5.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FLAG® M2 antibody | Millipore Sigma | Cat # F1804; RRID:AB_262044 |
| VHL mouse polyclonal | Santa Cruz Biotechnology | Cat # SC135657; RRID:AB_2215955 |
| Ub mouse monoclonal (mAb) | Santa Cruz Biotechnology | Cat # SC8017; RRID:AB_628423 |
| NSD3 rabbit mAb | Cell Signaling Technology | Cat # 92056; RRID:AB_2800178 |
| HA tag rabbit mAb | Cell Signaling Technology | Cat # 3724; RRID:AB_1549585 |
| IFITM1 rabbit mAb | Cell Signaling Technology | Cat # 13126; RRID:AB_2798126 |
| Cleaved caspase-3 rabbit mAb | Cell Signaling Technology | Cat # 9661; RRID:AB_2341188 |
| Cleaved caspase-7 rabbit mAb | Cell Signaling Technology | Cat # 8438; RRID:AB_11178377 |
| α-Tubulin rabbit mAb | Cell Signaling Technology | Cat # 2144; RRID:AB_2210548 |
| GAPDH rabbit mAb | Cell Signaling Technology | Cat # 5174; RRID:AB_10622025 |
| cMyc rabbit mAb | Cell Signaling Technology | Cat # 9402; RRID:AB_2151827 |
| CBLB rabbit mAb | Cell Signaling Technology | Cat # 9498; RRID:AB_2797707 |
| Normal rabbit IgG | Cell Signaling Technology | Cat # 2729; RRID:AB_1031062 |
| Anti-mouse IgG, HRP-linked antibody | Cell Signaling Technology | Cat # 7076; RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked antibody | Cell Signaling Technology | Cat # 7074; RRID:AB_2099233 |
| Bacterial and Virus Strains | ||
| DH5a competent cells | Thermo Fisher Scientific | Cat# 18265017 |
| One Shot Stbl3 competent cells | Thermo Fisher Scientific | Cat# C737303 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MS9715 | This study | NA |
| MS9715N | This study | NA |
| BI-9321 | (Bottcher et al., 2019) | NA |
| MLN4924 | Selleck Chemical | Cat # S7109 |
| MLN9708 | MedChemExpress | Cat # HY–10452 |
| Ac-VHL | This study | NA |
| Ac-VHL-me | This study | NA |
| Critical Commercial Assays | ||
| RNeasy Plus Mini Kit (250) | Qiagen | Cat # 74136 |
| Turbo DNA-free kit | Thermo Fisher | Cat # AM1907 |
| iSCRIPT cDNA Synthesis KIT | Biorad | Cat # 1708891 |
| iTaq Universal SYBR Green Supermix | Biorad | Cat # 1725125 |
| MycoAlert™ PLUS Mycoplasma Detection Kit | Lonza | Cat # LT27–286 |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher | Cat # L3000150 |
| NEBNext Ultra II RNA library Prep kit | New England Biolabs | Cat # E7770 |
| Nextseq 550 High Output Kit v2.5 | New England Biolabs | Cat # 20024906 |
| NEBNext Multiplex Oligos for Illumina | New England Biolabs | Cat # E7335S |
| Annexin V-FITC Apoptosis Detection Kit | BD Biosciences | Cat # 556570; RRID:AB_2869085 |
| SureBeads™ immunoprecipitation Kit with protein A and G conjugated magnetic beads | Biorad | Cat # 161–4833 |
| Deposited Data | ||
| Raw and analyzed mass spectrometry proteomics data | This study | PXD021901 |
| Raw and analyzed dataset of RNA-seq | This study | GEO: GSE158296 |
| Experimental Models: Cell Lines | ||
| K562 | ATCC | Cat # CCL–243; RRID:CVCL_0004 |
| RS4;11 | ATCC | Cat # CRL–1873; RRID:CVCL_0093 |
| MM1.S | ATCC | Cat # CRL–2974; RRID:CVCL_8792 |
| 293FT | Thermo Fisher | Cat # R70007 |
| MOLM-13 | DSMZ | Cat # ACC–554; RRID:CVCL_2119 |
| EOL-1 | DSMZ | Cat # ACC–386; RRID:CVCL_0258 |
| Oligonucleotides | ||
| RT-qPCR oligos and sgRNAs used | This study, Table S8 | NA |
| Recombinant DNA | ||
| pCDH-EF1 with HA-tagged cMyc | This study | NA |
| pLenti LRG-2.1_Neo with sgRNA of NSD3 (human) | This study | NA |
| Software and Algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/ |
| FlowJo | FlowJo LLc | https://www.flowjo.com/ |
| ImageJ | ImageJ | https://imagej.nih.gov/ij/index.html |
| GSEA | (Subramanian et al., 2005) | https://software.broadinstitute.org/gsea/index.jsp |
| Perseus (V1.6.2.2) | Perseus | https://maxquant.net/perseus/ |
| Other | ||
Highlights.
We discover the NSD3 PROTAC, MS9715, which effectively degrades NSD3.
MS9715 suppresses both NSD3- and cMyc-related oncogenic nodes in tumor cells.
MS9715 is superior to the reported NSD3 antagonist BI-9321 in targeting NSD3.
MS9715, but not BI-9321, is effective in suppressing the growth in tumor cells.
ACKNOWLEDGEMENTS
This work was supported in part by the US National Institutes of Health grants R01GM122749 (to J.J.), P30CA196521 (to J.J.), R01CA211336 (to G.G.W), R01CA215284 (to G.G.W), R24GM137786 (to A.J.T.), P20GM121293 (to A.J.T.), R01CA236209 (to A.J.T.), P20GM103429 (to A.J.T.) and an NIH/Office of the Director Grant S10OD018445 (to S.G.M.), an endowed professorship from the Icahn School of Medicine at Mount Sinai (to J.J.), and grants/awards from Gabrielle’s Angel Foundation for Cancer Research (to G.G.W.), When Everyone Survives (WES) Leukemia Research Foundation (to G.G.W.) and UNC Lineberger Cancer Center UCRF Stimulus Initiative Grants (to G.G.W. and L.C.). G.G.W. is an American Cancer Society (ACS) Research Scholar, a Leukemia and Lymphoma Society (LLS) Scholar, and an American Society of Hematology (ASH) Scholar in Basic Science. This work utilized the AVANCE NEO 600 MHz NMR Spectrometer System that was upgraded with funding from a National Institutes of Health SIG grant 1S10OD025132–01A1. We thank UNC’s facilities, including High-throughput Sequencing Facility (HTSF), Bioinformatics Core, and Flow Cytometry Core, for their professional assistance of this work. The cores affiliated to the UNC Cancer Center are supported in part by the UNC Lineberger Comprehensive Cancer Center Core Support Grant P30CA016086.
Footnotes
DECLARATION OF INTERESTS
The Jin laboratory received research funds from Celgene Corporation, Levo Therapeutics, and Cullgen, Inc. The Wang laboratory received research funds from the Deerfield Management/Pinnacle Hill Company. J.J. is an equity shareholder and consultant of Cullgen, Inc.
Supplemental information can be found online at
PDF containing Data S1 and Figure S1–S5 and Tables S1 and S8
Non-PDF format items include:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RL, Swaroop A, Troche C, and Licht JD (2017). The Role of Nuclear Receptor-Binding SET Domain Family Histone Lysine Methyltransferases in Cancer. Cold Spring Harb Perspect Med 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher J, Dilworth D, Reiser U, Neumuller RA, Schleicher M, Petronczki M, Zeeb M, Mischerikow N, Allali-Hassani A, Szewczyk MM, et al. (2019). Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3. Nat Chem Biol 15, 822–829. [DOI] [PubMed] [Google Scholar]
- Cai L, Tsai YH, Wang P, Wang J, Li D, Fan H, Zhao Y, Bareja R, Lu R, Wilson EM, et al. (2018). ZFX Mediates Non-canonical Oncogenic Functions of the Androgen Receptor Splice Variant 7 in Castrate-Resistant Prostate Cancer. Molecular cell 72, 341–354 e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau NG, Ma C, Danga K, Al-Sayegh H, Nardi V, Barrette R, Lathan CS, DuBois SG, Haddad RI, Shapiro GI, et al. (2020). An Anatomical Site and Genetic-Based Prognostic Model for Patients With Nuclear Protein in Testis (NUT) Midline Carcinoma: Analysis of 124 Patients. JNCI Cancer Spectr 4, pkz094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawade A, Alexandersson E, and Levander F (2014). Normalyzer: a tool for rapid evaluation of normalization methods for omics data sets. J Proteome Res 13, 3114–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Yu X, Lu K, Xie L, Wang L, Meng F, Han X, Chen X, Liu J, Xiong Y, et al. (2020). Discovery of Potent and Selective Epidermal Growth Factor Receptor (EGFR) Bifunctional Small-Molecule Degraders. J Med Chem 63, 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, and Wang GG (2010). Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromm PM, Samarasinghe KTG, Hines J, and Crews CM (2018). Addressing Kinase-Independent Functions of Fak via PROTAC-Mediated Degradation. Journal of the American Chemical Society 140, 17019–17026. [DOI] [PubMed] [Google Scholar]
- Dale et al. , M. BC; Park K-S; Kaniskan HÜ; Xiong Y; Jin J (2021). Advancing targeted protein degradation for cancer therapy. Nat Rev Cancer, in press. DOI: 10.1038/s41568-021-00365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T, and Huntly BJ (2012). Targeting epigenetic readers in cancer. N Engl J Med 367, 647–657. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. (2011). Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Lu J, Guo Y, Li D, Zhang ZM, Tsai YH, Pi WC, Ahn JH, Gong W, Xiang Y, et al. (2020). BAHCC1 binds H3K27me3 via a conserved BAH module to mediate gene silencing and oncogenesis. Nature genetics 52, 1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang C, Qin C, Xiang W, Fernandez-Salas E, Yang CY, Wang M, Zhao L, Xu T, Chinnaswamy K, et al. (2019). Discovery of ARD-69 as a Highly Potent Proteolysis Targeting Chimera (PROTAC) Degrader of Androgen Receptor (AR) for the Treatment of Prostate Cancer. J Med Chem 62, 941–964. [DOI] [PubMed] [Google Scholar]
- Hu J, Hu B, Wang M, Xu F, Miao B, Yang CY, Wang M, Liu Z, Hayes DF, Chinnaswamy K, et al. (2019). Discovery of ERD-308 as a Highly Potent Proteolysis Targeting Chimera (PROTAC) Degrader of Estrogen Receptor (ER). J Med Chem 62, 1420–1442. [DOI] [PubMed] [Google Scholar]
- Huang H, Howard CA, Zari S, Cho HJ, Shukla S, Li H, Ndoj J, Gonzalez-Alonso P, Nikolaidis C, Abbott J, et al. (2020). Covalent inhibition of NSD1 histone methyltransferase. Nat Chem Biol 16, 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, and Vingron M (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 Suppl 1, S96–104. [DOI] [PubMed] [Google Scholar]
- Khan S, He Y, Zhang X, Yuan Y, Pu S, Kong Q, Zheng G, and Zhou D (2020). PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics. Oncogene 39, 4909–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupperman E, Lee EC, Cao Y, Bannerman B, Fitzgerald M, Berger A, Yu J, Yang Y, Hales P, Bruzzese F, et al. (2010). Evaluation of the proteasome inhibitor MLN9708 in preclinical models of human cancer. Cancer Res 70, 1970–1980. [DOI] [PubMed] [Google Scholar]
- Lai AC, and Crews CM (2017). Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov 16, 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ahn JH, and Wang GG (2019). Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol Life Sci 76, 2899–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Galbo PM Jr., Gong W, Storey AJ, Tsai YH, Yu X, Ahn JH, Guo Y, Mackintosh SG, Edmondson RD, et al. (2021). ZMYND11-MBTD1 induces leukemogenesis through hijacking NuA4/TIP60 acetyltransferase complex and a PWWP-mediated chromatin association mechanism. Nat Commun 12, 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ivanov AA, Su R, Gonzalez-Pecchi V, Qi Q, Liu S, Webber P, McMillan E, Rusnak L, Pham C, et al. (2017). The OncoPPi network of cancer-focused protein-protein interactions to inform biological insights and therapeutic strategies. Nat Commun 8, 14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang P, Parton T, Zhou Y, Chrysovergis K, Rockowitz S, Chen WY, Abdel-Wahab O, Wade PA, Zheng D, et al. (2016). Epigenetic Perturbations by Arg882-Mutated DNMT3A Potentiate Aberrant Stem Cell Gene-Expression Program and Acute Leukemia Development. Cancer cell 30, 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A, Stratikopoulos E, Park KS, Wei J, Martin TC, Yang X, Schwarz M, Leshchenko V, Rialdi A, Dale B, et al. (2020). Discovery of a first-in-class EZH2 selective degrader. Nat Chem Biol 16, 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalawansha DA, and Crews CM (2020). PROTACs: An Emerging Therapeutic Modality in Precision Medicine. Cell Chem Biol 27, 998–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, Wang J, Chen X, Dong H, Siu K, et al. (2016). PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A 113, 7124–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Ahn JH, Liu H, Tsai YH, Bhanu NV, Koss B, Allison DF, Ma A, Storey AJ, Wang P, et al. (2019). PHF19 promotes multiple myeloma tumorigenicity through PRC2 activation and broad H3K27me3 domain formation. Blood 134, 1176–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati R, La Starza R, Veronese A, Aventin A, Schwienbacher C, Vallespi T, Negrini M, Martelli MF, and Mecucci C (2002). NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15). Blood 99, 3857–3860. [DOI] [PubMed] [Google Scholar]
- Schapira M, Calabrese MF, Bullock AN, and Crews CM (2019). Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov 18, 949–963. [DOI] [PubMed] [Google Scholar]
- Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, and Vakoc CR (2015). NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Mol Cell 60, 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Gao G, Yu X, Kim H, Wang L, Xie L, Schwarz M, Chen X, Guccione E, Liu J, et al. (2020). Discovery of First-in-Class Protein Arginine Methyltransferase 5 (PRMT5) Degraders. J Med Chem 63, 9977–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Morishita M, Lee D, Kim S, Lee T, Mevius D, Roh Y, and di Luccio E (2019). Identification of LEM-14 inhibitor of the oncoprotein NSD2. Biochem Biophys Res Commun 508, 102–108. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. (2009). An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736. [DOI] [PubMed] [Google Scholar]
- Su S, Yang Z, Gao H, Yang H, Zhu S, An Z, Wang J, Li Q, Chandarlapaty S, Deng H, et al. (2019). Potent and Preferential Degradation of CDK6 via Proteolysis Targeting Chimera Degraders. J Med Chem 62, 7575–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Gao H, Yang Y, He M, Wu Y, Song Y, Tong Y, and Rao Y (2019). PROTACs: great opportunities for academia and industry. Signal Transduct Target Ther 4, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner-Ivey B, Smith EL, Rutkovsky AC, Spruill LS, Mills JN, and Ethier SP (2017). Development of mammary hyperplasia, dysplasia, and invasive ductal carcinoma in transgenic mice expressing the 8p11 amplicon oncogene NSD3. Breast Cancer Res Treat 164, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Cai L, Pasillas MP, and Kamps MP (2007). NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 9, 804–812. [DOI] [PubMed] [Google Scholar]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. (2010). MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic acids research 38, e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Hu J, Wang L, Xie L, Jin MS, Chen X, Liu J, and Jin J (2019). Discovery of a First-in-Class Mitogen-Activated Protein Kinase Kinase 1/2 Degrader. J Med Chem 62, 10897–10911. [DOI] [PubMed] [Google Scholar]
- Xu BW, On DM, Ma AQ, Parton T, Konze KD, Pattenden SG, Allison DF, Cai L, Rockowitz S, Liu SC, et al. (2015). Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood 125, 346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Allis CD, and Wang GG (2021). The language of chromatin modification in human cancers. Nat Rev Cancer 21, 413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. (2011). RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S2. RNAseq identifies differentially expressed genes (DEGs) showing significant up-regulation in the EOL-1 cells treated with 2.5 uM of MS9715, in comparison to DMSO, for 4 days. Related to Figure 4.
Supplemental Table S3. RNAseq identifies differentially expressed genes (DEGs) showing significant up-regulation in the EOL-1 cells with NSD3 knockout (KO) (sgNSD3#1), in comparison to empty vector control (sgEV). Related to Figure 4.
Supplemental Table S4. RNAseq identifies differentially expressed genes (DEGs) showing significant down-regulation in the EOL-1 cells treated with 2.5 μM of BI-9321, in comparison to control (DMSO), for 4 days. Related to Figure 5.
Supplemental Table S5. RNAseq identifies differentially expressed genes (DEGs) showing significant up-regulation in the EOL-1 cells treated with 2.5 μM of MS9715N, in comparison to control (DMSO), for 4 days. Related to Figure 5.
Supplemental Table S6. RNAseq identifies differentially expressed genes (DEGs) showing significant down-regulation in the EOL-1 cells treated with 2.5 μM of MS9715, in comparison to a negative control (MS9715N lacking VHL binding activity), for 4 days. Related to Figure 5.
Supplemental Table S7. RNAseq identifies differentially expressed genes (DEGs) showing significant down-regulation in the EOL-1 cells treated with MS9715, in comparison to BI-9321, for 4 days. Related to Figure 5.
Data Availability Statement
All transcriptomic profiling datasets have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under accession number GSE158296. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Perez-Riverol et al., 2019) with the dataset identifier PXD021901. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.