Abstract
Background
Vancomycin-resistant Enterococcus faecium (VREfm) is an important agent of hospital-acquired infection. VanA phenotype is characterized by resistance to high levels of vancomycin and teicoplanin and is encoded by the vanA gene, whereas VanD phenotype is characterized by resistance to vancomycin and susceptibility or intermediate resistance to teicoplanin; however, some isolates carry a VanD phenotype with a vanA genotype, but there are many gaps in the knowledge about the genetic mechanisms behind this pattern.
Objective
To characterize the genetic structure, clonality, and mobile genetic elements of VRE isolates that display a VanD-vanA phenotype.
Results
All vanA VRE-fm isolates displayed minimum inhibitory concentration (MIC) for vancomycin > 32µg/mL and intermediate or susceptible MIC range for teicoplanin (8–16µg/mL). The isolates were not clonal, and whole-genome sequencing analysis showed that they belonged to five different STs (ST478, ST412, ST792, ST896, and ST1393). The absence of some van complex genes were observed in three isolates: Ef5 lacked vanY and vanZ, Ef2 lacked vanY, and Ef9 lacked orf1 and orf2; moreover, another three isolates had inverted positions of orf1, orf2, vanR, and vanS genes. IS1542 was observed in all isolates, whereas IS1216 in only five. Moreover, presence of other hypothetical protein-encoding genes located downstream the vanZ gene were observed in six isolates.
Conclusion
VRE isolates can display some phenotypes associated to vanA genotype, including VanA and VanB, as well as VanD; however, further studies are needed to understand the exact role of genetic variability, rearrangement of the transposon Tn1546, and presence of insertion elements in isolates with this profile.
Keywords: Enterococcus faecium, Whole-genome sequence, Resistance, Molecular characterization
Introduction
Vancomycin-resistant Enterococcus faecium (VREfm) is an important agent of hospital-acquired infection [1]. Among six glycopeptide-resistant phenotypes described, VanA is the most common, characterized by resistance to high levels of both vancomycin and teicoplanin, and usually encoded by the vanA gene [2]. VanD phenotype is characterized by resistance to vancomycin and susceptibility or intermediate resistance to teicoplanin [3].
Genetic variability can determine the phenotype. The VanD phenotype in VREfm is usually justified by the presence of vanD. However, some isolates presenting VanD phenotype carry the vanA gene, and some genetic phenomena have been associated with this pattern, such as impairment of accessory genes (vanY or vanZ) due to the insertion of IS16 [4]. However, studies investigating VanD phenotype vanA genotype are rare and explore but a few strains. Thus, there are many gaps in the knowledge about genetic mechanisms behind this pattern, such as the mobile genetic elements and bacterial strains involved.
The aim of this study was to characterize the genetic structure, clonality, and mobile genetic elements of the VRE isolates that display a VanD-vanA phenotype.
Material and methods
Nine clinical VREfm isolates from different patients, described in a previous study, were selected for molecular characterization based on their glycopeptide susceptibility profile (vancomycin-resistant and teicoplanin-susceptible, or teicoplanin-intermediate), confirmed by broth microdilution methods, interpreted according to Clinical and Laboratory Standards Institute [5], and based on the presence of the vanA gene after polymerase chain reaction (PCR) confirmation [6]. They were isolated from clinical samples of inpatients of a Bone Marrow Transplantation Unit in a teaching hospital in São Paulo, Brazil, between 2005 and 2014. These isolates were submitted to pulsed field gel electrophoresis (PFGE) to evaluate the clonality using enzyme FastDigest SmaI (Thermo Fisher Scientific, Waltham, MA, USA), and a tolerance of 1.5% and optimization of 0.5% were used for comparison. Isolates with similarity < 80% were considered different [7]. Whole-genome shotgun project was deposited at the DDBJ/EMBL/GenBank: QHLB00000000 (Ef1), MXAT00000000 (Ef2), QHLC00000000 (Ef3), QNUP00000000 (Ef4), MVGF00000000 (Ef5), MVGH00000000 (Ef6), QNUQ00000000 (Ef7), QHLD00000000 (Ef8), and MVGJ00000000 (Ef9).
Whole-genome sequencing of all VREfm isolates was performed using MiSeq Illumina™ (Illumina Inc., San Diego, CA, USA) or Ion Torrent technologies [8, 9]. The Enterococcus faecium BM4147 (GenBank accession number M97297) strain was used as a reference for molecular characterization. MLST, virulome, and resistome analysis were performed using MLSTFinder 2.0 [10], VirulenceFinder 2.0 [11], and ResFinder 4.0 [12], respectively. Additionally, the genes related to van transposon were searched in the genomes using local BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [13]. The draft genomes were used for alignment of the transposon region and visualized using EasyFig [14].
Results
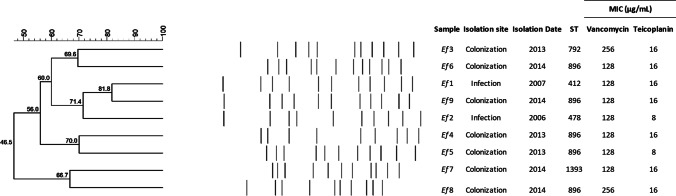
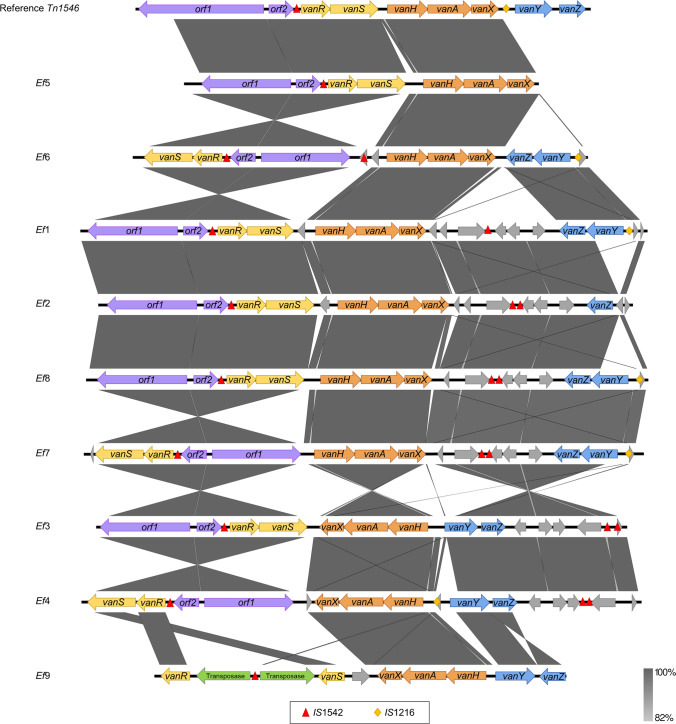
All vanA-positive VRE-fm isolates displayed minimum inhibitory concentration (MIC) for vancomycin of 128 µg/mL (Ef1, Ef2, Ef4, Ef5, Ef6, Ef7, Ef9) or 256 µg/mL (Ef3, Ef8). They were intermediate (MIC 16 µg/mL: Ef3, Ef6, Ef1, Ef9, Ef4, Ef7, Ef8) or susceptible (MIC 8 µg/mL: Ef2, Ef5) for teicoplanin, compatible with VanD phenotype. PFGE analysis showed that they are not clones and belong to nine different clusters (Fig. 1). Whole-genome sequencing analysis showed that they belong to five different lineages: ST478 (n = 1), ST412 (n = 1), ST792 (n = 1), ST896 (n = 5), and ST1393 (n = 1). Virulome analysis showed that the isolates harbor genes related to adherence (acm, efaAfm, esp, and asa1) and colonization (gelE), but the asa1 gene is present in only five isolates (Ef3, Ef6, Ef1, Ef9, and Ef2), and gelE in only three isolates (Ef3, Ef6, and Ef1). Resistome analysis identified that the isolates harbor resistance genes related to aminoglycoside, glycopeptide, macrolide, lincosamide, and streptogramin (Table 1). Three isolates did not harbor some of the vanA genotype complex genes: Ef5 lacked vanY and vanZ, Ef2 lacked vanY, and Ef9 lacked orf1 and orf2 (Fig. 2). Three isolates (Ef3, Ef4, and Ef7) had inverted positions of orf1, orf2, vanR, and vanS genes. The insertion sequence IS1542 was present in all isolates, and in seven of them, it was duplicated in different regions. Isolate Ef9 (ST896) presented a different pattern compared with others, with IS1542 between two transposases that were situated between vanR and vanS genes. The IS1216 was also present in five isolates (Ef6, Ef1, Ef8, Ef7, and Ef4), located downstream of the vanY gene. When compared to the reference, six isolates (Ef1, Ef2, Ef8, Ef7, Ef3, and Ef4) presented insertion of other hypothetical protein-encoding genes located downstream the vanZ gene (Fig. 2).
Fig. 1.
Clonality of nine VanD phenotype Enterococcus faecium
Table 1.
Phenotypic and genotypic features of the isolates from this study
| Sample | Isolation site | Isolation date | Susceptible profile | Resistome | Virulome | ||||
|---|---|---|---|---|---|---|---|---|---|
| Vancomycin MIC µg/mL | Teicoplanin MIC µg/mL | Aminoglycoside | Glycopeptide | MLS | Adherence | Colonization | |||
| Ef1 | Blood | 2007 | 128 | 16 | aph(3')-III, ant(6)-Ia |
vanH-A, vanA, vanX-A, vanZ-A, vanY-A, vanR-A, vanS-A |
msr(C), erm(B) | acm, efaAfm, esp, asa1 | gelE |
| Ef2 | Blood | 2006 | 128 | 8 | aph(3')-III, ant(6)-Ia |
vanH-A, vanA, vanX-A, vanZ-A, vanR-A, vanS-A |
msr(C), erm(B), lnu(B) | acm, efaAfm, esp, asa1 | gelE |
| Ef3 | Rectal swab | 2013 | 256 | 16 | aph(3')-III, ant(6)-Ia |
vanH-A, vanA, vanX-A, vanZ-A, vanY-A, vanR-A, vanS-A, |
msr(C), erm(B) | acm, efaAfm, esp, asa1 | gelE |
| Ef4 | Feces | 2013 | 128 | 16 | aph(3')-III, aac(6')-aph(2'') |
vanH-A, vanA, vanX-A, vanZ-A, vanY-A, vanR-A, vanS-A |
msr(C), erm(B), lnu(B) | acm, efaAfm, esp, asa1 | - |
| Ef5 | Feces | 2013 | 128 | 8 | aph(3')-III, ant(6)-Ia | vanH-A, vanA, vanX-A,vanR-A, vanS-A | msr(C), erm(B) | acm, efaAfm, esp, asa1 | - |
| Ef6 | Feces | 2014 | 128 | 16 | aph(3')-III, ant(6)-Ia |
vanH-A, vanA, vanX-A, vanZ-A, vanY-A, vanR-A, vanS-A |
msr(C), erm(B) | acm, efaAfm, esp | - |
| Ef7 | Nasal swab | 2014 | 128 | 16 | aph(3')-III |
vanH-A, vanA, vanX-A, vanZ-A, vanY-A, vanR-A, vanS-A |
msr(C), erm(B) | acm, efaAfm | - |
| Ef8 | Feces | 2014 | 256 | 16 | aph(3')-III, ant(6)-Ia |
vanH-A, vanA, vanX-A, vanZ-A, vanY-A, vanR-A, vanS-A |
msr(C), erm(B) | acm, efaAfm, esp | - |
| Ef9 | Feces | 2014 | 128 | 16 | aph(3')-III, ant(6)-Ia |
vanH-A, vanA, vanX-A, vanZ-A, vanY-A, vanR-A, vanS-A |
msr(C), erm(B) | acm, efaAfm, esp | - |
MIC minimum inhibitory concentration, MLS macrolide, lincosamide, and streptogramin
Fig. 2.
Structures and diversity of van transposons in nine VanD phenotype Enterococcus faecium
Discussion
This study describes the phenotypic and genotypic characteristics of nine Enterococcus faecium clinical isolates with vancomycin resistance profile and susceptibility or intermediate to teicoplanin; however, this profile has been previously described in other studies in Brazil [15, 16].
The isolates were not clonal, harbored vanA gene, and, although the isolates belonged to clonal complex (CC) 17, the STs were distinct, despite ST896 being the most prevalent among them. However, the STs are different to previous studies that reported isolates with VanD-vanA profile and belonged to the ST203, ST192, ST17, ST80, ST78, ST18, ST205, and ST206 [17, 18].
The most common mechanism of vancomycin and teicoplanin resistance is through the vanA gene that confers a VanA phenotype and is carried by transposon Tn1546. However, studies have reported isolates with genotype vanA, resistant to vancomycin, but exhibit susceptibility or intermediate to teicoplanin characterizing a VanD phenotype and were previously described in Asia and Europe [2, 4, 17, 19]. In the present study, all nine isolates carried the vanA gene and had the susceptibility profile consistent with previous reports.
Studies suggest that some characteristics may be related to the VanD-vanA phenotype, such as point mutations in vanS, presence of insertion elements along the transposon or in the vanY, and impairment of the accessory genes vanY and vanZ [17–19]. In our isolates, we did not observe mutations in vanS or the presence of insertion elements in the vanY gene, but rather, presence of the insertion sequence IS1216 was found in three isolates, and IS1542, in different positions along of transposons. Moreover, absence and rearrangement of accessory genes vanY and vanZ as well as the presence of uncharacterized proteins in most of the isolates were observed. Although inversions were found in three genomes (Ef3, Ef4, and Ef7), we could not find data in the literature supporting this phenomenon. All together, these results are in agreement with other studies that describe that ISs have an important role in the transposon formation, which could lead to genetic alterations as well as the deletion of vanY or vanZ or both in the loss of teicoplanin resistance [20–22].
Conclusion
Some VRE isolates can display phenotypes associated to vanA genotype, including the most commonly observed VanA and VanB, as well as VanD. However, further studies are needed to understand the exact role of genetic variability, rearrangement of the transposon Tn1546, and presence of insertion elements in isolates that exhibit the VanD-vanA profile.
Author contribution
All authors have reviewed and approved the manuscript.
Funding
FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) (São Paulo Research Foundation) and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior) (Coordination for the Improvement of Higher Education Personnel).
Declarations
Ethics approval
These experiments were approved by the Ethical Committee of Hospital das Clínicas of the University of São Paulo, Brazil.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhou X, Willems RJL, Friedrich AW, Rossen JWA, Bathoorn E. Enterococcus faecium: from microbiological insights to practical recommendations for infection control and diagnostics. Antimicrob Resist Infect Control. 2020;9(1):130. doi: 10.1186/s13756-020-00770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim D, Yoon EJ, Hong JS, et al. Impact of vanA-positive enterococcus faecium exhibiting diverse susceptibility phenotypes to glycopeptides on 30-day mortality of patients with a bloodstream infection. Antimicrob Agents Chemother. 2020;64(7):e02180–e2219. doi: 10.1128/AAC.02180-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato T, Wada T, Shinagawa M, et al. (2020) Emergence of vancomycin- and teicoplanin-resistant Enterococcus faecium via vanD5-harbouring large genomic island. J Antimicrob Chemother. 2020;75(9):2411–2415. doi: 10.1093/jac/dkaa220. [DOI] [PubMed] [Google Scholar]
- 4.Song JY, Cheong HJ, Seo YB, et al. Clinical and microbiological characteristics of vancomycin-resistant enterococci with the VanD phenotype and vanA genotype. Jpn J Infect Dis. 2013;66(1):1–5. doi: 10.7883/yoken.66.1. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Document M100-S30. Wayne, PA: CLSI, 2020.
- 6.Marchi AP, Perdigão Neto LV, Martins RCR, et al. Vancomycin-resistant enterococci isolates colonizing and infecting haematology patients: clonality, and virulence and resistance profile. J Hosp Infect. 2018;99:346–355. doi: 10.1016/j.jhin.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Faria NA, Carrico JA, Oliveira DC, et al. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Clin Microbiol. 2008;46(1):136–144. doi: 10.1128/JCM.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogaty C, Mataseje L, Gray A, et al. Investigation of a Carbapenemase-producing Acinetobacter baumannii outbreak using whole genome sequencing versus a standard epidemiologic investigation. Antimicrob Resist Infect Control. 2018;7:140. doi: 10.1186/s13756-018-0437-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothberg JM, Hinz W, Rearick TM, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;75(7356):348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 10.Larsen MV, Cosentino S, Rasmussen S, et al (2012) Multilocus sequence typing of total genome sequenced bacteria. Clin. Micobiol. 50(4): 1355–1361. 10.12.0/JCM.06094–11. [DOI] [PMC free article] [PubMed]
- 11.Joensen KG, Scheutz F, Lund O, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52(5):1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bortolaia V, Kaas RS, Ruppe E, et al (2020). ResFinder 4.0 for predictions of phenotypes from genotypes J Antimicrob Chemother. 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed]
- 13.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics. 1;27(7):1009–10. 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed]
- 15.Dalla Costa LM, Reynolds PE, Souza HA. Characterization of a divergent vanD-type resistance element from the first glycopeptide-resistant strain of Enterococcus faecium isolated in Brazil. Antimicrob Agents Chemother. 2000;44(12):3444–3446. doi: 10.1128/aac.44.12.3444-3446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camargo IL, Dalla Costa LM, Woodford N, et al (2006) Sequence analysis of Enterococcus faecium strain 10/96A (VanD4), the original vancomycin-resistant E. faecium strain in Brazil. J Clin Microbiol. 44(7):2635–7. 10.1128/JCM.00509-06. [DOI] [PMC free article] [PubMed]
- 17.Song JH, Ko KS, Suh JY, Oh WS, Kang CI, et al. Clinical implications of vancomycin-resistant Enterococcus faecium (VRE) with VanD phenotype and vanA genotype. J Antimicrob Chemother. 2008;61:838–844. doi: 10.1093/jac/dkn025. [DOI] [PubMed] [Google Scholar]
- 18.Cha JO, Yoo JI, Kim HK, et al. Diversity of Tn1546 in vanA-positive Enterococcus faecium clinical isolates with VanA, VanB, and VanD phenotypes and susceptibility to vancomycin. J Appl Microbiol. 2013;115(4):969–976. doi: 10.1111/jam.12300. [DOI] [PubMed] [Google Scholar]
- 19.Naas T, Fortineau N, Snanoudj R, Spicq C, Durrbach A, et al. First nosocomial outbreak of vancomycin-resistant Enterococcus faecium expressing a VanD-like phenotype associated with a vanA genotype. J Clin Microbiol. 2005;43:3642–3649. doi: 10.1128/JCM.43.8.3642-3649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wardal E, Kuch A, Gawryszewska I, et al. Diversity of plasmids and Tn1546-type transposons among VanA Enterococcus faecium in Poland. Eur J Clin Microbiol Infect Dis. 2017;36(2):313–328. doi: 10.1007/s10096-016-2804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee WG, Huh JY, Cho SR, Lim YA. Reduction in glycopeptide resistance in vancomycin-resistant enterococci as a result of vanA cluster rearrangements. Antimicrob Agents Chemother. 2004;48(4):1379–1381. doi: 10.1128/aac.48.4.1379-1381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagnon S, Lévesque S, Lefebvre B, et al. vanA-containing Enterococcus faecium susceptible to vancomycin and teicoplanin because of major nucleotide deletions in Tn1546. J Antimicrob Chemother. 2011;66(12):2758–2762. doi: 10.1093/jac/dkr379. [DOI] [PubMed] [Google Scholar]