Wood formation is a complex developmental process, exhibiting a continuous and iterative growth habit because of the existence of stem cells in the vascular cambium (Smetana et al., 2019). The process is controlled by hierarchical gene regulatory networks that regulate specific pathways. Stem‐differentiating xylem (SDX), a wood‐forming tissue that is part of the inner vascular cambium, differentiates into different types of cells. However, we currently lack distinct genetic markers defining sub‐stages or cell types of the SDX differentiation processes. An understanding of the networks and key genes in specific cell types is important for improved feedstock sustainability.
To decipher gene markers and the networks in specific cell types of wood formation tissue, protoplasts were prepared for scRNA‐seq, which are obtained from the surface of a freshly debarked stem segment of Populus trichocarpa (Figure 1a–e; Lin et al., 2014). Data were pre‐filtered at the cell and gene level, resulting in a pool of 12 466 cells with a median of 1566 genes per cell. The uniform manifold approximation and projection (UMAP) algorithm yielded largely overlapping distributions of cells from each of the biological replicates, indicating a high degree of reproducibility (Figure 1f). These unsupervised analyses grouped SDX cells into 12 cell clusters (Figure 1g). The analysis of cell cycle genes revealed that the clustering was not dominated by cell cycle status (Figure 1h). Looking through the clustering profiles, we found that the specialized cells exhibited relatively distinct transcriptome profiles (Figure 1g). Notably, our gene expression and signature analyses provide fresh insights into cluster‐specific properties.
Figure 1.
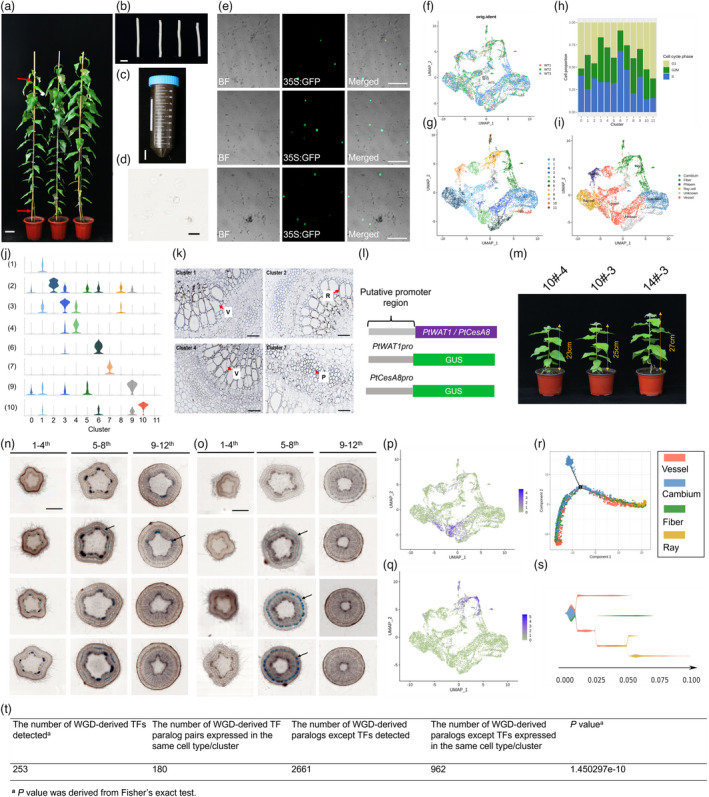
Isolation and cluster analysis of single‐cell transcriptomes from poplar stem‐differentiating xylem. (a) To generate protoplasts from the wood‐forming cells or DX cells, stems from a 6‐month‐old greenhouse‐grown Populus trichocarpa were cut into (b) 10 cm segments (~0.5–0.8 cm in diameter), and four such segments are submerged into the cell wall digestion solution in a (c) 50 ml conical tube. The red arrow shows the position of cutting. (d) Protoplasts (cell without cell wall), cell types include primary xylem, mature xylem, cambium and a few phloem cells. (e) Transfected protoplasts, the bright GFP label covers the whole cell, demonstrating that the cells are alive. The red arrow shows the position of cutting. (f) The UAMP visualization of the three replicates in SDX. (g) Twelve clusters were obtained using UMAP algorithms. (h) UMAP projection plot showing the combined transcript accumulation from marker genes, organized by the tissue/cell type of the marker gene group (cambium, vessel and fibre marker gene sets). (i) Proportion of inferred cell cycle phases (G1, S and G2/M) in each cell cluster. (j) Violin graph showing expression levels for 10 selected genes for RNA in situ hybridization. The violin represents the proportion of cells expressing in the cluster. (1) POPTR_007G044500; (2) POPTR_005G150300; (3) POPTR_005G070900; (4) POPTR_009G101900; (6) POPTR_009G139600; (7) POPTR_002G104600; (9) POPTR_013G009800; (10) POPTR_013G083300. (k) RNA in situ hybridization of four selected genes with obvious signals. V: vessel; R: ray; P: phloem. (l) Schematic diagram of PtWAT1 and PtCesA8 promoter‐GUS. (m) The transgenic plants of PtWAT1pro‐GUS and PtCesA8pro‐GUS. Serial transverse sections of the first to twelfth stem internode after GUS staining of PtWAT1pro‐GUS (n) and PtCesA8pro‐GUS (o) transgenic plants. Arrowheads left indicate phloem cells and arrowheads right indicate vessel cells. UMAP graph showing the distribution and expression level of (p) PtWAT1 and (q) PtCesA8. Blue represents low expression levels, and dark blue represents high expression levels. (r) Pseudotime analysis of all cell clusters except phloem and unknown cells. (s) Pseudotime analysis of all the clusters except phloem and unknown cells. (t) Comparison of the expression patterns of TFs and other genes. Scale bars (a) 10 cm; (b, c) 2 cm; (d) 50 μm; (e) 200 μm; (k) 100 μm; (n, o) 1 mm.
We then used the following three strategies to annotate cell types in the SDX. First, we used marker genes identified from several well‐established high‐resolution transcriptome analyses of poplar. Second, we performed RNA in situ hybridization assays (RISH) for eight cluster‐specific genes. Third, marker genes were selected to perform transgenic experiment to assign some cell clusters. The single‐cell transcriptome atlas enabled us to identify most of the major cell types and several markers in the cluster cloud (Figure 1i,j). The expression pattern of these genes in RISH not only confirmed the above annotations but also enabled us to assign unknown cell clusters (Figure 1k). POPTR_007G044500 (cluster 1) and POPTR_009G101900 (cluster 4) displayed a higher RNA abundance in vessel cells and approximately no signals in other cells. Obvious RNA signals were detected for POPTR_005G150300 (cluster 2) in ray cells, indicating the cluster 2 belongs to ray cells. RNA signals for POPTR_002G104600 (cluster 7) were mainly observed in the cells that belong to phloem cells. Cluster 0 is composed of cambium cells and expressed marker genes such as PtCRF4 (POPTR‐015G023200), PtLBD1 (POPTR‐008G043900) and PtAIL5 (POPTR‐018G091600); clusters 5, 8, 11 are composed of fibre cells and contained marker genes such as PtWPP2 (POPTR‐014G021900) and PtNEV (POPTR‐001G406300); clusters 1, 3 and 4 are composed of vessel cells and contained marker genes PtCSLC12 (POPTR‐005G146900), PtNAC82 (POPTR‐007G099400) and PtWAT1 (POPTR‐002G029100). PtWAT1 (POPTR‐002G029100) and PtCesA8 (POPTR‐004G059600) were identified to be upregulated and expressed in vessel and fibre cells in our clustering analysis. To test the applicability of their promoter for cluster‐specific expression, we generated transgenic P. tomentosa expressing β‐glucuronidase (GUS) under the control of the PtWAT1 and PtCesA8 promoter (PtWAT1pro::GUS, PtCesA8pro::GUS) and examined GUS staining patterns in various tissues (Figure 1l,m). Generally, GUS staining of the PtWAT1pro::GUS and PtCesA8pro::GUS lines displayed vessel and fibre specificity, respectively (Figure 1n,o). This is consistent with their transcriptional signals of cell clusters (Figure 1p,q). We also identified several novel marker genes that were highly and specifically enriched in their respective clusters.
To order cells along a reconstructed “trajectory” of SDX cell differentiation, Monocle 2 and STREAM algorithms were employed to disentangle and visualize complex branching trajectories. The vascular cambium is composed of two cell types including fusiform initials and ray initials, and cambium cells divide periclinally inwards to xylem mother cells and outwards to phloem mother cells. The SDX tissue differentiation is essential to understand the wood formation process. It is interesting to note that when considering all clusters of SDX together (phloem cells and unknown clusters were not included), the result revealed that cluster 0 (cambium cells) topologically bifurcated into two trajectories (Figure 1r). A similar output was observed by employing the STREAM software (Figure 1s). The vascular cambium has typically two morphologically distinct types of initials: the axillary elongated fusiform initials that will lead to the formation of the axial system (including vessel and fibre cells) and the smaller isodiametrical ray initials giving rise to the radially orientated parenchymatous rays, supporting that, one trajectory demonstrated gradual transitions from cells of cambium, to fibre and vessel cells and the other from cambium, to fibre and vessel cells and ray cells.
To study the evolutionary effects of polyploidy on the transcriptional network underlying wood formation, we reanalysed the functional genomic and transcriptome data for a large number of duplicated gene pairs formed by ancient polyploidy events in poplar (Sundell et al., 2017). Of the 2932 WGD pairs in our marker gene set, we identified 1142 paralogs that were expressed in the same cell type/cluster, suggesting the regulatory divergence contributes to the process of cambial growth and wood formation. TFs were more central in the network than other genes and may serve essential functional roles in cambial growth and wood formation. Notably, compared with the WGD‐derived TFs, other WGD paralogous pairs showed more narrow transcription profiles with expression restricted to the same cell type/cluster (Figure 1t; P = 1.450297e‐10; Fisher’s exact test). This may reflect the existence of strong genetic control over the TFs, as they perform central roles in regulatory networks.
Altogether, the profiling of distinct cell‐type‐specific transcription in multicellular organisms is essential for elucidating unique developmental regulators and functional genes that give cells their distinctive forms and functions. The data presented here show that profiling of SDX using high‐throughput scRNA‐seq of thousands of cells offers an unparalleled view of the high heterogeneity and provide key regulators in the wood formation process.
Conflict of interest
The authors declare no competing financial interest.
Author contributions
D.Z. designed the experiments; J.X., M.L. and Y.J. performed the research; J.X. wrote the paper; X.L. revised the manuscript and D.Z. obtained funding and is responsible for this article. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Project of the National Natural Science Foundation of China (Nos. 31872671, 32022057 and 31972954) and the 111 Project (No. B20050).
Xie, J. , Li, M. , Zeng, J. , Li, X. and Zhang, D. (2022) Single‐cell RNA sequencing profiles of stem‐differentiating xylem in poplar. Plant Biotechnol. J., 10.1111/pbi.13763
References
- Lin, Y.‐C. , Li, W. , Chen, H. , Li, Q. , Sun, Y.‐H. , Shi, R. , Lin, C.‐Y. et al. (2014) A simple improved‐throughput xylem protoplast system for studying wood formation. Nat. Protoc. 9, 2194–2205. [DOI] [PubMed] [Google Scholar]
- Smetana, O. , Mäkilä, R. , Lyu, M. , Amiryousefi, A. , Sánchez Rodríguez, F. , Wu, M.‐F. , Solé‐Gil, A. et al. (2019) High levels of auxin signalling define the stem‐cell organizer of the vascular cambium. Nature, 565, 485–489. [DOI] [PubMed] [Google Scholar]
- Sundell, D. , Street, N.R. , Kumar, M. , Mellerowicz, E.J. , Kucukoglu, M. , Johnsson, C. , Kumar, V. et al. (2017) AspWood: high‐spatial‐resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula . Plant Cell, 29, 1585–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]


