Abstract
The STA8 locus of Chlamydomonas reinhardtii was identified in a genetic screen as a factor that controls starch biosynthesis. Mutations of STA8 cause a significant reduction in the amount of granular starch produced during nutrient limitation and accumulate phytoglycogen. The granules remaining in sta8 mutants are misshapen, and the abundance of amylose and long chains in amylopectin is altered. Mutations of the STA7 locus, which completely lack isoamylase activity, also cause accumulation of phytoglycogen, although sta8 and sta7 mutants differ in that there is a complete loss of granular starch in the latter. This is the first instance in which mutations of two different genetic elements in one plant species have been shown to cause phytoglycogen accumulation. An analytical procedure that allows assay of isoamylase in total extracts was developed and used to show that sta8 mutations cause a 65% reduction in the level of this activity. All other enzymes known to be involved in starch biosynthesis were shown to be unaffected in sta8 mutants. The same amount of total isoamylase activity (approximately) as that present in sta8 mutants was observed in heterozygous triploids containing two sta7 mutant alleles and one wild-type allele. This strain, however, accumulates normal levels of starch granules and lacks phytoglycogen. The total level of isoamylase activity, therefore, is not the major determinant of whether granule production is reduced and phytoglycogen accumulates. Instead, a qualitative property of the isoamylase that is affected by the sta8 mutation is likely to be the critical factor in phytoglycogen production.
Plant mutants defective in α-1,6 glucanohydrolase (starch debranching enzyme) substitute the synthesis of insoluble granular starch with that of small size hydrosoluble glycogen-like particles. Because only plant cells accumulate semi-crystalline α-1,4-linked α-1,6 branched glucans in the form of large insoluble granules, it is presumed that starch debranching enzyme may constitute one of, if not the major, biochemical step distinguishing plant starch metabolism from that of bacterial or animal glycogen. Two types of starch debranching enzymes have been detected in plants (for review, see Manners, 1997; Myers et al., 2000). Limit-dextrinase, also known as plant pullulanase, can digest pullulan, a bacterial polysaccharide made of a regular succession of maltotriose chains linked together by α-1,6 linkages at the ends of each maltotriosyl residue. Plant isoamylases, on the other hand, cannot debranch such branches and thus will not hydrolyze pullulan. However, only isoamylases will rapidly debranch glycogen, whereas both types of enzymes will readily debranch amylopectin to completion. Both families of debranching enzyme can also be clearly distinguished at the level of their protein sequences (Myers et al., 2000). Limit-dextrinase and isoamylases are present at the time of starch biosynthesis in all tissues and organs analyzed so far and could therefore be involved in starch biosynthesis. Phytoglycogen-producing mutants have been reported in maize (Correns, 1901), rice (Matuo et al., 1987), sorghum (Watson and Hirata, 1960), Arabidopsis (Zeeman et al., 1998), and the unicellular green alga Chlamydomonas reinhardtii (Mouille et al., 1996). In vascular plants most mutants accumulate semi-crystalline insoluble granules and phytoglycogen, whereas in C. reinhardtii, the substitution of starch by phytoglycogen was complete. In all cases the mutants have been shown to lack an isoamylase. In addition, the defective gene was shown to encode an isoamylase type of debranching enzyme (James et al., 1995; Zeeman et al., 1998; Kubo et al., 1999). In rice and maize the mutant endosperm was shown to simultaneously display a reduction in pullulanase activity, whereas in Arabidopsis and C. reinhardtii no such decrease was ever recorded (James et al., 1995; Mouille et al., 1996; Zeeman et al., 1998; Kubo et al., 1999; Dauvillée et al., 2000). The reasons for these differences are presently unclear. In the case of cereals the resulting phenotype must thus be analyzed in the light of the absence of isoamylase and of the reduction in pullulanase. The disappearance of starch in the sta7 phytoglycogen-producing mutants of C. reinhardtii lead us to propose that polysaccharide debranching (pre-amylopectin trimming) was mandatory to obtain amylopectin synthesis in plants (Ball et al., 1996). We, and others, have explained this by assuming that starch-debranching enzymes selectively hydrolyze those branches that prevent proper alignment and crystallization of the polysaccharide (Ball et al., 1996; Myers et al., 2000). We also proposed that more functions may be involved in the trimming pathway and we have recently shown that malto-oligosaccharide metabolism must be functional to ensure proper processing of those chains released by debranching enzymes during amylopectin maturation (Mouille et al., 1996; Colleoni et al., 1999a, 1999b).
In all plants analyzed in sufficient detail it was shown that the isoamylase consists of a high mass enzyme complex (Ishizaki et al., 1983; Beatty et al., 1999; Fujita et al., 1999; Dauvillée et al., 2000). We now report a novel locus (STA8), which when defective, leads to the simultaneous production of high amylose starch and phytoglycogen. We show that despite the maintenance of the 88-kD debranching enzyme subunit the total isoamylase activity has decreased by 65% in the mutant strains. We demonstrate that it is not the quantitative change per se, but the modification in enzyme specificity or structure that is responsible for the defect in amylopectin biosynthesis.
RESULTS
STA8 Defines a Novel C. reinhardtii Locus Required for Normal Starch Synthesis
Transformants (5 × 104) were selected through complementation of the arg7 mutation by the wild-type argininosuccinate lyase gene and were screened individually through our standard iodine staining procedure (Fig. 1). Among 16 mutants defective for various aspects of starch biosynthesis and structure, two strains complemented all previously characterized mutant loci during trans complementation tests performed in vegetative diploid strains. The two mutants (BafV13 and BafO6) displayed similar phenotypes. They accumulated between 15% to 30% of the normal starch amounts during starch storage (Table I). The residual starch contained between 40% to 60% amylose, whereas the wild-type controls were characterized by amylose contents ranging between 12% to 30% (Table I; Fig. 2). Expressivity of the mutant phenotype was slightly reduced in nitrogen supplied cultures. This reduction in expressivity is often observed in mutants defective for starch biosynthesis (Libessart et al., 1995; Colleoni et al., 1999) and can be understood through the relief in nitrogen-supplied cultures of the severe energy limitations imposed on the nitrogen-starved cells. These limitations are due to the decrease in photosynthesis observed during nitrogen starvation. Both mutations were selected during distinct transformation experiments and therefore define two different mutant alleles. Southern analysis performed with a probe representing part of the bacterial sequences used in the transformation experiments demonstrate different hybridization patterns that characterize the two sta8 alleles. In addition, the integration of pARG7 proved to involve modifications at multiple sites, whereas only part of the Southern profile cosegregated with the mutant sta8-1::ARG7 allele (Fig. 3). BafV13 was subjected to extensive genetic analysis and sta8-1::ARG7 proved to segregate as a single Mendelian defect. As expected the sta8-1::ARG7 and sta8-2::ARG7 mutations were recessive in heterozygous diploids and did not complement with each other, but complemented the isoamylase defective sta7 mutations. This was not surprising since strains carrying a sta7 mutation display a very different phenotype and typically lack starch. We further proved that STA8 segregated independently from STA7 after meiosis. This was done by crossing BafV13 (mt+ arg7 cw15 nit1 nit2 sta8-1::ARG7) with strain GM7.27 (mt− pab2 sta7-1::ARG7). On a total of 208 haploid progeny obtained after meiosis, 50 colonies displayed a wild-type phenotype with respect to starch, whereas 46 clones accumulated low amounts of high amylose starch as BafV13 and 112 recombinants failed to accumulate starch as GM7.27. These results were consistent with the segregation of two independent genes if we assume full epistasis of sta7 on sta8. This assumption was further proven by performing transcomplementation tests on a sample of 10 starchless recombinants with sta7 and sta8 carrying reference strains of suitable genotypes. We found three double-mutant sta7 sta8 recombinants and seven single sta7 mutants in this sample, thereby confirming our epistasis hypothesis.
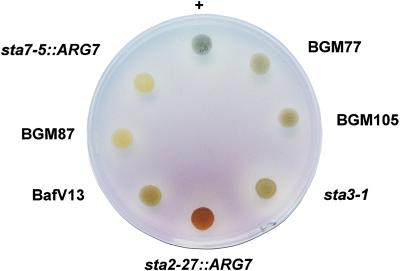
Figure 1.
Wild-type and mutant iodine-staining phenotype. Iodine stain of cell patches incubated for 7 d on solid nitrogen-deprived medium. Genotypes with respect to starch are indicated for our reference strains. sta2-27::ARG7, sta3-1, and sta7-5::ARG7 correspond to defects, respectively, in GBSS, SS, and debranching enzyme (Delrue et al., 1992; Fontaine et al., 1993; Mouille et al., 1996). +, Wild type. The original mutant strain BafV13 (sta8-1) and two recombinants, BGM77 and BGM105, display the dark green stain of sta8-1 mutants, whereas the BGM87 strain shows a typical yellow stain of low-starch mutants and were proved by complementation test to carry mutations sta7 and sta8.
Table I.
Phenotype of wild-type and mutant strains during storage (−N) or transitory starch (+N) synthesis
| Strain | Genotype | λmaxa
|
Starchb
|
WSPc
|
Amd
|
||||
|---|---|---|---|---|---|---|---|---|---|
| +N | −N | +N | −N | +N | −N | +N | −N | ||
| % | |||||||||
| BGM13 | + | 569 | 571 | 1.71 | 30 | 0.006 | 0.02 | 3 | 13 |
| BGM34 | + | 565 | 570 | 1.60 | 26 | 0.003 | 0.07 | 4 | 16 |
| 330 | + | 570 | 565 | 0.80 | 29 | 0.010 | 0.10 | 1 | 14 |
| BafV13 | sta8-1 | 573 | 591 | 0.93 | 7 | 0.018 | 1.05 | 3 | 46 |
| BafO6 | sta8-2 | 571 | 595 | 1.11 | 9 | 0.025 | 1.20 | 6 | 53 |
| BGM77 | sta8-1 | – | 600 | 1.20 | 11 | 0.028 | 0.81 | – | 42 |
Values listed are averages of three separate measures in a single experiment.
Wavelength of maximal absorbance of the iodine-polysaccharide complex of amylopectin purified by gel filtration.
Amount of insoluble polysaccharides, expressed in micrograms × 10−6 cells, purified through sedimentation as measured by the standard amyloglucosidase assay.
Amount of water-soluble polysaccharides expressed in micrograms × 10−6 cells.
The percentage of amylose in the purified starch was calculated after gel filtration of the dispersed polysaccharides.
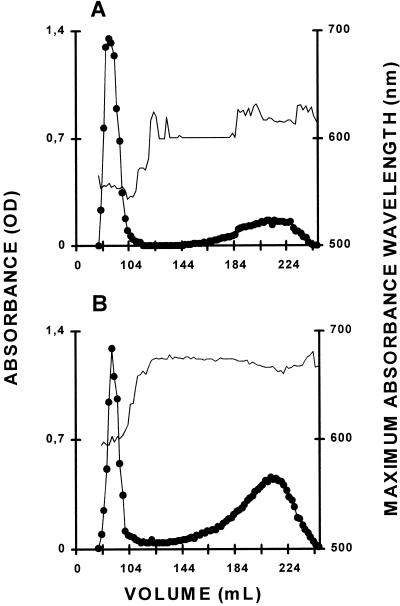
Figure 2.
Separation of amylopectin and amylose by CL2B-sepharose chromatography. The optical density (●) was measured for each 2-mL fraction at λmax (unbroken thin line). The sample was loaded on the same column setup described by Delrue et al. (1992). Starches from the wild-type strain 330 (A) and the mutant strain BafV13 (B) were extracted from nitrogen-deprived cultures. Quantification of amylose and amylopectin ratios (see Table I) was obtained by pooling amylopectin and amylose fractions separately and measuring the amount of Glc through the standard amyloglucosidase assay.
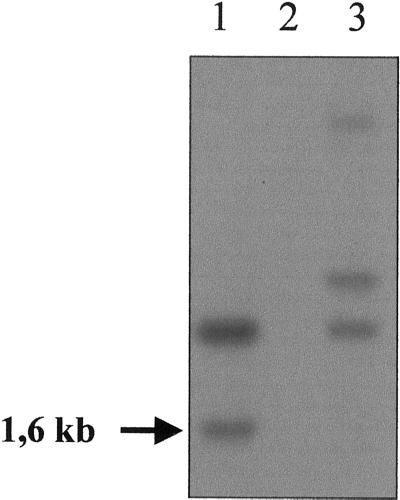
Figure 3.
Southern-blot analysis. The genomic DNAs from reference strains were digested by PstI and separated by electrophoresis. The hybridization patterns were obtained with a 323-bp probe corresponding to the bacterial part of the pARG7 plasmid that was used for mutagenesis. The original sta8-1::ARG7 mutant strain (BafV13) profile is displayed in lane 1. The wild-type strain profile 330 used for the insertional mutagenesis (lane 2) shows no signal. Strain BafO6 (sta8-2::ARG7) profile is displayed in lane 3. The arrow corresponds to the 1.6-kb signal cosegregating with the sta8-1::ARG7 mutation.
From all these analyses we have defined a novel C. reinhardtii locus (STA8) required for normal starch biosynthesis. Strains BafV13 and BafO6 defined, respectively, the sta8-1::ARG7 and sta8-2::ARG7 mutant alleles.
Mutants of the STA8 Locus Accumulate High Amylose Starch and Phytoglycogen
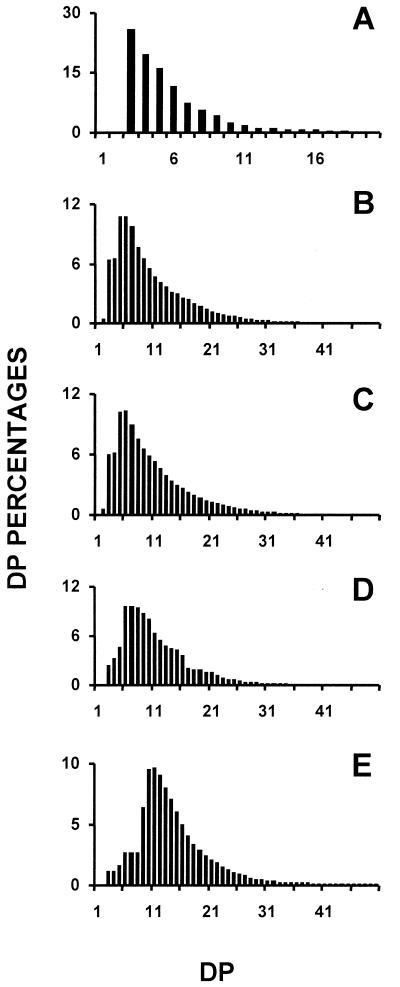
In addition to high amylose starch we detected the accumulation of 0.5 to 2 μg 10−6 cells of water-soluble polysaccharide (WSP). This material proved to contain high- and low-mass glucans, as evidenced by gel permeation chromatography of the purified WSP. The size distribution of the oligosaccharide fraction is displayed in Figure 4A, whereas the chain-length distribution of the debranched polysaccharide (Fig. 4B) is compared with those of amylopectin (Fig. 4E), glycogen (Fig. 4D), and phytoglycogen extracted from the isoamylase deficient (sta7) mutants of C. reinhardtii (Fig. 4C). The degree of branching was ascertained by quantifying the amount of reducing ends generated after debranching the polysaccharide fractions. The high-mass WSP contained 8.4% ± 0.3%, whereas branching ratios of 9% ± 0.5%, 8.3% ± 0.2%, and 5% ± 0.3% were measured, respectively, for bovine liver glycogen, phytoglycogen from the isoamylase deficient mutants of C. reinhardtii, and amylopectin. In addition, proton nuclear magnetic resonance was performed on the high-mass WSP fractions and compared with the spectra of glycogen and amylopectin. The spectra produced were identical to those previously displayed for phytoglycogen (Mouille et al., 1996). It is clear from all these results that the sta8 mutants accumulate glycogen-like polymers (phytoglycogen). We performed transmission electron microscopy (TEM) analysis of wild-type and mutant cells after staining of the polysaccharides with periodic acid thiosemicarlazid argent (PATAg). The glycogen granules located within the plastid were undistinguishable from those produced in the previously reported isoamylase-deficient (sta7) mutants of C. reinhardtii (Dauvillée et al., 1999).
Figure 4.
Determination of the chain-length distribution of sta8 high and low molecular mass WSPs. A, Histogram of chain-length distribution of low molecular mass WSP separated by TSK-HW-50 gel permeation chromatography (Fig. 4). B, Chain-length distribution of debranched high molecular mass WSP from strain BafV13 carrying sta8-1::ARG7. C, D, and E display chain length distributions of debranched reference polysaccharides, respectively, high molecular mass WSP (phytoglycogen) from a sta7 mutant strain, bovine liver glycogen, and maize amylopectin. The results are displayed as percentages of chains of degrees of polymerization (DP) between 1 and 50. The x scale displays a DP scale, and the y axis represents the relative frequencies of the chains expressed as percentages.
Mutants of the STA8 Locus Accumulate Starch Granules with a Change in Size, Shape, X-Ray Diffraction Pattern, and a Modification of Amylopectin Structure
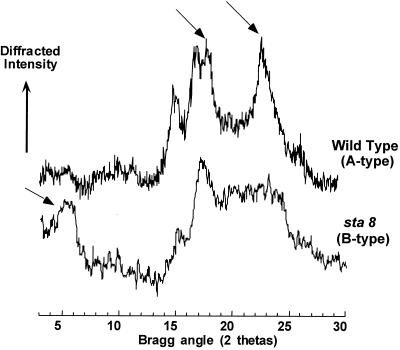
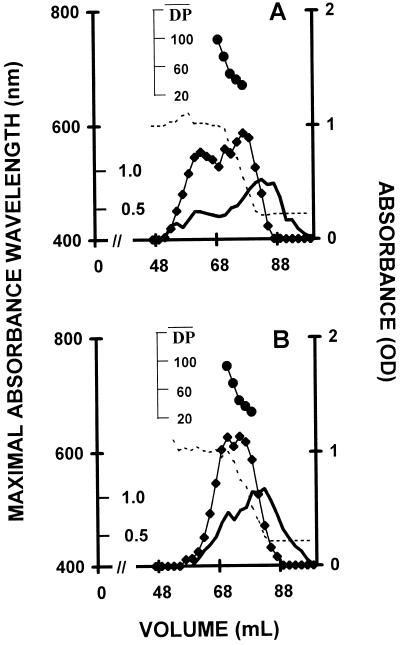
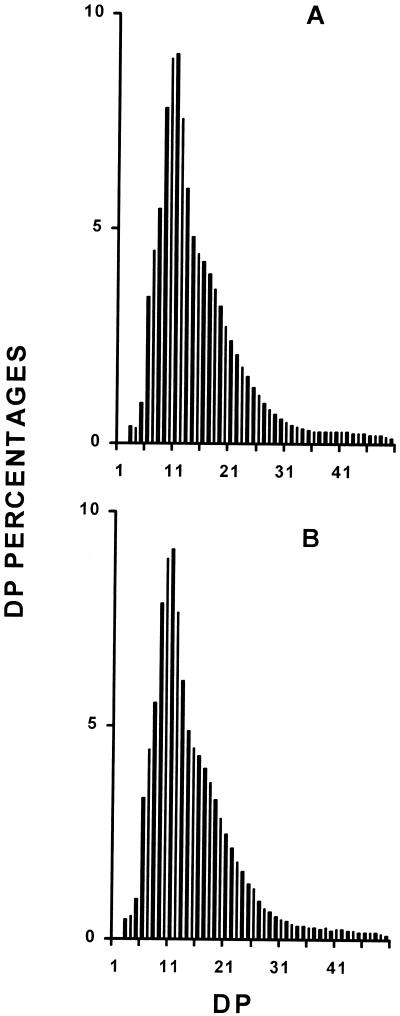
Starch granules were purified from wild-type and mutant sta8-1::ARG7 strains. The granules were directly subjected to scanning electron microscopy (SEM) or included in agar, sliced and subjected to PATAg staining. The granules of the mutant were significantly larger with distorted shapes, as is often the case for high-amylose starches (Fig. 5). In addition, the x-ray diffraction type of the starch has switched from the A to the B type (Fig. 6). The amylopectin from the wild-type and mutant starches were purified after gel permeation chromatography (GPC) on a Sepharose CL2B column. The debranched polysaccharide was subjected to GPC on a TSK-HW50 column to detect the long-chain fraction (Fig. 7, A and B) or to capillary electrophoresis after labeling of the reducing ends with 8-amino-1,3,6-pyrenetriolsulfonic acid (APTS) to detect the chain-length distribution of chains up 50 Glc residues in length (Fig. 8, A and B). It is clear that the mutant accumulates a significantly higher amount of long glucans (Fig. 7, A and B), whereas the small and average size chains did not differ significantly (Fig. 8, A and B). We believe that all of these results point to a selective defect in the amylopectin biosynthetic pathway.
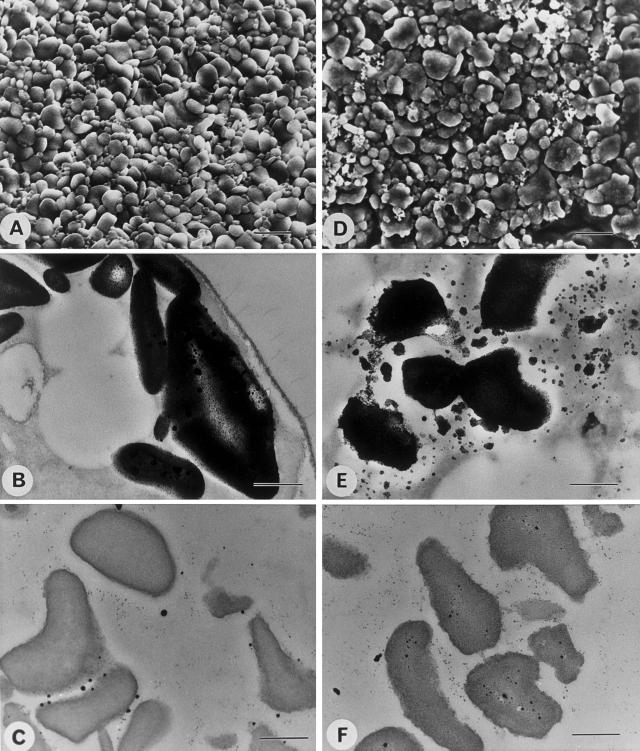
Figure 5.
SEM and TEM of starches from wild-type and mutant sta8-1::ARG7 strains. Electron micrographs of purified starches and in cells from nitrogen-starved wild-type (137C) and mutant sta8-1::ARG7 (BafV13) C. reinhardtii strains. A through C, Wild-type strain 137C; D through F, mutant sta8-1::ARG7 strain. A and D, SEM of purified starches (bar = 2 μm); B and E, TEM of starch-containing cells after PATAg staining (bar = 0.5 μm); C and F, TEM of purified starches after PATAg staining (bar = 0.5 μm).
Figure 6.
X-ray diffraction of wild-type and mutant starches. Powder x-ray diffractograms of the starches extracted from the wild type (strain 330) and mutant (strain BafV13). Crystalline lattices of vascular plant starches fall into three types. The A type defines cereal endosperm starches, the B type is found in tuber starches and some high amylose mutant starches (for review, see Buléon et al., 1998). The C type is found in pea embryos and is a mix of the A and B type. The organization of the glucan double helical structures are completely different in A and B type starches. The arrows display the individual reference peaks that define the A type or the B type evidenced, respectively, in the wild-type and mutant starches.
Figure 7.
Average chain length distribution of wild-type and mutant amylopectins. A and B, Average chain length distributions of amylopectin purified from a sta8-1 (strain BafV13) and wild-type (strain 330) after debranching by P. amyloderamosa isoamylase. Ten milligrams of purified amylopectins were loaded on a TSK-HW-50(F) GPC column. The optical density (♦) of the iodine-polysaccharide complex was measured for each 2-mL fraction at λmax. The optical density scale is on the right y axis. The amount (milligrams) of Glc for the whole 2-mL fractions is scaled on the inner side of the left y axis. The outer y axis on the left represents the λmax wavelength scale (in nanometers). The x axis shows the elution volume scale (in milliliters). λmax values are displayed for all fractions for which it could be determined (broken line). The DP scale (●) was generating by using the λmax values of the debranched glucans as internal standards according to Banks et al. (1971).
Figure 8.
Chain length distribution of wild-type and mutant amylopectins. A and B, Chain length distributions of wild-type and sta8-1-purified amylopectin, respectively, after debranching by P. amyloderamosa isoamylase. The results are displayed as percentages of chains of DP 1 to 50. The x scale displays a DP scale, and the y axis represents the relative frequencies of the chains expressed as percentages.
Devising a Specific Crude Extract Assay for Isoamylase in C. reinhardtii
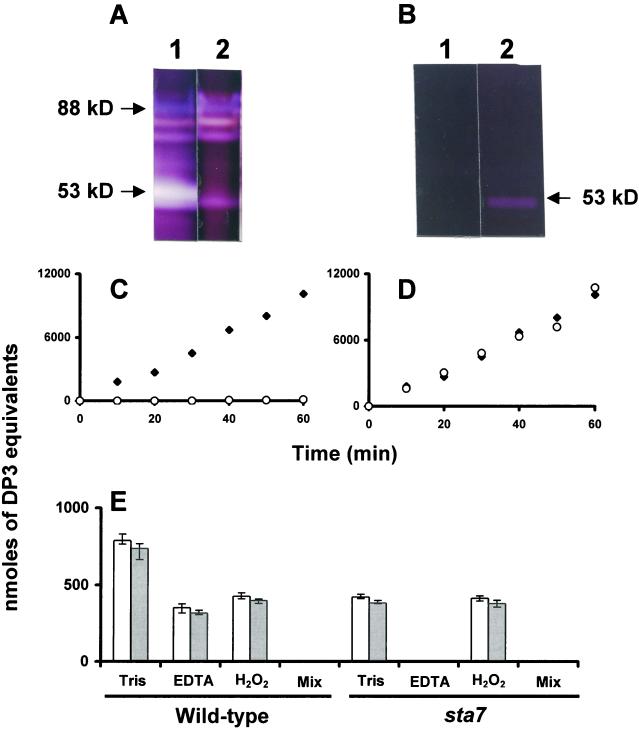
All phytoglycogen producing mutants presently described are specifically defective for a debranching enzyme of isoamylase specificity. Before embarking into the biochemical characterization of the sta8 mutants we needed to establish a specific assay for this enzyme, which would discriminate its activity from other starch hydrolases in crude extracts. In all plants, isoamylase and α-amylases will generate branched or linear oligosaccharides that will be assayed without discrimination through the appearance of reducing ends from the breakdown products of glycogen or amylopectin. Although branching enzymes and α-1,4 glucanotransferases will not produce soluble dextrins from polysaccharides under our experimental conditions, both limit-dextrinase (pullulanase) and α-glucosidase will readily produce oligosaccharides or Glc. However, even if these enzymes are given their optimal substrates (pullulan or maltose, respectively) they account for less than 2% of the reducing ends generated from amylopectin and glycogen using the crude extract mixture of C. reinhardtii (Table II; Dauvillée et al. 2000). We have previously reported that the purified C. reinhardtii isoamylase is highly sensitive to oxidation through pretreatment with H2O2 (Dauvillée et al., 2000). The purified enzyme remains, however, insensitive to concentrations of up to 10 mm EDTA. We have recently noted that the 50- to 53-kD α-amylase activities of C. reinhardtii display a strong requirement for calcium. Concentrations of EDTA as low as 5 mm were sufficient to inhibit the bulk of the enzyme activities detected on zymograms (Fig. 9A) performed with both denatured-renatured extracts (Mouille et al., 1996) or with native proteins (Kakefuda and Duke, 1984). We further confirmed the sensitivity of α-amylase to EDTA by purifying the enzyme over 50-fold and obtaining fractions lacking isoamylase activity (Fig. 9B). Over 99% inhibition was achieved through the use of 10 mm EDTA (Fig. 9C) in these purified extracts, whereas the enzyme was insensitive to pretreatment with 1 mm H2O2 (Fig. 9D). We proceeded to establish that production of reducing ends from glycogen breakdown in the presence of 10 mm EDTA or after treatment with 1 mm H2O2-defined quantitative crude extract assays, respectively, for isoamylase and α-amylase. We, therefore, compared crude extracts of wild-type and sta7 mutants for the production of reducing ends resulting from glycogen or amylopectin breakdown (Fig. 9E). In the presence of 10 mm EDTA, reducing end production in wild-type extracts decreased by 55%. The sta7 mutants lack isoamylase with no concomitant modification of amylase (Mouille et al., 1996). As predicted, this resulted in the complete absence of reducing end production (<1%) in the presence of 10 mm EDTA. Moreover, the activity measured in the sta7 mutant without EDTA precisely matched that of the amount of activity inhibited by 10 mm EDTA in wild-type extracts. In addition, the wild-type reducing end production decreased by 45% after treatment with 1 mm H2O2, whereas the sta7 extracts remained insensitive to H2O2. These results prove beyond doubt that reducing end production in the presence of 10 mm EDTA or after treatment with 1 mm H2O2 do define specific crude extract assays, respectively, for isoamylase and α-amylases in C. reinhardtii.
Table II.
Enzyme activities in wild-type and mutant sta8-1 progeny
| Enzyme | Wild-Type Strainsa | Mutant Strainsa | Zymogramb |
|---|---|---|---|
| AGPase | 25 ± 5 (6) | 22 ± 6 (6) | No |
| STP | 20 ± 3 (6) | 19 ± 2 (6) | Yes (7, 8) |
| PGM | 5.8 ± 2.1 (6) | 5.6 ± 1.7 (6) | Yes (8, 8) |
| SS | 2.7 ± 0.4 (6) | 2.8 ± 0.7 (6) | Yes (22, 24) |
| BE | 0.7 ± 0.1 (6) | 0.9 ± 0.2 (6) | Yes (12, 16) |
| GBSS | 43 ± 4 (4) | 42 ± 6 (4) | No |
| α-Glucosidase | 4.51 ± 0.37 (6) | 4.68 ± 0.61 (6) | No |
| Limit-dextrinase | 0.25 ± 0.1 (21) | 0.24 ± 0.1 (23) | Yes (45, 47) |
| D-enzyme | 86 ± 7 (6) | 84 ± 8 (6) | Yes (45, 47) |
| Isoamylase | 367 ± 34 (21) | 122 ± 12 (23) | Yes (45, 47) |
ADP-glucose pyrophosphorylase (AGPase; assayed in direction of pyrophosphorolysis in the presence of 1.5 mm 3-PGA), starch phosphorylase (STP), and phosphoglucomutase (PGM) units are expressed in nanomoles of Glc-1-P produced min−1 mg−1 protein. Soluble starch synthase (SS) and granule-bound starch synthase (GBSS) are expressed in nanomoles of ADP-Glc incorporated into polysaccharide min−1 mg−1 protein (SS) or milligrams of starch (GBSS). Branching enzymes (BE) is expressed as nanomoles of Glc-1-P incorporated into polysaccharide min−1 mg−1 protein (phosphorylase amplification assay). Limit dextrinase and D-enzyme are expressed in nanomoles of maltotriose formed from pullulan min−1 mg−1 protein and nanomoles of glucose formed from maltotriose min−1 mg−1 protein, respectively. α-Glucosidase activities are expressed in nanomoles of glucose formed from maltose min−1 mg−1 protein. The isoamylase activities were monitored by measuring the amount of reducing ends produced during incubation with glycogen in the presence of 10 mm EDTA. The activities are expressed as nanomoles of maltotriose equivalents liberated from glycogen per hour and per milligrams of protein.
The nos. between parentheses correspond to the no. of different strains examined.
The two nos. between parentheses correspond respectively to the no. of wild-type and mutant strains examined, respectively.
Figure 9.
Isoamylase and α-amylase inhibition experiments. A, Two hundred micrograms of wild-type crude extract was denatured and loaded on a starch containing zymogram (Mouille et al., 1996). After renaturation, part of the gel was incubated with or without 5 mm EDTA. Lane 1 displays the gel segment incubated in the incubation mix without EDTA, and lane 2 displays another gel segment containing the same sample, but incubated in a mix with 5 mm EDTA. B, Twenty micrograms of proteins from semi-purified α-amylase was denatured and loaded on a starch-containing zymogram according to Mouille et al. (1996). After renaturation, part of the gel was incubated with or without 5 mm EDTA. Lane 1 displays the gel segment incubated in the incubation mix containing 5 mm EDTA, and lane 2 displays another gel segment containing the same sample, but incubated in a mix without EDTA. C, Twenty microliters of the semipurified α-amylase (purified 50 times) was used to measure the increase in reducing ends from glycogen (see “Materials and Methods”) with (○) or without (♦) 10 mm EDTA. D, The same procedure was followed, but the enzyme was pretreated with 1 mm H2O2 (○) (see “Materials and Methods”). E, Wild-type and sta7 crude extracts (100 μg of proteins) were used to assay the increase in reducing power from bovine liver glycogen (white bars) or maize amylopectin (gray bars) in the presence of different inhibitors. Tris stands for the buffer used in standard assay (20 mm Tris, pH 7, containing 5 mg mL−1 of polysaccharide). EDTA stands for the presence of 10 mm EDTA in the buffer described above and H2O2 stands for for 1 mm hydrogen peroxide pretreatment. In C and D, the x axis represents the incubation time expressed in minutes. In C through E, the y axis scale displays maltotriose nanomoles equivalents liberated during incubation for the whole semi-purified α-amylase fraction (C and D) or for 100 μg of crude extract (E).
Mutants of the STA8 Locus Display a Selective 65% Quantitative Reduction in Isoamylase Activity, But Still Retain the 88-kD Debranching Enzyme Subunit
We have assayed the wild-type and mutant extracts for the presence of all enzymes suspected to be involved in starch biosynthesis or degradation. Results listed in Table II demonstrate that all these enzymes are unaffected by the presence of the sta8-1::ARG7 mutation. A 65% ± 5% reduction in isoamylase activity cosegregated with the mutant gene in 46 recombinants analyzed from a cross involving a wild-type and a mutant C. reinhardtii strain. Moreover, in contrast to the previously described sta7 mutants (Mouille et al., 1996), the sta8 strains still contained the 88-kD isoamylase subunit in seemingly normal amounts as far as could be evidenced by zymograms performed after denaturation-renaturation.
Gene Dosage Experiments
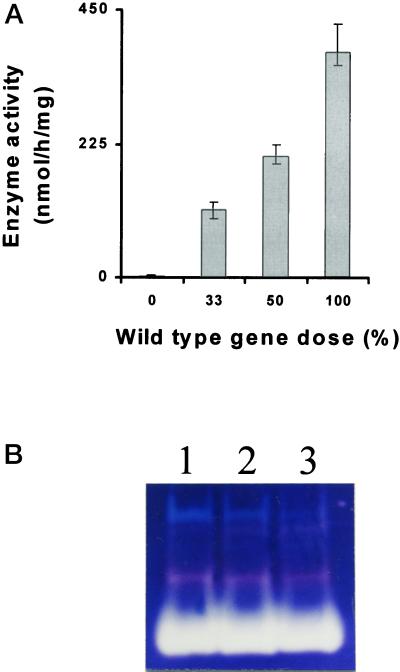
Establishing a specific crude extract assay for isoamylase enabled us to perform gene dosage experiments in diploid and triploid strains homozygous or heterozygous for STA7 or STA8. In addition to the three haploid standards containing sta7, sta8, and the wild-type reference (+), we have prepared (see “Materials and Methods”) the following three diploid and three triploid genotypes: sta7/sta7, sta7/+, +/+, sta7/sta7/sta7, sta7/sta7/+, and +/+/+. The results displayed in Figure 10A demonstrate a linear relationship between wild-type gene dose and isoamylase enzyme activity for the STA7 (+) locus. The results obtained with the STA8 locus are more complex and the relationship is not strictly linear with respect to the wild-type allele dose. In addition, although zymograms do not offer precise quantifications, we can clearly observe an increase of the 88-kD debranching enzyme subunit zymogram stain as a function of the wild-type STA7 (+) allele dose (Fig. 10B). No such relation was found with the STA8 locus, suggesting that STA7 could encode the 88-kD debranching enzyme subunit. The experiments described above provided us with two distinct genotypes (sta7/sta7/+, and sta8 haploid) containing the same amount of isoamylase specific activity. We observed in the homozygous sta8/sta8 the same mutant phenotype as that described for the haploid sta8-1::ARG7. However, despite the presence of a 65% decrease in isoamylase activity, the sta7/sta7/+ triploid displayed a fully wild-type phenotype. These results establish that the defect in amylopectin synthesis recorded in the sta8 mutants is not due to a decrease in isoamylase activity or to a modification of the branching-to-debranching enzyme ratio. It is, therefore, the residual enzyme's quality that is chiefly responsible for the dysfunction in amylopectin synthesis.
Figure 10.
Gene dosage. A, Gene dosages ranging from 0% wild-type alleles (homozygous mutant) to 100% wild type (homozygous wild-type); 50% corresponds to the heterozygous diploids, whereas 33% corresponds to a sta7/sta7/+ triploid. Histograms representing means and sds (n = 3) were calculated for each gene dose. One hundred micrograms of proteins from the different genotypes was used to assay isoamylase in the presence of 10 mm EDTA. Results are displayed as nanomoles of maltotriose equivalents produced from glycogen per hour and per milligram of protein. Total phosphoglucomutase-specific activities were monitored as internal controls and proved similar in all constructs (2.3 ± 0.5 nmol Glc-6-P formed from Glc-1-P min−1 mg−1 protein). B, Starch-containing zymograms. Fifty micrograms of crude extracts proteins from wild-type haploid strain A35 (lane 1), heterozygous diploid sta7/+ (lane 2), and heterozygous triploid sta7/sta7/+ (lane 3), were denatured and loaded on the same starch-containing zymogram according to the procedure described in Mouille et al. (1996). After renaturation, enzymes activities were revealed after incubation by staining the gel with a freshly made solution of iodine (0.2% [w/v] KI and 0.02% [w/v] I2).
DISCUSSION
A fundamental aspect of plant physiology is the storage of Glc units produced initially during photosynthesis in large, insoluble starch granules. Because this carbon source storage mechanism is in stark contrast to the soluble glycogen molecules found in animals, fungi, and prokaryotes, it has been of interest to seek mutant plants in which glycogen-like polymers accumulate at the expense of starch grains. Mutants of C. reinhardtii (Mouille et al., 1996), maize (James et al., 1995), rice (Kubo et al., 1999), and Arabidopsis (Zeeman et al., 1998) have been identified that display this phenotype. A striking observation is that in all these instances phytoglycogen accumulation results from a mutation that controls production of an isoamylase, and in at least three of the species the mutation directly alters the gene that codes for the enzyme.
The relation between the isoamylase and the form of the storage glucan, i.e. soluble or granular, appears to be very specific. Phytoglycogen-accumulating mutants in maize are easy to detect owing to the resultant kernel phenotype, and in the past century dozens of such mutations have been isolated in the course of the extensive genetic analysis of this species. All of these are allelic mutations of the su1 locus, indicating the high level of saturation of genetic screens for phytoglycogen-accumulating mutants. Mutation of an isoamylase coding sequence, therefore, may be the sole means of altering starch biosynthesis such that glycogen-like polymers are formed in plants.
The genetic analysis reported here indicates that at least two different genes in a plant species can determine the ability to produce storage glucans in the form of insoluble granules. This is the first description of two genes controlling phytoglycogen production, so further characterization of the STA8 gene is likely to be of interest in understanding how granular starch is produced. The sta8 mutations cause significant reductions in the activity of isoamylase in total extracts. The genetic analysis, therefore, has not identified a completely different biochemical activity involved in phytoglycogen production at the expense of starch, and the specific relationship between phytoglycogen and isoamylase activity remains. Identification of STA8, however, is likely to provide novel insights into the nature of the isoamylase. Only one isoamylase in C. reinhardtii cell extracts is detected in activity gels. This activity migrates as an 88-kD polypeptide, and it is completely eliminated by the sta7 mutations (Mouille et al., 1996; Dauvillée et al., 1999, 2000). The sta8 mutations, in contrast, reduce this activity, but do not eliminate it completely. Several possibilities may be considered to explain how STA7 and STA8 can affect the same enzyme. One of the two genes may directly code for a polypeptide that possesses isoamylase activity, whereas the second gene may control expression of the enzyme. In an alternate manner, STA7 and STA8 could code for polypeptides within the isoamylase, with the STA7 product as an essential component and the STA8 product as a contributing factor that is not absolutely necessary for activity. The gene dosage experiments reported in this paper strongly suggest that STA7 encodes a catalytic subunit within the enzyme complex.
Plant isoamylases are known to be multisubunit enzymes of high Mr, so the possibility of heteromultimeric compounds must be considered. This suggestion seemingly is contrary to nature of the rice endosperm isoamylase, which was defined as homomultimeric complex (Fujita et al., 1999). In potato, however, two polypeptides copurified with the isoamylase activity (Ishizaki et al., 1983). Furthermore, at least two genes coding for polypeptides highly homologous to known isoamylases are present in Arabidopsis and potato. Further characterization of the precise molecular nature of the isoamylase in various plant species is necessary to understand the role of this enzyme in amylopectin biosynthesis, and we expect that the genetic characterization of two loci controlling the enzyme in C. reinhardtii will be useful in this regard.
Mutations that affect isoamylase in various species can have different effects on amylopectin biosynthesis. This study revealed that in sta8 mutants the chain-length distribution of the small and medium size amylopectin chains (up to a length of 40 Glc residues) in the residual starch is similar to that of wild-type amylopectin, and the same result was obtained in Arabidopsis dbe1 mutants (Zeeman et al., 1998). In contrast, mutations of isoamylase polypeptides in maize and rice cause alterations in the chain length distribution in the remaining amylopectin (Nakamura et al., 1997; Dinges et al., 2001). A third phenotype is observed in the C. reinhardtii sta7 mutants, as well as in rice plants bearing specific alleles of the su1 locus. In these plants granular starch is completely lacking. A possible explanation for these varying results is that there are various means of altering isoamylase activity and that different types of mutation have different effects on amylopectin biosynthesis. These considerations raise the question of whether the total activity of isoamylase determines if storage glucans are converted into soluble or insoluble polymers or, alternatively, whether specific qualitative aspects such as substrate specificity also are involved in the process.
The different phenotypes caused by sta7 and sta8 mutations offered an opportunity to address this question. Comparison of a sta8/sta8 strain to a sta7/sta7/+ heterozygous triploid revealed two distinct phenotypes. In the latter instance starch production is normal, whereas in the former phytoglycogen is present and granular starch is strongly reduced. Yet the two strains possess the same total amount of isoamylase activity measured in total extracts. To explain these results we suggest that a qualitative aspect of isoamylase activity is critical in amylopectin biosynthesis. One possibility is that the substrate specificity of the isoamylase is different in the enzyme found in homozygous sta8 strains compared with that present in the sta7 heterozygotes. A second possibility is that the interactions of the multimeric isoamylase complex with other polypeptides are altered in the sta8 mutant.
A technical advance described here is the ability to measure total isoamylase activity in vitro in crude cell extracts. The presence of several different amylolytic enzymes that can produce new reducing ends from polysaccharide substrates is a significant complication in measurement of debranching enzyme activity. The fact that sta7 mutant extracts assayed in the presence of EDTA contain undetectable amounts of amylolytic activity indicates that the assay used here is effective in eliminating all extraneous enzyme activity above the isoamylase that is dependent on STA7. EDTA treatment, therefore, is an effective means of measuring isoamylase in C. reinhardtii. Whether this method is applicable to other species has, however, to be thoroughly checked and may vary according to the tissue and species. Three requirements that were met in the C. reinhardtii have to be ascertained in each case. First, the contributions of the many EDTA insensitive starch hydrolases have to account for less than 1% of the crude extract mixture activity to be considered negligible. Second, mutants lacking isoamylase must be available to confirm that in the presence of EDTA no activity can be detected in the assay. Third, the amount of activity assayed in the absence of EDTA in the mutant should match precisely the decrease in activity measured in the presence of EDTA in the wild-type controls.
MATERIALS AND METHODS
Materials
Pseudomonas amyloderamosa isoamylase was from Hayashibara Biochemical Laboratories, (Okayama, Japan). Rabbit or bovine liver glycogen and maize amylopectin were from Sigma (St. Louis). Boehringer-Mannheim (GmbH) provided Glc-1-P, yeast hexokinase, yeast Glc-6-P dehydrogenase, and rabbit muscle phosphorylase.
Chlamydomonas reinhardtii Strains, Insertional Mutagenesis, Growth Conditions, Cytological Observations, and Media
The mutant strains BafV13 and BafO6 were obtained by transformation of the cell wall-deficient Arg requiring strain 330 (mt+ arg7 cw15 nit1 nit2) with 1 μg of pARG7.8 carrying the wild-type argininosuccinate lyase gene. Transformants were selected by complementation of the Arg auxotrophy and were screened by spraying iodine on replica plates (Maddelein et al., 1994). All putative mutant strains were subjected to routine complementation tests.
The reference strains used in this study were 330, Baf V13 (mt+ cw15 nit1 nit2 sta8-1::ARG7), BafO6 (mt+ cw15 nit1 nit2 sta8-2::ARG7), and GM7.27 (mt− pab2 sta7-1::ARG7). BGM strains from a cross performed between BafV13 and GM7.27 were used throughout this work. SJ and SN strains were used to construct the diploid and triploid strains used for gene dosage experiments and were selected from a cross between the wild-type strain A35 (mt− ac14 pab2) and the mutant strain S (mt+ sta7-4::ARG7 nit1 nit2).
All experiments were carried out in continuous light (40 μE m−2 s−1) in the presence of acetate at 24°C in liquid cultures that were shaken without air or CO2 bubbling. Late-log phase cultures were inoculated at 105 cells mL−1 and harvested at 2 × 106 cells mL−1. Nitrogen-starved cultures were inoculated at 5 × 105 cells mL−1 and were harvested after 5 d at a final density of 1 to 2 × 106 cells mL−1. Recipes for media and genetic techniques can be found in Harris (1989a and 1989b). Fixation and embedding protocols were as described in Dauvillée et al. (1999).
Structural Analysis of Polysaccharides
A full account of amyloglucosidase assays, starch purification on Percoll gradients, and λmax, the wavelength of the maximal absorbance of the iodine-polysaccharide complex, can be found in Delrue et al. (1992). Amylopectin and amylose were separated through a CL2B gel permeation chromatography (Pharmacia Biotech, Piscataway, NJ) equilibrated in 10 mm NaOH as described in Delrue et al. (1992). Phytoglycogen and oligosaccharides found in the mutant strains were separated on a TSK-HW-50 GPC column (Merck, Darmstadt, Germany) eluted in 10% (w/v) dimethyl sulfoxide as described by Maddelein et al. (1994). The WSP fraction purification can be found in Dauvillée et al. (1999).
GPC-purified amylopectin and phytoglycogen were debranched by P. amyloderamosa isoamylase. The APTS-tagged chains produced by isoamylase-mediated debranching and the APTS-tagged oligosaccharide fraction were separated by capillary electrophoresis carried out as previously described (O'Shea et al., 1998). Wide-angle x-ray diffraction and TEM studies were as detailed in Buléon et al. (1997).
Crude Extract Preparation, Enzyme Assays, and Zymograms
Soluble crude extracts were always prepared from late-log phase cells (2 × 106 cells mL−1) grown in high-salt acetate medium under continuous light (40 μE m−2 s−1). All assays were conducted in conditions of linearity with respect to time and amount of crude extract. Phosphoglucomutase, ADP-Glc pyrophosphorylase, and phosphorylase activities were monitored by using the standard assays described in Ball et al. (1991) and Van den Koornhuyse et al. (1996). The SS and branching enzymes assays were those described by Fontaine et al. (1993) and Libessart et al. (1995). GBSSI was monitored as previously described in Delrue et al. (1992) or Van den Koornhuyse et al. (1996), from the starch purified from nitrogen-supplied cultures. The α-glucosidase and D-enzyme activities were monitored by measuring the Glc produced from maltose or maltotriose, respectively, as detailed in Colleoni et al. (1999a). The analysis was completed by zymograms as detailed in Buléon et al. (1997) and Mouille et al. (1996).
Amylase and isoamylase activities were monitored by measuring the appearance of reducing ends from glycogen or amylopectin as follows. Amounts of crude extract corresponding to 20 to 200 μg of total protein buffered in 2 mm dithiothreitol, 20 mm Tris, pH7, containing glycogen or amylopectin at 5 mg mL−1 in a 1-mL final volume were incubated from 10 to 120 min at 30°C in the presence of 1 mm of hydrogen peroxide (amylase assay, see below) or 10 mm EDTA (isoamylase assay). Hydrogen peroxide inactivation was achieved by pretreatment of the extract for 15 min in 1 mm H2O2 followed by extensive dialysis. Enzyme activity was subsequently monitored. Reducing ends produced during incubation were monitored by using the standard dinitrosolicyclote solution assay or the Nelson (1944) and Somogyi (1952) method using maltotriose as a standard as described in Dauvillée et al. (2000).
C. reinhardtii DNA Purification and Southern-Blot Analysis
Algal DNA was purified as described by Rochaix et al. (1991). Southern-blot analysis was performed using 10 μg of PstI-digested algal DNA using a NruI/SalI 321-bp fragment covering a small part of the bacterial tetracycline resistance gene contained in the plasmid pARG7.8 used for mutagenesis.
Gene Dosage Experiments
Diploid and triploid strains were constructed as follows. To obtain the homozygous mutant we crossed SJ6 (mt− ac14 nit1 nit2 sta7-4::ARG7) and SJ16 (mt+ pab2 nit1 nit2 sta7-4::ARG7) and selected the diploid after 4 d of growth on minimal medium supplied with ammonium. The wild-type homozygous diploid was obtained by crossing the strain SN25 (mt+ pab2 nit1 nit2) with A35 (mt− ac14 pab2) and selecting diploids on minimum medium supplemented with paraminobenzoic acid. The heterozygous diploid were selected on the same medium after crossing the strains SJ23 (mt+ pab2 nit1 nit2 sta7-4::ARG7) and A35. Vegetative diploid strains heterozygous for mating type display an mt− mating type. After checking the phenotype, cellular volume, protein content, and mating type, we crossed the heterozygous diploid with the strain S (mt+ sta7-4::ARG7 nit1 nit2) to obtain the sta7-4/sta7–4/+ triploid selected on minimal medium. After selection, on the appropriate medium, the haploid, diploid, and triploid nature of the clones was confirmed by retesting the phenotypes and measuring the average cell volume distribution and the cell protein content from unsynchronized cultures. For each construct we selected three independent clones. Gene dosages are thus averages from three separate colonies for each construct. Phosphoglucomutase activity was assayed (Ball et al., 1991) and used as an internal standard during these experiments. Isoamylase assays with crude extracts obtained from the different genotypes were performed as described above.
ACKNOWLEDGMENT
The authors thank A. Decq for excellent technical assistance.
Footnotes
This work was supported by the Ministère de l'Education Nationale, by the Centre National de la Recherche Scientifique, by the Institut National de la Recherche Agronomique, by Biogemma UK, and by the U.S. Department of Agriculture.
LITERATURE CITED
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J. From glycogen to amylopectin: a model explaining the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- Ball S, Marianne T, Dirick L, Fresnoy M, Delrue B, Decq A. A Chlamydomonas reinhardtii low-starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta. 1991;185:17–26. doi: 10.1007/BF00194509. [DOI] [PubMed] [Google Scholar]
- Banks W, Greenwood CT, Khan KM. The interaction of linear amylose oligomers with iodine. Carbohydr Res. 1971;17:25–33. [Google Scholar]
- Beatty MK, Rahman A, Cao H, Woodman W, Lee M, Myers AM, James MG. Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch debranching enzyme from maize. Plant Physiol. 1999;119:255–266. doi: 10.1104/pp.119.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buléon A, Ball S, Planchot V, Colonna P. Starch granules: Structure and biosynthesis. Int J Biol Macromol. 1998;23:85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- Buléon A, Gallant DJ, Bouchet B, Mouille G, D'Hulst C, Kossman J, Ball SG. Starches from A to C: Chlamydomonas reinhardtii as a model microbial system to investigate the biosynthesis of the plant amylopectin crystal. Plant Physiol. 1997;115:949–957. doi: 10.1104/pp.115.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni C, Dauvillee D, Mouille G, Buléon A, Gallant D, Bouchet B, Morell M, Samuel M, Delrue B, d'Hulst C. Genetic and biochemical evidence for the involvement of α-1,4 glucanotransferases in amylopectin synthesis. Plant Physiol. 1999a;120:993–1004. doi: 10.1104/pp.120.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni C, Dauvillee D, Mouille G, Morell M, Samuel M, Slomiany M-C, Lienard L, Wattebled F, d'Hulst C, Ball S. Biochemical characterization of the Chlamydomonas reinhardtii α-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiol. 1999b;120:1005–1014. doi: 10.1104/pp.120.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correns C. Bastarde zwischen maisrassen, mit besonder berucksichtung der xenien. Bibl Bot. 1901;53:1–161. [Google Scholar]
- Dauvillée D, Colleoni C, Shaw E, Mouille G, D'Hulst C, Morell M, Samuel MS, Bouchet B, Gallant DJ, Sinskey A. Novel starch-like polysaccharides are synthesized by a soluble form of granule-bound starch synthase in glycogen accumulating mutants of Chlamydomonas reinhardtii. Plant Physiol. 1999;119:321–330. doi: 10.1104/pp.119.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D, Mestre V, Colleoni C, Slomianny MC, Mouille G, Delrue B, d'Hulst C, Bliard C, Nuzillard J-M, Ball S. The debranching enzyme complex missing in glycogen accumulating mutants of Chlamydomonas reinhardtii displays an isoamylase-type specificity. Plant Sci. 2000;157:145–156. doi: 10.1016/s0168-9452(00)00256-9. [DOI] [PubMed] [Google Scholar]
- Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski JM, Van Den Koornhuyse N, Maddelein M-L, Fournet B, Ball S. Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase accumulate a structurally modified amylopectin. J Bacteriol. 1992;174:3612–3620. doi: 10.1128/jb.174.11.3612-3620.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, Myers AM, James MG. Molecular structure of three mutations of the maize sugary 1 locus and their allele-specific phenotypic effects. Plant Physiol. 2001;125:1406–1418. doi: 10.1104/pp.125.3.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine T, D'Hulst C, Maddelein M-L, Routier F, Marianne-Pepin T, Decq A, Wieruszeski JM, Delrue B, Van Den Koornhuyse N, Bossu J-P. Toward an understanding of the biogenesis of the starch granule: evidence that Chlamydomonas-soluble starch synthase II controls the synthesis of intermediate size glucans of amylopectin. J Biol Chem. 1993;268:16223–16230. [PubMed] [Google Scholar]
- Fujita N, Kubo A, Francisco PB, Nakakita M, Harada K, Minaka N, Nakamura Y. Purification, characterization, and cDNA structure of isoamylase form developing endosperm of rice. Planta. 1999;208:283–293. doi: 10.1007/s004250050560. [DOI] [PubMed] [Google Scholar]
- Harris EH. Culture and storage methods. In: Harris E, editor. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989a. pp. 25–63. [DOI] [PubMed] [Google Scholar]
- Harris EH. Genetic analysis. In: Harris E, editor. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989b. pp. 399–446. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Taniguchi H, Maruyama Y, Nakamura M. Debranching enzymes of potato tuber (Solanum tuberosum L.): I. Purification and some properties of potato isoamylase. Agric Biol Chem. 1983;47:771–779. [Google Scholar]
- James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakefuda G, Duke SH. Electrophoretic transfer as a technique for the detection and identification of the plant amylolytic enzymes in polyacrylamide gels. Plant Physiol. 1984;75:278–280. doi: 10.1104/pp.75.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y. The starch-debranching enzyme isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 1999;121:399–410. doi: 10.1104/pp.121.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libessart N, Maddelein M-L, Van Den Koornhuyse N, Decq A, Delrue B, Ball SG. Storage, photosynthesis and growth: the conditional nature of mutations affecting starch synthesis and structure in Chlamydomonas reinhardtii. Plant Cell. 1995;7:1117–1127. doi: 10.1105/tpc.7.8.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddelein M-L, Libessart N, Bellanger F, Delrue B, D'Hulst C, Van Den Koornhuyse N, Fontaine T, Wieruszeski JM, Decq A, Ball SG. Toward an understanding of the biogenesis of the starch granule: determination of granule-bound and soluble starch synthase functions in amylopectin synthesis. J Biol Chem. 1994;269:25150–25157. [PubMed] [Google Scholar]
- Manners D. Observations on the specificity and nomenclature of starch debranching enzymes. J Appl Glycosci. 1997;44:83–85. [Google Scholar]
- Matuo T, Yano M, Satoh H, Omura T. Effects of sugary and shrunken mutant genes on carbohydrates in rice endosperm during ripening period. Jpn J Breed. 1987;37:17–21. [Google Scholar]
- Mouille G, Maddelein M-L, Libessart N, Talaga P, Decq A, Delrue B, Ball S. Phytoglycogen processing: a mandatory step for starch biosynthesis in plants. Plant Cell. 1996;8:1353–1366. doi: 10.1105/tpc.8.8.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–997. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Kubo A, Shimamune T, Matsuda T, Harada K, Satoh H. Correlation between activities of starch debranching enzymes and α-polyglucan structure in endosperms of sugary-1 mutants of rice. Plant J. 1997;12:143–153. [Google Scholar]
- Nelson N. A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem. 1944;153:375–380. [Google Scholar]
- O'Shea MG, Samuel MS, Konik CM, Morell MK. Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labeling and high-resolution separation. Carbohydr Res. 1998;105:1–12. [Google Scholar]
- Rochaix J, Mayfield S, Goldschlmidt-Clermont M, Erickson J. Molecular biology of Chlamydomonas. In: Shaw C, editor. Plant Molecular Biology: A Practical Approach. Oxford: IRL Press; 1991. pp. 253–275. [Google Scholar]
- Somogyi M. Notes on sugar determination. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- Van den Koornhuyse N, Libessart N, Delrue B, Zabawinski C, Decq A, Iglesias A, Preiss J, Ball S. Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J Biol Chem. 1996;271:16281–16288. doi: 10.1074/jbc.271.27.16281. [DOI] [PubMed] [Google Scholar]
- Watson SA, Hirata Y. Sorghum Newslett . 1960;3:6. [Google Scholar]
- Zeeman SC, Umemoto T, Lue WL, Au-Yeung P, Martin C, Smith AM, Chen J. A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell. 1998;10:1699–1712. doi: 10.1105/tpc.10.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]