Abstract
Widespread application of hexaconazole for agriculture purpose poses a threat to human health by disrupting normal endocrine homeostasis. To avoid adverse health effects on human, it is crucial to identify the effects of hexaconazole on key enzymes responsible for steroidal hormone synthesis. In view of this, present study was conducted to investigate the interaction mechanisms of hexaconazole with key enzymes in comparison with their food drug administration (FDA) approved inhibitor by molecular docking and molecular dynamics simulations. Results indicate that hexaconazole contacts with the active site of the key enzymes required for steroidal hormonal synthesis. Results pertaining to root-mean-square deviation, root-mean-square calculation, radius of gyration, hydrogen bonding and solvent accessible surface area exhibited that the interaction pattern and stability of interaction of hexaconazole was similar to enzyme specific inhibitor. In addition, ligand and enzyme complex interaction energy of hexaconazole was almost similar to key enzyme and FDA-approved enzyme specific inhibitor complex. This study offers a molecular level of understanding of hexaconazole with different enzymes required for steroidal hormonal synthesis. Findings of the study clearly suggest that hexaconazole has efficacy to stably interact with various enzyme required to progress the pathway of hormonal synthesis. If incessant exposure of hexaconazole occurs during agricultural work it may lead to ravage hormonal synthesis or potent endocrine disruption. The result of binding energy and complex interaction energy is depicted in the graphical abstract.
Keywords: hexaconazole, biosynthesis, pathway, molecular, dynamics simulation, interaction energy, endocrine disruption
Graphical Abstract
Graphical Abstract.

Pictorial representation of stability and interactive efficacy of hexaconazole with different enzymes involved in the steroidogenic hormone pathway in comparison with their specific inhibitor.
Introduction
Hexaconazole [(RS)-2-(2,4-dichlorophenyl)-1-(1H-1,2,4- triazol-1-yl) hexaconazolean-2-ol] is a fungicide Fig 1. According to US EPA, hexaconazole belongs to Group C-category of human carcinogen. It is incredibly stubborn, and its nondiscernible degradation was observed in river water incubated at 20°C for 3 weeks [1]. In addition, hexaconazole has been shown to influence gene transcription in developmental toxicity pathways [2]. The dissipation time 50% (DT50) for hexaconazole field soil degradation was 225 days [2]. Due to their high biological activity, presence of hexaconazole has been reported in tomato and percent dissipation of hexaconazole residue in baked bread has been found around 46% [3, 4].
Figure 1.
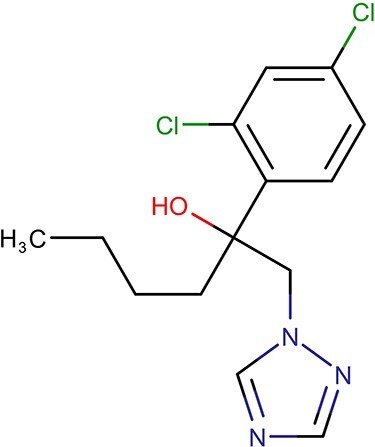
Chemical structure of Hexaconazole (PUB Chem ID 66461).
According to Jia et al. (2019), hexaconazole exposure can cause vacuolization and swelling in the livers of female and male zebrafish [5]. However, endocrine disruptive potential of hexaconazole in zebra fish has also been reported [6]. It is already known that steroidogenic enzymes in the pathway from cholesterol to active steroid hormones play important roles in hormone synthesis and their proper regulation. Nevertheless, the behaviour of hexaconazole for the steroidogenic enzymes responsible to proceed the pathway of steroid hormones synthesis is still unclear. However, Wang et al. (2015) reported bioaccumulation ability of hexaconazole [7]. To fully assess the risks of hexaconazole to humans and other higher animals, a careful research on its metabolism, toxicity and the possible mechanisms by which this compound affects normal body function is necessary, so that it can be remediated from the environment effectively after use. It is also one of the important aspects to know the ability of hexaconazole’s to modulate steroidogenic enzymes in the pathway of steroid hormones synthesis.
Use of hexaconazole for agriculture purposes is rising on daily basis. Therefore, in this work, we evaluated effects of hexaconazole in pathway of steroid hormones synthesis to know its efficacy to modulate the key enzyme responsible to proceed the step of hormone synthesis. The active sites of key enzyme having potential role in the pathway of steroid hormones synthesis and ability of hexaconazole to acquire their active site were explored by molecular interaction and the biophysical movement and stability of the enzyme–hexaconazole complex was confirmed using molecular dynamics (MD) simulation [8–12]. Present study was, therefore, conducted with aim to establish a deep molecular understanding of hexaconazole to interfere in the pathway of steroid hormones synthesis. Findings of this study are useful to provide an understanding that how incessant exposure of hexaconazole may be a risk factor for altered steroidal hormonal synthesis leading to endocrine disruption and disease manifestation.
Materials and Methods
Preparation of the enzymes and ligand
All the enzyme which is required on different step of steroid hormones biosynthesis were downloaded from protein data bank depending on their availability [13–18]. Selected enzyme with their protein databank id and their role in steroid hormones synthesis including their US food and drug administration (FDA)-approved inhibitor is given in Table 1. The inhibitors chosen were used to compare the binding pattern and biophysical movement of hexaconazole with the enzyme involved to proceed the step of hormone synthesis.
Table 1.
Selected enzymes, along with their protein databank id and roles in steroid hormone synthesis, as well as their FDA-approved inhibitors
| SN | Name of enzyme and protein data Bank ID | Role of enzyme in steroid hormones synthesis | Inhibitor ligands and their therapeutic uses |
|---|---|---|---|
| 1 | CYP11A | Converts cholesterol to pregnenolone | Aminoglutethimide (PUB Chem ID 2145) |
| 3N9Z | Therapeutic use— | ||
| • Anticonvulsant in the treatment of petit mal epilepsy | |||
| • Steroidogenesis inhibitor in the treatment of Cushing’s syndrome | |||
| 2 | CYP17A1 | Converts pregnenolone to dehydroepiandrosterone | Abiraterone (PUB Chem ID 9821849) |
| 4NKY | Therapeutic use— | ||
| • Prostate cancer | |||
| 3 | Hydroxysteroid dehydrogenase | Converts pregnenolone to progesterone | Trilostane (PUB Chem ID 656583) |
| 1HDC | Therapeutic use— | ||
| • Cushing’s syndrome | |||
| • Conn’s syndrome | |||
| • Postmenopausal breast cancer | |||
| 4 | CYP11B1 | Converts progesterone to corticosterone | Metyrapone (PUB Chem ID 4174) |
| 6M7X | Therapeutic use— | ||
| • Cushing’s syndrome | |||
| 5 | CYP19A1 | Converts androstenedione to testosterone and estradiol | Anastrozole (PUB Chem ID 2187) |
| 3S79 | Therapeutic use— | ||
| • Breast cancer | |||
| 6 | CYP11B2 | Converts testosterone to aldosterone | Metyrapone (PUB Chem ID 4174) |
| 4DVQ | Therapeutic use— | ||
| • Cushing’s syndrome |
Molecular docking analysis.
The fungicide hexaconazole was docked with different enzymes involved in the biosynthesis pathway of steroid hormones by AutoDock program version 4.2 (Morris et al., 1998). To avoid the hindrance of the ionic property of water molecules in interaction efficiency each enzyme was optimized by removing crystallographic water molecules. In addition, Kollman combined charges, hydrogens and solvation parameters, were added to the protein in ideal geometry. For the compound, torsions were fixed. Before saving the respective protein in PDBQT file format the initial parameters and van der Waals well depth was assigned. The grid was centered on the active site as determined by the PDBsum ligand interaction details. The grid parameter files (GPF) and docking parameter files (DPF) were generated using AutoDock tools after grid box setting. Finally, possible protein–ligand binding conformations were generated using the Lamarckian Genetic Algorithm by using command prompt autogrid -p protein. Gpf-l protein and autodock4-p ligand. Dpf-l ligand. Dlg [19]. The binding energy was calculated using 50 different poses. The complex with the lowest binding energy was chosen for MD studies. The molecular visualization software Discovery Studio 16 was used to analyse all of the complex compound conformations obtained [20].
Molecular dynamics analyses
The structural and dynamic changes in enzyme responsible to proceed the biosynthesis of steroid hormones in comparison with inhibitor were analysed by MD simulation for a period of 10 ns using the GROMACS-2019 software. To build topological structures of respective enzyme and ligand the respective enzyme and ligand complex were parameterized by Charmm 36 force field for all atoms [21–26]. The enzyme and ligand complex was solvated by simple point charge (SPC) water molecules and addition of counter ions (Cl or Na) to neutralize the protein, details is given in supplementary Table 1 [21–26]. The energy minimization was used to remove van der waals contacts between the atoms. There was two phase to perform equilibration of complex for a period of 10 ns. The first phase was a constant number of particles, volume, and temperature (NVT) ensemble with endothermic and exothermic processes which was exchanged with the thermostat. The second phase was with constant number of particles, pressure, and temperature (NPT) ensemble at 300 K along with constant pressure, following that, the Linear Constraint Solver (LINCS) algorithm was used to constrain covalent bonds. Finally, MD was run for 10 ns to evaluate the stability of each system [21–26]. The protein and ligand complex potential energy exhibited that each complex was energetically stable and the details for this are given in supplementary Figure 1. Visual molecular dynamics (VMD) video also represents that protein was in the centre of ligand during simulation run. Video clip for the same is attached as supplementary data.
To analyse the enzyme–substrate complex and its behaviour throughout the simulation, simply root-mean-square deviation (RMSD), root-mean-square calculation (RMSF), radius of gyration (RG), interaction energy and solvent accessible surface area (SASA) calculations were used in this study. In addition, hydrogen bond estimation was also performed in this MD simulation to robust the generated findings. The following command was used to compute g_rmsf, g_gyrate, g_hbond g _sasa plugins of GROMACS, respectively. In, addition “ignh” option of gromacs plugin was applied to ignore hydrogen atoms if required to generate topology file. [21–26]. The MDs parameters that were considered in depth are listed in the supplementary text 1.
ADME analysis
The pharmacokinetics properties of hexaconazole was evaluated by Swiss ADME online tool (http://www.swissadme.ch, governed by Swiss Institute of Bioinformatics [SIB], Lausanne, Switzerland).
Results and Discussion
Molecular docking analysis
Molecular docking analysis was performed for CYP11A, CYP17A1, Hydroxysteroid dehydrogenase, CYP11B1, CYP19A1, CYP11B2 with hexaconazole and for each enzyme their FDA-approved inhibitor was used as a comparator. The binding energy score of hexaconazole with enzyme CYP11A was −6.73 kcal/mol and with its inhibitor aminoglutethimide was −6.86 kcal/mol, respectively. Residues ARG, LEU was associated with hydrogen bond with hexaconazole and Alkyl, Pi Alkyl bond was associated with residue ARG, LEU, MET with hexaconazole. Only one residue CYS was found to be associated with sulphur bond formation with hexaconazole (Fig. 2). The residue ARG, GLN, LEU, TRP was associated with hydrogen bond with aminoglutethimide which was used as a comparator ligand for CYP11A in respect with hexaconazole. Alkyl bond was associated with LEU in aminoglutethimide (Fig. 3). As given in graphical abstract CYP11A is responsible to convert cholesterol to pregnenolone such similar interaction may be responsible for altered pathway of steroidal hormone synthesis. Albeit, altered energy metabolism, lipid metabolism and amino acid metabolism of zebrafish has been reported due to hexaconazole exposure [5].
Figure 2.
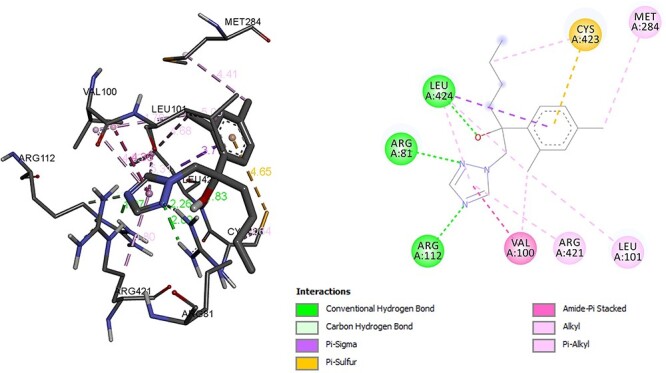
Interaction of hexaconazole with CYP11A.
Figure 3.

Interaction of aminoglutothimide with CYP11A.
The CYP17A1 enzyme is another steroidal hormonal synthesis enzyme that is responsible for transforming pregnenolone to dehydroepiandrosterone. Various therapeutic activities of this enzyme have been reported in cancer, specifically prostate cancer. Overall, because CYP17A1 produces androgen, which again is required for tumour cell growth, CYP17A1 suppression is an attractive objective for in the treatment of prostate cancer [6]. Molecular docking of CYP17A1 with hexaconazole reveals that hexaconazole has almost similar molecular interaction with CYP17A1 as abiraterone which is specific inhibitor of CYP17A1. The residue associated with hydrogen bond formation with hexaconazole are ARG, ILE and restudies associated with Alkyl, Pi Alkyl bond are ALA, LEU. In addition, hexaconazole forms Pi donor hydrogen bond formed with CYS and ILE (Fig. 4). However, abiraterone interactive residue with CYP17A1was ILE, CYS, GLY (hydrogen bond interaction) ALA, LEU, VAL (Alkyl, Pi Alkyl interaction). Albeit, LEU was also associated with Pi-Lone pair interaction (Fig. 5). The hexaconazole and abiraterone had binding energy of −6.58 kcal/mol and −9.22 kcal/mol, respectively. Such similar interaction with common interactive residue ILE, ALA, LEU, CYS between hexaconazole and abiraterone endorse the finding of [27] that azole fungicide may have potency to inhibit CYP17A1 if expose incessantly leading to reprotoxcity [27].
Figure 4.
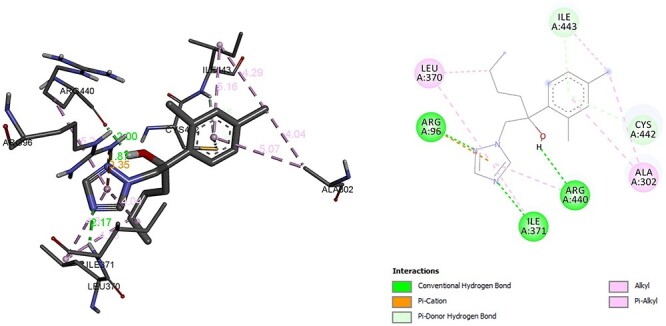
Interaction of hexaconazole with CYP17A1.
Figure 5.
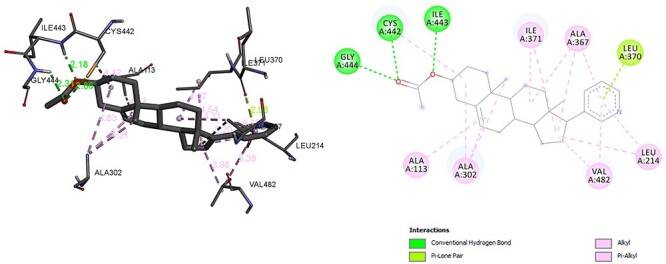
Interaction of abiraterone with CYP17A1.
In other aspect to converts pregnenolone to progesterone enzyme hydroxysteroid dehydrogenase play important role in this step of steroidal pathway. We have also done molecular interaction analysis of hexaconazole with hydroxysteroid dehydrogenase enzyme and its FDA-approved inhibitor trilostane. The hydrogen bond associated residues was THR, SER and residue ALA was associated with Alkyl, Pi Alkyl bond for hexaconazole (Fig. 6). The calculated binding energy for enzyme hydroxysteroid dehydrogenase and hexaconazole complex was −6.53 kcal/mol. However, calculated binding energy for inhibitor trilostane was −9.24 kcal/mol. The residue for hydrogen bond was THR, LEU, GLY and Alkyl, Pi Alkyl bond was associated with residue TYR, PRO, MET, ALA and LEU (Fig. 7). Similarly, Cao et al. has been concluded that fungicide may disrupt normal steroidogenesis through altering the hydroxysteroid dehydrogenase [28].
Figure 6.
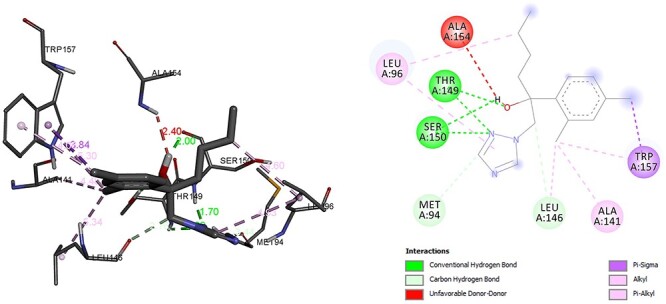
Interaction of hydroxysteroid dehydrogenase with hexaconazole.
Figure 7.

Interaction of hydroxysteroid dehydrogenase with trilostane.
Another important enzyme in the steroidal enzyme biosynthesis pathway is CYP11B1 which converts progesterone to corticosterone. CYP11B1 deficiency leads to elevated levels of adrenocorticotropic hormone, 11-deoxycorticosterone, 11-deoxycortisol. In addition, deficiency of 11-deoxycorticosterone, may be a factor for expansion in the extracellular fluid volume, hypertension and hypokalemia, with a concomitant suppression of the renin-angiotensin-aldosterone system [29–30]. However, CYP11B1 has to inhibited in the treatment of Cushing’s syndrome. In this molecular interaction analysis of CYP11B1 and hexaconazole, it was observed that hexaconazole was able to form hydrogen bond with ARG, Alkyl, Pi Alkyl bond with LEU, PHE, CYS, VAL, ARG and Pi-cation with ARG with efficient binding energy of −7.32 kcal/mol (Fig. 8). In comparison of molecular interaction analysis of CYP11B1 with comparator metyrapone. The binding energy of enzyme CYP11B1 with metyrapone was found to be −6.55 kcal/mol. The interacting residue to form hydrogen bond was ARG, LEU, PHE and Pi Alkyl with residue LEU. In addition, Pi-sulphur and Pi-cation bond was formed with residue CYS and ARG, respectively (Fig. 9).
Figure 8.
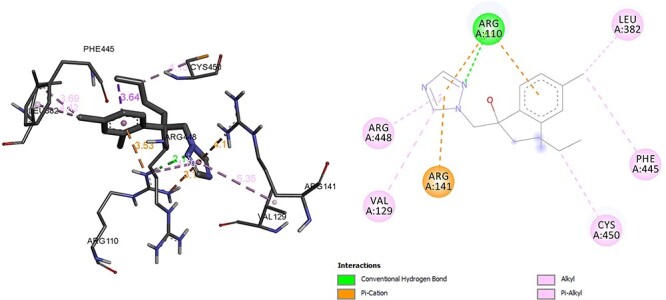
Interaction of hexaconazole with CYP11B1.
Figure 9.
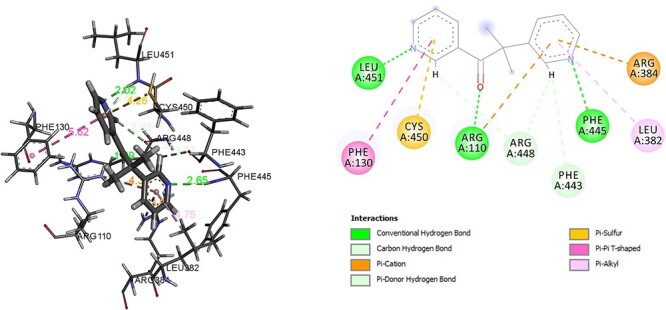
Interaction of metyrapone with CYP11B1.
In the final step of steroid hormones synthesis enzyme CYP19A1, CYP11B2 is responsible to transfer androstenedione to testosterone, estradiol and corticosterone to aldosterone, respectively. The ability of fungicide and fungicide contaminated food to disrupt endocrine system has been highlighted by different study [31, 32]. It has been observed that hexaconazole and inhibitor of CYP19A1 anastrozole have shown binding energy of −6.15 kcal/mol and −8.36 kcal/mol, respectively. The common interaction residues between hexaconazole and inhibitor of CYP19A1 anastrozole were ARG, MET, PHE, VAL, ILE. The residues associated with the formation of hydrogen in CYP19A1 and hexaconazole complex was ARG and Alkyl, Pi Alkyl bond with MET, PHE, VAL, CYS. In addition, hexaconazole also formed Pi-cation, Pi-sulphur with residue MET (Fig. 10). However, anastrozole formed hydrogen bond with MET, ARG and Alkyl and Pi Alkyl bond with ALA, CYS, PHE, VAL (Fig. 11). The influence of hexaconazole and azole containing fungicide on the enzyme of steroidal biosynthesis, cell cycle, and arachidonic acid metabolism pathways in earthworm has been also observed [27, 33].
Figure 10.
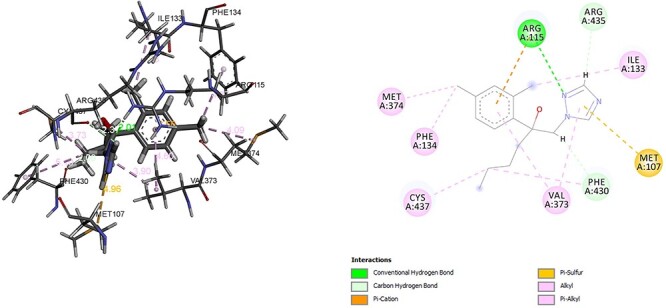
Interaction of hexaconazole with CYP19A1.
Figure 11.
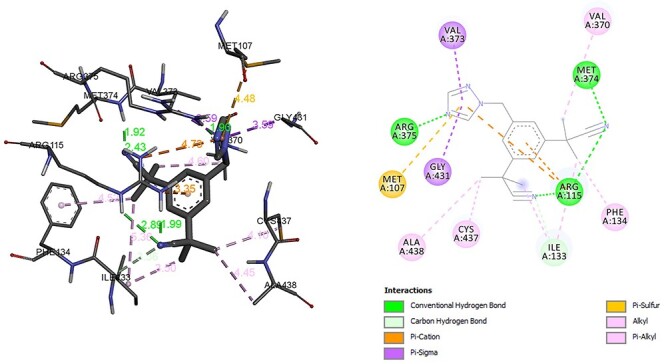
Interaction of anastrozole with CYP19A1.
The enzyme CYP11B2 is responsible to converts testosterone to aldosterone molecular interaction found that residue GLY was associated with hydrogen bonding and Alkyl and Pi Alkyl bond with CYS, ALA. Pi sigma with VAL, LEU and Pi–Pi was associated with PHE. The binding energy of hexaconazole with CYP11B2 was −6.67 kcal/mol (Fig. 12). However, binding energy of CYP11B2 inhibitor metyrapone was −6.61 kcal/mol which was almost similar to hexaconazole reflecting interactive ability of hexaconazole similar with its known inhibitor metyrapone. The residue LEU and CYS was associated with hydrogen bonding similar to hexaconazole CYS was associated with hydrogen bonding. In addition, similar to hexaconazole, metyrapone forms Pi sigma with LEU. The residue VAL was associated with Amide-Pi bond (Fig. 13). The molecular interactions details are given in Table 2.
Figure 12.
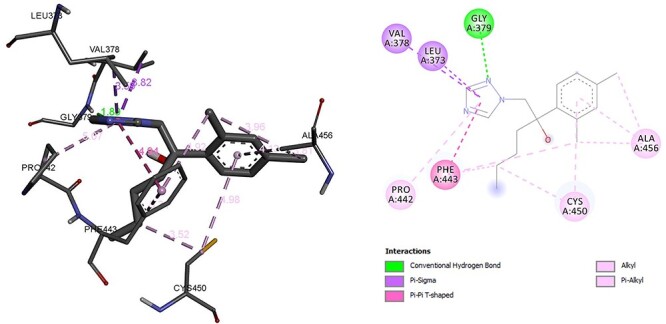
Interaction of hexaconazole with CYP11B2.
Figure 13.
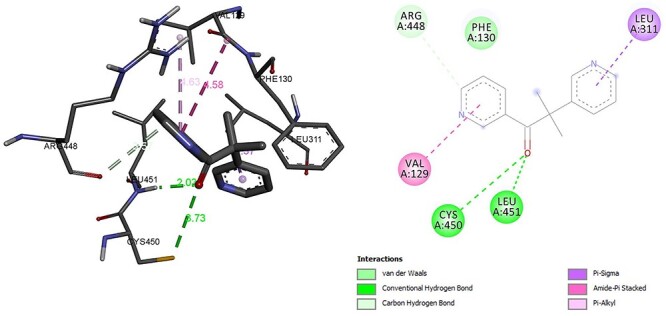
Interaction of metyrapone with CYP11B2.
Table 2.
Molecular interaction analyses
| H bond Distance (Å) | |||||||
|---|---|---|---|---|---|---|---|
| Ligands | Hydrogen bonds | Between hydrogen and acceptor atom | Between donor and acceptor atom | Amino acid residues involved in Hydrophobic interactions | Docking Final intermolecular Energy (Δ G) = vdW + Hbond + desolv Energy (kcal/mol) | Inhibition Constant (Ki) | Protein |
| Hexaconazole | ARG 81 | 2.09 | 3.07 | VAL100 | −8.68 | 11.71 | 3N9Z |
| ARG112 | 1.97 | 2.79 | LEU 424 | μM | CYP11A | ||
| LEU 424 | 1.83 | 2.72 | |||||
| Aminoglutothimide | TRP 108 | 2 | 2.95 | VAL 100 | −7.55 | 9.32 | |
| ARG 112 | 2.05 | 2.79 | LEU 424 | μM | |||
| ARG 421 | 2.43 | 3.25 | |||||
| GLN 422 | 2.42 | 3.31 | |||||
| LEU 424 | 1.79 | 2.68 | |||||
| Hexaconazole | ARG 96 | 1.87 | 2.73 | ALA 367 | −8.59 | 15 | 4NKY |
| ARG 440 | 2 | 2.58 | LEU 370 | μM | CYP17A1 | ||
| ILE 371 | 2.17 | 2.93 | ILE 443 | ||||
| Abiraterone | ILE 443 | 3.01 | 3.71 | ILE 443, ALA 113, 302, 367 | −1.07 | 174.43 | |
| GLY 444 | 2.31 | 2.85 | PHE 114, VAL 482 | μM | |||
| CYS 442 | 2.18 | 2.86 | |||||
| Hexaconazole | THR 149 | 2.42 | 3.36 | LEU 964 | −8.46 | 16.47 | 1HDC |
| SER 150 | 2.21 | 3.14 | PHE 104 | μM | Hydroxysteroid dehydrogenase | ||
| LEU 146 | |||||||
| ALA 154 | |||||||
| TRP 157 | |||||||
| Trilostane | THR 92 | 1.97 | 2.94 | ALA 141 | −9.73 | 167.87 | |
| THR 185 | 2.89 | 3.56 | LEU 146 | μM | |||
| GLY 183 | 1.17 | 2.61 | TYR 152 | ||||
| LEU 148 | 1.97 | 2.78 | |||||
| Hexaconazole | ARG 110 | 2.17 | 3.03 | LEU 382 | −8.56 | 13.56 | 6M7X |
| μM | CYP11B1 | ||||||
| Metyrapone | ARG 110 | 1.89 | 2.42 | LEU 382 | −7.02 | 22.05 | |
| LEU 451 | μM | ||||||
| PHE 445 | |||||||
| Hexaconazole | ARG 115 | 2.91 | 3.64 | ILE 133 A | −8.15 | 32.46 | 3S79 |
| PHE 134 | μM | CYP19A1 | |||||
| VAL 370373 | |||||||
| MET 374 | |||||||
| PHE 430 | |||||||
| Anastrozole | ARG 115 | 2.43 | 2.87 | PHE 134 | −9.25 | 0.742 | |
| MET 374 | 1.92 | 2.9 | VAL 370 | μM | |||
| Hexaconazole | GLY 379 | 1.89 | 2.89 | THR 318 | −8.74 | 12.88 | 4DVQ |
| PHE 443 | μM | CYP11B2 | |||||
| ALA 456 | |||||||
| Metyrapone | CYS 450 | 2.03 | 2.99 | VAL 129 | −7.33 | 14.16 | |
| LEU 451 | 2.03 | 2.73 | PHE 130 | μM | |||
| LEU 451 | |||||||
Molecular dynamics simulation
To discern the ability of the enzyme and fungicide complex in comparison with their FDA-approved inhibitor the MD simulation was performed. The MD simulation was used to analyse the physical movement of the atoms and molecules, and to predict the conformational changes at the molecular level. In this study, we observed the structural changes of CYP11A with hexaconazole and inhibitor aminoglutethimide, CYP17A1 with hexaconazole and inhibitor abiraterone. Hydroxysteroid dehydrogenase with hexaconazole and inhibitor trilostane, CYP11B1 and inhibitor metyrapone. CYP19A1 with hexaconazole and inhibitor anastrozole. A total of six key enzyme essential in the different stages of the steroidal biosynthesis were subjected to MD simulation analysis. The MD approach is successfully used for the identification of endocrine disruptive action of xenobiotic [34].
The variation in the enzyme–substrate complex was determined using RMSD at 10 ns. The RMSD of the enzyme–substrate complex is presented in Fig. 14. The RMSD deviation for CYP11A and hexaconazole complex was exhibited between the range of ~0.05 and 0.1 nm (0.5–1 Å). The RMSD of hexaconazole was almost similar to the RMSD of aminoglutethimide, and CYP11A [~0.05 to 0.05 nm (0.5–0.5 Å)] which was superimposed during MD run (Fig. 14A). The RMSD range of CYP17A1 and hexaconazole complex was in the range of ~0.05 and 0.15 nm (0.5–1.5 Å) and abiraterone which is inhibitor of CYP17A1demonstras RMSD in the range of ~0.1 to 0.2 nm (1–2 Å, Fig. 14A). However, hydroxysteroid dehydrogenase with hexaconazole exhibited RMSD of ~0.05 and 0.15 nm (0.5–1.5 Å) and with inhibitor trilostane ~0.05 and 0.1 nm (0.5–1 Å, Fig. 12B). The enzyme CYP11B1 with hexaconazole and metyrapone showed the RMSD of ~0.05 nm (0.5 Å, Fig. 14B). The RMSD of hexaconazole was superimposed with metyrapone vice versa, reflecting the stability of hexaconazole with enzyme CYP11B1 in similar to inhibitor metyrapone. Albeit, RMSD of CYP19A1 with hexaconazole and anastrozole exhibited the variation of ~0.06 nm to 0.6 Å (Fig. 14C). In the last step of steroidogenic enzyme CYP11B2 which is responsible for the generation of aldosterone. The CYP11B2 and hexaconazole, CYP11B2 and inhibitor metyrapone RMSD were in the range of ~0.06 nm to 0.6 Å (Fig. 14C). The RMSD is an important parameter in MD simulations which is used to measure the equilibration and the flexibility of the proteins/enzymes, as well as to assess the distance between the backbone and atoms of the protein [35]. Albeit, the RMSD for protein and ligand have been assessed to understand the fluctuation in the complex [36, 37].
Figure 14.
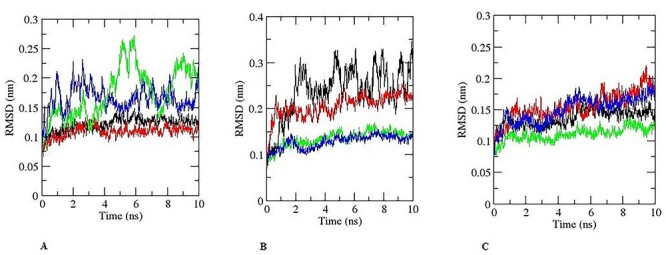
Pictorial representation of RMSD for 10 ns during MD simulation. (A). Black colour is for hexaconazole-CYP11A, red for aminoglutethimide -CYP11A. Green colour for CYP17A1- hexaconazole, blue colour for CYP17A1-abiraterone. (B) Black colour is for hexaconazole- hydroxysteroid dehydrogenase, red for hydroxysteroid dehydrogenase-trilostane. Green colour for CYP11B1- hexaconazole, blue colour for CYP11B1- metyrapone. (C) Black colour is for hexaconazole- CYP19A1, red for anastrozole- CYP19A1. Green colour for CYP11B2- hexaconazole, blue colour for CYP11B2-abiraterone.
This is factually correct that RMSF analysis is important to predict the movement of the position of an atom at a specific temperature and pressure. In addition, RMSF confirms flexible regions of protein and fluctuation in the protein during the MD simulation. It has been also confirmed that RMSF value towards the lower range is reflection of stability of the enzyme–ligand complex, while a higher value reflects the high flexibility in MD [38]. The fluctuations of enzyme–substrate complexes were observed during 10 ns trajectory time. The RMSF fluctuation of the MD simulation is represented in Fig. 15. The result exhibited with little fluctuation in the enzyme–ligand complex of steroidogenic pathway responsible to proceed the step of hormone synthesis one after another as presented in graphical abstract. The enzyme CYP11A with hexaconazole, aminoglutethimide and CYP17A1 with hexaconazole, abiraterone exhibited very less fluctuation pattern demonstrating the restricted movements during the simulation (Fig. 15A). The enzyme hydroxysteroid dehydrogenase and CYP11B1with hexaconazole and their inhibitor trilostane, metyrapone respectively exhibited RMFS fluctuation similar to their FDA-approved inhibitor (Fig. 15B). However, enzyme CYP19A1 and CYP11B2 showed higher fluctuation (Fig. 13C) than other enzyme responsible for progression of steroidogenic synthesis (Fig. 15A and B). RMSF analyses were also performed on the salicylaldehyde dehydrogenases, which revealed high RMSF values, indicating flexible protein residues. It has been shown that the RMSF is associated with cofactor and substrate binding [39]. The findings of the present study suggest that RMSF of hexaconazole—enzyme was similar to FDA-approved inhibitors.
Figure 15.
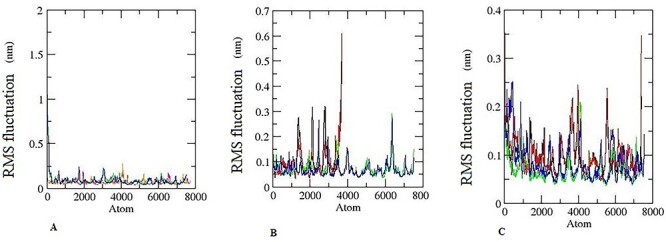
Pictorial representation of RMSF for 10 ns during MD simulation. (A) Black colour is for hexaconazole-CYP11A, red for aminoglutethimide -CYP11A. Green colour for CYP17A1- hexaconazole, blue colour for CYP17A1-abiraterone. (B) Black colour is for hexaconazole- hydroxysteroid dehydrogenase, red for hydroxysteroid dehydrogenase-trilostane. Green colour for CYP11B1- hexaconazole, blue colour for CYP11B1- metyrapone. (C) Black colour is for hexaconazole- CYP19A1, red for anastrozole- CYP19A1. Green colour for CYP11B2- hexaconazole, blue colour for CYP11B2-abiraterone.
The radius of gyration (Rg) is used to describe the compactness changes of an enzyme–substrate complex. It denotes the folding and unfolding of the proteins during MD simulations [22–26, 40]. The Rgs fluctuation for the CYP11A–hexaconazole and CYP11A–aminoglutethimide (near 2.28 nm and decreased to a minimum value of 2.27 nm, Fig. 16A). CYP17A1–hexaconazole and CYP17A1–abiraterone (near 2.23 nm and decreased to a minimum value of 2.28 nm, Fig. 15A), hydroxysteroid dehydrogenase–hexaconazole, hydroxysteroid dehydrogenase–trilostane (near 1.8 nm and decreased to a minimum value of 1.8 nm, Fig. 14B), CYP11B1–hexaconazole and CYP11B1–metyrapone (near 2.29 nm and decreased to a minimum value of 2.29 nm, Fig. 16B), CYP19A1–hexaconazole and CYP19A1– anastrozole (near 2.24 nm and decreased to a minimum value of 2.24 nm, Fig. 16C). CYP11B2–hexaconazole and CYP11B2–metyrapone (near 2.26 nm and decreased to a minimum value of 2.25 nm, Fig. 16B), respectively. From the graph, it is clear that enzyme–ligand complex of hexaconazole showed lower Rg values or it was similar to inhibitor of respective steroidogenic enzyme inhibitor complex. In addition, SASA denotes the interaction of enzyme–ligand complex with solvents. It indicates the interaction of enzyme–ligand complexes with solvents and also denotes the conformational changes which occurs during the time of interactions. The average SASA value for each of the complexes was calculated during MD simulation most of the complex exhibited SASA similar to its inhibitor. The SASA findings reveals that hexaconazole and different steroidogenic enzyme complexes spontaneously form in aqueous solution driven by the charm force field of GROMACS-2019 software. It is reasonable to explain the behaviour of hexaconazole for steroidogenic enzyme is to form a stable binding interaction leading to endocrine disruption (Fig. 17). In addition, interaction energy of the enzyme–substrate predicts the strength of the complex. The interaction energy was also calculated by GROMACS-2019 software. These results of hexaconazole–substrate interaction energy suggest that hexaconazole has a higher potential to inhibit steroidogenic enzyme and the interaction energy findings endorse the interactive ability of hexaconazole exhibited during molecular docking. Nevertheless, the interaction energy for the protein–ligand complexes were calculated and analysed previously and linked with their catalytic and binding affinity [36–41].
Figure 16.
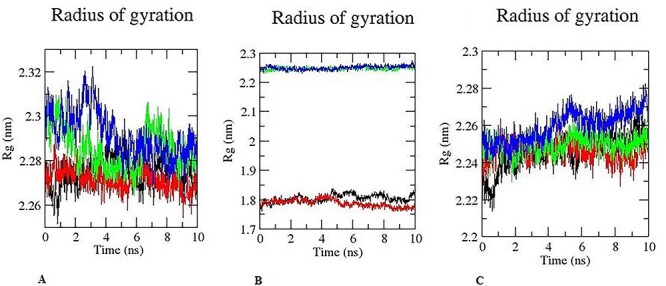
Pictorial representation of Gyration for 10 ns during MD simulation. (A) Black colour is for hexaconazole-CYP11A, red for aminoglutethimide -CYP11A. Green colour for CYP17A1- hexaconazole, blue colour for CYP17A1-abiraterone. (B) Black colour is for hexaconazole- hydroxysteroid dehydrogenase, red for hydroxysteroid dehydrogenase-trilostane. Green colour for CYP11B1- hexaconazole, blue colour for CYP11B1- metyrapone. (C) Black colour is for hexaconazole- CYP19A1, red for anastrozole- CYP19A1. Green colour for CYP11B2- hexaconazole, blue colour for CYP11B2-abiraterone.
Figure 17.
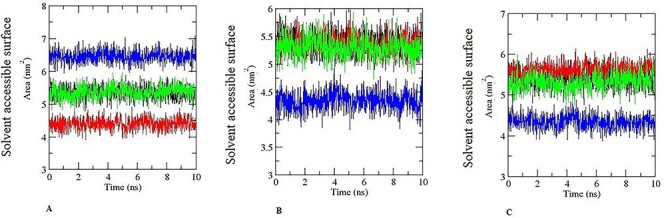
Pictorial representation of SASA for 10 ns during MD simulation. (A) Black colour is for hexaconazole-CYP11A, red for aminoglutethimide -CYP11A. Green colour for CYP17A1- hexaconazole, blue colour for CYP17A1-abiraterone. (B) Black colour is for hexaconazole- hydroxysteroid dehydrogenase, red for hydroxysteroid dehydrogenase-trilostane. Green colour for CYP11B1- hexaconazole, blue colour for CYP11B1- metyrapone. (C) Black colour is for hexaconazole- CYP19A1, red for anastrozole- CYP19A1. Green colour for CYP11B2- hexaconazole, blue colour for CYP11B2-abiraterone.
For enzyme–substrate binding, the hydrogen bond is essential [43]. It is critical for the specificity, metabolism, and catalysis of the substrate. The six enzyme–ligand complex of hexaconazole along with their specific inhibitor were investigated for hydrogen bond. The number of hydrogen bond was observed after 10 ns are depicted in Fig. 18. The results indicate that hexaconazole tightly bind with the active site of the enzyme responsible for the progression of steroidogenic pathway similar to its FDA-approved inhibitor. Previously, it has been found that hydrogen bonds coincide with enzyme–ligand binding of the substrate with the enzyme possessing more hydrogen bonds being associated with the strong interaction of the enzyme–substrate complex [42]. Hydrogen bonding ensures stability of biomolecules as well as the catalytic reactions of enzyme–substrate complexes [43–45]. Earlier researchers used phenol degrading enzymes to study hydrogen bonding [42]. Such hydrogen bonding represented the interaction of the enzyme and substrate after docking. The Stability analysis of the hexaconazole with steroidogenic enzyme is given in Table 3.
Figure 18.
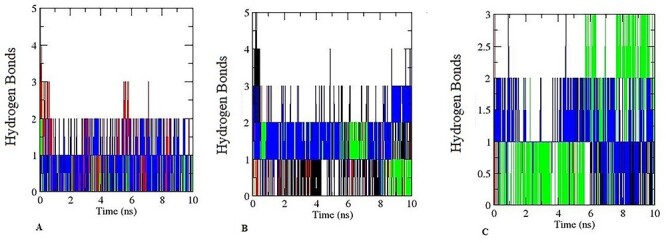
Pictorial representation of hydrogen bonds for 10 ns during MD simulation. (A) Black colour is for hexaconazole-CYP11A, red for aminoglutethimide -CYP11A. Green colour for CYP17A1- hexaconazole, blue colour for CYP17A1-abiraterone. (B) Black colour is for hexaconazole- hydroxysteroid dehydrogenase, red for hydroxysteroid dehydrogenase-trilostane. Green colour for CYP11B1- hexaconazole, blue colour for CYP11B1- metyrapone. (C) Black colour is for hexaconazole- CYP19A1, red for anastrozole- CYP19A1. Green colour for CYP11B2- hexaconazole, blue colour for CYP11B2-abiraterone.
Table 3.
Stability analyses of hexaconazole with different key enzymes involved in the steps of steroidal hormone synthesis in comparison with FDA approved inhibitor of respective enzymes
| SN | Enzyme substrate complex | RMSD (nm) | RMSF (nm) | RG | SASA (nm2) | Interaction energy (kj/mol) |
|---|---|---|---|---|---|---|
| (nm) | ||||||
| 1 | CYP11A–hexaconazole | 0.05 ± 0.01 | 0.23 ± 0.01 | 2.28 ± 0.01 | 5.8 ± 0.1 | −142.95 |
| CYP11A–aminoglutethimide | 0.05 ± 0.01 | 0.23 ± 0.01 | 2.28 ± 0.01 | 4.9 ± 0.1 | −111.86 | |
| 2 | CYP17A1–hexaconazole | 0.05 ± 0.15 | 0.23 ± 0.01 | 2.23 ± 0.02 | 5.8 ± 0.1 | −155.14 |
| CYP17A1–abiraterone | 0.05 ± 0.15 | 0.23 ± 0.01 | 2.23 ± 0.02 | 6.9 ± 0.9 | −209.89 | |
| 3 | Hydroxysteroid dehydrogenase–hexaconazole | 0.05 ± 0.15 | 0.1 ± 0.01 | 1.8 ± 0.01 | 5.8 ± 0.8 | −149.04 |
| Hydroxysteroid dehydrogenase–trilostane | 0.05 ± 0.15 | 0.1 ± 0.01 | 1.8 ± 0.01 | |||
| 4 | CYP11B1–hexaconazole | 0.05 ± 0.01 | 0.1 ± 0.01 | 2.22 ± 0.02 | 5.8 ± 0.1 | −178.59 |
| CYP11B1–metyrapone | 0.05 ± 0.01 | 0.1 ± 0.01 | 2.22 ± 0.02 | 4.6 ± 0.6 | −179.23 | |
| 5 | CYP19A1–hexaconazole | 0.06 ± 0.02 | 0.2 ± 0.1 | 2.23 ± 0.01 | 5.8 ± 0.81 | −14.21 |
| CYP19A1–anastrozole | 0.06 ± 0.02 | 0.2 ± 0.1 | 2.24 ± 0.01 | 5.8 ± 0.41 | −198.04 | |
| 6 | CYP11B2–hexaconazole | 0.06 ± 0.05 | 0.2 ± 0.1 | 2.25 ± 0.01 | 5.6 ± 0.60 | −152.25 |
| CYP11B2–metyrapone | 0.06 ± 0.05 | 0.2 ± 0.1 | 2.25 ± 0.01 | 4.8 ± 0.60 | −14.23 |
ADME analysis
Hexaconazole’s ADMET properties were predicted using the Swiss ADME server. Hexaconazole had significant values in many attributes. The log kp for skin permeation was −5.45 cm/s, and it was also effective in crossing the blood–brain barrier. Hexaconazole, surprisingly, does not violate Lipinski’s or Pfizer’s rule of five, and it may easily mimic the pharmacokinetic properties of FDA-approved inhibitor which is indicated for the treatment of various type pathological manifestation including cancer. Details of ADME are given in Table 4.
Table 4.
Important ADME and toxicological properties of hexaconazole
| SN | Parameters | Results |
|---|---|---|
| 1 | GI absorption | Yes |
| 2 | BBB permeant | Yes |
| 3 | Log Kp (skin permeation) | −5.45 cm/s |
| 4 | Lipinski violation | 0 violation |
| 5 | Molecular weight | 314.21 g/mol |
| 6 | Num. H-bond acceptors | 3 |
| 7 | Num. H-bond donors | 1 |
| 8 | CYP1A2 inhibitor | Yes |
| 9 | CYP2C19 inhibitor | Yes |
| 10 | CYP2D6 inhibitor | Yes |
| 11 | CYP3A4 inhibitor | No |
| 12 | Water solubility | 1.74e−02 mg/ml; 5.54e−05 mol/l |
Hexaconazole’s ADMET properties describe that hexaconazole has gastrointestinal absorption potency with a skin permeation coefficient (Log Kp) of −5.45 cm/s. Hexaconazole can cross blood brain barrier. Combining all findings, it is possible to say that hexaconazole can stably interact with key steroidogenic enzyme by accruing the sites of binding proteins similar to steroidogenic enzyme inhibitor leading to ravage steroidal hormonal synthesis depending upon duration and amount of exposure. The findings of this study are promising, and there is a requirement of further preclinical and clinical studies on hexaconazole.
Conclusion
In this study, we explored the molecular interactions of hexaconazole with key enzyme responsible for the progression of steroidal hormone synthesis. Hexaconazole acquires similar binding pattern in comparison with key enzyme inhibitor indicated for the amelioration of different disease. The protein–ligand complex interaction energy for hexaconazole was also similar or near to similar in comparison with the respective inhibitor. Incessant exposure of hexaconazole may intensify altered steroidal hormone synthesis leading to endocrine disruption.
Authors’ contribution
All the authors of the manuscript have contributed significantly in the manuscript to make it in its final stage. The lead author Dr Sayed Aliul Hasan Abdi has designed the study and has contributed by ideas; formulation or evolution of overarching research goals and aims. Dr Abidi has conducted a research and investigation process, specifically performing the experiments, or data/evidence collection, designed the methodology, created models, done Programming, software development, designed computer programmes etc. He has leadership responsibility for the research activity planning and execution.
Dr Abdulaziz Alzahrani, Dr Saleh Alghamdi, Dr Ali Alquraini, and Dr Adel Alghamdi were involved in verification, whether as a part of the activity or separate, of the overall replication/reproducibility of results/experiments and other research outputs.
Supplementary Material
Contributor Information
Sayed Aliul Hasan Abdi, Department of Pharmaceutical Chemistry, Faculty of Clinical Pharmacy, Albaha University, 1988, Saudi Arabia.
Abdulaziz Alzahrani, Department of Pharmaceutical Chemistry, Faculty of Clinical Pharmacy, Albaha University, 1988, Saudi Arabia.
Saleh Alghamdi, Department of Clinical Pharmacy, Faculty of Clinical Pharmacy, Albaha University, 1988, Saudi Arabia, Saudi Arabia.
Ali Alquraini, Department of Pharmaceutical Chemistry, Faculty of Clinical Pharmacy, Albaha University, 1988, Saudi Arabia.
Adel Alghamdi, Department of Pharmaceutical Chemistry, Faculty of Clinical Pharmacy, Albaha University, 1988, Saudi Arabia.
Conflict of interest statement
None declared.
Funding
There is no funding source for this study.
References
- 1. Yu M, Chen Y, Liu Wet al. Thyroid endocrine disruption in zebrafish larvae following exposure to hexaconazole and tebuconazole. Aquat Toxicol 2013;138:35–42. [DOI] [PubMed] [Google Scholar]
- 2. Hermsen SA, Pronk TE, Brandhof EJet al. Triazole-induced gene expression changes in the zebrafish embryo. Reprod Toxicol 2012;34:216–24. [DOI] [PubMed] [Google Scholar]
- 3. Han J, Jiang J, Su Met al. Bioactivity, toxicity and dissipation of hexaconazole enantiomers. Chemosphere 2013;93:2523–7. [DOI] [PubMed] [Google Scholar]
- 4. Randhawa M A, Ahmed A, Javed, M S. Wheat contaminants (pesticides) and their dissipation during processing. In Wheat and Rice in Disease Prevention and Health (pp. 263–77). 2014. United State: Academic Press. [Google Scholar]
- 5. Jia M, Wang Y, Wang Det al. The effects of hexaconazole and epoxiconazole enantiomers on metabolic profile following exposure to zebrafish (Danio rerio) as well as the histopathological changes. Chemosphere 2019;1:520–33. [DOI] [PubMed] [Google Scholar]
- 6. Yu L, Chen M, Liu Yet al. Thyroid endocrine disruption in zebrafish larvae following exposure to hexaconazole and tebuconazole. Aquat Toxicol 2013;138:35–42. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y, Xu L, Li Det al. Enantioselective bioaccumulation of hexaconazole and its toxic effects in adult zebrafish (Danio rerio). Chemosphere 2015;138:798–805. [DOI] [PubMed] [Google Scholar]
- 8. Bhardwaj VK, Purohit R, Kumar S. Himalayan bioactive molecules as potential entry inhibitors for the human immunodeficiency virus. Food Chem 2021; 15;347:128932. [DOI] [PubMed] [Google Scholar]
- 9. Singh R, Bhardwaj VK, Sharma Jet al. Identification of selective cyclin-dependent kinase 2 inhibitor from the library of pyrrolone-fused benzosuberene compounds: an in silico exploration. J Biomol Struct Dyn 2021;9:1–9. [DOI] [PubMed] [Google Scholar]
- 10. Sharma J, Bhardwaj VK, Das Pet al. Identification of naturally originated molecules as γ-aminobutyric acid receptor antagonist. J Biomol Struct Dyn 2021;39:911–22. [DOI] [PubMed] [Google Scholar]
- 11. Singh R, Bhardwaj V, Purohit R. Identification of a novel binding mechanism of Quinoline based molecules with lactate dehydrogenase of plasmodium falciparum. J Biomol Struct Dyn 2021;39:348–56. [DOI] [PubMed] [Google Scholar]
- 12. Singh R, Bhardwaj VK, Sharma Jet al. Discovery and in silico evaluation of aminoarylbenzosuberene molecules as novel checkpoint kinase 1 inhibitor determinants. Genomics 2021;1:707–15. [DOI] [PubMed] [Google Scholar]
- 13. Strushkevich N, MacKenzie F, Cherkesova Tet al. Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. PNAS 2011; 1; 25; 10139–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petrunak EM, DeVore NM, Porubsky PRet al. Structures of human steroidogenic cytochrome P450 17A1 with substrates. J Biol Chem 2014;47:32952–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghosh D, Erman M, Wawrzak Zet al. Mechanism of inhibition of 3α, 20β-hydroxysteroid dehydrogenaseby a licorice-derived steroidal inhibitor. Structure 1994;10:973–80. [DOI] [PubMed] [Google Scholar]
- 16. Brixius-Anderko S, Scott EE. Structure of human cortisol-producing cytochrome P450 11B1 bound to the breast cancer drug fadrozole provides insights for drug design. J Biol Chem 2019;2:453–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghosh D, Lo J, Morton Det al. Novel aromatase inhibitors by structure-guided design. J Med Chem 2012;19:8464–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strushkevich N, Gilep AA, Shen Let al. Structural insights into aldosterone synthase substrate specificity and targeted inhibition. J Mol Endocrinol 2013;2:315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morris G M, Goodsell DS, Halliday R Set al. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 1998; 19; 1639–62. [Google Scholar]
- 20. Biovia DS. Dassault Systemes BIOVIA, discovery studio, 2019. Dassault Syst_emes. https://www.3dsbiovia.com/about/ citations-references. 2019.
- 21. Ajao AT, Kannan M, Yakubu SEet al. Homology modeling, simulation and molecular docking studies of catechol-2, 3-dioxygenase from Burkholderia cepacia: involved in degradation of petroleum hydrocarbons. Bioinformation 2012;18:848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J, Cheng X, Swails JMet al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput 2016;1:405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abdi SA, Alzahrani A, Asad Met al. Molecular docking and dynamics simulation to screen interactive potency and stability of fungicide tebuconazole with thyroid and sex hormone-binding globulin: implications of endocrine and reproductive interruptions. J Appl Toxico 2021;10:1649–59. [DOI] [PubMed] [Google Scholar]
- 24. Thirumal Kumar D, Lavanya P, George Priya Doss Cet al. A molecular docking and dynamics approach to screen potent inhibitors against fosfomycin resistant enzyme in clinical klebsiella pneumoniae. J Cell Biochem 2017;11:4088–94. [DOI] [PubMed] [Google Scholar]
- 25. Rafi MO, Al-Khafaji K, Tok TTet al. Computer-based identification of potential compounds from Salviae miltiorrhizae against Neirisaral adhesion a regulatory protein. J Biomol Struct Dyn 2020;1:1–3. [DOI] [PubMed] [Google Scholar]
- 26. Kumar DT, Eldous HG, Mahgoub ZAet al. Computational modelling approaches as a potential platform to understand the molecular genetics association between Parkinson’s and Gaucher diseases. Metab Brain Dis 2018;6:1835–47. [DOI] [PubMed] [Google Scholar]
- 27. Bonomo S, Hansen CH, Petrunak EMet al. Promising tools in prostate cancer research: selective non-steroidal cytochrome P450 17A1 inhibitors. Sci Rep 2016;6:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roelofs MJ, Temming AR, Piersma AHet al. Conazole fungicides inhibit Leydig cell testosterone secretion and androgen receptor activation in vitro. Toxicol Rep 2014;1:271–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cao S, Ye L, Wu Yet al. The effects of fungicides on human 3β-hydroxysteroid dehydrogenase 1 and aromatase in human placental cell line JEG-3. Pharmacology 2017; 100; 139–147. [DOI] [PubMed] [Google Scholar]
- 30. White PC 2001. Steroid 11β-hydroxylase deficiency and related disorders. Endocrinol Metab Clin 2001; 1; 61–79. [DOI] [PubMed] [Google Scholar]
- 31. Simone GIOVANNI, Tommaselli AP, Rossi RICCARDOet al. Partial deficiency of adrenal 11-hydroxylase. A possible cause of primary hypertension. Hypertension 1985;2:204–10. [DOI] [PubMed] [Google Scholar]
- 32. Marx-Stoelting P, Knebel C, Braeuning Aet al. The connection of azole fungicides with xeno-sensing nuclear receptors, drug metabolism and hepatotoxicity. Cell 2020;5:1192–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zubrod JP, Bundschuh M, Arts Get al. Fungicides: an overlooked pesticide class? Environ Sci Technol 2019;7:3347–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu T, Fang K, Liu Yet al. Enantioselective residues and toxicity effects of the chiral triazole fungicide hexaconazole in earthworms (Eisenia fetida). Environ Pollut 2021;270:116269–75. [DOI] [PubMed] [Google Scholar]
- 35. Wahl J, Smiesko M. 2018. Endocrine disruption at the androgen receptor: employing molecular dynamics and docking for improved virtual screening and toxicity prediction. Int J Mol Sci 2018; 19;1784–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sargsyan K, Grauffel C, Lim Cet al. How molecular size impacts rmsd applications in molecular dynamics simulations. J Chem Theory Comput 2017;13:1518–24. [DOI] [PubMed] [Google Scholar]
- 37. Bhatt K, Maheshwari DK. Insights into zinc-sensing metalloregulator ‘Zur’deciphering mechanism of zinc transportation in bacillus spp. by modeling, simulation and molecular docking. J Biomol Struct Dyn 2020;14:1–16. [DOI] [PubMed] [Google Scholar]
- 38. Joshi T, Joshi T, Sharma Pet al. Molecular docking and molecular dynamics simulation approach to screen natural compounds for inhibition of Xanthomonas oryzae pv. Oryzae by targeting peptide deformylase. J. Biomol. Struct. Dyn 2020;12:1–18. [DOI] [PubMed] [Google Scholar]
- 39. Dong YW, Liao ML, Meng XLet al. Structural flexibility and protein adaptation to temperature: molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proc Natl Acad Sci USA 2018;115:1274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jia B, Jia X, Kim KHet al. Evolutionary, computational, and biochemical studies of the salicylaldehyde dehydrogenases in the naphthalene degradation pathway. Sci Rep 2017;7:43489–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adams MA, Luo Y, Hove-Jensen Bet al. Crystal structure of PhnH: an essential component of carbon-phosphorus lyase in Escherichia coli. J Bacteriol 2008;190:1072–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Y, Liu Z, Zeng Get al. Effect of surfactants on the interaction of phenol with laccase: molecular docking and molecular dynamics simulation studies. J Hazard Mater 2018;357:10–8. [DOI] [PubMed] [Google Scholar]
- 43. Singh S, Kumar V, Datta Set al. Glyphosate uptake, translocation, resistance emergence in crops, analytical monitoring, toxicity and degradation: a review. Environ Chem Lett 2020;18:663–702. [Google Scholar]
- 44. Maguire JB, Boyken SE, Baker Det al. Rapid sampling of hydrogen bond networks for computational protein design. J Chem Theory Comput 2018;14:2751–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen M, Zeng G, Xu Pet al. Interaction of carbon nanotubes with microbial enzymes: conformational transitions and potential toxicity. Environ Sci Nano 2017;4:1954–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


