Abstract
Chlamydomonas reinhardtii mutants of the STA8 gene produce reduced amounts of high amylose starch and phytoglycogen. In contrast to the previously described phytoglycogen-producing mutants of C. reinhardtii that contain no residual isoamylase activity, the sta8 mutants still contained 35% of the normal amount of enzyme activity. We have purified this residual isoamylase and compared it with the wild-type C. reinhardtii enzyme. We have found that the high-mass multimeric enzyme has reduced its average mass at least by one-half. This coincides with the disappearance of two out of the three activity bands that can be seen on zymogram gels. Wild-type and mutant enzymes are shown to be located within the plastid. In addition, they both act by cleaving off the outer branches of polysaccharides with no consistent difference in enzyme specificity. Because the mutant enzyme was demonstrated to digest phytoglycogen to completion in vitro, we propose that its inability to do so in vivo supports a function of the enzyme complex architecture in the processing of pre-amylopectin chains.
Mutants lacking isoamylase, an enzyme that catalyzes the breakdown of α-1,6 branches, are unable to synthesize normal starch in maize, rice, and Chlamydomonas reinhardtii (for review, see Nakamura, 1996; Myers et al., 2000). In maize and rice the so-called “sugary” mutations that map in an isoamylase structural gene lead to the disappearance of all detectable isoamylase activity. Mutants defective in the STA7 locus of C. reinhardtii equally lack starch, but the relation to the isoamylase structural gene still needs to ascertained (Mouille et al., 1996). This has led to the contention that debranching of a precursor of amylopectin known as pre-amylopectin is an integral part of the amylopectin synthesis pathway (Ball et al., 1996; Myers et al., 2000). It was proposed that the release of improperly positioned α-1,6 branches allowed proper alignment of the α-1,4-linked glucans within the growing polysaccharide. In turn, this alignment facilitated crystallization within large-size starch granules. Mutations in another gene (STA8) that leads to some partial defect in isoamylase activity have recently been documented in C. reinhardtii. These mutations lead to a two-third reduction in total isoamylase activity, together with a substantial decrease in amylopectin content and the concomitant appearance of glycogen-like polymers (phytoglycogen; Dauvillée et al., 2001). In C. reinhardtii, heterozygous triploids with a sta7/sta7/STA7 genotype contain the same wild-type isoamylase specific activity as the mutant sta8 haploids (Dauvillée et al., 2001). Yet these heterozygous triploids accumulate wild-type amounts of normal starch and no phytoglycogen. These results suggest that it is the quality of the residual isoamylase in the sta8 mutant rather than the reduced amount of activity that is responsible for the dysfunctions in amylopectin synthesis. Because the wild-type isoamylase activity is not present in rate controlling amounts in C. reinhardtii, explanations have to be sought that are consistent with the finding of a substantial amount of residual activity in the sta8 mutants. The first obvious explanation is that the enzyme's substrate specificity has changed. The second is that the mutant enzyme is mislocated and that only tiny and insufficient amounts of activity are transported within the plastid. The third is that although significant enzyme activity amounts can be measured in vitro, the mutant enzyme is nearly completely inactive in vivo. The fourth is that the known multimeric architecture of plant isoamylases per se might be of particular relevance for amylopectin synthesis. We now report the partial purification of this residual isoamylase present in the sta8 mutant and its comparison with the wild-type enzyme. We provide evidence that the multimeric organization of the isoamylase per se is of paramount importance in vivo during amylopectin synthesis
RESULTS
Mutants of the STA8 Locus Lack Two out of the Three Native Isoamylase Zymogram Bands
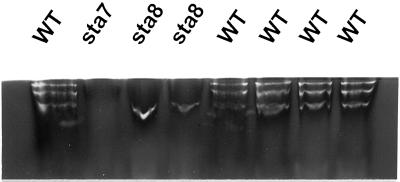
In C. reinhardtii zymogram, procedures have been devised that allow migration of denatured proteins and their renaturation in the presence of substrate (Mouille et al., 1996). This technique enabled us to visualize an 88-kD debranching enzyme with isoamylase specificity. The presence of this polypeptide is clearly under control of the STA7 locus and gene dosage experiments are consistent with STA7 encoding this catalytic isoamylase subunit. STA8 has no detectable influence on the quality or quantity of 88-kD debranching enzyme as detected in these zymograms. Because we know the isoamylase to be substantially reduced in sta8 mutants, we embarked in a more detailed zymogram investigation by looking at native proteins. We used the set-up detailed in Kakefuda and Duke (1984) and a novel procedure devised by us for glycogen containing gels (see “Materials and Methods”). In this system three clear, white-staining isoamylase bands of similar intensity were detected in all wild-type strains (Fig. 1). All three bands disappeared in sta7 mutants. It is interesting that the sta8 mutant clearly lacked the two slow migrating bands (Fig. 1). To ensure that it was specifically due to the presence of sta8-1::ARG7 we examined cosegregation of the zymogram defect with the sta8 mutation on 26 wild-type and 27 mutant recombinants from a cross involving the BafV13 mutant and the A35 wild-type strain. Cosegregation was observed on all 53 meiotic recombinants. In addition, we observed full epistasis of sta7 on sta8 and no activity bands could be scored in the double mutants.
Figure 1.
Detection of isoamylase complexes on glycogen-containing zymograms. Native crude extracts (100 μg of protein) were loaded on a rabbit liver glycogen containing zymogram (see “Materials and Methods”). C. reinhardtii isoamylase complexes appears as white bands after incubation at the top of the gel. The isoamylase defective mutants carrying the sta7-1::ARG7 mutation show no band on this zymogram. Wild-type strains always display three distinct bands of similar intensity, whereas sta8-1::ARG7 strains display only one lower white band on over 50 meiotic segregants (27 mutant strains and 26 wild type).
Partial Purification and Comparison of the Wild-Type and Mutant Isoamylase Architecture
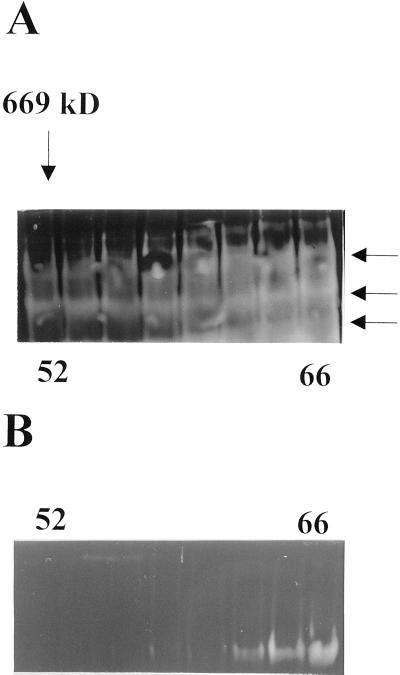
To better understand the qualitative modification of isoamylase activity present in the sta8 mutants we partly purified the enzyme activity according to a pre-established procedure (Dauvillée et al., 2000) and we compared the elution patterns of the mutant enzymes with those of the wild-type activities. Purification of the wild-type enzyme relied on a S-300 fast-protein liquid chromatography (FPLC) gel filtration followed by anion-exchange on a UNOQ FPLC column. Yields and purification factors are summarized in Table I. Figure 2 exemplifies results obtained with the S-300 FPLC. We used the zymogram procedure developed by Kakefuda and Duke (1984) to visualize the isoamylase activities within the protein mixture. We also sized the proteins by using commercially available mass standards (thyroglobulin, apoferritin, and catalase, respectively, of 669, 443, and 240 kD). Figure 2 displays the gel filtration step performed on the same amount of protein extract with the relevant molecular mass standard (thyroglobulin, 669 kD) position for wild-type (Fig. 2A) and mutant (Fig. 2B) strains of isogenic background. Three blue activity bands can again be seen in zymograms from the wild-type strain, whereas only one such band is witnessed in the sta8 mutant. As a control we checked that all three bands disappeared in sta7 mutants. In addition, we calculated the average mass of the debranching activities from three distinct purification experiments (see “Materials and Methods”). We find values of 500 ± 100 kD for the wild type and 150 ± 50 kD for the mutant activities. It is interesting that the presence of the 88-kD DBE subunit visualized after denaturation on zymogram gels displays an equivalent shift, proving that this catalytic subunit is present within a protein complex of much larger size in the wild type than in the mutant. We found no clear correlation in the wild-type strain chromatogram between molecular mass and the presence of any of the three distinct zymogram bands, suggesting as was recently observed in rice that the three separate bands are probably generated by the experimental procedures from an otherwise intact multimeric complex (Fujita et al., 1999). To confirm that molecular mass of the complex and presence of a distinct subset of zymogram bands were direct consequences of the sta8 mutation we partly purified the DBEs from five wild-type (strains BGM13, BGM15, BGM80, BGM200, and BGM202) and five mutant (strains BGM6, BGM11, BGM12, BGM97, and BGM201) meiotic recombinants and found identical results.
Table I.
Isoamylase purification
| Extract Origin | Yield
|
Purification
Factor
|
Total Activitya
|
|||
|---|---|---|---|---|---|---|
| sta8 | + | sta8 | + | sta8 | + | |
| % | ||||||
| Crude extract | 100 | 100 | 0 | 0 | 225 | 322 |
| Protamine sulfate | 74 | 65 | 1, 7 | 1, 3 | 167 | 210 |
| Ammonium sulfate | 46 | 40 | 2, 4 | 1, 6 | 105 | 130 |
| Gel permeation (52–65) | – | 17 | – | 9, 9 | – | 55 |
| Gel permeation (58–66) | 19 | – | 9, 9 | – | 42 | – |
| Anion exchange (19–20) | – | 2, 5 | – | 140 | – | 8, 2 |
| Anion exchange (15–16) | 4, 5 | – | 77 | – | 10, 2 | – |
Purification was from 4 × 1010 cells. Debranching activity corresponds to micromoles of maltotriose equivalents produced per hour from amylopectin for the whole fraction in study. After the gel permeation step, wild-type isoamylase was free of interfering starch hydrolytic activity, whereas trace amounts of α-amylase can be detected by zymograms in the sta8 semipurified fractions.
Figure 2.
Semipurification of wild-type and sta8 isoamylases. Crude extracts from 20-L cultures of wild-type strain A35 or the sta8-1 BafV13 strain were subjected to the purification procedure described previously by Dauvillée et al. (2000). The undenatured samples (80 μL from the fractions 52, 54, 56, 58, 60, 62, 64, and 66 of the S300 GPC) separated by PAGE under native conditions were blotted onto starch-containing gels according to Kakefuda and Duke (1984). The color of the bands observed after staining the gel with iodine will change according to the enzyme specificities. Branching enzymes will stain white or light red, amylases and glucosidases will stain white, and debranching enzymes (pullulanases or isoamylases) will stain blue. A, Zymogram obtained from wild-type isoamylase semi-purification. B, The same fractions analyzed from the reference sta8-1 mutant strain.
Substrate Specificity and Mode of Action of Wild-Type and Mutant Enzymes
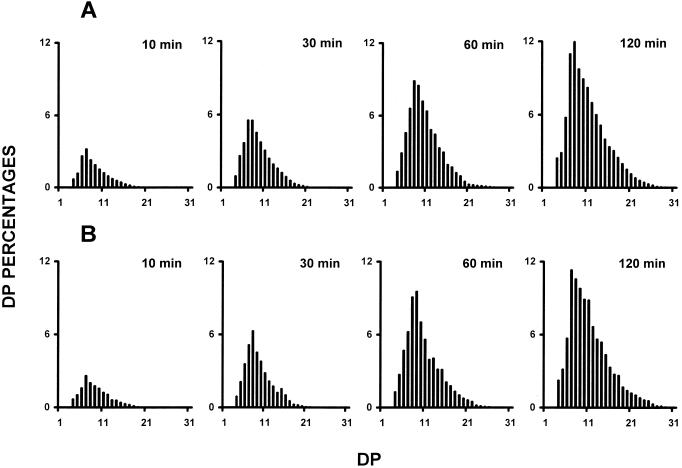
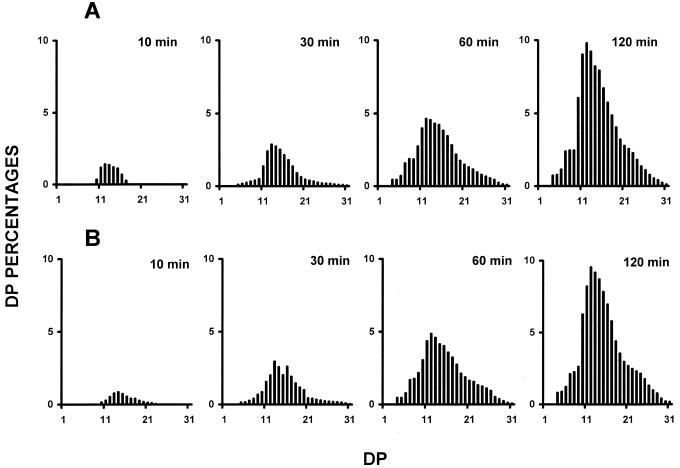
The trimming function that we envision for the plant isoamylases in amylopectin synthesis implies that the DBEs splice out only those branches that would otherwise prevent amylopectin crystallization (Mouille et al., 1996; Myers et al., 2000). This suggests that any modification of the substrate specificity of the isoamylases will interfere with polysaccharide crystallization and therefore with amylopectin synthesis. Modification of the isoamylase complex through the sta8 mutation is thus expected to generate some change in substrate specificity in the DBE complex that would conveniently explain the mutant phenotype. We, therefore, embarked on experiments designed to probe the specificity of the wild-type and mutant enzyme not only in semi-pure fractions, but also in crude extracts. This was done to avoid selecting any subset of the activities displayed in Figure 1 that might differ with the bulk of the isoamylase activities and therefore lead us to biased conclusions. We used two different analytical procedures (fluoro-phore-assisted carbohydrate electrophoresis and high performance anion-exchange with pulsed amperometric detection [HPAED-PAD]) to characterize the chain-length distributions of the glucans liberated through enzymatic debranching from bovine liver glycogen (Fig. 3) and maize amylopectin (Fig. 4). In addition, we quantified the amount of branches present in the malto-oligosaccharides produced upon incubation with the enzyme extracts. We also checked the chain-length distribution within the undigested polysaccharide fraction. This was achieved by treatment of this fraction with the Pseudomonas amyloderamosa isoamylase. We never released branched malto-oligosaccharides during these experiments and the branching percentage always fell below our level of detection (<1%). Moreover, the polysaccharide fraction remaining after digestion with the wild-type or mutant C. reinhardtii isoamylase displayed similar chain-length distribution. These results prove that the plant isoamylases are only able to digest the outer branches of the substrate polysaccharides. Figures 3 and 4 exemplify the type of results we got through all these experiments. Although in some cases we did witness some small differences, the latter vanished upon reproducing the experiment and displayed no constant pattern distinguishing the wild-type and mutant enzyme. We conclude that both enzymes display apparently the same or a very similar specificities during the debranching of amylopectin and glycogen. These conclusions hold within the limits afforded by the presently available analytical tools.
Figure 3.
Debranching kinetics of bovine liver glycogen by wild-type and mutant isoamylases. Semipurified isoamylases were incubated in the presence of 5 mg mL−1 of bovine liver glycogen (see “Materials and Methods”). Aliquots were boiled after 10, 30, 60, and 120 min and chain-length distribution of the glucans liberated were analyzed by HPAED-PAD. A, Chain length distribution of glucans liberated by wild-type semipurified isoamylase (fraction from 52–65, the gel filtration FPLC). B, Chain length distribution of glucans produced by semipurified isoamylase from sta8 defective strains (fraction from 58– 66, the gel filtration FPLC). The results are displayed as percentages of chains of degrees of polymerization (DP) 1 to 32. The x scale displays a DP scale, and the y axis represents the relative frequencies of the chains expressed as percentages.
Figure 4.
Debranching kinetics of maize amylopectin by wild-type and mutant enzymes. Semipurified isoamylases were incubated in the presence of 5 mg mL−1 of maize amylopectin (see “Materials and Methods”). Aliquots were boiled after 10, 30, 60, and 120 min and chain length distributions were analyzed through HPAED-PAD. A, Chain length distribution of glucans liberated by wild-type semipurified isoamylase. B, Chain length distribution of glucans liberated by semipurified isoamylase from the sta8 defective strain. The results are displayed as percentages of chains of DP between 1 to 32. The x scale displays a DP scale, and the y axis represents the relative frequencies of the chains expressed as percentages.
Because glycogen and amylopectin do not define the true in vivo substrates for the C. reinhardtii enzyme we reasoned that the phytoglycogen synthesized in the sta8 mutants should be specifically enriched in chains selectively resistant to the mutant enzyme, thereby explaining accumulation of the latter in the mutant plastid. We were thus surprised to find that the wild-type and mutant isoamylase are equally effective for debranching phytoglycogen in vitro (initial rates measured for phytoglycogen debranching were, respectively, of 370 ± 12 and 359 ± 21 nmol equivalents maltotriose protein mg−1 h−1); moreover, both enzymes were equally effective in debranching phytoglycogen to completion.
We measured the Km for glycogen and amylopectin and the optimum pH and temperature values for mutant and wild-type and we were unable to find any convincing difference.
Localization of the Wild-Type and Mutant Isoamylase and in Vivo Activity
Another possible explanation for the presence of an altered phenotype within the sta8 mutants can be easily found if the mutant isoamylase was not efficiently transported within the plastid compartment. An older study by Levi and Gibbs (1984) established α-amylase as a plastidial enzyme. However, the enzyme assays used by Levi and Gibbs would not distinguish amylases in general from isoamylases and glucosidases. Therefore, we embarked in cell fractionation experiments to revisit this issue with the aim of probing the localization of the wild-type and mutant isoamylase. The results listed in Table II establish wild-type and mutant isoamylases as entirely (>95%) plastidial, whereas the amylase assay attributes only part of this family of enzymes to the plastid compartment.
Table II.
Enzyme activities of cell homogenates and chloroplasts from Chlamydomonas reinhardtii wild-type and sta8 strains
| Enzymes | Wild Type
|
sta8
|
||
|---|---|---|---|---|
| Homogenate | Chloroplasts | Homogenate | Chloroplasts | |
| UGPase | 609 ± 36 | 116 ± 15 | 567 ± 38 | 126 ± 14 |
| Phosphoenolpyruvate carboxylase | 12 ± 0.7 | 2.2 ± 0.3 | 10 ± 1 | 1.6 ± 0.2 |
| NADP GAP DH | 138 ± 6 | 184 ± 11 | 120 ± 6 | 159 ± 4 |
| Isoamylase | 9.4 ± 0.3 | 10.2 ± 0.8 | 2.6 ± 0.1 | 3.7 ± 0.1 |
Activities are expressed in micromoles per milligram of chlorophyll per hour and are the average of three separate measures.
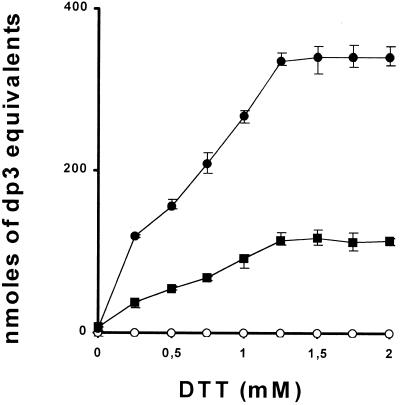
All of our measures of enzyme activity are performed in the presence of 2 mm dithiothreitol (DTT) to obviate enzyme oxidation and inactivation. The wild-type C. reinhardtii enzyme is exquisitely sensitive to oxidation and no enzyme activity can be measured in its absence. This property is shared by many other plant isoamylases. In addition, once oxidized, we are unable to recover any significant enzyme activity if we reintroduce DTT in the assays. It thus remains possible that in vivo the residual activity present in the sta8 mutants remains insufficiently reduced. In this specific case the addition of 2 mm DTT might restore some activity to the complex and therefore lead to an overestimation of the biologically relevant activity left within the mutant strains. To tackle this difficult question we designed an experimental procedure aimed at maximizing our chances of observing some activity in the natural reduced state occurring within the concentrated crude extract without the addition of 2 mm DTT (see “Materials and Methods”). We reasoned that if the ratio of mutant-to-wild-type activities remained constant within a full range of DTT concentrations, it would suggest that the residual isoamylase present within the mutants is not less active in vivo because of its decreased ability to be reduced. Results displayed in Figure 5 show that wild-type and mutant enzyme are equally sensitive to reduction by DTT, suggesting that the one-third ratio observed by in vitro measures between mutant and wild-type activities reflects the in vivo situation within the plastid at the site of starch biosynthesis.
Figure 5.
DTT sensitivity of C. reinhardtii isoamylases. The fresh crude extracts from synchronized cultures were used to assay isoamylase activities in wild-type and sta8 strains using bovine liver glycogen as substrate (see “Materials and Methods”) at various DTT concentrations. Results are displayed as nanomoles of maltotriose formed by hour and per milligram of protein. The x axis represents the DTT concentrations used in the incubation buffer. Means (●, ▪, ○ for wild-type [330], sta8-1::ARG7 [BafV13], sta7-4::ARG7 [Bafj6] strains, respectively) and sd from three distinct measures were calculated for each DTT concentration.
Mutants of the STA7 and STA8 Locus Display Identical Rates of Phytoglycogen Degradation in Vivo
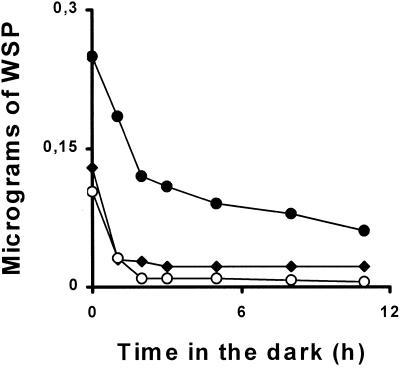
Starch and water-soluble polysaccharide degradation can be induced by simply switching C. reinhardtii cultures to darkness. In the experiments depicted in Figure 6 we induced accumulation of phytoglycogen from three different genotypes carrying the sta7, sta7 sta8, and sta8 mutations, respectively. Despite differences in the initial phytoglycogen content of the strains used in these experiments, the rates of polysaccharide degradation were similar in all three mutants. It is remarkable that strains that contain respectively 35% and 0% of residual isoamylase activity display similar amounts of phytoglycogen and identical polysaccharide degradation kinetics in vivo. We believe these results prove that isoamylase does not play a rate controlling role in the breakdown of phytoglycogen in vivo.
Figure 6.
Kinetics of WSP degradation. One week TAP-N cultures (6 L) inoculated at 5.105 cells mL−1 in the light were switched to darkness. Five-hundred-milliliter samples were harvested and the WSP was assayed by the amyloglucosidase assay (see “Materials and Methods”). ●, ♦, and ○ represent strains harboring the sta7-1::ARG7 (strain GM7.27), the sta8-1::ARG7 (strain BafV13), and both mutations (strain BGM87, respectively).
The Residual Mutant Isoamylase Activity Becomes Rate-Controlling for Amylopectin Synthesis in a sta8 Background
Mutants of the sta8 locus display phytoglycogen and a reduced amount of amylopectin with normal small- and medium-size (up to DP50) chain-length distributions. One possible explanation for this observation is that the mutant isoamylase activity has become rate controlling for the production of amylopectin. The presence of the mutant enzyme would simply slow down the process of amylopectin crystallization to the point where pre-amylopectin can be transformed into phytoglycogen by other enzymes of the starch pathway. To test this hypothesis we constructed a diploid strain homozygous for the sta8-1::ARG7 mutation, but heterozygous for sta7-1::ARG7. Such a sta8-1::ARG7/sta8-1::ARG7 sta7-1::ARG7/+ diploid strain contains a mutant isoamylase amounting to one-half of the activity present in haploid or homozygous mutant diploid sta8 strains. We reasoned that if the residual mutant isoamylase has become rate controlling for amylopectin synthesis, we should observe a significant reduction in starch content in these diploids when compared with the reference sta8 homozygous diploids. Measures of starch and phytoglycogen levels performed in a series of three distinct diploid clones of identical genotype for diploid classes confirm these speculations. We have measured levels of 3 ± 0.5 μg starch × 10−6 cells and 3 ± 2 μg phytoglycogen × 10−6 cells in the sta8-1::ARG7/sta8-1::ARG7 sta7-1::ARG7/+ diploids, whereas the homozygous mutant diploid reference sta8-1::ARG7/sta8-1::ARG7 +/+ contained 10 ± 3 μg starch and 1 ± 0.5 μg phytoglycogen. We, therefore, conclude that the presence of sta8 renders the residual isoamylase rate controlling for amylopectin biosynthesis.
DISCUSSION
The Rate of Semicrystalline Amylopectin Formation Decreases in Strains Harboring a Defect in the Assembly of the Isoamylase Multimer Complex
Mutants of the STA8 locus in C. reinhardtii accumulate restricted amounts of high amylose starch and phytoglycogen. This coincides with a drop of 65% in isoamylase activity, whereas no other enzyme known to interfere with starch biosynthesis seems modified (Dauvillée et al., 2000). In this work we prove that this residual activity is exported normally to the plastid compartment and that the substrate specificity has not been significantly altered. Moreover, we provide suggestive evidence that this enzyme remains functional in vivo. Because we know that triploids displaying one-third of the wild-type isoamylase activity display a fully wild-type phenotype, the mutant enzyme must differ by some other property essential in vivo for normal amylopectin synthesis. This work establishes that the high-mass multimeric complex has collapsed from a minimum of one-half to one-fourth of its normal size in the mutants. This coincides with a simplification of the zymogram profile from three to one fast-migrating activity band, although only one form might in fact exist in vivo in wild-type and mutant strains. The modification of the isoamylase multimeric architecture is presently the only convincing qualitative difference that we can reasonably offer to explain the mutant phenotype. We, therefore, propose that the multimeric assembly of isoamylase subunits into a large size complex per se is essential to proper enzyme function.
If we do not take into account the increase in amylose content, the residual starch synthesized in the sta8 mutants displays a structure quite similar to that of wild-type polysaccharide. The increase in amylose and in the long chain content of amylopectin can be attributed to a relative decrease of the rate of amylopectin synthesis or to an increase in malto-oligosaccharide and/or ADP-Glc levels, which define mechanisms documented to control amylose content and synthesis by granule-bound starch synthase I (Leloir et al., 1961; Denyer et al., 1996; Van den Koornhuyse et al., 1996; van de Wal et al., 1998). In line with the absence of modification that we now report for the mutant enzyme's substrate specificity is the observation that the distribution in size of the small and intermediate chains of amylopectin has not significantly changed. This result is at variance with that reported for maize and rice where mutations in a gene encoding an isoamylase subunit essential for catalytic activity are reported to significantly modify the residual amylopectin. It is also at variance with the deeply modified structure of the granular polysaccharide found in sta7 mutants of C. reinhardtii that equally lack the major isoamylase catalytic subunit (Dauvillée et al., 1999)
We believe that the presence of a significantly reduced amount of otherwise normal amylopectin points to a reduction in the rate of normal amylopectin synthesis due to a severe dysfunction of the isoamylase complex. We do not believe that the normal function of isoamylase is impeded completely in the mutants, but rather that the reaction simply slows down to the point where it becomes limiting for amylopectin synthesis, whereas simultaneously allowing the formation of phytoglycogen. This suggestion is confirmed by the observation that the mutant isoamylase has become rate controlling for amylopectin synthesis in a mutant sta8 background.
Isoamylase Does Not Directly Control the Steady-State Levels and Structure of Phytoglycogen
We have proven that the steady-state levels and phytoglycogen structure are identical in sta7 mutants lacking all isoamylase activities and in sta8 mutants that still contain a significant amount of biologically active enzyme. In addition, we have proven that the isoamylase activity remaining in the sta8 mutant debranches phytoglycogen in vitro with an efficiency that is similar to that of the wild-type enzyme. Taken together, these results make a very strong case against isoamylase being a rate-controlling factor responsible for down-regulating phytoglycogen production. This conclusion is also supported by the measures of identical rates of phytoglycogen breakdown in vivo in strains lacking or displaying reduced amounts of isoamylase. Because the isoamylase of the sta8 mutant has become rate controlling for starch synthesis, we believe these results suggest a function of the wild-type isoamylase in amylopectin synthesis rather than in phytoglycogen breakdown.
The Nature of the sta7 and sta8 Gene Products
C. reinhardtii is presently the only plant system where two loci have been described to control phytoglycogen production. The exact molecular nature of both loci still needs to be established. Gene dosage experiments performed with the STA7 locus together with the absence the 88-kD isoamylase subunit documented in such mutants clearly point to STA7 as a structural gene that would encode the 88-kD catalytic subunit of the enzyme. This hypothesis is further supported by the discovery of the molecular nature of analogous mutations in rice and maize (James et al., 1995; Kubo et al., 1999). The nature of the STA8 gene products remains, however, elusive. Two equally viable assumptions can be made with respect to this locus. First, the phenotype of the C. reinhardtii sta8 mutants matches precisely that which was reported for a mutant of Arabidopsis defective in a gene known to encode an isoamylase-like subunit (Zeeman et al., 1998). One might thus be tempted to believe STA8 encodes another isoamylase-like subunit in a heteromultimer enzyme. This subunit would have more a regulatory than a catalytic function within the enzyme complex in a fashion that is reminiscent of the catalytic and regulatory subunits of ADP-Glc pyrophosphorylase. The potato isoamylase complex has been previously purified to homogeneity and was demonstrated to contain at least two distinct subunits (Ishizaki et al., 1983). However, only one isoamylase subunit has been demonstrated to exist within the pure rice isoamylase complex (Fujita et al., 1999). It remains, however, equally possible that the leaf enzyme complexes contain several distinct isoamylases, whereas the cereal endosperm complex would only contain one such subunit. Another possibility would be that STA8 encodes a protein required for the assembly of the isoamylase complex. In line with such a hypothesis is the copurification of the maize isoamylase sugary 1 subunit together with bacterial chaperonins upon expression in Escherichia coli (A.M. Myers, personal communication). It must be stressed that although investigations concerning the molecular nature of STA7 and STA8 clearly define our next research priorities, the conclusions that we propose in this paper do not require that STA8 encodes a functional isoamylase subunit.
MATERIALS AND METHODS
Materials
The apoferritin and thyroglobulin mass standards, the maize amylopectin, the soluble potato starch, and the bovine or rabbit liver glycogen were supplied by Sigma Chemical (St. Louis). The catalase mass standard was from Boehringer-Mannheim (GmbH).
Chlamydomonas reinhardtii Strains, Growth Conditions, Water-Soluble Polysaccharide (WSP) Assays, and Media
The wild-type reference strain used in this work is 330 (mt+ arg7 cw15 nit1 nit2), whereas strains BafV13 (mt+ cw15 nit1 nit2 sta8-1::ARG7) and GM7.27 (mt− pab2 sta7-1::ARG7) were used as our reference sta8 and sta7 mutant strains, respectively. Wild-type and mutant segregants from a cross between BafV13 and GM7.27 were used throughout this work (strains called BGM). The genotype of the segregants was checked by standard complementation tests and starch-containing zymograms. WSP was assayed as follows. One liter of Tris-acetate phosphate- (TAP) N cultures inoculated at 5.105 cells mL−1 were harvested after 5 d and were ruptured by one passage in a French Press (10,000 psi). Standard TAP medium was fully detailed in Harris (1989b). TAP-N defines TAP medium where NH4Cl was substituted by an equivalent concentration of NaCl. The lysate was kept at 4°C and was immediately cleared by centrifugation at 3,000g for 10 min. The supernatant was immediately frozen (−20°C) and thawed after 2 h. The thawed extract was boiled for 5 min in a water bath and was further cleared by spinning at 10,000g for 15 min at 4°C. The WSP was immediately assayed by the amyloglucosidase assay (Delrue et al., 1992).
All experiments were carried out in continuous light (40 μE m−2 s−1) in the presence of acetate at 24°C in liquid cultures that were shaken without air or CO2 bubbling. Late-log phase cultures were inoculated at 105 cells mL−1 and were harvested at 2 × 106 cells mL−1. Genetic techniques were as described by Harris (1989a).
Enzyme Purification and Zymograms
Crude extract preparation and isoamylase purification from C. reinhardtii crude extracts were fully detailed in Dauvillée et al. (2000). In brief, the thawed extracts were subjected to successive protamine sulfate and ammonium sulfate precipitations. Proteins obtained in the 35% (w/v) ammonium sulfate pellet were loaded on a FPLC gel permeation chromatography Sephacryl S-300 HR. The fractions containing the isoamylase activity were detected by starch containing zymogram and were pooled and loaded on a FPLC UnoQ1 (Bio-Rad, Hercules, CA) column. Gel permeation columns were precalibrated by subjecting 5 mg of thyroglobulin (669 kD), apoferritin (443 kD), and catalase (240 kD) to the same chromatographic procedure.
Zymograms in starch-containing gels (0.3% [w/v] soluble potato starch) allowing the detection of most starch hydrolases have been described for undenaturated samples by Kakefuda and Duke (1984). To detect C. reinhardtii isoamylase isoforms, 100 μg of crude extract proteins were loaded on a 29:1 (acrylamide:bisacrylamide) 7.5% (w/v; 1.5 mm thick) native polyacrylamide gel containing 0.6% (w/v) of rabbit liver glycogen (Sigma) and were run at 20 V cm−1 for 120 min at 4°C in 25 mm Tris Gly, pH 8.3, and 1 mm DTT. Gels were incubated for 1 to 12 h in the same buffer containing 20 mm DTT and were finally stained by using a freshly prepared 0.02% (w/v) I2 and 0.2% (w/v) KI stock solution.
Chloroplast Isolation and Characterization
Synchronized C. reinhardtii cultures on a 12-h light/12-h dark cycle were used to extract chloroplasts. The cells were harvested in the middle of the third light period. The method used to isolate chloroplasts has been fully detailed in Mason et al. (1991). The chloroplast fractions obtained were used to measure marker enzyme activities including UDP-Glc pyrophosphorylase and phosphoenolpyruvate carboxylase as cytosolic markers and NADP glyceraldehyde 3-phosphate dehydrogenase as plastidial markers. The corresponding enzyme assays have been previously described in Borchert et al. (1993). Activities were assayed in the plastidial fraction and the homogenate from the same cultures. Chloroplast contamination by cytosolic enzymes in all preparations was estimated to be around 20%.
Isoamylase Assays and Debranching Kinetics
Isoamylase was always assayed at 30°C in 20 mm Tris, 10 mm EDTA, and 2 mm DTT, pH 7, unless otherwise specified, using bovine liver glycogen or amylopectin at 5 mg mL−1. Activities were measured as previously detailed (Dauvillée et al., 2000) through the increase in reducing power generated by the debranching reaction. This increase was translated into maltotriose equivalents by using the latter as standard (Dauvillée et al., 2000). After stopping the reaction by boiling for 5 min, the determination of the chain-length distribution of the glucans released by the C. reinhardtii isoamylases were obtained by fluorophore-assisted carbohydrate electrophoresis or HPAED-PAD chromatography as fully detailed in O'Shea et al. (1998) and Libessart et al. (1995).
To check the sensitivity of the wild-type and mutant enzyme to DTT, we used the same protocol, but with several concentrations of DTT including 0, 0.25, 0.5, 0.75, 1, 1.25, 1.5, 1.75, and 2 mm. To obtain cells in the same redox state we used 3-d-old synchronized cultures undergoing 12-h light/12-h dark cycles. The cells were harvested rapidly for 5 min at 3,000g. All subsequent steps were carried out under high light and on ice to avoid oxidation within the extract. The crude extract was obtained by breaking the cells with a tissue grinder tube on ice and was used immediately to assay isoamylase with the procedure described above.
ACKNOWLEDGMENT
The authors thank A. Decq for excellent technical assistance.
Footnotes
This work was supported by the Ministère de l'Education Nationale, by the Centre National de la Recherche Scientifique, by Biogemma UK, and by the U.S. Department of Agriculture.
LITERATURE CITED
- Ball S, Guan H-P, James M, Myers A, Keeling P, Mouille G, Buléon A, Colonna P, Preiss J. From glycogen to amylopectin: a model explaining the biogenesis of the plant starch granule. Cell. 1996;86:349–352. doi: 10.1016/s0092-8674(00)80107-5. [DOI] [PubMed] [Google Scholar]
- Borchert S, Harborth J, Schünemann D, Hoferichter P, Heldt HW. Studies of the enzymic capacities and transport properties of pea root plastids. Plant Physiol. 1993;101:303–312. doi: 10.1104/pp.101.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D, Colleoni C, Mouille G, Buléon A, Gallant DJ, Bouchet B, Morell M, D'Hulst C, Myers AM, Ball SG (2001) Two loci control phytoglycogen production in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol (in press) [DOI] [PMC free article] [PubMed]
- Dauvillée D, Colleoni C, Shaw E, Mouille G, D'Hulst C, Morell M, Samuel MS, Bouchet B, Gallant DJ, Sinskey A. Novel starch-like polysaccharides are synthesized by a soluble form of granule-bound starch synthase in glycogen accumulating mutants of Chlamydomonas reinhardtii. Plant Physiol. 1999;119:321–330. doi: 10.1104/pp.119.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D, Mestre V, Colleoni C, Slomianny MC, Mouille G, Delrue B, d'Hulst C, Bliard C, Nuzillard J-M, Ball S. The debranching enzyme complex missing in glycogen accumulating mutants of Chlamydomonas reinhardtii displays an isoamylase-type specificity. Plant Sci. 2000;157:145–156. doi: 10.1016/s0168-9452(00)00256-9. [DOI] [PubMed] [Google Scholar]
- Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski JM, Van Den Koornhuyse N, Maddelein M-L, Fournet B, Ball S. Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase accumulate a structurally modified amylopectin. J Bacteriol. 1992;174:3612–3620. doi: 10.1128/jb.174.11.3612-3620.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer K, Clarke B, Hylton C, Tatge H, Smith A. The elongation of amylose and amylopectin chains in isolated starch granules. Plant J. 1996;10:1135–1143. [Google Scholar]
- Fujita N, Kubo A, Francisco PB, Nakakita M, Harada K, Minaka N, Nakamura Y. Purification, characterization, and cDNA structure of isoamylase form developing endosperm of rice. Planta. 1999;208:283–293. doi: 10.1007/s004250050560. [DOI] [PubMed] [Google Scholar]
- Harris EH. Culture and storage methods. In: Harris E, editor. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989a. pp. 25–63. [DOI] [PubMed] [Google Scholar]
- Harris EH. Genetic analysis. In: Harris E, editor. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989b. pp. 399–446. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Taniguchi H, Maruyama Y, Nakamura M. Debranching enzymes of potato tuber (Solanum tuberosum L.): I. Purification and some properties of potato isoamylase. Agric Biol Chem. 1983;47:771–779. [Google Scholar]
- James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakefuda G, Duke SH. Electrophoretic transfer as a technique for the detection and identification of the plant amylolytic enzymes in polyacrylamide gels. Plant Physiol. 1984;75:278–280. doi: 10.1104/pp.75.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y. The starch-debranching enzyme isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 1999;121:399–410. doi: 10.1104/pp.121.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloir LF, de Fekete MAR, Cardini CE. Starch and oligosaccharide synthesis from uridine diphosphate glucose. J Biol Chem. 1961;236:636–641. [PubMed] [Google Scholar]
- Levi C, Gibbs M. Starch degradation in synchronously grown Chlamydomonas reinhardtii and characterization of the amylase. Plant Physiol. 1984;74:459–463. doi: 10.1104/pp.74.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libessart N, Maddelein M-L, Van Den Koornhuyse N, Decq A, Delrue B, Ball SG. Storage, photosynthesis and growth: the conditional nature of mutations affecting starch synthesis and structure in Chlamydomonas reinhardtii. Plant Cell. 1995;7:1117–1127. doi: 10.1105/tpc.7.8.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason CB, Matthews S, Bricker TM, Moroney JV. Simplified procedure for the isolation of intact chloroplasts. Plant Physiol. 1991;97:1576–1580. doi: 10.1104/pp.97.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Maddelein M-L, Libessart N, Talaga P, Decq A, Delrue B, Ball S. Phytoglycogen processing: a mandatory step for starch biosynthesis in plants. Plant Cell. 1996;8:1353–1366. doi: 10.1105/tpc.8.8.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–997. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. Some properties of starch debranching enzymes and their possible role in amylopectin biosynthesis. Plant Sci. 1996;121:1–18. [Google Scholar]
- O'Shea MG, Samuel MS, Konik CM, Morell MK. Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labeling and high-resolution separation. Carbohydr Res. 1998;307:1–12. [Google Scholar]
- van de Wal M, D'Hulst C, Vincken J-P, Buléon A, Visser R, Ball S. Amylose is synthesized in vitro by extension of and cleavage from amylopectin. J Biol Chem. 1998;273:22232–22240. doi: 10.1074/jbc.273.35.22232. [DOI] [PubMed] [Google Scholar]
- Van den Koornhuyse N, Libessart N, Delrue B, Zabawinski C, Decq A, Iglesias A, Preiss J, Ball S. Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J Biol Chem. 1996;271:16281–16288. doi: 10.1074/jbc.271.27.16281. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Umemoto T, Lue WL, Au-Yeung P, Martin C, Smith AM, Chen J. A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell. 1998;10:1699–1712. doi: 10.1105/tpc.10.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]