Abstract
Background: Long non-coding RNAs (lncRNAs) are key regulators of pancreatic cancer development and are involved in ferroptosis regulation. LncRNA transcript levels serve as a prognostic factor for pancreatic cancer. Therefore, identifying ferroptosis-related lncRNAs (FRLs) with prognostic value in pancreatic cancer is critical.
Methods: In this study, FRLs were identified by combining The Cancer Genome Atlas (TCGA) and FerrDb databases. For training cohort, univariate Cox, Lasso, and multivariate Cox regression analyses were applied to identify prognosis FRLs and then construct a prognostic FRLs signature. Testing cohort and entire cohort were applied to validate the prognostic signature. Moreover, the nomogram was performed to predict prognosis at different clinicopathological stages and risk scores. A co-expression network with 76 lncRNA-mRNA targets was constructed.
Results: Univariate Cox analysis was performed to analyze the prognostic value of 193 lncRNAs. Furthermore, the least absolute shrinkage and selection operator and the multivariate Cox analysis were used to assess the prognostic value of these ferroptosis-related lncRNAs. A prognostic risk model, of six lncRNAs, including LINC01705, AC068620.2, TRAF3IP2-AS1, AC092171.2, AC099850.3, and MIR193BHG was constructed. The Kaplan Meier (KM) and time-related receiver operating characteristic (ROC) curve analysis were performed to calculate overall survival and compare high- and low-risk groups. There was also a significant difference in survival time between the high-risk and low-risk groups for the testing cohort and the entire cohort, with AUCs of .723, .753, respectively. Combined with clinicopathological characteristics, the risk model was validated as a new independent prognostic factor for pancreatic adenocarcinoma through univariate and multivariate Cox regression. Moreover, a nomogram showed good prediction.
Conclusion: The signature of six FRLs had significant prognostic value for pancreatic adenocarcinoma. They may be a promising therapeutic target in clinical practice.
Keywords: pancreatic adenocarcinoma, ferroptosis, long non-coding RNA, risk model, nomogram
1 Introduction
Pancreatic adenocarcinoma (PAAD) is an aggressive gastrointestinal malignancy, with a high mortality rate (Mizrahi et al., 2020). Because of its early subclinical symptoms, the vast majority of PAAD patients were diagnosed in the middle to late stages, even with distant metastasis. Worse still, the level of acceptance of surgical treatment is relatively low at 20% (Singhi et al., 2019). Improving the prognosis of PAAD remains a challenge, with less than 5% of patients surviving for 5 years (Hidalgo, 2010; Hidalgo et al., 2015). In recent years, neoadjuvant therapy, targeted therapy, and immunotherapy have gained widespread application and yielded good clinical outcomes (Neoptolemos et al., 2018; Tempero, 2019).
Ferroptosis id a new type of cell death regulated by multiple metabolic pathways, lipid peroxide accumulation and iron dependence and, thus, may be involved in the mechanism of PAAD progression (Gupta et al., 2017; Wei et al., 2019; Badgley et al., 2020; Zhou et al., 2020). Emerging evidence suggests that ferroptosis provides a link between the pathophysiological disease mechanism and the altered human health status through its involvement in metabolic and redox reactions and negatively affects (Zheng and Conrad, 2020). It is promoted by BRCA1-associated protein 1 (BAP1) through SLC7A11 repression, and inhibited by glutathione-dependent phospholipid peroxidase 4, preventing lipid peroxide accumulation. Promoting ferroptosis circumvents therapy-resistant cancer cells (Hassannia et al., 2019). Therefore, a validated predictive model is needed to assess patient prognosis and guide patients to personalized targeted therapy.
Long non-coding RNAs (lncRNAs) are transcribed from genes but do not undergo translation into proteins (Bhan et al., 2017). They mediate the regulation of ferroptosis pathways; for instance, the cytoplasmic lncRNA P53RRA can promote ferroptosis through nuclear segregation of p53 (Mao et al., 2018). Furthermore, lncRNAs can interact with mRNA, thus modulating mRNA stability. Therefore, they are a crucial factor in regulating cholesterol metabolism processes in cancer.
Whether or not targeted therapies can have anti-cancer effects on patients needs to be further explored. A validated predictive model is required to accurately assess patient prognosis, account for individual differences, and guide individualized treatment to prolong survival.
The aim of the present study was to identify ferroptosis-related lncRNAs (FRLs) that have prognostic value in PAAD. We utilized The Cancer Genome Atlas (TCGA) dataset for lncRNA expression in PAAD to create a FRLs signature and elucidate its ability to predict overall survival (OS) in PAAD. Further, we summarize six previous reports of FRLs and briefly describe their potential mechanisms of action to provide a reference for further research.
2 Materials and Methods
2.1 Gene Expression and Clinical Data Access to Pancreatic Adenocarcinoma
Gene expression data and clinical information were downloaded from the TCGA-PAAD cohort FPKM file from the TCGA Information Portal (https://portal.gdc.cancer.gov/). After excluding 14 patients without RNA-seq or clinical information, 171 patients were included in this study. The 171 patients were divided into training cohort (n = 86) and testing cohort (n = 85). The training cohort were used for risk score model building, and the testing cohort and entire cohort were used for validation of the risk score model to test the robustness of the risk model.
TCGA data is open to the community and our research is based on the TCGA Data Access Policy and Publication Guidelines.
2.2 Screening for Ferroptosis-Related lncRNAs
The ferroptosis-related mRNAs were obtained from the FerrDb database (http://www.zhounan.org/ferrdb), which comprised a total of 239 genes. Then, Pearson’s correlation analysis was performed for FRLs identified using the “limma” package. LncRNAs with R2 > .6 and p < .001 were considered to be FRLs.
2.3 Construction of a Prognostic Ferroptosis-Related lncRNAs Signature
First, the training cohort was used to construct a prognostic FRLs signature. Co-expressed FRLs were tested using the univariate Cox analysis to determine the prognostic value. Subsequently, based on the candidate lncRNAs with p < .01 in the univariate Cox screen analysis, a least absolute shrinkage and selection operator (LASSO) regression model was constructed by “glmnet” package. Morever, the resulting lncRNAs were introduced into a multivariate Cox model to obtain hazard ratios (HR) and regression coefficients for each lncRNA used in the construction of the final prognostic FRLs signature using “survival” package. Risk scores were calculated using the following equation. The risk score was calculated using the following formula:
where is the expression of the ith selected lncRNA, and is its regression coefficient.
Subsequently, patients were divided into high- and low-risk groups based on the median risk score. The “survival” package was used to perform the Kaplan Meier (KM) survival and receiver operating characteristic (ROC) curve analysis of the performance of prognostic factors in terms of overall survival (OS). The testing cohort and the entire cohort were applied to validate the prognosis signature constructed based on the training cohort.
2.4 Assessment of Prognostic Factors
The KM analysis was performed in high-risk and low-risk groups to elucidate differences between subgroups based on gender (male and female), age (<65 and ≥65 years), grade (G1, G2 and G3, G4), T stage (T1, T2 and T3, T4) and N stage (N0 and N1). Univariate and multivariate Cox regression analysis of clinical characteristics and risk scores were performed to assess whether or not the risk score was an independent prognostic factor as well as to identify clinical characteristics that were independent prognostic factors.
2.5 Clinical Value of a Prognostic Ferroptosis-Related lncRNAs Signature
In the classification of clinicopathological characteristics, the age, sex, grade, American Joint Committee on Cancer (AJCC) stage, T-stage, and N-stage were used to elucidate the independent risk factors associated with the prognosis. However, the M-stage was excluded from the study because of the high rate of missing data on the M-stage.
2.6. Construction of a Predictive Nomogram
A nomogram was used to predict the 12-, 18-, and 24-month survival. The stability of the model was assessed using the consistency index (C-index) and calibration curves.
2.7 Ferroptosis-Related LncRNA-mRNA Co-Expression Network Construction
LncRNA-mRNA co-expression pairs with R 2 > .4 and p < 0.05 were considered to be potential regulatory pathways. Subsequently, the co-expression network was visualized by Cytoscape software (version 3.8.2, http://www.cytoscape.org/).
2.8 Functional Analysis
To elucidate potential biological functions of FRLs, Gene Ontology (GO) (Gene Ontology Consortium, 2015) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000) was performed on lncRNA-mRNAs. Then, significantly related biological functions and pathways, which may be potential pathways for ferroptosis-related lncRNA regulation, were listed.
2.9 Statistical Analysis
Conversion of gene names and merging of the patient gene expression data with the clinical data were performed using PERL (version 5.30.2, http://www.perl.org/). All statistical analyses were performed in the R software (version 4.1.0).
3 Results
3.1 Patient Data Sets
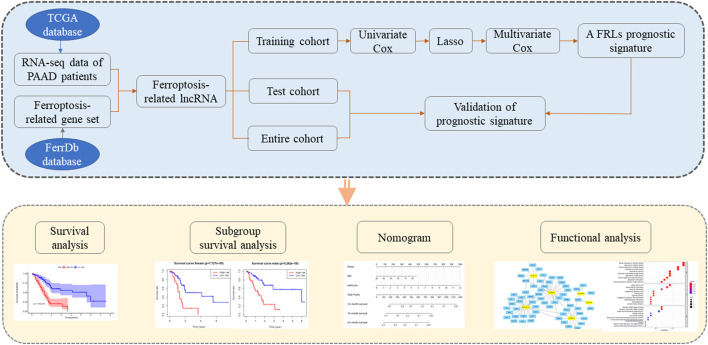
Data from the TCGA-PAAD cohort were downloaded and processed, as reported previously. Inclusion criteria were: 1) a histological diagnosis of pancreatic adenocarcinoma; 2) available data on gene expression and clinicopathological characteristics; and 3) complete survival information and a follow-up duration >30 days. When a patient had two or more samples then the first sample was selected. After applying inclusion and exclusion criteria, 171 patients were enrolled. Table 1 summarizes the basic patient profile. Then, the 171 PAAD patients were randomly divided into a training cohort (n = 86) and a testing cohort (n = 85) at a 1:1 ratio. The flow chart of the study is shown in Figure 1.
TABLE 1.
Baseline table for TCGA-PAAD.
| Clinical/Pathological features | Training cohort (n = 86) | Testing cohort (n = 85) | Entire cohort (n = 171) |
|---|---|---|---|
| Age | |||
| <65 | 36 | 42 | 78 |
| ≥65 | 50 | 43 | 93 |
| Gender | |||
| Male | 49 | 44 | 93 |
| Female | 37 | 41 | 78 |
| Grade | |||
| G1 | 15 | 13 | 28 |
| G2 | 44 | 48 | 92 |
| G3 | 25 | 22 | 47 |
| G4 | 1 | 1 | 2 |
| GX | 1 | 1 | 2 |
| Stage | |||
| Stage I | 1 | 0 | 1 |
| Stage IA | 3 | 2 | 5 |
| Stage IB | 8 | 5 | 13 |
| Stage IIA | 19 | 9 | 28 |
| Stage IIB | 52 | 62 | 114 |
| Stage III | 1 | 2 | 3 |
| Stage IV | 1 | 3 | 4 |
| Unknown | 1 | 2 | 3 |
| T | |||
| T1 | 4 | 3 | 7 |
| T2 | 10 | 11 | 21 |
| T3 | 70 | 68 | 138 |
| T4 | 1 | 2 | 3 |
| TX | 0 | 1 | 1 |
| Unknown | 1 | 0 | 1 |
| M | |||
| M0 | 37 | 40 | 77 |
| M1 | 1 | 3 | 4 |
| MX | 48 | 42 | 90 |
| N | |||
| N0 | 30 | 17 | 47 |
| N1 | 50 | 65 | 115 |
| N1b | 3 | 1 | 4 |
| NX | 2 | 2 | 4 |
| Unknown | 1 | 0 | 1 |
FIGURE 1.
Flow chart of the study.
3.2 Identification of Ferroptosis-Related lncRNAs in Pancreatic Adenocarcinoma
First, 13,830 lncRNAs were identified by analyzing gene expression data from TCGA-PAAD. Altogether 239 ferroptosis-related genes (Supplementary Table S1) were expressed in patients with PAAD. Finally, 435 FRLs were selected. (R 2 > 0.6 and p < .001).
3.3 Construction of a Prognostic Ferroptosis-Related lncRNAs Signature
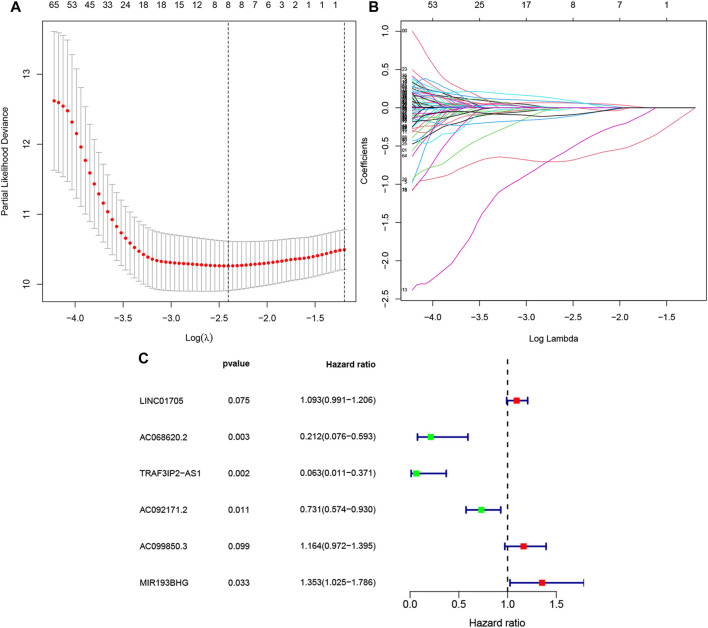
The univariate Cox regression performed on 86 training PAAD samples with OS revealed 193 FRLs to be relevant with the PAAD prognosis (p < .01, Supplementary Table S2). Subsequently, LASSO regression of these genes, and eight FRLs were identified after LASSO regression (Figures 2A,B). Finally, multivariate Cox regression revealed six FRLs (LINC01705, AC068620.2, TRAF3IP2-AS1, AC092171.2, AC099850.3, and MIR193BHG, Figure 2C). A FRLs signature was created (Table 2) with the following risk score formula:
FIGURE 2.
Selection of FRLs using LASSO and multivariate Cox regression. (A)Eight FRLs based LASSO cross validation plot. (B) LASSO coefficient of eight FRLs in PAAD. (C) Multivariate Cox regression showing six FRLs (LINC01705, AC068620.2, TRAF3IP2-AS1, AC092171.2, AC099850.3, and MIR193BHG) associated with OS in TCGA-PAAD.
TABLE 2.
Multivariate Cox results for FRLs based on TCGA-PAAD.
| Id | Coef | HR | HR.95L | HR.95H | p-value | Risk |
|---|---|---|---|---|---|---|
| LINC01705 | .0892 | 1.0933 | .9911 | 1.2060 | .0748 | High |
| AC068620.2 | −1.5521 | .2118 | .0756 | .5932 | .0031 | Low |
| TRAF3IP2-AS1 | −2.7649 | .0630 | .0107 | .3707 | .0022 | Low |
| AC092171.2 | −.3138 | .7307 | .5742 | .9298 | .0107 | Low |
| AC099850.3 | .1522 | 1.1644 | .9719 | 1.3951 | .0988 | High |
| MIR193BHG | .3023 | 1.3529 | 1.0246 | 1.7863 | .0330 | High |
Table 3 summarizes previous reports on the six FRLs. To the best of our knowledge, two lncRNAs (AC068620.2 and AC092171.2) have not been previously reported.
TABLE 3.
Summary statistics of available studies on FRLs.
| Gene symbol | Evidence types | References |
|---|---|---|
| LINC01705 | Cell culture; Mouse models; Clinical samples | Xie et al. (2018), Du et al. (2020), Lei Yang et al. (2021) |
| TRAF3IP2-AS1 | Cell culture; Mouse models; Clinical samples; review; Bioinformatics Analysis | Zou et al. (2014), Bannon et al. (2015), Fabbri and Serretti, (2016), Zan and Li, (2019), He et al. (2021), Shuai Yang et al. (2021) |
| AC099850.3 | Bioinformatics Analysis | Jia et al. (2020), Hao Wu et al. (2020), Jiang et al. (2021), Wu et al. (2021), Xu et al. (2021), Zhang et al. (2021), Zheng et al. (2021a), Junliang Zhou et al. (2021) |
| MIR193BHG | Cell culture; Bioinformatics Analysis; Clinical samples | Chan Meng et al. (2020), Xue Wu et al. (2020), Zhang et al. (2020), Zheng et al. (2021b), Qiwei Zhou et al. (2021) |
3.4 Assessment of a Prognostic Ferroptosis-Related lncRNAs Signature
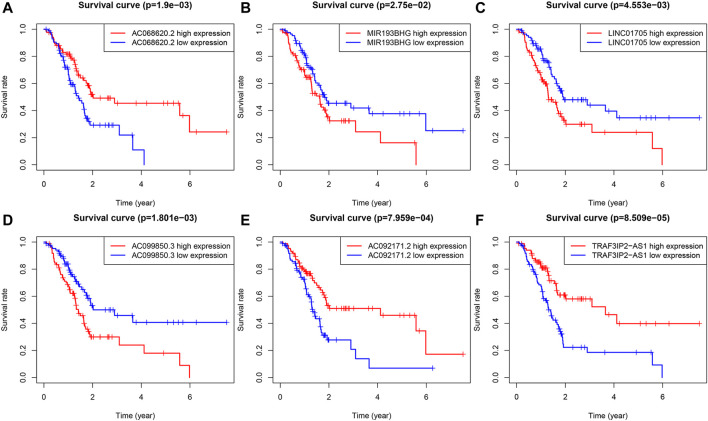
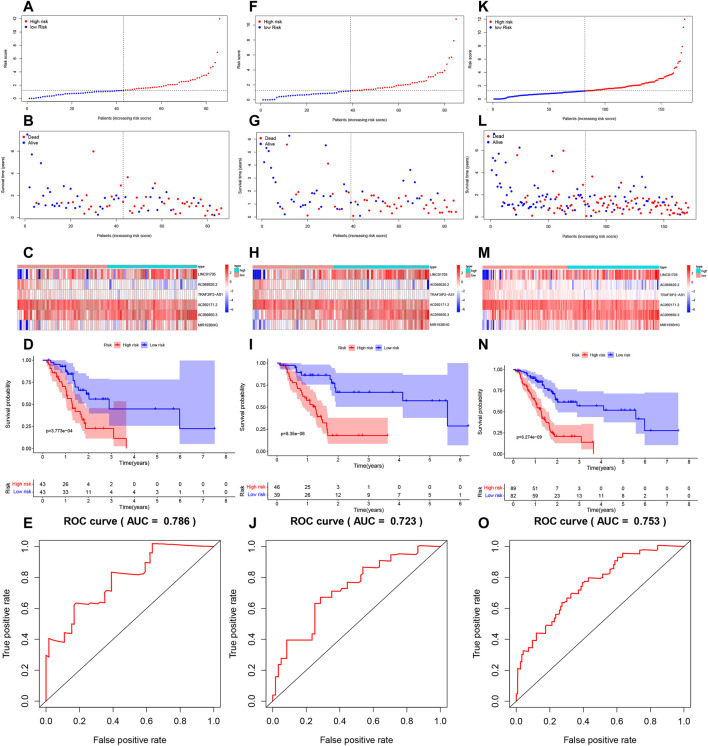
LINC01705, AC099850.3, and MIR193BHG were risk factors for PAAD, while three lncRNAs (AC068620.2, TRAF3IP2-AS1, and AC092171.2) were protective factors for PAAD (Figures 3A–F). The risk score formula applied to gene expression data of 86 training patients revealed a median risk score of 1.231. Accordingly, all cases could be classified as low-risk (n = 43) or high-risk (n = 43) (Figure 4A). The increased risk score was accompanied by gradually decreasing survival time and increased mortality (Figure 4B). Finally, the expression level of OS-related FRLs was presented in the form of heatmaps (Figure 4C). Regrettably, none of the patients in the high-risk group survived longer than 4 years, whereas many of those in the low-risk group survived longer than 7 years. After log-rank test, there was a significant difference in OS between the two groups (p = 1.135e-08, Figure 4D). Time-dependent ROC curves showed prediction of lncRNA biomarkers in the prognosis, with AUC of 0.786, respectively (Figure 4E). We then validated the a prognostic FRLs signature on the testing cohort and the entire cohort (Figures 4F–O). Survival analysis showed, significant differences in survival time between both the testing cohort and the entire cohort of patients, with significantly higher OS in the low-risk group than in the high-risk group. In addition, the AUCs for the testing cohort and the entire cohort were .723, .753, respectively, showing that the six FRLs prognostic signature has good robustness.
FIGURE 3.
KM survival curves for the six prognostic FRLs. Three FRLs (LINC01705, AC099850.3 and MIR193BHG) were independent unfavorable factors and three lncRNAs (AC068620.2, TRAF3IP2-AS1, and AC092171.2) were independent favorable factors for PAAD.
FIGURE 4.
Risk score model development and validation. (A) Trianing cohort risk score distribution of a patient with PAAD based on FRLs. (B) Trianing cohort scatter plots showing the association between the OS and the risk score in PAAD patients according to prognostic features of FRLs. (C) Trianing cohort heatmap showing three unfavorable genes (LINC01705, AC099850.3, and MIR193BHG) with high expression in high-risk patients, contray to the expression of the three favorable genes (AC068620.2, TRAF3IP2-AS1, and AC092171.2). (D) Trianing cohort KM survival curve analysis. (E) Testing cohort area under the ROC curve based on FRLs-based prognostic features at 12 months. (F) Testing cohort risk score distribution of a patient with PAAD based on FRLs. (G) Testing cohort scatter plots showing the association between the OS and the risk score in PAAD patients according to prognostic features of FRLs. (H) Testing cohort heatmap showing three unfavorable genes (LINC01705, AC099850.3, and MIR193BHG) with high expression in high-risk patients, contray to the expression of the three favorable genes (AC068620.2, TRAF3IP2-AS1, and AC092171.2). (I) Testing cohort KM survival curve analysis. (J) Testing cohort area under the ROC curve based on FRLs-based prognostic features at 12 months. (K) Entire cohort risk score distribution of a patient with PAAD based on FRLs. (L) Entire cohort scatter plots showing the association between the OS and the risk score in PAAD patients according to prognostic features of FRLs. (M) Entire cohort heatmap showing three unfavorable genes (LINC01705, AC099850.3, and MIR193BHG) with high expression in high-risk patients, contray to the expression of the three favorable genes (AC068620.2, TRAF3IP2-AS1, and AC092171.2). (N) Entire cohort KM survival curve analysis. (O) Entire cohort area under the ROC curve based on FRLs-based prognostic features at 12 months.
3.5Clinical Value of a Prognostic Ferroptosis-Related lncRNAs Signature
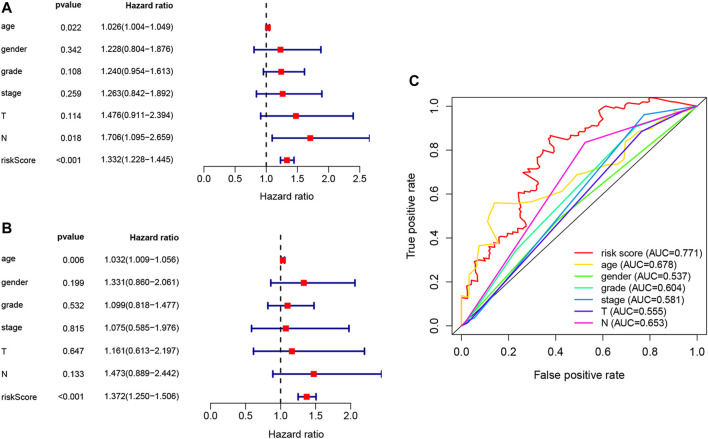
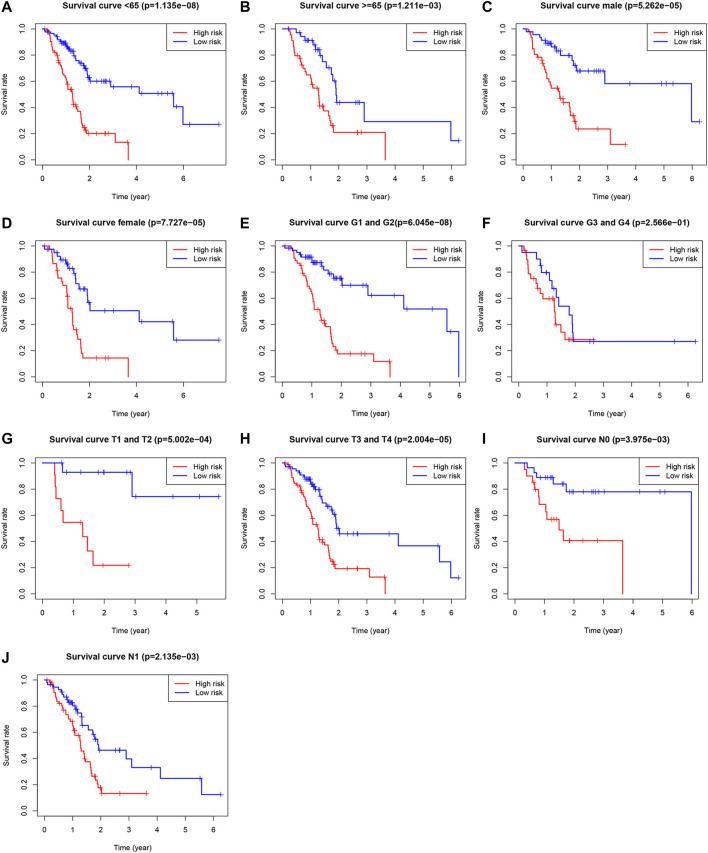
Univariate Cox, regression revealed the risk score to be a risk factor significantly associated with the prognosis of patients with PAAD (95% confidence interval (CI): 1.228–1.445, p < .001), with a higher risk score suggesting a worse prognosis. In addition, the N-stage (95% CI: 1.095–2.659, p = .018) and patient age (95% CI: 1.004–1.049, p = .022) were also closely associated with the prognosis. (Figure 5A). After controlling for clinical characteristics, risk score remained an independent indicator (hazard ratio = 1.372, 95% CI: 1.250–1.506, p < .001, Figure 5B, Table 4). However, the N-stage was not significantly associated with the prognosis in multivariate Cox regression. Moreover, higher accuracy of the risk score in predicting patients compared to other clinicopathological features was also confirmed in the ROC analysis. The prognostic value of FRLs (.771) was higher than that of age (.678), sex (.537), grade (.604), the AJCC stage (.581), the T-stage (.555), and the N-stage (.653; Figure 5C). Similarly, across all age, sex, T-stage, grade, and AJCC stage subgroups, patients with PAAD showed a lower OS rate in the high-risk group than in the low-risk group (Figure 6). All data suggested that the prognostic FRLs signature is significantly associated with the prognosis of patients with PAAD.
FIGURE 5.
Estimated prognostic accuracy of FRLs in patients with PAAD. (A) Univariate Cox regression showing that the age, N-stage, and risk score were associated with OS (p < .05). (B) Multivariate Cox regression showing that the age and the risk score (p < .01) were independent prognostic indicators of OS in patients with PAAD. (C) ROC curve showing that the risk score has the highest prognostic accuracy.
TABLE 4.
Multivariate Cox regression analysis of clinical characteristics and the risk score for PAAD.
| Id | HR | HR.95L | HR.95H | p-value |
|---|---|---|---|---|
| Age | 1.032 | 1.009 | 1.056 | .006 |
| Gender | 1.328 | .856 | 2.060 | .206 |
| Grade | 1.099 | .818 | 1.476 | .532 |
| Stage | 1.076 | .586 | 1.978 | .813 |
| T | 1.155 | .607 | 2.199 | .661 |
| M | .987 | .792 | 1.229 | .903 |
| N | 1.477 | .890 | 2.450 | .132 |
| RiskScore | 1.372 | 1.250 | 1.506 | .000 |
FIGURE 6.
Survival rates of PAAD patients with high- and low-risk patients with PAAD in the subgroups based on clinicopathological characteristics. (A) Subgroup of age <65 years. (B) Subgroup of age ≥65 years. (C) Male subgroup. (D) Female subgroup. (E) G1 and G2 subgroups. (F) G3 and G4 subgroups. (G) T1 and T2 subgroups. (H) T3 and T4 subgroups. (I) N0 subgroup. (J) N1 subgroup.
3.6 Construction and Assessment of Nomogram
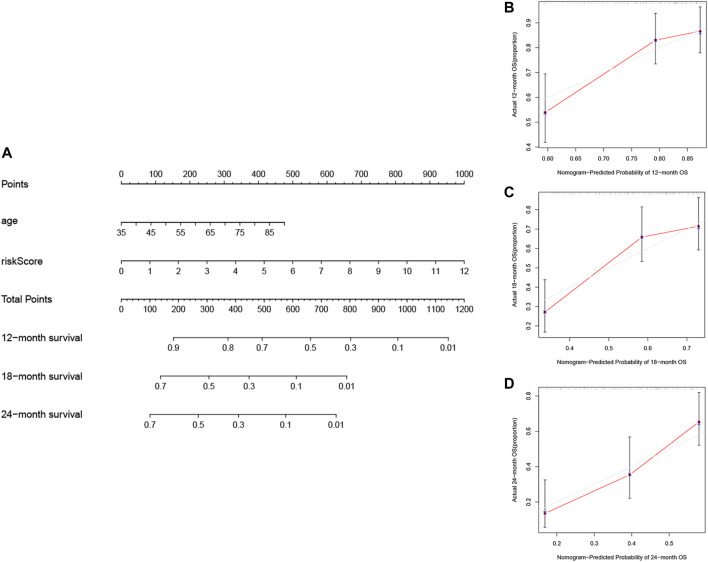
The nomogram, a common prognostic visualization tool in oncology, allows for the quantification of patient survival based on the inclusion of an index score. Based on multivariate Cox regression, the indicators, age and risk score, were included in the construction of the nomogram. Total scores were obtained by adding the individual indicator scores for age and risk score and predicting the probability of survival at 12, 18, and 24 months (Figure 7A). The nomogram had good stability at a consistency index of .697. Furthermore, calibration curves showed that predicted survival times at 12, 18, and 24 months were consistent compared to the reference line (Figures 7B–D). Furthermore, Table 5 lists other studies in which lncRNA predicted the prognosis of patients with PAAD.
FIGURE 7.
Construction and validation of the nomogram. (A) Prognostic nomogram based on the risk score using the prognostic FRLs signature and the age to predict 12-, 18-, and 24-month survival in patients with PAAD. Calibration curves corrected for deviations in agreement between the predicted and observed survival rates at (B) 12, (C) 18 and (D) 24-months.
TABLE 5.
Comparative studies on available lncRNA signatures in PAAD.
| Feature | Database | Gene list | OS | AUC | Refer |
|---|---|---|---|---|---|
| All lncRNA | TCGA | AC009014.3, RP11-48O20.4, UCA1 | OS | .689 | Li et al. (2020) |
| ICGC | |||||
| Immune-related lncRNA | TCGA | MANCR, LINC01655, UCA1, LINC01940, FIRRE, LINC01776, LINC01436, LINC00242 | OS | .74 | Qi et al. (2021) |
| M6A-related lncRNA | TCGA | AC099850.3, UCA1, AP005233.2, AL513165.1, PTOV1-AS2 | OS | .682 | Yuan et al. (2021) |
| ICGC | |||||
| GTEx | |||||
| All lncRNA | GEO | lA2M-AS1, DLEU2, LINC01133, LINC00675, MIR155HG, SLC25A25-AS1, LINC01857, LOC642852(LINC00205), ITGB2-AS1, TSPOAP1-DACAS1, PSMB2-AS1 | — | — | Giulietti et al. (2018) |
| All lncRNA | TCGA | AL137789.1, MIR600HG, AC079015.1 | OS | .742 | Wu et al. (2018) |
| All lncRNA | ICGC | AC009014.3, RP11-48O20.4, UCA1 | — | — | Li et al. (2020) |
| CeRNA | GEO | SCAMP1, HCP5, MAL2, LINC00511 | — | — | Wang et al. (2019) |
3.7 Construction of the Prognostic Ferroptosis-Related lncRNAs Associated LncRNA-mRNA Co-Expression Network and the Functional Enrichment Analysis
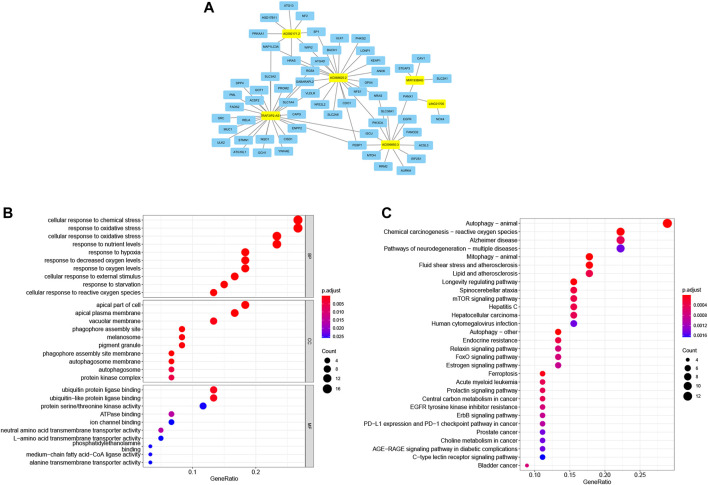
The co-expression analysis of FRLs revealed a total of 61 mRNAs involved in formation of the co-expression network of 76 lncRNA-mRNA targets with R 2 > .4 and p < .05 (Figure 8A; Supplementary Table S3). The GO analysis showed that it was involved in 476 biological processes, such as cellular responses to chemical stress and responses to oxidative stress, constituted 25 cellular components, including the phagosome assembly site and the apical portion of cells, and mediated 35 molecular functions, such as ubiquitin protein ligase binding and ubiquitin-like protein ligase binding (Figure 8B; Table 6). KEGG showed ferroptosis to be a significantly enriched pathway (Figure 8C; Table 7).
FIGURE 8.
Ferroptosis-related lncRNA-mRNA co-expression network and functional enrichment. (A) LncRNA-mRNA network showing 76 lncRNA-mRNA co-expression pairs formed between six FRLs and 61 mRNAs. The yellow rectangles denote FRLs, and the blue rectangles denote mRNAs. (B) GO analysis showing enriched biological function of these mRNAs co-expressed with six FRLs. (C) KEGG pathway analysis showing enriched signaling pathways.
TABLE 6.
Significantly enriched GO terms of co-expression mRNA with PAAD with an adjusted p < .05
| Category | Term | Count | p-value | Adj p-value |
|---|---|---|---|---|
| BP | GO:0062197∼ cellular response to chemical stress | 16 | <.001 | <.001 |
| BP | GO:0006979∼ response to oxidative stress | 16 | <.001 | <.001 |
| BP | GO:0034599∼ cellular response to oxidative stress | 14 | <.001 | <.001 |
| BP | GO:0031667∼ response to nutrient levels | 14 | <.001 | <.001 |
| BP | GO:0001666∼ response to hypoxia | 11 | <.001 | <.001 |
| BP | GO:0042594∼ response to starvation | 9 | <.001 | <.001 |
| BP | GO:0036293∼ response to decreased oxygen levels | 11 | <.001 | <.001 |
| BP | GO:0070482∼ response to oxygen levels | 11 | <.001 | <.001 |
| BP | GO:0034614∼ cellular response to reactive oxygen species | 8 | <.001 | <.001 |
| BP | GO:0071496∼ cellular response to external stimulus | 10 | <.001 | <.001 |
| CC | GO:0000407∼phagophore assembly site | 5 | <.001 | <.001 |
| CC | GO:0045177∼apical part of cell | 11 | <.001 | <.001 |
| CC | GO:0016324∼apical plasma membrane | 10 | <.001 | <.001 |
| CC | GO:0034045∼phagophore assembly site membrane | 4 | <.001 | <.001 |
| CC | GO:0000421∼autophagosome membrane | 4 | <.001 | <.001 |
| CC | GO:0042470∼melanosome | 5 | <.001 | <.001 |
| CC | GO:0048770∼pigment granule | 5 | <.001 | <.001 |
| CC | GO:0005774∼vacuolar membrane | 8 | <.001 | .001 |
| CC | GO:0005776∼autophagosome | 4 | <.001 | .005 |
| CC | GO:1902911∼protein kinase complex | 4 | <.001 | .006 |
| MF | GO:0031625∼ubiquitin protein ligase binding | 8 | <.001 | .001 |
| MF | GO:0044389∼ubiquitin-like protein ligase binding | 8 | <.001 | .001 |
| MF | GO:0015175∼neutral amino acid transmembrane transporter activity | 3 | <.001 | .016 |
| MF | GO:0051117∼ATPase binding | 4 | <.001 | .016 |
| MF | GO:0004674∼protein serine/threonine kinase activity | 7 | .001 | .026 |
| MF | GO:0008429∼phosphatidylethanolamine binding | 2 | .001 | .026 |
| MF | GO:0031956∼medium-chain fatty acid-CoA ligase activity | 2 | .001 | .026 |
| MF | GO:0022858∼alanine transmembrane transporter activity | 2 | .001 | .026 |
| MF | GO:0044325∼ion channel binding | 4 | .001 | .026 |
| MF | GO:0015179∼L-amino acid transmembrane transporter activity | 3 | .001 | .026 |
TABLE 7.
Signifcantly enriched KEGG terms of co-expression mRNA with PAAD with an adjusted p < .05
| KEGG term | Count | p-value | Adj p-value |
|---|---|---|---|
| hsa04140∼Autophagy—animal | 13 | <.001 | <.001 |
| hsa04137∼Mitophagy—animal | 8 | <.001 | <.001 |
| hsa04136∼Autophagy—other | 6 | <.001 | <.001 |
| hsa05208∼Chemical carcinogenesis—reactive oxygen species | 10 | <.001 | <.001 |
| hsa04211∼Longevity regulating pathway | 7 | <.001 | <.001 |
| hsa05418∼Fluid shear stress and atherosclerosis | 8 | <.001 | <.001 |
| hsa04216∼Ferroptosis | 5 | <.001 | <.001 |
| hsa05017∼Spinocerebellar ataxia | 7 | <.001 | <.001 |
| hsa01522∼Endocrine resistance | 6 | <.001 | <.001 |
| hsa05417∼Lipid and atherosclerosis | 8 | <.001 | <.001 |
| hsa04150∼mTOR signaling pathway | 7 | <.001 | <.001 |
| hsa05160∼Hepatitis C | 7 | <.001 | <.001 |
| hsa05221∼Acute myeloid leukemia | 5 | <.001 | <.001 |
| hsa05163∼Human cytomegalovirus infection | 7 | <.001 | .001 |
| hsa04933∼AGE-RAGE signaling pathway in diabetic complications | 5 | <.001 | .001 |
| hsa05022∼Pathways of neurodegeneration—multiple diseases | 10 | <.001 | .001 |
| hsa04625∼C-type lectin receptor signaling pathway | 5 | <.001 | .002 |
4 Discussion
PAAD is increasing in incidence every year and is highly malignant. It has a low early clinical detection rate and a 5-year survival rate <5%. Previous studies have elucidated prognostic factors associated with PAAD, such as the tumor grade, stage, size, and number. PAAD involves complex biological processes, some of which are closely related to its prognosis, such as autophagy (Yue et al., 2020), immunity, and ferroptosis. LncRNAs have been identified as key regulators of biological processes that could be used as potential prognostic biomarkers and might provide insights into clinically targeted therapies. In our study, the combination analysis of Cox and LASSO regression was applied to establish a ferroptosis-related lncRNA signature. The signature showed good predictive performance, with patients in the low-risk group having higher OS than those in the high-risk group.
Six prognosis-associated lncRNAs finally obtained in the risk model were LINC01705, AC068620.2, TRAF3IP2-AS1, AC092171.2, AC099850.3, and MIR193BHG. These FRLs could be prognostic marker molecules, potential markers for PAAD, and potential therapeutic targets.
Among the six ferroptosis-related gene risk models, AC084018.1 and AC092171.2 have not been reported to date. Regulations of AC084018.1 and AC092171.2 should be investigated in future studies. The biological mechanism of the other FRLs has been previously reported. LINC01705 positively regulates the translocation promoter region nuclear basket protein by competitively binding to miR-186-5p, thereby promoting the aggressiveness of breast cancer cells (Du et al., 2020). Moreover, miR-223-5p-LINC01705 is involved in pulmonary metastasis of osteosarcoma as a microRNA–lncRNA target pair (Lei Yang et al., 2021). In addition, LINC01705 has high expression during the pathogenesis of proliferative vitreoretinopathy (Xie et al., 2018). Shuai Yang et al. (2021) investigated the inhibitory effect of the tumor growth suppressor TRAF3IP2-AS1 on the progression of NONO-TFE3-translocated renal carcinoma and found that the overexpression of TRaf3IP2-AS1 could stimulate N6-methyladenosine of PARP1 mRNA and downregulate PTEN, further inhibiting the progression of renal carcinoma. He et al. (2021) showed that TRAF3IP2-AS1 may be an attractive therapeutic target for IL-17-related autoimmune diseases, such as psoriasis and multiple sclerosis. Bannon et al. (2015) analyzed clinical samples and showed that TRAF3IP2-AS1 exhibits high expression in cocaine abusers, positively correlating with the opposite chain protein encoding transcript TRAF3IP2. In the study by Zou et al. (2014), TRAF3IP2-AS1 expression was altered in gastric cancer cells after 125I irradiation, providing a target for future drug development. AC099850.3 has been repeatedly extracted as a prognosis-related gene in the bioinformatics analysis of cancer, including squamous cell carcinoma of the tongue, hepatocellular carcinoma, and non-small cell lung cancer (Hao Wu et al., 2020; Jia et al., 2020; Zheng et al., 2021a; Junliang Zhou et al., 2021; Jiang et al., 2021; Wu et al., 2021; Xu et al., 2021; Zhang et al., 2021). The MIR193BHG motif can fine-tune cellular sterol/steroid biosynthesis by producing lincNORS to repress the expression of multiple pathway components (Xue Wu et al., 2020). MIR193BHG was found to be significantly associated with the prognosis in both autophagy-related lncRNA in ovarian cancer and ferroptosis-related lncRNA in lung adenocarcinoma (Chan Meng et al., 2020; Zheng et al., 2021b). In the analysis of competing endogenous RNAs in lung and renal cell carcinomas, MIR193BHG-miR-140-3p may be a promising upstream regulatory pathway for GPRIN1 (Qiwei Zhou et al., 2021). Furthermore, MIR193BHG has diagnostic value for placental tissue in patients with early-onset pre-eclampsia (Zhang et al., 2020).
KEGG and GO enrichment results indicated that these screened lncRNAs were related to regulation of reactive oxygen species (ROS), which can cause ptosis. The effect of ROS on ferroptosis in pancreatic cancer is not fully understood. However, many regulators of ROS related to ferroptosis, such as irisin, deerskin, QD394, and QD394-me, have been found. ROS metabolism, iron metabolism, and the cysteine/glutamate reverse transport system (SYSTEM Xc-) play important roles in the regulatory process (Hu et al., 2020; Song et al., 2020; Yang and Leung, 2020). In terms of environment, many studies have shown that arsenic may stimulate ferroptosis in cancer cells through oxidative stress. Our enrichment analysis revealed that the co-expressed mRNA was significantly enriched in the autophagy pathway (Pan Meng et al., 2020; Wei et al., 2020). To date, the genetic link between autophagy and ferroptosis remains unclear. Autophagy has been shown to promote ferroptosis by degrading ferritin in cancer cells (Hou et al., 2016). Therefore, identifying lncRNAs associated with iron sagging in pancreatic cancer may provide guidance for the search for regulatory factors.
Ours was the first study to combine patient prognostic analysis with FRLs to preliminarily explore the molecular mechanism of ferroptosis affecting the prognosis of patients with PAAD. Six FRLs signatures were established to further supplement the traditional clinical prognostic factors, guiding the search for therapeutic targets and the selection of prognostic decisions for pancreatic cancer. In addition, we provided a columnar line graph correlating FRLs with clinical factors to predict OS in PAAD in a validated and quantitative way.
5 Conclusion
We created a ferroptosis-related lncRNAs signature that can be used as a new biomarker to predict PAAD progression. This signature could help provide insights into the correlation between ferroptosis and tumorigenesis for clinical ferroptosis-related targeted therapies.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://portal.gdc.cancer.gov/.
Author Contributions
JL designed the study and conducted data analysis. JH, ST, and JZ designed the manuscript. JL, WC, and YZ revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81774230) and the Natural Science Foundation of Zhejiang Province (No. LY21H270012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.838021/full#supplementary-material
Abbreviations
AUC, area under the receiver operating characteristic curve; FRLs, ferroptosis-related lncRNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; LASSO, least absolute shrinkage and selection operator; OS, overall survival; PAAD, pancreatic adenocarcinoma; ROC, receiver operating characteristic; TCGA, The Cancer Genome Atlas.
References
- Badgley M. A., Kremer D. M., Maurer H. C., DelGiorno K. E., Lee H.-J., Purohit V., et al. (2020). Cysteine Depletion Induces Pancreatic Tumor Ferroptosis in Mice. Science 368 (6486), 85–89. 10.1126/science.aaw9872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon M. J., Savonen C. L., Jia H., Dachet F., Halter S. D., Schmidt C. J., et al. (2015). Identification of Long Noncoding RNAs Dysregulated in the Midbrain of Human Cocaine Abusers. J. Neurochem. 135 (1), 50–59. 10.1111/jnc.13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A., Soleimani M., Mandal S. S. (2017). Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 77 (15), 3965–3981. 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Zhang J.-l., Wang Y., Zhang Y.-y., Zhang J.-h., Zhang L.-f., et al. (2020). The Long Non-coding RNA LINC01705 Regulates the Development of Breast Cancer by Sponging miR-186-5p to Mediate TPR Expression as a Competitive Endogenous RNA. Front. Genet. 11, 779. 10.3389/fgene.2020.00779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri C., Serretti A. (2016). Genetics of Long-Term Treatment Outcome in Bipolar Disorder. Prog. Neuro-Psychopharmacology Biol. Psychiatry 65, 17–24. 10.1016/j.pnpbp.2015.08.008 [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2015). Gene Ontology Consortium: Going Forward. Nucleic Acids Res. 43 (Database issue), D1049–D1056. 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulietti M., Righetti A., Principato G., Piva F. (2018). LncRNA Co-expression Network Analysis Reveals Novel Biomarkers for Pancreatic Cancer. Carcinogenesis 39 (8), 1016–1025. 10.1093/carcin/bgy069 [DOI] [PubMed] [Google Scholar]
- Gupta R., Amanam I., Chung V. (2017). Current and Future Therapies for Advanced Pancreatic Cancer. J. Surg. Oncol. 116 (1), 25–34. 10.1002/jso.24623 [DOI] [PubMed] [Google Scholar]
- Hassannia B., Vandenabeele P., Vanden Berghe T. (2019). Targeting Ferroptosis to Iron Out Cancer. Cancer Cell 35 (6), 830–849. 10.1016/j.ccell.2019.04.002 [DOI] [PubMed] [Google Scholar]
- He R., Wu S., Gao R., Chen J., Peng Q., Hu H., et al. (2021). Identification of a Long Noncoding RNA TRAF3IP2-AS1 as Key Regulator of IL-17 Signaling through the SRSF10-IRF1-Act1 Axis in Autoimmune Diseases. J.I. 206 (10), 2353–2365. 10.4049/jimmunol.2001223 [DOI] [PubMed] [Google Scholar]
- Hidalgo M., Cascinu S., Kleeff J., Labianca R., Löhr J.-M., Neoptolemos J., et al. (2015). Addressing the Challenges of Pancreatic Cancer: Future Directions for Improving Outcomes. Pancreatology 15 (1), 8–18. 10.1016/j.pan.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Hidalgo M. (2010). Pancreatic Cancer. N. Engl. J. Med. 362 (17), 1605–1617. 10.1056/nejmra0901557 [DOI] [PubMed] [Google Scholar]
- Hou W., Xie Y., Song X., Sun X., Lotze M. T., Zeh H. J., 3rd, et al. (2016). Autophagy Promotes Ferroptosis by Degradation of Ferritin. Autophagy 12 (8), 1425–1428. 10.1080/15548627.2016.1187366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Sechi M., Singh P. K., Dai L., McCann S., Sun D., et al. (2020). A Novel Redox Modulator Induces a GPX4-Mediated Cell Death that Is Dependent on Iron and Reactive Oxygen Species. J. Med. Chem. 63 (17), 9838–9855. 10.1021/acs.jmedchem.0c01016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Chen Y., Liu J. (2020). Prognosis-Predictive Signature and Nomogram Based on Autophagy-Related Long Non-coding RNAs for Hepatocellular Carcinoma. Front. Genet. 11, 608668. 10.3389/fgene.2020.608668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Xue D., Shi F., Qiu J. (2021). Prognostic Significance of an Autophagy-Related Long Non-coding RNA Signature in Patients with Oral and Oropharyngeal Squamous Cell Carcinoma. Oncol. Lett. 21 (1), 29. 10.3892/ol.2020.12290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28 (1), 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Li H., Chen Q., Wu W., Chen X., Ran L., et al. (2020). A Novel and Robust Long Noncoding RNA Panel to Predict the Prognosis of Pancreatic Cancer. DNA Cel Biol. 39 (7), 1282–1289. 10.1089/dna.2019.5241 [DOI] [PubMed] [Google Scholar]
- Mao C., Wang X., Liu Y., Wang M., Yan B., Jiang Y., et al. (2018). A G3BP1-Interacting lncRNA Promotes Ferroptosis and Apoptosis in Cancer via Nuclear Sequestration of P53. Cancer Res. 78 (13), 3484–3496. 10.1158/0008-5472.CAN-17-3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C., Zhou J. Q., Liao Y. S. (2020). Autophagy-related Long Non-coding RNA Signature for Ovarian Cancer. J. Int. Med. Res. 48 (11), 300060520970761. 10.1177/0300060520970761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng P., Zhang S., Jiang X., Cheng S., Zhang J., Cao X., et al. (2020). Arsenite Induces Testicular Oxidative Stress In Vivo and In Vitro Leading to Ferroptosis. Ecotoxicology Environ. Saf. 194, 110360. 10.1016/j.ecoenv.2020.110360 [DOI] [PubMed] [Google Scholar]
- Mizrahi J. D., Surana R., Valle J. W., Shroff R. T. (2020). Pancreatic Cancer. The Lancet 395 (10242), 2008–2020. 10.1016/s0140-6736(20)30974-0 [DOI] [PubMed] [Google Scholar]
- Neoptolemos J. P., Kleeff J., Michl P., Costello E., Greenhalf W., Palmer D. H. (2018). Therapeutic Developments in Pancreatic Cancer: Current and Future Perspectives. Nat. Rev. Gastroenterol. Hepatol. 15 (6), 333–348. 10.1038/s41575-018-0005-x [DOI] [PubMed] [Google Scholar]
- Qi B., Liu H., Zhou Q., Ji L., Shi X., Wei Y., et al. (2021). An Immune-Related lncRNA Signature for the Prognosis of Pancreatic Adenocarcinoma. Aging 13 (14), 18806–18826. 10.18632/aging.203323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhi A. D., Koay E. J., Chari S. T., Maitra A. (2019). Early Detection of Pancreatic Cancer: Opportunities and Challenges. Gastroenterology 156 (7), 2024–2040. 10.1053/j.gastro.2019.01.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xiang X., Li J., Deng J., Fang Z., Zhang L., et al. (2020). Ruscogenin Induces Ferroptosis in Pancreatic Cancer Cells. Oncol. Rep. 43 (2), 516–524. 10.3892/or.2019.7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell B. R., Jiang X., Gu W. (2020). Emerging Mechanisms and Disease Relevance of Ferroptosis. Trends Cel Biol. 30 (6), 478–490. 10.1016/j.tcb.2020.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Huang Z., Sheng W., Xu M.-d. (2018). Emerging Roles of Long Non-coding RNAs in Tumor Metabolism. J. Hematol. Oncol. 11 (1), 106. 10.1186/s13045-018-0648-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A. Cancer J. Clin. 71 (3), 209–249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Tempero M. A. (2019). NCCN Guidelines Updates: Pancreatic Cancer. J. Natl. Compr. Canc Netw. 17 (55), 603–605. 10.6004/jnccn.2019.5007 [DOI] [PubMed] [Google Scholar]
- Wang W., Lou W., Ding B., Yang B., Lu H., Kong Q., et al. (2019). A Novel mRNA-miRNA-lncRNA Competing Endogenous RNA Triple Sub-network Associated with Prognosis of Pancreatic Cancer. Aging 11 (9), 2610–2627. 10.18632/aging.101933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Liang Q., Li X., Li H., Liu Y., Huang X., et al. (2019). Bioinformatics Profiling Utilized a Nine Immune‐related Long Noncoding RNA Signature as a Prognostic Target for Pancreatic Cancer. J. Cel Biochem 120 (9), 14916–14927. 10.1002/jcb.28754 [DOI] [PubMed] [Google Scholar]
- Wei S., Qiu T., Yao X., Wang N., Jiang L., Jia X., et al. (2020). Arsenic Induces Pancreatic Dysfunction and Ferroptosis via Mitochondrial ROS-Autophagy-Lysosomal Pathway. J. Hazard. Mater. 384, 121390. 10.1016/j.jhazmat.2019.121390 [DOI] [PubMed] [Google Scholar]
- Wu B., Wang K., Fei J., Bao Y., Wang X., Song Z., et al. (2018). Novel three-lncRNA S-ignature P-redicts S-urvival in P-atients with P-ancreatic C-ancer. Oncol. Rep. 40 (6), 3427–3437. 10.3892/or.2018.6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Wei H., Liu G., Zhang Y. (2021). Bioinformatics Profiling of Five Immune-Related lncRNAs for a Prognostic Model of Hepatocellular Carcinoma. Front. Oncol. 11, 667904. 10.3389/fonc.2021.667904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Liu T., Qi J., Qin C., Zhu Q. (2020). Four Autophagy-Related lncRNAs Predict the Prognosis of HCC through Coexpression and ceRNA Mechanism. Biomed. Res. Int. 2020, 3801748. 10.1155/2020/3801748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Niculite C. M., Preda M. B., Rossi A., Tebaldi T., Butoi E., et al. (2020). Regulation of Cellular Sterol Homeostasis by the Oxygen Responsive Noncoding RNA lincNORS. Nat. Commun. 11 (1), 4755. 10.1038/s41467-020-18411-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Yao Z., Zhang Y., Li D., Hu F., Liao Y., et al. (2018). Deep RNA Sequencing Reveals the Dynamic Regulation of miRNA, lncRNAs, and mRNAs in Osteosarcoma Tumorigenesis and Pulmonary Metastasis. Cell Death Dis 9 (7), 772. 10.1038/s41419-018-0813-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B.-H., Jiang J.-H., Luo T., Jiang Z.-J., Liu X.-Y., Li L.-Q. (2021). Signature of Prognostic Epithelial-Mesenchymal Transition Related Long Noncoding RNAs (ERLs) in Hepatocellular Carcinoma. Medicine (Baltimore) 100 (30), e26762. 10.1097/md.0000000000026762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B. C., Leung P. S. (2020). Irisin Is a Positive Regulator for Ferroptosis in Pancreatic Cancer. Mol. Ther. - Oncolytics 18, 457–466. 10.1016/j.omto.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Chen Y., Liu N., Shi Q., Han X., Gan W., et al. (2021). Low Expression of TRAF3IP2-AS1 Promotes Progression of NONO-TFE3 Translocation Renal Cell Carcinoma by Stimulating N6-Methyladenosine of PARP1 mRNA and Downregulating PTEN. J. Hematol. Oncol. 14 (1), 46. 10.1186/s13045-021-01059-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Li H., Yao H., Zhang Y., Bao H., Wu L., et al. (2021). Long Noncoding RNA ERLR Mediates Epithelial-Mesenchymal Transition of Retinal Pigment Epithelial Cells and Promotes Experimental Proliferative Vitreoretinopathy. Cell Death Differ 28 (8), 2351–2366. 10.1038/s41418-021-00756-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Ren J., Li L., Li S., Xiang K., Shang D. (2021). Development and Validation of a Novel N6-Methyladenosine (m6A)-Related Multi- Long Non-coding RNA (lncRNA) Prognostic Signature in Pancreatic Adenocarcinoma. Bioengineered 12 (1), 2432–2448. 10.1080/21655979.2021.1933868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P., Zhu C., Gao Y., Li Y., Wang Q., Zhang K., et al. (2020). Development of an Autophagy-Related Signature in Pancreatic Adenocarcinoma. Biomed. Pharmacother. 126, 110080. 10.1016/j.biopha.2020.110080 [DOI] [PubMed] [Google Scholar]
- Zan X. Y., Li L. (2019). Construction of lncRNA-Mediated ceRNA Network to Reveal Clinically Relevant lncRNA Biomarkers in Glioblastomas. Oncol. Lett. 17 (5), 4369–4374. 10.3892/ol.2019.10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Cheng M., Fan Z., Jin Q., Cao P., Zhou G. (2021). Identification of Cancer Cell Stemness-Associated Long Noncoding RNAs for Predicting Prognosis of Patients with Hepatocellular Carcinoma. DNA Cel Biol. 40 (8), 1087–1100. 10.1089/dna.2021.0282 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wang P., Zhang L., Huang C., Gao J., Li Y., et al. (2020). Identification of Key Genes and Long Noncoding RNA-Associated Competing Endogenous RNA (ceRNA) Networks in Early-Onset Preeclampsia. Biomed. Res. Int. 2020, 1673486. 10.1155/2020/1673486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Conrad M. (2020). The Metabolic Underpinnings of Ferroptosis. Cel Metab. 32 (6), 920–937. 10.1016/j.cmet.2020.10.011 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Lai C., Li W., Zhang C., Ma K., Yao Y. (2021). Identification of a Novel Glycolysis-Related LncRNA Signature for Predicting Overall Survival in Patients with Bladder Cancer. Front. Genet. 12, 720421. 10.3389/fgene.2021.720421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Zhang Q., Wu W., Xue Y., Liu S., Chen Q., et al. (2021). Identification and Validation of a Ferroptosis-Related Long Non-coding RNA Signature for Predicting the Outcome of Lung Adenocarcinoma. Front. Genet. 12, 690509. 10.3389/fgene.2021.690509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Liu J., Kang R., Klionsky D. J., Kroemer G., Tang D. (2020). Ferroptosis Is a Type of Autophagy-dependent Cell Death. Semin. Cancer Biol. 66, 89–100. 10.1016/j.semcancer.2019.03.002 [DOI] [PubMed] [Google Scholar]
- Zhou J., Zhang M., Dong H., Wang M., Cheng Y., Wang S., et al. (2021). Comprehensive Analysis of Acetylation-Related lncRNAs and Identified AC099850.3 as Prognostic Biomarker in Non-small Cell Lung Cancer. J. Oncol. 2021, 4405697. 10.1155/2021/4405697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Li D., Zheng H., He Z., Qian F., Wu X., et al. (2021). A Novel lncRNA‐miRNA‐mRNA Competing Endogenous RNA Regulatory Network in Lung Adenocarcinoma and Kidney Renal Papillary Cell Carcinoma. Thorac. Cancer 12 (19), 2526–2536. 10.1111/1759-7714.14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R.-S., Zhang E.-X., Sun Q.-F., Ye Z.-J., Liu J.-W., Zhou D.-H., et al. (2019). Integrated Analysis of lncRNA-miRNA-mRNA ceRNA Network in Squamous Cell Carcinoma of Tongue. BMC Cancer 19 (1), 779. 10.1186/s12885-019-5983-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Luo K., Qiao O., Xu J. (2014). Global Gene Expression Responses to Iodine-125 Radiation in Three Human Gastric Cancer Cell Lines. Zhonghua Wai Ke Za Zhi 52 (8), 612–616. 10.3760/cma.j.issn.0529-5815.2014.08.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://portal.gdc.cancer.gov/.