Abstract
UDP-Glycosyltransferases (UGTs) catalyze the transfer of nucleotide-activated sugars to specific acceptors, among which the GT1 family enzymes are well-known for their function in biosynthesis of natural product glycosides. Elucidating GT function represents necessary step in metabolic engineering of aglycone glycosylation to produce drug leads, cosmetics, nutrients and sweeteners. In this review, we systematically summarize the phylogenetic distribution and catalytic diversity of plant GTs. We also discuss recent progress in the identification of novel GT candidates for synthesis of plant natural products (PNPs) using multi-omics technology and deep learning predicted models. We also highlight recent advances in rational design and directed evolution engineering strategies for new or improved GT functions. Finally, we cover recent breakthroughs in the application of GTs for microbial biosynthesis of some representative glycosylated PNPs, including flavonoid glycosides (fisetin 3-O-glycosides, astragalin, scutellarein 7-O-glucoside), terpenoid glycosides (rebaudioside A, ginsenosides) and polyketide glycosides (salidroside, polydatin).
Keywords: Glycosyltransferases, Glycosylated plant natural products, Enzyme mining, Protein engineering, Biosynthesis
1. Introduction
More than 200,000 natural products are known to be synthesized by plants (hereafter, plant natural products; PNPs), collectively constituting a large, structural diverse library of compounds with widely varying biological activities [1]. A considerable proportion of PNPs are glycosylated with diverse sugar moieties attached to the aglycones, thus greatly increasing the variety and complexity of structures. These glycosylated PNPs have been used as medicines, sweeteners, nutrients, cosmetics, and health products, consequently drawing considerable research attention to their biosynthesis and modification [2].
The biosynthesis of glycosylated compounds involves multiple complex biological processes that are orchestrated by many enzymatic systems in plants. Glycosyltransferases (GTs) (EC 2.4.x.y) are crucial for the biosynthesis of glycosylated PNPs and commonly perform the final step in their biosynthesis pathway, mediating the regio- and stereo-specific glycosidic bond formation via transfer of nucleotide-diphosphate-activated sugar moieties to a variety of biomolecules [3]. GTs involved in the biosynthesis of glycosylated PNPs are universally classified into the GT1 family in the Carbohydrate Active Enzyme database (CAZy, http://www.cazy.org/). Currently, nearly 30,000 GTs are included in the GT1 family, and an increasing number of GT sequences are reported from different organisms with the development of the high-throughput sequencing and advances in deep-learning-based analysis [[4], [5], [6]]. However, only 1% of GTs have been functionally characterized, which has limited the clarification of glycosylated PNPs biosynthetic pathways and validation of enzymatic mechanisms [7].
Using enzymatic catalysis or metabolic engineering methods, scientists can produce glycosylated PNPs at industrial scale, thus providing a reliable and scalable alternative to conventional production methods based on extraction from natural resources [[8], [9], [10], [11]]. However, wild-type GTs are often accompanied by unfavorable properties for the synthesis of target products, such as low expression levels and low catalytic activity. Thus, these obstacles persist as limiting factors in glycosylated PNP biosynthesis which require protein engineering through rational design and/or directed evolution [[12], [13], [14]].
In recent years, the rapid development of synthetic biology has accelerated the metabolic engineering of complex synthetic pathways and increased efficiency during production of limited natural resources [15,16]. Systematic optimization of redirecting metabolic fluxes in microbial hosts to produce desired products through the combined heterologous expression of plant pathways and enzymes makes it possible to construct new and efficient PNP biosynthetic routes [17]. Significant progress has been made in producing glycosylated PNPs via different hosts using synthetic biology approaches [8]. Once a microbial factory has been developed that can produce even low quantities of a target product, strategies are then employed to engineer that strain for production of industrial-scale titers [18,19].
Recently, the structure-function relationship and glycosylation mechanisms of terpenoid GTs were summarized [3,20]. The evolution and substrates coverage of GT1 family were also overviewed to pave the way for the future exploration of GT proteins [12,21]. However, there is still a lack of comprehensive review on the catalytic diversity of plant GTs in the synthesis of glycosylated PNPs. Moreover, considerable progress has been made in recent years in the mining, engineering and applications of GTs. A timely systematic review will help in constructing synthetic pathways of glycosylated PNPs. In this review, we focus on recent advances in the mining, engineering, and applications of GTs in glycosylated PNP biosynthesis. We systematically summarize the phylogenetic distribution and catalytic diversity of the characterized GTs from plant sources, as well as advanced methods for identification of candidate GTs for glycosylated PNP synthesis. In addition, we also discuss current and ongoing efforts to engineer GTs for new or improved functions by rational design and directed evolution. Finally, this review also covers recent progress in GT applications in the microbial biosynthesis of glycosylated PNPs.
2. GT diversity in glycosylated PNP synthetic pathways
In plants, GTs can glycosylate almost all major classes of secondary metabolites, such as phenylpropanoids (flavanoids, coumarins, lignans, etc.) [[22], [23], [24]], alkaloids (indole alkaloids, steroidal alkaloids, cytokinins, etc.) [[25], [26], [27]], terpenoids (monoterpenoids, diterpenoids, triterpenoids, etc.) [[28], [29], [30]], and polyketides (phenol polyketides, polycyclic aromatic polyketides, etc.) (Fig. 1) [[31], [32], [33], [34]].
Fig. 1.
Representative substrates of functionally characterized GTs in the synthesis of glycosylated PNPs. Chemical groups that are typically glycosylated were colored red.
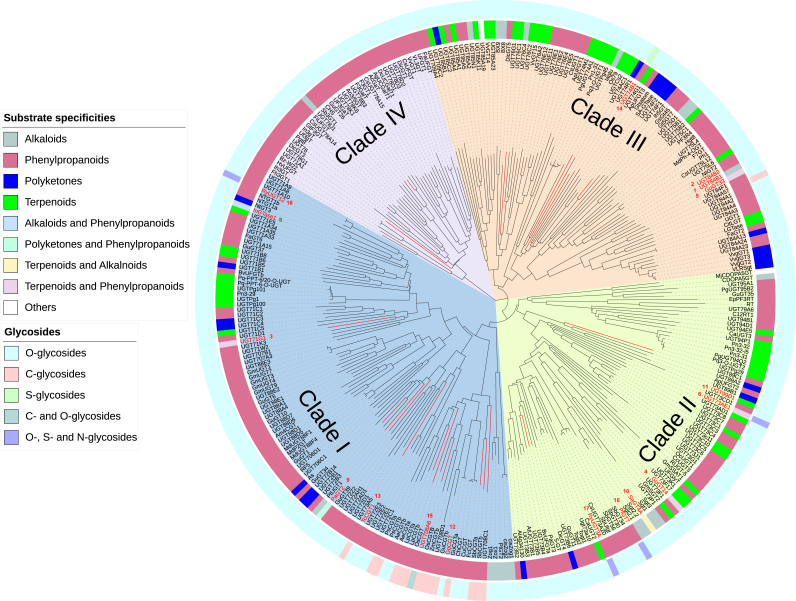
In the present review, we phylogenetically summarized 303 characterized plant GTs (215 were collected in CAZy database, and 88 were collected from recent studies), and analyzed the diversity of their natural substrates and different glycosidic bonds they catalyze (Fig. 2). Phylogenetic trees were generated using the neighbor-joining method with the poisson model applied and using a bootstrap replication of 1000 using MEGAX64. The trees were then imported into the online software iTOL (https://itol.embl.de/) for further optimization. The 303 selected plant GTs could be phylogenetically classified into four major clades. Among the four clades, GTs associated with phenylpropanoid glycoside synthesis comprise the largest family, followed by those that synthesize terpenoid glycosides and polyketide glycosides, while GTs involved in alkaloid glycosides production are relatively rare.
Fig. 2.
Phylogenetic tree analysis of the characterized plant GTs from the GT1 family. The substrate specificities of the characterized plant GTs were depicted by different colors in peripheral circle. The glycosidic bond catalytic by the characterized plant GTs were depicted by different colors in interlayer circle. The characterized plant GTs with co-crystal structures reported were highlighted by red lines. GTs mentioned in the text were marked and numbered in red.
The different clades showed no strict catalytic boundaries for four classes of substrates (including phenylpropanoids, alkaloids, terpenoids, and polyketides). At the clade level, GTs in clades I-III collectively provide broad substrate selectivity, typically accepting almost all of the compound classes listed in Fig. 1. By contrast, clade IV GTs have a relatively narrow substrate spectrum, e.g., with obvious preference for phenylpropanoids or alkaloids.
Occasionally, functional characterizations reveal that evolutionarily close GTs may glycosylate disparate chemical aglycones. For instance, UGT84B1 accepts alkaloids [35], while UGT84B2 (78.3% similarity with UGT84B1) transforms phenylpropanoids (1 and 2 in Fig. 2, respectively) [36]. The reasons for this phenomenon may be attributable to either incomplete characterization of catalytic function, or that specific mutations in key residues, often outside of the active site region, have altered their substrate spectrum [37].
Moreover, despite the diversity in substrates at the clade level, plant GTs have high substrate specificity at the individual level, that they frequently will only accommodate one class of acceptors (Fig. 2). However, some GTs show remarkable promiscuity, showing the ability to catalyze different classes of compounds. This flexibility is key for the success of many glycosylation engineering strategies aimed at facilitating the production of diverse glycosylated PNPs. In particular, UGT71G1 (Medicago truncatula) [36,38], GuGT14, GuGT33 (Glycyrrhiza uralensis) [39] and UGT73AE1 (Carthamus tinctorius) [40] have been shown to catalyze glycosylation of phenylpropanoids (coumarins, coumarones, flavones, isoflavones, and others) and terpenoids (glycyrrhetinic acid and glycyrrhizic acid) (3–6 in Fig. 2, respectively), while InGTase1 (Ipomoea nil) [41] can catalyze phenylpropanoids and alkaloids (7 in Fig. 2), PaGT2 (Phytolacca americana) [42] can catalyze phenylpropanoids and polyketides (9 in Fig. 2), and SaGT4A (Solanum aculeatissimum) [43] can catalyze terpenoids and alkaloids (10 in Fig. 2).
In addition to handling diverse substrates, GTs can also mediate sugar moiety transfer to a diversity of acceptor atoms, O-, C-, N- and even S-atoms in PNP aglycons [3]. O-glycosides are the most common natural products of glycosylation, while the C-, N- and S-glycosides are relatively rare (Fig. 2) [21,44]. The first triterpene arabinosyl-O-GT (UGT99D1, 11 in Fig. 2) was recently discovered and characterized from Avena strigosa. This enzyme selectively adds l-arabinoside to the triterpene scaffold at the C-3 position, a modification critical for disease resistance [45]. Another O-GT (UGT84B1, 1 in Fig. 2) from Arabidopsis thaliana can transfer a glucosyl moiety to the –COOH of indole-3-acetic acid and phenylacetic acid, two primary natural auxins. In addition, 11 O-GTs from Glycyrrhiza uralensis, including isoflavone 7-O-GTs, flavonol 3-O-GTs, and promiscuous O-GTs catalyzing flavones, chalcones, and triterpenoids have been characterized [39].
C-glycoside secondary metabolites and bioactive molecules are widely distributed in plants, and are metabolically more stable than O-, N-, S-glycosides [46,47]. Interestingly, the vast majority of plant C-GTs are found in Clade I, which may indicate their evolutionary conservation and unique catalytic function for C-glycoside synthesis (Fig. 2). In particular, Ye et al. reported a di-C-glycosyltransferase GgCGT (12 in Fig. 2) from Glycyrrhiza glabra, which catalyzes a two-step di-C-glycosylation of flopropione-containing substrates with conversion rates of >98%. GgCGT is the first di-C-GT with a crystal structure containing a sugar acceptor [48]. Recently, TcCGT1 (13 in Fig. 2) from A. thaliana have been identified as an 8-C-GT, which can efficiently and regio-specifically catalyze the 8-C-glycosylation of 36 flavones and other flavonoids. This broad substrate promiscuity of TcCGT1 is enabled by a spacious binding pocket and provides a basis for efficient directed biosynthesis of valuable and diverse bioactive flavonoid C-glycosides [49].
In contrast, few examples of S- and N-GTs from plants have been described in the literature. Historically, glucosinolates were the first identified S-glycosides, which were found in cruciferous vegetables. UGT74B1 (14 in Fig. 2) from A. thaliana was reported to efficiently glycosylate thiohydroximate in glucosinolate biosynthesis [50].
A few plant GTs were shown to perform multiple catalytic functions for different glycosides. For example, a bifunctional maize glycosyltransferase (UGT708A6, 15 in Fig. 2) can produce both C- and O-glycosylated flavonoids, a property not previously described for any other GTs [51]. Additionally, PlGT7 (Pueraria lobata) [52], UGT73AE1 (Carthamus tinctorius) [40], RyUGT3A (Rubia yunnanensis), and RyUGT12 (Rubia yunnanensis) [53] (16, 6, 17, 18 in Fig. 2, respectively) can even transfer various sugars to three different nucleophilic groups (OH, NH2, and SH) of diverse compounds, thus producing O-, N-, and S-glycosides.
3. Mining GTs for biosynthesis of glycosylated PNPs
The discovery of GTs with novel functions is a necessity for advancing their practical applications. Recently, the development of high-throughput sequencing and deep learning analysis have enabled considerable advances in enzyme mining, leading to progress in discovery of novel GTs for glycosylated PNP production (Fig. 3) [[54], [55], [56], [57], [58]].
Fig. 3.
The workflow for GT mining and application in biosynthesis of glycosylated PNPs.
Genomic analysis of whole genome sequence data can provide a multi-level resource for mining novel enzymes that can be greatly informative for researchers seeking to identify genes involved in glycosylated PNP synthesis [[59], [60], [61]]. Huang et al. demonstrated that plant GTs harbor a highly conserved 44 amino acid C-terminus motif (the plant secondary product glycosyltransferase box, PSPG box), which has been proposed to serve as a nucleotide diphosphate sugar binding site [62,63]. A recent genome-wide analysis of soybean by Rehman et al. identified 149 putative UGTs based on the PSPG box [64]. Similar approaches were used for GT identification in the genomes of chickpea, cotton, maize, and flax [[65], [66], [67], [68]]. In addition, genomic data can also provide insights into glycosylated PNP biosynthetic pathways evolution, regulation, and production [69]. Despite the exponential increase in genomic sequence data, the use of glycosylated PNPs for engineering biosynthetic pathways is still restricted to experimentally characterized GTs [70]. For example, to identify the 2′-O-GT (P2′GT) responsible for phloretin production, Zhou et al. performed genome-wide analysis in domesticated apple (Malus x domestica Borkh), which identified two P2′GTs (MdUGT88F1 and MdUGT88F4) that were validated by in vitro activity assays and relative expression analysis (Table 1) [4].
Table 1.
The recent five years characterized GTs identified by high-throughput sequencing strategies.
| Enzyme Name | Accession No. | Taxonomy | Systems biology tool | Reference |
|---|---|---|---|---|
| UGT75D1 | AAB58497.1 | Arabidopsis thaliana | Genomic | [25] |
| UGT71C3 | AAF82195.1 | Arabidopsis thaliana | Genomic | [71] |
| UGT1 | QGI57841.1 | Atropa belladonna | Genomics | [72] |
| MdUGT88F1 | ARV88476.1 | Malus domestica | Genomics | [4] |
| UGT84A23 | ANN02875.1 | Punica granatum | Genomics | [73] |
| MdPh-4-OGT | AAX16493.1 | Malus domestica Brokh | Genomics | [74] |
| DcUCGalT1 | AKI23632.1 | Daucus carota | Genomics | [75] |
| GuCGTa, GuCGTb | QLF98865.1, QLF98866.1 | Glycyrrhiza uralensis | Genomic | [76] |
| UGT72AD1, UGT72AH1, UGT72Z2 |
AP009657.1, AOG18241.1, AKK25344.1 |
Lotus japonicus | Genomic | [77] |
| MdP2GT | AMA68117.1 | Malus domestica | Genomic | [78] |
| MdUGT88F, MdUGT88F4 | ARV88476.1 | Malus domestica | Genomic | [4] |
| MdPh-4-OGT | AAX16493.1 | Malus x domestica Brokh | Genomic | [79] |
| AgUCGalT1 | AXU98426.1 | Apium graveolens | Transcriptomic | [80] |
| UGT71B5 | ANM66102.1 | Arabidopsis thaliana | Transcriptomic | [81] |
| UGT76C1, UGT76C2 | BAB10792.1, BAB10791.1 | Arabidopsis thaliana | Transcriptomic | [82] |
| UGT76E12 | AAK82559.1 | Arabidopsis thaliana | Transcriptomic | [83] |
| UGT85A1 | AAF18537.1 | Arabidopsis thaliana | Trancsriptomic | [83] |
| AeCGTb, AeCGTa | QLF98868.1, QLF98867.1 | Arisaema erubescens | Transcriptomic | [84] |
| CsUGT73A20, CsUGT75L12, CsUGT78A14, CsUGT78A15 | ALO19886.1, ALO19892.1 ALO19888.1, ALO19889.1 | Camellia sinensis | Transcriptomic | [85] |
| UGT84A57 | BBI55602.1 | Eutrema japonicum | Transcriptomic | [86] |
| GgCGT | QGL05036.1 | Glycyrrhiza glabra | Transcriptomic | [87] |
| Pn1-31, Pn3-29, Pn3-31, Pn3-32 | QOJ43864.1, QOJ43865.1, QOJ43866.1, QOJ43868.1 | Panax notoginseng | Transcriptomic | [88] |
| RyUGT3A | QSB46663.1 | Rubia yunnanensis | Transcriptomic | [53] |
| RsUGT75L20, RsUGT75T4 | AWU66063.1, AWU66062.1 | Rubus suavissimus | Transcriptomic | [76] |
| UGT76E2 | BAA97493.1 | Arabidopsis thaliana | Transcriptomic | [89] |
| TcCGT1 | QCZ42162.1 | Trollius chinensis | Transcriptomic | [49] |
| GuGT1 | QDM38894.1 | Glycyrrhiza uralensis | Transcriptomic | [39] |
| ApUFGT1 | QDA11331.1 | Andrographis paniculata | Transcriptomic | [90] |
| CcUGT3 | QDH43895.1 | Crocosmia | Transcriptomic | [91] |
| Sb3GT1 | QBL54224.1 | Scutellaria baicalensis | Transcriptomic | [92] |
| PgUGT95B2 | AZB52139.1 | Punica granatum | Transcriptomic | [93] |
| OcUGT1 | AWD73588.1 | Ornithogalum caudatum | Transcriptomic | [94] |
| Pq3-O-UGT1 | ALE15279.1 | Panax quinquefolius | Transcriptomic | [95] |
| Pq3-O-UGT2 | ALE15280.1 | Panax quinquefolius | Transcriptomic | [96] |
| UGT72B3 | AAF97321.1 | Arabidopsis thaliana | Transcriptomic | [97] |
| UGT73F17 | AXS75258.1 | Glycyrrhiza uralensis | Transcriptomic | [98] |
| EpPF3RT | MG264429.1 | Epimedium pseudowushanense | Transcriptomic | [99] |
| UGT76E11 | CAB62337.1 | Arabidopsis thaliana | Transcriptomic | [100] |
| PgUGT71A27 | A0A0A7HB61.1 | Panax ginseng | Transcriptomic | [101] |
| UGT75B2 | AAF79732.1 | Arabidopsis thaliana | Genomic & Trancsriptomic | [102] |
| ChCGT, CuCGT, FcCGT | BBA18064.1, BA18063.1, BBA18062.1 | Citrus hanaju | Genomic & Trancsriptomic | [103] |
| CtUGT1 | MW629113 | Cistanche tubulosa | Genomic and transcriptomic | [104] |
| GmSGT2 | BAI99584.1 | Glycine max | Genomic & Transcriptomic | [105] |
| UGT71K3 | XP_004294260.1 | Fragaria ananassa | Genomics & Transcriptomic | [106] |
| Pq-PPT-6/20-O-UGT, Pq-PPT-6-O-UGT | QEV87497.1, QEV87498.1 | Panax quinquefolius | Genomic and Transcriptomic | [107] |
| PsCGTa, PsCGTb | QLF98871.1, QLF98872.1 | Pistia stratiotes | Genomic and Transcriptomic | [76] |
| SbGT30, SbGT34 SbGT56 |
AMK52071.1,AMK52072.1 AMK52073.1 |
Scutellaria baicalensis | Genomic & Trancsriptomic | [108] |
| UGT90F1, UGT73B26, UDPG1 | MF417497.1, MF417498.1, HQ259620.1 | Siraitia grosvenorii | Genomic and Transcriptomic | [109] |
| ZmCGTb | QLF98873.1 | Zea mays | Genomic and Transcriptomic | [76] |
| UGT99D1 | AZQ26921.1 | Avena strigosa | Transcriptomic & Protemic | [45] |
| UGT74H5, UGT74H6 | ACD03250.1, ACD03261.1 | Avena strigosa | Transcriptomic & Protemic | [110] |
| UGT73AD1 | ALD84259.1 | Centella asiatica | Transcriptomic and Proteomic | [35] |
| GmSSAT1 | XP_003532274.1 | Glycine max | Transcriptomic & Proteomic | [111] |
In addition to genomic data, whole transcriptome sequencing (RNA-seq) can provide the cDNA sequence and tissue-specific expression levels of specific genes not available through genomic data [112]. Thus, many studies opt for this approach to mine for GTs relevant to the synthesis of glycosylated PNPs [113,114]. For example. Murukarthick et al. carried out transcriptome sequencing on four root samples type from Panax ginseng including whole roots of from one-year-old plants, and the main root bodies, rhizomes, and lateral roots of six-year-old plants. This analysis ultimately identified 189 GT-derived transcripts involved in ginsenoside biosynthesis [115]. Similarly, a comprehensive analysis of the transcriptome landscape of three genotypes of Stevia (SR-1, SR-2, and SR-3) revealed 143 total GT unigenes, some of which were determined to contribute to steviol glycoside biosynthesis [116].
Indeed, comparative transcriptome analysis of different organs or tissues can provide more information than genomic analysis that is relevant to screening novel candidate GTs [117]. For instance, Fan et al. showed that the anthraquinone glycosides accumulated to higher levels in roots of R. yunnanensis than in the stems or leaves [118]. Based on this finding, Yi et al. compared the transcriptomes of R. yunnanensis roots, stems and leaves and identified 32 novel candidate GT genes with high expression in root tissue from 499 putative GTs found in transcriptomic data [53]. These findings largely guided subsequent screening of candidate GTs for glycosylated PNP biosynthesis.
Taking advantage of advances in LC-MS/MS technology, a recent study developed a proteomics workflow to identify candidate GTs involved in glycosylated PNP biosynthesis [119]. For instance, Suliman et al. used tandem mass spectrometry (LC-MS/MS) to identify proteins extracted from the Golgi-enriched fractions of wheat endosperm. This analysis revealed 1135 proteins in the wheat endosperm, which identified 64 GTs by searching mass data against four databases, including UniProt, Gene Index Databases Wheat release 12.0, an in-house glycosyltransferase databank, and a contaminant database (i.e., keratins and trypsin) [120].
Moreover, comprehensive gene mining has realized through the integration of genomic, transcriptomics, and proteomics data. As an example, in depth metabolic fingerprinting and LC-MS profiling of various parts of Asparagus racemosus led to the identification of a significant number of steroidal saponins exclusively present in roots. Transcriptome sequencing from three different tissues led to the identification of 321 different genes involved in saponin biosynthesis [6]. Similarly, two homologous GTs, itUGT1 and itUGT2 (86% similarity) have been identified based on peptide mass fingerprinting and previously described transcriptomics data of Indigofera tinctoria leaves [121].
A large number of GT genes have been obtained by using the above gene mining methods, however, there are still some challenges in narrowing down the scope of target GTs mining for specific PNPs glycosylation owing to the labor-intensive function characterization of individual enzymes [20]. Therefore, development of efficient and flexible enzyme function detection methods is crucial for obtaining comprehensive enzyme functional data. Some progress has been made recently, such as the development of a high-throughput screening method by using mass spectrometry [122]. Another publication has recently highlighted the potential for a fluorescence-based method to universally monitor the activity of GTs by detecting the nucleotides generated in a biochemical reaction [123].
In addition, the development of artificial intelligence provides a unique avenue for the identification of target enzymes through rapidly expanding sequence databases. Recent advances in deep learning models for feature extraction and patter recognition for sequence classification and functional prediction of enzymes in large datasets have also facilitated the discovery of novel GTs [124,125]. Yang et al. developed a chemical-bioinformatic model for functional prediction of uncharacterized GT1 family GTs based on sequence data of 54 GTs of A. thaliana and structural information of 91 candidate substrates. The model successfully identified novel substrates for GTs and enabled functional annotation of GTs from other sources including alfalfa, oats, and bacteria. Using enzyme sequences that did not rely on experimental data, this analysis provided meaningful biological insights that guided subsequent directed evolution and mechanistic studies of GT enzymes [126].
4. Engineering of GTs for biosynthesis of glycosylated PNPs
Wild-type GTs often lack properties desirable for the synthesis of glycosylated PNPs, such as high activity, high stereo- and regio-selectivity, and minimal undesirable activities and promiscuity towards valuable unnatural substrate [12]. However, structure-based rational design and directed evolution can efficiently improve the properties of GTs for industrial applications.
As of October 20, 2021, a total of 26 GT1 family GTs involved PNP glycosylation with solved crystal structures (among nearly 30,000 total GT sequences) are available in the CAZy database (Table 2). Increasing the availability of GT structures, especially in the presence of both donor and acceptor analogues, is essential for the continued success of guided rational engineering of GT mutants with improved or altered functions [13,14]. As an alternative strategy for GT engineering, directed evolution may offer the potential for altering GT specificity and/or fine-tuning the activity of rational GT chimeras [12,127,128]. Here, we summarize several recently reported protein engineering strategies to increase the catalytic activity, broaden the substrate spectrum, and alter the regioselectivity of GTs.
Table 2.
The structure-based rational design and directed evolution of GTs that involved in the biosynthesis of glycosylated plant natural products.
| Protein name | Organism | Genbank | PDB code | Representative schematic reaction | Key residues | Engineering | Reference |
|---|---|---|---|---|---|---|---|
| YjiC | Bacillus subtilis | NP_389104.1 | 7BOV |
|
|
[129] | |
| UGT72B1 | Arabidopsis thaliana | CAB80916.1 | 2VCE | 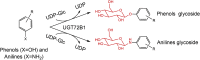 |
|
– | [130] |
| UGT74F2 | Arabidopsis thaliana | AAB64024.1 | 5U6M | 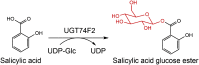 |
|
– | [131] |
| UGT89C1 | Arabidopsis thaliana | AAF80123.1 | 6IJ7 |
|
|
[132] | |
| UGT78K6 | Clitoria ternatea | BAF49297.1 | 3WC4 | 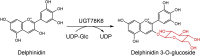 |
|
– | [133,134] |
| UGT708C1 | Fagopyrum esculentum | BAP90360.1 | 6LLG |
|
– | [135] | |
| GgCGT | Glycyrrhiza glabra | QGL05036.1 | 6L5P |  |
|
|
[48] |
| UGT73P12 | Glycyrrhiza uralensis | BBN60799.1 | 7C2X |  |
|
– | [136] |
| LpCGTa | Landoltia punctata | QLF98869.1 | 6LG1 | 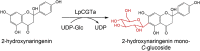 |
– | – | [76] |
| LpCGTb | Landoltia punctata | QLF98870.1 | 6LFN | 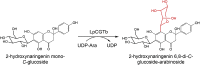 |
– | – | [76] |
| SbCGTa | Scutellaria baicalensis | QLF98861.1 | 6LG0 |  |
|
– | [76] |
| SbCGTb | Scutellaria baicalensi | QLF98862.1 | 6LFZ | 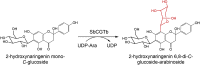 |
|
|
[76] |
| UGT708A6 | Zea mays | ACF81582.1 | 6LF6 | 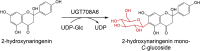 |
– | – | [76] |
| UGT71G1 | Medicago truncatula | AAW56092.1 | 2ACV | 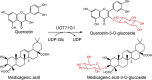 |
|
– | [2] |
| UGT85H2 | Medicago truncatula | ABE87250.1 | 2PQ6 |
|
– | [137] | |
| UGT78G1 | Medicago truncatula | ABI94025.1 | 3HBF |
|
– | [138] | |
| Os79 | Oryza sativa | BAF14158.1 | 5TMB |
|
– | [139] | |
| PtUGT1 | Persicaria tinctoria | BBB06426.1 | 5NLM | 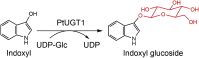 |
|
– | [140] |
| PaGT2 | Phytolacca americana | BAG71125.1 | 6JEL |
|
|
[42] | |
| PaGT3 | Phytolacca americana | BAG71127.1 | 6LZX |
|
– | [141] | |
| UGT51 | Saccharomyces cerevisiae | AAB67475.1 | 5GL5 | 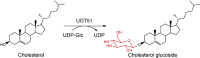 |
|
|
[142,143] |
| UGT74AC1 | Siraitia grosvenorii | AEM42999.1 | 6L8W |  |
|
|
[144] |
| UGT74AC2 | Siraitia grosvenorii | AXK92493.1 | 7BV3 | 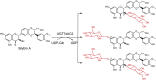 |
|
|
[145] |
| UGT76G1 | Stevia rebaudiana | AAR06912.1 | 6INF |
|
– | [146] | |
| TcCGT1 | Trollius chinensis | QCZ42162.1 | 6JTD | 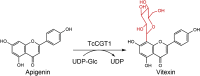 |
|
|
[49] |
| VvGT1 | Vitis vinifera | AAB81683.1 | 2C1X | 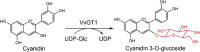 |
|
– | [147] |
| UGTPg45 | Panax ginseng | AKA44586.1 | – | – |
|
[148] |
Wild-type GTs often show low catalytic activity when expressed in heterogeneously engineered hosts, which in many cases can restrict the industrial-scale production of glycosylated PNPs. Recently, to address the inefficiency of the key ginseng GT, our group selected fifteen mutation hotspots in the receptor binding site of Saccharomyces cerevisiae UGT51 based on its crystal structure in complex with UDP-glucose. One mutant (S81A/L82A/V84A/K92A/E96K/S129A/N172D) showed an ∼1,800-fold increase catalytic activity in the conversion of protopanaxadiol (PPD) to ginsenoside Rh2 in vitro (Table 2) [143]. Similarly, Liu et al. engineered the GT Yjic from Bacillus subtilis for Rh2 synthesis. Using a semi-rational design that included structure-guided alanine scanning and saturation mutations, mutant M315F was found to efficiently synthesize Rh2 (∼99%) and block the further glycosylation of C12–OH [149].
An in vivo directed evolution strategy in which mutations were directly introduced into the chassis was also recently developed in an effort to enhance catalytic activity. The mutants were screened based on in vivo yield of target products, resulting mutated bioparts with improved enzymatic characteristics and performance in compatibility with chassis. Using this method, the poor performance of UGTPg45 in catalyzing the conversion of its unnatural substrate, PPD to ginsenoside Rh2, was improved. As a result, the UGT mutant UGTPg45-HV was acquired which carried two missense mutations (Q222H and A322V) that conferred a 70% increase in ginsenoside Rh2 yield [148]. Recently, our group developed an ultrahigh-throughput dual-channel microfluidic droplet screening system and a fluorescence-activated cell sorting system that both enabled the high-throughput screening (>107 mutants/hour) of mutants, which can contribute to the effective engineering of high activity GTs [150,151].
Recently, the promiscuous substrate specificity of GTs has inspired studies exploring how to effectively design biocatalysts for efficient and directed biosynthesis of bioactive glycosides. Structure-guided mutagenesis was conducted to alter the catalytic specificity of C-/O-glycosylation by TcCGT1. As a C-glycosyltransferase (CGT) from the medicinal plant Trollius Chinensis, TcCGT1 can catalyze the 8-C-glycosylation of 36 different flavonoids and the O-glycosylation of diverse phenolics. The spacious binding pocket characterized using its crystal structure in complex with uridine diphosphate explains its substrate promiscuity, with the substrate binding pose determining its C- or O-glycosylation activity. Site-directed mutagenesis at two residues (I94E and G284K) enabled the conversion from C- to O-glycosylation [49]. Other studies have also successfully switched sugar donor preference and acceptor substrates using either single- or multiple-point mutations not exclusively located within the binding site (Table 2) [48,76,132]. Based on the successful GT engineering cases, the specificity for the donor substrate, especially the nucleotide residue, is largely determined by the highly conserved PSPG motif in the C-terminus. Besides, mutations in the residues involved in substrate recognition may change the preference of glycosylation.
Regioselectivity is also a typical problem that must be considered in the synthesis of structurally diverse glycosides [152]. Recent studies of plant GTs have shown that point mutations can alter regioselectivity due to flexibility in the GT substrate binding pocket [153]. Fan et al. demonstrated the successful switching of regioselectivity by UGTBL1 from Bacillus licheniformis to yield polydatin (resveratrol 3-O-β-glucoside) instead of resveratrol 4′-O-β-glucoside polydatin, a compound used to relieve the toxic side effects of cisplatin and treat acute severe hemorrhagic shock. To this end, a 3D model of UGTBL1 was constructed, and residue Ile62 was found to significantly influence its regioselectivity. Mutation I62G ultimately led to the switch in regioselectivity from 4′-OH to 3-OH of resveratrol, with a roughly sevenfold increase in the formation of the preferred polydatin over that of 4′-O-glucoside compared to wild type [154]. Recently, Sun and co-workers tuned a newly identified GT from Siraitia grosvenorii (UGT74AC2) to serve as the catalyst of targeted regioselective glycosylation of the polyhydroxy substrate silybin and derivatives. Three single-site mutants (P12Y, L200W and Y145W) showed 94%, >99%, and >99% selectivity on the 3-OH, 7-OH and 3,7-O-diglycoside of the substrates, respectively, compared with that of wild type, which produced a 22%:39%:39% product mixture [132].
5. GT applications in glycosylated PNP biosynthesis
Emerging synthetic biology strategies are rapidly expanding the application of GTs in the synthesis of glycosylated PNPs, or their desirable precursors, in microbial fermentation systems [[155], [156], [157], [158]]. Here, we discuss some breakthrough of GT-based biosynthesis of glycosylated PNPs at industrial scale in model hosts like Escherichia coli and S. cerevisiae (Table 3). These representative products include flavonoid glycosides (fisetin 3-O-glycosides, astragalin, scutellarein 7-O-glucoside), terpenoid glycosides (rebaudioside A, ginsenosides), and polyketide glycosides (salidroside, polydatin).
Table 3.
The recent glycosylated plant natural products synthesis by microbial sources.
| Compound | Microbial sources | GTs | Titer | Reference |
|---|---|---|---|---|
| Cyanidin 3-O-glucoside | E. Coli | 3 GT | 0.35 g/L | [159] |
| Luteolin-7-O-glucuronide, Quercetin-3-O-glucuronide, Quercetin 3-O-galactoside | E. Coli | AmUGT10, VvUGT, PhUGT | 0.30, 0.69, 0.28 g/L | [160] |
| Fisetin 3-O-glucoside, Fisetin 3-O-rhamnoside | E. Coli | UGT78K1, ArGt-3 |
0.39, 0.34 g/L | [161] |
| Quercetin 3-O-galactoside, Quercetin 3-O-rhamnoside | E. Coli | RhaGT | 0.94, 1.12 g/L | [162] |
| Scutellarein 7-O-glucoside | S. cerevisiae | SbGT34 | 1.20 g/L | [108] |
| Kaempferol 3-O-glucoside | E. Coli | AtUGT78D2 | 3.60 g/L | [163] |
| Rebaudioside A | S. cerevisiae | UGT76G1 | 1.16 g/L | [164] |
| Tyrosol glucoside | E. Coli | UGT72B14 | 6.7 mg/L | [165] |
| Ginsenoside Rh1, Ginsenoside F1 | S. cerevisiae | UGTPg1, UGTPg100 | 0.10, 0.04 g/L | [166] |
| Ginsenoside Rh2, Ginsenoside Rg3 | S. cerevisiae | UGTPg29, UGTPg45 | 0.02, 0.05 g/L | [30] |
| Ginsenoside Rh2 | S. cerevisiae | UGT51 | 0.30 g/L | [143] |
| 3β,12β-Di-O-Glc-PPD, -PPT, DM | S. cerevisiae | UGT109A1 | 9.05, 4.57, 11.5 mg/L | [167] |
| 3β-O-Glc-DM | S. cerevisiae | UGT74AE2 | 5.60 g/L | [168] |
| Ginsenoside Rh2, PPD, DM | S. cerevisiae | UGTPn50 | 2.25, 9.05, 8.09 g/L | [148] |
| Ginsenoside Compound K | Yarrowia lipolytica | UGTPg1 | 0.16 g/L | [169] |
| Ginsenoside Compound K | S. cerevisia | UGTPg1 | 5.74 g/L | [170] |
| Ginsenoside Rg1, Notoginsenoside R1, Notoginsenoside R2 | S. cerevisia | PgUGT71A53, PgUGT94Q13, PgUGT71A54 | 1.95, 1.62, 1.25 g/L | [171] |
| Crocetin | E. coli | YjiC, YdhE, YojK | 4.42 mg/L | [172] |
| Kaempferol, astragalin | E. coli | AtUGT78D2 | 1.18, 1.74 g/L | [173] |
| Geranyl glucoside | E. coli | VvGT14a | 0.93 g/L | [174] |
| Glycyrrhizin and Glycyrrhetic acid 3-O-mono-β-D-glucuronide | S. cerevisiae | UGT1A1 | 5.98, 2.31 mg/L | [175] |
| Polydatin | S. cerevisiae | PcR3GAT | 0.55 g/L | [176] |
Fisetin glycoside is a medicinally important flavonoid glycoside produced by various plants that has been reported to exhibit diverse medicinal effects such as prevention of cardiovascular diseases, antioxidant activity, anti-diabetic activity, and anticancer activity [[177], [178], [179]]. The regiospecific GT (UGT78K1) from Glycine max or ArGt-3 from A. thaliana were introduced into E. coli BL21 (DE3), along with the respective UDP-glucose and TDP-rhamnose biosynthetic genes from different bacterial sources, in order to achieve the bioconversion of fisetin. Approximately 1.18 g of fisetin 3-O-glucoside and 1.03 g of fisetin 3-O-rhamnoside were produced in a 3 L bioreactor [161].
Similar to this accomplishment, the GT (AtUGT78D2) from A. thaliana and a highly efficient UDP-glucose synthesis pathway were introduced into E. coli BL21 (DE3) to produce another flavonoid glycoside, astragalin, which resulted in maximal astragalin production of 1.74 g/L [173]. Other than E. coli, S. cerevisiae has also been used for flavonoid glycoside biosynthesis. Successful deletion of glucosidases in S. cerevisiae in conjunction with overexpression of the flavonoid GT SbGT34 from Scutellaria baicalensis enabled production of the medicinal compound scutellarein 7-O-glucoside. The feasibility of scaling in vivo glycosylation was demonstrated by large-scale production of 1.20 g/L scutellarein 7-O-glucoside by optimization of the appropriate fermentation conditions [108].
As the sweetest terpenoid glycoside from Stevia rebaudiana, rebaudioside A is commercially significant as a natural sweetener used in the food and beverage industry [180,181]. To produce rebaudioside A in yeast, UGT76G1 from S. rebaudiana was overexpressed under the control of the PGK1 promoter. At the same time, the Nocardia farcinica phosphoglucomutase gene, nfa44530, which participates in UDP-glucose synthesis, and the E. coli K12 glucose-1-phosphate-1 uridylyltransferase gene, galU, were also co-expressed in the recombinant S. cerevisiae. By optimizing the availability of UDP-glucose, rebaudioside A production reached 1.16 g/L [164].
As a group of glycosylated triterpenes found in Panax species, ginsenosides are synthesized from 2,3-oxidosqualene through the universal precursors, dimethylallyl diphosphate (DMAPP) and isopentenyl diphosphate (IPP) [182]. An approach for ginsenoside production was recently developed for S. cerevisiae. Following the chromosomal integration of a PPD biosynthetic pathway in yeast, UGTPg45 and UGTPg29 from P. ginseng were introduced into the recombinant cells, enabling production of the Rh2 and Rg3 ginsenosides. However, Rh2 production was relatively low (16.95 mg/L in shaken flasks) for commercialization due to the poor performance of the GT (UGTPg45) [148]. Based on these findings, our group built an efficient ginsenoside Rh2 biosynthetic cell factory by repurposing an inherently promiscuous GT (UGT51 mutant) from S. cerevisiae with an 1800-fold increase in catalytic efficiency over wild type. This strain harboring the engineered GT could produce 0.3 g/L of ginsenoside Rh2 in a 5 L fed-batch fermentation system [143]. Notably, Zhou's group successfully constructed yeast strains that could produce ginsenoside CK, ginsenoside Rg1, notoginsenoside R1, and notoginsenoside R2 by introducing a group of GTs including UGTPg1, PgUGT71A53, PgUGT94Q13, and PgUGT71A54. De novo production of these ginsenosides reached 5.74, 1.95, 1.62, and 1.25 g/L, respectively [170,171].
As one of the major polyketide glycosides in Rhodiola, salidroside (the 8-O-β-glucoside of tyrosol) has been purported to confer adaptogenic and ergogenic effects [183]. Xue et al. reported salidroside production through the expression of Rhodiola UGT72B14 in E. coli. Codon optimization resulted in significantly enhanced salidroside accumulation, reaching 6.7 mg/L, a 3.2-fold increase over that of wild-type GT [170].
Polydatin is a well-known pharmaceutical polyketide glycoside that provides anticancer, antiaging, and anti-inflammatory effects [[184], [185], [186]]. Recently, Liu et al. explored the development of a microbial chassis for polydatin production that could potentially replace plant extraction in future systems. This work also identified a key enzyme for polydatin biosynthesis, resveratrol GT, PcR3GAT. Polydatin production thus reached 0.54 g/L through the incorporation of a resveratrol biosynthesis module, UDP-glucose supply module, and GT expression module and subsequent optimization of fermentation conditions [176].
6. Conclusion and future perspectives
Glycosylation is one of the most important physiological and biochemical reactions in nature, given its crucial roles in a multitude of essential processes. This intrinsic importance has attracted long-standing and wide research attention into the characteristics of GTs to facilitate their applications in glycosylation reactions for metabolic engineering of natural product biosynthesis. While several GTs have been found to be suitable for altering glycosylation patterns, generally low catalytic activity and stringent substrate specificity remains a limiting factor in the diversification of PNPs for industrial fermentation systems. The data mining and engineering GTs are still the most promising approaches for discovering and developing novel enzymes with well-defined characteristics. High-throughput sequencing has enabled efforts to comprehensively profile the genomes, transcriptomes, and proteomes of plant species with important medicinal, industrial, or scientific applications. Furthermore, this sequencing data corroborate with functional data that have fed artificial intelligence-based computational approaches to GT discovery.
A series of discoveries have been made recently, using principles of synthetic biology, in the field of PNP glycoside biosynthesis catalyzed by GTs. However, there remains some challenges to engineering industrial hosts [187]. The metabolic engineering needed to produce a particular PNP glycoside relies on the biosynthetic routes of PNP-precursors, however these details are frequently unavailable or incomplete. In this case, candidate pathway design, enzyme selection, and pathway testing all bring different challenges [8]. More efforts should be made to reveal the complexity of natural PNP pathways.
Another obstacle present in GT application in the synthesis of PNP glycosides is that when introducing GTs into a heterologous host, they may function sub-optimally or not at all for reasons that include low expression or activity, improper folding, and mislocalization. Therefore, modifying the function of GTs need for glycosylated PNP synthesis, and tuning the biosynthetic systems to improve the yields of these natural products, is an increasing research priority, given the urgent need for affordable, effective drugs that are inefficiently produced in nature. Since traditional GT activity assays are not suitable for rapid detection, a central goal of future research on GTs is to develop universal high-throughput detection methods, such as qualitative mass spectrometry-based assays or fluorescence-based assays for rapid screening of target GTs. In addition, further structural elucidation of GTs will help to increase our understanding of the catalytic mechanism of these enzymes. The crystal structures validated by experimental methods, or predicted by high accurate artificial intelligence methods, e.g. AlphaFold2 [188] and RoseTTAFold [189], will accelerate the elucidation of the structure-function relationship. In particular, this involves identifying which hot domains or motifs of the protein affects its structure and the associated enzymatic activity, which will further guide the rational design of GTs.
Moving forward, synthetic biology-driven metabolic engineering of different desirable GT characteristics combine with relevant components of synthesis pathways will reinvigorate current efforts to increase the diversity of PNP glycosides used in industrial production for medicine, functional foods, and cosmetics.
CRediT authorship contribution statement
Bo He: Conceptualization, Writing – original draft, Data curation, Validation. Xue Bai: Writing – original draft, Data curation. Yumeng Tan: Investigation, Data curation. Wentao Xie: Investigation, Data curation. Yan Feng: Supervision. Guang-Yu Yang: Funding acquisition, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no conflict of interests.
Acknowledgement
This study was financially supported by the National Key R&D Program of China (2020YFA0907900, 2018YFE0200501), and the National Natural Science Foundation of China (Grant number 32030063).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Atanasov A.G., et al. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol Adv. 2015;33:1582–1614. doi: 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shao H., et al. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell. 2005;17:3141–3154. doi: 10.1105/tpc.105.035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang D.M., et al. Glycosyltransferases: mechanisms and applications in natural product development. Chem Soc Rev. 2015;44:8350–8374. doi: 10.1039/c5cs00600g. [DOI] [PubMed] [Google Scholar]
- 4.Zhou K., et al. Genome-wide identification of glycosyltransferases converting phloretin to phloridzin in Malus species. Plant Sci. 2017;265:131–145. doi: 10.1016/j.plantsci.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Shen C.J., et al. Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNA-Seq. Sci Rep. 2017;7:187. doi: 10.1038/s41598-017-00292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srivastava P.L., Shukla A., Kalunke R.M. Comprehensive metabolic and transcriptomic profiling of various tissues provide insights for saponin biosynthesis in the medicinally important Asparagus racemosus. Sci Rep. 2018;8:9098. doi: 10.1038/s41598-018-27440-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombard V., et al. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cravens A., Payne J., Smolke C.D. Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat Commun. 2019;10:2142. doi: 10.1038/s41467-019-09848-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C.L., et al. Microbial platform for terpenoid production: Escherichia coli and yeast. Front Microbiol. 2018;9:2460. doi: 10.3389/fmicb.2018.02460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muhammad A., et al. Production of plant natural products through engineered Yarrowia lipolytica. Biotechnol Adv. 2020;43:107555. doi: 10.1016/j.biotechadv.2020.107555. [DOI] [PubMed] [Google Scholar]
- 11.Wang L.X., Huang W. Enzymatic transglycosylation for glycoconjugate synthesis. Curr Opin Chem Biol. 2009;13:592–600. doi: 10.1016/j.cbpa.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McArthur J.B., Chen X. Glycosyltransferase engineering for carbohydrate synthesis. Biochem Soc Trans. 2016;44:129–142. doi: 10.1042/BST20150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lairson L.L., et al. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 14.Breton C., et al. Recent structures, evolution and mechanisms of glycosyltransferases. Curr Opin Struct Biol. 2012;22:540–549. doi: 10.1016/j.sbi.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Nemhauser J.L., Torii K.U. Plant synthetic biology for molecular engineering of signalling and development. Native Plants. 2016;2:16010. doi: 10.1038/nplants.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng F.K., Ellis T. The second decade of synthetic biology: 2010-2020. Nat Commun. 2020;11:5174. doi: 10.1038/s41467-020-19092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J.W., et al. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol. 2012;8:536–546. doi: 10.1038/nchembio.970. [DOI] [PubMed] [Google Scholar]
- 18.Liu H., et al. Enhanced β-amyrin synthesis in Saccharomyces cerevisiae by coupling an optimal acetyl-CoA supply pathway. J Agric Food Chem. 2019;67:3723–3732. doi: 10.1021/acs.jafc.9b00653. [DOI] [PubMed] [Google Scholar]
- 19.Ajikumar P.K., et al. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurze E., et al. Structure-function relationship of terpenoid glycosyltransferases from plants. Nat Prod Rep. 2021 doi: 10.1039/d1np00038a. Advance article. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P., et al. Glycosyltransferase GT1 family: phylogenetic distribution, substrates coverage, and representative structural features. Comput Struct Biotechnol J. 2020;18:1383–1390. doi: 10.1016/j.csbj.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer C.M., et al. Cloning and regiospecificity studies of two flavonoid glucosyltransferases from Allium cepa. Phytochemistry. 2003;64:1069–1076. doi: 10.1016/s0031-9422(03)00507-7. [DOI] [PubMed] [Google Scholar]
- 23.Ono E., et al. Yellow flowers generated by expression of the aurone biosynthetic pathway. Proc Natl Acad Sci U S A. 2006;103:11075–11080. doi: 10.1073/pnas.0604246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim C.E., Ahn J.H., Lim J. Molecular genetic analysis of tandemly located glycosyltransferase genes, UGT73B1, UGT73B2, and UGT73B3, in Arabidopsis thaliana. J Plant Biol. 2006;49:309–314. [Google Scholar]
- 25.Zhang G.Z., et al. Ectopic expression of UGT75D1, a glycosyltransferase preferring indole-3-butyric acid, modulates cotyledon development and stress tolerance in seed germination of Arabidopsis thaliana. Plant Mol Biol. 2016;90:77–93. doi: 10.1007/s11103-015-0395-x. [DOI] [PubMed] [Google Scholar]
- 26.Eudes A., et al. Metabolism of the folate precursor p-aminobenzoate in plants: glucose ester formation and vacuolar storage. J Biol Chem. 2008;283:15451–15459. doi: 10.1074/jbc.M709591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veach Y.K., et al. O-glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol. 2003;131:1374–1380. doi: 10.1104/pp.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y.Q., et al. Discovery of Arabidopsis UGT73C1 as a steviol-catalyzing UDP-glycosyltransferase with chemical probes. Chem Commun. 2018;54:7179–7182. doi: 10.1039/c7cc09951g. [DOI] [PubMed] [Google Scholar]
- 29.Smehilova M., et al. Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front Plant Sci. 2016;7:1264. doi: 10.3389/fpls.2016.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P.P., et al. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Pandey R.P., Parajuli P., Sohng J.K. Metabolic engineering of glycosylated polyketide biosynthesis. Emerg Top Life Sci. 2018;2:389–403. doi: 10.1042/ETLS20180011. [DOI] [PubMed] [Google Scholar]
- 32.Lee H., Raskin I. Purification, cloning, and expression of a pathogen inducible UDP-glucose: salicylic acid glucosyltransferase from tobacco. J Biol Chem. 1999;274:36637–36642. doi: 10.1074/jbc.274.51.36637. [DOI] [PubMed] [Google Scholar]
- 33.Mittasch J., et al. Identification of UGT84A13 as a candidate enzyme for the first committed step of gallotannin biosynthesis in pedunculate oak (Quercus robur) Phytochemistry. 2014;99:44–51. doi: 10.1016/j.phytochem.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 34.Ma L.Q., et al. Molecular cloning and overexpression of a novel UDP-glucosyltransferase elevating salidroside levels in Rhodiola sachalinensis. Plant Cell Rep. 2007;26:989–999. doi: 10.1007/s00299-007-0317-8. [DOI] [PubMed] [Google Scholar]
- 35.de Costa F., et al. Molecular cloning of an ester-forming triterpenoid: UDP-glucose 28-O-glucosyltransferase involved in saponin biosynthesis from the medicinal plant Centella asiatica. Plant Sci. 2017;262:9–17. doi: 10.1016/j.plantsci.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Modolo L.V., et al. A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol. 2007;64:499–518. doi: 10.1007/s11103-007-9167-6. [DOI] [PubMed] [Google Scholar]
- 37.Peng M., et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat Commun. 2017;8:1975. doi: 10.1038/s41467-017-02168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Achnine L., et al. Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 2005;41:875–887. doi: 10.1111/j.1365-313X.2005.02344.x. [DOI] [PubMed] [Google Scholar]
- 39.Chen K., et al. Diversity of O-glycosyltransferases contributes to the biosynthesis of flavonoid and triterpenoid glycosides in Glycyrrhiza uralensis. ACS Synth Biol. 2019;8:1858–1866. doi: 10.1021/acssynbio.9b00171. [DOI] [PubMed] [Google Scholar]
- 40.Xie K.B., et al. Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius. Org Lett. 2014;16:4874–4877. doi: 10.1021/ol502380p. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki H., et al. Identification and characterization of an Ipomoea nil glucosyltransferase which metabolizes some phytohormones. Biochem Biophys Res Commun. 2007;361:980–986. doi: 10.1016/j.bbrc.2007.07.147. [DOI] [PubMed] [Google Scholar]
- 42.Maharjan R., et al. An ambidextrous polyphenol glycosyltransferase PaGT2 from Phytolacca americana. Biochemistry. 2020;59:2551–2561. doi: 10.1021/acs.biochem.0c00224. [DOI] [PubMed] [Google Scholar]
- 43.Kohara A., et al. A novel glucosyltransferase involved in steroid saponin biosynthesis in Solanum aculeatissimum. Plant Mol Biol. 2005;57:225–239. doi: 10.1007/s11103-004-7204-2. [DOI] [PubMed] [Google Scholar]
- 44.Salcedo R.G., et al. Elucidation of the glycosylation steps during biosynthesis of antitumor macrolides PM100117 and PM100118 and engineering for novel derivatives. Microb Cell Factories. 2016;15:187. doi: 10.1186/s12934-016-0591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louveau T., et al. Analysis of two new arabinosyltransferases belonging to the Carbohydrate-Active Enzyme (CAZY) glycosyl transferase family1 provides insights into disease resistance and sugar donor specificity. Plant Cell. 2018;30:3038–3057. doi: 10.1105/tpc.18.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lalitha K., et al. Recent developments in β-C-glycosides: synthesis and applications. Carbohydr Res. 2015;402:158–171. doi: 10.1016/j.carres.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 47.Thibodeaux C.J., Melancon C.E., Liu H.W. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 48.Zhang M., et al. Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra. J Am Chem Soc. 2020;142:3506–3512. doi: 10.1021/jacs.9b12211. [DOI] [PubMed] [Google Scholar]
- 49.He J.B., et al. Molecular and structural characterization of a promiscuous C-glycosyltransferase from Trollius chinensis. Angew Chem Int Ed Engl. 2019;58:11513–11520. doi: 10.1002/anie.201905505. [DOI] [PubMed] [Google Scholar]
- 50.Grubb C.D., et al. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 2004;40:893–908. doi: 10.1111/j.1365-313X.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- 51.Ferreyra M.L.F., et al. Identification of a bifunctional maize C- and O-Glucosyltransferase. J Biol Chem. 2013;288:31678–31688. doi: 10.1074/jbc.M113.510040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun L., et al. Exploring the aglycon promiscuity of a new glycosyltransferase from Pueraria lobata. Tetrahedron Lett. 2016;57:1518–1521. [Google Scholar]
- 53.Yi S.Y., et al. Discovery and characterization of four glycosyltransferases involved in anthraquinone glycoside biosynthesis in Rubia yunnanensis. Org Chem Front. 2020;7:2442–2448. [Google Scholar]
- 54.Samuels D.C., et al. Finding the lost treasures in exome sequencing data. Trends Genet. 2013;29:593–599. doi: 10.1016/j.tig.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo Y., et al. Large scale comparison of gene expression levels by microarrays and RNAseq Using TCGA data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinger M.E., et al. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Briefings Funct Genomics Proteomics. 2009;8:407–423. doi: 10.1093/bfgp/elp038. [DOI] [PubMed] [Google Scholar]
- 57.Saliba A.E., et al. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42:8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jain M., et al. De novo transcriptome assembly and comprehensive expression profiling in Crocus sativus to gain insights into apocarotenoid biosynthesis. Sci Rep. 2016;6:22456. doi: 10.1038/srep22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y., et al. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.) BMC Plant Biol. 2018;18:67. doi: 10.1186/s12870-018-1286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shang J.Z., et al. The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol. 2020;21:200. doi: 10.1186/s13059-020-02088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X.Y., et al. The chromosome-level Stevia genome provides insights into steviol glycoside biosynthesis. Hortic Res. 2021;8:129. doi: 10.1038/s41438-021-00565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hughes J., Hughes M.A. Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq. 1994;5:41–49. doi: 10.3109/10425179409039703. [DOI] [PubMed] [Google Scholar]
- 63.Vogt T., Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5:380–386. doi: 10.1016/s1360-1385(00)01720-9. [DOI] [PubMed] [Google Scholar]
- 64.Rehman H.M., et al. Genome-wide analysis of Family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J Plant Physiol. 2016;206:87–97. doi: 10.1016/j.jplph.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 65.Barvkar V.T., et al. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genom. 2012;13:175. doi: 10.1186/1471-2164-13-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Y.J., et al. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays) Planta. 2014;239:1265–1279. doi: 10.1007/s00425-014-2050-1. [DOI] [PubMed] [Google Scholar]
- 67.Huang J., et al. Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol Genet Genom. 2015;290:1805–1818. doi: 10.1007/s00438-015-1040-8. [DOI] [PubMed] [Google Scholar]
- 68.Schmutz J., et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet. 2014;46:707–713. doi: 10.1038/ng.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kellner F., et al. Genome-guided investigation of plant natural product biosynthesis. Plant J. 2015;82:680–692. doi: 10.1111/tpj.12827. [DOI] [PubMed] [Google Scholar]
- 70.Bateman A., et al. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen L., et al. Methyl salicylate glucosylation regulates plant defense signaling and systemic acquired resistance. Plant Physiol. 2019;180:2167–2181. doi: 10.1104/pp.19.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu F., et al. Functional genomics analysis reveals two novel genes required for littorine biosynthesis. New Phytol. 2020;225:1906–1914. doi: 10.1111/nph.16317. [DOI] [PubMed] [Google Scholar]
- 73.Ono N.N., et al. Two UGT84 family glycosyltransferases catalyze a critical reaction of hydrolyzable tannin biosynthesis in pomegranate (Punica granatum) PLoS One. 2016;11 doi: 10.1371/journal.pone.0156319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yahyaa M., et al. Identification and characterization of UDP-glucose: phloretin 4'-O-glycosyltransferase from Malus x domestica Borkh. Phytochemistry. 2016;130:47–55. doi: 10.1016/j.phytochem.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 75.Xu Z.S., et al. Identification and characterization of DcUCGalT1, a galactosyltransferase responsible for anthocyanin galactosylation in purple carrot (Daucus carota L.) taproots. Sci Rep. 2016;6:27356. doi: 10.1038/srep27356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Z.L., et al. Dissection of the general two-step di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants. Proc Natl Acad Sci U S A. 2020;117:30816–30823. doi: 10.1073/pnas.2012745117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin Q.G., et al. Involvement of three putative glucosyltransferases from the UGT72 family in flavonol glucoside/rhamnoside biosynthesis in Lotus japonicus seeds. J Exp Bot. 2017;68:594–609. doi: 10.1093/jxb/erw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang T.J., et al. Purification and characterization of a novel phloretin-2'-O-glycosyltransferase favoring phloridzin biosynthesis. Sci Rep. 2016;6:35274. doi: 10.1038/srep35274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yahyaa M., et al. Identification and characterization of UDP-glucose: phloretin 4'-O-glycosyltransferase from Malus x domestica Borkh. Phytochemistry. 2016;130:47–55. doi: 10.1016/j.phytochem.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 80.Kai F., et al. Isolation, purification, and characterization of AgUCGalT1, a galactosyltransferase involved in anthocyanin galactosylation in purple celery (Apium graveolens L.) Planta. 2018;247:1363–1375. doi: 10.1007/s00425-018-2870-5. [DOI] [PubMed] [Google Scholar]
- 81.Chen X., et al. Tandem UGT71B5s catalyze lignan glycosylation in Isatis indigotica with substrates promiscuity. Front Plant Sci. 2021;12:637695. doi: 10.3389/fpls.2021.637695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smehilova M., et al. Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front Plant Sci. 2016;7:1264. doi: 10.3389/fpls.2016.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen L., et al. Overexpression of the UGT76E12 gene modulates seed germination, growth, and response to NaCl, mannitol, and abscisic acid. Biol Plant (Prague) 2019;63:328–334. [Google Scholar]
- 84.Sun Y.W., et al. Diterpenoid UDP-glycosyltransferases from Chinese sweet tea and Ashitaba complete the biosynthesis of rubusoside. Mol Plant. 2018;11:1308–1311. doi: 10.1016/j.molp.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 85.Jiang X.L., et al. Four flavonoid glycosyltransferases present in tea overexpressed in model plants Arabidopsis thaliana and Nicotiana tabacum for functional identification. J Chromatogr B. 2018;1100:148–157. doi: 10.1016/j.jchromb.2018.09.033. [DOI] [PubMed] [Google Scholar]
- 86.Mashima K., et al. Identification and characterization of apigenin 6-C-glucosyltransferase involved in biosynthesis of isosaponarin in wasabi (Eutrema japonicum) Plant Cell Physiol. 2019;60:2733–2743. doi: 10.1093/pcp/pcz164. [DOI] [PubMed] [Google Scholar]
- 87.Zhang M., et al. Functional characterization and structural basis of an efficient di-C-glycosyltransferase from Glycyrrhiza glabra. J Am Chem Soc. 2020;142:3506–3512. doi: 10.1021/jacs.9b12211. [DOI] [PubMed] [Google Scholar]
- 88.Wang D., et al. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab Eng. 2020;61:131–140. doi: 10.1016/j.ymben.2020.05.007. [DOI] [PubMed] [Google Scholar]
- 89.Haroth S., et al. The glycosyltransferase UGT76E1 significantly contributes to 12-O-glucopyranosyl-jasmonic acid formation in wounded Arabidopsis thaliana leaves. J Biol Chem. 2019;294:9858–9872. doi: 10.1074/jbc.RA119.007600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y., et al. Functional characterization of three flavonoid glycosyltransferases from Andrographis paniculata. R Soc Open Sci. 2019;6:190150. doi: 10.1098/rsos.190150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Irmisch S., et al. Biosynthesis of the anti-diabetic metabolite montbretin A: glucosylation of the central intermediate mini-MbA. Plant J. 2019;100:879–891. doi: 10.1111/tpj.14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Z.L., et al. Highly promiscuous flavonoid 3-O-glycosyltransferase from Scutellaria baicalensis. Org Lett. 2019;21:2241–2245. doi: 10.1021/acs.orglett.9b00524. [DOI] [PubMed] [Google Scholar]
- 93.Wilson A.E., Wu S., Tian L. PgUGT95B2 preferentially metabolizes flavones/flavonols and has evolved independently from flavone/flavonol UGTs identified in Arabidopsis thaliana. Phytochemistry. 2019;157:184–193. doi: 10.1016/j.phytochem.2018.10.025. [DOI] [PubMed] [Google Scholar]
- 94.Yuan S., et al. Isolation and characterization of a multifunctional flavonoid glycosyltransferase from Ornithogalum caudatum with glycosidase activity. Sci Rep. 2018;8:5886. doi: 10.1038/s41598-018-24277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu C., Zhao S.J., Wang X.S. Functional regulation of a UDP-glucosyltransferase gene (Pq3-O-UGT1) by RNA interference and overexpression in Panax quinquefolius. Plant Cell Tiss Org. 2017;129:445–456. [Google Scholar]
- 96.Lu C., et al. Functional regulation of ginsenoside biosynthesis by RNA interferences of a UDP-glycosyltransferase gene in Panax ginseng and Panax quinquefolius. Plant Physiol Biochem. 2017;111:67–76. doi: 10.1016/j.plaphy.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 97.Lin J.S., et al. UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 2016;88:26–42. doi: 10.1111/tpj.13229. [DOI] [PubMed] [Google Scholar]
- 98.He J.B., et al. UGT73F17, a new glycosyltransferase from Glycyrrhiza uralensis, catalyzes the regiospecific glycosylation of pentacyclic triterpenoids. Chem Commun. 2018;54:8594–8597. doi: 10.1039/c8cc04215b. [DOI] [PubMed] [Google Scholar]
- 99.Feng K.P., et al. A regiospecific rhamnosyltransferase from Epimedium pseudowushanense catalyzes the 3-O-rhamnosylation of prenylflavonols. Org Biomol Chem. 2018;16:452–458. doi: 10.1039/c7ob02763j. [DOI] [PubMed] [Google Scholar]
- 100.Li Q., et al. Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis. Plant Biol. 2018;20:10–19. doi: 10.1111/plb.12627. [DOI] [PubMed] [Google Scholar]
- 101.Yan X., et al. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carvalho D.R.A., et al. Gene expression and physiological responses associated to stomatal functioning in Rosa x hybrida grown at high relative air humidity. Plant Sci. 2016;253:154–163. doi: 10.1016/j.plantsci.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 103.Ito T., et al. C-Glycosyltransferases catalyzing the formation of di-C-glucosyl flavonoids in citrus plants. Plant J. 2017;91:187–198. doi: 10.1111/tpj.13555. [DOI] [PubMed] [Google Scholar]
- 104.Xu X.P., et al. Molecular cloning and biochemical characterization of a new coumarin glycosyltransferase CtUGT1 from Cistanche tubulosa. Fitoterapia. 2021;153:104995. doi: 10.1016/j.fitote.2021.104995. [DOI] [PubMed] [Google Scholar]
- 105.Gao Y.N., et al. Galactosylation of monosaccharide derivatives of glycyrrhetinic acid by UDP-glycosyltransferase GmSGT2 from Glycine max. J Agric Food Chem. 2020;68:8580–8588. doi: 10.1021/acs.jafc.0c03842. [DOI] [PubMed] [Google Scholar]
- 106.Song C.K., et al. A UDP-glucosyltransferase functions in both acylphloroglucinol glucoside and anthocyanin biosynthesis in strawberry (Fragaria x ananassa) Plant J. 2016;85:730–742. doi: 10.1111/tpj.13140. [DOI] [PubMed] [Google Scholar]
- 107.Feng P.C., et al. Identification and RNAi-based gene silencing of a novel UDP-glycosyltransferase from Panax quinquefolius. Plant Cell Tissue Organ Cult. 2021;144:567–576. [Google Scholar]
- 108.Wang H.M., et al. Engineering Saccharomyces cerevisiae with the deletion of endogenous glucosidases for the production of flavonoid glucosides. Microb Cell Factories. 2016;15:134. doi: 10.1186/s12934-016-0535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Itkin M., et al. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc Natl Acad Sci U S A. 2016;113:E7619–E7628. doi: 10.1073/pnas.1604828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Owatworakit A., et al. Glycosyltransferases from oat (Avena) implicated in the acylation of avenacins. J Biol Chem. 2013;288:3696–3704. doi: 10.1074/jbc.M112.426155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Louveau T., et al. Analysis of two new arabinosyltransferases belonging to the carbohydrate-active enzyme (CAZY) glycosyl transferase family1 provides insights into disease resistance and sugar donor specificity. Plant Cell. 2018;30:3038–3057. doi: 10.1105/tpc.18.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lowe R., et al. Transcriptomics technologies. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sarangi B.K., Minami Y., Thul S.T. RNA-seq analysis for indigo biosynthesis pathway genes in Indigofera tinctoria and Polygonum tinctorium. Genom Data. 2015;6:212–213. doi: 10.1016/j.gdata.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang J., et al. Functional characterization of a novel glycosyltransferase (UGT73CD1) from Iris tectorum Maxim. for the substrate promiscuity. Mol Biotechnol. 2021;63:1030–1039. doi: 10.1007/s12033-021-00364-1. [DOI] [PubMed] [Google Scholar]
- 115.Jayakodi M., et al. Comprehensive analysis of Panax ginseng root transcriptomes. BMC Plant Biol. 2015;15:138. doi: 10.1186/s12870-015-0527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen J.W., et al. RNA-seq for gene identification and transcript profiling of three Stevia rebaudiana genotypes. BMC Genom. 2014;15:571. doi: 10.1186/1471-2164-15-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li Z.Y., et al. Comparative transcriptome analysis of Anthurium “Albama” and its anthocyanin-loss mutant. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fan J.T., et al. Biologically active arborinane-type triterpenoids and anthraquinones from Rubia yunnanensis. J Nat Prod. 2011;74:2069–2080. doi: 10.1021/np2002918. [DOI] [PubMed] [Google Scholar]
- 119.Chandramouli K., Qian P.Y. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genom proteomics. 2009;2009:239204. doi: 10.4061/2009/239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Suliman M., et al. Identification of glycosyltransferases involved in cell wall synthesis of wheat endosperm. J Proteonomics. 2013;78:508–521. doi: 10.1016/j.jprot.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 121.Inoue S., et al. Characterization of UDP-glucosyltransferase from Indigofera tinctoria. Plant Physiol Biochem. 2017;121:226–233. doi: 10.1016/j.plaphy.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 122.de Rond T., et al. A high-throughput mass spectrometric enzyme activity assay enabling the discovery of cytochrome P450 biocatalysts. Angew Chem Int Ed. 2019;58:10114–10119. doi: 10.1002/anie.201901782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Engel L., et al. Utility of bioluminescent homogeneous nucleotide detection assays in measuring activities of nucleotide-sugar dependent glycosyltransferases and studying their inhibitors. Molecules. 2021;26:6230. doi: 10.3390/molecules26206230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Taujale R., et al. Mapping the glycosyltransferase fold landscape using interpretable deep learning. Nat Commun. 2021;12:5656. doi: 10.1038/s41467-021-25975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Albuquerque-Wendt A., et al. Membrane topological model of glycosyltransferases of the GT-C superfamily. Int J Mol Sci. 2019;20:4842. doi: 10.3390/ijms20194842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang M., et al. Functional and informatics analysis enables glycosyltransferase activity prediction. Nat Chem Biol. 2018;14:1109–1117. doi: 10.1038/s41589-018-0154-9. [DOI] [PubMed] [Google Scholar]
- 127.Packer M.S., Liu D.R. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 128.Williams G.J., Thorson J.S. A high-throughput fluorescence-based glycosyltransferase screen and its application in directed evolution. Nat Protoc. 2008;3:357–362. doi: 10.1038/nprot.2007.538. [DOI] [PubMed] [Google Scholar]
- 129.Liu B., et al. Structural and biochemical studies of the glycosyltransferase Bs-YjiC from Bacillus subtilis. Int J Biol Macromol. 2021;166:806–817. doi: 10.1016/j.ijbiomac.2020.10.238. [DOI] [PubMed] [Google Scholar]
- 130.Brazier-Hicks M., et al. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci U S A. 2007;104:20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thompson A.M.G., et al. Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci Rep. 2017;7:46629. doi: 10.1038/srep46629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zong G.N., et al. Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-β-l-rhamnose and rhamnosylation mechanism. Plant J. 2019;99:257–269. doi: 10.1111/tpj.14321. [DOI] [PubMed] [Google Scholar]
- 133.Hiromoto T., et al. Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. J Synchrotron Radiat. 2013;20:894–898. doi: 10.1107/S0909049513020712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hiromoto T., et al. Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. Protein Sci. 2015;24:395–407. doi: 10.1002/pro.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu M., et al. Crystal structures of the C-glycosyltransferase UGT708C1 from buckwheat provide insights into the mechanism of C-glycosylation. Plant Cell. 2020;32:2917–2931. doi: 10.1105/tpc.20.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nomura Y., et al. Functional specialization of UDP-glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J. 2019;99:1127–1143. doi: 10.1111/tpj.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li L.N., et al. Crystal structure of Medicago truncatula UGT85H2 - insights into the structural basis of a multifunctional (Iso)flavonoid glycosyltransferase. J Mol Biol. 2007;370:951–963. doi: 10.1016/j.jmb.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 138.Modolo L.V., et al. Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (Iso)flavonoids. J Mol Biol. 2009;392:1292–1302. doi: 10.1016/j.jmb.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 139.Wetterhorn K.M., et al. Crystal structure of Os79 (Os04g0206600) from Oryza sativa: a UDP-glucosyltransferase involved in the detoxification of deoxynivalenol. Biochemistry. 2016;55:6175–6186. doi: 10.1021/acs.biochem.6b00709. [DOI] [PubMed] [Google Scholar]
- 140.Hsu T.M., et al. Employing a biochemical protecting group for a sustainable indigo dyeing strategy. Nat Chem Biol. 2018;14(3):256–261. doi: 10.1038/nchembio.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maharjan R., et al. Crown-ether-mediated crystal structures of the glycosyltransferase PaGT3 from Phytolacca americana. Acta Crystallogr D Struct Biol. 2020;76:521–530. doi: 10.1107/S2059798320005306. [DOI] [PubMed] [Google Scholar]
- 142.Chen L.Q., Zhang Y., Feng Y. Structural dissection of sterol glycosyltransferase UGT51 from Saccharomyces cerevisiae for substrate specificity. J Struct Biol. 2018;204:371–379. doi: 10.1016/j.jsb.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 143.Zhuang Y., et al. Biosynthesis of plant-derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme. Metab Eng. 2017;42:25–32. doi: 10.1016/j.ymben.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 144.Li J., et al. Efficient O-glycosylation of triterpenes enabled by protein engineering of plant glycosyltransferase UGT74AC1. ACS Catal. 2020;10:3629–3639. [Google Scholar]
- 145.Li J., et al. Near-perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases. Green Syn Catal. 2021;2:45–53. [Google Scholar]
- 146.Yang T., et al. Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations. Nat Commun. 2019;10:3214. doi: 10.1038/s41467-019-11154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Offen W., et al. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006;25:1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang P.P., et al. Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency. Cell Discov. 2019;5:5. doi: 10.1038/s41421-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma W., et al. Oriented efficient biosynthesis of rare ginsenoside Rh2 from PPD by compiling UGT-Yjic mutant with sucrose synthase. Int J Biol Macromol. 2019;146:853–859. doi: 10.1016/j.ijbiomac.2019.09.208. [DOI] [PubMed] [Google Scholar]
- 150.Tan Y.M., et al. Directed evolution of an α1,3-fucosyltransferase using a single-cell ultrahigh-throughput screening method. Sci Adv. 2019;5 doi: 10.1126/sciadv.aaw8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ma F.Q., et al. Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform. Nat Commun. 2018;9:1030. doi: 10.1038/s41467-018-03492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Flitsch S.L. Chemical and enzymatic synthesis of glycopolymers. Curr Opin Chem Biol. 2000;4:619–625. doi: 10.1016/s1367-5931(00)00152-6. [DOI] [PubMed] [Google Scholar]
- 153.Wang J.B., Li G.Y., Reetz M.T. Enzymatic site-selectivity enabled by structure-guided directed evolution. Chem Commun. 2017;53:3916–3928. doi: 10.1039/c7cc00368d. [DOI] [PubMed] [Google Scholar]
- 154.Fan B., et al. Switching glycosyltransferase UGT(BL)1 regioselectivity toward polydatin synthesis using a semi-rational design. Org Biomol Chem. 2018;16:2464–2469. doi: 10.1039/C8OB00376A. [DOI] [PubMed] [Google Scholar]
- 155.Wilson S.A., Roberts S.C. Metabolic engineering approaches for production of biochemicals in food and medicinal plants. Curr Opin Biotechnol. 2014;26:174–182. doi: 10.1016/j.copbio.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 156.Paddon C.J., et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 157.Wang Q., et al. Metabolic engineering of terpene biosynthesis in plants using a trichome-specific transcription factor MsYABBY5 from spearmint (Mentha spicata) Plant Biotechnol J. 2016;14:1619–1632. doi: 10.1111/pbi.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Smanski M.J., et al. Synthetic biology to access and expand nature's chemical diversity. Nat Rev Microbiol. 2016;14:135–149. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lim C.G., et al. Development of a recombinant Escherichia coli strain for overproduction of the plant pigment anthocyanin. Appl Environ Microbiol. 2015;81:6276–6284. doi: 10.1128/AEM.01448-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kim S.Y., et al. Metabolic engineering of Escherichia coli for the biosynthesis of flavonoid-O-glucuronides and flavonoid-O-galactoside. Appl Microbiol Biotechnol. 2015;99:2233–2242. doi: 10.1007/s00253-014-6282-6. [DOI] [PubMed] [Google Scholar]
- 161.Parajuli P., et al. Synthetic sugar cassettes for the efficient production of flavonol glycosides in Escherichia coli. Microb Cell Factories. 2015;14:76. doi: 10.1186/s12934-015-0261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.De Bruyn F., et al. Metabolic engineering of Escherichia coli into a versatile glycosylation platform: production of bio-active quercetin glycosides. Microb Cell Factories. 2015;14:138. doi: 10.1186/s12934-015-0326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pei J.J., et al. Modulating heterologous pathways and optimizing fermentation conditions for biosynthesis of kaempferol and astragalin from naringenin in Escherichia coli. J Ind Microbiol Biotechnol. 2019;46:171–186. doi: 10.1007/s10295-018-02134-6. [DOI] [PubMed] [Google Scholar]
- 164.Li Y., et al. Production of rebaudioside A from stevioside catalyzed by the engineered Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2016;178:1586–1598. doi: 10.1007/s12010-015-1969-4. [DOI] [PubMed] [Google Scholar]
- 165.Xue F.Y., et al. Expression of codon-optimized plant glycosyltransferase UGT72B14 in Escherichia coli enhances salidroside production. BioMed Res Int. 2016;2016:9845927. doi: 10.1155/2016/9845927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wei W., et al. Characterization of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts. Mol Plant. 2015;8:1412–1424. doi: 10.1016/j.molp.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 167.Liang H.C., et al. Production of a bioactive unnatural ginsenoside by metabolically engineered yeasts based on a new UDP-glycosyltransferase from Bacillus subtilis. Metab Eng. 2017;44:60–69. doi: 10.1016/j.ymben.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 168.Hu Z.F., et al. Construction and optimization of microbial cell factories for sustainable production of bioactive dammarenediol-II glucosides. Green Chem. 2019;21:3286–3299. [Google Scholar]
- 169.Li D.S., et al. Production of triterpene ginsenoside compound K in the non-conventional yeast Yarrowia lipolytica. J Agric Food Chem. 2019;67:2581–2588. doi: 10.1021/acs.jafc.9b00009. [DOI] [PubMed] [Google Scholar]
- 170.Wang P.P., et al. Systematic optimization of the yeast cell factory for sustainable and high efficiency production of bioactive ginsenoside compound K. Synth Syst Biotechnol. 2021;6:69–76. doi: 10.1016/j.synbio.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li X.D., et al. High-level sustainable production of the characteristic protopanaxatriol-type saponins from Panax species in engineered Saccharomyces cerevisiae. Metab Eng. 2021;66:87–97. doi: 10.1016/j.ymben.2021.04.006. [DOI] [PubMed] [Google Scholar]
- 172.Wang W., et al. Construction of Escherichia coli cell factories for crocin biosynthesis. Microb Cell Factories. 2019;18:120. doi: 10.1186/s12934-019-1166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pei J.J., et al. Metabolic engineering of Escherichia coli for astragalin biosynthesis. J Agric Food Chem. 2016;64:7966–7972. doi: 10.1021/acs.jafc.6b03447. [DOI] [PubMed] [Google Scholar]
- 174.Priebe X., et al. Byproduct-free geraniol glycosylation by whole-cell biotransformation with recombinant Escherichia coli. Biotechnol Lett. 2021;43:247–259. doi: 10.1007/s10529-020-02993-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xu K., Zhao Y.-J., Ahmad N., et al. O-glycosyltransferases from Homo sapiens contributes to the biosynthesis of glycyrrhetic acid 3-O-mono-β-D-glucuronide and glycyrrhizin in Saccharomyces cerevisiae. Synth Syst Biotechnol. 2021;6:173–179. doi: 10.1016/j.synbio.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liu T., et al. De novo biosynthesis of polydatin in Saccharomyces cerevisiae. J Agric Food Chem. 2021;69:5917–5925. doi: 10.1021/acs.jafc.1c01557. [DOI] [PubMed] [Google Scholar]
- 177.Veitch N.C., Grayer R.J. Flavonoids and their glycosides, including anthocyanins. Nat Prod Rep. 2011;28:1626–1695. doi: 10.1039/c1np00044f. [DOI] [PubMed] [Google Scholar]
- 178.Juergenliemk G., et al. In vitro studies indicate that miquelianin (quercetin 3-O-β-D-glucuronopyranoside) is able to reach the CNS from the small intestine. Planta Med. 2003;69:1013–1017. doi: 10.1055/s-2003-45148. [DOI] [PubMed] [Google Scholar]
- 179.Soundararajan R., et al. Quercetin 3-glucoside protects neuroblastoma (SH-SY5Y) cells in vitro against oxidative damage by inducing sterol regulatory element-binding protein-2-mediated cholesterol biosynthesis. J Biol Chem. 2008;283:2231–2245. doi: 10.1074/jbc.M703583200. [DOI] [PubMed] [Google Scholar]
- 180.Carakostas M.C., et al. Overview: the history, technical function and safety of rebaudioside A, a naturally occurring steviol glycoside, for use in food and beverages. Food Chem Toxicol. 2008;46:S1–S10. doi: 10.1016/j.fct.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 181.Wu X.L., et al. The non-cytotoxicity characterization of rebaudioside A as a food additive. Food Chem Toxicol. 2014;66:334–340. doi: 10.1016/j.fct.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 182.Christensen L.P. Ginsenosides: chemistry, biosynthesis, analysis, and potential health effects. Adv Food Nutr Res. 2009;55:1–99. doi: 10.1016/S1043-4526(08)00401-4. [DOI] [PubMed] [Google Scholar]
- 183.Chiang H.M., et al. Rhodiola plants: chemistry and biological activity. J Food Drug Anal. 2015;23:359–369. doi: 10.1016/j.jfda.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang C.M., et al. Polydatin induces apoptosis and inhibits growth of acute monocytic leukemia cells. J Biochem Mol Toxicol. 2016;30:200–205. doi: 10.1002/jbt.21779. [DOI] [PubMed] [Google Scholar]
- 185.Wang H.L., et al. Comparative studies of polydatin and resveratrol on mutual transformation and antioxidative effect in vivo. Phytomedicine. 2015;22:553–559. doi: 10.1016/j.phymed.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 186.Lanzilli G., et al. Anti-inflammatory effect of resveratrol and polydatin by in vitro IL-17 modulation. Inflammation. 2012;35:240–248. doi: 10.1007/s10753-011-9310-z. [DOI] [PubMed] [Google Scholar]
- 187.Zhang C.Z., et al. Production of terpenoids by synthetic biology approaches. Front Bioeng Biotechnol. 2020;8:347. doi: 10.3389/fbioe.2020.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tunyasuvunakool K., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pennisi E. Protein structure prediction now easier, faster. Science. 2021;373:262–263. doi: 10.1126/science.373.6552.262. [DOI] [PubMed] [Google Scholar]