Take Home Message
The results from this study indicate that among patients with prostate cancer undergoing active surveillance, those with higher Decipher scores were more likely to have higher-grade disease found over time. These findings indicate that the Decipher test might be useful for guiding the intensity of monitoring during active surveillance, such as more frequent biopsy for patients with higher scores.
Keywords: Prostate cancer, Active surveillance, Decipher
Abstract
Background
Although the Decipher genomic classifier has been validated as a prognostic tool for several prostate cancer endpoints, little is known about its role in assessing the risk of biopsy reclassification for patients on active surveillance, a key event that often triggers treatment.
Objective
To evaluate the association between Decipher genomic classifier scores and biopsy Gleason upgrading among patients on active surveillance.
Design, setting, and participants
This was a retrospective cohort study among patients with low- and favorable intermediate-risk prostate cancer on active surveillance who underwent biopsy-based Decipher testing as part of their clinical care.
Outcome measurements and statistical analysis
We evaluated the association between the Decipher score and any increase in biopsy Gleason grade group (GG) using univariable and multivariable logistic regression. We compared the area under the receiver operating characteristic curve (AUC) for models comprising baseline clinical variables with or without the Decipher score.
Results and limitations
We identified 133 patients for inclusion with a median age of 67.7 yr and median prostate-specific of 5.6 ng/ml. At enrollment, 75.9% had GG1 and 24.1% had GG2 disease. Forty-three patients experienced biopsy upgrading. On multivariable logistic regression, the Decipher score was significantly associated with biopsy upgrading (odds ratio 1.37 per 0.10 unit increase, 95% confidence interval [CI] 1.05–1.79; p = 0.02). The Decipher score was associated with upgrading among patients with biopsy GG 1 disease, but not GG2 disease. The discriminative ability of a clinical model (AUC 0.63, 95% CI 0.51–0.74) was improved by integration of the Decipher score (AUC 0.69, 95% CI 0.58–0.80).
Conclusions
The Decipher genomic classifier score was associated with short-term biopsy Gleason upgrading among patients on active surveillance.
Patient summary
The results from this study indicate that among patients with prostate cancer undergoing active surveillance, those with higher Decipher scores were more likely to have higher-grade disease found over time. These findings indicate that the Decipher test might be useful for guiding the intensity of monitoring during active surveillance, such as more frequent biopsy for patients with higher scores.
1. Introduction
Active surveillance is the recommended initial management strategy for most patients with low-grade prostate cancer and an option for selected patients with favorable intermediate-risk disease and is now adopted by the majority of eligible patients [1]. Evidence from randomized trials and institutional cohort studies supports the long-term safety of active surveillance and its effectiveness as a strategy to avoid or defer definitive treatment [2], [3]. Nonetheless, 20–60% of patients who are initially enrolled in active surveillance ultimately experience reclassification of their disease according to changes in biopsy Gleason grade, prostate-specific antigen (PSA) levels, or cancer volume [4], [5]. As a result, up to half of patients undergo definitive treatment in the near term, most frequently because of Gleason upgrading [6]. A smaller number of patients with clinically low-risk features ultimately experience clinically significant progression over time, underscoring the need for close monitoring to detect early signs of reclassification [7]. Estimating the risk of disease reclassification during active surveillance on the basis of standard clinical parameters is imperfect, leading to patient anxiety, avoidable treatment, and imprecision in monitoring (eg, overuse or underuse of surveillance testing) [8], [9], [10].
Genomic classifiers measuring features associated with prostate cancer aggressiveness developed largely in patients with high-risk disease provide robust predictions of disease outcome, yet little is known about their role in estimating the trajectory of untreated favorable-risk prostate cancer [11]. The Decipher classifier (GenomeDx Biosciences, Vancouver, BC, Canada) is a tissue-based platform evaluating the expression of 22 genes selected from whole-transcriptome analysis and reflecting pathways involved in cellular proliferation, differentiation, immune modulation, and androgen-receptor signaling. The test has been widely validated as both a prognostic and a predictive marker associated with several clinical outcomes, including adverse pathology at prostatectomy, biochemical recurrence, metastasis, and prostate cancer mortality after treatment [12], [13]. However, less information is available regarding its utility in predicting the outcome for patients being managed with active surveillance. Such information would be useful as a means for tailoring the approach to clinical management, with potential for moderating surveillance protocols for those at lowest risk and for intensifying or foregoing surveillance for patients most likely to experience reclassification or disease progression [8].
In this study we evaluated the association between Decipher genomic classifier scores and biopsy outcomes among patients with favorable-risk prostate cancer managed with initial active surveillance [14]. Analytic and clinical validation of commercially available genomic tests were largely conducted using archival tissue obtained in the era before widespread use of prostate magnetic resonance imaging (MRI), an approach that significantly improves the accuracy of sampling and reduces the risk of initial misclassification [5], [15], [16]. Therefore, commensurate with current clinical practice, we conducted our study in a contemporary cohort of patients managed with active surveillance following a prostate biopsy guided by MRI-ultrasound fusion.
2. Patients and methods
2.1. Study design and patient selection
We performed a retrospective cohort study of patients enrolled on active surveillance for prostate cancer who underwent Decipher testing. We identified subjects from a prospectively maintained institutional repository of patients with known or suspected prostate cancer undergoing prostate MRI and prostate biopsy at a single tertiary care center. The primary study objective was to examine the association between a patient’s baseline Decipher score (scale 0–1.0 units) and Gleason upgrading during active surveillance, defined as an increase in Gleason grade group (GG) on subsequent biopsy. The secondary objectives were to evaluate the performance of clinical prediction models with or without the genomic classifier score, and to identify a clinical threshold for the Decipher score in predicting Gleason upgrading. In addition, we evaluated the association between Gleason upgrading and the clinically reported Decipher risk groups: low (score <0.45), intermediate (score 0.45–0.60), and high (score >0.60).
Of 1432 patients undergoing prostate MRI-ultrasound fusion biopsy, we identified 133 who elected for initial active surveillance and underwent at least one additional biopsy and Decipher testing between July 2016 and November 2020. Patients with low-risk prostate cancer (Gleason score ≤ 3 + 3, cT1 stage, PSA ≤10 ng/ml) and select patients with favorable intermediate-risk prostate cancer (Gleason score ≤ 3 + 4 with no more than 1 core with Gleason pattern 4, ≤cT2 stage, PSA 10–20 ng/ml) detected on combined systematic and MRI-ultrasound fusion targeted biopsy (MRF-TB) were enrolled in the active surveillance program and included in an institutional review board–approved prospective data registry. The institutional surveillance protocol consisted of semi-annual PSA testing, a confirmatory prostate biopsy within 1 yr of diagnosis, and subsequent prostate MRI and prostate biopsy on a yearly or biennial basis. Protocols for MRI and MRF-TB were as previously described [17]. Genomic testing was routinely offered to patients considering active surveillance without restriction according to disease characteristics. We compiled clinical, pathology, and sociodemographic information, including prostate MRI findings and the Decipher score.
2.2. Statistical analysis
We compiled clinicopathologic variables, Decipher scores, and biopsy upgrading status for each patient. Categorical variables are reported as the frequency and proportion; continuous variables are reported as the median and interquartile range (IQR). We used McNemar’s test for statistical analysis of proportions, and the Kruskal-Wallis test was for continuous variables. We constructed multivariable logistic regression models to evaluate the association between baseline characteristics, including the Decipher score, and biopsy Gleason upgrading. Variables that were significantly associated with upgrading on univariable analysis were included in the model, as well as a priori variables shown to be associated with Gleason upgrading in prior studies (age, PSA density, number of biopsy cores positive for cancer, and prostate MRI findings). We compared the performance of a baseline clinical model with the Decipher classifier alone and a combined model consisting of clinical parameters and Decipher score. We used Youden’s index to identify a potential threshold for the Decipher score that could be clinically used to identify patients at greater risk of reclassification during active surveillance. All statistical analyses were performed using SPSS v27 (IBM, Armonk, NY, USA).
3. Results
The study sample consisted of 133 patients initially managed with active surveillance who underwent Decipher testing. The median age at enrollment was 67.7 yr (IQR 62.4–71.4) and the median PSA at diagnosis was 5.6 ng/ml (IQR 4.3–7.1; Table 1). In this cohort, 66 men (49.6%) had Decipher testing performed on their initial diagnostic biopsy and 67 (50.4%) had testing on a subsequent biopsy. The biopsy Gleason grade at enrollment was GG1 for 75.9% and GG2 for 24.1%. The median interval between biopsies was 13.6 mo (IQR 11.9–16.9), and the median Decipher score was 0.39 (IQR 0.25–0.48). The Decipher risk group reported was low for 64.4%, intermediate for 25.3%, and high for 10.3% of patients. A change in prostate MRI PI-RADS score occurred in 41 patients (30.7%).
Table 1.
Baseline characteristics of patients with favorable-risk prostate cancer undergoing active surveillance and Decipher genomic testing
| Variable | Result |
|---|---|
| Median age, yr (IQR) | 67.7 (62.4–71.4) |
| Median body mass index, kg/m2 (IQR) | 27.1 (25.0–30.5) |
| Median prostate-specific antigen, ng/ml (IQR) | 5.6 (4.3–7.1) |
| Biopsy Gleason grade group 1, n (%) | 101 (75.9) |
| Biopsy Gleason grade group 2, n (%) | 32 (24.1) |
| Race/ethnicity, n (%) | |
| White | 120 (90.2) |
| Black/African American | 8 (6.0) |
| Latino | 3 (2.3) |
| Other | 2 (1.5) |
| Median prostate volume, ml (IQR) | 46.0 (32.6–59.0) |
| Median Decipher score (IQR) | 0.39 (0.39–0.48) |
| Decipher risk category, n (%) | |
| Low (score <0.45) | 94 (64.4) |
| Intermediate (score 0.45–0.60) | 37 (25.3) |
| High (score >0.60) | 15 (10.3) |
IQR = interquartile range.
In total, 43 patients (32.3%) experienced biopsy upgrading. The median Decipher score was 0.39 (IQR 0.25–0.46) among patients with upgrading and 0.41 (IQR 0.32–0.54) among those without upgrading (p = 0.06; Table 2). The distribution of upgrading events did not differ significantly by Decipher risk group (28.6% in the low-risk, 34.3% in the intermediate-risk, and 50.0% in the high-risk group; p = 0.27). On univariable analysis, increasing Decipher score was associated with greater odds of upgrading (odds ratio [OR] 1.24 per 0.10 unit; p = 0.045). When stratified by diagnostic Gleason grade group, the Decipher score was associated with upgrading among patients with GG1 (OR 1.29 per 0.10 unit; p = 0.047) but not among those with GG2 disease (p = 0.41). On multivariable logistic regression analysis, the Decipher score remained significantly associated with the odds of biopsy upgrading (OR 1.37 per 0.10 units; p = 0.02; Table 3).
Table 2.
Comparison of characteristics for patients who did and did not experience biopsy Gleason upgrading during active surveillance
| Variable | No biopsy upgrading (n = 90) |
Biopsy upgrading (n = 43) |
p value |
|---|---|---|---|
| Median age, yr (IQR) | 68.0 (62.4–70.9) | 66.3 (61.9–73.2) | 0.93 |
| Median PSA, ng/ml (IQR) | 5.6 (4.1–7.0) | 5.6 (4.4–7.2) | 0.97 |
| Median body mass index, kg/m2 (IQR) | 26.8 (24.7–30.4) | 28.1 (25.3–30.9) | 0.20 |
| Median PSA density, ng/ml/ml (IQR) | 0.12 (0.08–- .18) | 0.12 (0.08–0.16) | 0.69 |
| Median prostate volume, ml (IQR) | 45.0 (33.5–57.1) | 47.0 (31.0–60.0) | 0.81 |
| Decipher risk category, n (%) | 0.27 | ||
| Low (score <0.45) | 60 (66.7) | 24 (55.8) | |
| Intermediate (score 0.45–0.60) | 23 (25.6) | 12 (27.9) | |
| High (score >0.60) | 7 (7.8) | 7 (16.3) | |
| Median Decipher score (IQR) | 0.39 (0.25–0.46) | 0.41 (0.32– 0.54) | 0.06 |
| Median positive SBx cores, n (IQR) | 2 (1–4) | 3 (1–4) | 0.53 |
| PI-RADS score, n (%) | 0.57 | ||
| 1–2 | 22 (25.0) | 8 (18.6) | |
| 3 | 7 (7.9) | 5 (11.7) | |
| 4 | 39 (44.4) | 20 (46.5) | |
| 5 | 20 (22.7) | 10 (23.2) | |
| Increase in PI-RADS score, n (%) | 22 (28.2) | 10 (32.2) | 0.68 |
IQR = interquartile range; PI-RADS = Prostate Imaging-Reporting and Data System; PSA = prostate-specific antigen; SBx = systematic biopsy.
Table 3.
Multivariable logistic regression analysis of factors associated with biopsy Gleason upgrading during active surveillance
| Variable | Odds ratio (95% CI) | p value |
|---|---|---|
| Age | 1.02 (0.97–1.07) | 0.44 |
| Baseline PI-RADS score | 0.77 | |
| 1–2 | Reference | |
| 3 | 0.63 (0.14–2.82) | 0.55 |
| 4–5 | 1.02 (0.36–2.89) | 0.97 |
| Prostate-specific antigen density (per 0.1 unit) | 0.83 (0.50–1.44) | 0.52 |
| Decipher score (per 0.1 unit) | 1.37 (1.05–1.79) | 0.02 |
| Three or more positive systematic biopsy cores | 2.55 (1.04–6.29) | 0.04 |
CI = confidence interval; PI-RADS = Prostate Imaging-Reporting and Data System.
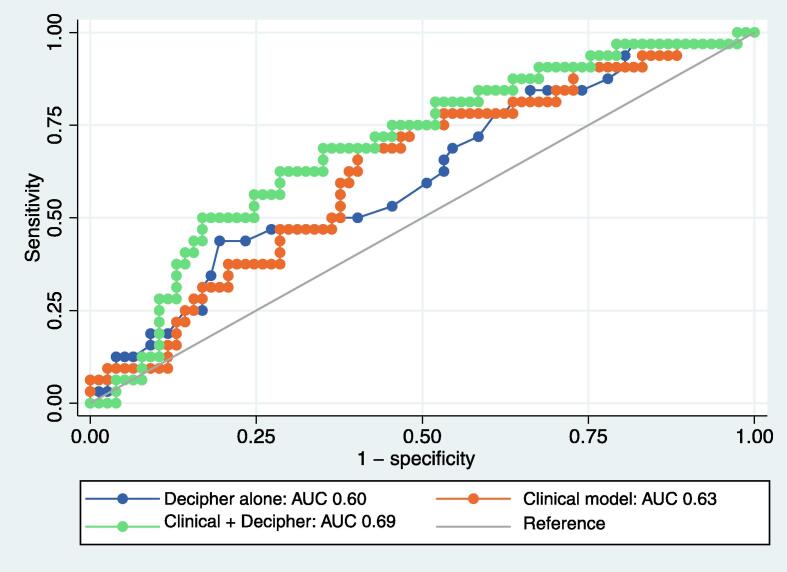
The baseline clinical model showed modest discrimination of biopsy upgrading (area under the receiver operating characteristic curve [AUC] 0.63, 95% CI 0.51–0.74). The AUC for Decipher alone was 0.60 (95% CI 0.49–0.70). A combined model including the Decipher score and clinical variables improved the AUC to 0.69 (95% CI 0.58–0.80; Fig. 1). A Decipher score cutoff of 0.475 maximized the sensitivity and specificity for prediction of biopsy upgrading while on AS. At a dichotomous threshold of 0.475, the sensitivity and specificity for biopsy upgrading were 41.9% and 78.9%, respectively, with modest discrimination for biopsy upgrading (AUC 0.60, 95% CI 0.52–0.69). Among patients with Decipher scores of <0.475 versus ≥0.475, the incidence of biopsy upgrading was 26.0% vs 48.6% (p = 0.02). On univariable analysis, Decipher scores ≥0.475 were associated with higher odds of biopsy upgrading (OR 2.69, 95% CI 1.22–5.92; p = 0.01). On multivariable logistic regression analysis, Decipher scores ≥0.475 were independently associated with higher odds of biopsy upgrading (OR 3.71, 95% CI 1.45–9.50; p = 0.01).
Fig. 1.
Discriminative performance of clinical models for prediction of biopsy upgrading during active surveillance with and without integration of the Decipher genomic score. AUC = area under the receiver operating characteristic curve.
4. Discussion
We found that the Decipher genomic classifier score was associated with subsequent biopsy upgrading among patients enrolled on active surveillance for low-risk or favorable intermediate-risk prostate cancer. In this contemporary cohort of patients undergoing prostate MRF-TB, the Decipher score stratified the risk of reclassification independently of clinical features, including PSA density and MRI findings. Integration of the Decipher score improved the discriminative performance of a model based on baseline clinical parameters and prostate MRI, although the overall performance remained modest. We further found that, according to the lower distribution of Decipher scores in the active surveillance population, reported risk groupings were not informative for predicting Gleason upgrading. As a result, distinct cut points or consideration of the classifier as a continuous measure of risk may have the most utility in active surveillance. These data provide novel quantitative information regarding the possibility of integrating this baseline genomic classifier information in clinical counseling.
Among patients with GG1 but not GG2 prostate cancer choosing active surveillance, Decipher scores were independently associated with Gleason upgrading on a subsequent biopsy. This additional predictive information may have greater utility for patients with low risk than for those with low to intermediate risk, who have a two- to fourfold higher risk of reclassification on the basis of clinical and pathology parameters [18], [19]. Although a large body of evidence has been accumulated on the associations between the Decipher score and clinical and pathology outcomes, there is little direct evidence concerning its short-term prognostic significance for patients managed with active surveillance. The findings from this study suggest that the Decipher classifier may be useful in identifying patients whose initial biopsies may have been misclassified or will experience progression of their disease in the short term. However, we did not identify a significant association between the Decipher score and biopsy upgrading among the subset of patients with GG2 disease. This may reflect the smaller sample size relative to GG1 and lower power for this comparison, the contributions of biopsy sampling leading by chance to detection of a higher proportion of Gleason pattern 4 disease, or the possibility that the Decipher score is indeed not associated with further biopsy upgrading in this group.
The setting of this study in the contemporary era of MRF-TB increases the generalizability of the results, as MRI is increasingly used to improve the initial assessment of cancer grade. However, it is important to note that prostate MRI does not eliminate the possibility of misclassification [5]. In this context, we found that baseline clinical parameters—including PSA density, number of cores positive for cancer, MRI findings, and age—offered only marginal discriminative ability for prediction of biopsy upgrading but were improved by the addition of the Decipher classifier. Therefore, further optimization of prediction tools for active surveillance outcomes remains an important and still unmet clinical need [20], [21].
Our findings build on prior studies of surrogate endpoints for active surveillance candidacy. For example, Herlemann and colleagues [12] evaluated 647 patients diagnosed with National Comprehensive Cancer Network (NCCN) very low-, low-, and favorable intermediate-risk prostate cancer treated with initial prostatectomy. In this cohort, the Decipher score was an independent predictor of adverse pathology (high grade and/or high stage at prostatectomy; OR 1.34 per 0.1 unit increase, 95% CI 1.11–1.63) [12]. Similarly, Kim and colleagues [22] analyzed Decipher scores for biopsies from 266 patients with NCCN very low-, low-, and favorable intermediate-risk prostate cancer and reported that the Decipher score was an independent predictor of adverse pathology on prostatectomy (odds ratio 1.29 per 10% increase, 95% CI 1.03–1.61 per 0.1 unit increase). However, by directly evaluating outcomes for patients managed with active surveillance, our findings fill an important gap in knowledge and indicate reveal an association between Decipher scores and biopsy upgrading. Furthermore, as reclassification events constitute the most significant triggers for conversion to active treatment, our results may have implications for questions on health-related quality of life and cost-effectiveness in future studies.
The reclassification events in our study were assessed over a relatively short interval after enrollment in active surveillance. Nearly one-third of patients in this study experienced biopsy Gleason upgrading, which is a larger proportion than reported for large institutional cohorts such as the multi-institutional Canary Prostate Active Surveillance Study, in which 27% of patients experienced Gleason reclassification at median follow-up of 4.1 yr [23]. This suggests that the upgrading observed was largely because of initial biopsy sampling error rather than disease progression [24]. Serial molecular profiling of prostate biopsies using immunohistochemistry and next-generation sequencing has also identified the potential contributions of short-term clonal progression of low-grade disease [25]. Regardless of the cause of upgrading, the potential role of a genomic classifier in enhancing estimates of a patient’s trajectory at the time of active surveillance addresses an important clinical need. The overall modest performance of even a refined clinical model incorporating prostate MRI and genomic testing in this study underscores the need to improve risk estimation for patients enrolled on active surveillance.
We found that the distribution of Decipher scores among active surveillance patients was narrow and, as expected, clustered at the lower end of the risk distribution. As a result, different groupings may be required for distinguishing risk among active surveillance patients, as the existing reporting classifications may be better suited for the wider spectrum of genomic risk. Although the Decipher score as a continuous variable (per 0.1 unit) was associated with Gleason upgrading, significant differences could not be appreciated when using the Decipher standard risk groups generated in clinical reporting (low, intermediate, and high) that are applied in the setting of more advanced disease. Assessing a putative clinical cut point that would maximize sensitivity and specificity in this select group of patients yielded a binary classification value of 0.475; scores above this threshold were associated with a nearly fourfold higher odds of biopsy upgrading. A theoretical clinical application of these findings would include offering Decipher testing broadly to patients with Gleason 3 + 3 disease for whom disease reclassification would lead to actionable differences in management, such as those with a stronger inclination for undergoing definitive treatment because of younger age, less comorbidity, and preference. For patients with the lowest risk of disease reclassification, as assessed via clinical and genomic features such as a Decipher score <0.475, reducing the intensity of surveillance by increasing the intervals between biopsies, or avoiding biopsy altogether, might be feasible. However, these findings require further study in larger cohorts and over longer periods of surveillance, or explicit investigation in a randomized trial.
The study has several limitations. Selection of patients for Decipher testing may not have occurred at random and could potentially favor use in patients at higher risk of disease reclassification for whom testing was undertaken to confirm suitability for active surveillance. However, Decipher testing was routinely offered without known systematic preference for those at higher risk, and patient baseline disease characteristics are consistent with widely accepted criteria for adoption of active surveillance [6]. A higher incidence of short-term reclassification was also reported in studies of patients receiving genomic testing that may relate to the preferential use of genomic testing in higher-risk populations [26]. In addition, we defined biopsy upgrading as any increase in biopsy Gleason score, an approach used in prior studies, but this may fail to account for more substantial changes in risk such as a simultaneous increase in tumor volume [27]. Although all prostate MRI scans were reviewed by expert genitourinary radiologists at our institution, central re-review of scans to apply the PRECISE criteria for MRI progression [28] was not conducted for this study. Lastly, the sample size and follow-up are insufficient for assessment of meaningful distant longitudinal outcomes. Despite these limitations, the strengths of the study include novel data on Decipher testing with outcomes for patients enrolled in a contemporary active surveillance program.
5. Conclusions
The Decipher genomic classifier score was associated with biopsy Gleason upgrading among patients with low-risk prostate cancer enrolled in active surveillance who had undergone MRI-enhanced biopsy procedures.
Author contributions: Michael S. Leapman had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Press, Sprenkle, Leapman.
Acquisition of data: Sprenkle, Leapman.
Analysis and interpretation of data: Press, Sprenkle, Leapman, Lin.
Drafting of the manuscript: Press, Jones, Olawoyin, Lokeshwar, Rahman, Khajir.
Critical revision of the manuscript for important intellectual content: Leapman, Sprenkle, Press, Loeb, Seibert, Catalona.
Statistical analysis: Press, Leapman, Khajir, Leapman.
Obtaining funding: None.
Administrative, technical, or material support: Lin, Cooperberg, Loeb, Darst, Zheng, Chen, Seibert, Catalona, Witte.
Supervision: Sprenkle, Leapman.
Other: None.
Financial disclosures: Michael S. Leapman certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Ronald C. Chen is a consultant for Myovant, AbbVie, Accuray, Blue Earth, and Janssen. Matthew R. Cooperberg has received personal fees from Astellas, Bayer, MDx Health, Myriad Genetics, Dendreon, Steba Biotech, Astra Zeneca, and AbbVie outside of the submitted work. Stacy Loeb has equity in Gilead, unrelated to the current research. The remaining authors have nothing to disclose.
Funding/Support and role of the sponsor: None.
Associate Editor: Guillaume Ploussard
References
- 1.Chen R.C., Rumble R.B., Loblaw D.A., et al. Active surveillance for the management of localized prostate cancer (Cancer Care Ontario guideline): American Society of Clinical Oncology Clinical Practice Guideline Endorsement. J Clin Oncol. 2016;34:2182–2190. doi: 10.1200/JCO.2015.65.7759. [DOI] [PubMed] [Google Scholar]
- 2.Cooperberg M.R., Carroll P.R. Trends in management for patients with localized prostate cancer, 1990–2013. JAMA. 2015;314:80–82. doi: 10.1001/jama.2015.6036. [DOI] [PubMed] [Google Scholar]
- 3.Womble P.R., Montie J.E., Ye Z., Linsell S.M., Lane B.R., Miller D.C. Contemporary use of initial active surveillance among men in Michigan with low-risk prostate cancer. Eur Urol. 2015;67:44–50. doi: 10.1016/j.eururo.2014.08.024. [DOI] [PubMed] [Google Scholar]
- 4.Timilshina N., Komisarenko M., Martin L.J., et al. Factors associated with discontinuation of active surveillance among men with low-risk prostate cancer: a population-based study. J Urol. 2021;206:903–913. doi: 10.1097/JU.0000000000001903. [DOI] [PubMed] [Google Scholar]
- 5.Williams C., Khondakar N.R., Daneshvar M.A., et al. The risk of prostate cancer progression in active surveillance patients with bilateral disease detected by combined magnetic resonance imaging-fusion and systematic biopsy. J Urol. 2021;206:1157–1165. doi: 10.1097/JU.0000000000001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klotz L., Zhang L., Lam A., Nam R., Mamedov A., Loblaw A. Clinical results of long-term follow-up of a large, active surveillance cohort with localized prostate cancer. J Clin Oncol. 2010;28:126–131. doi: 10.1200/JCO.2009.24.2180. [DOI] [PubMed] [Google Scholar]
- 7.Hamdy F.C., Donovan J.L., Lane J.A., et al. 10-Year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med. 2016;375:1415–1424. doi: 10.1056/NEJMoa1606220. [DOI] [PubMed] [Google Scholar]
- 8.Cooperberg M.R., Zheng Y., Faino A.V., et al. Tailoring intensity of active surveillance for low-risk prostate cancer based on individualized prediction of risk stability. JAMA Oncol. 2020;6:e203187. doi: 10.1001/jamaoncol.2020.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsson H., Nordström T., Clements M., Grönberg H., Lantz A.W., Eklund M. Intensity of active surveillance and transition to treatment in men with low-risk prostate cancer. Eur Urol Oncol. 2020;3:640–647. doi: 10.1016/j.euo.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Rajwa P., Sprenkle P.C., Leapman M.S. When and how should active surveillance for prostate cancer be de-escalated? Eur Urol Focus. 2021;7:297–300. doi: 10.1016/j.euf.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leapman M.S., Wang R., Ma S., Gross C.P., Ma X. Regional adoption of commercial gene expression testing for prostate cancer. JAMA Oncol. 2021;7:52–58. doi: 10.1001/jamaoncol.2020.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herlemann A., Huang H.C., Alam R., et al. Decipher identifies men with otherwise clinically favorable-intermediate risk disease who may not be good candidates for active surveillance. Prostate Cancer Prostat Dis. 2020;23:136–143. doi: 10.1038/s41391-019-0167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jairath N.K., Dal Pra A., Vince R., Jr., et al. A systematic review of the evidence for the Decipher genomic classifier in prostate cancer. Eur Urol. 2021;79:374–383. doi: 10.1016/j.eururo.2020.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Washington S.L., 3rd, Jeong C.W., Lonergan P.E., et al. Regional variation in active surveillance for low-risk prostate cancer in the US. JAMA Netw Open. 2020;3:e2031349. doi: 10.1001/jamanetworkopen.2020.31349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloom J.B., Daneshvar M.A., Lebastchi A.H., et al. Risk of adverse pathology at prostatectomy in the era of MRI and targeted biopsies; rethinking active surveillance for intermediate risk prostate cancer patients. Urol Oncol. 2021;39:729.e721–729.e726. doi: 10.1016/j.urolonc.2021.02.018. [DOI] [PubMed] [Google Scholar]
- 16.O’Connor L.P., Wang A.Z., Yerram N.K., et al. Combined MRI-targeted plus systematic confirmatory biopsy improves risk stratification for patients enrolling on active surveillance for prostate cancer. Urology. 2020;144:164–170. doi: 10.1016/j.urology.2020.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsiang W., Ghabili K., Syed J.S., et al. Outcomes of serial multiparametric magnetic resonance imaging and subsequent biopsy in men with low-risk prostate cancer managed with active surveillance. Eur Urol Focus. 2021;7:47–54. doi: 10.1016/j.euf.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Balakrishnan A.S., Cowan J.E., Cooperberg M.R., Shinohara K., Nguyen H.G., Carroll P.R. Evaluating the safety of active surveillance: outcomes of deferred radical prostatectomy after an initial period of surveillance. J Urol. 2019;202:506–510. doi: 10.1097/JU.0000000000000247. [DOI] [PubMed] [Google Scholar]
- 19.Godtman R.A., Holmberg E., Khatami A., Pihl C.G., Stranne J., Hugosson J. Long-term results of active surveillance in the Göteborg randomized, population-based prostate cancer screening trial. Eur Urol. 2016;70:760–766. doi: 10.1016/j.eururo.2016.03.048. [DOI] [PubMed] [Google Scholar]
- 20.Chu C.E., Lonergan P.E., Washington S.L., et al. Multiparametric magnetic resonance imaging alone is insufficient to detect grade reclassification in active surveillance for prostate cancer. Eur Urol. 2020;78:515–517. doi: 10.1016/j.eururo.2020.06.030. [DOI] [PubMed] [Google Scholar]
- 21.Rajwa P., Pradere B., Quhal F., et al. Reliability of serial prostate magnetic resonance imaging to detect prostate cancer progression during active surveillance: a systematic review and meta-analysis. Eur Urol. 2021;80:549–563. doi: 10.1016/j.eururo.2021.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Kim H.L., Li P., Huang H.C., et al. Validation of the Decipher test for predicting adverse pathology in candidates for prostate cancer active surveillance. Prostate Cancer Prostat Dis. 2019;22:399–405. doi: 10.1038/s41391-018-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liss M.A., Newcomb L.F., Zheng Y., et al. Magnetic resonance imaging for the detection of high grade cancer in the Canary Prostate Active Surveillance Study. J Urol. 2020;204:701–706. doi: 10.1097/JU.0000000000001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue L.Y., Trock B.J., Partin A.W., Carter H.B., Etzioni R. Modeling grade progression in an active surveillance study. Stat Med. 2014;33:930–939. doi: 10.1002/sim.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salami S.S., Tosoian J.J., Nallandhighal S., et al. Serial molecular profiling of low-grade prostate cancer to assess tumor upgrading: a longitudinal cohort study. Eur Urol. 2021;79:456–465. doi: 10.1016/j.eururo.2020.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cedars B.E., Washington S.L., 3rd, Cowan J.E., et al. Stability of a 17-gene genomic prostate score in serial testing of men on active surveillance for early stage prostate cancer. J Urol. 2019;202:696–701. doi: 10.1097/JU.0000000000000271. [DOI] [PubMed] [Google Scholar]
- 27.Leapman M.S., Ameli N., Cooperberg M.R., et al. Quantified clinical risk change as an end point during prostate cancer active surveillance. Eur Urol. 2017;72:329–332. doi: 10.1016/j.eururo.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 28.Moore C.M., Giganti F., Albertsen P., et al. Reporting magnetic resonance imaging in men on active surveillance for prostate cancer: the PRECISE recommendations—a report of a European School of Oncology task force. Eur Urol. 2017;71:648–655. doi: 10.1016/j.eururo.2016.06.011. [DOI] [PubMed] [Google Scholar]