Abstract
Background
To characterize genomic determinants of response to pembrolizumab in recurrent/metastatic (R/M) head and neck squamous cell carcinoma (HNSCC) in the KEYNOTE-012 study.
Methods
Associations between biomarkers (tumor mutational burden (TMB), neoantigen load (NL), 18-gene T-cell-inflamed gene expression profile (TcellinfGEP), and PD-L1 combined positive score (CPS)) and clinical outcomes with pembrolizumab were assessed in patients with R/M HNSCC (n=192). Tumor human papillomavirus (HPV) status was also evaluated with the use of p16 immunohistochemistry and whole exome sequencing (WES; HPV+, mapping >20 HPV reads) in pretreatment tumor samples (n=106).
Results
TMB, clonality-weighted TMB, and TcellinfGEP were significantly associated with objective response (p=0.0276, p=0.0201, and p=0.006, respectively), and a positive trend was observed between NL and PD-L1 CPS and clinical response (p=0.0550 and p=0.0682, respectively). No correlation was observed between TMB and TcellinfGEP (Spearman ρ=–0.026) or TMB and PD-L1 (Spearman ρ=0.009); a correlation was observed between TcellinfGEP and PD-L1 (Spearman ρ=0.511). HPV status by WES and p16 immunohistochemistry showed concordance (84% ҡ=0.573) among patients whose HPV results were available using both methods.
Conclusions
TMB and inflammatory biomarkers (TcellinfGEP and PD-L1) may represent distinct and complementary biomarkers predicting response to anti-programmed death 1 therapies in HNSCC; further study of these relationships in randomized clinical trials is needed.
Trial registration number
Keywords: gene expression profiling, head and neck neoplasms, immunotherapy, programmed cell death 1 receptor, tumor biomarkers
Introduction
Immune checkpoint inhibitors demonstrate antitumor activity in a variety of tumor types.1–4 Programmed death ligand 1 (PD-L1) expression can predict response to programmed death 1 (PD-1) inhibition and is an approved diagnostic for some cancers.5 6 Despite the predictive value of PD-L1 expression, some patients with PD-L1-negative tumors experience clinical benefit with PD-1-targeting/PD-L1-targeting regimens,7–9 and crossover of progression-free survival (PFS) or overall survival (OS) curves for patients with PD-L1-positive and PD-L1-negative tumors using various cutoffs has been observed,8 10 suggesting the existence of unidentified immunotherapy-responsive subpopulations. Greater understanding of the tumor microenvironment, beyond PD-L1 expression, is needed to predict clinical benefit more reliably.
Certain molecular signatures have been linked with clinical outcomes in several solid tumors.11 Tumor mutational burden (TMB), as determined by next-generation sequencing (NGS) or whole exome sequencing (WES), can predict response to immunotherapies (anti-cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) or anti-PD-1/PD-L1) as demonstrated in retrospective analyses across multiple tumor types.12–15 Additionally, the anti-PD-1 monoclonal antibody pembrolizumab is now approved as treatment for patients with TMB-high (TMB-H; ≥10 mutations/megabase) solid tumors.6 Neoantigen load (NL), a less validated biomarker, can also be determined by NGS and WES, but identifying criteria to further define neoantigens is needed.13 Some tumors exhibit a T-cell-inflamed phenotype, described by an 18-gene T-cell-inflamed gene expression profile (TcellinfGEP) composed of infiltrating T cells, chemokines, and an interferon gamma (IFN-γ) signature, and additional gene expression T-cell inflammation signatures have been developed.16 17 A recent study suggested that tumor antigenicity, whether originating from somatic mutations or viral epitopes, and T-cell infiltration provide complementary information that predicts pembrolizumab activity.16 18 Markers of tumor T-cell inflammation are related to response to anti-PD-1/PD-L1 therapies, suggesting that tumors exhibit adaptive and cytotoxic T-cell responses of variable intensity.16 19 The primary objective of the evaluations presented here is to characterize response to pembrolizumab in head and neck squamous cell carcinoma (HNSCC) according to the landscape of these key mutational and inflammation biomarkers.
Oncogenic viruses (eg, human papillomavirus (HPV), hepatitis B virus, Merkel cell polyomavirus, and Epstein-Barr virus) generate viral antigens distinct from somatic mutations.20 21 Response to immunotherapy is reportedly higher in some virus-associated cancers than in their virally unrelated counterparts.4 22 PD-L1 expression is increased in some virus-associated cancers, reflecting an inflamed tumor phenotype.21–23 However, genetic determinants of response to immunotherapy in virus-associated cancers are not well understood.21
HNSCC includes HPV-associated cancers that might be highly immune cell infiltrated; the level of immune infiltration and activation varies according to HPV status, molecular subtype, and genomic instability.24 HNSCC tumors, irrespective of HPV status, benefit from immunotherapy.24–26 Pembrolizumab was well tolerated and conferred durable antitumor activity in patients with HNSCC in the non-randomized phase 1b multicohort KEYNOTE-012 and phase 2 KEYNOTE-055 (NCT02255097) studies.2 4 27 28 The efficacy of pembrolizumab was associated with PD-L1 expression and IFN-γ-related gene expression.4 29 Although HPV-mediated HNSCC has a distinct natural history, biomolecular signature, and response to cytotoxic therapy,30 31 the association of HPV status with pembrolizumab efficacy has been equivocal. In CheckMate-141, response rates were higher in nivolumab-treated patients with p16-positive HNSCC (15.9%) than in those with p16-negative HNSCC (8.0%).32 In KEYNOTE-012, patients with HPV-associated (defined as p16-positive) HNSCC achieved a higher response to pembrolizumab than their p16-negative counterparts; this was not statistically significant given the small number of patients with p16-positive disease.2 Conversely, in KEYNOTE-055, patient response to pembrolizumab was not impacted by HPV status though response rate alone may not be a sensitive outcome measure for patient benefit.28 By contrast, survival has consistently been more favorable in patients with HPV-associated than HPV-negative HNSCC across treatment regimens (eg, radiotherapy, chemotherapy, and immunotherapy across anti-PD-1/PD-L1 studies).33 34
Although HPV is often determined indirectly using p16 immunohistochemistry (IHC) as a surrogate marker with a 5% false-positive rate in HPV-negative tumors,35 direct genomic methods such as in situ hybridization, PCR, and WES are available.36 37 In the current study, the key secondary objective beyond characterizing genomic determinants of response to pembrolizumab was to descriptively assess the distribution of mutational load and inflammatory biomarkers (TcellinfGEP and PD-L1) according to HPV status and to assess the role of HPV status as an explanatory variable for pembrolizumab response beyond the key genomic determinants of mutation and inflammation in patients with recurrent/metastatic (R/M) HNSCC enrolled in KEYNOTE-012, for whom long-term data have been detailed.29
Methods
Study design
The design and patient population of KEYNOTE-012 have been reported.4 In brief, adult patients with histologically or cytologically confirmed R/M HNSCC were enrolled. Initially, only patients with PD-L1-positive tumors (expression in stroma or ≥1% of tumor cells38) were included (cohort B1); after a protocol amendment, patients were enrolled regardless of PD-L1 status (cohort B2). Cohort B1 received pembrolizumab 10 mg/kg every 2 weeks intravenously; cohort B2 received pembrolizumab 200 mg every 3 weeks intravenously.
Assessments
In this exploratory analysis, the predictive value of TMB and inflammatory biomarkers (TcellinfGEP, PD-L1) were assessed in both KEYNOTE-012 cohorts (B1 and B2). The methods used to analyze TMB, NL, and TcellinfGEP have been reported.16 18 The cut-off of −0.318 used to define GEPlow and GEPnonlow is synonymous with GEPlo and GEPhi used in Cristescu et al.18 Tumor PD-L1 expression was assessed by IHC combined positive score (CPS); CPS ≥1 was considered positive.
HPV status was confirmed by p16 IHC using the CINtec p16 Ventana assay on the BenchMark Ultra using formalin-fixed paraffin-embedded pretreatment clinical specimens and by WES of germline and tumor DNA. For HPV status determined by p16 IHC, HPV-positive status (defined as ≥70% tumor cells with positive nuclear and/or cytoplasmic diffuse staining and H score of 210) included patients with primary tumor locations in the oropharynx, and HPV-negative status included patients with non-HPV-associated oropharyngeal cancers and primary tumor locations outside the oropharynx. Additional details are described in online supplemental file 1.
jitc-2021-003026supp001.pdf (110.8KB, pdf)
Statistical analysis
Logistic regression modeling was used to test the association between TMB (and/or TcellinfGEP/PD-L1) and best overall response. A Cox model was used to assess the association between TMB (and/or TcellinfGEP/PD-L1) and PFS and OS; log or square root transformation was used when needed for TMB/PD-L1. Regression models were adjusted for Eastern Cooperative Oncology Group (ECOG) performance status and cohort. One-sided p values were calculated according to the hypothesized positive association between these exploratory biomarkers and improved clinical outcomes. The area under the receiver operating characteristic (AUROC) curve was used to measure discriminatory ability. A TMB cut-off of 175 mutations/exome (mut/exome) was used for illustrative purposes and was identified in previous exploratory analyses as the threshold with strong support for predicting response to pembrolizumab across multiple tumor types.18 39 Additionally, this WES-based TMB cut-off of 175 mut/exome proved to be the most concordant with the FoundationOne®CDx (Foundation Medicine) 10 mutations/megabase cut-off,40 the current assay and cut-off for the tumor-agnostic indication for which pembrolizumab is approved.6 The cut-off for TcellinfGEP was −0.318 (TcellinfGEPlow <–0.318; TcellinfGEPnonlow ≥–0.318); this was associated with the Youden Index in an ROC analysis of a pan-cancer data set using TcellinfGEP to predict whether a tumor was inflamed, as defined by observations on the dendrogram from unsupervised clustering of the pan-cancer data and supported as an enriching cut-off across multiple tumor types. Correlations between TMB and inflammatory biomarkers were assessed using Spearman correlation. Nominal p values were reported for signature testing. No adjustment was made for multiplicity. The concordance of HPV status, defined by WES versus p16 IHC, was evaluated using a contingency table. The distribution of each biomarker (TMB/TcellinfGEP/PD-L1) by HPV status was illustrated using boxplots, and the mean difference of each biomarker in HPV-positive versus HPV-negative subgroups was tested using a two-sample t-test, and the adjusted p values are reported for multiple testing across the three biomarkers (TMB/TcellinfGEP/PD-L1). The Hochberg Step-up procedure was used for multiplicity to control the family-wise error rate. Testing for differential biomarker relationships according to HPV status was performed with an interaction term between the biomarker (TMB/TcellinfGEP/PD-L1) and HPV status in a logistic regression model (other terms were ECOG, cohort, the biomarker itself (TMB/TcellinfGEP/PD-L1), and HPV status) and was similarly adjusted for multiplicity.
Results
One hundred and ninety-two patients were included in this analysis; 106 (55.2%; 33 from cohort B1, 73 from cohort B2) had available WES data. Patient baseline characteristics are listed in online supplemental table S1.
Association between TMB and clinical outcomes
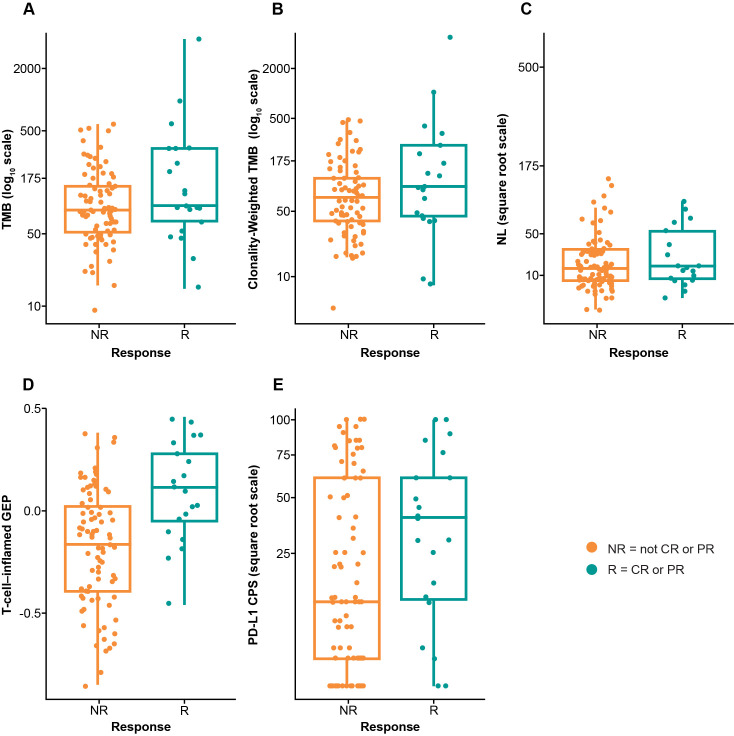
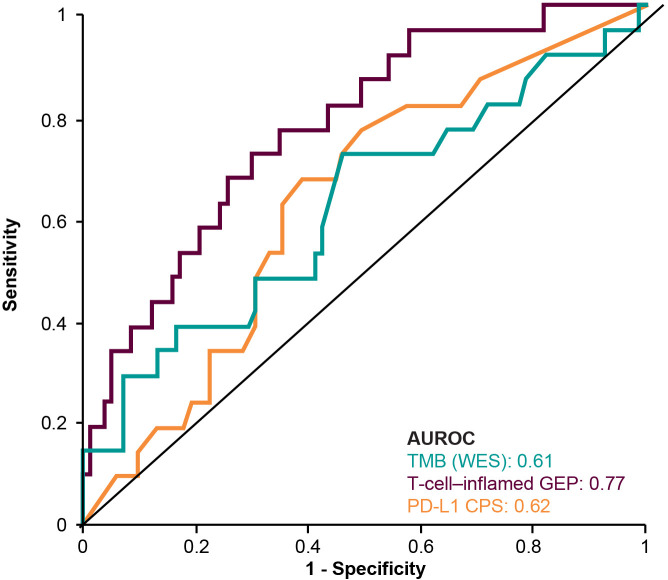
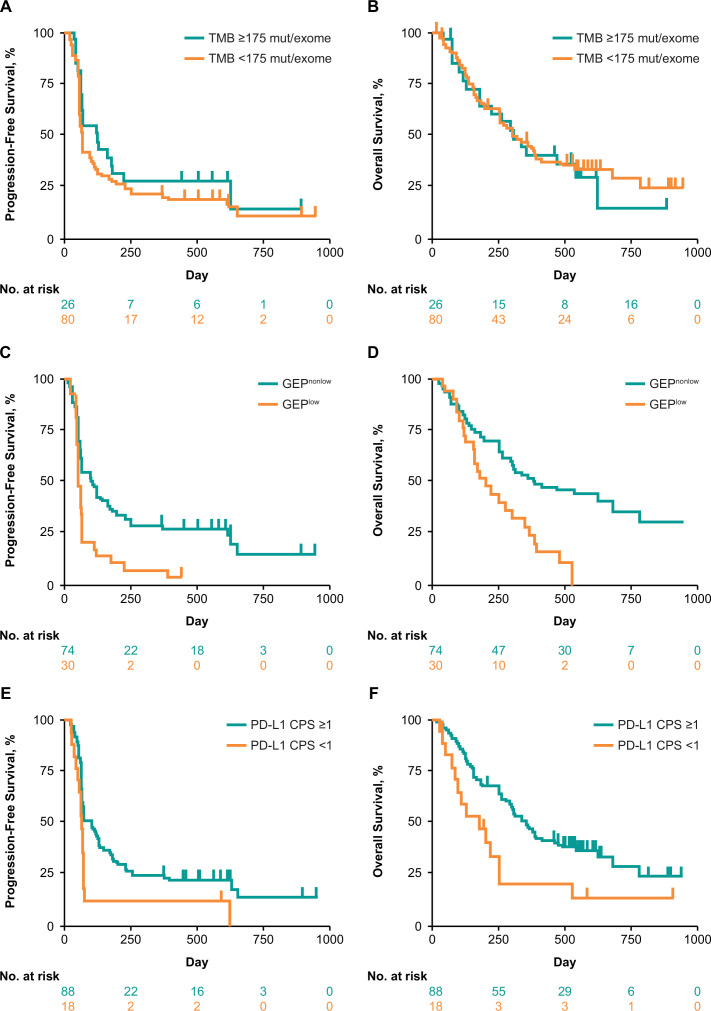
TMB was significantly associated with objective response (p=0.0276) (figure 1A).18 The AUROC curve for TMB and response was 0.61 (figure 2). Median PFS was longer in the TMB ≥175 mut/exome subgroup than the TMB <175 mut/exome subgroup (121 vs 64 days) (figure 3A); median OS times were similar regardless of TMB cut-off (301 vs 303 days, respectively) (figure 3B).
Figure 1.
Association between biomarkers and response in the overall patient population. (A) TMB, (B) TMB weighted by clonality, (C) NL, (D) T-cell-inflamed GEP, and (E) PD-L1 CPS. CPS, combined positive score; CR, complete response; GEP, gene expression profile; NL, neoantigen load; NR, non-responder; PD-L1, programmed death ligand 1; PR, partial response; R, responder; TMB, tumor mutational burden.
Figure 2.
AUROC curve by biomarker. AUROC, area under the receiver operating characteristic; CPS, combined positive score; GEP, gene expression profile; PD-L1, programmed death ligand 1; TMB, tumor mutational burden; WES, whole exome sequencing.
Figure 3.
Association between biomarkers and PFS and OS in the overall patient population at prespecified cut-offs. (A) TMB and PFS, (B) TMB and OS, (C) T-cell-inflamed GEP and PFS, (D) T-cell-inflamed GEP and OS, (E) PD-L1 CPS and PFS, and (F) PD-L1 CPS and OS. CPS, combined positive score; GEP, gene expression profile; OS, overall survival; PD-L1, programmed death ligand 1; PFS, progression-free survival; TMB, tumor mutational burden.
Clonality-weighted TMB, an estimate of TMB restricted to clonal mutations, was highly correlated with TMB (Spearman ρ=0.91). Similarly, clonality-weighted TMB was significantly associated with response (p=0.0201) (figure 1B). NL was also highly correlated with TMB (Spearman ρ=0.83) and tended to associate with higher response rates (p=0.0550) (figure 1C).
Association between TcellinfGEP and clinical outcomes
TcellinfGEP was significantly associated with response (p=0.0006) (figure 1D).18 The AUROC curve for TcellinfGEP and response was 0.77 (figure 2). Of importance, the TcellinfGEP was initially identified with a training data set including cohort B1 and later independently validated using data from cohort B2.16 Median PFS was longer in the TcellinfGEPnonlow subgroup than the TcellinfGEPlow subgroup (106 vs 57 days) (figure 3C). Similarly, median OS was longer in the TcellinfGEPnonlow subgroup than the TcellinfGEPlow subgroup (385 vs 199 days) (figure 3D).
Association between PD-L1 and clinical outcomes
PD-L1 tended to associate with higher response rates (p=0.0682) (figure 1E). The AUROC curve for PD-L1 response was 0.62 (figure 2). Median PFS was longer in the PD-L1 CPS≥1 subgroup than the CPS<1 subgroup (97 vs 60 days) (figure 3E). Similarly, median OS was longer in the CPS≥1 subgroup than the CPS<1 subgroup (353 vs 173 days) (figure 3F).
Joint assessment of biomarkers
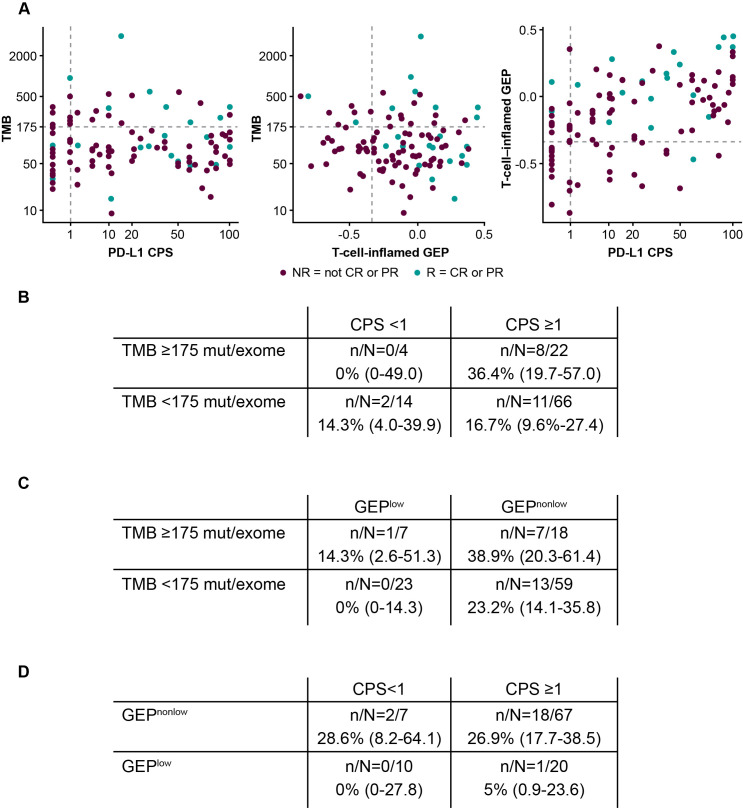
There was no correlation between TMB and TcellinfGEP (Spearman ρ=−.026) or between TMB and PD-L1 (Spearman ρ=0.009) (figure 4). TcellinfGEP and PD-L1 were correlated with Spearman ρ=0.511. When TMB, TcellinfGEP, and PD-L1 were evaluated as continuous variables for their independent predictive values in a multivariate model with any two included into a model simultaneously, TMB and TcellinfGEP showed independent predictive value with p<0.05 after adjusting for another biomarker (TMB/TcellinfGEP/PD-L1); PD-L1 was not statistically significant after adjusting for TcellinfGEP or TMB (p>0.05). Responses were enriched in the TMB ≥175 mut/exome subgroup (8/26 patients; 30.8%; 95% CI 16.5% to 50.0%), the CPS ≥1 subgroup (19/88 patients; 21.6%; 95% CI 14.3% to 31.3%), and the TcellinfGEPnonlow subgroup (20/74 patients; 27.0%; 95% CI 18.2% to 38.1%). The TMB ≥175 mut/exome and PD-L1 CPS ≥1 (8/22 patients; 36.4%; 95% CI 19.7% to 57.0%) (figure 4B) and TMB ≥175 mut/exome and TcellinfGEPnonlow (7/18 patients; 38.9%; 95% CI 20.3% to 61.4%) (figure 4C) subgroups had the highest response rate.
Figure 4.
Correlation between (A) TMB and T-cell-inflamed GEP or (B) TMB and PD-L1 CPS and the response rate (95% CI) of the dual biomarkers (C) TMB and PD-L1 CPS, (D) TMB and T-cell-inflamed GEP, and (E) T-cell-inflamed GEP and PD-L1 CPS in the overall patient population. Dashed horizontal lines represent clinically applicable TMB threshold of ≥175 mut/exome. Dashed vertical line represents discovery cut-off for the T-cell-inflamed GEP (≥−.318) selected through analysis of data across multiple tumor types16 or the PD-L1 cut-off of CPS 1. CPS, combined positive score; CR, complete response; GEP, gene expression profile; mut/exome, mutations/exome; NR, non-responder; PD-L1, programmed death ligand 1; PR, partial response; R, responder; TMB, tumor mutational burden.
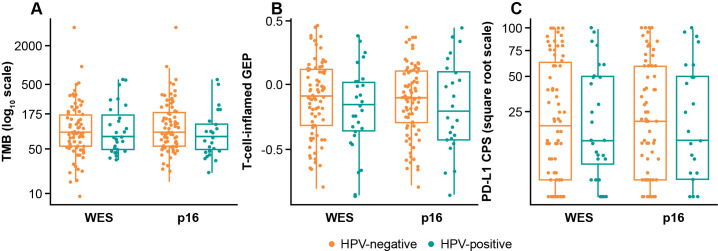
Distribution of biomarkers by HPV status
Of 192 patients, 106 (55%) had evaluable WES data. There was agreement between p16 IHC and WES for HPV status (89/106 (84%); ҡ=0.573) (online supplemental table S2). Both WES-defined and p16 IHC-defined HPV status demonstrated mutual exclusivity with TP53 mutation (online supplemental table S3).
The distribution of biomarkers (TMB/TcellinfGEP/PD-L1) was comparable in HPV-positive and HPV-negative subgroups defined by WES or p16 IHC (figure 5). Two-sample t-testing showed no significant difference between HPV-positive and HPV-negative subgroups defined by WES or p16 IHC for any biomarkers (adjusted p>0.6).
Figure 5.
Biomarker distribution by HPV status for (A) TMB, (B) T-cell-inflamed GEP, and (C) PD-L1 CPS. CPS, combined positive score; GEP, gene expression profile; HPV, human papillomavirus; TMB, tumor mutational burden; WES, whole exome sequencing.
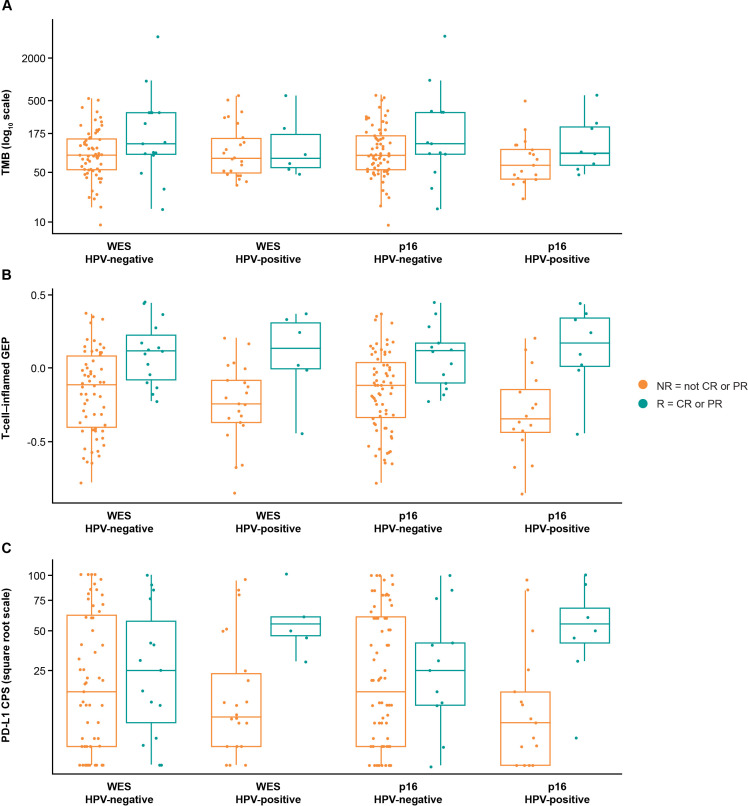
Biomarker relationships with clinical outcomes by HPV status
Evaluating trends for TMB, TcellinfGEP, and PD-L1 within HPV status subtypes suggested an association between each biomarker and response in HPV-positive and HPV-negative subgroups (figure 6A–C). Trends for association between PD-L1 and response and TcellinfGEP and response in HPV subgroups were observed regardless of methodology used to evaluate HPV status. In contrast, though a trend for association between TMB and response was seen in HPV subgroups analyzed by p16 IHC, a trend was observed only for the HPV-negative subgroup defined by WES.
Figure 6.
Association between biomarkers and response by HPV status for (A) TMB, (B) T-cell-inflamed GEP, and (C) PD-L1. CPS, combined positive score; CR, complete response; GEP, gene expression profile, HPV, human papillomavirus; NR, non-responder; PD-L1, programmed death ligand 1; PR, partial response; R, responder; TMB, tumor mutational burden; WES, whole exome sequencing.
Interaction testing did not confirm evidence of unique relationships for these biomarkers with clinical outcome, depending on HPV status. A test of the interaction effect, TMB by HPV, showed no difference of TMB association with response in HPV-positive versus HPV-negative subgroups defined by WES (TMB*HPV interaction, adjusted p=0.8184) or p16 IHC (adjusted p=0.4879). Similarly, a test of the interaction effect, TcellinfGEP by HPV, showed no difference of TcellinfGEP association with response in HPV-positive versus HPV-negative subgroups defined by WES (TcellinfGEP*HPV interaction, adjusted p=0.8184) or p16 IHC (adjusted p=0.4879). Last, a test of the interaction effect, PD-L1 by HPV, showed no difference of PD-L1 association with response in HPV-positive versus HPV-negative subgroups defined by WES (PD-L1*HPV interaction, adjusted p=0.3129) or p16 IHC (adjusted p=0.3069).
Discussion
Pembrolizumab is approved in the USA for patients with R/M HNSCC6; however, not all patients respond to treatment. Establishing patient-specific and tumor-specific factors that predict response can help identify patients likely to achieve clinical benefit and may accelerate the study of novel strategies for patients less likely to benefit from therapy. In the current study, we explored genomic determinants of response, including by HPV status, to pembrolizumab; this is a comprehensive analysis reporting the impact of TMB and inflammatory biomarkers (TcellinfGEP and PD-L1) on response to pembrolizumab in patients with HNSCC. Results indicate that higher mutation and inflammation levels are associated with improved propensity to respond.
In the overall population, our data show that TMB and TcellinfGEP were orthogonal, independently significant predictors of response. Responses were higher in patients with TMB≥175 mut/exome or TcellinfGEPnonlow tumors; a similar pattern was observed for PFS. OS was prolonged in the TcellinfGEPnonlow compared with the TcellinfGEPlow subgroups; no significant associations between TMB and OS were observed. TMB-H is associated with response and prolonged survival in patients receiving immune checkpoint inhibitors.13 15
The distributions of TMB, TcellinfGEP, and PD-L1 were not dramatically different between HPV-positive and HPV-negative subgroups, and no significant statistical evidence supporting HPV-specific relationships between these biomarkers and clinical outcome was observed. In the small subset of patients with WES-defined HPV-positive tumors, TMB did not appear to be associated with response, possibly because of the dominance of viral neoepitopes not measured by WES compared with somatic neoepitopes captured by WES and potentially reflected by that fact that HPV-positive tumors exhibit a characteristic somatic mutation signature (apolipoprotein B editing catalytic polypeptide, potentially virally induced) more frequently than do HPV-negative tumors.31 Nevertheless, the general statistical shortcomings of evaluating biomarker distributions across small HPV subgroups must be acknowledged. The small sample size of the HPV-positive subgroup in this study precludes reliable conclusions, but emerging evidence on establishing HPV infection status to inform treatment decisions highlights its importance in HNSCC. HPV-positive HNSCC is associated with improved survival and response to cytotoxic therapy33; it is now recommended that HPV status be reported before therapy is chosen.35 41 Similarly, in a recent pooled analysis, HPV-positive tumors were associated with greater clinical benefit from PD-1/PD-L1 inhibitors than were HPV-negative tumors.34 HPV infection promotes T-cell infiltration, immune effector cell activation, and T-cell receptor diversity in HNSCC,34 suggesting that HPV-positive tumors may be more amenable to immunotherapy. The current analysis, though limited by sample size, highlights the importance of evaluating genomic signatures in HNSCC and of trying to understand whether those genomic correlates of response operate similarly in their associations regardless of HPV status.
Genomic correlates of response to immune checkpoint inhibitors have been studied in several tumor types.13 14 16 42–45 An 18-gene TcellinfGEP predicted response to pembrolizumab across multiple solid tumor types, including HNSCC, and other T-cell inflammation/IFN-γ-related signatures are being developed.16 In metastatic melanoma, TMB, NL, and expression of cytolytic markers (genes encoding granzyme A and perforin) were predictors of clinical benefit with ipilimumab (anti-CTLA-4).13 In patients with advanced melanoma, TcellinfGEP predicted best overall response with anti-PD-1 but not with anti-CTLA-4 therapy.43 46
The widely used method of detecting HPV infection—p16 IHC47—is readily available and evaluable in pathology laboratories.35 47 p16 positivity is commonly defined as strong and diffuse nuclear and cytoplasmic staining in ≥70% of the tumor specimen,48 49 although there has been some debate with regard to the cut-off and staining patterns.50 Potential problems exist with p16 IHC detection of HPV, which include p16 expression in 5% of HPV-negative head and neck cancers as well as p16 staining heterogeneity, false-positive staining, and lack of a standardized cut-off for positivity.51 Moreover, WES-defined and p16-defined HPV status demonstrated mutual exclusivity with TP53 mutation in this data set; such mutual exclusivity has been reported in major head and neck molecular studies, which include The Cancer Genome Atlas (TCGA).52 Increased sensitivity of NGS methods for HPV detection over p16 IHC was demonstrated for HNSCC, but no direct comparison was made.36 In the first study to describe large-scale WES in HNSCC, Stransky et al36 identified more HPV-positive cases by use of WES than by use of p16/CDKN2A detection. Further validation of the usefulness of WES could be achieved by determining whether the natural history of patients with WES-defined HPV-positive disease is the same as that for p16 IHC-defined patients. Because WES HPV genetic material detection is accurate,52 the absence of HPV genetic material is considered reliable for determining HPV-negative status; however, the presence of HPV DNA is considered less accurate than HPV RNA (the gold standard for HPV determination). Nevertheless, DNA-based and RNA-based methods showed similar accuracy in a TCGA analysis in HNSCC,52 and discrepancies between HPV DNA-based methods and p16 IHC similar to those found in our analysis have been observed.53
Sample size is an important limitation of this study, particularly for subset analysis based on HPV status and attendant lack of power for exploring HPV-specific biomarker relationships. Further evaluation in larger studies will provide greater insight into potential interactions between mutation, inflammation, and HPV status in their association with response to pembrolizumab monotherapy.
Conclusion
TMB and inflammatory markers (TcellinfGEP and PD-L1) in HNSCC appear to represent distinct, fairly uncorrelated measures within the tumor microenvironment, providing complementary information for propensity to respond to pembrolizumab monotherapy. In this limited data set, TMB and inflammatory measures appeared to follow similar distributions in HPV-positive and HPV-negative tumors, and no evidence of HPV-specific relationships with clinical outcome for these biomarkers was observed. Evaluation in larger, randomized HNSCC studies will facilitate further understanding of the role TMB and inflammatory markers have as biomarkers of response to anti-PD-1 therapies in HNSCC and, when assessed separately or jointly, potentially aid in the identification of patients who will benefit from treatment.
Acknowledgments
We thank the patients and their families and caregivers for participating in the KEYNOTE-012 study. Critical review of the manuscript was provided by Joanne Tomassini. Medical writing and/or editorial assistance was provided by Holly C. Cappelli, PhD, CMPP, Matthew Grzywacz, PhD, and Dana Francis, PhD, of ApotheCom (Yardley, PA). This assistance was funded by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA. Funding for this research was provided by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA.
Footnotes
Contributors: Conception, design, or planning of study: AA, MA, BB, RIH, LH and RC. Acquisition of data: AA, BB, JC, LQMC, JPE, IG, SG, RIH, HK, BK, RMe, RMo, KM, RC, TYS and JW. Analysis of data: LQMC, RIH, LH, JL, RC and TYS. Interpretation of results: BB, JC, LQMC, RIH, BK, LH, JL, RMe, RMo, RC, TYS, MT and JW. Drafting of manuscript: LQMC, LH, JL, RC, TYS and MT. Critically review/revision of manuscript: all authors. Approval to submit for publication: all authors. Guarantor of the study: RIH.
Funding: This study was funded by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co. Inc., Kenilworth, New Jersey, USA.
Competing interests: RIH: research grant (to institution) from Merck; consultant/ad board member for Merck, BMS, Astra Zeneca, Pfizer, Genentech, Kura, Celgene, Eisai, Loxo, Immunomic, GSK, Gilead, Vaccinex, EMD Serono, BioNTech, Achilles; royalties from Up to Date; data safety monitoring board for Nanobiotix, ISA. TYS: grant (to institution) from Merck, Nanobiotix, Regeneron, Bristol Myers Squibb, AstraZeneca; honorarium from Merck, Nanobiotix, Regeneron, Innate Pharma, AstraZeneca, eTheRNA, Nektar. LQMC: grants from Merck, Lily/Imclone, Bristol Myers Squibb, AstraZeneca/MedImmune, Pfizer, Seattle Genetics, Dynavax, Alkermes, Novartis; personal fees from Merck, Pfizer, Dynavax, Synthrox, Alkermes, Cullinan, Elicio, Genentech, Novartis, Daiichi Sankyo, Gilead, Regeneron/Sanofi Genzyme. SG: consultant to Seattle Genetics. JW: research grant (to institution) from Merck; research grant from Merck, AstraZeneca, Celgene, G1; personal fees from AstraZeneca, EMD Serono, Genentech, Inivata, Celgene, G1, Jounce, AbbVie, Rakuten. IG: has nothing to disclose. JPE: has nothing to disclose. BB: grants from Merck, Bristol Myers Squibb, Fox Chase Cancer Center; personal fees from Merck, AstraZeneca, Fox Chase Cancer Center. MT: research grant from MSD, Ono Pharmaceutical, Bristol Myers Squibb, Bayer, Eisai, Merck Biopharma, Pfizer, Rakuten Medical, Novartis; personal fees from MSD, Ono Pharmaceutical, Bristol Myers Squibb, Bayer, Eisai, Merck Biopharma, LOXO, Pfizer, Celgene, Rakuten Medical, Amgen, Novartis. BK: grants from Ono Pharmaceutical, MSD Oncology, AstraZeneca; personal fees from MSD Oncology, AstraZeneca, Genexis, Handok. HK: research grant from Merck, Kura Oncology, Lilly, Exelixis, Elevar Therapeutics, Ayala Pharmaceuticals, Novartis; consulting for PIN Therapeutics, Mitoimmune; advisory board for Bayer, GlaxoSmithKline, Prelude Therapeutics, Achilles Therapeutics. KM: research grant from Solasio Pharma, Pfizer, Amgen, Daiichi Sankyo, Parexel International, MSD, Merck Serono; research grant and honorarium from Sanofi, Ono, Taiho, consulting and honorarium from Eli Lilly, Chugai, honorarium from Takeda, Bristol Myers Squibb, Bayer. AA: employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and stockholder of Merck & Co., Inc., Kenilworth, NJ, USA. RM: employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and stockholder of Merck & Co., Inc., Kenilworth, NJ, USA. MA: employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and stockholder of Merck & Co., Inc., Kenilworth, NJ, USA; patent (US20180327848) issued for RNA Gene Signatures (inventor of 18-gene T-cell inflamed signature). LH: employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and stockholder of Merck & Co., Inc., Kenilworth, NJ, USA. JL: employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and stockholder of Merck & Co., Inc., Kenilworth, NJ, USA; patent (US20180327848) issued for RNA Gene Signatures (inventor of 18-gene T-cell inflamed signature). RC: employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and stockholder of Merck & Co., Inc., Kenilworth, NJ, USA; patent pending for angiogenesis and mMDSC gene expression-based biomarker of tumor response to PD-1 antagonists (patent 2020/167619). JC: former employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and stockholder of Merck & Co., Inc., Kenilworth, NJ, USA. RM: research grant from AstraZeneca, Merck; consulting/advisory board for Rakuten Medical.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not required
Ethics approval
The KEYNOTE-012 study was conducted in accordance with the Declaration of Helsinki and the International Conference on Harmonization Good Clinical Practice guidelines and was approved by the institutional review boards or ethics committees at all sites. All patients provided written informed consent.
References
- 1.Herbst RS, Baas P, Kim D-W, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540–50. 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 2.Chow LQM, Haddad R, Gupta S, et al. Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE-012 expansion cohort. J Clin Oncol 2016;34:3838–45. 10.1200/JCO.2016.68.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 2013;369:134–44. 10.1056/NEJMoa1305133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1B trial. Lancet Oncol 2016;17:956–65. 10.1016/S1470-2045(16)30066-3 [DOI] [PubMed] [Google Scholar]
- 5.Garon EB, Rizvi N, Hui R. Efficacy of pembrolizumab (MK-3475) and validation of PD-L1 expression as a biomarker in patients with non-small cell lung cancer (NSCLC): findings from KEYNOTE-001. presented at: AACR annual meeting; April 18-22, 2015; Philadelphia, PA.
- 6.KEYTRUDA (pembrolizumab) injection, for intravenous use. 08/2021. Merck Sharp & Dohme Corp.: Whitehouse Station, NJ, USA 2021.
- 7.Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255–65. 10.1016/S0140-6736(16)32517-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med 2015;373:123–35. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulley JL, Rajan A, Spigel DR, et al. Avelumab for patients with previously treated metastatic or recurrent non-small-cell lung cancer (JAVELIN Solid Tumor): dose-expansion cohort of a multicentre, open-label, phase 1b trial. Lancet Oncol 2017;18:599–610. 10.1016/S1470-2045(17)30240-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018–28. 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 11.Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–56. 10.1038/nm.3850 [DOI] [PubMed] [Google Scholar]
- 12.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 2017;16:2598–608. 10.1158/1535-7163.MCT-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N Engl J Med 2018;378:2288–301. 10.1056/NEJMoa1716948 [DOI] [PubMed] [Google Scholar]
- 18.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayers M, Nebozhyn M, Cristescu R, et al. Molecular profiling of cohorts of tumor samples to guide clinical development of pembrolizumab as monotherapy. Clin Cancer Res 2019;25:1564–73. 10.1158/1078-0432.CCR-18-1316 [DOI] [PubMed] [Google Scholar]
- 20.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69–74. 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 21.Topalian SL, Taube JM, Anders RA, et al. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275–87. 10.1038/nrc.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nghiem PT, Bhatia S, Lipson EJ, et al. PD-1 blockade with pembrolizumab in advanced Merkel-cell carcinoma. N Engl J Med 2016;374:2542–52. 10.1056/NEJMoa1603702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyford-Pike S, Peng S, Young GD, et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res 2013;73:1733–41. 10.1158/0008-5472.CAN-12-2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandal R, Şenbabaoğlu Y, Desrichard A, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight 2016;1:e89829. 10.1172/jci.insight.89829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burtness B, Harrington KJ, Greil R, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 2019;394:1915–28. 10.1016/S0140-6736(19)32591-7 [DOI] [PubMed] [Google Scholar]
- 26.Cohen EEW, Soulières D, Le Tourneau C, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet 2019;393:156–67. 10.1016/S0140-6736(18)31999-8 [DOI] [PubMed] [Google Scholar]
- 27.Mehra R, Seiwert TY, Mahipal A, et al. Efficacy and safety of pembrolizumab in recurrent/metastatic head and neck squamous cell carcinoma (r/m HNSCC): pooled analyses after long-term follow-up in KEYNOTE-012. JCO 2016;34:6012. 10.1200/JCO.2016.34.15_suppl.6012 [DOI] [Google Scholar]
- 28.Bauml J, Seiwert TY, Pfister DG, et al. Pembrolizumab for platinum- and cetuximab-refractory head and neck cancer: results from a single-arm, phase II study. J Clin Oncol 2017;35:1542–9. 10.1200/JCO.2016.70.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehra R, Seiwert TY, Gupta S, et al. Efficacy and safety of pembrolizumab in recurrent/metastatic head and neck squamous cell carcinoma: pooled analyses after long-term follow-up in KEYNOTE-012. Br J Cancer 2018;119:153–9. 10.1038/s41416-018-0131-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posner MR, Lorch JH, Goloubeva O, et al. Survival and human papillomavirus in oropharynx cancer in TAX 324: a subset analysis from an international phase III trial. Ann Oncol 2011;22:1071–7. 10.1093/annonc/mdr006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannataro VL, Gaffney SG, Sasaki T, et al. APOBEC-induced mutations and their cancer effect size in head and neck squamous cell carcinoma. Oncogene 2019;38:3475–87. 10.1038/s41388-018-0657-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferris RL, Blumenschein G, Fayette J, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 2016;375:1856–67. 10.1056/NEJMoa1602252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fakhry C, Westra WH, Li S, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 2008;100:261–9. 10.1093/jnci/djn011 [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Sun H, Zeng Q, et al. HPV-positive status associated with inflamed immune microenvironment and improved response to anti-PD-1 therapy in head and neck squamous cell carcinoma. Sci Rep 2019;9:13404. 10.1038/s41598-019-49771-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Comprehensive Cancer Network . NCCN clinical practice guidelines in oncology (NCCN guidelines): head and neck cancer (version 1.2021). Available: https://www.nccn.org/store/login/login.aspx?ReturnURL=https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf [Accessed 7 Apr 2021].
- 36.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157–60. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandrani P, Kulkarni V, Iyer P, et al. NGS-based approach to determine the presence of HPV and their sites of integration in human cancer genome. Br J Cancer 2015;112:1958–65. 10.1038/bjc.2015.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nanda R, Chow LQM, Dees EC, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. JCO 2016;34:2460–7. 10.1200/JCO.2015.64.8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herbst RS, Lopes G, Kowalski DM, et al. Association between tissue TMB (tTMB) and clinical outcomes with pembrolizumab monotherapy (pembro) in PD-L1-positive advanced NSCLC in the KEYNOTE-010 and -042 trials. Ann Oncol 2019;30:v916–7. 10.1093/annonc/mdz394.077 [DOI] [Google Scholar]
- 40.Aurora-Garg D, Albright A, Qiu P. Large-Scale evaluation of concordance of genomic scores in whole exome sequencing and Foundation medicine comprehensive genomic platform across cancer types. J Immunother Cancer 2019;7:172–3.31291990 [Google Scholar]
- 41.Brandwein-Gensler M, Smith RV. Prognostic indicators in head and neck oncology including the new 7th edition of the AJCC staging system. Head Neck Pathol 2010;4:53–61. 10.1007/s12105-010-0161-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson DB, Lovly CM, Flavin M, et al. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol Res 2015;3:288–95. 10.1158/2326-6066.CIR-14-0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ribas A, Robert C, Schachter J. T cell inflamed gene expression profile (GEP) analysis of Pembrolizumab-and Ipilimumab-treated patients wtih advanced melanoma in the multicenter, randomized, open-label phase 3 KEYNOTE 006 study. J Immunother Cancer 2017;5:Abstract P79. [Google Scholar]
- 44.Maia MC, Almeida L, Bergerot PG, et al. Relationship of tumor mutational burden (TMB) to immunotherapy response in metastatic renal cell carcinoma (mRCC). JCO 2018;36:662. 10.1200/JCO.2018.36.6_suppl.662 [DOI] [Google Scholar]
- 45.Hellmann MD, Ciuleanu T-E, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018;378:2093–104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribas A, Robert C, Schachter J. Tumor mutational burden (TMB), T cell-inflamed gene expression profile (GEP) and PD-L1 are independently associated with response to pembrolizumab (Pembro) in patients with advanced melanoma in the KEYNOTE (KN)-006 study. Cancer Res 2019;79:4217. [Google Scholar]
- 47.Schlecht NF, Brandwein-Gensler M, Nuovo GJ, et al. A comparison of clinically utilized human papillomavirus detection methods in head and neck cancer. Mod Pathol 2011;24:1295–305. 10.1038/modpathol.2011.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer 2010;116:2166–73. 10.1002/cncr.25033 [DOI] [PubMed] [Google Scholar]
- 49.Fakhry C, Lacchetti C, Rooper LM, et al. Human papillomavirus testing in head and neck carcinomas: ASCO clinical practice guideline endorsement of the College of American pathologists guideline. J Clin Oncol 2018;36:3152–61. 10.1200/JCO.18.00684 [DOI] [PubMed] [Google Scholar]
- 50.Chen ZW, Weinreb I, Kamel-Reid S, et al. Equivocal p16 immunostaining in squamous cell carcinoma of the head and neck: staining patterns are suggestive of HPV status. Head Neck Pathol 2012;6:422–9. 10.1007/s12105-012-0382-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahajan A. Practical issues in the application of p16 immunohistochemistry in diagnostic pathology. Hum Pathol 2016;51:64–74. 10.1016/j.humpath.2015.12.021 [DOI] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Network . Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ndiaye C, Mena M, Alemany L, et al. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol 2014;15:1319–31. 10.1016/S1470-2045(14)70471-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003026supp001.pdf (110.8KB, pdf)