Abstract
Introduction:
Increasing occurrence of infections caused by multidrug-resistant Gram-negative bacteria resulted in colistin being the last agent for treatment. Apart from plasmid-mediated mcr genes, mutations involving several genes like mgrB, phoP/phoQ, pmrA, pmrB, pmrC, and crrABC genes, are leading causes of colistin resistance. Four colistin susceptibility testing methods were compared against broth microdilution (BMD) and determined the presence of the mcr1-5 gene.
Methodology:
A total of 100 carbapenem-resistant Enterobacterales isolates were tested for colistin susceptibility by commercial broth microdilution (cBMD), E-test, VITEK-2, and rapid polymyxin NP assay (RPNP) and compared with BMD. The presence of the mcr1-5 gene was determined by modified RPNP and PCR. Two non-mcr colistin-resistant XDR isolates were subjected to whole-genome sequencing using Illumina MiSeq sequencing platform.
Results:
Among 100 carbapenem-resistant Enterobacterales isolates, 15% were resistant to colistin. Essential agreement, categorical agreement, major error, and very major error for cBMD/E-test/VITEK-2/RPNP were 96%/73%/82%/NA; 99%/86%/88%/91%, 1.2%/9.4%/11.8%/8.2% and 0%/40%/13.3%/13.3%, respectively. Only one Klebsiella pneumoniae isolate harbored the mcr-1 gene, observed by both methods. Whole-genome sequencing of two non-mcr XDR Klebsiella pneumoniae showed multiple mutations in 10 genes responsible for lipopolysaccharide biosynthesis.
Conclusions:
The performance of cBMD was excellent, whereas the E-test was unacceptable. VITEK-2 and RPNP performed better but remained unreliable due to high error rates. Multiple mutations in the target proteins involving lipopolysaccharide formation, modification, and regulation were seen, resulting in colistin resistance.
Keywords: broth microdilution, carbapenem-resistant Enterobacterales, colistin, mcr-1, Whole-genome sequencing
Introduction
The increasing incidence of multidrug-resistant (MDR) Gram-negative bacilli (GNB) such as Enterobacterales, Acinetobacter baumannii, and Pseudomonas aeruginosa infections leads to the resurrection and repurposing of old antibiotics, among which polymyxins have shown promising activity against these pathogens.1–5
As part of the routine antibiogram panel, the susceptibility of colistin must be tested in settings where these MDR GNBs are prevalent to avoid delays in reporting. For appropriate therapeutic decision-making, rapid and reliable colistin susceptibility testing methods is the need of the hour in routine diagnostic microbiology laboratories. To date, few studies around the globe have elaborately studied the performance of different colistin susceptibility methods, showing opposing results; therefore, more studies are needed to establish the most accurate method. 6
The addition of L-Ara4-N and phosphoethanolamine (PEtN) molecules causing an alteration in the lipopolysaccharide region of bacterial outer membrane is the most common way leading to colistin resistance. 7 Mutations involving the two-component regulatory systems like phoP/phoQ, pmrA, pmrB, pmrC, and crrABC genes, or their regulator like mgrB leading to Lipid A modifications have been reported in the literature.7,8 Recently, plasmid-mediated colistin resistance genes, that is, mcr-1-5, which encodes for PEtN transferase, were demonstrated in Klebsiella pneumoniae (K. pneumoniae), Escherichia coli (E. coli), and Salmonella spp.9,10
Disk diffusion is the most prevalent antimicrobial susceptibility testing method used in routine diagnostic microbiology laboratories. But this method is not recommended for detection of colistin resistance like other antibiotics because of high error rates leading to discordant and unreliable results compared to minimum inhibition concentration (MIC) based methods.11,12
Some studies have demonstrated excellent correlations between the results of the E-test and broth microdilution (BMD) methods for colistin,12–16 while others questioned its reliability.17,18 Although E-test strips are convenient in determining MIC levels but because of conflicting performance results, routine colistin susceptibility testing using E-test is not recommended. In addition, limited studies have tested the performance of automated methods like VITEK-2 for colistin susceptibility and could not reach any conclusive decision.12,15,18
BMD is mostly considered the gold standard method for determining antibiotic MICs, although not convenient for routine clinical laboratories. 19 It is sometimes becomes impractical to include BMD as a part of the routine antibiogram panel for laboratories specifically in resource-poor settings due to its extensive requirement of human resources, high cost, and lengthy test timing. Many laboratories perform BMD either due to specific requests for colistin susceptibility or after detecting resistance to other drugs like carbapenems, causing a delay in reporting.20,21 In addition, Colistin shows a property of adherence of varying degrees to different surfaces used to make BMD plates, which may cause a reduction in antibiotic concentrations affecting colistin susceptibility testing. 22 Surfactant polysorbate 80’s addition to BMD tray minimized colistin adhesion leading to a significant decrease in colistin MICs, affecting mainly bacteria with relatively low MICs.17,20,23 Nevertheless, surfactant polysorbate 80 is not recommended by the Clinical and Laboratory Standards Institute (CLSI) for colistin susceptibility by BMD. 24
‘Colistin Heteroresistance’ is defined as colistin‑resistant subpopulations that emerge from a colistin‑susceptible population under colistin pressure and may be evidenced by the presence of skip wells in BMD. 7 Due to all these issues, commercial BMD (cBMD) methods have come up recently. These methods do not need hectic standardization steps, do not have cumbersome logistic problems and show excellent correlation with BMD. 25 Apart from cBMD platforms, two other methods, namely colistin broth disk elution method and colistin agar test, have performed at par with BMD and are now recommended as standard tests for colistin susceptibility testing by CLSI.24,26
The breakpoints for colistin have changed every year, and different organizations have differences among them, which ultimately complicates the interpretation of colistin susceptibility results.24,27 In this study, the performance of various colistin MIC testing methods, such as cBMD, E-test, and VITEK-2, along with the colorimetric test, that is, RPNP, were tested against BMD using a collection of non-repeating carbapenem-resistant Enterobacterales (CRE) isolates. Also, we performed genetic profiling of extremely drug-resistant (XDR) non-mcr colistin-resistant isolates by whole-genome sequencing (WGS).
Material and methods
Isolate collection
A total of 100 CRE isolates comprising only E. coli, K. pneumoniae, and Enterobacter cloacae from 263 Enterobacterales isolates were collected. These non-repeating strains were isolated from blood, sterile fluids, and respiratory samples of patients treated in a tertiary care hospital in Delhi, India. The study was conducted from 2018–2019 and all these 100 CRE isolates were tested for colistin susceptibility using colistin breakpoints published in European Committee on Antimicrobial Susceptibility Testing (EUCAST) and CLSI guidelines.24,27 VITEK-2 automated AST system was used to determine CRE status based on MIC level against carbapenems. Species identification was performed using MALDI-TOF (VITEK-MS system, bioMérieux, Marcy-l'Étoile, France). The isolates were stored at -80ºC and sub-cultured before testing. The MIC of these 100 isolates out of 263 Enterobacterales isolates was determined by VITEK-2.
Colistin susceptibility
Various phenotypic methods used to determine colistin MICs are cBMD (Mikrolatest® MIC colistin), E-test (Himedia, India), and VITEK-2 (bioMérieux, Marcy-l'Étoile, France) following which comparison of the results were made with BMD results according to CLSI breakpoints (intermediate, ⩽ 2 µg /mL; resistant, ⩾ 4 µg/mL). 24 For the purpose of this study, the ‘Intermediate’ category mentioned in CLSI guideline is considered as ‘Susceptible’. In the interpretation of colistin MIC, as EUCAST breakpoints (resistant > 2µg/mL; susceptible ⩽ 2 µg/mL) were similar to that of CLSI, we used both the guidelines in CLSI/EUCAST format for the study. 27 In case of disagreement, the BMD method was used for reporting susceptibility. cBMD is a ready-to-use BMD test kit coated with different colistin concentrations to determine colistin MIC. Rapid Polymyxin NP (RPNP) (ELITech Group, Puteaux, France) is a colorimetry-based method detecting bacterial growth indicated by glucose metabolism in the presence of 2 μg/mL of colistin. Colistin resistance is indicated by pH shift due to the formation of acid metabolites resulting from glucose metabolism, which in turn causes a color shift of phenol red indicator from orange to yellow. 28 VITEK-2 AST susceptibility cards AST- GN-280 (bioMérieux, Marcy-l'Étoile, France) were used as per the instructions mentioned by the manufacturer, where colistin MIC is reported in the range between ⩽ 0.5 to ⩾ 16 μg/mL. E. coli ATCC 25922 (colistin-susceptible, colistin MIC, 0.25 µg/mL to 2 µg/mL) was used as negative quality control strains, whereas E. coli NCTC 13846 (colistin-resistant harboring mcr-1 gene, colistin MIC, 4 µg/mL) was used as positive quality control strains in the study.24,29
Evaluation of minimum inhibitory concentration correlations
Essential agreement (EA) was defined as an E-test/ cBMD/ VITEK-2 MIC equal to or within ± 1 dilution of the MIC result of BMD. Categorical agreement (CA) was met when E-test/ cBMD/ RPNP/ VITEK- 2 interpretive criteria agreed (susceptible /resistant) with BMD interpretive criteria. A major error (ME) occurred when cBMD/ RPNP/ VITEK-2 results were resistant, and BMD was susceptible and was calculated only for susceptible isolates. A very major error (VME) occurred when cBMD/ RPNP/ VITEK-2 results were susceptible, and BMD was resistant and was calculated only for resistant isolates. 30 Colistin susceptibility performance was considered acceptable if both the CA and EA were ⩾ 90%. In addition, MEs and VMEs rates were benchmarked as ⩽ 10% and ⩽ 3%, respectively, for acceptable performance. 31
All the discordant results (MEs and VMEs) were retested in triplicate to reconfirm the results. In the repeat testing, if the error was resolved, the repeat results were kept as final. If repeat MIC values were within ± 2 log dilution and categorical error in the form of MEs and VMEs remained, the categorical error was accepted. Skip well phenomenon was defined as the absence of growth of an isolate at a lower antimicrobial concentration(s). A single skip well did not affect the MIC interpretation, while multiple skip wells were considered uninterpretable according to CLSI guidelines. 19 For the skip well phenomenon in BMD panels, the isolates were retested.
Modified rapid polymyxin NP
RPNP solution was prepared by adding 6.25 g of cation-adjusted Mueller-Hinton II broth powder and 0.0125 g of phenol red (Sigma-Aldrich) in 225 mL of distilled water. 32 The pH of the resultant solution was adjusted at 6.7 with 1 mol/L HCl, following which sterilization was carried out by autoclaving the solution at 121°C for 15 min at 15 lbs. After sterilization, the RPNP solution was cooled at room temperature, and 25 mL of filter-sterilized 10% anhydrous D(+)-glucose (Sigma-Aldrich) was added.
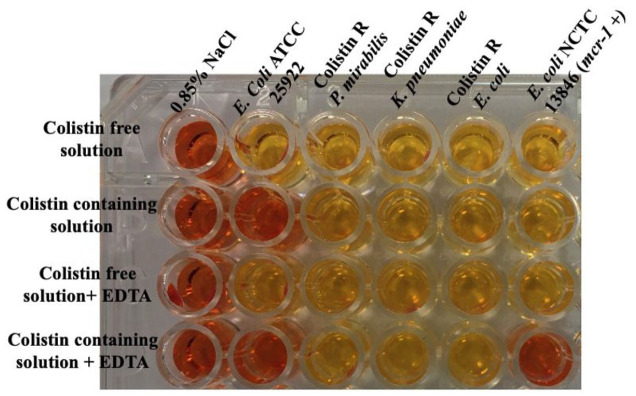
The RPNP test was modified by incorporating two additional wells, one containing colistin-free RPNP solution plus EDTA (80 µg/mL) [EDTA is an inhibitor of PEtN transferase] and the other having RPNP solution containing colistin (2 µg/mL) along with EDTA (80 µg/mL). In the modified scheme (Figure 3), row A wells were filled with 150 µL of colistin-free RPNP solution. In row B wells, 150 µL of RPNP solution containing colistin sulfate was filled. Next, row C wells were added with 150 µL of colistin-free RPNP solution plus EDTA only. Finally, row D wells were added with 150 µL of RPNP solution containing both colistin sulfate and EDTA. In the next step, to all wells of Column 1, 50 µL of 0.85% NaCl (negative sterility control) was added. In other respective columns for each strain, 50 µL of a 3.0 to 3.5 McFarland bacterial suspension corresponding to ~109 CFU/mL was added and adequately mixed with 150 µL reaction solution in each well. Incubation of the 96 -well plate was done at 35 ± 2°C under aerobic conditions for 4 h.
Figure 3.
Modified rapid polymyxin NP test (mRPNP). Wells A1-A6 were filled with colistin-free RPNP solution. Wells B1 to B6 were filled with RPNP solution + colistin sulfate. Wells C1 to C6 were filled with colistin-free NP + EDTA. Wells D1 to D6 were added with RPNP solution + colistin sulfate + EDTA. Wells in column 1 were filled with 0.85% NaCl (negative sterility control), whereas Columns 2 to 7 represent the mRPNP test performed for E. coli ATCC 25922, P. mirabilis ATCC 25933, colistin-resistant mcr-1-5 negative K. pneumoniae, colistin-resistant mcr-1-5 negative E. coli strain, and mcr-1-positive E. coli NCTC 13846, respectively. The plates were incubated at 35 ± 2°C under aerobic conditions for 4 h, and visual changes in the color of the wells were monitored each hour. In wells, B1 to B6, a color change from orange to yellow was considered positive to colistin resistance, whereas the mRPNP test was considered positive to MCR-1 phosphoethanolamine transferase production when the colistin-containing solution supplemented with EDTA (wells D1 to D6) remained orange (i.e. absence of glucose metabolization); this shows that growth of the colistin-resistant E. coli (mcr-1-positive) in the well containing colistin solution (well D6) was inhibited by EDTA. Since mcr-1 translates to PEtN transferase, which is a zinc enzyme, exposure to chelators like EDTA could reduce colistin resistance in mcr-1-producing strains.
At every 1 h, change of color of the 200 µL reaction mixture in each well was monitored. Any color change from orange to yellow in wells containing colistin sulfate was considered positive for colistin resistance. When the wells with colistin-containing solution supplemented with EDTA remained orange, signifying the absence of glucose metabolism due to inhibition of mcr-1 positive colistin-resistant strains. mRPNP was performed with all the 100 CRE isolates and ten intrinsically colistin-resistant Morganellaceae isolates ( that is, 4 Proteus mirabilis; 3 Proteus vulgaris; 3 Morganella morganii). E. coli ATCC 25922 (colistin-susceptible, colistin MIC, 0.25 µg/mL to 2 µg/mL), P. mirabilis ATCC 25933 (non-mcr negative intrinsic colistin-resistant, colistin MIC, > 2 µg/mL) was used as negative quality control strains, whereas E. coli NCTC 13846 (colistin-resistant harboring mcr-1 gene, colistin MIC, 4 µg/mL) was used as positive quality control strains.
DNA extraction and conventional multiplex PCR for the mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 genes
Extraction of the genomic DNA of the 100 CRE and 10 intrinsically colistin-resistant Morganellaceae isolates was done by the boiling method, following which conventional multiplex PCR was performed using the Taq PCR Premix (Thermo-scientific) to amplify the mcr-1(mcr1_320bp_fw: AGTCCGTTTGTTCTTGTGGC, mcr1_320bp_rev: AGATCCTTGGTCTCGGCTTG; 320 bp), mcr-2(mcr2_700bp_fw: CAAGTGTGTTGGTCGCAGTT, mcr2_700bp_rev: TCTAGCCCGACAAGCATACC; 715 bp), mcr-3(mcr3_900bp_fw: AAATAAAAATTGTTCCGCTTATG, mcr3_900bp_rev: AATGGAGATCCCCGTTTTT; 929 bp), mcr-4(mcr4_1100bp_fw: TCACTTTCATCACTGCGTTG, mcr4_1100bp_rev: TTGGTCCATGACTACCAATG; 1116 bp), and mcr-5(MCR5_fw: ATGCGGTTGTCTGCATTTATC, MCR5_rev: TCATTGTGGTTGTCCTTTTCTG; 1644 bp) genes. Amplification of mcr-1-5 genes was done with a 25 µl PCR reaction mixture with primers using the following PCR cycling conditions [ i.e. pre-denaturation: 94℃ for 15 minutes; amplification: 30 seconds at 94℃, 90 seconds at 58℃, and 60 seconds at 72℃ for 25 cycles; and final amplification: 72℃ for10 minutes]. Gel electrophoresis to visualize the respective amplified DNA PCR product was done using 2% agarose gel and ethidium bromide. 33
Whole genome sequencing, sequence assembly, annotation, and analysis
The only two colistin-resistant XDR [non-susceptibility to at least one agent in all but two or fewer antimicrobial categories] 30 non-mcr K. pneumoniae isolates mentioned as H-53, and H-62 in the present study were subjected to WGS in a high-throughput Illumina MiSeq sequencing platform (Illumina, Inc., USA) at Translational Health Science and Technology Institute, Faridabad. The rest of the 13 colistin-resistant isolates were not included for WGS due to economic constraints. The quality and quantity of the isolated genomic DNA of each of the isolates were monitored by Thermo Scientific NanoDrop 2000 and resolving the nucleic acid in 0.8% agarose gel, respectively. Any contamination of phenolic compounds in the nucleic acid preparation was monitored by BioSpectrometer (Eppendorf, Germany). Approximately 0.2 ng of pure genomic DNA from each isolate was used for shotgun sequencing. DNA fragmentation, library preparation, and pair-end sequencing were done using the Nextera XT DNA Library preparation kit (Illumina, Inc., USA). FastQC and Trimmomatic programs measured the quality of raw reads. The Unicycler pipeline was used for the assembly of cleaned pair-end reads. 34 Rapid Annotation Subsystem Technology (RAST) automated annotation pipeline was used for genome annotation of both the assembled genomes. 35 The publicly available protein database was used to confirm the annotated proteins. The amino acid sequences were analyzed with Protein Variation Effect Analyzer (PROVEAN, http://provean.jcvi.org/index.php) allowing prediction by algorithm of the functional impact for all classes of sequence variations. 36 The change in the alignment score was considered as a measure of change in similarity caused by variation and thus to protein functionality.
Nucleotide sequence accession numbers
The whole-genome sequences of K. pneumoniae H-53 and H-62 have been deposited in National Center for Biotechnology Information (NCBI) under BioProject database IDPRJNA685993 (Accession: SAMN17103367) and PRJNA685995 (Accession: SAMN17103397), respectively.
Statistical analyses
SPSS software v.20.0 (SPSS Inc., Chicago, IL) was used to analyze the data, and χ2 test was done. Fisher's exact test was used when data were scarce. The significance was set at p < 0.05 using two-sided comparisons. The correlation between cBMD, E-test, VITEK-2, and RPNP against BMD was calculated using the Pearson method.
Results
Out of 100 CRE isolates most common organism to be isolated was E. coli 50% (50/100), followed by K. pneumoniae. 48% (48/100), and finally, Enterobacter cloacae 2% (2/100).
Colistin susceptibility by different methods
Among these 100 CRE isolates, according to CLSI/EUCAST guidelines, total colistin resistance by standard BMD method was observed to be 15%. Species wise, E. coli had 16% (8/50) colistin resistance, followed by K. pneumoniae. 14.6% (7/48). Both the isolates of Enterobacter cloacae were found to be susceptible to colistin, although one isolate showed skip well phenomenon at MIC value of 0.25 µg/mL. The result of cBMD, VITEK-2, and E-test following CLSI/EUCAST breakpoints had been shown in the table (Table 1). The percentage of resistant isolates by the cBMD method was 16%. E-test showed similar results whereas VITEK-2 showed a higher rate of colistin resistance, that is, 23% (Table 1). On the other hand, the colorimetric method, RPNP, showed colistin resistance of 20%.
Table 1.
Colistin resistance profile among different species of CRE by different phenotypic methods along with their MIC50/90 values.
| Organism | Colistin MIC50/90
(µg/ml)(MIC range) (µg/ml) |
Colistin resistance(%) by CLSI/EUCAST | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cBMD | E-test | VITEK-2 | RPNP | |||||||||
| BMD | cBMD | E-test | VITEK-2 | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Negative | Positive | |
| E. coli (50) | 0.5/4(0.125-32) | 0.5/4(0.125-32) | 0.5/16(0.125-256) | 0.5/8(0.5-16) | 82 | 18 | 82 | 18 | 82 | 18 | 82 | 18 |
| Klebsiella pneumoniae (48) | 0.5/8(0.125-16) | 0.5/8(0.125-32) | 0.5/16(0.125-128) | 0.5/16(0.5-16) | 85.4 | 14.6 | 85.4 | 14.6 | 70.8 | 29.2 | 77.1 | 22.9 |
| Enterobacter cloacae (2) | 1/2(0.25-2) | 0.5/1(0.5-1) | 0.25/0.25(0.25) | 0.5/0.5(0.5) | 100 | 0 | 100 | 0 | 100 | 0 | 100 | 0 |
| Total (100) | 0.5/4(0.125-32) | 0.5/16(0.125-32) | 0.5/4(0.125-256) | 0.5/4(0.5-16) | 84 | 16 | 84 | 16 | 77 | 23 | 80 | 20 |
BMD, Broth microdilution; CLSI, Clinical and Laboratory Standards Institute; E. coli, Escherichia coli; EUCAST, European Committee on Antimicrobial Susceptibility Testing; MIC, Minimum inhibition concentration; RPNP, Rapid Polymyxin NP.
Evaluation of minimum inhibitory concentration correlations and error rates
Colistin had the same MIC50 and MIC90 (0.5 µg/mL) in the colistin susceptible strain. In colistin-resistant strains, overall MIC50 of 8 µg/mL and MIC90 of 16 µg/mL were observed. The MIC50/90 rates of cBMD were similar to BMD, whereas both E-test and VITEK-2 had higher MIC90 values (Table 1). The rate of colistin resistance was identical for all the methods with respect to E. coli, 36% (18/50), and Enterobacter cloacae 0% (0/2). Whereas in the case of K. pneumoniae RPNP and VITEK-2 showed higher rate of colistin resistance than cBMD and E-test (Table 1).
The overall sensitivity (SN) for cBMD was 100%, whereas specificity (SP) was 98.8%.VITEK-2 showed SN ad SP, of 86.7% and 88.2%, respectively. In comparison, RPNP had SN similar to VITEK-2 but with a higher SP of 91.8%. Among all the phenotypic tests compared in the study, E-test fared poorly with SN and SP of 60% and 90.6%, respectively (Table 2).
Table 2.
In-vitro antibacterial activity of colistin against 100 CRE isolates using cBMD, E-test, VITEK-2, and RPNP and categorization of errors compared with BMD method.
| cBMD (%) | E-test (%) | VITEK-2 (%) | RPNP (%) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EA | CA | ME | VME | SN | SP | EA | CA | ME | VME | SN | SP | EA | CA | ME | VME | SN | SP | CA | ME | VME | SN | SP | |
| E. coli (50) | 94 | 98 | 2.3 | 0 | 100 | 97.6 | 74 | 86 | 9.5 | 37.5 | 62.5 | 88.1 | 86 | 94 | 4.8 | 14.3 | 87.5 | 95.2 | 96 | 2.4 | 12.5 | 100 | 97.6 |
| Klebsiella pneumoniae. (48) | 97.9 | 100 | 0 | 0 | 100 | 100 | 89.5 | 79.2 | 17.1 | 42.9 | 57.1 | 92.7 | 77.1 | 89.6 | 8.9 | 14.3 | 85.7 | 80.5 | 83.3 | 14.6 | 28.6 | 71.4 | 85.4 |
| Enterobacter cloacae (2) | 100 | 100 | 0 | 0 | 100 | 100 | 50 | 100 | 0 | 0 | 100 | 100 | 50 | 100 | 0 | 0 | 100 | 100 | 100 | 0 | 0 | 100 | 100 |
| Total (100) | 96 | 99 | 1.2 | 0 | 100 | 98.8 | 72 | 86 | 9.4 | 40 | 60 | 90.6 | 81 | 88 | 11.8 | 13.3 | 86.7 | 88.2 | 91 | 8.2 | 13.3 | 86.7 | 91.8 |
CA, Categorical agreement; cBMD, Commercial Broth microdilution; EA, Essential agreement; E. coli, Escherichia coli; ME, Major error; MIC, Minimum inhibition concentration; RPNP, Rapid Polymyxin NP; SN, Sensitivity; SP, Specificity; VME, Very major error.
According to CLSI/ EUCAST breakpoints, EA, CA, ME, and VME for cBMD/E-test/VITEK-2 were 96%/72%/81%; 99%/86%/88%, 1.2%/9.4%/11.8% and 0%/40%/13.3%, respectively. In RPNP, CA, ME, and VME rates were 91%, 8.2%, and 13.3%, respectively (Table 2).
The performance of cBMD for K. pneumoniae was found to be excellent in comparison to E. coli. VITEK-2 and RPNP had similar error rates, with their performance better in E. coli than K. pneumoniae. However, E-test fared poorly, having the highest ME and VME rates compared to other methods (Table 2).
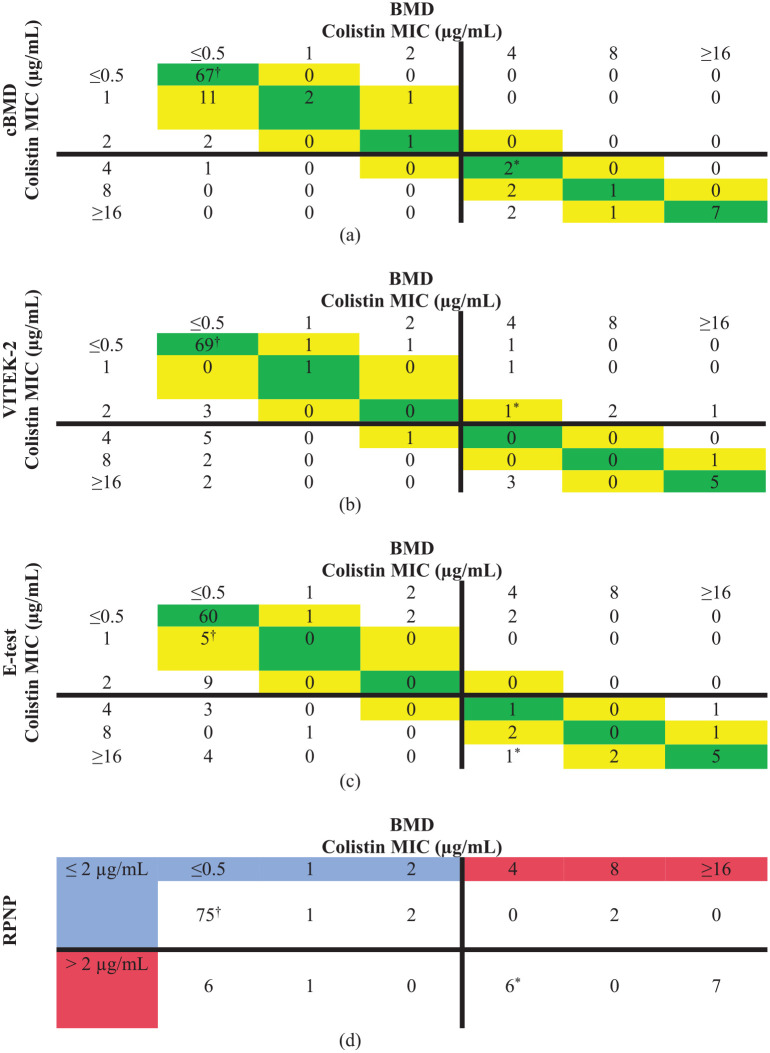
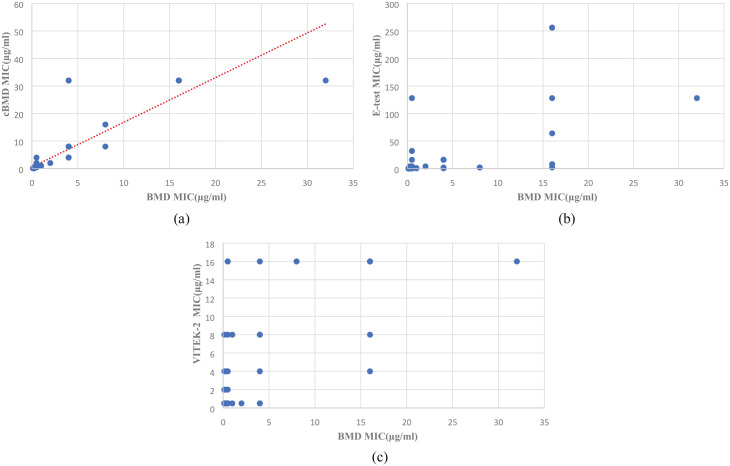
A Scatter diagram was plotted using colistin MIC results of 100 isolates by cBMD, VITEK-2, E-test, and RPNP and compared with the BMD test result. Ninety-five isolates showed an excellent correlation in the case of cBMD when compared with BMD results (Figure 1(a)). VITEK-2 showed excellent correlation with 79% isolate lying with ± 1 log2 dilution of BMD (Figure 1(b)). However, a relatively poor correlation was seen between the E-test and BMD results with 77% isolates having E-test MIC lying between ± 1 log2 dilution of BMD MIC. Sixteen percent of isolates having BMD MIC of ⩽ 0.5 µg/mL had E-test MIC of ⩾ 2 µg/mL (Figure 1(c)). Comparing the RPNP method results with BMD, a good categorical agreement was observed with isolates having MIC value ⩽ 2 µg/mL, showing 78/85(91%) isolates susceptible to colistin. Few isolates, that is, 2 out of 15 (13.3%) with MIC > 2 µg/mL by BMD, were wrongly classified as susceptible by RPNP (Figure 1(d)). The correlation plot between MIC results of cBMD and BMD values was drawn for the comparative analysis. Pearson R2 value was found at 0.8736, which indicates excellent concordance between both methods. On the other hand, both VITEK-2 and E-test showed fair concordance compared to BMD (Figure 2).
Figure 1.
Scatter diagram of colistin MICs obtained by BMD compared with a) cBMD, b) VITEK-2, c) E-test, and d) CNP for 100 isolates. Essential agreement is highlighted in green (perfect agreement) and yellow (±1 log2 dilution). The solid horizontal and vertical lines represent the clinical breakpoint value established by CLSI/ EUCAST. Blue color signifies susceptible MIC, whereas Red color signifies resistance MIC range. Escherichia coli NCTC 13846 (mcr-1 positive) and Escherichia coli ATCC 25922 (colistin susceptible) are represented by * and †, respectively.
Figure 2.
Correlation plot of colistin MICs obtained by BMD compared with a) cBMD, b) E-test, c) VITEK-2 for 100 isolates. Pearson correlation values show an excellent correlation between BMD and cBMD, whereas it shows a fair correlation for E-test and VITEK-2.
Modified rapid polymyxin NP and conventional multiplex PCR for mcr 1-5 genes
Only one K. pneumoniae isolate out of 15 (6.7%) colistin-resistant strains confirmed by BMD harbored mcr-1 gene, which was observed by conventional mcr 1-5 gene multiplex PCR and as well as predicted by mRPNP. Both PCR and mRPNP were negative for mcr1-5 genes in all 10 intrinsic colistin-resistant isolates of family Morganellaceae. mRPNP showed a SN and SP of 100% when compared with PCR.
Whole-genome sequencing and analysis
The WGS analysis revealed that the closest neighbor of H-53 and H-62 strains is K. pneumoniae MGH 78578. Our analysis of the amino acid sequences of all the 15 protein molecules associated with colistin resistance identified the accumulation of multiple mutations (Table 3). Out of these 15 proteins, such as sensor proteins PmrB, UDP-glucose 6-dehydrogenase PmrE, Lipid A phosphoethanolamine transferase PmrC, bifunctional polymyxin resistance protein ArnA_FT, UDP-4-amino-4-deoxy-L-arabinose-oxoglutarate aminotransferase ArnB, undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase ArnC, and undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase ArnT except PhoP/PhoQ regulator MgrB regulator MgrB had multiple amino acid changes in their protein sequences. In the case of Lipid A palmitoyl transferase PagP, 2 amino acid deletion, and addition of 17 amino acids were found at N-terminal of H-53 and H-62, respectively. Both isolates also harbored efflux pump AcrAB and spermidine export protein MdtI/KpnF, which are also known to confer resistance to many antimicrobials, including colistin.
Table 3.
Cumulative results of amino acid substitutions in various genes contributing to colistin resistance found upon WGS analysis of two non-MCR colistin-resistant XDR K. pneumoniae strains.
| Genes | Isolate-53 | Isolate-62 | ||||
|---|---|---|---|---|---|---|
| Mutation | PROVEAN score | Prediction (cut off = −2.5) | Mutation | PROVEAN score | Prediction (cut off = −2.5) | |
| pmrA (transcriptional regulatory protein PmrA) | G53V | -7.575 | Deleterious | A217V | -0.729 | Neutral |
| A217V | -0.729 | Neutral | ||||
| pmrB (Sensor protein PmrB) | A246T | -1.132 | Neutral | A246T | -1.132 | Neutral |
| M213L | -0.623 | Neutral | T157P | -5.787 | Deleterious | |
| M213L | -0.623 | Neutral | ||||
| pmrC/ eptA (Lipid A phosphoethanolamine transferase) | S25G | -1.726 | Neutral | S25G | -1.726 | Neutral |
| Q319R | -1.735 | Neutral | Q319R | -1.735 | Neutral | |
| F27C | -2.411 | Neutral | F27C | -2.411 | Neutral | |
| pmrE/ugd (UDP-glucose 6-dehydrogenase) | I17V | -0.008 | Neutral | I17V | -0.008 | Neutral |
| mgrB (PhoP/PhoQ regulator) | G37C | -9.000 | Deleterious | G37C | -9.000 | Deleterious |
| arnB (UDP-4-amino-4-deoxy-L-arabinose--oxoglutarate | A112D | 0.683 | Neutral | A112D | 0.683 | Neutral |
| S150P | 1.430 | Neutral | S150P | 1.430 | Neutral | |
| aminotransferase) | V168I | 0.203 | Neutral | V168I | 0.203 | Neutral |
| S280T | -1.149 | Neutral | S280T | -1.149 | Neutral | |
| arnC (Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase) | A32T | -1.050 | Neutral | A32T | -1.050 | Neutral |
| A115E | -1.351 | Neutral | A115E | -1.351 | Neutral | |
| T139S | -1.383 | Neutral | T139S | -1.383 | Neutral | |
| I286V | -0.461 | Neutral | I286V | -0.461 | Neutral | |
| arnT (Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase) | L114M I117V | -0.942 | Neutral | L114M I117V | -0.942 | Neutral |
| S164G | 0.037 | Neutral | S164G | 0.037 | Neutral | |
| M177I | 2.646 | Neutral | M177I | 2.646 | Neutral | |
| K372R | -3.810 | Deleterious | K372R | -3.810 | Deleterious | |
| 0.301 | Neutral | 0.301 | Neutral | |||
| arnA_FT (Bifunctional polymyxin resistance protein ArnA) | H363N | -3.981 | Deleterious | H363N | -3.981 | Deleterious |
| T185A | 0.119 | Neutral | T185A | 0.119 | Neutral | |
| pagP (Lipid A palmitoyltransferase PagP) | Addition of 2 amino acid at N-terminal, F to I at C-terminal | Deletion of 17 amino acid at N-terminal, F to I at C-terminal | ||||
| Efflux pump AcrAB | + | + | ||||
| Spermidine export protein MdtI)/KpnF | + | + | ||||
PROVEAN score is a measure of the change in protein structure: if the score is equal or below to the predefined threshold (cut off = −2.5), the variant is predicted to have a ‘deleterious’ effect; if above, the variant is predicted to have a ‘neutral’ effect. In the latter column, there is a prediction of the mutation effect on the protein functionality.
Discussion
Colistin therapy is one of the last viable options while treating serious life-threatening infections caused by MDR GNBs, especially CRE. 5 However, a worldwide trend of increasing colistin MIC levels has been noted,37,38 underlining the need for a robust and precise colistin susceptibility testing method.
The final verdict for an optimal method for colistin susceptibility testing is an ongoing debatable and developing field. Therefore, the performances of four colistin susceptibility methods against CRE isolates were evaluated in this study.
While false colistin susceptible result is considered as a serious error, false colistin resistant results should be viewed as an equally gravely. Therefore, excellent essential agreement regarding colistin susceptibility testing is needed.
In our study, overall colistin resistance among CRE was 15% with standard BMD which is similar to findings presented in other studies worldwide, with colistin resistance among Enterobacterales lying between 20% and 30%.7,39–42 Colistin resistance was slightly more in E. coli isolates than K. pneumoniae contrary to observations published in previous literature.41,42 Skip well phenomenon was only observed in only one isolate of Enterobacter cloacae, although the strain was susceptible to colistin which may be due to presence of heteroresistant subpopulation. Heteroresistance can only be confirmed after a population analysis profile of the isolate, similarly shown by previous studies.42,43 By both guidelines, all the methods considered in this study showed higher colistin resistance, especially VITEK-2, compared to BMD (Table 1).
MIC50/90 of E. coli (0.5/4 µg/mL) by BMD method was found to be lower than MIC50/90 of K. pneumoniae (0.5/8 µg/mL) and much lower than data mentioned in other studies.44,45 Thus, the increased MIC values against colistin among these isolates might be due to the patient population chosen from the hospital setting.
In this study, cBMD meet all the CLSI recommendations regarding VMEs and MEs when compared with the MIC results of BMD with EA and CA lying around 96% and 99%, respectively, with a high concordance with BMD (R2, 0.8736; Figure 2a). Similar results were found in Singh et al. 46 2019’s study, with 100% EA and both ME and VME at 0%. VITEK-2 and E-test have VMEs more than the recommended percentages laid down the CLSI. 31 Our study result showed more discrepancy by the E-test method in comparison to VITEK-2. The CA for VITEK-2 and E-test was 88% and 86%, respectively. Very major error was more frequent for the E-test (40%) compared to VITEK-2 (13.3%). The ME for all methods (except cBMD) exceeded the CLSI recommendation of ⩽ 3.0%. 31 In the case of RPNP, CA was 91%, with ME and VME rates at 8.2% and 13.3%, respectively (Table 2). The percentage of VME exceeding CLSI recommendations might be due to fewer resistant isolates included in the current study.
All the phenotypic methods did appear to perform well against E. coli in comparison to K. pneumoniae, except for cBMD (Table 2). Similar to our current study, a poor agreement was shown by various previous investigations between E-test and BMD for isolates of Enterobacterales with lower rates of agreements (<50%) and high rates of errors (i.e. VME, > 40%; ME, > 10%).17,18,44,46,47 The current study also showed EA of 72%, CA of 86%, and VME of 40%, reflecting the E‑test's unreliability among Enterobacterales. In addition, poor concordance (R2 = 0.6630; Figure 2) was observed between BMD and E‑test MICs among Enterobacterales, in line with the previously reported studies.45,48 In contrast to most studies, using agar dilution as a reference method, Maalej et al. 2011 reported good concordance of E‑test among Enterobacterales without any VME. 13 However, E‑test is also prone to error in terms of MIC determination resulting in more than 2‑fold difference with respect to BMD, thus giving the wrong impression regarding the level of colistin resistance depending on the quality of Mueller-Hinton agar. 17
In the case of VITEK-2, previous studies have a controversial and ambiguous conclusion for Enterobacterales, with some showing a high rate of EA and CA without any VME.12,47 Still, other researchers strictly advise not to use VITEK-2 due to VME as high as 36% in their studies.18,45 VITEK-2 system was also observed to overestimate resistance (i.e. ME), as previously reported. 7 Despite fair concordance with BMD (R = 0.6884; Figure 2) and fair CA and EA rates, this study found VITEK-2 to be unreliable due to unacceptable rates of VME (13.3%) as well as ME (11.8%).
RPNP showed high CA (91%), but it also suffered from high ME (8.2%) and VME (13.3%), which is similar to previously done studies.28,49 There are several possible explanations for the higher VME rates observed in general for the RPNP method. Primary reasons for higher error rates may be due to difficulty in interpreting the color changes, inoculum effect, and borderline MICs. 49 Previous studies have noted this same limitation, with both in-house methods and the commercial kits.50,51
BMD is a very laborious and expensive method for MIC detection. However, the cBMD method in the present study is easy to use without the requirement of additional equipment or great technical expertise. Therefore, after internal validation, cBMD can be utilized for colistin MIC testing in clinical microbiology laboratories. Furthermore, as per cost comparison, cBMD and standard BMD costs are comparable, while the costs of both RPNP and VITEK-2 are higher.
This present study also looked for the presence of plasmid-mediated mcr-1-5 genes by conventional multiplex PCR and mRPNP methods. Only one isolate out of 100 isolates, showed presence of mcr-1 gene by PCR which was also predicted by mRPNP method. mRPNP showed SN and SP of 100% compared to PCR, similar to earlier literature.32,52
In this study, extensive analysis of the genome of both the XDR K. pneumoniae, H-53, and H-62 strains was done. The WGS analysis identified multiple mutations in the target proteins which take part in pathways of lipopolysaccharide formation, modification, and regulation (Table 3), resulting in colistin resistance. Similar amino acid changes have previously been reported in the similar target molecules of colistin-resistant K. pneumoniae isolates.53,54 Previous literature mentions various mutation in arnB(A112D), arnT(L114M, I117 V), pmrA(A217 V), pmrB(A246 T, T157 P) and pmrC/eptA(S25G, Q319R) genes as important mutations conferring colistin resistance in K. pneumoniae.53,55 Apart from these mutations mentioned, many new novel mutations were detected, which need further research to determine which are responsible for colistin resistance. PROVEAN score predicted to have a ‘deleterious’ effect on the protein functionality of arnA_FT(H363 N), arnT(M177I), pmrA(G53 V), pmrB(T157 P) and mgrB(G37 C) mutations. Both the strains also had efflux pumps AcrAB and MdtI/KpnF (Table 3), which confer resistance to many antimicrobials, including colistin. 56
This study has several limitations. The isolates were taken from a single center with fewer numbers contributed by Enterobacter cloacae, and no isolates from Acinetobacter spp. or Pseudomonas aeruginosa were included in the study. The susceptible isolates were disproportionately more than resistant strains. Further studies are needed based on the correlation between clinical outcomes and MIC breakpoints to better guide treatment for a good prognosis. Ongoing research should focus on the significance of heteroresistance in colistin susceptibility testing and the clinical importance of the acquisition of mcr genes. Suitable colistin breakpoints can be implemented confidently in routine testing by clinical microbiology laboratories only when these questions are answered.
Conclusion
In conclusion, the present study's findings showed that cBMD performed well with excellent correlation compared to BMD, whereas the performance of gradient tests like the E-test was unacceptable. This unacceptable performance of the E-test is probably related to the poor and unpredictable diffusion of colistin sulfate. VITEK-2 and RPNP performed better than E‑test but remained unreliable due to high ME and VME. Further studies correlating MICs with the clinical outcome are needed to determine the precise breakpoint to lead patient management. The present study highlights the importance of optimal colistin susceptibility amid CRE isolates. Although there is a substantial colistin resistance among the CRE isolates mediated mainly by the accumulation of mutation in lipopolysaccharide formation and regulation pathway, only one isolate with a plasmid-mediated mcr-1 gene was observed. The genome sequences of both the XDR K. pneumoniae strains indicate lateral gene transfer and spontaneous mutation accumulations are the major drivers of colistin resistance. Globally colistin susceptibility testing should preferably shift to commercial BMD platforms and improved automated systems in clinical microbiology laboratories where BMD can hardly be implemented.
Supplemental Material
Supplemental material, sj-docx-1-tai-10.1177_20499361221080650 for Analysis of colistin resistance in carbapenem-resistant Enterobacterales and XDR Klebsiella pneumoniae by Raunak Bir, Hitender Gautam, Nazneen Arif, Priyanka Chakravarti, Jyoti Verma, Sayantan Banerjee, Sonu Tyagi, Sarita Mohapatra, Seema Sood, Benu Dhawan, Rama Chaudhry, Arti Kapil, Bimal Kumar Das and Bhabatosh Das in Therapeutic Advances in Infectious Disease
Acknowledgments
We thank the Laboratory staff, Bacteriology Section, All India Institute of Medical Sciences, New Delhi, India, and Translational Health Science and Technology Institute, Faridabad, for helping us carry out this study.
Footnotes
Author contributions: Raunak Bir: Formal analysis; Methodology; Writing – original draft.
Hitender Gautam: Conceptualization; Funding acquisition; Project administration; Visualization; Writing – review & editing.
Nazneen Arif: Data curation; Software.
Priyanka Chakravarti: Formal analysis; Methodology.
Jyoti Verma: Formal analysis; Methodology.
Sayantan Banerjee: Conceptualization.
Sonu Tyagi: Methodology; Validation.
Sarita Mohapatra: Data curation; Investigation.
Seema Sood: Project administration; Supervision.
Benu Dhawan: Project administration; Supervision.
Rama Chaudhry: Writing – review & editing.
Arti Kapil: Supervision.
Bimal Kumar Das: Project administration; Supervision; Writing – review & editing.
Bhabatosh Das: Methodology; Resources; Writing – review & editing.
Conflict of interest statement: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Intramural Research Grant from All India Institute of Medical Sciences, New Delhi with Project Code No.: A-597.
Ethics statement: Ethical approval for this study has been granted by the Institutional Ethical Committee, AIIMS, New Delhi (Ref. No.: Institutional Ethical Committee-283/01.06.2018), and all the protocols were followed as per the ethical standards. As the inclusion criteria was CRE isolates from routinely sent samples, the requirement of informed consent was exempted.
ORCID iD: Hitender Gautam  https://orcid.org/0000-0002-1409-1543
https://orcid.org/0000-0002-1409-1543
Data transparency: All relevant data related to this scientific work is available.
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Raunak Bir, Department of Microbiology, All India Institute of Medical Sciences, New Delhi, India.
Hitender Gautam, Department of Microbiology, All India Institute of Medical Sciences, New Delhi 110029, India.
Nazneen Arif, Department of Microbiology, All India Institute of Medical Sciences, New Delhi, India.
Priyanka Chakravarti, Translational Health Science and Technology Institute, Faridabad, India.
Jyoti Verma, Translational Health Science and Technology Institute, Faridabad, India.
Sayantan Banerjee, 1Day Sooner.
Sonu Tyagi, Department of Microbiology, All India Institute of Medical Sciences, New Delhi, India.
Sarita Mohapatra, Department of Microbiology, All India Institute of Medical Sciences, New Delhi, India.
Seema Sood, Department of Microbiology, All India Institute of Medical Sciences, New Delhi, India.
Benu Dhawan, Department of Microbiology, All India Institute of Medical Sciences, New Delhi, India.
Rama Chaudhry, Department of Microbiology, All India Institute of Medical Sciences, New Delhi, India.
Arti Kapil, Department of Microbiology, All India Institute of Medical Sciences, New Delhi, India.
Bimal Kumar Das, Department of Microbiology, All India Institute of Medical Sciences, New Delhi, India.
Bhabatosh Das, Translational Health Science and Technology Institute, Faridabad, India.
References
- 1. Stein A, Raoult D. Colistin: an antimicrobial for the 21st century? Clin Infect Dis 2002; 35: 901–902. [DOI] [PubMed] [Google Scholar]
- 2. Falagas ME, Rafailidis PI, Kasiakou SK, et al. Effectiveness and nephrotoxicity of colistin monotherapy vs. Colistin-meropenem combination therapy for multidrug-resistant Gram-negative bacterial infections. Clin Microbiol Infect 2006; 12: 1227–1230. [DOI] [PubMed] [Google Scholar]
- 3. Landman D, Georgescu C, Martin DA, et al. Polymyxins revisited. Clin Microbiol Rev 2008; 21: 449–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J, Nation RL, Turnidge JD, et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 2006; 6: 589–601. [DOI] [PubMed] [Google Scholar]
- 5. Karaiskos I, Giamarellou H. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother 2014; 15: 1351–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Humphries RM. Susceptibility testing of the polymyxins: where are we now. Pharmacotherapy 2015; 35: 22–27. [DOI] [PubMed] [Google Scholar]
- 7. Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 2017; 30: 557–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wright MS, Suzuki Y, Jones MB, et al. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 2015; 59: 536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu Y, Liu F, Lin IY, et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 146–147. [DOI] [PubMed] [Google Scholar]
- 10. Kluytmans J. Plasmid-encoded colistin resistance: mcr-one, two, three and counting. Euro Surveill 2017; 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gales AC, Reis AO, Jones RN. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J Clin Microbiol 2001; 39: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lo-Ten-Foe JR, de Smet AM, Diederen BM, et al. Comparative evaluation of the VITEK 2, disk diffusion, etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob Agents Chemother 2007; 51: 3726–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maalej SM, Meziou MR, Rhimi FM, et al. Comparison of disc diffusion, Etest and agar dilution for susceptibility testing of colistin against Enterobacteriaceae. Lett Appl Microbiol 2011; 53: 546–551. [DOI] [PubMed] [Google Scholar]
- 14. Arroyo LA, Garcia-Curiel A, Pachon-Ibanez ME, et al. Reliability of the E-test method for detection of colistin resistance in clinical isolates of Acinetobacter baumannii. J Clin Microbiol 2005; 43: 903–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee SY, Shin JH, Lee K, et al. Comparison of the Vitek 2, MicroScan, and Etest methods with the agar dilution method in assessing colistin susceptibility of bloodstream isolates of Acinetobacter species from a Korean university hospital. J Clin Microbiol 2013; 51: 1924–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goldstein FW, Ly A, Kitzis MD. Comparison of Etest with agar dilution for testing the susceptibility of Pseudomonas aeruginosa and other multidrug-resistant bacteria to colistin. J Antimicrob Chemother 2007; 59: 1039–1040. [DOI] [PubMed] [Google Scholar]
- 17. Hindler JA, Humphries RM. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol 2013; 51: 1678–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tan TY, Ng SY. Comparison of Etest, Vitek and agar dilution for susceptibility testing of colistin. Clin Microbiol Infect 2007; 13: 541–544. [DOI] [PubMed] [Google Scholar]
- 19. Clinical Laboratory Standard Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard. Wayne, PA: CLSI, 2015. [Google Scholar]
- 20. Albur M, Noel A, Bowker K, et al. Colistin susceptibility testing: time for a review. J Antimicrob Chemother 2014; 69: 1432–1434. [DOI] [PubMed] [Google Scholar]
- 21. Nation RL, Li J, Cars O, et al. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis 2015; 15: 225–234. [DOI] [PubMed] [Google Scholar]
- 22. Karvanen M, Malmberg C, Lagerback P, et al. Colistin is extensively lost during standard in vitro experimental conditions. Antimicrob Agents Chemother 2017; 61: e00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sader HS, Rhomberg PR, Flamm RK, et al. Use of a surfactant (polysorbate 80) to improve MIC susceptibility testing results for polymyxin B and colistin. Diagn Microbiol Infect Dis 2012; 74: 412–414. [DOI] [PubMed] [Google Scholar]
- 24. Clinical Laboratory Standard Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Wayne, PA: CLSI, 2020. [Google Scholar]
- 25. Matuschek E, Ahman J, Webster C, et al. Antimicrobial susceptibility testing of colistin – evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect 2018; 24: 865–870. [DOI] [PubMed] [Google Scholar]
- 26. Humphries RM, Green DA, Schuetz AN, et al. Multicenter evaluation of colistin broth disk elution and Colistin Agar Test: a report from the Clinical and Laboratory Standards Institute. J Clin Microbiol 2019; 57: e01269-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Växjö: EUCAST, 2020. [Google Scholar]
- 28. Jayol A, Kieffer N, Poirel L, et al. Evaluation of the Rapid Polymyxin NP test and its industrial version for the detection of polymyxin-resistant Enterobacteriaceae. Diagn Microbiol Infect Dis 2018; 92: 90–94. [DOI] [PubMed] [Google Scholar]
- 29. Wattal C, Goel N, Oberoi JK, et al. Performance of three commercial assays for colistin susceptibility in clinical isolates and Mcr-1 carrying reference strain. Indian J Med Microbiol 2019; 37: 488–495. [DOI] [PubMed] [Google Scholar]
- 30. Clinical Laboratory Standard Institute (CLSI). Development of in-vitro susceptibility testing criteria and quality control parameters; approved guideline. Wayne, PA: CLSI, 2008. [Google Scholar]
- 31. Clinical Laboratory Standard Institute (CLSI). Verification of commercial microbial identification and susceptibility test systems. Wayne, PA: CLSI, 2017. [Google Scholar]
- 32. Poirel L, Larpin Y, Dobias J, et al. Rapid polymyxin NP test for the detection of polymyxin resistance mediated by the mcr-1/mcr-2 genes. Diagn Microbiol Infect Dis 2018; 90: 7–10. [DOI] [PubMed] [Google Scholar]
- 33. Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill 2018; 23: 17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wick RR, Judd LM, Gorrie CL, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012; 7: e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sader HS, Farrell DJ, Flamm RK, et al. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009-2011). Diagn Microbiol Infect Dis 2014; 78: 443–448. [DOI] [PubMed] [Google Scholar]
- 38. Control ECfDPa. Surveillance of antimicrobial resistance in Europe – Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017. Stockholm: ECDC, 2018. [Google Scholar]
- 39. Gandra S, Mojica N, Klein EY, et al. Trends in antibiotic resistance among major bacterial pathogens isolated from blood cultures tested at a large private laboratory network in India, 2008-2014. Int J Infect Dis 2016; 50: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chandy M, Das A, Bhattacharyya A, et al. Colistin-Resistant Klebsiella Infections Among Pediatric Oncology and Hematopoietic Stem Cell Transplantation Patients in Eastern India. Infect Control Hosp Epidemiol 2018; 39: 118–121. [DOI] [PubMed] [Google Scholar]
- 41. Manohar P, Shanthini T, Ayyanar R, et al. The distribution of carbapenem- and colistin-resistance in Gram-negative bacteria from the Tamil Nadu region in India. J Med Microbiol 2017; 66: 874–883. [DOI] [PubMed] [Google Scholar]
- 42. Das S, Roy S, Roy S, et al. Colistin susceptibility testing of gram-negative bacilli: better performance of vitek2 system than E-test compared to broth microdilution method as the gold standard test. Indian J Med Microbiol 2020; 38: 58–65. [DOI] [PubMed] [Google Scholar]
- 43. Hong YK, Lee JY, Ko KS. Colistin resistance in Enterobacter spp. Isolates in Korea. J Microbiol 2018; 56: 435–440. [DOI] [PubMed] [Google Scholar]
- 44. Galani I, Adamou P, Karaiskos I, et al. Evaluation of ComASP Colistin (formerly SensiTest Colistin), a commercial broth microdilution-based method to evaluate the colistin minimum inhibitory concentration for carbapenem-resistant Klebsiella pneumoniae isolates. J Glob Antimicrob Resist 2018; 15: 123–126. [DOI] [PubMed] [Google Scholar]
- 45. Chew KL, La MV, Lin RTP, et al. Colistin and polymyxin B susceptibility testing for Carbapenem-resistant and mcr-positive enterobacteriaceae: comparison of Sensititre, MicroScan, Vitek 2, and Etest with Broth Microdilution. J Clin Microbiol 2017; 55: 2609–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh RI, Bhatia M, Anusha KR, et al. Comparative evaluation of microscan walkaway 96 plus ID/AST system and mikrolatest broth microdilution kit in assessing In vitro colistin susceptibility of carbapenem-resistant clinical gram-negative bacterial isolates: experience from a tertiary care teaching hospital in Rishikesh, Uttarakhand. Indian J Med Microbiol 2019; 37: 502–508. [DOI] [PubMed] [Google Scholar]
- 47. Dafopoulou K, Zarkotou O, Dimitroulia E, et al. Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible klebsiella pneumoniae and acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother 2015; 59: 4625–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simar S, Sibley D, Ashcraft D, et al. Colistin and polymyxin B minimal inhibitory concentrations determined by etest found unreliable for gram-negative Bacilli. Ochsner J 2017; 17: 239–242. [PMC free article] [PubMed] [Google Scholar]
- 49. Kon H, Abramov S, Amar Ben Dalak M, et al. Performance of rapid polymyxin NP and Rapid Polymyxin Acinetobacter for the detection of polymyxin resistance in carbapenem-resistant Acinetobacter baumannii and Enterobacterales. J Antimicrob Chemother 2020; 75: 1484–1490. [DOI] [PubMed] [Google Scholar]
- 50. Belda-Orlowski A, Pfennigwerth N, Gatermann SG, et al. Evaluation and readout optimization of the rapid polymyxin NP test for the detection of colistin-resistant Enterobacteriaceae. J Med Microbiol 2019; 68: 1189–1193. [DOI] [PubMed] [Google Scholar]
- 51. Karatuna O, Matuschek E, Ahman J, et al. Evaluation of rapid polymyxin NP, Pseudomonas and Acinetobacter tests for rapid determination of colistin susceptibility in Escherichia coli, Klebsiella pneumoniae, P. Aeruginosa and A. Baumannii. In: Twenty-ninth European congress of clinical microbiology & infectious diseases, Amsterdam, 13–16 April, 2019. [Google Scholar]
- 52. Esposito F, Fernandes MR, Lopes R, et al. Detection of colistin-resistant MCR-1-positive escherichia coli by use of assays based on inhibition by EDTA and Zeta potential. J Clin Microbiol 2017; 55: 3454–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pragasam AK, Shankar C, Veeraraghavan B, et al. Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India – a first report. Front Microbiol 2016; 7: 2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jaidane N, Bonnin RA, Mansour W, et al. Genomic insights into Colistin-Resistant Klebsiella pneumoniae from a Tunisian Teaching Hospital. Antimicrob Agents Chemother 2018; 62: e01601-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Elias R, Duarte A, Perdigao J. A molecular perspective on colistin and Klebsiella pneumoniae: mode of action, resistance genetics, and phenotypic susceptibility. Diagnostics (Basel) 2021; 11: 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aghapour Z, Gholizadeh P, Ganbarov K, et al. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist 2019; 12: 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tai-10.1177_20499361221080650 for Analysis of colistin resistance in carbapenem-resistant Enterobacterales and XDR Klebsiella pneumoniae by Raunak Bir, Hitender Gautam, Nazneen Arif, Priyanka Chakravarti, Jyoti Verma, Sayantan Banerjee, Sonu Tyagi, Sarita Mohapatra, Seema Sood, Benu Dhawan, Rama Chaudhry, Arti Kapil, Bimal Kumar Das and Bhabatosh Das in Therapeutic Advances in Infectious Disease