Abstract
The opaque-2 mutation in maize (Zea mays) is associated with an increased level of free amino acids (FAA) in the mature endosperm. In particular, there is a high concentration of lysine, the most limiting essential amino acid. To investigate the basis for the high-FAA phenotype of opaque-2 maize, we characterized amino acid accumulation during endosperm development of several wild-type and opaque-2 inbreds. Oh545o2 was found to have an exceptionally high level of FAA, in particular those derived from aspartate (Asp) and intermediates of glycolysis. The FAA content in Oh545o2 is 12 times greater than its wild-type counterpart, and three and 10 times greater than in Oh51Ao2 and W64Ao2, respectively. We crossed Oh545o2 to Oh51Ao2 and analyzed the F2:3 progeny to identify genetic loci linked with the high FAA level in these mutants. Quantitative trait locus mapping identified four significant loci that account for about 46% of the phenotypic variance. One locus on the long arm of chromosome 2 is coincident with genes encoding a monofunctional Asp kinase 2 and a bifunctional Asp kinase-homo-Ser dehydrogenase-2, whereas another locus on the short arm of chromosome 3 is linked with a cytosolic triose phosphate isomerase 4. The results suggest an alternation of amino acid and carbon metabolism leads to overproduction and accumulation of FAA in opaque-2 mutants.
The opaque-2 (o2) mutation nearly doubles the Lys content of maize (Zea mays) endosperm, and thereby significantly improves the protein quality of the grain (Mertz et al., 1964). The increased level of Lys in o2 mutants results from several factors: (a) The synthesis of zein storage proteins is reduced, and zeins contain no Lys; (b) there is an increased synthesis of a number of other proteins (non-zeins), many of which contain Lys; and (c) there is a general increase in the level of free amino acids (FAA), including Lys. The extent to which the Lys content of an o2 mutant is increased depends on the genetic background. For example, the protein concentration can range nearly 2-fold (between 6.5% and 11.9%), as does the percent of Lys in the flour (between 0.24–0.48 mg 100 mg−1; Moro et al., 1996). Most of the Lys is protein bound, but free Lys can also make a significant contribution. For example, in Oh545o2, free Lys accounts for nearly one-third of the total Lys in the endosperm (Moro et al., 1996).
The mechanism by which the o2 mutation increases the Lys content of maize endosperm is only partially understood. The O2 gene encodes a transcriptional activator that regulates the expression of a number of genes (Schmidt, 1993), including the 22-kD α-zeins (Kodrzycki et al., 1989; Schmidt et al., 1990, 1992). The reduction in α-zein synthesis is associated with varying degrees of increased accumulation of several non-zein proteins, which accounts for most of the higher percentage of Lys (Damerval and de Vienne, 1993; Habben et al., 1993). Habben et al. (1995) showed that elongation factor 1α (eEF1A) is increased in o2 mutants, and its concentration is highly correlated with the Lys content of endosperm flour. A survey of a large number of normal and o2 inbreds showed a consistently high correlation (r = 0.9) between the concentration of eEF1A and endosperm Lys content (Moro et al., 1996). Only in genotypes with high levels of free Lys was this relationship less consistent.
The high level of FAA in o2 mutants was recognized many years ago (Mertz et al., 1974; Misra et al., 1975), and there has been a great deal of research to try to explain it. O2 has features of GCN4 (Mauri et al., 1993), which serves as a general transcription factor regulating amino acid biosynthesis in yeast (Saccharomyces cerevisiae; Hinnebusch, 1990). However, the level of FAA is increased in o2 mutants. As a consequence, it seems unlikely that O2 acts through a mechanism similar to GCN4 to increase the levels of FAA. Several studies have shown that certain key enzymes involved in amino acid and carbon metabolism are altered in o2 mutants. The activity of Asp kinase (AK) is up-regulated by o2 (Brennecke et al., 1996). AK is an important enzyme involved in the synthesis of several amino acids, including Thr, Lys, Met, and Leu (Bryan, 1990). However, the effect of o2 on AK must be indirect, as there is no evidence that O2 inhibits expression of the gene. The activity of bifunctional Lys ketoglutarate reductase-saccaropine dehydrogenase, which regulates Lys degradation in maize endosperm, is down-regulated by the o2 mutation as a consequence of reduced levels of mRNA (Kemper et al., 1999) and enzyme (Brochetto-Braga et al., 1992; Yunes et al., 1994). It is thought that the low level of Lys ketoglutarate reductase- saccaropine dehydrogenase is an important factor responsible for the high level of free Lys in mature endosperm. Damerval and Le Guilloux (1998) found that acetohydroxyacid synthase, the enzyme catalyzing the first common step in the synthesis of branched amino acids derived from pyruvate and Thr, is down-regulated by o2, and this could decrease the level of Val and Leu. In addition, several investigators demonstrated that cytosolic pyruvate phosphate dikinase (cyPPDK) is activated by the O2 gene (Gallusci et al., 1996; Maddaloni et al., 1996; Damerval and Le Guilloux, 1998). This enzyme is a major regulator of the glycolytic pathway, which could be linked to carbon and amino acid metabolism by converting pyruvate to phosphoenol pyruvate. Thus, o2 could increase FAA levels by influencing a number of enzymatic steps in glycolysis, the tricarboxylic acid cycle, and amino acid synthesis and degradation.
We used a genetic approach to identify loci influencing the pools of FAA in o2 mutants. We characterized the FAA levels in developing and mature Oh545+/o2 and W64A+/o2, two inbreds with significantly different levels of FAA in mature endosperm. The Oh545+/o2 inbreds were found to contain much higher than average levels of FAAs, including Lys, which causes them to deviate from the relationship between eEF1A concentration and Lys content (Moro et al., 1996). Oh545o2 contains 12 times more FAAs than its wild-type counterpart, and almost all amino acids in Oh545o2, especially those derived from the Asp pathway, are greatly increased compared with other inbreds. To identify loci influencing FAA content in Oh545o2, we crossed it to Oh51Ao2, an inbred with a smaller increase in FAA, and characterized the F2:3 progeny. Quantitative trait locus (QTL) mapping of loci associated with the high FAA trait identified in four regions on chromosome 2, 3, and 7. Asp kinase 2 (Ask2) and/or AK-homo-Ser dehydrogenase (HSDH) 2 appear to be good candidate genes for the QTL on the long arm of chromosome 2.
RESULTS
To evaluate phenotypic variation in FAA composition of o2 endosperm, we analyzed mature kernels of the near-isogenic normal and o2 maize inbreds, W64A+/o2 and Oh545A+/o2. These materials were grown at different times and locations. Although there was variation in total level of FAA, depending on the season and location in which the plants were grown, the relative concentrations of FAAs and their ratio in wild-type and o2 inbreds were consistent (Table I). (Asn was not measured in the analysis of mature W64A+/o2 endosperm, but it was determined for 50-d after pollination (DAP) developing endosperm [Table II], which is essentially a mature stage.) It is common that o2 mutants have a higher content of FAAs compared with their wild-type counterparts (Sodek and Wilson, 1971; Misra et al., 1975; Moro et al., 1996), but there is striking variation between these two inbreds. W64Ao2 has nearly 3.5 times more FAA than W64A+ and Oh545o2 has nearly 12 times more FAA than Oh545+. In W64A, the o2 mutation results in a general 2- to 3-fold increase in most amino acids, with Lys showing the greatest (nearly 12-fold) increase. W64Ao2 also has a large increase in Glu-derived amino acids: Gln, His, and Arg. The effect of the o2 mutation on FAA composition is very pronounced in Oh545. Compared with W64A, on average there is a 10-fold increase in most FAAs, and there is a dramatic increase (18–24-fold) in amino acids derived from the Asp pathway (Lys, Thr, Met, and iso-Leu). Other amino acids significantly elevated above average are Ser, Leu, Ala, and Val, several of which are derived from glycolytic intermediates.
Table I.
FAA composition of W64A+/o2, Oh545+/o2, and Oh51Ao2 mature endosperm
| Amino Acid | W64A+
|
W64Ao2
|
Oh545+
|
Oh545o2
|
Oh51Ao2a
|
W64Ao2/W64A+ | Oh545o2/Oh545+ | Oh545+/W64A+ | Oh545o2/W64Ao2 | Oh545o2/ Oh51Ao2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | se | Mean | se | Mean | se | Mean | se | Mean | se | ||||||
| Asp | 0.63 | 0.17 | 1.56 | – | 2.89 | 0.03 | 9.67 | 0.62 | 15.76 | 3.55 | 2.5 | 3.3 | 4.6 | 6.2 | 0.6 |
| Asn | – | – | – | – | 1.32 | 0.42 | 11.89 | 2.51 | 20.24 | 0.87 | – | 9.0 | – | – | 0.6 |
| Glu | 1.35 | 0.25 | 3.45 | 0.76 | 3.43 | 0.96 | 26.58 | 5.24 | 11.48 | 3.31 | 2.6 | 7.7 | 2.5 | 7.7 | 2.3 |
| Gln | 0.27 | 0.02 | 1.74 | 0.04 | 0.79 | 0.38 | 14.00 | 4.18 | 9.56 | 5.81 | 6.5 | 17.8 | 3.0 | 8.1 | 1.5 |
| Ala | 0.87 | 0.21 | 2.98 | 1.15 | 4.82 | 1.63 | 86.30 | 16.52 | 7.24 | 4.97 | 3.4 | 17.9 | 5.6 | 29.0 | 11.9 |
| Lys | 0.12 | 0.01 | 1.43 | 0.28 | 0.39 | 0.08 | 6.93 | 0.79 | 1.11 | 0.34 | 11.8 | 17.7 | 3.2 | 4.8 | 6.2 |
| Thr | 0.14 | 0.05 | 0.49 | 0.13 | 0.54 | 0.14 | 7.47 | 0.82 | 1.19 | 0.49 | 3.6 | 13.9 | 3.9 | 15.3 | 6.3 |
| Met | 0.04 | 0.01 | 0.12 | 0.05 | 0.08 | 0.03 | 1.88 | 0.39 | 0.18 | 0.18 | 2.7 | 23.1 | 1.9 | 16.3 | 10.5 |
| Ile | 0.04 | 0.01 | 0.15 | 0.04 | 0.15 | 0.02 | 3.69 | 0.97 | 0.10 | 0.08 | 3.4 | 24.0 | 3.6 | 24.9 | 37.6 |
| Leu | 0.08 | 0.02 | 0.28 | 0.10 | 0.25 | 0.06 | 5.75 | 0.71 | 0.60 | 0.32 | 3.5 | 23.1 | 3.1 | 20.4 | 9.5 |
| Val | 0.12 | 0.02 | 0.66 | 0.20 | 0.40 | 0.11 | 7.13 | 0.87 | 1.11 | 0.45 | 5.6 | 17.8 | 3.4 | 10.9 | 6.4 |
| Ser | 0.52 | 0.23 | 0.71 | 0.27 | 1.76 | 0.65 | 22.70 | 2.91 | 2.22 | 1.00 | 1.4 | 12.9 | 3.4 | 32.0 | 10.2 |
| Gly | 0.28 | 0.03 | 0.72 | 0.22 | 0.93 | 0.30 | 7.66 | 0.95 | 0.85 | 0.40 | 2.6 | 8.2 | 3.4 | 10.7 | 9.0 |
| Tyr | 0.08 | 0.01 | 0.53 | 0.05 | 0.21 | 0.01 | 2.71 | 0.29 | 0.67 | 0.17 | 6.9 | 12.7 | 2.8 | 5.1 | 4.1 |
| Phe | 0.07 | 0.03 | 0.21 | 0.05 | 0.18 | 0.01 | 2.54 | 0.28 | 0.41 | 0.17 | 2.9 | 13.9 | 2.5 | 11.8 | 6.2 |
| Arg | 0.13 | 0.01 | 0.87 | 0.25 | 0.23 | 0.06 | 2.22 | 0.33 | 1.03 | 0.29 | 6.7 | 9.8 | 1.8 | 2.6 | 2.2 |
| His | 0.07 | 0.01 | 0.45 | 0.04 | 0.30 | 0.09 | 1.41 | 0.20 | 0.44 | 0.05 | 6.0 | 4.6 | 4.1 | 3.1 | 3.2 |
| Total | 4.81b | 1.22 | 16.33b | 4.45 | 18.68 | 4.52 | 220.53 | 24.39 | 74.20 | 19.64 | 3.4 | 11.8 | 3.9b | 13.5b | 3.0 |
Values are nanomoles per milligram dry endosperm. Measurements for Oh545+/o2 and W64A+/o2 are the mean of duplicate samples from two locations.
Samples for Oh51Ao2 were from three different seasons.
Asn content was not determined.
Table II.
FAA composition of W64A+/o2 and Oh545+/o2 developing endosperm
| Amino Acid | 20 DAP
|
30
DAP
|
40 DAP
|
50 DAP
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| W64A+ | W64Ao2 | OH545+ | OH545o2 | W64A+ | W64Ao2 | OH545+ | OH545o2 | W64A+ | W64Ao2 | OH545+ | OH545o2 | W64A+ | W64Ao2 | OH545+ | OH545o2 | |
| Asp | 5.39 | 11.59 | 3.97 | 8.29 | 3.51 | 13.94 | 5.00 | 12.90 | 2.31 | 17.22 | 2.19 | 16.36 | 1.33 | 14.72 | 2.89 | 14.56 |
| Asn | 3.24 | 12.34 | 4.93 | 11.10 | 1.87 | 12.06 | 2.82 | 7.72 | 1.53 | 14.86 | 1.53 | 9.64 | 0.53 | 11.32 | 1.27 | 9.91 |
| Glu | 10.55 | 27.27 | 24.30 | 24.22 | 7.41 | 21.17 | 11.24 | 26.28 | 5.05 | 24.76 | 3.73 | 31.91 | 0.56 | 10.35 | 4.30 | 22.21 |
| Gln | 10.04 | 78.85 | 4.30 | 59.13 | 4.00 | 50.07 | 3.81 | 53.64 | 1.89 | 26.00 | 0.88 | 22.34 | 0.06 | 1.53 | 0.99 | 14.66 |
| Ala | 12.72 | 40.42 | 25.29 | 59.36 | 7.91 | 24.41 | 17.42 | 53.76 | 5.74 | 26.08 | 6.22 | 64.51 | 0.21 | 2.07 | 9.42 | 37.70 |
| Lys | 0.97 | 1.62 | 1.63 | 1.22 | 0.52 | 2.06 | 0.93 | 3.31 | 0.30 | 2.72 | 0.24 | 3.81 | 0.08 | 1.70 | 0.27 | 4.95 |
| Thr | 1.45 | 4.63 | 2.25 | 6.16 | 0.80 | 3.25 | 1.35 | 6.29 | 0.48 | 3.34 | 0.47 | 5.66 | 0.04 | 1.01 | 0.56 | 4.12 |
| Met | 0.62 | 1.91 | 1.05 | 1.41 | 0.39 | 1.90 | 0.48 | 1.85 | 0.25 | 1.49 | 0.18 | 1.21 | 0.02 | 0.28 | 0.25 | 1.35 |
| Ile | 0.26 | 0.70 | 0.42 | 1.46 | 0.12 | 0.41 | 0.28 | 1.17 | 0.05 | 0.55 | 0.07 | 1.39 | 0.02 | 0.26 | 0.08 | 1.14 |
| Leu | 0.50 | 2.87 | 0.68 | 2.35 | 0.20 | 1.73 | 0.50 | 2.76 | 0.11 | 2.08 | 0.10 | 3.46 | 0.03 | 0.67 | 0.18 | 2.55 |
| Val | 1.48 | 5.23 | 1.98 | 4.49 | 0.51 | 2.88 | 1.00 | 3.89 | 0.26 | 2.79 | 0.15 | 4.13 | 0.04 | 1.14 | 0.27 | 3.78 |
| Ser | 2.86 | 7.50 | 6.17 | 10.41 | 1.21 | 5.09 | 3.08 | 9.32 | 0.70 | 6.07 | 0.98 | 8.78 | 0.09 | 1.25 | 1.41 | 10.27 |
| Gly | 1.84 | 4.48 | 1.72 | 3.95 | 0.65 | 3.41 | 1.24 | 4.20 | 0.48 | 2.66 | 0.36 | 4.64 | 0.05 | 0.40 | 0.62 | 1.62 |
| Tyr | 0.33 | 0.81 | 0.60 | 0.54 | 0.20 | 0.59 | 0.36 | 1.19 | 0.14 | 1.18 | 0.08 | 1.85 | 0.05 | 1.30 | 0.13 | 2.06 |
| Phe | 0.34 | 0.80 | 0.69 | 1.34 | 0.25 | 0.65 | 0.40 | 1.15 | 0.12 | 0.99 | 0.10 | 1.75 | 0.02 | 0.64 | 0.10 | 1.35 |
| Arg | 0.35 | 0.85 | 0.50 | 0.59 | 0.20 | 0.44 | 0.30 | 0.92 | 0.06 | 0.57 | 0.05 | 1.16 | 0.03 | 0.76 | 0.09 | 1.45 |
| His | 0.39 | 0.67 | 0.59 | 0.34 | 0.13 | 0.40 | 0.29 | 0.64 | 0.06 | 0.49 | 0.01 | 0.75 | 0.02 | 0.58 | 0.06 | 1.09 |
| Pro | 0.59 | 1.38 | 1.31 | 6.26 | 0.16 | 1.36 | 0.83 | 3.09 | 0.17 | 5.82 | 0.24 | 11.44 | 0.10 | 4.54 | 0.38 | 19.27 |
| Gaba | 0.63 | 2.42 | 3.08 | 5.66 | 0.63 | 1.80 | 3.11 | 7.33 | 0.76 | 5.01 | 1.69 | 19.77 | 0.38 | 2.77 | 3.50 | 14.95 |
| Urea | 0.13 | 2.56 | 0.28 | 0.97 | 0.06 | 1.11 | 0.21 | 1.22 | 0.04 | 1.02 | 0.05 | 1.06 | 0.05 | 0.43 | 0.09 | 0.71 |
| Others | 1.84 | 9.63 | 3.01 | 6.50 | 0.78 | 7.68 | 1.63 | 7.85 | 0.54 | 6.23 | 0.36 | 9.23 | 0.33 | 1.52 | 0.93 | 7.37 |
| Total | 56.58 | 218.54 | 88.76 | 215.82 | 31.51 | 156.42 | 56.24 | 210.47 | 21.02 | 151.99 | 19.70 | 224.84 | 4.03 | 59.35 | 27.78 | 177.04 |
Values are nanomoles per milligram dry endosperm; measurements are the mean of three independent extractions.
At least part of the differences in elevated FAAs in the o2 inbreds is related to their genetic background. The level of FAA in mature endosperm of Oh545+ is generally three to four times greater than in W64A. When we subtracted the difference (fold increase) between an FAA in the high and low normal inbreds from that of the o2 versions, it was evident that the o2 mutation in Oh545 has a more pronounced effect on amino acids derived from the Asp pathway. Lys content is only 1.6 times greater in Oh545o2 compared with W64Ao2, whereas iso-Leu (21.5 times), Met (14.4 times), and Thr (11.4 times) are much more dramatically increased.
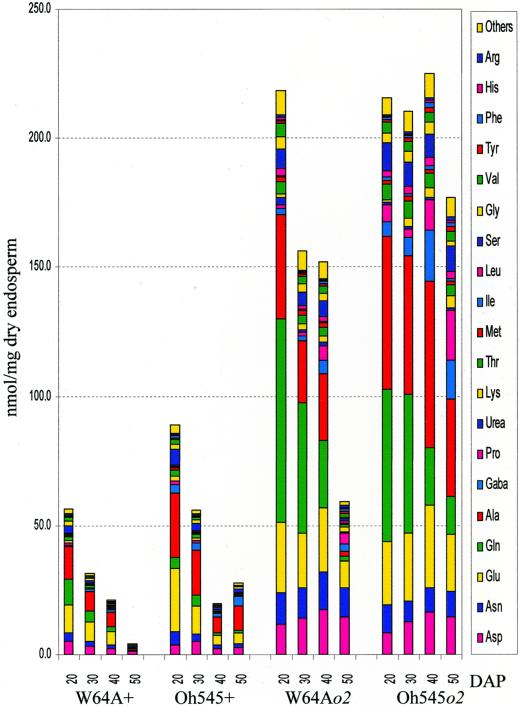
To understand the dynamic changes that contribute to final concentration of FAAs in mature endosperm, we characterized the level and composition of FAAs during W64A+/o2 and Oh545+/o2 kernel development. Ears were harvested at 20, 30, 40, and 50 DAP; kernel samples from the center of well-filled ears were pooled, frozen, and lyophilized; and the amino acids were extracted as with the mature endosperm. The data in Table II show the amino acid composition at each developmental stage, and these results are illustrated graphically in Figure 1.
Figure 1.
FAA in developing endosperm of Oh545+/o2 and W64A+/o2. FAA analysis was performed with pooled kernel samples as described in “Materials and Methods.” The values are the means of three independent extractions and measurements.
To a large extent, the results of this analysis are similar to those obtained with mature endosperm. There is a much higher level of FAAs in developing endosperm of o2 than wild-type inbreds, and there is a significantly higher level of FAA in Oh545+/o2 compared with W64A+/o2 throughout development. The o2 mutants have nearly four times more FAAs compared with their wild-type counterparts at 20 DAP, and in general their concentrations of FAA drop much more slowly than in wild-type endosperm. For example, in W64Ao2 the concentration of FAA at 50 DAP is nearly the same as in the wild type at 20 DAP. It is striking that W64Ao2 and Oh545o2 have similar concentrations of FAAs at 20 DAP, but although the FAA level declines in W64Ao2 endosperm as it matures, there is only a slight reduction in FAA concentration in Oh545o2.
Although there is some fluctuation in the pool size of various amino acids during endosperm development, to a large extent the concentration at maturity reflects that during development (Table II). Glu, Gln, Asp, Asn, and Ala are present in the highest concentrations throughout most of endosperm development for both normal and o2 genotypes. There is a significant reduction in Gln concentration as the endosperm matures, but with the exception of Pro, which increases in concentration in o2 endosperm (particularly in Oh545o2), there is not a dramatic increase in concentration of any particular amino acid. The higher concentrations of Lys, Thr, Met, and iso-Leu in o2 endosperm as it matures comes as a consequence of the reduction in concentration of the more abundant amino acids, such as Gln.
We investigated the genetic basis for the enhanced level of FAAs in Oh545o2 by crossing it with Oh51Ao2. The FAA content in mature endosperm of Oh545o2 is three times higher than Oh51Ao2, with the amino acids derived from the AK pathway being more highly elevated than most others. The level of FAA in Oh51Ao2 is four times higher than W64Ao2 (Table I), and with the exceptions of Lys and iso-Leu, the concentration of each amino acid is higher in mature endosperm of Oh51Ao2 compared with W64Ao2 (Table I). However, with this particular cross we were able to simultaneously analyze segregation for high levels of free Lys and Lys-containing proteins, and investigate the interaction between these two traits (Wang et al., 2001).
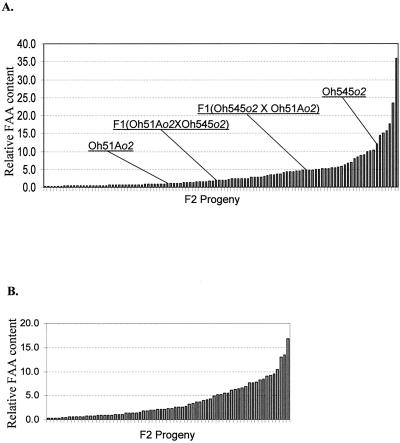
We used a ninhydrin assay as a rapid method to phenotype the level of FAAs in Oh545o2, Oh51Ao2, and their F1 and F2:3 progeny (Fig. 2). The phenotypic segregation of this trait suggests both recessive and dosage-dependent effects because the relative FAA content in the F1 is closer to the low-FAA parent (Oh51Ao2); the FAA level in each F1 is directly related to its ear parent. In the spring and fall of 1996, we obtained 106 and 69 F2:3 progeny, respectively, and there was continuous phenotypic variation in the endosperm FAA content both seasons. The F2:3 progeny generally had an FAA level intermediate between the two parents, but most of them have an amino acid level more like the low FAA parent, Oh51Ao2 (Fig. 2, A and B). Only a few F2:3 progeny had an FAA level close to the high FAA parent, Oh545o2. Because the F2:3 population from the spring of 1996 was significantly larger than the fall season, we selected it for QTL mapping.
Figure 2.
Relative concentration of FAA in mature endosperm of Oh545o2, Oh51Ao2, their reciprocal F1 crosses, and F2:3 progeny. Flour was prepared from a pool of 20 kernels from the middle of well-filled ears and assayed with ninhydrin to determine the FAA content. The relative content of FAA in Oh51Ao2 (the low-FAA parent) was defined as “1.” The relative FAA content of each F2:3 individual is the mean of two measurements from two independent extractions. Plants were grown in the spring (A) and fall (B) of 1996.
A linkage map for the Oh545o2 × Oh51Ao2 cross was created with 83 polymorphic simple sequence repeat (SSR) markers (Wang et al., 2001). To reduce the labor of genotyping this population with informative SSR markers, we analyzed 40 F2 progeny, including the 20 individuals with the highest and 20 individuals with the lowest FAA content. We conducted interval mapping with a simple regression model to identify a few potential QTLs using the Map Manager QXTb03 program (http://mcbio. med.buffalo.edu/mapmgr.html). Additional SSRs in these regions were then tested and the informative markers were then used to genotype the entire population. All markers flanking potential QTLs were used to genotype the entire F2:3 population.
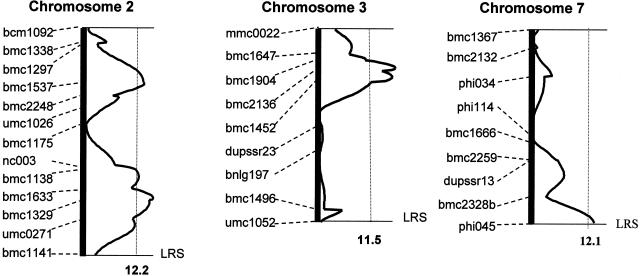
Using the genotypic and phenotypic analysis of the F2:3 progeny, 1,000 permutation tests were performed on individual chromosomes to determine a significant (95%) threshold value of the likelihood ratio statistic (LRS). Four significant QTLs were identified that account for about 46% of the phenotypic variation in FAA content (Fig. 3; Table III). Each of these QTLs has both additive and recessive genetic effects, consistent with the phenotypic variation observed in the F1 and F2:3 progeny. The QTL on the short arm of chromosome 2 has an LRS value of 12.7, which is slightly larger than the threshold value of 12.2 given by permutation tests. It contributes 10% of variance in FAA content. A second QTL on the long arm of chromosome 2 has an LRS value of 14.8, which is much larger than the significant threshold value of 12.2, and it explains 11% of the total phenotypic variance. The third QTL on the short arm of chromosome 3 has the largest effect among the four QTLs. It accounts for about 14% of the variance in FAA level, and has an LRS value of 15.6, which is much higher than the significant threshold value of 11.5 given by the permutation test. The fourth QTL is near the telomere of the long arm of chromosome 7 and has an LRS value of 12.6. This QTL primarily has an additive effect, with a minor recessive effect.
Figure 3.
Interval mapping of QTLs influencing FAA content in maize endosperm. The plots are derived from the interval mapping of loci associated with FAA content in the F2 population of Oh51Ao2 × Oh545o2. A free regression model was used to perform interval mapping. The solid curve illustrates the LRS; SSR DNA markers are indicated on the left. The significant threshold values of LRS for each chromosome were estimated with 1,000 permutations using Map Manager QTXb03. The LRS considered as a significant threshold value (95%) for chromosomes 2, 3, and 7 are 12.2, 11.5, and 12.1, respectively (shown with dashed lines).
Table III.
Characteristics of QTLs influencing free amino acid content
| QTL | Chromosome | Flanking Markers | LRS | Variance | Additive | Dominant |
|---|---|---|---|---|---|---|
| % | ||||||
| 1 | 2L | bmc1633, bmc1329 | 14.8 | 11 | 2.32 | −1.35 |
| 2 | 2S | bmc1537, bmc2248 | 12.8 | 10 | 2.59 | −1.63 |
| 3 | 3S | bmc1904, bmc2136, bmc1452 | 17.9 | 15 | 2.68 | −3.40 |
| 4 | 7L | bmc2328b, phi045 | 12.6 | 10 | 2.37 | −0.93 |
To evaluate the significance of the linkage between the SSRs flanking QTLs and FAA content, a chi-square test was performed using the 20 individuals with extreme high and 20 individuals with extreme low FAA content (Table IV). The flanking markers, bmc1633 and bmc1329, which define the QTL on the long arm of chromosome 2, show significant linkage with this trait, based on their occurrence in the high- and low-FAA individuals. The flanking marker of the QTL on the short arm of chromosome 2, bmc1537, is significantly linked to this QTL, with a chi-square value of 6.4 among the high FAA individuals; there is no linkage among low FAA individuals. There is only one flanking marker tightly linked to the QTL on the long arm of chromosome 7, phi045. It is surprising that based on the chi-square test we did not find significant linkage of the markers flanking the QTL on chromosome 3, which has the greatest phenotypic effect and LRS value. This could result from the five individuals with the highest FAA content having a SSR genotype identical to Oh545o2 (data not shown), whereas the marker genotypes are randomly distributed in other high-FAA individuals. This analysis shows that for all four QTLs, the alleles contributed by Oh545o2 are responsible for the high FAA level.
Table IV.
Segregation of flanking markers among F2 individuals with extreme FAA content
| Marker | Chromosome | High-FAA
Individuals
|
Low-FAA Individuals
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | H | B | X2 | p | A | H | B | X2 | p | ||
| bmc1537 | 2S | 1 | 10 | 9 | 6.4 | ** | 3 | 12 | 5 | 1.2 | NS |
| bmc2248 | 2S | 1 | 12 | 7 | 5.4 | * | 5 | 11 | 4 | 0.3 | NS |
| bmc1633 | 2L | 0 | 13 | 6 | 6.3 | ** | 8 | 11 | 1 | 5.1 | * |
| bmc1329 | 2L | 1 | 9 | 9 | 6.5 | ** | 8 | 11 | 1 | 5.1 | * |
| bmc1904 | 3S | 6 | 8 | 6 | 0.8 | NS | 2 | 13 | 4 | 2.9 | NS |
| bmc2136 | 3S | 5 | 7 | 7 | 1.7 | NS | 2 | 13 | 5 | 2.7 | NS |
| bmc1452 | 3S | 5 | 6 | 8 | 3.4 | NS | 4 | 9 | 5 | 0.3 | NS |
| phi045 | 7L | 2 | 8 | 10 | 7.2 | ** | 8 | 10 | 2 | 3.6 | NS |
A, No. of plants homozygous for the Oh51Ao2 allele. H, No. of plants heterozygous. B, No. of plants homozygous for the Oh545o2 allele. p, Significance level: 0.1, *; 0.05, **; and no significance, NS.
DISCUSSION
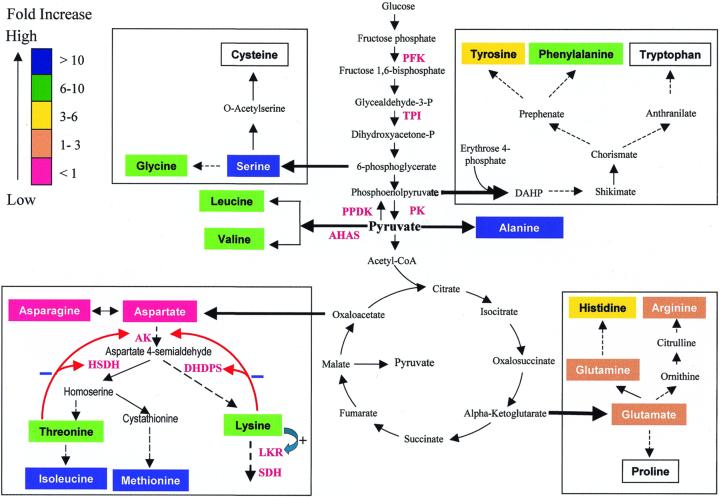
We found significant variability in the FAA content of different maize inbred lines that is accentuated by the o2 mutation. FAAs typically constitute about 1% to 3% of the non-protein nitrogen in wild-type endosperm, and this is increased several-fold in o2 mutants (Sodek and Wilson, 1971; Misra et al., 1975; Moro et al., 1996). The levels of FAA in W64A and Oh51A are fairly typical in this regard, but Oh545 appears to have a genetic predisposition for overaccumulating FAAs, especially amino acids derived from the Asp pathway and glycolytic intermediates (Tables I and II; Fig. 1). Figure 4 illustrates the relative differences in FAA concentration in Oh51Ao2 and Oh545o2. The concentration ratios are somewhat larger when W64Ao2 and Oh545o2 are compared, but the same amino acids, hence biosynthetic pathways, are affected.
Figure 4.
Metabolic pathways showing the major points of regulation for amino acid biosynthesis and Lys degradation. The color scale shows the relative increase (blue, >10-fold; green, 6–10-fold; yellow, 3–6-fold; orange, 1–3-fold; red, <1-fold) in FAA concentration in mature endosperm of Oh545o2 compared with Oh51Ao2. AHAS, Acetohydroxyacid synthase; DAHP, 7-phospho-2-keto-3-deoxy-d-arabino-heptonate; DHDPS, dihydrodipicolinate synthase; FPK, Fru phosphate kinase; LKR, Lys ketoglutarate reductase; PK, pyruvate kinase; PPDK, pyruvate phosphate dikinase; SDH, sacchropine dehydrogenase; TPI, triose phosphate isomerase.
The high concentration of FAAs in o2 endosperm is a consequence of both high levels of synthesis/accumulation during development and the apparent inability to incorporate these amino acids, or turn them over (Arruda et al., 2000), before endosperm desiccation. It is unfortunate that our data do not provide insight regarding the mechanisms responsible for the rapid decline of FAAs in maturing wild-type endosperm, nor why this fails to occur in o2 mutants. The concentration of all amino acids is increased in o2 mutants, but Asp/Asn, Glu/Gln, and Ala are by far the most abundant early in endosperm development. The reduction of these abundant amino acids, especially Gln, is largely responsible for the relative increase in concentration of minor amino acids, especially Lys, Met, and Thr, as the endosperm matures. We found significant variation in the relative concentration of FAAs in the endosperm of the wild-type and o2 inbreds when they were grown at different times and locations. However, the amount of each amino acid (percent of total) was consistently affected for each genotype, regardless of the growing conditions (Table I; data not shown).
Although the results of our study do not address the source of the high level of FAAs in Oh545o2 endosperm, several types of evidence, including what is known about the molecular action of O2 (Schmidt, 1993), suggest that alterations in amino acid biosynthesis are responsible. In theory, the high level of FAA in Oh545o2 could result from higher rates of transport into the endosperm, reduced rates of incorporation into proteins, higher rates of synthesis, and lower rates of turnover, or a combination of these. However, we would not expect the differences in FAA content to result from variation in amino acid transport (Arruda and Silva, 1979; Bush, 1999). The amino acid composition of the vascular sap from the ear peduncle is distinct from that of the endosperm (Arruda and Silva, 1979). Furthermore, the endosperm has been shown to contain many of the enzymes necessary to synthesize its own amino acids (Sodek and Wilson, 1970; Sodek, 1976; Gengenbach et al., 1978). For example, Pro is not present in vascular sap (Arruda and Silva, 1979), but it is abundant in the endosperm (Table II; Fig. 1).
There could be an association between high levels of FAAs and reduced amounts of storage protein synthesis, but our data are incomplete in this regard. The Oh545+/o2 inbreds have a lower protein content than W64A+/o2 and Oh51A+/o2, i.e. 8% and 7% compared with 11% and 9% and 12% and 10%, respectively (Moro et al., 1996). The progeny of the Oh545o2 × Oh51Ao2 cross are all o2 mutants with a reduced capacity for storage protein synthesis. A partial analysis of the F2:3 progeny revealed protein concentrations ranging from 8.5% to 11.7%, with common values of 9%. This would not be considered a particularly low protein concentration. We are developing recombinant inbred lines from these materials, and they will be characterized with regard to protein and FAA content. It is noteworthy that the level of eEF1A, which provides an index of the protein-bound Lys (Moro et al., 1996), segregated independently from the high FAA phenotype among the F2:3 individuals (Wang et al., 2001). Thus, a high level of free Lys did not correlate with the level of Lys-containing proteins in this population.
The four QTLs we identified account for approximately 46% of the variability for FAA content in the progeny of the Oh545o2 × Oh51Ao2 cross. Although this represents only a small proportion of the variability, this outcome is not uncommon for QTL mapping studies, which frequently only can explain about 50% of the variability (Kearsey and Farquhar, 1998). As previously described (Wang et al., 2001), we encountered difficulties developing the Oh545o2 × Oh51Ao2 mapping population, which was half the intended size. We also recognize there are limitations in our mapping data as a consequence of environmental effects. We hope to address the deficiencies of the sample size through a genetic analysis of the recombinant inbred lines that are being developed from the F2:3 individuals.
The QTLs on the short arm of chromosome 2 and the long arm of chromosome 7 have LRS values of 12.8 and 12.6, respectively, which slightly exceed the significant threshold value of 12.1 provided by the permutation tests (Fig. 3). The flanking markers showed significant linkage with the high FAA trait, and additional genotyping with the recombinant inbred progeny lines will help resolve the validity of these QTLs.
The QTL on the short arm of chromosome 3 has the largest phenotypic effect and LRS value, but the chi-square test with the 40 individuals showing the extreme FAA phenotypes failed to show linkage of the flanking markers. Nevertheless, the five individuals with the highest FAA content contain the Oh545o2 allele of the SSR markers flanking this QTL: bmc2136, bmc1904, and bmc1452. As a consequence, it is premature to dismiss this QTL on the basis of a random combination of marker genotypes and FAA phenotypes. It should also be possible to address this question by analyzing the recombinant inbred lines.
O2 encodes a transcription factor that regulates zein gene expression (Hartings et al., 1989; Kodrzycki et al., 1989; Schmidt et al., 1990), but it also has several activities related to carbon and amino acid metabolism. An alteration of one or more of the enzymes involved in amino acid synthesis/degradation or glycolysis could significantly change the steady-state level of FAAs. Alterations in enzymes affecting amino acid metabolism have been shown to have pleiotropic affects on FAA levels in plant tissues. For example, the levels of Thr, Met, and other amino acids are increased in AK mutants that are less sensitive to Lys feedback inhibition (Hibberd and Green, 1982; Diedrick et al., 1990; Dotson et al., 1990). A feedback-insensitive AK mutant in tobacco, LT 70, not only has a higher level of amino acids derived from the Asp pathway, but other pathways as well (Frankard et al., 1991, 1992). Alteration of Trp and Tyr levels in transgenic tobacco leaves affected the level of Trp, as well as the nonaromatic amino acids Met, Val, and Leu (Guillet et al., 2000).
The large amount of Ala and Ser in Oh545o2 could be caused by an increased level of pyruvate and 3-phosphohydroxypyruvate (Huppe and Turpin, 1994; Plaxton 1996; Bourguignon and Rebeille, 1999), which are intermediates or end products of gylcolysis. It was shown that a cyPPDK is down-regulated by the o2 mutation (Gallusci et al., 1996; Maddaloni et al., 1996; Damerval and Le Guilloux, 1998). This enzyme, which catalyzes the conversion of pyruvate to phosphoenol pyruvate, is a key regulator of the glycolytic pathway and is linked to carbon and amino acid metabolism. Thus, the o2 mutation may lead to an increased level of pyruvate by down-regulating cyPPDK, resulting in higher levels of Ala.
Mitchell-Olds and Pedersen (1998) suggested that both regulatory and structural genes influence the glycolytic pathway. Because regulators of glycolysis have not been mapped in maize, it is of interest to compare the activity of several key enzymes in this pathway. Preliminary results show that pyruvate kinase activity is higher in Oh545o2 than W64Ao2 endosperm at 15 and 25 DAP (data not shown). However, a systematic characterization of such enzymes will be necessary before any inferences are warranted. We determined that a locus encoding cytosolic triose phosphate isomerase 4, which encodes a catalytic step of glycolysis (Wendel et al., 1989), is located near the QTL on the short arm of chromosome 3. However, there is no evidence that this enzyme influences the flux rate of glycolysis.
Several amino acids of the Asp pathway are significantly increased in Oh545o2 (Tables I and II; Fig. 4), and it was interesting to note that monofunctional Ask2 and the bifunctional AK-HSDH2 both map to the same region as the QTL on the long arm of chromosome 2 (Galili, 1995; Azevedo et al., 1997). The maize monofuctional Ask2 presumably encodes a Lys-sensitive AK, and one mutant, ask2, overproduces Thr, Lys, and Met (Hibberd and Green, 1982; Dotson et al., 1990; Muehlbauer et al., 1994a), similar to the phenotype of Oh545o2. AK-HSDH, which is Thr sensitive, is encoded by three genes (Azevedo et al., 1992; Muehlbauer et al., 1994b). One of them was mapped to the long arm of chromosome 2, and one was mapped to the short arm of chromosome 4 using maize cDNA clones; however, the location of the third is unknown (Muehlbauer et al., 1994b). Ask2 and AK-HSDH2 are both located a few centimorgans apart on the long arm of chromosome 2. The ask1 mutant, which also encodes a Lys-insensitive AK, results in high levels of Lys, Thr, and Met, and a double mutant of o2 and ask1 has higher AK activity and a higher level of FAA than the single mutants (Azevedo et al., 1990; Brennecke et al., 1996). Thus, if the AK in Oh545o2 has a higher total activity and/or is less sensitive to Lys or Thr feedback inhibition, it could explain why the ratio of FAA is so much larger in Oh545o2/W64Ao2 compared with Oh545+/W64A+. Whether the monofunctional Ask2 and/or bifunctional AK-HSDH2 are the candidate genes on the long arm of chromosome 2 will be addressed by investigating total activity and feedback inhibition properties of these enzymes.
MATERIALS AND METHODS
Plant Materials
We obtained seed samples of the maize (Zea mays) inbreds W64A+/o2, Oh545+/o2, and Oh51Ao2 from Crows Hybrid Corn Company (Milford, IL). One set of FAA data was obtained using flour from these samples. W64A+/o2, Oh545+/o2, and Oh51Ao2 were also grown at the University of Arizona West Campus Agricultural Center or in the greenhouse. Oh545o2 (high FAA) and Oh51Ao2 (low FAA) and their F1 and F2:3 progeny were planted in the spring and fall seasons of 1996 in 6-m rows, 75 cm apart, with 15 plants per row. The plants were self-pollinated or crossed, as appropriate, and well-filled ears were collected and air dried. For FAA analysis of developing endosperm, single plants were grown in pots in the greenhouse between November, 1996, and January, 1997. Developing kernels from well-filled ears were collected at 20, 30, 40, and 50 DAP. The endosperm and embryo were separated by dissection, frozen with liquid nitrogen, and stored at −80°C.
Preparation of Samples from Mature and Developing Endosperm for FAA Analysis
Twenty kernels were collected from well-filled, mature parental lines, F1 and F2:3 ears. The kernels were degermed with a variable-speed dental-type drill (model 750, Dremel, Racine, WI), and the endosperms were combined and pulverized to a fine flour with a Wig-l-Bug amalgamator (Crescent Dental Manufacturing Co., Chicago). A modification of the ninhydrin assay described by Mertz et al. (1974) was used to estimate the relative content of FAA. Twenty milligrams of endosperm flour was defatted by adding 1 mL of petroleum ether and incubating the samples for 1 h on a rocking platform. The samples were then centrifuged at 16,000g for 10 min. The ether was decanted and a second 1 mL of petroleum ether was added to the flour. The samples were briefly vortexed, centrifuged, and then the ether was removed. The flour was resuspended in 1 mL of water and placed on a rocking platform for 20 min at 25°C. Particulate matter was pelleted by centrifugation and 600 μL of the supernatant was transferred to a fresh 1.5-mL microfuge tube and stored at 4°C as a working sample. One hundred microliters of the working sample was added to a 1.5-mL microfuge tube containing 250 μL of ninhydrin reagent (Sigma, St. Louis), and the tube was placed in a boiling water bath for 20 min. An aliquot of 200 μL was removed, added to an ELISA plate, and three 2-fold serial dilutions were made into adjacent wells containing water. Absorbance was read at 590 nm with an ELISA plate reader (RR 700, Dynatech Laboratories Inc., Chantilly, VA). Two samples from two independent extractions were used to measure amino acid content; a regression was used to calculate the absorbance of the original sample. The slope of the relationship between A590 (absorbance at 590 nm) and flour weight for Oh51Ao2 was used to calculate the relative FAA content in endosperm samples.
For qualitative analysis, the extracted amino acids were filtered through a C18 reverse phase mini-cartridge (Vydac, Hesperia, CA) to remove soluble proteins. An aliquot of 500 μL of the filtrate was dried in a speed vacuum drier (Southwest Instruments Biomedical Instrumentation, Tucson, AZ). The pellet was resuspended in 50 μL of sterile double-distilled water for amino acid analysis.
A similar procedure was used to analyze FAA in developing endosperms. Two or three well-filled ears were collected at different developmental stages, and 10 to 20 fresh kernels were degermed and the embryos and endosperms pooled. These tissues were lyophilized and stored at −80°C until analyzed. Flour was prepared as described above, and amino acids were extracted from 20-mg samples. In this case, the flour was resuspended in 1 mL of sterile water containing 0.1 mm phenylmethylsulfonyl fluoride. After extraction at 4°C for 20 min, the soluble extract was filtered through a C18 reverse phase minicartidge (Vydac) to remove soluble proteins. Five hundred micoliters of supernatant was lyophilized with a speed vacuum drier (Southwest Instruments Biomedical Instrumentation). The pellet was dissolved in 50 μL of sterile, double-distilled water for amino acid analysis. Assays were made of triplicate samples and ses were calculated (data not shown).
Amino Acid Analysis
Amino acid analysis was performed by the Laboratory for Protein Sequencing and Analysis at the University of Arizona using a post column Amino Acid Analyzer (model 7300, Beckman Instruments, Palo Alto, CA; ninhydrin method). Amino acids were separated by ion-exchange chromatography, using citrate buffer of increasing ionic strength and pH, at varying temperatures. Amino acids were detected with ninhydrin; the reaction was monitored with a colorimeter at 570 nm for primary amino acids and 440 nm for secondary amino acids.
Interval Mapping of QTLs Influencing Amino Acid Content
SSR DNA marker selection, conditions for PCR, linkage map creation, and interval mapping were described by Wang et al. (2001). Twenty F2 individuals with extreme high and 20 F2 individuals with extreme low levels of FAAs were genotyped with 83 SSR markers that are polymorphic between Oh51Ao2 and Oh545o2 (Wang et al., 2001). The first round of interval mapping (Lander and Botstein, 1989) was performed with these selected individuals, and regions with an LRS (Haley and Knott, 1992; Kearsey and Farquhar, 1998) of more than 10 were considered potential QTLs. Additional SSRs near these loci subsequently were tested for polymorphism, and the entire population was genotyped with flanking markers. Interval mapping was conducted with a free regression model and permutation tests (1,000 shuffles) on individual chromosomes were done to establish the significant threshold value of LRS (Churchill and Doerge, 1994; Doerge and Churchill, 1996).
Statistical Analyses
Variance and linear regression analysis were performed with the statistical package in Excel (Microsoft, Redwood, CA). Regressions were always significant (F test, P < 0.01) and had an r2 value greater than 0.9. The correlation was compared using a standard t test. Map Manager QTXb03 was used to detect QTLs (http://mcbio.med.buffalo.edu/mapmgr.html). The LRS threshold values for significant QTLs (95%) were based on 1,000 permutation tests with a 1-cM step for an individual chromosome.
ACKNOWLEDGMENT
We thank Dr. Richard Jorgensen for the use of his PCR machine.
Footnotes
This research was supported by Pioneer Hi-Bred (grant to B.A.L.).
LITERATURE CITED
- Arruda P, Kemper EL, Papes F, Leite S. Regulation of lysine catabolism in higher plants. Trends Plant Sci. 2000;5:324–330. doi: 10.1016/s1360-1385(00)01688-5. [DOI] [PubMed] [Google Scholar]
- Arruda P, Silva WJ. Amino acid composition of vascular sap of maize ear peduncle. Phytochemistry. 1979;18:409–410. [Google Scholar]
- Azevedo RA, Arana JL, Arruda P. Biochemical genetics of the interaction of the lysine plus threonine-resistant mutant Ltr*1 with opaque2 maize mutant. Plant Sci. 1990;70:81–90. [Google Scholar]
- Azevedo RA, Arruda P, Turner WL, Lea PJ. The biosynthesis and metabolism of the aspartate derived amino acids in higher plants. Phytochemistry. 1997;46:395–419. doi: 10.1016/s0031-9422(97)00319-1. [DOI] [PubMed] [Google Scholar]
- Azevedo RA, Smith RJ, Lea PJ. Aspartate kinase regulation in maize: evidence for co-purification of threonine-sensitive aspartate kinase and homoserine dehydrogenase. Phytochemistry. 1992;31:3731–3734. [Google Scholar]
- Bourguignon J, Rebeille F. Serine and glycine metabolism in higher plants. In: Singh BK, editor. Plant Amino Acids. New York: Marcel Dekker Inc.; 1999. pp. 111–146. [Google Scholar]
- Brennecke K, Souza Neto AJ, Lugli J, Lea PJ, Azevedo RA. Aspartate kinase in maize mutants ask1-lt19 and opaque-2. Phytochemistry. 1996;41:707–712. [Google Scholar]
- Brochetto-Braga MR, Leite A, Arruda P. Partial purification and characterization of lysine-ketoglutarate reductase in normal and opaque-2 maize endosperm. Plant Physiol. 1992;98:1139–1147. doi: 10.1104/pp.98.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan JK. Advances in the biochemistry of amino acid biosynthesis. In: Milfin BJ, Lea PJ, editors. The Biochemistry of Plants. Vol. 16. New York: Academic Press; 1990. pp. 161–195. [Google Scholar]
- Bush DR. Amino acid transport. In: Singh BK, editor. Plant Amino Acids. Princeton, NJ: American Cyanamid Company; 1999. pp. 305–318. [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C, de Vienne D. Quantification of dominance for proteins pleiotropically affected by opaque-2 in maize. Heredity. 1993;70:38–51. [Google Scholar]
- Damerval C, Le Guilloux M. Characterization of novel proteins affected by the o2 mutation and expressed during maize endosperm development. Mol Gen Genet. 1998;257:354–361. doi: 10.1007/s004380050657. [DOI] [PubMed] [Google Scholar]
- Diedrick TL, Frisch DA, Gengenbach BG. Tissue culture isolation of a second mutant locus for increased threonine accumulation in maize. Theor Appl Genet. 1990;79:209–215. doi: 10.1007/BF00225953. [DOI] [PubMed] [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson SB, Frish DA, Somers DA, Gengenbach BG. Lysine-insensitive aspartate kinase in two threonine-overproducing mutants in maize. Planta. 1990;182:546–552. doi: 10.1007/BF02341030. [DOI] [PubMed] [Google Scholar]
- Frankard V, Ghislain M, Jacobs M. Two feedback-insensitive enzymes of the aspartate pathway in Nicotiana sylvestris. Plant Physiol. 1992;99:1285–1293. doi: 10.1104/pp.99.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankard V, Ghislain M, Negrutiu I, Jacobs M. High threonine producer mutant of Nicotiana sylvestris (Spegg. and Comes) Theor Appl Genet. 1991;82:273–282. doi: 10.1007/BF02190612. [DOI] [PubMed] [Google Scholar]
- Galili G. Regulation of lysine and threonine synthesis. Plant Cell. 1995;7:899–906. doi: 10.1105/tpc.7.7.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallusci P, Varotto S, Matsuoko M, Maddaloni M, Thompson RD. Regulation of cytosolic pyruvate, orthophosphate dikinase expression in developing maize endosperm. Plant Mol Biol. 1996;31:45–55. doi: 10.1007/BF00020605. [DOI] [PubMed] [Google Scholar]
- Gengenbach BG, Walter TJ, Green CE, Hibberd KA. Feedback regulation of lysine, threonine, and methionine biosynthetic enzymes in corn. Crop Sci. 1978;18:472–576. [Google Scholar]
- Guillet G, Poupart J, Basurco J, De Luca V. Expression of tryptophan decarboxylase and tyrosine decarboxylase genes in tobacco results in altered biochemical and physiological phenotypes. Plant Physiol. 2000;122:933–343. doi: 10.1104/pp.122.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habben JE, Kirlies AW, Larkins BA. The origin of lysine containing proteins in opaque2 maize endosperm. Plant Mol Biol. 1993;23:825–838. doi: 10.1007/BF00021537. [DOI] [PubMed] [Google Scholar]
- Habben JE, Moro GL, Hunter BG, Hamaker BR, Larkins BA. Elongation factor -1α concentration is highly correlated with the lysine content of maize endosperm. Pro Natl Acad Sci USA. 1995;92:8640–8644. doi: 10.1073/pnas.92.19.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Hartings H, Maddaloni M, Lazzaroni N, di Fonzo N, Motto M, Salamini F, Thompson R. The o2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J. 1989;8:2795–2801. doi: 10.1002/j.1460-2075.1989.tb08425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd KA, Green CE. Inheritance and expression of lysine plus threonine resistance selected in maize tissue culture. Proc Natl Acad Sci USA. 1982;79:559–563. doi: 10.1073/pnas.79.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Transcriptional and translational regulation of gene expression in the amino acid biosynthesis in Saccharomyces cerevisiae. Prog Nucleic Acids Res Mol Biol. 1990;89:292–299. doi: 10.1016/s0079-6603(08)60712-6. [DOI] [PubMed] [Google Scholar]
- Huppe HC, Turpin DH. Intergration of carbon and nitrogen metabolism in plant and algal cells. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:577–607. [Google Scholar]
- Kearsey MJ, Farquhar AG. QTL analysis in plants: where are we now? Heredity. 1998;80:137–142. doi: 10.1046/j.1365-2540.1998.00500.x. [DOI] [PubMed] [Google Scholar]
- Kemper EL, Cord Neto G, Papes F, Martinez Moraes KC, Leite A. The role of Opaque 2 in the control of lysine-degrading activities in developing maize endosperm. Plant Cell. 1999;11:1981–1993. doi: 10.1105/tpc.11.10.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodrzycki R, Boston R, Larkins BA. The opaque-2 mutation of maize defferentially reduces zein gene transcription. Plant Cell. 1989;1:105–114. doi: 10.1105/tpc.1.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaloni M, Donini G, Balconi C, Rizzi E, Gallusci P, Forlani F, Lohmer S, Thompson R, Salamini F, Motto M. The transcriptonal activator Opaque-2 controls the expression of a cytosolic form of pyruvate orthophosphate dikinase-1 in maize endosperms. Mol Gen Genet. 1996;250:647–654. doi: 10.1007/BF02174452. [DOI] [PubMed] [Google Scholar]
- Mauri I, Maddaloni M, Lohmer S, Motto M, Salamini F, Thompson R, Martegani E. Functional expression of the transcriptional activator Opaque-2 of Zea mays in transformed yeast. Mol Gen Genet. 1993;241:319–326. doi: 10.1007/BF00284684. [DOI] [PubMed] [Google Scholar]
- Meer JM, Cudmore RH, Mauly KF (1999) Map Manager QTX. http://mcbio.med.buffalo.edu/mmQTX.html
- Mertz ET, Bates LS, Nelson OE. Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science. 1964;145:279. doi: 10.1126/science.145.3629.279. [DOI] [PubMed] [Google Scholar]
- Mertz ET, Misra PS, Jambunathan R. Rapid ninhydrin color test for screening high-lysine mutants of sorghum, barley, and other cereal grains. Cereal Chem. 1974;51:304–307. [Google Scholar]
- Misra PS, Mertz ET, Glover DV. Studies on corn proteins: VIII. Free amino acid content of opaque-2 and double mutants. Cereal Chem. 1975;52:844–848. [Google Scholar]
- Mitchell-Olds T, Pedersen D. The molecular basis of quantitative genetic variation in central and secondary metabolism in Arabidopsis. Genetics. 1998;149:739–747. doi: 10.1093/genetics/149.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro GL, Habben JE, Hamaker BR, Larkins BA. Characterization of the variability for lysine content in normal and opaque-2 maize endosperm. Crop Sci. 1996;36:1651–1659. [Google Scholar]
- Muehlbauer GJ, Gengenbach BG, Somers DA, Donovan CM. Genetic and amino acid analysis of two maize threonine-overproducing, lysine-insensitive aspartate kinase mutants. Theor Appl Genet. 1994a;89:767–774. doi: 10.1007/BF00223717. [DOI] [PubMed] [Google Scholar]
- Muehlbauer GJ, Somers DA, Matthews BF, Gengenbach BG. Molecular genetics of the maize (Zea mays L.) aspartate kinase-homoserine dehydrogenase gene family. Plant Physiol. 1994b;106:1303–1312. doi: 10.1104/pp.106.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC. The organization and regulation of plant glycolysis. Annul Rev Plant Physiol Plant Mol Biol. 1996;47:185–214. doi: 10.1146/annurev.arplant.47.1.185. [DOI] [PubMed] [Google Scholar]
- Schmidt RJ. Opaque-2 and zein gene expression. In: Verma DPS, editor. Control of Plant Gene Expression. Boca Raton, FL: CRC Press; 1993. pp. 337–355. [Google Scholar]
- Schmidt RJ, Burr FA, Aukerman MJ, Burr B. Maize regulatory gene opaque-2 encodes a protein with a “leucine zipper” motif that binds to zein DNA. Proc Natl Acad Sci USA. 1990;87:46–50. doi: 10.1073/pnas.87.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Ketudat M, Aukerman MJ, Hoschek G. Opaque-2 is a transcription activator that recognizes a specific target site in 22-kD zein genes. Plant Cell. 1992;4:689–700. doi: 10.1105/tpc.4.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodek L. Biosynthesis of lysine and other amino acids in the developing maize endosperm. Phytochemistry. 1976;15:1903–1906. [Google Scholar]
- Sodek L, Wilson CM. Incorporation of leucine-14C and lysine-14C into protein in the developing endosperm of normal and opaque-2 corn. Arch Biochem Biophys. 1970;140:29–38. doi: 10.1016/0003-9861(70)90006-8. [DOI] [PubMed] [Google Scholar]
- Sodek L, Wilson CM. Amino acid composition of proteins isolated from normal, opaque-2, and floury-2 corn endosperms by a modified Osborne procedure. J Agric Food Chem. 1971;19:1144–1149. [Google Scholar]
- Wang X-L, Woo Y, Kim C, Larkins BA (2001) QTL mapping of loci influencing eEF1A content in maize endosperm. Plant Physiol (in press) [DOI] [PMC free article] [PubMed]
- Wendel JF, Stuber CW, Goodman MM, Beckett JB. Duplicated plastid and triplicated cytosolic isozymes of triose phosphate isomerase in maize (Zea mays L.) J Hered. 1989;80:218–228. doi: 10.1093/oxfordjournals.jhered.a110839. [DOI] [PubMed] [Google Scholar]
- Yunes JA, Cord Neto G, Leite A, Ottoboli LMM, Arruda P. The role of the Opaque-2 transcriptional factor in the regulation of protein accumulation and amino acid metabolism in maize seeds. An Acad Bras Ciencias. 1994;66:227–238. [PubMed] [Google Scholar]