Abstract
Epidermal growth factor receptor (EGFR) has critical roles in epithelial cell physiology. Over-expression and over-activation of EGFR have been implicated in diverse cancers, including triple-negative breast cancers (TNBCs), prompting anti-EGFR therapies. Therefore, developing potent therapies and addressing the inevitable drug resistance mechanisms necessitates deciphering of EGFR related networks. Here, we describe Sorting Nexin 3 (SNX3), a member of the recycling retromer complex, as a critical player in the epidermal growth factor (EGF) stimulated EGFR network in TNBCs. We show that SNX3 is an immediate and sustained target of EGF stimulation initially at the protein level and later at the transcriptional level, causing increased SNX3 abundance. Using a proximity labeling approach, we observed increased interaction of SNX3 and EGFR upon EGF stimulation. We also detected colocalization of SNX3 with early endosomes and endocytosed EGF. Moreover, we show that EGFR protein levels are sensitive to SNX3 loss. Transient RNAi models of SNX3 downregulation have a temporary reduction in EGFR levels. In contrast, long-term silencing forces cells to recover and overexpress EGFR mRNA and protein, resulting in increased proliferation, colony formation, migration, invasion in TNBC cells, and increased tumor growth and metastasis in syngeneic models. Consistent with these results, low SNX3 and high EGFR mRNA levels correlate with poor relapse-free survival in breast cancer patients. Overall, our results suggest that SNX3 is a critical player in the EGFR network in TNBCs with implications for other cancers dependent on EGFR activity.
INTRODUCTION
EGFR (Epidermal Growth Factor Receptor) regulates signal transduction pathways that control diverse cell phenotypes, including proliferation, migration, and survival [1]. Overexpression and over-activation of EGFR in breast tumors are associated with poor clinical outcomes, including early recurrence, increased risk of metastasis, and decreased survival [2–6]. Therefore, it is essential to understand mechanisms regulating EGFR levels and activity. Among other critical processes, internalization and recycling steps regulate surface EGFR levels and activity in normal and in cancer cells.
SNX3 belongs to sorting nexins and is a critical player in the recycling pathway. SNX3 is involved in an early step of endocytosis by binding to phosphatidylinositol-3-phosphate (PI3P) of early endosomes. Multiple lines of evidence established that SNX3 is an essential component of the VPS35:VPS26:VPS29 retromer complex. Retromer complexes recycle the endocytosed receptors such as Wnt receptor Wntless (Wls) and EGFR back to the trans-Golgi network or plasma membrane, respectively [7–16]. If not recycled, endosomes mature into late endosomes, which later fuse with the lysosomes, and the cargo is degraded [17, 18].
SNX proteins, SNX1, SNX2, SNX5, SNX6, SNX12, and SNX16, have been implicated in lysosomal targeting of internalized receptors, including EGFR [19–24]. SNX3, on the other hand, plays a direct role in multivesicular body formation and has a cargo recognition function within the retromer [25, 26]. Hence, SNX3 is involved in the recycling of receptors rather than directly targeting them to lysosomes. Only in the absence of SNX3 function, endocytosed receptors are degraded in the lysosomes. For example, genetic loss of Snx3 leads to lysosomal degradation of internalized Wls receptors in C. elegans and Drosophila [27, 28]. Likewise, lysosomal degradation of EGFR increases upon siRNA silencing of SNX3 in epidermoid carcinoma cells [14].
Despite the role of SNX3 in recycling, knowledge on its contribution to cancer phenotypes is limited [29]. The objective of this study was to investigate SNX3 regulation with EGF stimulation in EGFR positive mammary cells and determine whether SNX3 has a role in breast tumorigenesis.
RESULTS
SNX3 is a target of EGF stimulation in EGFR positive cells, first at the protein level and later at transcript level
To begin investigating the role of SNX3 concerning EGFR in breast cancers, we first asked whether SNX3 is an EGF/EGFR induced gene. We used the MCF10A cell line, an EGFR positive, non-tumorigenic, and hormone receptor-negative mammary cell line. MCF10A is also a well-established model for studying gene expression patterns in response to EGF stimulation [30–32]. We stimulated cells with EGF in a time-course experiment to detect dynamic changes in gene expression and collected lysates and RNA.
Shortly after EGF (15–30 min) stimulation, the abundance of SNX3 protein was increased and remained high throughout the EGF treatment (i.e., 12 h) (Fig. 1A). This increase in SNX3 following EGF treatment was also seen in HEK293 (EGFR positive human embryonic kidney cells) (Supplementary Fig. S1). These results showed that the EGF responsiveness of SNX3 was not MCF10A specific.
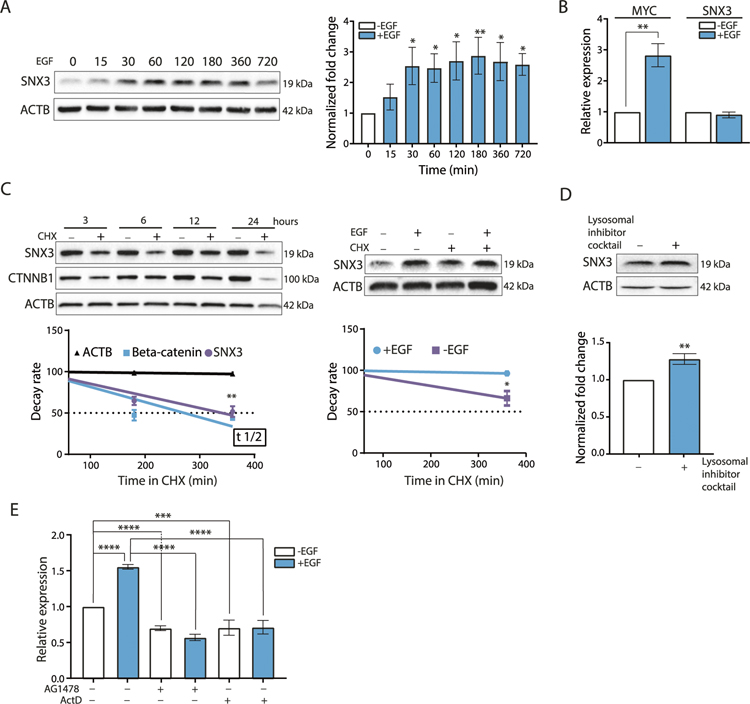
Fig. 1. EGF induction of SNX3.
A Western blotting analysis for SNX3 in MCF10A cells treated with EGF (20 ng/ml) for the indicated time points (minutes). ACTB was used as a loading control. Blots are representative of 3 independent treatments. Graphs show densitometric quantification of bands. The data represent the mean (SD) of 3 experiments. One-way ANOVA with Tukey’s multiple comparison test was used, *p < 0.05, **p < 0.01. B RT-qPCR for the expression of MYC and SNX3 upon EGF stimulation (20 ng/ml, for 30 min) relative to untreated MCF10A cells. The data represent the mean (SD) of 3 independent treatments. Unpaired t-test was performed for statistical significance, **p < 0.01. C Western blotting analysis for the decay rate of SNX3 protein in MCF10A cells treated with CHX (30 μg/ml) with or without EGF (20 ng/ml) for the indicated time points. Cell lysates were immunoblotted for SNX3, for ACTB as a loading control, and for CTNNB1 (Beta-catenin) as a positive control for CHX treatments. The data represent the mean (SD) of 3 independent treatments. Unpaired t-test with Welch’s correction was performed for statistical significance, *p < 0.05, **p < 0.01. D MCF10A cells were treated with a lysosomal inhibitor cocktail (10 μM Leupeptin, 1.5 μM Pepstatin A, 1 μM E-64) for 24 h, and lysates were collected. Blots are representative of 3 independent treatments. Graphs show densitometric quantification of bands. Unpaired t-test was performed for statistical significance, ** indicates p < 0.01, E RT-qPCR for SNX3 mRNA. MCF10A cells were pre-treated with actinomycin D (10 μg/ml) for 1 h and with (EGF 20 ng/ml) for 12 h. The data represent the mean (SD) of 3 independent treatments. One-way ANOVA with Tukey’s multiple comparison test was used ***p < 0.001, ***p < 0.0001.
To understand how SNX3 protein abundance increased in response to EGF, we first tested whether regulation was transcriptional. Transcriptional upregulation of MYC mRNA was evident as an EGF responsive gene [32]. However, we did not observe transcriptional upregulation of SNX3 mRNA upon EGF stimulation at early time points (Fig. 1B, Supplementary Fig. S2). Next, we turned to protein stability as a possible mechanism to explain increased SNX3 protein levels at early time points of EGF stimulation. In a time-course experiment, we measured SNX3 protein half-life after blocking translation with cycloheximide (CHX) treatment alone or with EGF stimulation and collected cell lysates for western blotting. CTNNB1 (Beta-catenin) was used as a control for the CHX treatment [33]. We also confirmed that CHX treatment did not impair the lysosomal degradation of EGF-induced EGFR (Supplementary Fig. S3). Inhibition of translation by CHX caused a reduction in SNX3 protein levels (Fig. 1C, left panel). Unlike SNX3, EGFR protein decay was not rapid (Supplementary Fig. S3). Interestingly, EGF treatment enhanced the abundance of SNX3 protein levels in CHX treated cells (Fig. 1C, right panel). EGF inflicted increase in protein levels suggested post-translational regulation of SNX3 protein, possibly through degradation. Accordingly, blocking proteasomal degradation did not recover SNX3 protein levels (Supplementary Fig. S4), whereas lysosomal inhibition enhanced SNX3 abundance (Fig. 1D).
On the other hand, EGF stimulation for 12 h resulted in increased levels of SNX3 mRNA (Fig. 1E). To test whether the increase in SNX3 mRNA levels was transcriptional or due to enhanced mRNA stability, we treated cells with actinomycin D, a transcription inhibitor, or AG1478, a selective inhibitor of EGFR tyrosine kinase activity [34]. Both actinomycin D and AG1478 treatments reversed the EGF-induced increase of the SNX3 mRNA (Fig. 1E). In addition, we measured the decay rate of SNX3 mRNA by blocking transcription for up to 12 h. We showed that the SNX3 transcript was not rapidly degraded (Supplementary Fig. S5). Therefore, it is unlikely that SNX3 mRNA stability is enhanced further by EGF stimulation.
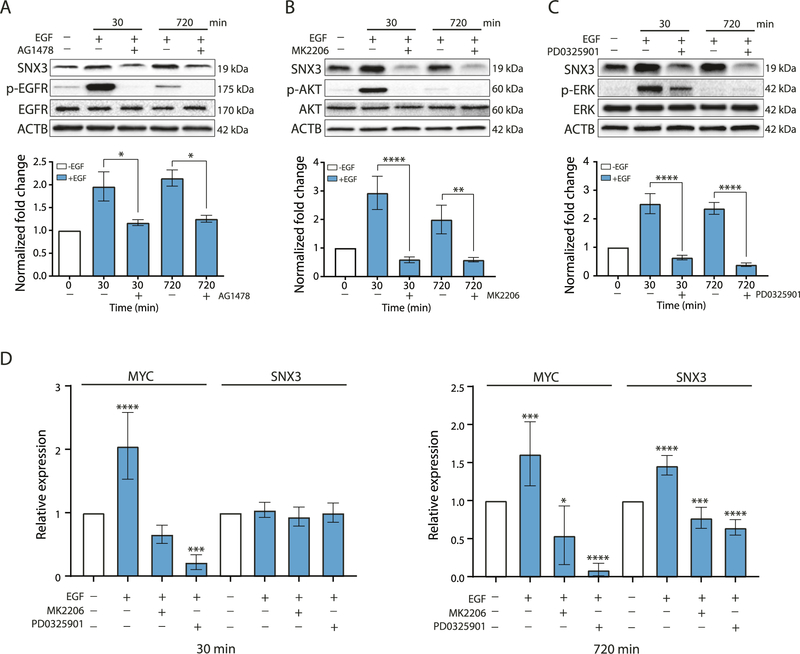
Furthermore, upregulation of SNX3 protein upon EGF stimulation was reversible by AG1478 [34] (Fig. 2A), by MK2206, an Akt inhibitor [35] (Fig. 2B), and by PD0325901, a MEK inhibitor [36] (Fig. 2C). These results showed that inhibiting EGFR and downstream pathways blocked the upregulation of SNX3. Treating EGF stimulated cells with MK2206 and PD0325901 further decreased SNX3 protein levels (Supplementary Fig. S6). As part of the late response, transcriptional regulation of SNX3 was also inhibited after blocking EGFR and downstream signaling pathways, preventing the increase of SNX3 protein (Fig. 2D). In support of these results, using the same inhibitors prevented upregulation of SNX3 in HEK293 cells (Supplementary Fig. S7).
Fig. 2. Effect of blocking of EGFR and downstream pathways on SNX3 protein levels.
Western blotting for SNX3, p-EGFR, p-AKT, p-ERK, EGFR, AKT, and ERK in MCF10A cells pre-treated with (A) AG1478 (25 μM) for 12 h, (B) with MK2206 (1 μM), (C) with PD0325901 (1 μM) for 4 h. Pre-treated cells were stimulated with EGF (20 ng/ml) for the indicated time points. Blots are representative of 3 independent experiments. Bars show densitometric quantification of SNX3 levels (mean (SD) of 3 experiments). One-way ANOVA with Tukey’s multiple comparison test was used, *p < 0.05, **p < 0.01, **** indicates p < 0.0001. (D) RT-qPCR for the mRNA levels of SNX3 in MCF10A cells upon EGF stimulation (30 min or 720 min) and inhibitor treatments compared with untreated cells. MYC expression was used as a positive control. The fold change for SNX3 mRNA was calculated as described. The data represent the mean (SD) of 3 independent treatments. One-way ANOVA with Dunnett’s multiple comparison test was used, *p < 0.05, ***p < 0.001, ****p < 0.0001.
In addition, we tested whether any other proliferative signal would induce SNX3 mRNA or protein in a similar pattern. However, estradiol stimulation of estrogen receptor (ER) positive MCF7 breast cells did not induce SNX3 at the mRNA or protein level (Supplementary Fig. S8).
Taken altogether, SNX3 is a specific target of EGF stimulation in EGFR positive cells, first at the protein level and later at the transcriptional level.
SNX3 interacts with EGFR, co-localizes with early endosomes and endocytosed EGF
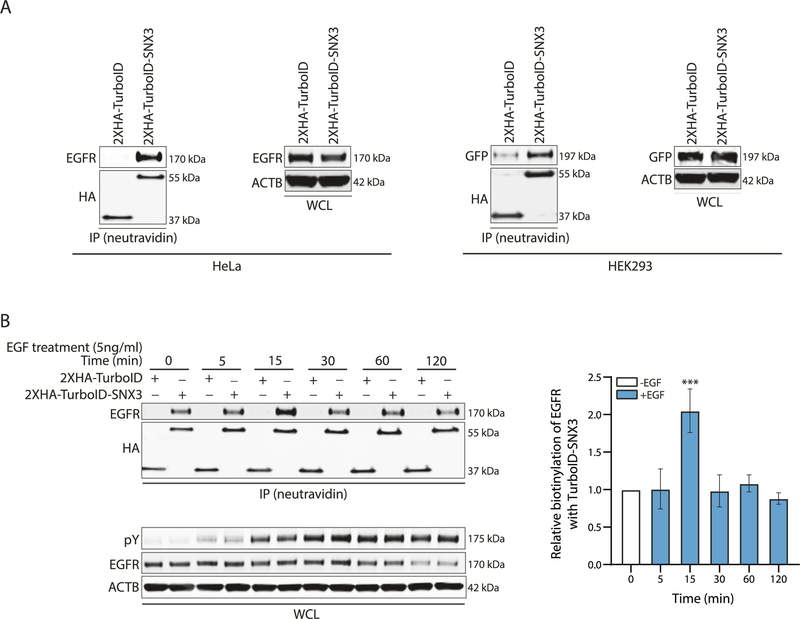
After establishing EGF/EGFR specific upregulation of SNX3, we began examining how SNX3 might, in turn, regulate EGFR. First, we took the proximity ligation approach to confirm the dynamic interaction between SNX3 and EGFR [37, 38]. We fused SNX3 protein to the C-terminus of 2XHA-tagged TurboID (2X HA-TurboID-SNX3). We transfected this construct to HeLa cells. In parallel, we transfected HEK293 cells with the same TurboID constructs and an EGFR-GFP fusion construct. The reasons for choosing these cells were their high transfection rates, EGFR positivity, and previous literature that established the functionality of the retromer and the endocytic pathways [25, 39, 40].
Following biotin incubation and neutravidin-mediated pulldown,immuno-blotting showed TurboID-SNX3 biotinylated endogenous EGFR and EGFR-GFP protein in HeLa and HEK293 cells, respectively (Fig. 3A). Immuno-blotting with whole-cell lysates (WCL) showed equal EGFR levels in TurboID vector control and 2XHA-TurboID-SNX3 transfected cells.
Fig. 3. Proximity-based labeling by SNX3-TurboID.
A Western blotting analysis on lysates from cells transfected with 2XHA-TurboID or 2XHA-TurboID-SNX3 constructs for proximity-based labeling assay. For HeLa cells, immune-precipitated (IP) samples were immunoblotted with EGFR and HA antibodies. In parallel, the 2XHA-TurboID or 2XHA-TurboID-SNX3 and EGFR-GFP constructs were tranfected to HEK293 cells. IP samples were immunoblotted with HA and GFP antibodies. GFP-antibody was used to detect the EGFR-GFP in the IP samples. HA-antibody was used to verify the expression of TurboID or TurboID-SNX3 fusion protein for both cells. Whole-cell lysates (WCL) were used to show the equal loading of proteins. Blots are representative of 3 independent experiments. B Western blotting analysis on lysates from HeLa cells transfected with 2XHA-TurboID or 2XHA-TurboID-SNX3 constructs and treated with EGF as indicated. Biotinylated EGFR levels were detected. HA-antibody was used to verify the expression of TurboID or TurboID-SNX3 fusion protein. WCL shows EGFR, phosphotyrosine (pY), and ACTIN levels. Blots and the graph for densitometry analysis represent 3 independent experiments. One-way ANOVA and Dunnett’s multiple comparison tests were used to measure statistical significance, ***p < 0.001.
Next, we stimulated HeLa cells with EGF (5 ng/ml) for a period of 5, 15, 30, 60, and 120 min. EGFR is known to be recycled back to the plasma membrane upon low-dose EGF treatment [41]. Accordingly, biotinylation of EGFR by SNX3-TurboID was highest at 15 min of low-dose EGF stimulation (Fig. 3B). In contrast, following long-term EGF stimulation, EGFR is degraded in lysosomes to circumvent the over-activation of downstream pathways [41]. In agreement, total EGFR levels did not change until EGF stimulation was extended to 60 min when EGFR levels were downregulated (Fig. 3B). Biotinylation of EGFR by SNX3-TurboID was also decreased after 15 min of EGF stimulation. Of note, EGFR was also biotinylated in unstimulated cells, albeit at a reduced level. We may explain this by the low-level internalization of EGFR upon non-EGF ligand interactions [42]. It is also plausible that overexpressing SNX3 may have enhanced the interaction with EGFR.
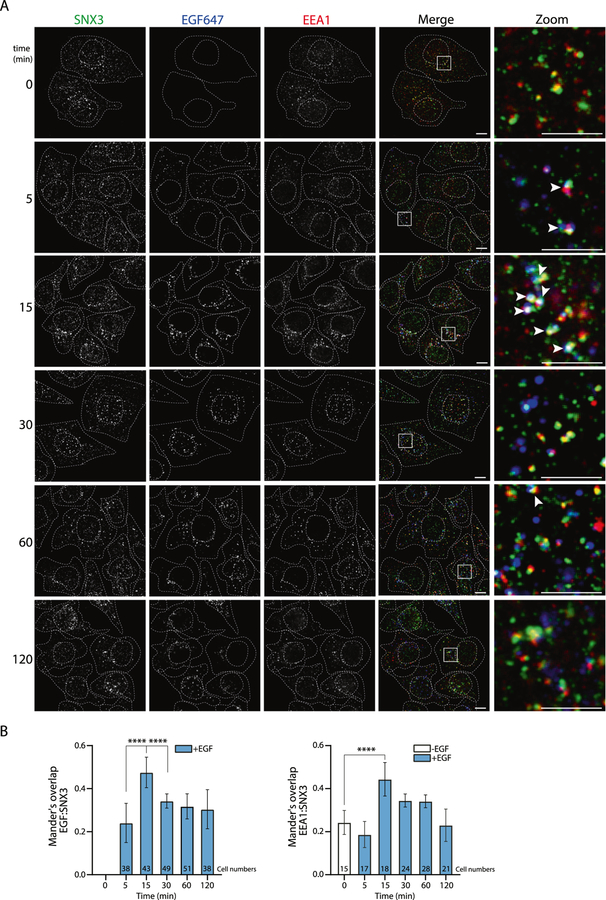
Next, we further examined the dynamic interaction of SNX3 and EGFR with confocal microscopy. We used Alexa Fluor™ labeled-EGF (EGF647) to follow endocytosis of EGF-induced EGFR. SNX3 colocalization with early endosomes (EEA1) and endocytosed EGF was highest at 15 min of EGF stimulation, consistent with the biotinylation results (Fig. 4A, B).
Fig. 4. Time-dependent interactions of SNX3.
A Representative images of confocal microscopy show SNX3, EEA1, and EGF in HeLa cells. Boxed regions in the merge images indicate zoomed areas. The colocalization of SNX3 (green), EGF (blue), and EEA1 (red) are highlighted with white arrows in the zoom images. Dashed white lines indicate cell and nuclear boundaries. Scale bars are 10 μm. B Graphs of colocalization analyses were plotted using Mander’s overlap values, which were quantified from multi-cell images. Cell numbers are shown in the bar graphs. Data are from 3 independent experiments. Statistical significance was calculated using one-way ANOVA with Tukey’s multiple comparison test. ****p < 0.0001.
Effects of SNX3 silencing on EGFR levels in TNBC cells
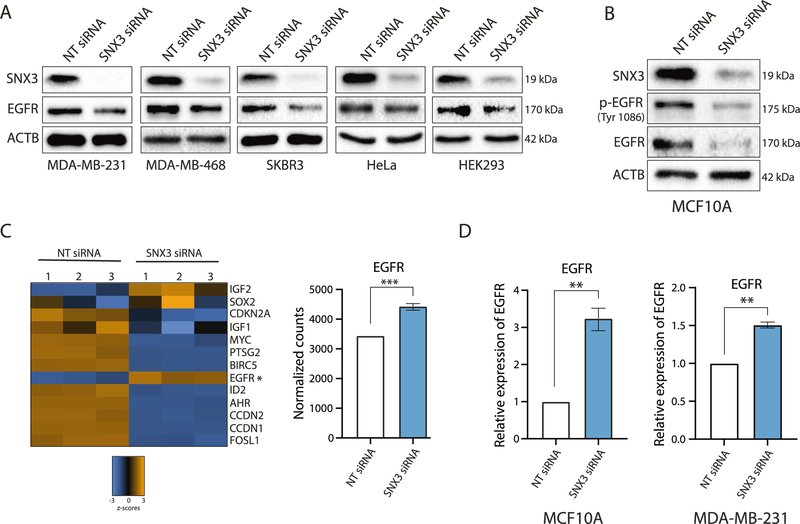
Next, we wanted to ascertain further the functional relationship between SNX3 and EGFR in TNBCs. For this, we examined whether SNX3 silencing would affect EGFR protein levels. We tested MDA-MB-231, MDA-MB-468 TNBC cell lines [43], and additional cell lines that are EGFR positive (SK-Br-3, HeLa, and HEK293). All of the cell lines transfected with a siRNA pool targeting SNX3 had decreased levels of EGFR protein compared to non-targeting siRNA transfected controls within 48–72 h of silencing (Fig. 5A). SNX3-siRNA transfected MCF10A cells also had decreased total EGFR levels and decreased p-EGFR (Fig. 5B).
Fig. 5. Effects of transient SNX3 silencing.
A Western blotting analysis for SNX3 and EGFR on lysates from indicated cell lines treated with SNX3 siRNA pool or non-targeting siRNAs (NT) for 48–72 h. Blots are representative of at least two independent experiments. B Western blotting analysis for SNX3, p-EGFR (Tyr1086), and EGFR on lysates from MCF10A cells transfected with NT siRNA and SNX3 siRNA for 72 h. Blots are representative of 2 independent experiments. C Transcription level changes in NT siRNA and SNX3 siRNA transfected MCF10A cells using the NanoString Vantage 3D RNA cartridge. Blue and orange colors on the heat map represent z scores. The asterisk shows EGFR mRNA. The graph shows normalized mRNA counts for EGFR. Unpaired t-test was performed for statistical significance, ***p < 0.001. D RT-qPCR for the expression of EGFR relative to RPLP0 in MCF10A and MDA-MB-231 cells transfected with SNX3 siRNA compared to NT siRNA transfected cells. Unpaired t-test was performed for statistical significance, **p < 0.01.
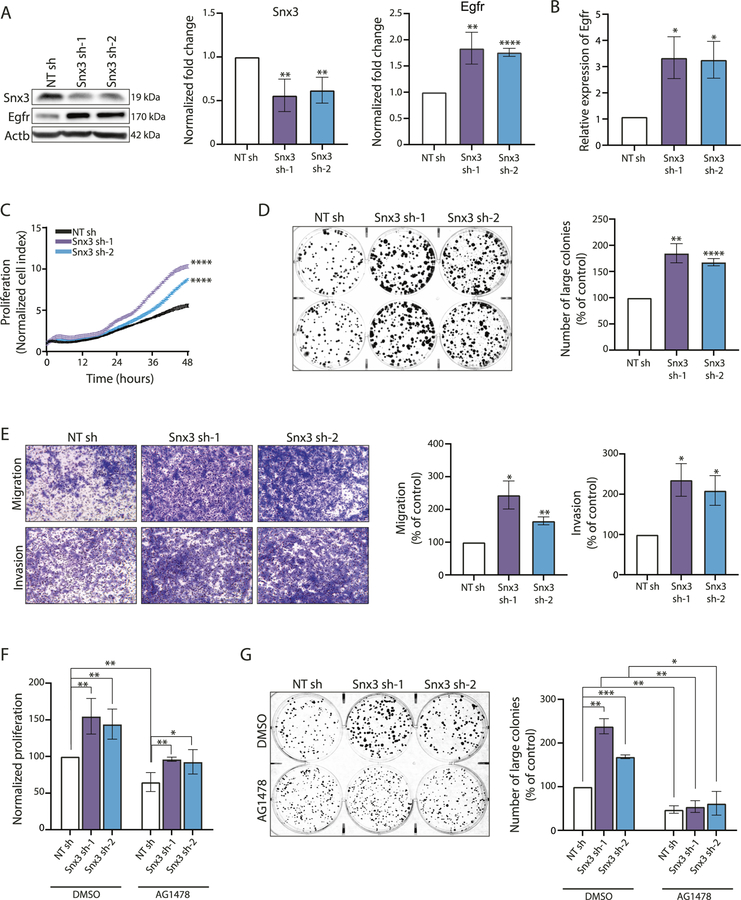
Moreover, a targeted transcriptomic analysis in SNX3-siRNA transfected MCF10A cells using NanoString technology showed decreased expression of cell cycle and survival-related genes including MYC, CCDN1, CCDN2, and BIRC5 (Survivin) (Fig. 5C). Downregulation of these genes was consistent with the downregulation of EGFR in SNX3 silenced cells. However, while transient SNX3 silencing caused decreased EGFR availability and activity, to our surprise, EGFR mRNA was upregulated in SNX3-targeted siRNA transfected cells (Fig. 5C). RT-qPCR also confirmed upregulation of the EGFR mRNA in SNX3 silenced MCF10A and MDA-MB-231 cells (Fig. 5D). Upregulation of EGFR mRNA suggested a compensatory response to rescue downregulated EGFR levels upon SNX3 silencing. To test this in another cancer cell line and investigate this further in vivo, we first generated shRNA silenced SNX3 (Snx3sh) models in the 4T1 cell line, a well-established syngeneic model for TNBCs, mimicking tumor growth and metastatic spread of human breast cancers [44]. We observed upregulated levels of EGFR protein (Fig. 6A) upon long-term silencing of SNX3. EGFR mRNA was also upregulated in these Snx3sh cells (Fig. 6B).
Fig. 6. Effects of long-term silencing of SNX3.
A 4T1 cells were stably transfected with shRNA vectors (NTsh and Snx3sh). Immuno-blotting shows sustained silencing of SNX3 and overexpression of EGFR in two monoclonal populations (Snx3sh-1 and Snx3sh-2) compared with 4T1 control (NTsh) cells. Bars show densitometry analysis of 3 independent experiments (**p < 0.001, ****p < 0.0001, unpaired t-test). B Egfr mRNA levels in NTsh and Snx3sh cells, detected by RT-qPCR. (*p < 0.05, unpaired t-test, n = 3). C Real-time proliferation analysis of Snx3sh 4T1 cells with xCELLigence RTCA. NTsh and Snx3sh-1 and Snx3sh-2 cells were monitored for 48 h with three technical replicates (****p < 0.0001, two-way ANOVA). D Colony formation of NTsh and Snx3sh cells in seven days of seeding. Large colonies (≥1000 pixels) were counted using CountPHICS software (n = 3 independent experiments, *p < 0.05, **p < 0.001, ***p < 0.0001, unpaired t-test). E Transwell migration and invasion of Snx3sh cells. Cells were allowed to migrate for 8 h. For invasion assay, cells were allowed to invade Matrigel-coated chambers for 12 h. Pictures were taken at ×4 magnification. Three independent experiments were analyzed with unpaired t-test (*p < 0.05, **p < 0.001). F Effect of EGFR blockage on the proliferation of Snx3sh 4T1 cells by AG1478 treatment for 72 h, detected by MTT (n = 3 independent experiments, *p < 0.05, **p < 0.001, unpaired t-test). G Colony-forming potential upon AG1478 treatment. NTsh and Snx3sh cells were treated with AG1478 or DMSO for seven days. Bars show the number of large colonies (≥1000 pixels) counted using the CountPHICS software (n = 3 independent experiments, unpaired t-test, *p < 0.05, **p < 0.001, ***p < 0.0001).
Long-term silencing of SNX3 enhances neoplastic phenotypes in vitro and promotes tumor progression and metastasis in vivo
Next, we wanted to determine whether cellular phenotypes were altered in Snx3sh 4T1 cells due to increased EGFR protein levels. SNX3sh cells had increased cell proliferation, measured by xCELLigence live proliferation system (Fig. 6C), increased number of large colonies (Fig. 6D), increased migration, and invasion detected by transwell assays compared to control cells (Fig. 6E). Because SNX3 may recycle other receptors, we tested whether EGFR overexpression was responsible for these phenotypes. Inhibition of EGFR kinase activity with AG1478 treatment in Snx3sh 4T1 cells reversed increased proliferative rate and colony-forming potential, confirming the role of EGFR activity in these in vitro phenotypes (Fig. 6F, G).
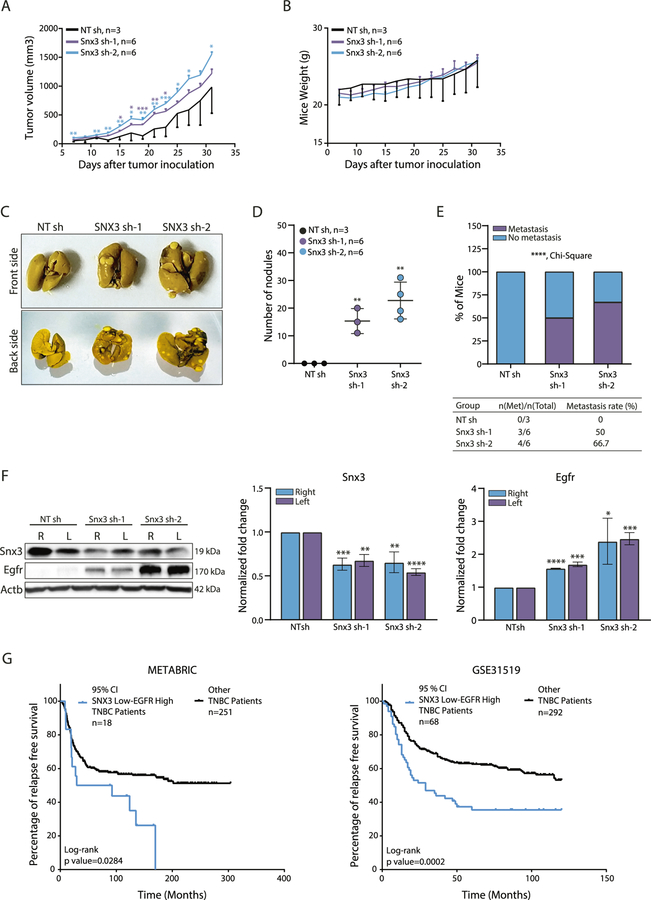
When we moved to an orthotopic mouse model and injected Snx3sh-4T1 cells subcutaneously near mammary fat pads (MFP) of BALB/c mice without surgery, tumors were larger in Snx3sh 4T1 injected mice compared with control animals. There were no significant changes to body weight (Fig. 7A, B). Although we did subcutaneous injection near the MFP (instead of injection to MFPs with surgery), there was gross metastasis in the lungs of mice bearing Snx3sh tumors compared with controls (p < 0.0001) (Fig. 7C–E). Later, tumors isolated from animals were lysed and examined for Snx3 and Egfr status. Snx3 silencing was maintained in the primary tumors, and Egfr protein was significantly overexpressed (Fig. 7F).
Fig. 7. Effect of Snx3 loss on tumor progression and metastasis in vivo.
A Changes in tumor volume of 4T1 syngeneic model upon Snx3 knockdown. Significance was calculated using multiple t-test for each data point for each day compared with control. B Mice body weight (g) of 4T1 syngeneic tumors. C Representative images of lungs collected from each group. Mice were sacrificed when one of the groups reached 1500 mm3. D The frequency of lung metastasis. Significance was determined with Chi-square analysis (****p < 0.0001). E Number of nodules on the lung surface of each group (*p < 0.05, **p < 0.01; ***p < 0.001, Student’s t test). F Primary tumors were collected, and lysates were used for Snx3 and Egfr protein expression analysis. Actb antibody was used as the loading control. Bars show the densitometry analysis of three independent experiments. Expression values of Snx3sh tumor samples were normalized to the average of NTsh tumor samples (*p < 0.05, **p < 0.001, ***p < 0.0001, unpaired t-test) (R: right side, L: left side). G Expression of SNX3 (ILMN_1740180) and EGFR (ILMN_1696521) mRNAs were determined using ±95% (Z = 1.96) confidence interval in the METABRIC dataset. Relapse-free survival (%) of low SNX3 and high EGFR expressing TNBC breast cancer patients (n = 18) was compared to other TNBC patients (n = 251). For the GSE31519 dataset, expression values of SNX3 (2160648_x_at) and EGFR (201983_s_at) mRNAs were determined using ±95% (Z = 1.96) confidence interval. Relapse-free survival of low SNX3 and high EGFR expressing TNBC patients (n = 68) was compared to other TNBC patients (n = 292).
Of note, SNX3sh-2 tumors with higher EGFR expression had higher numbers of metastatic nodules.
These results showed that 4T1 cells became further aggressive due to overexpression of EGFR, inflicted by SNX3 loss. To investigate the clinical relevance of these observations, we analyzed mRNA expressions of SNX3 and EGFR in TNBC patients in the METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) dataset [45]. We found that low SNX3 and high EGFR mRNA levels correlated with poor relapse-free survival in patients (Fig. 7G). Similarly, low SNX3 and high EGFR mRNA levels correlated with poor relapse-free survival in TNBC patients in an independent dataset (GSE31519) (Fig. 7G, right panel).
Collectively, our results showed SNX3 to be a critical player in the EGFR network as an EGF responsive target and a regulator of EGFR protein levels. Based on these results, we speculate that SNX3 loss may be linked to EGFR overexpression in not only TNBCs but possibly other cancers where gene-level amplification and/or mutations do not exist.
DISCUSSION
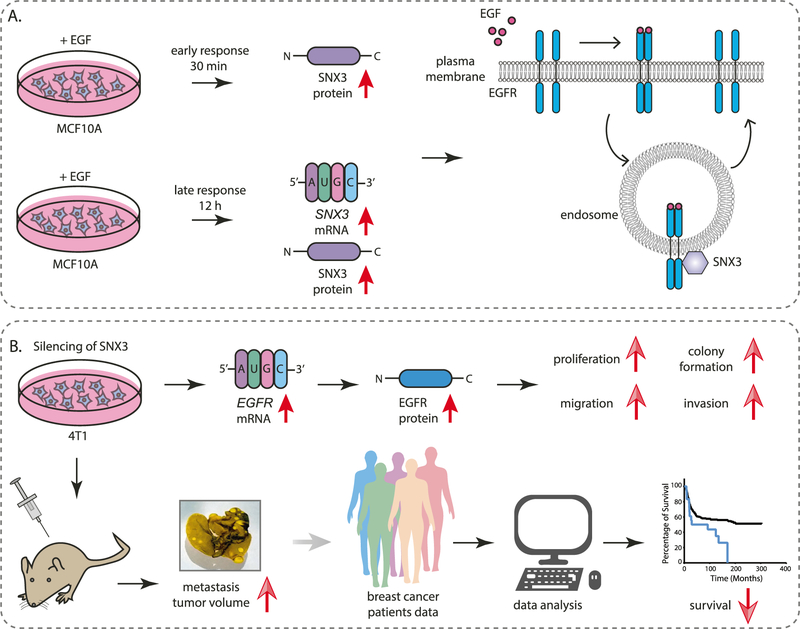
We report multiple lines of evidence suggesting that the EGF-SNX3-EGFR axis drives tumor progression and metastasis in triple-negative breast cancers (Fig. 8).
Fig. 8. Schematic diagram showing the EGF-SNX3-EGFR axis.
A SNX3 protein levels are upregulated during the early response to EGF stimulation. Prolonged exposure to EGF causes transcriptional upregulation. In turn, SNX3, as a retromer protein, interacts with EGFR and endocytosed EGF in early endosomes. B Sustained silencing of SNX3 causes EGFR mRNA and protein upregulation, causing increased neoplastic phenotypes in vitro and in vivo. In support of these, TNBC patients with low SNX3 and high EGFR mRNA have poor overall survival.
First, we identified SNX3 as an immediate and sustained target of EGF stimulation. In EGFR positive mammary cells, SNX3 protein levels were upregulated quickly after EGF stimulation. The rapid upregulation of SNX3 protein was possibly due to the delayed turnover of SNX3 in lysosomes. SNX3 protein levels were high throughout EGF stimulation, with the contribution of transcriptional upregulation only at much later time points (i.e., 12 h). These results, showing multiple mechanisms to regulate SNX3 mRNA/protein abundance in response to EGF, highlight the role of SNX3 in EGFR biology.
Next, to provide insight into SNX3 function, we used a proximity-based labeling approach in live cells. We showed close proximity between SNX3 and EGFR, mainly when ligand-activated and endocytosed EGFR is recycled most [41]. These observations were consistent with increased colocalization of SNX3, early endosomes, and endocytosed EGF at 15 min of EGF stimulation.
After establishing SNX3 upregulation upon EGF stimulation and confirming its association with EGFR, we investigated the role of SNX3 in TNBCs. To this end, we performed short and long-term silencing of SNX3 and conducted a targeted gene expression analysis. This approach allowed us to capture dynamic changes of the EGFR mRNA and protein levels. As expected, short-term silencing of SNX3 in a panel of breast and non-breast cancer cell lines showed decreased EGFR protein levels. However, transcriptome level analysis showed increased levels of EGFR mRNA in SNX3 silenced cells, suggesting activation of a compensatory mechanism to restore EGFR protein levels. Indeed, when we investigated the long-term effects of SNX3 silencing, we observed significant upregulation of EGFR protein in TNBC cells. The same gene expression pattern (low SNX3, high EGFR) correlates with the shorter survival of TNBC patients. In agreement with the clinical data, in vivo studies showed increased tumor growth and metastasis upon SNX3 loss and EGFR overexpression. Importantly, oncogenic phenotypes in vitro were reversible by pharmacological inhibition of EGFR.
Overexpression of EGFR and its association with poor prognosis is known in many cancer types, including breast cancer [46–48]. However, EGFR gene amplifications and/or mutations can only partly explain cases of EGFR overexpression. Other mechanisms have been proposed to explain the upregulation of EGFR. For example, increased translation or decreased protein degradation leads to EGFR overexpression in the hypoxic microenvironment [49, 50]. Hypoxia also induces Early Growth Response Factor 1 (EGR1) and activates the basal transcriptional activity of the EGFR promoter [51]. In addition, changes in the local epigenetic landscape of the EGFR promoter modulate the accessibility of other transcription factors in different cellular contexts [52]. Despite these findings, mechanisms of EGFR overexpression and activation in cancers are not entirely understood. Our results suggest that deregulated retromer function may also drive EGFR overexpression. The molecular network behind compensatory upregulation of EGFR remains to be investigated.
It is also worth noting that SNX3 plays a role in recycling other receptors (e.g., transferrin receptor, Wls, and possibly other members of the ErbB family). We observed increased transferrin receptor levels in SNX3 silenced 4T1 cells but not in primary tumors (Supplementary Fig. S9). This result suggests that deregulated recycling activates compensatory mechanisms to repopulate the receptor levels. However, these mechanisms are possibly dynamic and context-dependent. As in the case of the transferrin receptor, protein levels were back to normal in SNX3 silenced tumors, whereas EGFR was still overexpressed. The differential upregulation of transferrin receptor and EGFR in tumors could depend on the biological context and the inherent complexity of signaling networks in TNBC cells. Hence, it is plausible that deregulated recycling causes diverse phenotypic outcomes in a cell context-dependent manner. This view may also clarify why SNX3 is linked to various diseases, including cancer, cardiac hypertrophy [53], and Alzheimer’s disease [54].
In closing, our results provide insight into SNX3 gene/protein regulation, with important implications for the overexpression of EGFR protein in a group of breast and possibly other cancers. This observation may partly explain EGFR overexpression cases with no genomic amplification or mutations. Thus, the EGF-SNX3-EGFR axis presented here may have future implications for developing potent therapies targeting the EGFR network and possibly understanding resistance mechanisms to anti-EGFR therapies.
METHODS
Cell lines and treatments
MCF10A, MDA-MB-231, HEK293, HeLa cells were grown as described [55]. 4T1 cells were grown in DMEM high glucose medium with 10% FBS (Biowest, France, 181T-500), 1% P/S, and 1% nonessential amino acids (Biological Industries, Israel, 03–031-1B). All cell lines were grown as monolayers and were incubated at 37 °C with 95% humidified air and 5% CO2. Cells were routinely monitored for mycoplasma contamination. 4T1 cells expressing stable shRNAs in pSUPER vector were maintained in G418 (175 μg/ml, Roche, Germany, 04727894001) containing medium. For EGF (Thermo Fisher Scientific, MA, USA, PHG0313) treatments, cells were serum-starved and then stimulated with EGF for the indicated time points. Cycloheximide, actinomycin D, and AG1478 were purchased from Tocris, UK (0970, 1299, 1276). Akt inhibitor MK2206 (Selleckchem, USA, 1078) and MEK inhibitor PD0325901 (Selleckchem, USA, 1036) were gifts from Dr. Onur Cizmecioglu (Bilkent University). E2 (β-Estradiol) was purchased from Sigma-Aldrich (E2257).
Plasmids and siRNAs
The EGFR-GFP vector was purchased from Addgene (Plasmid 32751) [56]. The coding sequence of SNX3 was cloned into 2X HA-TurboID (pEGFP-C1) vector (a gift from Dr. S. Neefjes, Leiden University) and sequenced. Mouse SNX3 short-hairpin RNA (shRNA) (5’-AACTTCCTCGAGATCGACG-3’) and non-targeting (NT) shRNA (5’-CGTACGCGGAATACTTCGATT-3’) were cloned into pSUPER retro.neo-GFP (a gift from Dr. U. Tazebay, Gebze Teknik University) and sequenced. SNX3 siRNA pool (Dharmacon, USA, SMARTpool siGENOME M-011521–01,) and NT siRNAs (Qiagen, Germany, 1027310) were used for transient silencing of SNX3. DharmaFect (Dharmacon, T-2004) was used to transfect siRNAs.
Proximity-based labeling assay
Cells were transfected with either pEGFP_2XHA_TurboID or pEGFP_2X-HA_TurboID_SNX3. After 24 h, cells were starved with serum-free media for 4 h. 5 ng/ml EGF was given to cells for the indicated time points. 50 μM biotin (Sigma-Aldrich, MO, USA, B4639) was given to cells for 5 min during the last 5 min of EGF treatment. Cell lysis and IP with high capacity neutravidin beads (Thermo Scientific, 29202) were performed as described [38]. Lysates were loaded to 4–12% SDS-PAGE for western blotting.
Confocal microscopy
4× 105 HeLa cells were seeded onto 24 well plates with glass coverslips (Menzel Gläser, Germany MENZCB00130RAC). Cells were kept in serum-free media for 4 h and induced with 100 ng/ml Alexa Fluor™ 647-tagged EGF for indicated times. Cells were stained as described [57]. For permeabilization and blocking, 0.1% Triton X-100 (Sigma-Aldrich, T8787) and 1% BSA (ChemCruz, TX, USA, sc-2323A) in PBS were added to cells and incubated for 30 min. 3 μg/ml primary antibodies in blocking buffer was given to cells for 1 h. Alexa Fluor™ tagged secondary antibodies were used (1:300). Following PBS washes, 4% paraformaldehyde was used for 30 min for post fixation. Cells were then mounted using ProLong Gold antifade Mounting medium with DAPI (Life Technologies, CA, USA, P36941). Imaging was performed using appropriate solid-state lasers on Leica SP8 microscope. HyD detectors and HCX PL 63X magnification oil emersion objectives were used. Images were taken with 2.5 digital zoom in 1024×1024 scanning format with line averaging. Mander’s overlap was used to indicate colocalization.
Expression analysis
RNA isolation, quantification, cDNA synthesis and expression analysis were conducted according to MIQE Guidelines [58] and as described previously [59]. Ssoadvanced Universal SYBR Green Supermix (BioRad, CA, USA, 1725271) and CFX Connect Real-Time PCR Detection System (BioRad, 1855201) were used for RT-qPCR. Fold changes were normalized against RPLP0 reference gene. For relative quantification, the reaction efficiency incorporated ΔΔCt formula was used [60].
Western blotting
M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific, 78501) with protease and phosphatase inhibitor cocktails (Roche, 05056489001, 4906845001) was used. Lysate concentrations were measured using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23227), and western blotting was performed as described [55]. WesternBright ECL (Advansta, USA, K12045-D50) and Chemidoc MP Imaging System (BioRad) were used for visualization.
Antibodies
SNX3 antibody (rabbit, 10772–1-AP) was from Proteintech (USA). P-EGFR (rabbit, 2220) and p-AKT (rabbit, 4060) antibodies were from Cell Signaling Technology (USA). EGFR (mouse, sc-373746), AKT (rabbit, sc-8312), p-ERK 1/2 (rabbit, sc-16982), ERK 1/2 (mouse, sc-514312), CTNNB1 (mouse, sc-133240), GFP (mouse, sc-9996), and HA (mouse, MMS-101R) antibodies were from Covance (US). EGFR (rabbit, 06–847) and phosphotyrosine (pY, 4G10, mouse, 05–321) antibodies were from Millipore (Germany). Anti-Transferrin receptor (TfR) was from Thermo Fisher Scientific (13–6890). ACTB (mouse, sc-47778, Santa Cruz Biotechnology) and ACTB (mouse, A5441, Sigma-Aldrich) antibodies were used. HRP-conjugated secondary antibodies (mouse, R-05071; rabbit, R-05072) were purchased from Advansta. Fluorescent secondary antibodies (IRDye 800CW goat anti-rabbit IgG (H + L) 926–32211, IRDye 800CW goat anti-mouse IgG (H + L) 926–32210, IRDye 680LT goat anti-rabbit IgG (H + L) 926–68021, IRDye 680LT goat anti-mouse IgG (H + L) 926–68020 were purchased from Li-COR (USA). The following antibodies were used in confocal microscopy: SNX3 (Abcam, UK, ab56078), EEA1 (BD Transduction Laboratories, US 610457), Alexa 568 (Thermo Fisher Scientific, A10037), and Alexa 488 (Thermo Fisher Scientific, A21206).
NanoString
Three independent siRNA transfections to MCF10A were performed, RNAs were pooled and loaded in three replicates. 100 ng of total RNA was loaded for each sample into the cartridge nCounter® Vantage 3D™ RNA Panel (VRXC-Wnt1–12). nSolver and advanced analysis softwares of NanoString Technologies were used to assess the quality of the run and process data. Two normalization analyses were done as recommended by the manufacturer to generate normalized counts. The first normalization included six internal positive controls whose geometric means were calculated, and a normalization factor was used to equilibrate counts. The same process was applied to each gene in the second normalization, using the geometric means of housekeeping genes (CC2D1B, COG7, EDC3, GPATCH3, HDAC3, MTMR14 NUBP1, PRPF38A, SAP130, SF3A3, TLK2, ZC3H14).
Proliferation assays
pSUPER-NTshRNA and pSUPER-Snx3shRNA stably transfected 4T1 cells were seeded at a density of 2 × 104 cells/well on E-plates placed to the RTCA plate station (xCELLigence Real-Time Cell Analyzer, ACEA Biosciences). Impedance was monitored live for 48 h with 15-min intervals. For MTT, 2 × 103 cells were plated in 96-well flat-bottom plates and were treated with AG1478 (15 μM) or vehicle control. MTT was performed as described [55].
Colony formation assay
Stably transfected 4T1 cells (500 cells/well) were grown in 6-well plates for seven days. Following methanol fixation, colonies were stained with 0.5% crystal violet as described [61]. Cells were treated with AG1478 (IC50 = 15 μM) or vehicle control DMSO for the same duration. Colonies (≥1000 pixels) were determined and counted using CountPHICS software [62].
Transwell migration and invasion assays
5× 104 cells in 100 μl of 1%FBS growth medium were seeded onto upper chamber of transwell inserts (Greiner, The Netherlands). 10%FBS-medium was added to bottom chambers. Cells were allowed to migrate for 8 h at 37 °C. For invasion assay, transwell chambers were coated with 100 μl Matrigel (300 μg/ml, Corning, NY, USA), and 8 × 104 cells were seeded per well for 12 h. Staining and counting of cells were performed as previously described [63].
In vivo tumor growth and metastasis experiments
Animal experiments were conducted following institutional ethical guidelines and regulations for animal experimentations of Bilkent University. Mice were housed in a temperature-controlled room with 12-h light/dark cycles and received a standard diet with water ad libitum. Six to eight-week-old female Balb/c mice were injected with 1 × 106 4T1 NTsh and two different Snx3sh monoclonal (Snx3sh-1, Snx3sh-2) cells into left and right mammary fat pads (MFP) subcutaneously. Cells were prepared in 1:1 PBS: Matrigel (Corning). Primary tumor growth was monitored by measuring tumor volume every other day with a caliper. Tumor volume was calculated using the formula (length × width2)/2. All mice were sacrificed when one of the groups reached the threshold of 1500 mm3 volume. Tumors were collected for subsequent analysis. Lung metastasis was evaluated with Bouin’s fixation [64]. Number of tumor nodules on lung surface was counted under a dissecting microscope, and metastasis rate was determined. One mouse from each injection group was sacrificed, and tumor samples were collected for western blot analysis.
Patient data analysis
The normalized mRNA expression data for SNX3 (ILMN_1740180) and EGFR (ILMN_1696521) were downloaded from METABRIC [39] (European Genome-Phenome Archive with EGAS00000000083), and the clinical data of the study were downloaded from cBioPortal [65, 66]. TNBC patients were grouped based on negativity of ER, HER2, and PR expression (n = 318). For disease-specific relapse-free survival (RFS) analyses, the patients who died of other causes were excluded from the analysis. The relapse-free survival was determined by comparing the low SNX3, high EGFR expressing tumors with other TNBC patients. Grouping was done by filtering the outlier normalized expression values of SNX3 and EGFR according to ±95% (Z = 1.96) confidence interval. Patients in the GSE31519 dataset were also grouped based on low SNX3 and high EGFR mRNA levels.
Supplementary Material
ACKNOWLEDGEMENTS
We thank current and past members of the laboratory for critical reading of the manuscript and discussions. We thank METABRIC Consortium for sharing the breast cancer data. TUBITAK 116Z257 (AEEB), and 119Z672 (EC). EMBO short term fellowship (MO). National Institutes of Health Grant 2P20GM109091-06 (OS), 1R01CA251374-01A1 (OS). The Turkish Ministry of Development project #KanSil 2016K121540 (CanSyL).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
ADDITIONAL INFORMATION
Supplementary information The online version contains supplementary material available at https..org/10.1038/s41388-021-02086-9.
Reprints and permission information is available at http://www.nature.com/reprints
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
REFERENCES
- 1.Wieduwilt MJ, Moasser MM. The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell Mol Life Sci. 2008;65:1566–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richard J, Sainsbury C, Needham GK, Farndon JR, Malcolm AJ, Harris AL. Epidermal-growth-factor receptor status as predictor of early recurrence of and death from breast cancer. Lancet. 1987;329:1398–402. [DOI] [PubMed] [Google Scholar]
- 3.Dickler MN, Cobleigh MA, Miller KD, Klein PM, Winer EP. Efficacy and safety of erlotinib in patients with locally advanced or metastatic breast cancer. Breast Cancer Res Treat. 2009;115:115–21. [DOI] [PubMed] [Google Scholar]
- 4.Rimawi MF, Shetty PB, Weiss HL, Schiff R, Osborne CK, Chamness GC, et al. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer. 2010;116:1234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ, Kim YJ, et al. High EGFR gene copy number predicts poor outcome in triple-negative breast cancer. Mod Pathol. 2014;27:1212–22. [DOI] [PubMed] [Google Scholar]
- 6.Ali R, Wendt MK. The paradoxical functions of EGFR during breast cancer progression. Signal Transduct Target Ther. 2017;2:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seaman MNJ, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with Ptdlns(3)P. Nat Cell Biol. 2001;3:658–66. [DOI] [PubMed] [Google Scholar]
- 9.Arighi CN, Harmell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seaman MNJ. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temkin P, Lauffer B, Jäger S, Cimermancic P, Krogan NJ, von Zastrow M. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol. 2011;13:715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Weering JRT, Sessions RB, Traer CJ, Kloer DP, Bhatia VK, Stamou D, et al. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 2012;31:4466–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiow KH, Tan Y, Chua RY, Huang D, Ng MLM, Torta F, et al. SNX3-dependent regulation of epidermal growth factor receptor (EGFR) trafficking and degradation by aspirin in epidermoid carcinoma (A-431) cells. Cell Mol Life Sci. 2012;69:1505–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucas M, Gershlick DC, Vidaurrazaga A, Rojas AL, Bonifacino JS, Hierro A. Structural mechanism for cargo recognition by the retromer complex. Cell. 2016;167:1623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinberg F, Gallon M, Winfield M, Thomas EC, Bell AJ, Heesom KJ, et al. A global analysis of SNX27 – retromer assembly and cargo specificity reveals a function in glucose and metal ion transport relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol. 2013;15:821–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burd C, Cullen PJ. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6:a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakker J, Spits M, Neefjes J, Berlin I. The EGFR odyssey - from activation to destruction in space and time. J Cell Sci. 2017;130:4087–96. [DOI] [PubMed] [Google Scholar]
- 19.Kurten RC, Cadena DL, Gill GN. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272:1008–10. [DOI] [PubMed] [Google Scholar]
- 20.Choi JH, Hong WP, Kim MJ, Kim JH, Ryu SH, Suh PG. Sorting nexin 16 regulates EGF receptor trafficking by phosphatidylinositol-3-phosphate interaction with the Phox domain. J Cell Sci. 2004;117:4209–18. [DOI] [PubMed] [Google Scholar]
- 21.Gullapalli A, Garrett TA, Paing MM, Griffin CT, Yang Y, Trejo JA. A role for sorting nexin 2 in epidermal growth factor receptor down-regulation: evidence for distinct functions of sorting nexin 1 and 2 in protein trafficking. Mol Biol Cell. 2004;15:2143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Liu ZQ, Chen CXQ, Magill S, Jiang Y, Liu YJ. Inhibitory regulation of EGF receptor degradation by sorting nexin 5. Biochem Biophys Res Commun. 2006;342:537–46. [DOI] [PubMed] [Google Scholar]
- 23.Cavet ME, Pang J, Yin G, Berk BC. An epidermal growth factor (EGF) -dependent interaction between GIT1 and sorting nexin 6 promotes degradation of the EGF receptor. FASEB J. 2008;22:3607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Priya A, Sugatha J, Parveen S, Lacas-Gervais S, Raj P, Gilleron J, et al. Essential and selective role of SNX12 in transport of endocytic and retrograde cargo. J Cell Sci. 2017;130:2707–21. [DOI] [PubMed] [Google Scholar]
- 25.Pons V, Luyet PP, Morel E, Abrami L, van der Goot FG, Parton RG, et al. Hrs and SNX3 functions in sorting and membrane invagination within multivesicular bodies. PLoS Biol. 2008;6:1942–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucas M, Gershlick DC, Vidaurrazaga A, Rojas AL, Bonifacino JS, Hierro A. Structural mechanism for cargo recognition by the retromer complex. Cell. 2016;167:1623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P, Wu Y, Belenkaya TY, Lin X. SNX3 controls Wingless/Wnt secretion through regulating retromer-dependent recycling of Wntless. Cell Res. 2011;21:1677–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harterink M, Port F, Lorenowicz MJ, McGough IJ, Silhankova M, Betist MC, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13:914–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan B, Zhang T, Yang W, Liu Y, Chen Y, Zhou Z, et al. SNX3 suppresses the migration and invasion of colorectal cancer cells by reversing epithelial-to-mesenchymal transition via the β-catenin pathway. Oncol Lett. 2019;18:5332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503–12. [DOI] [PubMed] [Google Scholar]
- 31.Zeisel A, Köstler WJ, Molotski N, Tsai JM, Krauthgamer R, Jacob-Hirsch J, et al. Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli. Mol Syst Biol. 2011;7:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golan-Lavi R, Giacomelli C, Fuks G, Zeisel A, Sonntag J, Sinha S, et al. Coordinated pulses of mRNA and of protein translation or degradation produce EGF-induced protein bursts. Cell Rep. 2017;18:3129–42. [DOI] [PubMed] [Google Scholar]
- 33.Hwang SG, Yu SS, Ryu JH, Jeon HB, Yoo YJ, Eom SH, et al. Regulation of β-catenin signaling and maintenance of chondrocyte differentiation by ubiquitin-independent proteasomal degradation of α-catenin. J Biol Chem. 2005;280:12758–65. [DOI] [PubMed] [Google Scholar]
- 34.Levitzki A, Gazit A. Tyrosine kinase inhibition: an approach to drug development. Science. 1995;67:1782–8. [DOI] [PubMed] [Google Scholar]
- 35.Hirai H, Sootome H, Nakatsuru Y, Miyama K, Taguchi S, Tsujioka K, et al. MK-2206, an allosteric akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Therapeutics. 2010;9:1956–67. [DOI] [PubMed] [Google Scholar]
- 36.Ciuffreda L, del Bufalo D, Desideri M, di Sanza C, Stoppacciaro A, Ricciardi MR, et al. Growth-inhibitory and antiangiogenic activity of the MEK inhibitor PD0325901 in malignant melanoma with or without BRAF mutations. Neoplasia. 2009;11:720–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, Montgomery JE, Eckert MA, Chang JW, Tienda SM, Lengyel E, et al. An activity-dependent proximity ligation platform for spatially resolved quantification of active enzymes in single cells. Nat Commun. 2017;8:1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sapmaz A, Berlin I, Bos E, Wijdeven RH, Janssen H, Konietzny R, et al. USP32 regulates late endosomal transport and recycling through deubiquitylation of Rab7. Nat Commun. 2019;10:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horibe T, Torisawa A, Akiyoshi R, Hatta-Ohashi Y, Suzuki H, Kawakami K. Transfection efficiency of normal and cancer cell lines and monitoring of promoter activity by single-cell bioluminescence imaging. Luminescence. 2014;29:96–100. [DOI] [PubMed] [Google Scholar]
- 40.McGough IJ, de Groot REA, Jellett AP, Betist MC, Varandas KC, Danson CM, et al. SNX3-retromer requires an evolutionary conserved MON2:DOPEY2:ATP9A complex to mediate Wntless sorting and Wnt secretion. Nat Commun. 2018;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bag N, Huang S, Wohland T. Plasma membrane organization of epidermal growth factor receptor in resting and ligand-bound states. Biophysical J. 2015;109:1925–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh B, Carpenter G, Coffey RJ. EGF receptor ligands: recent advances. F1000Res. 2016;5:F1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kathryn JC, Sireesha VG, Stanley L. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2012;32:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pulaski BA, Ostrand-Rosenberg S. Mouse 4T1 breast tumor model. Curr Protoc Immunol. 2000;39:1–16. [DOI] [PubMed] [Google Scholar]
- 45.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lurje G, Lenz HJ. EGFR signaling and drug discovery. Oncology. 2010;77:400–10. [DOI] [PubMed] [Google Scholar]
- 47.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakha EA, El-Sayed ME, Green AR, Lee AHS, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. [DOI] [PubMed] [Google Scholar]
- 49.Franovic A, Gunaratnam L, Smith K, Robert I, Patten D, Lee S. Translational upregulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proc Natl Acad Sci USA. 2007;104:13092–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinde SR, Maddika S. PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat Commun. 2016;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishi H, Nishi KH, Johnson AC. Early growth response-1 gene mediates upregulation of epidermal growth factor receptor expression during hypoxia. Cancer Res. 2002;62:827–34. [PubMed] [Google Scholar]
- 52.Erfani P, Tome-Garcia J, Canoll P, Doetsch F, Tsankova NM. EGFR promoter exhibits dynamic histone modifications and binding of ASH2L and P300 in human germinal matrix and gliomas. Epigenetics. 2015;10:496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu J, Xu S, Huo Y, Sun D, Hu Y, Wang J, et al. Sorting nexin 3 induces heart failure via promoting retromer-dependent nuclear trafficking of STAT3. Cell Death Differ. 2021;28:2871–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vardarajan BN, Bruesegem SY, Harbour ME, st. George-Hyslop P, Seaman MNJ, Farrer LA. Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol Aging. 2012;33:2231–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akman HB, Oyken M, Tuncer T, Can T, Erson-Bensan AE. 3’UTR shortening and EGF signaling: Implications for breast cancer. Hum Mol Genet. 2015;24:6910–20. [DOI] [PubMed] [Google Scholar]
- 56.Carter RE, Sorkin A. Endocytosis of functional epidermal growth factor receptor-green fluorescent protein chimera. J Biol Chem. 1998;273:35000–7. [DOI] [PubMed] [Google Scholar]
- 57.Miller HE, Larson CL, Heinzen RA. Actin polymerization in the endosomal pathway, but not on the Coxiella-containing vacuole, is essential for pathogen growth. PLoS Pathogens. 2018;14:e1007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. [DOI] [PubMed] [Google Scholar]
- 59.Akman BH, Can T, Elif Erson-Bensan A. Estrogen-induced upregulation and 3'-UTR shortening of CDC6. Nucleic Acids Res. 2012;40:10679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Asp Med. 2006;27:126–39. [DOI] [PubMed] [Google Scholar]
- 61.Crowley LC, Christensen ME, Waterhouse NJ. Measuring survival of adherent cells with the Colony-forming assay. Cold Spring Harbor Protocols. 2016;2016:721–4. [DOI] [PubMed] [Google Scholar]
- 62.Brzozowska B, Galecki M, Tartas A, Ginter J, Kazmierczak U, Lundholm L. Freeware tool for analysing numbers and sizes of cell colonies. Radiat Environ Biophys. 2019;58:109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akhavantabasi S, Sapmaz A, Tuna S, Erson-Bensan AE. MiR-125b targets ARID3B in breast cancer cells. Cell Struct Funct. 2012;37:27–38. [DOI] [PubMed] [Google Scholar]
- 64.Huang Y, Pan L, Helou K, Xia Q, Parris TZ, Li H, et al. Mechanical ventilation promotes lung metastasis in experimental 4T1 breast cancer lung-metastasized models. Cancer Manag Res. 2018;10:545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.