Abstract
Three new polyketide dimers named huoshanmycins A‒C (1–3) were isolated from a plant endophytic Streptomyces sp. HS-3-L-1 in the leaf of Dendrobium huoshanense, which was collected from the Cultivation base in Jiuxianzun Huoshanshihu Co., Ltd. The dimeric structures of huoshanmycins were composed of unusual polyketides SEK43, SEK15, or UWM4, with a unique methylene linkage. Their structures were elucidated through comprehensive 1D-/2D-NMR and HRESIMS spectroscopic data analysis. The cytotoxicity against MV4-11 human leukemia cell by the Cell Counting Kit-8 (CCK8) method was evaluated using isolated compounds with triptolide as positive control (IC50: 1.1 ± 0.4 μM). Huoshanmycins A and B (1, 2) displayed moderate cytotoxicity with IC50 values of 32.9 ± 7.2 and 33.2 ± 6.1 μM, respectively.
Keywords: huoshanmycin, polyketide, Streptomyces, Dendrobium huoshanense, endophyte
Introduction
Dendrobium huoshanense is a perennial epiphytic Orchidaceae herb with important medicinal and ornamental value. The leaves of D. huoshanense have long been utilized for dermatologic disorders, metabolic syndromes, nervous system disorders, and musculoskeletal system disorders (Wang, 2021). Modern pharmacological research has revealed that D. huoshanense has anti-inflammatory (Ge et al., 2018; Li et al., 2020), cytotoxic (Chen et al., 2022), hypoglycemic (Wang et al., 2019), anti-atherosclerosis (Fan et al., 2020), and antioxidant (Tian et al., 2013) activity. Dendrobium plant is well-known for its rich and diverse endophytic bacterial and fungal community (Chen et al., 2019; Chen et al., 2020). Previous studies have revealed the close relationship between Dendrobium and its endophytes, such as improving the seed germination rate (Tsavkelova et al., 2007) and supply of nutrients (Li et al., 2017). At present, research on D. huoshanense endophytes mainly focuses on the diversity and functions, while not much is known about their secondary metabolites, especially for bacteria. The main species of bacterial microorganisms of D. huoshanense are Sphingomonas, Acinetobacter, Enterococcus, Bacillus, and Methylobacterium (Chen et al., 2020). Streptomyces is the largest genus of Actinobacteria and characterized by producing complex secondary metabolites. They produce over two-thirds of the clinically useful antibiotics of natural origin (e.g., chloramphenicol, streptomycin, tetracycline, erythromycin, ivermectin, and rifamycin) (Raja and Prabakarana, 2011). The last four compounds all belong to polyketides, which are derived from a precursor molecule consisting of a chain of alternating ketone (or reduced forms of a ketone) and methylene groups.
Since the discovery of taxol and taxane produced by an endophytic fungus from the phloem (inner bark) of Pacific yew in 1993 (Stierle et al., 1993), endophytes have become an important resource in the field of bioactive natural products discovery (Newman and Cragg, 2015; Gómez and Luiz, 2018), as they can produce analogs or bioactivity-related compounds as their hosts did (Cui et al., 2012; Zhao et al., 2020c). As part of an effort to characterize novel natural products from medicinal plants (Wang et al., 2009; Ding et al., 2021; Hu et al., 2021) and their endophytes (Zhao et al., 2020b; Zhao et al., 2020a; Zhu et al., 2021), herein we report the isolation and characterization of three new polyketide dimers from an endophyte Streptomyces sp. HS-3-L-1 of the D. huoshanense leaf. The dimeric structures of new huoshanmycins A‒C (1–3) were composed of SEK43, SEK15, or UWM4 (Meurer et al., 1997), with a unique methylene linkage. Herein, we report the fermentation, extraction, isolation, structural elucidation, and cytotoxic activity of these secondary metabolites.
Materials and Methods
General Experimental Procedures
UV data were acquired on a Persee TU-1810 spectrophotometer (Persee analytics, Beijing, China). IR spectra were measured on a Thermo Scientific Nicolet iS5 FT-IR spectrometer (Thermo, United States). NMR spectra were obtained on a Bruker Advance AV500 spectrometer (Bruker, Germany). HRESIMS spectra were recorded on an Orbitrap Elite mass spectrometer (Thermo Scientific, United States). Liquid chromatography–mass spectrometry (LC-MS) was conducted with an Agilent 1290 system equipped with 6120 Quadrupole MSD mass spectrometer (Agilent Technologies, United States). HPLC analysis was performed on a Waters 2695 system equipped with 2998 PDA detector. Total component analysis was performed on an Agilent 1290 UHPLC-6520 Q-TOF/MS. Preparative HPLC separation was performed on a Waters 1525 EF LC system (Waters Company, United States). MCI GEL high-porous polymer (75–150 μm) was purchased from Mitsubishi Chemical Corporation (Japan). Sephadex LH-20 resin (25–100 μm) was purchased from GE Healthcare Company (Sweden). XAD16N resin (20–60 mesh) was obtained from Yuanye Company (Nanjing, Jiangsu, China). Chemicals were purchased from Juyou Company or Aldrich and used without further purification unless otherwise noted.
Strain Isolation
Plant samples of D. huoshanense were provided by Jiuxianzun Huoshanshihu Co., Ltd. (Liu-An City, Anhui Province, China) and identified by co-author Dr. Yang Hu. A voucher specimen (no. 20190309) was deposited at Jiangsu Key Laboratory for Functional Substances of Chinese Medicine, China. The roots, leaves, and stems of D. huoshanense were separated and cleaned with water and then rinsed in 0.1% Tween-20 for 30 s, sequentially immersed in 75% ethanol for 5 min and in 2% sodium hypochlorite for 5 min and rinsed with 10% NaHCO3 for 10 min to inhibit fungal growth. After each treatment, samples were rinsed three times in sterile water. The surface sterilized samples were aseptically dissected into small pieces; 0.5 g of each sample was suspended in 1.0 ml of sterile H2O, and heated at 75°C for 1 min to eliminate nonsporulating bacteria (Zhao et al., 2020a). A 100-μl aliquot of supernatant was streaked on oatmeal agar and on ISP4 agar plates supplemented with nalidixic acid (25 μg/ml) and amphotericin B (25 μg/ml). A number of sporulating bacterial colonies were observed after 1–2 months of incubation at 28°C, and each colony was subsequently purified on a M2 agar plate (Wang et al., 2013). Overall, 54 endophytic strains were isolated from plant samples. The endophytic strain HS-3-L-1 was isolated from the leaf of D. huoshanense.
Phylogenetic Analysis
Strain HS-3-L-1 was inoculated in a 20-ml test tube with 4 ml of TSB broth. After 3 days culture at 28°C with 160 rpm agitation, the partial 16S rRNA gene fragment was amplified using universal primers (27F 5′-AGAGTTTGATCMTGGCTCAG-3’; 1492R 5′-GGTTACCTTGTTACGACTT-3′). The amplified fragment (1,367 bp) was sent for sequencing analysis (Shanghai Sangong Company, China), which displayed 99.85% identity (BlastN, https://blast.ncbi.nlm.nih.gov/Blast.cgi) to Streptomyces polaris (MW164959.1). The sequence of 16S rRNA has been deposited in the NCBI nucleotide database with the accession number OK161010.
Fermentation, Extraction, and Isolation
Streptomyces sp. HS-3-L-1 was grown on M2 agar plate (glucose 4 g/L, malt extract 10 g/L, yeast extract 4 g/L, and agar 15 g/L) at 28°C for a week. Small pieces of agar with bacterial growth were added to eleven 250-ml Erlenmeyer flasks, each containing 50 ml of medium Bran [corn flour, 40.0 g/L; gluten powder, 5.0 g/L; K2HPO4•3H2O, 0.5 g/L; glucose, 10.0 g/L; bran, 10.0 g/L; CaCO3, 2.0 g/L; and (NH4)2SO4, 1.0 g/L]. After 3 days of incubation at 28°C with 200 rpm agitation, the seed cultures were used to inoculate 100 Erlenmeyer flasks (250 ml), each containing 100 ml of medium Bran (total 10 L). The fermentation was carried out on a rotary shaker (200 rpm) at 28°C for a week. All obtained culture broth was combined and centrifuged at 5,000 ×g for 30 min to separate the mycelium and supernatant. Mycelium was extracted with MeOH (3 × 2 L), and the organic phase was evaporated to afford 50.2 g of crude extract A. The supernatant was mixed with 4% (w/v) XAD-16 resin and stirred for 6 h, followed by filtration. The resin was washed with water (3 × 500 ml) and then eluted with MeOH until the eluant was colorless. The MeOH extract was subsequently evaporated to afford 13.3 g of crude extract B.
Crude extract A (50.2 g) was subjected to an MCI column (500 g, 10 × 80 cm) and eluted with a gradient of aqueous MeOH (20, 40, 60, 80, and 100%) to yield 16 fractions (Fr. 1-1 to Fr. 1-16). Fr. 1-9 (0.5 g) was subjected to a Sephadex LH-20 column (4 × 100 cm, 2 ml/min) eluted with 80% aqueous MeOH to obtain 11 subfractions (Fr. 1-9-1 to Fr. 1-9-11). Compound 5 (13.6 mg) was obtained from Fr. 1-9-4. Fr. 1-9-9 was further purified by semi-preparative HPLC (38% aqueous MeOH) to yield compound 3 (5.8 mg). Crude extract B (13.3 g) was subjected to an MCI column (200 g, 6 × 30 cm), using gradient elution from 20% to 100% aqueous MeOH to provide 11 fractions (Fr. 2-1 to Fr. 2–11). Fractions Fr. 2-5 (0.7 g) was subjected to a Sephadex LH-20 column (4 × 100 cm, 2 ml/min, 80% aqueous MeOH) to obtain 12 subfractions (Fr. 2-5-1 to Fr. 2-5-12). Fr. 2-5-6 (0.3 g) was further purified by semi-preparative HPLC (45% aqueous MeOH) to yield compounds 4 (7.6 mg) and 7 (9.4 mg). Fr. 2-10 (0.5 g) was also purified by a Sephadex LH-20 column (4 × 100 cm, 2 ml/min, 80% aqueous MeOH) to obtain seven subfractions (Fr. 2-10-1 to Fr. 2-10-7). Fr. 2-10-6 (0.2 g) was further purified by semi-preparative HPLC (63% aqueous MeOH) to yield compounds 1 (8.6 mg), 2 (16.8 mg), and 6 (12.3 mg).
Huoshanmycin A (1): Yellow amorphous powder; UV (MeOH) λmax (log ε) 294 nm (4.17); IR (KBr) ν max 3,164, 2,927, 1,680, 1,617, 1,437, 1,401, 1,384, 1,284, 1,207, 1,138, 1,027, 829, 803, and 724 cm−1; 13C and 1H NMR data, see Table 1; (+)-ESI-MS: m/z 749.0 [M + H]+; (‒)-ESI-MS: m/z 747.2 [M − H]‒; (−)-HR-ESI-MS m/z 747.17096 [M − H]− (calcd. for C41H31O14 747.1714).
TABLE 1.
1H (500 MHz) and 13C (125 MHz) NMR data of compounds 1‒3 in DMSO-d 6.
| No | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δ C, type | δ H, J in Hz | δ C, type | δ H, J in Hz | δ C, type | δ H, J in Hz | |
| 1 | 165.2, C | — | 165.2, C | — | 165.2, C | — |
| 2 | 100.0, C | — | 100.1, C | — | 100.1, C | — |
| 3 | 166.1, C | — | 166.1, C | — | 166.3, C | — |
| 4 | 101.8, CH | 5.76, s | 101.8, CH | 5.75, s | 101.6, CH | 5.74, s |
| 5 | 160.9, C | — | 160.9, C | — | 160.8, C | — |
| 6 | 36.5, CH2 | 3.54, s | 36.6, CH2 | 3.55, s | 36.5, CH2 | 3.53, s |
| 7 | 133.2, C | — | 133.2, C | — | 133.2, C | — |
| 8 | 121.3, CH | 6.74, dd (7.6, 2.1) | 121.3, CH | 6.75, d (7.6) | 121.2, CH | 6.75, d (7.7) |
| 9 | 130.5, CH | 7.22, t (7.7) | 130.5, CH | 7.22, t (7.9) | 130.5, CH | 7.21, t (7.9) |
| 10 | 115.0, CH | 6.80, t (8.7) | 115.0, CH | 6.80, d (8.2) | 114.9, CH | 6.80, d (8.2) |
| 11 | 154.2, C | — | 154.2, C | — | 154.2, C | — |
| 12 | 131.2, C | — | 131.3, C | — | 131.2, C | — |
| 13 | 200.4, C | — | 200.4, C | — | 200.6, C | — |
| 14 | 116.0, C | — | 116.0, C | — | 116.0, C | — |
| 15 | 165.5, C | — | 165.5, C | — | 165.5, C | — |
| 16 | 101.1, CH | 6.13, d (2.3) | 101.8, CH | 6.14, s | 101.1, CH | 6.13, d (2.1) |
| 17 | 163.7, C | — | 163.6, C | — | 163.6, C | — |
| 18 | 112.0, CH | 6.09, d (2.0) | 112.0, CH | 6.09, s | 112.0, CH | 6.09, d (1.7) |
| 19 | 143.4, C | 143.4, C | 143.4, C | |||
| 20 | 21.9, CH3 | 1.83, s | 21.9, CH3 | 1.84, s | 21.9, CH3 | 1.84, s |
| 1′ | 161.9, C | — | 165.2, C | — | 165.1, C | — |
| 2′ | 100.1, C | — | 100.1, C | — | 100.0, C | — |
| 3′ | 166.3, C | — | 166.1, C | — | 166.2, C | — |
| 4′ | 101.7, CH | 5.70, s | 101.8, CH | 5.75, s | 101.5, CH | 5.74, s |
| 5′ | 161.4, C | — | 160.9, C | — | 161.2, C | — |
| 6′ | 37.0, CH2 | 3.47, s | 36.6, CH2 | 3.55, s | 36.7, CH2 | 3.48, s |
| 7′ | 134.8, C | — | 133.2, C | — | 135.8, C | — |
| 8′ | 120.9, CH | 6.74, dd (7.6, 2.1) | 121.3, CH | 6.75, d (7.6) | 109.5, CH | 6.16, d (1.9) |
| 9′ | 129.1, CH | 7.17, t (7.9) | 130.5, CH | 7.22, t (7.9) | 160.1, C | |
| 10′ | 114.4, CH | 6.80, t (8.7) | 115.0, CH | 6.80, d (8.2) | 101.9, CH | 6.23, d (1.9) |
| 11′ | 154.7, C | — | 154.2, C | — | 157.8, C | — |
| 12′ | 129.6, C | — | 131.3, C | — | 121.5, C | — |
| 13′ | 141.1, C | — | 200.4, C | — | 200.3, C | — |
| 14′ | 111.3, CH | 5.95, d (2.4) | 116.0, C | — | 117.6, C | — |
| 15′ | 162.4, C | — | 165.5, C | — | 163.2, C | — |
| 16′ | 102.3, CH | 6.27, d (2.4) | 101.8, CH | 6.14, s | 100.9, CH | 6.11, s |
| 17′ | 163.6, C | — | 163.6, C | — | 162.4, C | — |
| 18′ | 115.9, C | — | 112.0, CH | 6.09, s | 111.3, CH | 6.06, s |
| 19′ | 203.6, C | — | 143.4, C | — | 142.1, C | — |
| 20′ | 30.3, CH3 | 1.87, s | 21.9, CH3 | 1.84, s | 21.4, CH3 | 1.83, s |
| 1″ | 17.8, CH2 | 3.25, s | 17.8, CH2 | 3.24, s | 17.7, CH2 | 3.24, s |
| 11-OH | — | 9.81, s | — | 9.82, s | — | 9.83, s |
| 15-OH | — | 12.66, s | — | 12.68, s | — | 12.68, s |
| 17-OH | — | 10.44, s | — | 10.45, s | — | 10.45, s |
| 9′-OH | — | — | — | — | — | 9.88, s |
| 11′-OH | — | 9.48, s | — | 9.82, s | — | 10.26, s |
| 15′-OH | — | 10.36, s | — | 12.68, s | — | 12.05, s |
| 17′-OH | — | 12.50, s | — | 10.45, s | — | 10.21, s |
Huoshanmycin B (2): Yellow amorphous powder; UV (MeOH) λmax (log ε) 294 nm (4.44); IR (KBr) ν max 3,184, 2,260, 2,129, 1,675, 1,587, 1,464, 1,284, 1,167, 1,158, 1,024, 995, 926, 826, 769, 722, 699, 656, 633, 611, 576, and 523 cm−1; 13C and 1H NMR data, see Table 1; (+)-ESI-MS: m/z 749.0 [M + H]+; (‒)-ESI-MS: m/z 747.2 [M − H]‒; (+)-HR-ESI-MS m/z 749.18768 [M + H]+ (calcd. for C41H33O14 749.1970).
Huoshanmycin C (3): Yellow amorphous powder; UV (MeOH) λmax (log ε) 294 nm (4.41); IR (KBr) ν max 3,172, 2,259, 2,129, 1,677, 1,616, 1,587, 1,464, 1,384, 1,271, 1,204, 1,169, 1,142, 1,024, 999, 926, 844, 800, 766, 722, 600, and 524 cm−1; 13C and 1H NMR data, see Table 1; (+)-ESI-MS: m/z 765.2 [M + H]+; (‒)-ESI-MS: m/z 763.2 [M − H]‒; (+)-HR-ESI-MS m/z 765.1819 [M + H]+ (calcd. for C41H33O15, 765.1819), 787.1640 [M + Na]+ (calcd. for C41H32O15Na, 787.1639).
Cell Culture and Proliferation Inhibition Assay
The human AML cell line MV4-11 (CRL-9591) was purchased from ATCC and cultured in IMDM (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin–streptomycin (Gibco). To conduct cell proliferation assay, cells (1.5 × 106 cells/well) in the logarithmic phase were seeded into 96-well plates simultaneously with various concentrations of different compounds (5 μl, final concentration of 50–0.023 µM for IC50 determination) or vehicle (0.5% DMSO) for 48 h. Cell viability of compounds 1‒4 was measured using Cell Counting Kit-8 (DoJINDO) according to the manufacturer’s instructions with triptolide as positive control (Table 2). The absorbance was measured at 450 nm using a microplate reader (Epoch, Bio-Tek, United States). The value of half maximal inhibitory concentration (IC50) was calculated using GraphPad Prism 7.
TABLE 2.
Antiproliferative activity of compounds 1‒4 against MV4-11 cell line.
| Compounds | IC50 ± SD, μM |
|---|---|
| 1 | 32.9 ± 7.2 |
| 2 | 33.2 ± 6.1 |
| 3 | >50 |
| 4 | >50 |
| Triptolide | 1.1 ± 0.4 |
Antimicrobial Assay
Standard strains of Staphylococcus aureus (ATCC 29213), Escherichia coli (ATCC 25922), Bacillus subtilis (A186), Pseudomonas aeruginosa (ATCC 27853), and Acinetobacter baumannii (ATCC 19606) were obtained from CICC (China Center of Industrial Culture Collection, China). Bacteria were inoculated in LB Broth media and incubated overnight at 37°C. The cultures were quantified via a spectrophotometer, and then diluted to A = 0.02 (OD600) and dispensed to 96-well black, clear-bottom assay plates (100 µl/well). Test compounds (final concentration 50 µM) and controls (positive control of 50 µM polymyxin and placebo control of DMSO) were then added. The plates were incubated for 16 h at 37°C, and then measured the absorbance at 600 nm using a microplate reader. Compound activity was calculated on a per-plate basis (Zhao et al., 2020a).
Results and Discussion
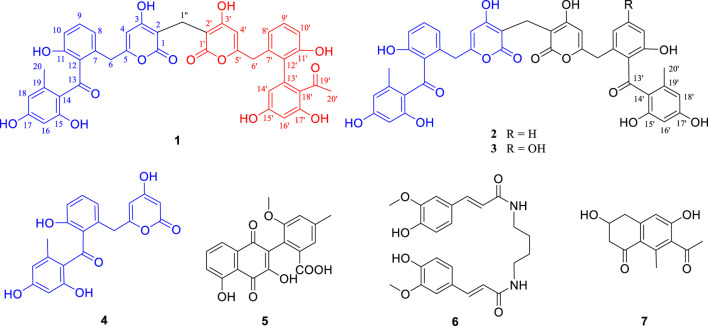
Preliminary HPLC-HRMS metabolic profiling of endophytic actinomycete strains isolated from D. huoshanense plant samples revealed that Streptomyces sp. HS-3-L-1 is capable of novel secondary metabolite production (Supplementary Figure S1). After scale-up fermentation (10 L) of Streptomyces sp. HS-3-L-1, and further extraction, fractionation, and standard chromatography, three new polyketide dimers were isolated and identified [huoshanmycins A (1, yield: 1.68 mg/L), B (2, yield: 0.86 mg/L), and C (3, yield: 0.58 mg/L)], together with four previously reported metabolites, SEK43 (4, yield: 0.76 mg/L), WS-5995 C (5, yield: 1.36 mg/L), JBIR-94 (6, yield: 12.3 mg/L), and GTRI-02 (7, yield: 0.94 mg/L)] (Figure 1).
FIGURE 1.
Chemical structures of compounds isolated from Streptomyces sp. HS-3-L-1.
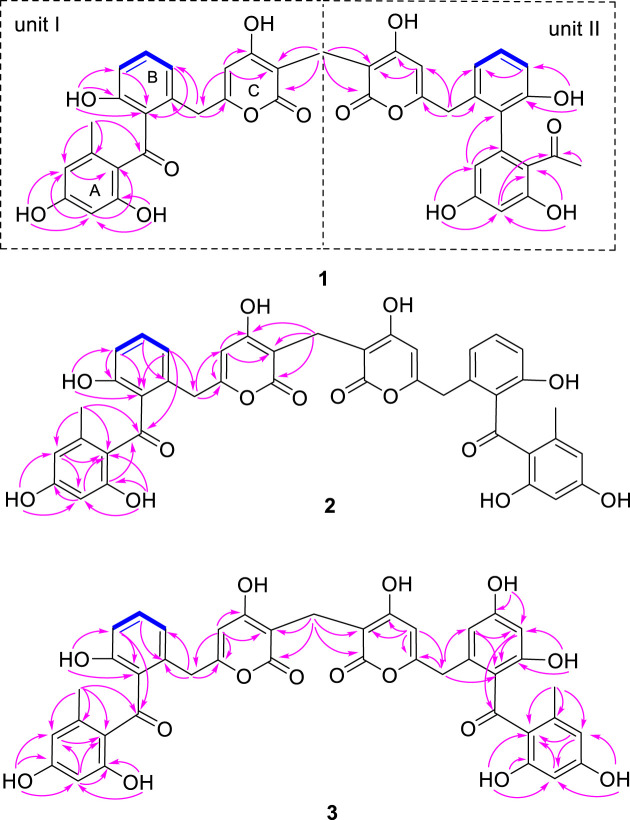
Compound 1 was obtained as a yellow amorphous powder. Its molecular formula was established as C41H32O14 based on HRESIMS data, which indicated 26 degrees of unsaturation. The 13C NMR of compound 1 showed 41 carbons, which can be sorted into two methyls, three methylenes, twelve aromatic methines, twenty sp2 quarternary carbons, and four carbonyls/carboxylic acids, with the aid of HSQC spectrum. The 1H-NMR and HSQC spectra of 1 indicated twelve aromatic protons [δ H 5.70 (1H, s), 5.76 (1H, s), 5.95 (1H, d, J = 2.4 Hz), 6.09 (1H, d, J = 2.0 Hz), 6.13 (1H, d, J = 2.3 Hz), 6.27 (1H, d, J = 2.4 Hz), 6.74 (2H, dd, J = 2.1, 7.6 Hz), 6.80 (2H, t, J = 8.7 Hz), 7.17 (1H, t, J = 7.9 Hz), and 7.22 (1H, t, J = 7.7 Hz)], two methyls [δ H 1.83 and 1.87 (each 3H, s)], three methylenes [δ H 3.25, 3.47, and 3.54 (each 2H, s)], and six hydroxyls showed at low field: δ H 9.48, 9.81, 10.36, 10.44, 12.50, and 12.66 (each 1H, s). Two spin systems of H-8 (δ H 6.74)/H-9 (δ H 7.22)/H-10 (δ H 6.80) and H-8′ (δ H 6.74)/H-9′ (δ H 7.17)/H-10′ (δ H 6.80) observed in COSY spectrum suggested the presence of two 1,2,3-trisubstiuted benzene rings. Two aromatic protons δ H 6.13 (H-16) and 6.09 (H-18), together with the HMBC correlations from H3-20 (δ H 1.83) to C-13 (δ C 200.4), C-18 (δ C 112.0), and C-19 (δ C 143.4), and from H-16 to C-14 (δ C 116.0) and C-18, and from H-18 to C-14, C-16 (δ C 101.1), and C-20 (δ C 21.9) suggested the presence of a 2,4-dihydroxy-6-methyl-1-keto-phenyl moiety (ring A in Figure 2). The two active hydrogens showed key HMBC correlations from 15-OH (δ H 12.66, s) to C-14/C-15/C-16, and from 17-OH (δ H 10.44, s) to C-16/C-17/C-18, indicating two hydroxyl groups located at C-15 and C-17. An unsaturated six-member lactone ring (ring C in Figure 2) was constructed by HMBC cross peaks from H-4 (δ H 5.76) to C-2 (δ C 100.0), C-3 (δ C 166.1), and C-6 (δ C 36.5), and from H-1″ to C-1 (δ C 165.2) and C-2. The obvious HMBC correlations from H2 -6 (δ H 3.54) to C-4 (δ C 101.8), C-5 (δ C 160.9), C-7 (δ C 133.2) and C-8 (δ C 121.3) permitted the assembly of ring B and ring C through a CH2 linkage. Analysis of the remaining NMR data revealed that unit I of 1 (Figure 2) as C2-substitued SEK43 (4) (Meurer et al., 1997). SEK43 was reported as an engineered biosynthesis product, which was isolated and identified likewise from this endophytic strain. In a similar manner, the other decaketide-related subunit II was elucidated as C2′-substitued UMW4 (Meurer et al., 1997) mainly through HMBC correlations (Figure 2), e.g., from H-4′ (δ H 5.70, s) to C-2′ (δ C 100.1) and C-3′ (δ C 166.3), from H2-6′ (δ H 3.47, s) to C-4′, C-5′ (δ C 161.4), C-7′ (δ C 134.8), and C-8′ (δ C 120.9), from H-14′ (δ H 5.95, d) to C-12′ (δ C 129.6) and C-13′ (δ C 141.1), and from H-16′ (δ H 6.27, d) to C-14′ (δ C 111.3), C-18′ (δ C 115.9), and C-19′ (δ C 203.6). The strong HMBC correlations from H2-1′′ (δ H 3.25) to C-1 (δ C 165.2), C-2 (δ C 100.0), C-1′ (δ C 161.9), and C-2′ (δ C 100.1) unambiguously connected SEK43 and UMW4 with a unique methylene bridge, to form the final dimeric structure of 1. Thus, compound 1 was identified as a novel polyketide dimer and named huoshanmycin A, to reflect the producing strain’s point of origin.
FIGURE 2.
Key COSY (bolds, blue), HMBC (arrows, pink) correlations of 1‒3.
Compound 2 was also obtained as a white amorphous powder and shared the same molecular formula (C41H32O14) with huoshanmycin A (1). Compound 2 was clearly recognized as a polyketide dimer from its NMR data (Table 1), which showed only 21 carbons in 13C NMR spectrum. Analysis of NMR data of compound 2 revealed that it was highly similar to SEK43 (4) and shared a same methylene linkage between C-2 and C-2′. This was confirmed by HMBC correlations from H2-1" (δ H 3.24) to C-1/C-1′ (δ C 165.2), C-2/C-2′ (δ C 100.1), and C-3/C-3′ (δ C 166.1). The remaining HMBC correlations (Figure 2) and NMR data (Table 1) were in full agreement with the new structure of compound 2, and it was named huoshanmycin B.
Compound 3 was obtained as a yellow amorphous powder and its molecular formula was determined as C41H32O15 from HRESIMS results. A detailed comparison of the NMR data of 3 and 2 indicated that their structures were highly similar. The significant differences observed in NMR spectra was that one of the 1,2,3-trisubstiuted benzene ring protons in 2 (δ H 6.75, 7.22, 6.80) was replaced with two olefinic methines (δ H 6.16, 6.23) and one hydroxyl (δ H 9.88) in compound 3. This tetra-substituted benzene moiety was confirmed by the HMBC correlations from H-6′ (δ H 3.48) to C-7, C-8′, and C-12′, from H-8′ (δ H 6.16) to C-6′, C-9′, C-10′, and C-12′, and from H-10′ (δ H 6.23) to C-8′, C-9′, C-11′, and C-12′, as well as the correlation signals from 9′-OH (δ H 9.88) to C-8′, C-9′, and C-10′. Therefore, the structure of huoshanmycin C (3) was elucidated as shown in Figure 1.
The other five known compounds were identified as SEK43 (4) (McDaniel et al., 1995), WS-5995 C (5) (Ikushima et al., 1983), JBIR-94 (6) (Taj and Sorensen, 2015), and GTRI-02 (7) (Yeo et al., 1998), through comparison with reported data. Although compounds 1–7 were inactive at or below 50 μM in a standard antimicrobial assay, huoshanmycins A and B (1, 2), the two isomers, showed antiproliferative activity against the MV4-11 cell line with IC50 values of 32.9 ± 7.2 and 33.2 ± 6.1 μM, respectively (Table 2).
In summary, the discovery of compounds 1–7 as metabolites of the D. huoshanense isolate Streptomyces sp. HS-3-L-1 further highlights the potential for novel microbial natural product discovery from medicinal plants. Among them, compounds 1–3 were unique dimers of SEK43 (4), SEK15, or UWM4, three decaketide-related shunt products discovered from minimal jadPKS constructs (Meurer et al., 1997). So far, only two similar natural products, strepolyketides B and C, were recently reported from a marine-derived Streptomyces (Jiang et al., 2020). Moreover, huoshanmycins A and B showed moderate cytotoxicity against MV4-11 human leukemia cell. The newly isolated 1–3 enriched the structural diversity of microbial source. Future investigation to explore the biosynthetic logic of these structurally unique dimers is ongoing.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, OK161010.
Author Contributions
YZ contributed to the chemical experiments and prepared the manuscript draft. YK performed the identification of strain and the cytotoxicity assay. YuH, LZ, and SL participated in the endophytic strain isolation and antimicrobial assay. SH and XC undertook the MS analysis. TX contributed to collection of plant samples and financial support. YaH and XW contributed to the design of experiment, financial support, and manuscript revision. All authors have approved the submission of this manuscript for publication.
Funding
This research was financially supported by the Jiangsu Provincial Innovation and Entrepreneurship Talent Plan (202010528), the Nature Science Foundation of Jiangsu Higher Education Institutions of China (No. 19KJA180008), the National Natural Science Foundation (81803391), and the Natural Science Foundation Youth Program of Nanjing University of Chinese Medicine (NZY81803391).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2021.807508/full#supplementary-material
References
- Chen S., Dai J., Song X., Jiang X., Zhao Q., Sun C., et al. (2020). Endophytic Microbiota Comparison of Dendrobium huoshanense Root and Stem in Different Growth Years. Planta Med. 86, 967–975. 10.1055/a-1046-1022 [DOI] [PubMed] [Google Scholar]
- Chen S. T., Dai J., Jiang X. P., Song X. W., Chen C. W., Chen N. F., et al. (2019). Diversity and Difference of Endophytes in Dendrobium huoshanense with Different Growth Years. Zhongguo Zhong Yao Za Zhi 44, 1145–1150. 10.19540/j.cnki.cjcmm.2019.0022 [DOI] [PubMed] [Google Scholar]
- Chen X., Hu J., Wu S., Zhao H., Peng D., Wu D., et al. (2022). Cytotoxic picrotoxane-type sesquiterpenoid lactones from Dendrobium huoshanense. Rec. Nat. Prod. 16, 144–149. 10.25135/rnp.260.21.04.2048 [DOI] [Google Scholar]
- Cui Y., Yi D., Bai X., Sun B., Zhao Y., Zhang Y. (2012). Ginkgolide B Produced Endophytic Fungus (Fusarium oxysporum) Isolated from Ginkgo Biloba . Fitoterapia 83, 913–920. 10.1016/j.fitote.2012.04.009 [DOI] [PubMed] [Google Scholar]
- Ding N., Wang J., Liu J., Zhu Y., Hou S., Zhao H., et al. (2021). Cytotoxic Guaianolide Sesquiterpenoids from Ainsliaea fragrans . J. Nat. Prod. 84, 2568–2574. 10.1021/acs.jnatprod.1c00587 [DOI] [PubMed] [Google Scholar]
- Fan X., Han J., Zhu L., Chen Z., Li J., Gu Y., et al. (2020). Protective activities of Dendrobium huoshanense C. Z. Tang et S. J. Cheng polysaccharide against high-cholesterol diet-induced atherosclerosis in zebrafish. Oxidative Med. Cell Longevity 2020, 1–10. 10.1155/2020/8365056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J.-C., Zha X.-Q., Nie C.-Y., Yu N.-J., Li Q.-M., Peng D.-Y., et al. (2018). Polysaccharides from Dendrobium huoshanense Stems Alleviates Lung Inflammation in Cigarette Smoke-Induced Mice. Carbohydr. Polym. 189, 289–295. 10.1016/j.carbpol.2018.02.054 [DOI] [PubMed] [Google Scholar]
- Gómez O. C., Luiz J. H. H. (2018). Endophytic Fungi Isolated from Medicinal Plants: Future Prospects of Bioactive Natural Products from Tabebuia/Handroanthus Endophytes. Appl. Microbiol. Biotechnol. 102, 9105–9119. 10.1007/s00253-018-9344-3 [DOI] [PubMed] [Google Scholar]
- Hu Y., Zhao H., Yang A., Lv Q., Ding N., Lu T.-L., et al. (2021). Jatrophacine, a 4,5-seco-Rhamnofolane Diterpenoid with Potent Anti-inflammatory Activity from Jatropha curcas . Nat. Product. Res. 35, 2748–2752. 10.1080/14786419.2019.1660656 [DOI] [PubMed] [Google Scholar]
- Ikushima H., Takase S., Kawai Y., Itoh Y., Okamoto M., Tanaka H. (1983). Structure and Synthesis of New Anticoccidial Antibiotics Isolated from Streptomyces auranticolor . Agric. Biol. Chem. 47, 2231–2235. 10.1080/00021369.1983.10865932 [DOI] [Google Scholar]
- Jiang Y., Huang Y., Chen S., Ji Y., Ding W., Ma Z. (2020). Strepolyketides A-C, Three Novel SEK15-Derived Polyketides from Streptomyces sp. HN2A53. Tetrahedron Lett. 61, 151996. 10.1016/j.tetlet.2020.151996 [DOI] [Google Scholar]
- Li O., Xiao R., Sun L., Guan C., Kong D., Hu X. (2017). Bacterial and Diazotrophic Diversities of Endophytes in Dendrobium catenatum Determined through Barcoded Pyrosequencing. PLoS One 12, e0184717. 10.1371/journal.pone.0184717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.-M., Jiang H., Zha X.-Q., Wu D.-L., Pan L.-H., Duan J., et al. (2020). Anti-inflammatory Bibenzyls from the Stems of Dendrobium huoshanense via Bioassay Guided Isolation. Nat. Product. Res. 34, 563–566. 10.1080/14786419.2018.1489394 [DOI] [PubMed] [Google Scholar]
- McDaniel R., Ebert-Khosla S., Hopwood D. A., Khosla C. (1995). Rational Design of Aromatic Polyketide Natural Products by Recombinant Assembly of Enzymatic Subunits. Nature 375, 549–554. 10.1038/375549a0 [DOI] [PubMed] [Google Scholar]
- Meurer G., Gerlitz M., Wendt-Pienkowski E., Vining L. C., Rohr J., Richard Hutchinson C. (1997). Iterative Type II Polyketide Synthases, Cyclases and Ketoreductases Exhibit Context-dependent Behavior in the Biosynthesis of Linear and Angular Decapolyketides. Chem. Biol. 4, 433–443. 10.1016/s1074-5521(97)90195-2 [DOI] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M. (2015). Endophytic and Epiphytic Microbes as sources of Bioactive Agents. Front. Chem. 3, 34. 10.3389/fchem.2015.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja A., Prabakaran P. (2011). Actinomycetes and Drug-An Overview. Am. J. Drug Discov. Develop. 1, 75–84. 10.3923/ajdd.2011.75.84 [DOI] [Google Scholar]
- Stierle A., Strobel G., Stierle D. (1993). Taxol and Taxane Production by Taxomyces Andreanae, an Endophytic Fungus of Pacific Yew. Science 260, 214–216. 10.1126/science.8097061 [DOI] [PubMed] [Google Scholar]
- Taj R., Sorensen J. L. (2015). Synthesis of Actinomycetes Natural Products JBIR-94, JBIR-125, and Related Analogues. Tetrahedron Lett. 56, 7108–7111. 10.1016/j.tetlet.2015.11.020 [DOI] [Google Scholar]
- Tian C.-C., Zha X.-Q., Pan L.-H., Luo J.-P. (2013). Structural Characterization and Antioxidant Activity of a Low-Molecular Polysaccharide from Dendrobium huoshanense . Fitoterapia 91, 247–255. 10.1016/j.fitote.2013.09.018 [DOI] [PubMed] [Google Scholar]
- Tsavkelova E. A., Cherdyntseva T. A., Klimova S. Y., Shestakov A. I., Botina S. G., Netrusov A. I. (2007). Orchid-associated Bacteria Produce Indole-3-Acetic Acid, Promote Seed Germination, and Increase Their Microbial Yield in Response to Exogenous Auxin. Arch. Microbiol. 188, 655–664. 10.1007/s00203-007-0286-x [DOI] [PubMed] [Google Scholar]
- Wang H.-Y., Li Q.-M., Yu N.-J., Chen W.-D., Zha X.-Q., Wu D.-L., et al. (2019). Dendrobium huoshanense Polysaccharide Regulates Hepatic Glucose Homeostasis and Pancreatic β-cell Function in Type 2 Diabetic Mice. Carbohydr. Polym. 211, 39–48. 10.1016/j.carbpol.2019.01.101 [DOI] [PubMed] [Google Scholar]
- Wang X.-C., Zheng Z.-P., Gan X.-W., Hu L.-H. (2009). Jatrophalactam, a Novel Diterpenoid Lactam Isolated from Jatropha curcas . Org. Lett. 11, 5522–5524. 10.1021/ol902349f [DOI] [PubMed] [Google Scholar]
- Wang X., Shaaban K. A., Elshahawi S. I., Ponomareva L. V., Sunkara M., Zhang Y., et al. (2013). Frenolicins C-G, Pyranonaphthoquinones from streptomyces sp. RM-4-15. J. Nat. Prod. 76, 1441–1447. 10.1021/np400231r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-H. (2021). Traditional Uses, Chemical Constituents, Pharmacological Activities, and Toxicological Effects of Dendrobium Leaves: A Review. J. Ethnopharmacology 270, 113851. 10.1016/j.jep.2021.113851 [DOI] [PubMed] [Google Scholar]
- Xu F., Chen X., Hu J., Zhao H., Peng D., Wu D., et al. (2022). Cytotoxic Picrotoxane-type Sesquiterpenoid Lactones from Dendrobium huoshanense . Rec. Nat. Prod. 16, 144–149. 10.25135/rnp.260.21.04.2048 [DOI] [Google Scholar]
- Yeo W.-H., Yun B.-S., Kim S.-S., Park E.-K., Kim Y.-H., Yoo I.-D., et al. (1998). GTRI-02, a New Lipid Peroxidation Inhibitor from Micromonospora sp. SA246. J. Antibiot. 51, 952–953. 10.7164/antibiotics.51.952 [DOI] [PubMed] [Google Scholar]
- Zhao H., Chen X., Chen X., Zhu Y., Kong Y., Zhang S., et al. (2020c). New Peptidendrocins and Anticancer Chartreusin from an Endophytic Bacterium of Dendrobium officinale . Ann. Transl. Med. 8, 455. 10.21037/atm.2020.03.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Yang A., Liu J., Bao S., Peng R., Hu Y., et al. (2020b). Chartspiroton, a Tetracyclic spiro-naphthoquinone Derivative from a Medicinal Plant Endophytic Streptomyces . Org. Lett. 22, 3739–3743. 10.1021/acs.orglett.0c00696 [DOI] [PubMed] [Google Scholar]
- Zhao H., Yang A., Zhang N., Li S., Yuan T., Ding N., et al. (2020a). Insecticidal Endostemonines A-J Produced by Endophytic Streptomyces from Stemona sessilifolia . J. Agric. Food Chem. 68, 1588–1595. 10.1021/acs.jafc.9b06755 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Li S., Kong Y., Zhao H., Hu Y., Meng J., et al. (2021). Terragines F-G Produced by Endophytic bacillus sp. SH-1.2-ROOT-18 from Dendrobium officinale . Nat. Product. Res. 10.1080/14786419.2021.1914614 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, OK161010.