Abstract
Membrane traffic in eukaryotic cells is mediated by transport vesicles that bud from a precursor compartment and are transported to their destination compartment where they dock and fuse. To reach their intracellular destination, transport vesicles contain targeting signals such as Rab GTPases and polyphosphoinositides that are recognized by tethering factors in the cytoplasm and that connect the vesicles with their respective destination compartment. The final step, membrane fusion, is mediated by SNARE proteins. SNAREs are connected to targeting signals and tethering factors by multiple interactions. However, it is still debated whether SNAREs only function downstream of targeting and tethering or whether they also participate in regulating targeting specificity. Here, we review the evidence and discuss recent data supporting a role of SNARE proteins as targeting signals in vesicle traffic.
Keywords: membrane fusion, snare proteins, trafficking
Introduction
Eukaryotic cells are compartmentalized into membrane-enclosed organelles. Since lipid bilayers form a hydrophobic barrier for all hydrophilic metabolites and macromolecules, compartments allow for the segregation of specific materials and biochemical reactions. Membrane vesicles are stable in the aqueous environment. They rarely fuse or split spontaneously [1], which is very useful for maintaining the identity of individual organelles. On the other hand, the compartments need to communicate with each other in order to exchange molecules for their homeostasis and to allow for stepwise processing and delivery of newly synthesized macromolecules or of material destined for remodeling or degradation. Therefore, a hallmark of all eukaryotic cells is the ability to generate vesicles from donor membranes and transport them to a specific acceptor membrane where they dock and fuse [2]. In fact, with the exception of mitochondria and, to some extent, also peroxisomes, all organelles are connected with each other by vesicular traffic, allowing for the directed transfer of soluble cargoes, membrane proteins, and lipids [2–4]. Importantly, vesicular traffic is highly organized in a network of central organelles serving as hubs such as the endoplasmic reticulum, the Golgi complex, the endosomal system, and, of course, the plasma membrane, which all generate and receive transport vesicles in a directed fashion.
Owing to recent progress in super-resolution microscopy and high-speed imaging it is now appreciated that vesicular traffic is stunningly complex. At any given time, organellar hubs are surrounded by thousands of vesicles that are constantly coming and going. In fact, despite its overall directionality transport is seemingly chaotic. Vesicles may undergo spurts of movement, pause, sometimes even move backwards a bit, and in addition to directed movement vesicle trafficking also seems to include diffusive components [5,6]. Thus, it is essential for cells to ‘know’ from where a given vesicle is originating (e.g. from the TGN or from the plasma membrane), and where it is to be delivered (e.g. to the late endosome or to the plasma membrane). Accordingly, vesicles must be equipped with molecular ‘zip codes’ that govern the subsequent trafficking step. Indeed, a robustly functioning zip code system is critical not only for cellular homeostasis but also for mitosis, differentiation and migration, with perturbations of vesicle targeting being associated with various diseases [7].
In the past decades, major progress was made in deciphering not only the basic molecular mechanisms of vesicle budding and fusion but also in the molecular foundation of organellar identity and targeting specificity [2,8]. In this review, we will attempt to discuss some of the mechanisms that explain how in this ‘chaotic’ environment vesicles released from a specific donor compartment find their target, focusing on the role of SNARE proteins.
Specific targeting of trafficking vesicles: Rab GTPases and polyphosphoinositides as central regulators
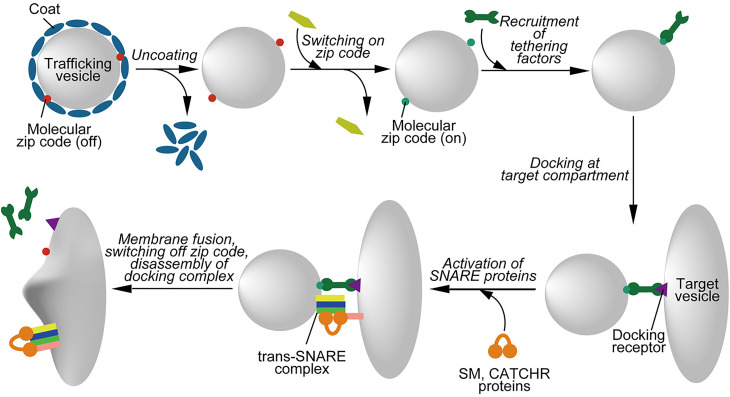
Vesicle trafficking generally proceeds in three consecutive steps: the generation of a vesicle from a precursor membrane which involves the selection of specific macromolecules (sorting), the motor protein-dependent transport of the vesicle to its destination followed by recognition/tethering (targeting), docking, and the fusion of the vesicle with the target membrane [2,9]. Here we define tethering as initial interaction at a distance, whereas docking is a closer interaction, probably involving trans-pairing of SNAREs between the membranes. In general, these processes are regulated by different proteins (see below). Formation of a new vesicle frequently requires the participation of protein coats. When a vesicle is set free from its donor compartment it needs to contain a built-in ‘navigation plan’ in order to reach the correct intracellular destination. In other words, molecular zip-codes are attached on the surface when the vesicles are generated (Figure 1). These membrane-anchored tags orchestrate the next step, which includes the ordered recruitment of multiple effector proteins from the cytoplasm. It is the specific combination of these effectors decorating the vesicle that in turn are been read by other molecules, thus allowing the vesicle to proceed to the next step. Such steps include the removal of coat proteins, the recruitment of motor proteins for directed transport, tethering factors, the attachment to the target membrane, or the activation of the fusion machine (Figure 1). While these steps are consecutive, their order may vary. For instance, coats may disassemble before arriving at the target membrane (i.e. COPII and clathrin coats), but in at least one other case the coat (COPI) is only disassembled upon contact with the preassembled fusion machine [10].
Figure 1. Schematic overview over the steps involved in the the targeting of a trafficking vesicle.
The figure shows a typical sequence of molecular steps governing the delivery and fusion of a trafficking vesicle to the correct target compartment. Formation of a trafficking vesicle by budding (not depicted here) frequently involves protein coats. Once these coats are disassembled, the molecular zip codes become “visible” to cytoplasmic proteins and are activated, for instance by GDP-GTP exchange or by phosphorylation/dephosphorylation of PtdIns variants. Active zip codes recruit tethering factors from the cytoplasm which, after transport along cytoskeletal tracks (not shown here), bind to a docking receptor on the target compartment. SNARE proteins are then activated by members of the conserved SM- and CATCHR protein families, followed by SNARE assembly and membrane fusion. After fusion, zip codes are switched off, tethering and activation factors dissociate, and the assembled SNARE complexes are disassembled (not shown). See text for details.
It is well established that Rab GTPases and phosphorylated variants of the membrane lipid phosphatidylinositol (PtdIns) serve as master organizers of vesicle traffic that confer vesicle identity and serve as zip codes for targeting [8]. Rab proteins belong to the Ras superfamily of small GTPases. The human genome contains ∼60 different Rab genes [11]. Apparently, each intracellular compartment contains a specific set of Rab GTPases that, sometimes in cooperation with small GTPases of other subfamilies (e.g. Arf GTPases), define the identity of the organelle, orchestrate membrane traffic, demarcate specific domains on the surface of organelles for tubulation or budding or directed transport, and undergo dynamic remodeling during progression to the next trafficking step [8]. Rabs are activated by guanine exchange factors (GEFs) and inactivated by GTPase activating proteins (GAPs). In the active (GTP-bound) form, Rab GTPases are anchored to the membrane by geranylgeranyl moieties and recruit specific sets of effector proteins including tethering factors onto membranes. After GTP is cleaved into GDP, the effector proteins bound to the active Rab protein disassemble, and most Rabs are then extracted from the membrane by forming a complex with GDP dissociation inhibitor (GDI), thus being returned to the cytoplasmic pool [11]. Progression along a specific trafficking route may involve the consecutive action of different Rab proteins that are regulated by both positive and negative feedback loops [12,13]. One of the best-studied examples involves the maturation of an early into a late endosome. While early endosomes are mainly governed by Rab5, one of its effectors (the Mon1–Ccz1 complex) recruits and activates Rab7 [14], thus changing the identity of the vesicle from a Rab5-containing early endosomes to a Rab7-containing late endosome.
Phosphorylated variants of PtdIns constitute a minor class of comparably short-lived membrane phospholipids. Seven different PtdInsPx-species are known that are phosphorylated at either one or several of the 3-, 4- and 5-OH groups of the inositol ring of phosphatidylinositol [15]. Switching between individual PtdInsPx variants is mediated by an array of specific PtdIns-kinases and phosphatases. In mammalian cells, there are 19 kinases and 28 phosphatases that coordinately work together in regulatory networks to control PtdInsPx levels [16]. Some kinases and phosphatases form a complex such as the 5-kinase PIKfyve and 5-phosphatase Fig4, which control PtdIns(3,5)P2 levels. Interestingly, deletion of the phosphatase Fig4 reduces rather than increases the overall level of PtdIns(3,5)P2 [17], suggesting that complex formation with the phosphatase is necessary for the kinase-function of PIKfyve.
Different PtdInsPx species are selectively localized on distinct subcellular compartments, thus contributing to defining the identity of the vesicles as molecular zip codes [8]. Similar to Rab proteins, different PtdInsPx recruit specific sets of effector proteins. In many cases, PtdInsPx function in a combinatorial fashion with Rab GTPases or with other signals (e.g. sorting signals in proteins) to recruit effector proteins and thus ‘sharpen’ the specific identity of a vesicle or a membrane domain [8,13]. Indeed, many effector proteins are only stably recruited to the membrane when two signals are present simultaneously such as a specific Rab GTPase and a specific PtdInsPx-species, termed coincidence detection [13,18]. Coincidence detection involving PtdInsPx-species also regulates the activity of sorting signals in integral membrane proteins. For instance, endocytotic signals such as tyrosine-based or dileucine-based motifs are only recognized by the clathrin adaptor complex AP2 when PtdIns(4,5)P2 is present in the membrane [19]. Furthermore, recent data suggest that the nature of the acyl-chains of PtdInsPx regulate clustering and spacing of lipids, thus creating hotspot microdomains to which effector proteins are recruited on the membrane [20].
In summary, both Rab GTPases and PtdInsPx exhibit common properties that are essential hallmarks of their zip code function. They serve as signals that are switched ‘on’ at a predefined step in membrane traffic, orchestrate the next step, and are then switched ‘off’. While the signals are generated on the membrane, all necessary components are recruited from the cytoplasm including not only the precursors and enzymes needed for generating the signal but also the effectors required to execute the trafficking step. When the signal is switched in the ‘off’ position, the entire machinery dissociates from the membrane, allowing the vesicle with its membrane constituents and cargo to proceed to the next trafficking step without being burdened with remnants of the regulators governing the preceding steps. While such a system is robust and versatile, it requires a tightly controlled ‘hand-over’ mechanisms, i.e. the initial components of the next step need to be firmly in place before the machinery of the past step is completely dismantled. As discussed above, several examples of such hand-over mechanisms have been identified (Rab cascades, ‘maturation’ of PtdInsPx signals [12,13]).
Vesicle targeting by controlling the specificity of SNARE pairing?
With the partial exception of homotypic fusion of ER membranes [21], SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins are responsible for catalyzing membrane fusion in the secretory pathway. SNARE proteins form a small and conserved protein family with 25 members in yeast and 36 members in humans. SNAREs contain characteristic heptad repeats of 60–70 amino acids, called SNARE motifs, which belong to four evolutionarily conserved subfamilies termed Qa, Qb, Qc, and R-SNAREs, respectively (Figure 2A). In most SNAREs, the SNARE motif is connected by a short linker to a C-terminal membrane anchor domain. Catalysis of membrane fusion by SNARE proteins is mediated by the progressive, zipper-like assembly of four SNARE motifs, one of each subfamily, residing in the two opposing membranes. Zippering is exergonic and results in the formation of a four-helix bundle with the transmembrane domains adjacent to each other, thus pulling the membranes together and overcoming the energy barrier for fusion (Figure 2B). Indeed, numerous in vitro experiments have shown that fusion is always associated with the formation of SNARE-complexes with a QabcR composition, confirming that complex formation constitutes the core engine responsible for every SNARE-mediated fusion reaction. After fusion, the ATPase NSF (N-ethylmaleimide sensitive factor), together with SNAPs (soluble NSF attachment protein), disassembles the SNARE complex with energy released in ATP hydrolysis, making individual SNAREs available for next rounds of fusion [22,23].
Figure 2. Domain structure of SNARE proteins.
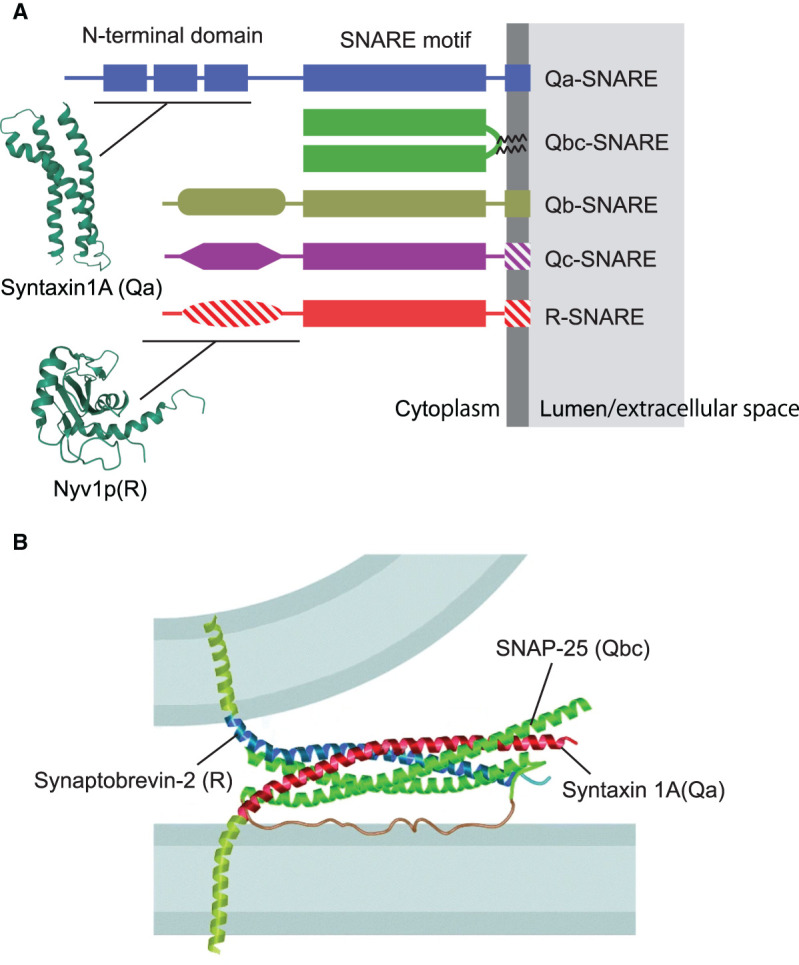
(A) Overview over the main subfamilies of SNARE proteins Note that there is some variability within the subfamilies: Hatched areas signify domains that are absent from some of the family members. Moreover, transmembrane domains may be substituted by domains binding to PtdInxPx variants. The structures of the N-terminal domains of syntaxin 1A and Nyv1p are based on the Protein Data Bank entries 1S94 and 2FZ0, respectively (wwPDB.org). (B) Crystal structure of the neuronal SNARE complex, modeled between the two membranes destined to fuse. Note that the N-terminal region of syntaxin 1 was removed. Reproduced with permission from [24].
SNAREs are differentially distributed among intracellular membranes, thus requiring specific sorting signals. Moreover, specific sets of SNAREs are responsible for individual fusion events in the secretory pathway [23]. Indeed, there are hundreds of theoretically possible combinations of SNAREs in QabcR-complexes, many more than there are distinct fusion steps in the secretory pathway. Accordingly, it was originally suggested that each individual fusion step is defined by a unique combinatorial code of SNARE pairing that ultimately controls docking and fusion of a trafficking vesicle with the correct target, with the intrinsic specificity of SNARE pairing serving as the final checkpoint for ensuring correct targeting and fusion [25,26]. Therefore, SNAREs were considered to serve as targeting signals that operate alongside with Rabs and PtdInsPx in defining the destination of trafficking vesicles. However, it is still debated to which extent SNAREs confer targeting specificity, thus functioning as a third class of zip-code molecules. SNAREs mediate fusion, which is the last step in trafficking and only occurs when correct targeting is already completed [8]. Moreover, in contrast with the temporally and spatially regulated Rabs and PtdInsPx most SNAREs are membrane-anchored and thus need to be recycled by vesicular membrane traffic to the point of origin as a passenger. Since as discussed above on-off switching is a mandatory feature for targeting molecules, this would require regulation of the protein's ability to interact with effectors while remaining anchored in the membrane, being ‘on’ when the SNARE is approaching its target and being ‘off’ while it is returned as passenger to its origin.
There are two possible mechanisms by which SNAREs may confer targeting specificity, which are not mutually exclusive. First, the topological specificity of SNARE function may be encoded in the combinatorial specificity of SNARE pairing. Second, individual SNAREs may contain targeting signals that function independent of SNARE pairing and involve the recruitment of targeting/tethering factors and/or factors regulating the activity of the SNARE motifs. These two mechanisms are encoded in different domains of the SNARE proteins (with some overlap) and are discussed in the following sections.
Specificity of SNARE pairing as observed in vitro and in artificial membranes
For many years, it was debated to which extent SNARE pairing is specific for individual SNAREs and thus functioning as one of the decisive and final steps in defining targeting specificity or at least, final proof-reading of correct targeting, thus preventing a wrongly targeted vesicle from fusion. Originally, it was assumed that each intracellular fusion event is mediated by a unique SNARE complex and that pairing specificity is encoded in the SNAREs themselves, without the need of additional regulatory proteins [27]. However, in some of the early in vitro experiments it was overlooked that only SNARE complexes with a ‘QabcR’ composition can function in fusion. Moreover, in vitro studies on mammalian exocytotic and endosomal SNAREs showed a high degree of promiscuity, with individual SNAREs of the same subfamily being able to substitute for each other in complex formation and fusion, particularly R-SNAREs. Indeed, the X-ray structures of diverse SNARE complexes revealed a striking structural conservation, despite considerable sequence divergence (see [23,26], for a more thorough discussion). More recently, a systematic analysis of SNARE complex formation and SNARE-mediated fusion in vitro was carried out using a panel of 14 different yeast SNARE proteins [28]. While the authors observed specificity in the formation of SNARE complexes operating in fusion events between the ER and the Golgi apparatus, they confirmed that SNAREs alone, in the absence of other regulatory factors, are not sufficient to determine targeting/fusion specificity in vacuolar, endosomal, and plasma membrane fusion reactions. Apparently, the mechanistic functioning of the SNARE engine requires a highly conserved core structure, which prevented diversification of an ancient SNARE machine into a multitude of specialized engines, even though there is considerable evolutionary sequence divergence within the four SNARE subfamilies.
Specificity of SNARE pairing in their native membrane environment
Considering the promiscuity of SNARE pairing in vitro, the question arises whether a similar flexibility also exists inside cells, with appropriate SNAREs being able to substitute for each other in individual fusion steps. Perhaps it is not surprising that there is no simple answer to this question. On one hand, genetic ablations of many SNAREs result in surprisingly mild phenotypes (only a minority of SNAREs in yeast are encoded by essential genes), revealing at least partial redundancy and confirming that different SNAREs of the same subfamily can substitute for each other in intact cells [23]. On the other hand, a large body of evidence shows that the specificity of SNARE-pairing mediating fusion in a native environment is higher than suggested by the promiscuity of SNARE assembly in vitro. For example, chemical neurotransmission involves exocytosis of synaptic vesicles which is mediated by a SNARE complex consisting of syntaxin 1 (Qa), SNAP-25 (Qbc), and synaptobrevin/VAMP-2 (R). Detailed electrophysiological experiments have revealed, however, that synaptic vesicles exist in separate pools exhibiting characteristic differences in their exocytotic behavior [29]. While signaling is dominated by a burst of rapid exocytosis in response to calcium influx, slower and scattered fusion activity is also observed, either as a tail of ‘asynchronous’ release following the exocytotic burst or occurring spontaneously, i.e. independent of action potential-mediated calcium influx. Apparently these differences are reflected by different sets of SNAREs. While synaptobrevin 2 (VAMP2) is governing rapid, Ca2+-dependent fusion, deletion of synaptobrevin 2 did not affect spontaneous release as dramatically as evoked release [30]. Similar observations were made when exocytotic SNAREs were inactivated by clostridial neurotoxins [31]. Indeed it appears that these fusion events involve other SNAREs such as the endosomal SNAREs VAMP4, VAMP7, and vti1a [32–34], although VAMP2 and VAMP4 can substitute for each other in vitro[35].
In vitro studies provide additional insights into the specificity of SNARE pairing in native membranes. As discussed in more detail below, it is becoming evident that the activity of many (but perhaps not all) SNAREs is tightly regulated. For instance, we showed earlier that the homotypic fusion of ‘native’ early endosomes isolated from cell extracts in vitro is mediated by a unique SNARE-complex involving the R-SNARE VAMP-4 [35]. This is remarkable when considering that endosomes contain a large diversity of R-SNAREs as ‘passengers’ en route to different destinations. Neuronal endosomes contain the exocytotic R-SNARE VAMP-2/synaptobrevin at a concentration that is 100-fold higher than that of VAMP4 [36]. While synaptobrevin is recycled via early endosomes, it does not participate in homotypic fusion of endosomes [37]. In contrast, the same early endosomal Q-SNAREs accept a much broader range of R-SNAREs as fusion partners, including VAMP-2/synaptobrevin, when these SNAREs are reconstituted in liposomes rather than being in their native environment [35]. Moreover, there appears to be a difference in specificity depending on whether the SNAREs assemble in ‘cis’ or ‘trans’: While specificity is high in the fusion reaction (‘trans’ assembly), it appears to be much lower when SNAREs residing in the same membrane form ‘cis’ QabcR-complexes, which appears to happen rather promiscuously [36].
Specificity of SNARE pairing by regulating SNARE activity
As discussed above, the SNARE motifs do not convey sufficient specificity to SNARE assembly to explain pairing specificity in intact cells and membranes. Rather, it is evident that the activity of SNAREs for entering trans-SNARE complexes is tightly regulated by a multitude of other factors that decide whether a SNARE is switched ‘on’ or ‘off’ for fusion. As far as we know, however, there are several rather diverse mechanisms for regulating SNARE activity, and they contribute to the specificity of SNARE pairing by different mechanisms.
After fusion SNAREs are in an inactive ‘cis’ conformation and need to be disassembled and thus re-activated by NSF and SNAP-proteins. Indeed, the continuous activity of NSF appears to be required to maintain SNAREs in an active form since they not only form complexes during fusion (trans-assembly) but also have a tendency to form cis-complexes in the membrane [38]. For instance, studies on yeast homotypic vacuolar fusion in vitro have revealed that Sec18 (the yeast homologue of mammalian NSF) and Sec17 (the yeast homologue of mammalian α-SNAP) are necessary for subsequent tethering and fusion [39–41]. NSF and SNAPs not only operate on fully assembled cis-complexes but also dissociate partial complexes. Moreover, recent evidence suggests that binding of α-SNAP, and perhaps also NSF, may also occur to SNARE trans-complexes and facilitate fusion without requiring ATP hydrolysis [42]. However, NSF operates on all SNAREs and thus is unlikely to contribute to pairing specificity.
SNARE-activity may also be regulated by the lateral organization of SNAREs in the plane of the membrane. For instance, it appears that at least in the plasma membrane SNAREs are concentrated in nanodomains of high density [43] that probably affect their ability to interact with partner SNAREs in fusion. Moreover, we have shown earlier that during fusion of neuronal endosomes, VAMP2 and VAMP4 are laterally segregated at endosomal contact sites, leading to a local enrichment of VAMP4, and a concomitant depletion of VAMP2, at the contact site although VAMP2 overall is much more abundant (see above) [36], thus contributing to pairing specificity. The mechanisms underlying such lateral segregation are not known, and it is not clear whether lateral segregation into nanodomans is a general mechanism by which the specificity of SNARE pairing at the membrane contact site is regulated.
Most importantly, SNARE activity is regulated by specific binding proteins that, while controlling assembly of the SNARE motifs, require additional features of the SNAREs for recognition. In many cases, this involves N-terminal domains present on many but not all SNAREs (Figure 2A) that are separated from the SNARE motif by short and flexible linkers. The sequences of the N-terminal domains are more divergent than those of the SNARE motifs and, as far as structurally characterized, are represented by at least two different folds. The first is termed Habc domain. It consists of three-helical bundles (Ha, Hb, and Hc helices) and is found on Qa, Qb, and some Qc-SNAREs [23]. The second is termed longin domain and is observed in some R-SNAREs including VAMP7, Sec22, and Ykt6 [44,45]. It is arranged in the form of α-β-α sandwich architecture and differs substantially from the Habc domain [45]. In some cases, N-terminal domains directly regulate the SNARE by intramolecular switching between a ‘closed’ conformation in which the domain reversibly binds to the SNARE motif and inhibits its ability to enter SNARE complexes, and an ‘open’ conformation in which there is no contact to the SNARE motif [46,47].
The best-characterized class of SNARE regulators includes the SM (Sec1/Munc18) proteins. They represent a small family of highly conserved proteins that appear to be essential for SNAREs to function. They are recruited by SNARE N-terminal domains of the Habc type, and in some cases binding stabilizes the closed inactive conformation [46]. Generally, however, the main function of SM proteins is to greatly facilitate trans-SNARE assembly, probably by providing a platform for the correct alignment of the SNARE motifs for SNARE zippering and thus promote fusion [46,48]. While individual SM-proteins specifically operate on specific intracellular fusion reactions, there are only four different SM-proteins in yeast and at least seven in mammals, i.e. fewer than fusion steps or SNAREs [49]. Indeed, several SM proteins are known to regulate multiple Qa-SNAREs. The yeast SM protein Vps45p binds to the SNAREs Pep12p and Tlg2p that participate in different SNARE complexes [50,51]. Similarly, human VPS45 interacts with multiple syntaxins present on early endosomes (e.g. syntaxin 4, 6, 13, and 16) [52,53].
Do SM proteins contribute to the specificity of SNARE pairing? While obviously there are too few of them to guarantee correct assembly of cognate complexes in all trafficking steps, they may restrict the range of theoretically possible complexes and define ‘domains’ of membrane traffic. Thus, in yeast Sec1p regulates exocytosis, Vps45p appears to operate in the endosomal system, Vp33p in the vacuole, and Sly1p in fusion events between the ER and the Golgi system [49].
SM proteins do not act alone but rather interact with various other proteins that affect their function. In neurons, the SM protein Munc18-1 is cooperating with the protein Munc-13, a conserved member of the CATCHR (Complex Associated with Tethering Containing Helical Rod) protein family, and with the specialized regulatory factors synaptotagmin and complexin to mediate docking and calcium-dependent fusion of synaptic vesicles [46]. More relevant for targeting appear to be various other binding partners that integrate SM-proteins into a network of Rab/PtdInsPx-effectors governing vesicle targeting, thus linking targeting to SNARE-mediated fusion. One of the best-studied examples includes the multisubunit HOPS tethering complex that regulates homotypic fusion of yeast vacuoles [54]. This complex contains two subunits (Vps39 and Vps41) that are effectors of the GTPase Rab7, and Vps33, the SM protein required for SNARE assembly, thus placing SNARE regulation downstream of Rab-dependent targeting. Detailed analysis of the function of the HOPS complex have identified several subsequent steps, with the initial recognition of the SNARE N-terminal domains being followed by catalysis of trans-assembly, associated with a weakening of the N-terminal interactions [46]. A second example is provided by the mammalian SM protein VPS45 that binds to Rabenosyn5, a protein recruited to the endosomal membrane by coincidence detection between two endosomal Rab GTPases (Rab4 and Rab5) and PtdIns(3)P [53,55–57]. Thus, SM-proteins may not only increase SNARE pairing specificity by restriction of the possible SNARE combinations but also by being selectively recruited to tethering factors, thus ensuring that they only operate at a defined intracellular fusion step.
Vesicle targeting by recruitment of tethering factors to SNARE proteins?
Tethering factors mediate vesicle targeting and define its specificity
Tethering factors are considered as the most important group of molecules defining targeting specificity. They mediate the initial recognition and contact of transport vesicles with their target membrane, with tethering preceding SNARE-dependent fusion [54,58,59] (Figure 1). So far, ∼20 different tethering factors are known that fall into two major groups: homodimeric coiled-coil proteins and multisubunit tethering complexes. Coiled-coil tethers are represented by large and hydrophilic proteins containing elongated coiled-coil domains. Owing to their large size (more than 200 nm), coiled-coil tethers can interact with vesicles over long distances [59]. After vesicles are captured by tethering factors at the target site, the homodimeric coiled-coil tethering factors shift from an extended to collapsed conformation [60,61]. As result, the captured vesicles are pulled closely toward the target membrane, thus preparing them for SNARE trans-assembly that requires no more than 8 nm distance between the membranes [62]. Multisubunit tethering complexes (MTCs) comprise a diverse family of large protein complexes ranging from 250 to 800 kDa overall mass [54,59,63]. MTCs also capture transport vesicles but they operate at closer ranges (30 nm) [59]).
Both classes of tethering factors share certain characteristics, most importantly the presence of multiple distinct binding sites, a prerequisite for ‘cross-linking’ two different membranes. All of them are recruited to the membrane by binding to specific Rab GTPases and to specific PtdInsPx, often in a combinatorial fashion, and they thus represent the main effectors of these zip code molecules [58,59]. Tethering factors are not only necessary for targeting but also appear to be sufficient as shown in elegant relocation assays. In these experiments, tethering factors were artificially mis-localized to the surface of mitochondria by fusing them to mitochondria-targeting signals, resulting in mis-targeting of the corresponding vesicles [64–67]. Therefore, capture of transport vesicles by a target membrane, mediated by cross-linking both membranes with specific tethering factors, appears to be the key step in ensuring targeting specificity in vesicle traffic.
Tethering factors bind to SNARE proteins
It is well established that most tethering factors bind to SNAREs [68–71], but the underlying mechanisms are diverse, particularly for MTCs. For instance, the Tip20 and Sec39 subunits of the Dsl1 complex, an MTC operating in retrograde traffic from the cis-Golgi to the endoplasmic reticulum, specifically bind to the N-terminal domains of Sec20 and Use1, respectively [72]. In contrast, subunits of COG complex interact with various SNAREs functioning in Golgi- and endosomal trafficking, but they bind mainly to the SNARE motifs rather than to the N-terminal domains [64,69]. In the GARP complex, an MTC governing the fusion of endosomal vesicles with the trans Golgi network, both binding modes were described for individual subunits: Whereas Vps51 interacts with the N-terminal Habc domain of Syntaxin6, Vps53 and Vps54 interact with the SNARE motifs of Syntaxin6, Syntaxin16, and VAMP4 [73–75].
These findings support the view that the function of MTCs is not limited to membrane recognition and tethering. Rather, evidence is increasing that they are directly involved in regulating SNAREs by facilitating trans-SNARE assembly and fusion. The Dsl complex recruits and stabilizes a pre-assembled SNARE acceptor complex, readying it for fusion with the incoming COPI-vesicle (see also below). Similarly, subunits of the COG complex, in conjunction with the SM-protein Vps45, enhance assembly of cognate SNARE complexes while preventing non-productive SNARE interactions [69,76]. As discussed above, a similar role in SNARE assembly was also shown for the HOPS complex that chaperones the SNAREs from initial assembly all the way towards fusion while protecting intermediate, pre-assembled SNARE complexes from the disassembly machinery before fusion is completed [77,78]. Indeed, the HOPS complex has recently been suggested to also control fusion itself: Apparently, it reduces the energy barrier for fusion by imposing steric constraints [79]. However, as mentioned, the main function of tethering factors is to recognize and then connect the membranes destined to fuse.
Do SNAREs function as ‘zip codes’ in vesicle targeting by recruiting tethering factors?
So far, we have discussed that some SNARE proteins are interacting with tethering factors in a highly specific manner, thus linking vesicle capture and proofreading directly to the subsequent fusion step. This, however, raises the question whether SNAREs are ‘effectors’ that in vesicle targeting and tethering operate downstream of the Rab and PtdInsP zip codes or whether SNAREs, on their own, have the capability of directing vesicles to specific subcellular localizations by recruiting tethering factors as a first step, with all other steps occurring downstream of this initial capturing reaction.
To address this question, we have recently introduced a novel approach: microinjection of artificial proteoliposomes with precisely defined compositions [80]. By introducing ‘naïve’ vesicles devoid of any intracellular history or imprinting, this new approach can identify the minimum requirements (‘zip-codes’) a vesicle must fulfil for specific targeting, which cannot be done by more conventional manipulations such as knock-outs of individual molecules or mistargeting experiments as those mentioned above. To become functional in trafficking, injected vesicles need to orchestrate their decoration and modification by recruiting the appropriate components from the surrounding cytoplasm such as tethering factors. Obviously, this implies that trafficking vesicles, once separated from their parent membrane, are autonomous, i.e. they are capable of following the correct trafficking route when introduced at a random site anywhere in the cytoplasm. Indeed, endosomes purified by cell fractionation from a donor cell, when injected into a different cell, were captured by cytoskeletal transport and correctly docked and fused with their respective intracellular target membrane (see below, [80]), thus providing proof of principle for the experimental approach.
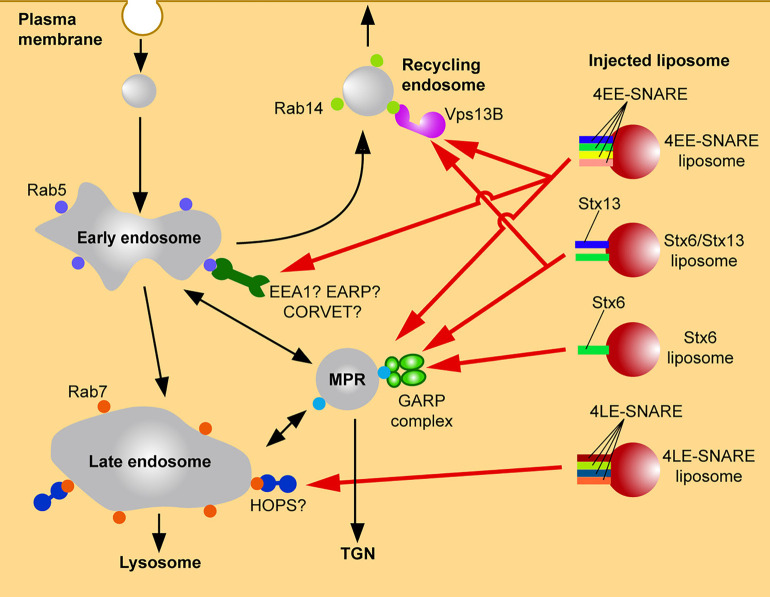
Our results show that artificial liposomes prepared with a standard membrane lipid composition and containing SNAREs as the only proteins were specifically targeted when microinjected into living cells. Indeed, proteoliposomes reconstituted with SNAREs known to mediate fusion of early endosomes (syntaxin 13 (Qa), vti1a (Qb), syntaxin 6 (Qc), and VAMP4 (R)) were captured by endogenous early endosomes (Figure 3). Conversely, liposomes reconstituted with late endosomal SNAREs (syntaxin 7 (Qa), vti1b (Qb), syntaxin 8 (Qc), and VAMP8 (R)) were specifically targeted to late endosomes [66] (Figure 3). In contrast, protein-free liposomes did not show any specific targeting with the lipids and accumulated in the Golgi area only after a very long delay period. These findings suggest that SNARE proteins alone, in the absence of Rab proteins or PtdInsPx, can mediate specific targeting of vesicles. The liposomes were not only targeted to the correct domain of the secretory pathway but also effectively fused with their respective target membrane [80].
Figure 3. Targeting of microinjected liposomes carrying early endosomal SNAREs to endosomal compartments.
Targeting of microinjected liposomes carrying early endosomal SNAREs to endosomal compartments. Liposomes containing early endosomal SNAREs but no other molecular zip codes are differentially targeted to early endosomal compartments where they dock and fuse, with the specificity being defined by a combinatorial interaction with tethering factors (GARP complex, Vps13B, possibly also additional tethering factors). Note that liposomes containing late endosomal SNAREs are correctly targeted to late endosomes, possibly via interaction with tethering complexes such as the HOPS complex. See text for details.
When we asked which of the four SNAREs are responsible for targeting, an interesting combinatorial interplay was discovered. Omission of vti1a and VAMP4 did not change the targeting specificity. Moreover, liposomes containing either of these SNAREs as the only protein showed no targeting, similar to protein-free liposomes. Liposomes containing only syntaxin 6 co-localized selectively with vesicles containing the mannose-6-phosphate receptor rather than with vesicles containing early endosomal markers whereas liposomes containing syntaxin 13 showed only very weak targeting activity. That means that the combination of syntaxin 6 and syntaxin 13 results in a targeting specificity that is clearly distinct from that of either SNARE alone [66] (Figure 3). Finally, we investigated how SNAREs exert targeting effects. Liposomes reconstituted with SNARE proteins did not show directed transport in cells after injection, similar to protein-free liposomes, suggesting that SNAREs are not directly involved in the recruitment of motor proteins (unpublished data). Surprisingly, we found that the N-terminal domain of SNAREs are recognized by tethering factors, thus allowing SNAREs to function as zip codes in a very similar manner as Rab GTPases and PtdInsPx [66].
We then identified the tethering factors responsible for the differential sorting of the two liposome populations. In our experiments, targeting of syntaxin 6 liposomes is mediated by the selective recruitment of Vps51, a component of the GARP (Golgi-associated retrograde protein) complex. The GARP complex regulates trafficking between endosomes and the trans Golgi network [59]. Our findings agree with earlier reports showing that Vps51 forms a specific complex with the N-terminal domain of syntaxin 6 [74,81,82]. Interestingly, it was also reported that the GARP complex interacts with the SNARE motifs of syntaxin 6, syntaxin 16, and VAMP4 [73,81], but it needs to be tested whether this interaction is also involved in targeting. We showed that the interaction between Vps51 and syntaxin 6 is necessary and sufficient for the targeting of Syntaxin 6 liposomes to M6PR-positive compartments [66] by mis-localization of Vps51 on mitochondrial membranes suggesting that the GARP complex can capture syntaxin 6-containing vesicles (Figure 3).
When we investigated targeting mechanism of liposomes containing both syntaxin6 and syntaxin 13, we unexpectedly discovered the protein Vps13B as novel tethering factor. Vps13B selectively binds only to syntaxin 6 and syntaxin 13 in combination. It is localized to recycling endosomes and the TGN in HeLa cells. Deletion of Vps13B delayed recycling of transferrin to plasma membrane [66], suggesting that Vps13B is required for the targeting of transferrin receptor-containing vesicles from early to recycling endosomes (Figure 3). Vps13B shares key properties with other tethering factors such as the ability to bind to Rab proteins, PtdIns phosphates, and SNAREs. Furthermore, EM imaging showed that yeast Vps13p has an elongated seahorse-like structure [83] that is similar to the HOPS tethering complex in shape and size [84], albeit HOPS has no significant sequence homology with Vps13p. Thus, Vps13B does not belong to one of the established classes of tethering proteins including long coiled-coil long proteins and multi-subunit complexes and may be the first representative of a novel and third class of tethering proteins.
Concluding remarks: multiple functions of SNAREs in trafficking
In summary, it is evident that SNAREs are integral components of vesicle trafficking that are participating not only in membrane fusion but also in sorting and targeting. First, SNAREs possess sorting signals that interact with the intracellular sorting machineries to deliver or return them to the correct compartment. Second, SNAREs interact with tethering factors that contribute to the targeting of the vesicle and then mediate docking by binding to the correct target membrane. Third, after docking SNARE-mediated fusion is regulated by proteins that activate SNAREs and guide them towards trans-assembly with the correct partners. Finally, after fusion SNARE complexes are disassembled to allow recycling of individual SNAREs and subsequent activation for the next fusion step. These steps are not only connected with each other and may involve overlapping sets of regulatory proteins, but they are also embedded in a network of regulatory interactions with Rab and PtdInsPx and their effector machineries.
Sorting
Specific sorting is a feature either of individual SNAREs independent of the pairing partners, or, in some cases, of partially assembled SNARE complexes. Sorting implies that the protein is recruited to a specific site of the precursor membrane where a trafficking vesicle is forming. Sorting is usually mediated by the interaction of a sorting signal with an appropriate adaptor complex. However, except of VAMP4 that contains a dileucine motif [85] most SNAREs do not appear to possess canonical sorting signals [86]. Rather, it appears that in many cases adaptor proteins interact with the N-terminal domains of SNAREs. For instance, the longin domain of VAMP7, also known as TI-VAMP (tetanus neurotoxin-insensitive SNARE) interacts with the ArfGAP Hrb and the δ subunit of the adaptor complex AP-3 [87–89]. Apparently, ArfGAP Hrb sorts VAMP7 into clathrin-coated vesicles at the plasma membrane. After endocytosis VAMP7 reaches recycling endosomes where AP3 mediates sorting of VAMP7 to late endosomes/lysosomes [88]. Similarly, the N-terminal domain of the SNARE vti1b binds to the clathrin adaptor EpsinR [90–92]. More complex mechanisms involving multiple factors have also emerged, which determine the intracellular localization of a given SNARE or mediate sorting of two SNAREs together [93,94].
In many cases, however, the molecular mechanisms of SNARE sorting are not known. SNAREs have to return to the donor compartment after fusion, thus utilizing vesicle trafficking routes as ‘passengers’, resulting the overlap of many different SNAREs in a given trafficking vesicle (see e.g. [95]), while they are en route to different destinations. Highly selective sorting mechanisms are thus required to distinguish individual SNAREs in the plane of the membrane. Using time-lapse imaging we have recently observed that the SNAREs syntaxin 6 and syntaxin 13 are differentially sorted, into trafficking vesicles at the early endosome, with one set containing only syntaxin 6 and a second set containing both SNAREs [66]. The underlying molecular mechanisms still need to be established.
Targeting and tethering
In contrast with sorting that recruits individual proteins or protein complexes to membrane subdomains and regulates their packaging into trafficking vesicles, targeting regulates the delivery of an entire trafficking organelle, with all of its components, to the correct intracellular destination. Targeting thus requires the recognition of, and bridging between, two distinct signals that are specific for the trafficking vesicle and for the acceptor membrane, respectively.
Our microinjection experiments have revealed that the SNARE-tethering factor axis is sufficient for the targeting of vesicles and for the recruitment of all factors needed for subsequent docking and fusion. They also have shown that the ability to direct vesicles to their target is restricted to subsets of SNAREs whereas others such as VAMP4 and vti1a, while clearly being specifically sorted, do not appear to participate in vesicle targeting [66]. Our findings confirm earlier observations showing that targeting is dependent on the N-terminal domains of the respective SNAREs. For example, overexpression of the N-terminal deletion mutant of syntaxin 3 that is necessary for trafficking of axonal targeting vesicles resulted in mis-targeting of the axonal cargos [96]. Similarly, deletion of Habc domain of syntaxin 1 reduced the number of the readily releasable pool of synaptic vesicles at a synapse [97]. Moreover, the fact that the tethering factor Vps13B only binds when both syntaxin 6 and syntaxin 13 are present shows that in this case coincidence detection is required, which may explain differential sorting of vesicle populations that both contain syntaxin 6 (Figure 4).
Figure 4. Interactions of SNAREs with tethering factors via their N-terminal domains, showing various levels of regulation.
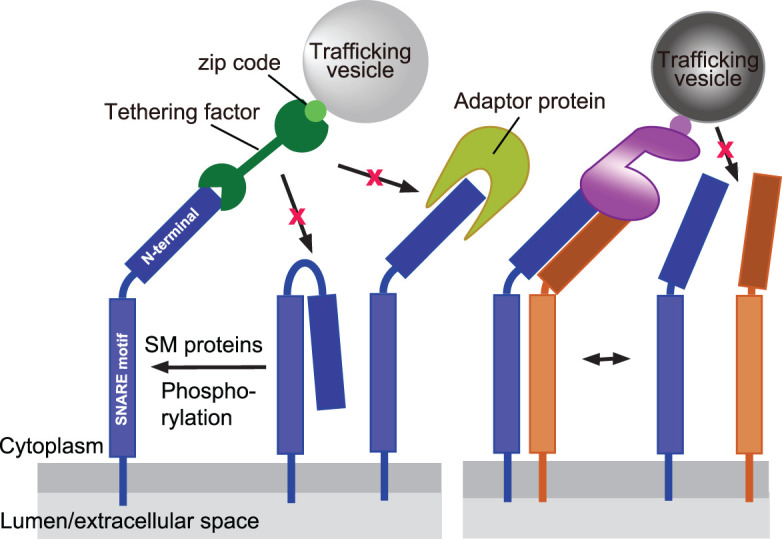
SNAREs may switch from an inactive closed conformation, with the N-terminal domain binding to the SNARE motif, to an active open conformation. This transition may be promoted by SM proteins, or in some cases also by posttranslational modifications. In the open conformation, the N-terminal domains may bind to tethering factors or to other adaptors in a mutually exclusive fashion. In some cases, association with another SNARE is required for the specific recruitment of a tethering factor. See text for details.
Although these data firmly establish that SNAREs on their own can function as ‘zip codes’ directing trafficking vesicles to the correct domain of the secretory pathway, they are not sufficient for achieving the required precision in targeting. Injected early endosomes were targeted to a more restricted population of endosomes than liposomes containing early endosomal SNAREs [80] that showed broader specificity. Most likely, the combination of Rab/PtdIns/SNARE-mediated recruitment of tethering factors is responsible for the final refinement of targeting.
Fusion
Although targeting and tethering are preceding fusion, it appears that these processes are generally linked, which may contribute to the targeting specificity in the secretory pathway. Above we have already discussed that SM-proteins that are needed for SNARE-mediated fusion are interacting with tethering factors or are even part of a tethering complex. Other examples include the Dsl tethering complex functioning in the docking and fusion of retrogradely transported COPI-coated vesicles with the endoplasmic reticulum. Here, the tethering proteins form a complex with the three Q-SNAREs required for fusion [10,98]. In this case, the tethering complex is formed on the target membrane, thus flagging the docking site for the incoming vesicle and ensuring that the SNAREs required for the subsequent fusion step are already in place. Such local recruitment of SNAREs at the prospective fusion site may also be needed since – except for neuronal exocytosis, the concentration of SNAREs in a given intracellular membrane is rather low; early endosomes, containing only few copies of the SNAREs needed for homotypic fusion [37].
Another example includes the protein Varp (Vps9 domain and ankyrin repeats containing protein), which is a Guanine Exchange Factor (GEF) for Rab21 [99]. Varp interacts with both the GDP-bound form of Rab21 and the longin domain of VAMP7 [99,100], yet another example for a direct link between SNAREs and Rabs. Varp binding to VAMP7 maintains VAMP7 in a closed and inactive conformation, preventing assembly with its partner SNAREs [101]. Knock-down of Varp decreases the colocalization of VAMP7 and Rab21 in the perinuclear area and also decreases neurite formation in PC12 cells [100]. Furthermore, in Rhodopsin transport carriers of Xenopus laevis photoreceptor cells, the longin domain of VAMP7 binds Rab11 and Rab8. Arf/Rab11 effector RAB11FIP3 did not directly bind to VAMP7, but completely inhibited Rab11a–VAMP7 interactions [102]. Apparently, Rab proteins regulate the conformation of VAMP7 and thus directly control its fusion activity.
Switching of SNAREs: by consecutive and/or competitive mechanisms?
Since SNAREs participate in several consecutive trafficking steps, it is evident that they must be tightly controlled by several layers of regulation in their various activities. But how are these different regulatory steps operating on a given SNARE coordinated? Or more precisely: how are SNAREs switched on or off as required for targeting signals? In contrast with the GTPases and PtdInsPx, there appears to be no common mechanism although the N-terminal domains seem to play a central role. It is conceivable that, depending on the position of a given SNARE in the trafficking pathway, different ‘effectors’ bind to a given SNARE. Interaction with a sorting adaptor may be favored when a SNARE is to be concentrated in a budding trafficking vesicle (e.g. associated with coat formation), which is followed by decoating and ‘freeing’ of the N-terminal domain for interaction with a targeting effector. We do not know how competition between effectors is controlled. It is conceivable that one effector blocks the accessibility of the N-terminal domains to other factors such as tethering proteins. Another possibility is to integrate proteins required for SNARE activation (SM proteins) into the tethering complex, thus making sure that the SNARE is only being able to become active once tethering has occurred between the correct membranes. Therefore, regulation between exposure and covering of N-terminal domain, and the regulation of assembly/disassembly of SNAREs on membrane would function as the switch of SNAREs during trafficking (Figure 4). Finally, it needs to be considered that SNAREs are conformationally dynamic and highly reactive molecules, and a high amount of energy is released upon SNARE assembly. Thus, capturing of SNAREs by effectors may also serve to stabilize them in a high-energy state until they are released for constructive trans-SNARE pairing at the correct time and the correct site to mediate fusion with the correct partners.
In conclusion, membrane targeting is controlled by multiple layers that are highly interconnected but exhibit a certain degree of redundancy, making the system robust and allow it to perform even in the presence of major disturbances. Indeed, a combination of consecutive and recursive steps, each of which being of moderate specificity and tolerant to variations in stoichiometry, with complexes being assembled when needed and disassembled when the task is completed may explain how complex functions such as vesicle trafficking in which thousands of molecules are participating are achieved with high fidelity.
Abbreviations
- GARP
Golgi-associated retrograde protein
- MTCs
multisubunit tethering complexes
- NSF
N-ethylmaleimide sensitive factor
- SM
Sec1/Munc18
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by JSPS KAKENHI Grant Number JP19K06559.
Open Access Statement
Open access for this article was enabled by the participation of Max Planck Digital Library in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with Max Planck Digital Library.
References
- 1.Cohen, F.S. and Melikyan, G.B. (2004) The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 199, 1–14 10.1007/s00232-004-0669-8 [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino, J.S. and Glick, B.S. (2004) The mechanisms of vesicle budding and fusion. Cell 116, 153–166 10.1016/S0092-8674(03)01079-1 [DOI] [PubMed] [Google Scholar]
- 3.Farré, J., Mahalingam, S.S., Proietto, M. and Subramani, S. (2019) Peroxisome biogenesis, membrane contact sites, and quality control. EMBO Rep. 20, e46864 10.15252/embr.201846864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellenrieder, L., Rampelt, H. and Becker, T. (2017) Connection of protein transport and organelle contact sites in mitochondria. J. Mol. Biol. 14, 2148–2160 10.1016/j.jmb.2017.05.023 [DOI] [PubMed] [Google Scholar]
- 5.Li, D., Shao, L., Chen, B.C., Zhang, X., Zhang, M., Mose, B.et al. (2015) ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349, aab3500 10.1126/science.aab3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valm, A.M., Cohen, S., Legant, W.R., Melunis, J., Hershberg, U., Wait, E.et al. (2017) Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 7656, 162–167 10.1038/nature22369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aridor, M. and Hannan, L.A. (2000) Tarffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic 1, 836–851 10.1034/j.1600-0854.2000.011104.x [DOI] [PubMed] [Google Scholar]
- 8.Behnia, R. and Munro, S. (2005) Organelle identity and the signposts for membrane traffic. Nature 7068, 597–604 10.1038/nature04397 [DOI] [PubMed] [Google Scholar]
- 9.Cai, H., Reinisch, K. and Ferro-Novick, S. (2007) Coats, tethers, rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 12, 671–682 10.1016/j.devcel.2007.04.005 [DOI] [PubMed] [Google Scholar]
- 10.Zink, S., Wenzel, D., Wurm, C.A. and Schmitt, H.D. (2009) A link between ER tethering and COP-I vesicle uncoating. Dev. Cell 17, 403–416 10.1016/j.devcel.2009.07.012 [DOI] [PubMed] [Google Scholar]
- 11.Homma, Y., Hiragi, S. and Fukuda, M. (2021) Rab family of small GTPases: an updated view on their regulation and functions. FEBS J. 288, 36–55 10.1111/febs.15453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuno-Yamasaki, E., Rivera-Molina, F. and Novick, P. (2012) GTPase networks in membrane traffic. Annu. Rev. Biochem. 81, 637–659 10.1146/annurev-biochem-052810-093700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jean, S. and Kiger, A.A. (2012) Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat. Rev. Mol. Cell Biol. 13, 463–470 10.1038/nrm3379 [DOI] [PubMed] [Google Scholar]
- 14.Poteryaev, D., Datta, S., Ackema, K., Zerial, M. and Spang, A. (2010) Identification of the switch in early-to-late endosome transition. Cell 141, 497–508 10.1016/j.cell.2010.03.011 [DOI] [PubMed] [Google Scholar]
- 15.Balla, T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki, T., Takasuga, S., Sasaki, J., Kofuji, S., Eguchi, S., Yamazaki, M.et al. (2009) Mammalian phosphoinositide kinases and phosphatases. Prog. Lipid Res. 48, 307–343 10.1016/j.plipres.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 17.Duex, J.E., Nau, J.J., Kauffman, E.J. and Weisman, L.S. (2006) Phosphoinositide 5-phosphatase Fig4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot. Cell 5, 723–731 10.1128/EC.5.4.723-731.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Paolo, G. and De Camilli, P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- 19.Traub, L.M. and Bonifacino, J.S. (2013) Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 5, a016790 10.1101/cshperspect.a016790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bone, L.N., Dayam, R.M., Lee, M., Kono, N., Fairn, G.D., Arai, H.et al. (2017) The acyltransferase LYCAT controls specific phosphoinositides and related membrane traffic. Mol. Biol. Cell 28, 161–172 10.1091/mbc.e16-09-0668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orso, G., Pendin, D., Liu, S., Tosetto, J., Moss, T.J., Faust, J.E., et al. (2009) Homotypic fusion of ER membranes requires the dynamin-like GTPase Atlastin. Nature 460, 978–983 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- 22.Ryu, J.K., Jahn, R. and Yoon, T.Y. (2016) Review: progresses in understanding N-ethylmaleimide sensitive factor (NSF) mediated disassembly of SNARE complexes. Biopolymers 105, 518–531 10.1002/bip.22854 [DOI] [PubMed] [Google Scholar]
- 23.Jahn, R. and Scheller, R.H. (2006) SNAREs: engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- 24.Sutton, R.B., Fasshauer, D., Jahn, R. and Brunger, A.T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolurion. Nature 395, 347–353 10.1038/26412 [DOI] [PubMed] [Google Scholar]
- 25.Rothman, J.E. (1994) Mechanisms of intracellular protein transport. Nature 372, 55–63 10.1038/372055a0 [DOI] [PubMed] [Google Scholar]
- 26.Pelham, H.R.B. (2001) SNAREs and the specificity of membrane fusion. Trends Cell Biol. 11, 99–101 10.1016/S0962-8924(01)01929-8 [DOI] [PubMed] [Google Scholar]
- 27.McNew, J.A., Parlatl, F., Fukuda, R., Johnston, R.J., Paz, K., Paumet, F.et al. (2000) Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature 407, 153–159 10.1038/35025000 [DOI] [PubMed] [Google Scholar]
- 28.Furukawa, N. and Mima, J. (2014) Multiple and distinct strategies of yeast SNAREs to confer the specificity of membrane fusion. Sci. Rep. 4, 4277 10.1038/srep04277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford, D.C. and Kavalali, E.T. (2015) Molecular underpinnings of synaptic vesicle pool heterogeneity. Traffic 16, 338–364 10.1111/tra.12262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoch, S., Deák, F., Königstorfer, A., Mozhayeva, M., Sara, Y., Südhof, T.C.et al. (2001) SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science 294, 1117–1122 10.1126/science.1064335 [DOI] [PubMed] [Google Scholar]
- 31.Hua, S.-Y., Raciborska, D.A., Trimble, W.S. and Charlton, M.P. (1998) Different VAMP/Synaptobrevin complexes for spontaneous and evoked transmitter release at the crayfish neuromuscular junction. J. Neurophysiol. 80, 3233–3246 10.1152/jn.1998.80.6.3233 [DOI] [PubMed] [Google Scholar]
- 32.Ramirez, D.M.O., Khvotchev, M., Trauterman, B. and Kavalali, E.T. (2012) Vti1a identifies a vesicle pool that preferentially recycles at rest and maintains spontaneous neurotransmission. Neuron 73, 121–134 10.1016/j.neuron.2011.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerona, R.R.L., Larsen, E.C., Kowalchyk, J.A. and Martin, T.F.J. (2000) The C terminus of SNAP25 is essential for Ca2+-dependent binding of synaptotagmin to SNARE complexes. J. Biol. Chem. 275, 6328–6336 10.1074/jbc.275.9.6328 [DOI] [PubMed] [Google Scholar]
- 34.Rossetto, O., Pirazzini, M. and Montecucco, C. (2014) Botulinum neurotoxins: genetic, structural and mechanistic insights. Nat. Rev. Microbiol. 12, 535–549 10.1038/nrmicro3295 [DOI] [PubMed] [Google Scholar]
- 35.Brandhorst, D., Zwilling, D., Rizzoli, S.O., Lippert, U., Lang, T. and Jahn, R. (2006) Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc. Natl Acad. Sci. U.S.A. 103, 2701–2706 10.1073/pnas.0511138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bethani, I., Lang, T., Geumann, U., Sieber, J.J., Jahn, R. and Rizzoli, S.O. (2007) The specificity of SNARE pairing in biological membranes is mediated by both proof-reading and spatial segregation. EMBO J. 26, 3981–3992 10.1038/sj.emboj.7601820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bethani, I., Werner, A., Kadian, C., Geumann, U., Jahn, R. and Rizzoli, S.O. (2009) Endosomal fusion upon SNARE knockdown is maintained by residual SNARE activity and enhanced docking. Traffic 10, 1543–1559 10.1111/j.1600-0854.2009.00959.x [DOI] [PubMed] [Google Scholar]
- 38.Sanyal, S., Tolar, L.A., Pallanck, L. and Krishnan, K.S. (2001) Genetic interaction between shibire and comatose mutations in Drosophila suggest a role for snap-receptor complex assembly and disassembly for maintenance of synaptic vesicle cycling. Neurosci. Lett. 311, 21–24 10.1016/S0304-3940(01)02125-5 [DOI] [PubMed] [Google Scholar]
- 39.Price, A., Seals, D., Wickner, W., Ungermann, C. and Wickner, B. (2000) The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J. Cell Biol. 148, 1231–1238 10.1083/jcb.148.6.1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer, A., Wickner, W. and Haas, A. (1996) Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell 85, 83–94 10.1016/S0092-8674(00)81084-3 [DOI] [PubMed] [Google Scholar]
- 41.Wickner, W. (2010) Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a rab, all dancing in a ring on yeast vacuoles. Annu. Rev. Cell Dev. Biol. 26, 115–136 10.1146/annurev-cellbio-100109-104131 [DOI] [PubMed] [Google Scholar]
- 42.Song, H., Torng, T.L., Orr, A.L., Brunger, A.T. and Wickner, W.T. (2021) Sec17/Sec18 can support membrane fusion without help from completion of SNARE zippering. eLife 10, e67578 10.7554/eLife.67578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang, T., Bruns, D., Wenzel, D., Riedel, D., Holroyd, P., Thiele, C.et al. (2001) SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 20, 2202–2213 10.1093/emboj/20.9.2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi, V., Banfield, D.K., Vacca, M., Dietrich, L.E.P., Ungermann, C., D'Esposito, M.et al. (2004) Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 29, 682–688 10.1016/j.tibs.2004.10.002 [DOI] [PubMed] [Google Scholar]
- 45.Daste, F., Galli, T. and Tareste, D. (2015) Structure and function of longin SNAREs. J. Cell Sci. 128, 4263–4272 10.1242/jcs.178574 [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y. and Hughson, F.M. (2021) Chaperoning SNARE folding and assembly. Annu. Rev. Biochem. 90, 581–603 10.1146/annurev-biochem-081820-103615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin, P., Gandasi, N.R., Arora, S., Omar-Hmeadi, M., Saras, J. and Barg, S. (2018) Syntaxin clusters at secretory granules in a munc18-bound conformation. Mol. Biol. Cell 29, 2700–2708 10.1091/mbc.E17-09-0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizo, J. and Südhof, T.C. (2012) The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices—guilty as charged? Annu. Rev. Cell Dev. Biol. 28, 279–308 10.1146/annurev-cellbio-101011-155818 [DOI] [PubMed] [Google Scholar]
- 49.Toonen, R.F.G. and Verhage, M. (2003) Vesicle trafficking: pleasure and pain from SM genes. Trends Cell Biol. 13, 177–186 10.1016/S0962-8924(03)00031-X [DOI] [PubMed] [Google Scholar]
- 50.Burd, C.G., Peterson, M., Cowles, C.R. and Emr, S.D. (1997) A novel sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol. Biol. Cell 8, 1089–1104 10.1091/mbc.8.6.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nichols, B.J., Holthuis, J.C.M. and Pelham, H.R.B. (1998) The Sec1p homologue Vps45p binds to the syntaxin Tlg2p. Eur. J. Cell Biol. 77, 263–268 10.1016/S0171-9335(98)80084-8 [DOI] [PubMed] [Google Scholar]
- 52.Dulubova, I., Yamaguchi, T., Gao, Y., Min, S.W., Huryeva, I., Südhof, T.C.et al. (2002) How Tlg2p/syntaxin 16 “snares” Vps45. EMBO J. 21, 3620–3631 10.1093/emboj/cdf381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen, E., Christoforidis, S., Uttenweiler-Joseph, S., Miaczynska, M., Dewitte, F., Wilm, M.et al. (2000) Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151, 601–612 10.1083/jcb.151.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ungermann, C. and Kümmel, D. (2019) Structure of membrane tethers and their role in fusion. Traffic 20, 479–490 10.1111/tra.12655 [DOI] [PubMed] [Google Scholar]
- 55.Peterson, M.R., Burd, C.G. and Emr, S.D. (1999) Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr. Biol. 9, 159–162 10.1016/S0960-9822(99)80071-2 [DOI] [PubMed] [Google Scholar]
- 56.Eathiraj, S., Pan, X., Ritacco, C. and Lambright, D.G. (2005) Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature 436, 415–419 10.1038/nature03798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Renzis, S., Sönnichsen, B. and Zerial, M. (2002) Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat. Cell Biol. 4, 124–133 10.1038/ncb744 [DOI] [PubMed] [Google Scholar]
- 58.Yu, I.M. and Hughson, F.M. (2010) Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 26, 137–156 10.1146/annurev.cellbio.042308.113327 [DOI] [PubMed] [Google Scholar]
- 59.Chia, P.Z.C. and Gleeson, P.A. (2014) Membrane tethering. F1000Prime Rep. 6, 74 10.12703/P6-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murray, D.H., Jahnel, M., Lauer, J., Avellaneda, M.J., Brouilly, N., Cezanne, A., et al. (2016) An endosomal tether undergoes an entropic collapse to bring vesicles together. Nature 537, 107–111 10.1038/nature19326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheung, P.Y.P., Limouse, C., Mabuchi, H. and Pfeffer, S.R. (2015) Protein flexibility is required for vesicle tethering at the Golgi. eLife 4, e12790 10.7554/eLife.12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li, F., Pincet, F., Perez, E., Giraudo, C.G., Tareste, D. and Rothman, J.E. (2011) Complexin activates and clamps SNAREpins by a common mechanism involving an intermediate energetic state. Nat. Struct. Mol. Biol. 18, 941–946 10.1038/nsmb.2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bröcker, C., Engelbrecht-Vandré, S. and Ungermann, C. (2010) Multisubunit tethering complexes and their role in membrane fusion. Curr. Biol. 20, R943–R952 10.1016/j.cub.2010.09.015 [DOI] [PubMed] [Google Scholar]
- 64.Willett, R., Kudlyk, T., Pokrovskaya, I., Schönherr, R., Ungar, D., Duden, R.et al. (2013) COG complexes form spatial landmarks for distinct SNARE complexes. Nat. Commun. 4, 1553 10.1038/ncomms2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong, M. and Munro, S. (2014) The specificity of vesicle traffic to the Golgi is encoded in the golgin coiled-coil proteins. Science 346, 1256898 10.1126/science.1256898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koike, S. and Jahn, R. (2019) SNAREs define targeting specificity of trafficking vesicles by combinatorial interaction with tethering factors. Nat. Commun. 10, 1608 10.1038/s41467-019-09617-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin, J.J., Crook, O.M., Borgeaud, A.C., Cattin-Ortolá, J., Peak-Chew, S., Breckels, L.M.et al. (2020) Spatial proteomics defines the content of trafficking vesicles captured by golgin tethers. Nat. Commun. 11, 5987 10.1038/s41467-020-19840-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong, W.J. and Lev, S. (2014) Tethering the assembly of SNARE complexes. Trends Cell Biol. 24, 35–43 10.1016/j.tcb.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 69.Laufman, O., Hong, W.J. and Lev, S. (2013) The COG complex interacts with multiple Golgi snares and enhances fusogenic assembly of SNARE complexes. J. Cell Sci. 126, 1506–1516 10.1242/jcs.122101 [DOI] [PubMed] [Google Scholar]
- 70.Shorter, J., Beard, M.B., Seemann, J., Barbara Dirac-Svejstrup, A. and Warren, G. (2002) Sequential tethering of golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J. Cell Biol. 157, 45–62 10.1083/jcb.200112127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dubuke, M.L. and Munson, M. (2016) The secret life of tethers: the role of tethering factors in SNARE complex regulation. Front. Cell Dev. Biol. 4, 42 10.3389/fcell.2016.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Travis, S.M., Damico, K., Yu, I.M., McMahon, C., Hamid, S., Ramirez-Arellano, G.et al. (2020) Structural basis for the binding of SNAREs to the multisubunit tethering complex Dsl1. J. Biol. Chem. 295, 10125–10135 10.1074/jbc.RA120.013654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pérez-Victoria, F.J. and Bonifacino, J.S. (2009) Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-Golgi network. Mol. Cell. Biol. 29, 5251–5263 10.1128/MCB.00495-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Conibear, E., Cleck, J.N. and Stevens, T.H. (2003) Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell 14, 1610–1623 10.1091/mbc.e02-10-0654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siniossoglou, S. and Pelham, H.R.B. (2002) Vps51p links the VFT complex to the SNARE Tlg1p. J. Biol. Chem. 277, 48318–48324 10.1074/jbc.M209428200 [DOI] [PubMed] [Google Scholar]
- 76.Shestakova, A., Suvorova, E., Pavliv, O., Khaidakova, G. and Lupashin, V. (2007) Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. J. Cell Biol. 179, 1179–1192 10.1083/jcb.200705145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu, H., Jun, Y., Thompson, J., Yates, J. and Wickner, W. (2010) HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 29, 1948–1960 10.1038/emboj.2010.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mima, J., Hickey, C.M., Xu, H., Jun, Y. and Wickner, W. (2008) Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 27, 2031–2042 10.1038/emboj.2008.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.D'Agostino, M., Risselada, H.J., Endter, L.J., Comte-Miserez, V. and Mayer, A. (2018) SNARE -mediated membrane fusion arrests at pore expansion to regulate the volume of an organelle. EMBO J. 37, e99193 10.15252/embj.201899193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koike, S. and Jahn, R. (2017) Probing and manipulating intracellular membrane traffic by microinjection of artificial vesicles. Proc. Natl Acad. Sci. U.S.A. 114, E9883–E9892 10.1073/pnas.1713524114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pérez-Victoria, F.J., Schindler, C., Magadán, J.G., Mardones, G.A., Delevoye, C., Romao, M.et al. (2010) Ang2/fat-free is a conserved subunit of the Golgi-associated retrograde protein complex. Mol. Biol. Cell 21, 3386–3395 10.1091/mbc.e10-05-0392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abascal-Palacios, G., Schindler, C., Rojas, A.L., Bonifacino, J.S. and Hierro, A. (2013) Structural basis for the interaction of the Golgi-associated retrograde protein complex with the t-SNARE Syntaxin 6. Structure 21, 1698–1706 10.1016/j.str.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De, M., Oleskie, A.N., Ayyash, M., Dutta, S., Mancour, L., Abazeed, M.E.et al. (2017) The Vps13p-Cdc31p complex is directly required for TGN late endosome transport and TGN homotypic fusion. J. Cell Biol. 216, 425–439 10.1083/jcb.201606078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bröcker, C., Kuhlee, A., Gatsogiannis, C., Kleine Balderhaar, H.J., Hönscher, C., Engelbrecht-Vandré, S.et al. (2012) Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl Acad. Sci. U.S.A. 109, 1991–1996 10.1073/pnas.1117797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peden, A.A., Park, G.Y. and Scheller, R.H. (2001) The Di-leucine motif of vesicle-associated membrane protein 4 is required for its localization and AP-1 binding. J. Biol. Chem. 276, 49183–49187 10.1074/jbc.M106646200 [DOI] [PubMed] [Google Scholar]
- 86.Bonifacino, J.S. and Traub, L.M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 10.1146/annurev.biochem.72.121801.161800 [DOI] [PubMed] [Google Scholar]
- 87.Pryor, P.R., Jackson, L., Gray, S.R., Edeling, M.A., Thompson, A., Sanderson, C.M.et al. (2008) Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134, 817–827 10.1016/j.cell.2008.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martinez-Arca, S., Rudge, R., Vacca, M., Raposo, G., Camonis, J., Proux-Gillardeaux, V., et al. (2003) A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc. Natl Acad. Sci. U.S.A. 100, 9011–9016 10.1073/pnas.1431910100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kent, H.M., Evans, P.R., Schäfer, I.B., Gray, S.R., Sanderson, C.M., Luzio, J.P.et al. (2012) Structural basis of the intracellular sorting of the SNARE VAMP7 by the AP3 adaptor complex. Dev. Cell 22, 979–988 10.1016/j.devcel.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chidambaram, S., Müllers, N., Wiederhold, K., Haucke, V. and Von Mollard, G.F. (2004) Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J. Biol. Chem. 279, 4175–4179 10.1074/jbc.M308667200 [DOI] [PubMed] [Google Scholar]
- 91.Hirst, J., Miller, S.E., Taylor, M.J., Von Mollard, G.F. and Robinson, M.S. (2004) EpsinR is an adaptor for the SNARE protein Vti1b. Mol. Biol. Cell 15, 5593–5602 10.1091/mbc.e04-06-0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miller, S.E., Collins, B.M., McCoy, A.J., Robinson, M.S. and Owen, D.J. (2007) A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature 450, 570–574 10.1038/nature06353 [DOI] [PubMed] [Google Scholar]
- 93.Bowman, S., Le, L., Zhu, Y., Harper, D.C., Sitaram, A., Theos, A.C.et al. (2021) A BLOC-1-AP-3 super-complex sorts a cis-SNARE complex into endosome-derived tubular transport carriers. J. Cell Biol. 220, e202005173 10.1083/jcb.202005173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao, G. and Banfield, D.K. (2020) Multiple features within the syntaxin Sed5p mediate its Golgi localization. Traffic 21, 274–296 10.1111/tra.12720 [DOI] [PubMed] [Google Scholar]
- 95.Takamori, S., Holt, M., Stenius, K., Lemke, E.A., Grønborg, M., Riedel, D., et al. (2006) Molecular anatomy of a trafficking organelle. Cell 127, 831–846 10.1016/j.cell.2006.10.030 [DOI] [PubMed] [Google Scholar]
- 96.Hoo, L.S., Banna, C.D., Radeke, C.M., Sharma, N., Albertolle, M.E., Low, S.H.et al. (2016) The SNARE protein syntaxin 3 confers specificity for polarized axonal trafficking in neurons. PLoS ONE 11, e0163671 10.1371/journal.pone.0163671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou, P., Pang, Z.P., Yang, X., Zhang, Y., Rosenmund, C., Bacaj, T.et al. (2013) Syntaxin-1 N-peptide and H abc -domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 32, 159–171 10.1038/emboj.2012.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Meiringer, C.T.A., Rethmeier, R., Auffarth, K., Wilson, J., Perz, A., Barlowe, C.et al. (2011) The Dsl1 protein tethering complex is a resident endoplasmic reticulum complex, which interacts with five soluble NSF (N-Ethylmaleimide-sensitive factor) attachment protein receptors (SNAREs): implications for fusion and fusion regulation. J. Biol. Chem. 286, 25039–25046 10.1074/jbc.M110.215327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang, X., He, X., Fu, X.Y. and Chang, Z. (2006) Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J. Cell Sci. 119, 1053–1062 10.1242/jcs.02810 [DOI] [PubMed] [Google Scholar]
- 100.Burgo, A., Sotirakis, E., Simmler, M.C., Verraes, A., Chamot, C., Simpson, J.C.et al. (2009) Role of Varp, a Rab21 exchange factor and TI-VAMP/VAMP7 partner, in neurite growth. EMBO Rep. 10, 1117–1124 10.1038/embor.2009.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schäfer, I.B., Hesketh, G.G., Bright, N.A., Gray, S.R., Pryor, P.R., Evans, P.R.et al. (2012) The binding of varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation. Nat. Struct. Mol. Biol. 19, 1300–1309 10.1038/nsmb.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kandachar, V., Tam, B.M., Moritz, O.L. and Deretic, D. (2018) An interaction network between the SNARE VAMP7 and Rab GTPases within a ciliary membrane-targeting complex. J. Cell Sci. 131, jcs222034 10.1242/jcs.222034 [DOI] [PMC free article] [PubMed] [Google Scholar]