Abstract
Flowering (inflorescence formation) of the grass Lolium temulentum is strictly regulated, occurring rapidly on exposure to a single long day (LD). During floral induction, L. temulentum differs significantly from dicot species such as Arabidopsis in the expression, at the shoot apex, of two APETALA1 (AP1)-like genes, LtMADS1 and LtMADS2, and of L. temulentum LEAFY (LtLFY). As shown by in situ hybridization, LtMADS1 and LtMADS2 are expressed in the vegetative shoot apical meristem, but expression increases strongly within 30 h of LD floral induction. Later in floral development, LtMADS1 and LtMADS2 are expressed within spikelet and floret meristems and in the glume and lemma primordia. It is interesting that LtLFY is detected quite late (about 12 d after LD induction) within the spikelet meristems, glumes, and lemma primordia. These patterns contrast with Arabidopsis, where LFY and AP1 are consecutively activated early during flower formation. LtMADS2, when expressed in transgenic Arabidopsis plants under the control of the AP1 promoter, could partially complement the organ number defect of the severe ap1-15 mutant allele, confirming a close relationship between LtMADS2 and AP1.
In Arabidopsis and snapdragon (Antirrhinum majus), two orthologous genes, LEAFY/FLORICAULA (LFY/FLO) and APETALA1/SQUAMOSA (AP1/SQUA), are important for the initiation of floral development. The flo mutants mostly lack flowers and produce secondary inflorescences in positions normally occupied by solitary flowers, with the secondary inflorescences repeating the pattern of the main inflorescence (Carpenter and Coen, 1990). The squa mutants have a similar phenotype as flo mutants, but flowers are produced more often than in flo mutants (Huijser et al., 1992). The lfy mutants also show flower-to-shoot conversions, but similarly to squa mutants produce flowers more often than flo mutants. These flowers lack petals and stamens and contain leaf-like organs, indicating partial shoot character (Schultz and Haughn, 1991). The weakest phenotypes are seen in ap1 mutants, in which flower-to-shoot conversions are relatively rare. Instead, flowers have leaf-like organs in the first whorl, with secondary flowers in their axils (Irish and Sussex, 1990). The increased proliferation of floral meristems is more pronounced when ap1 mutations are combined with mutations in the AP1 paralogs CAULIFLOWER (CAL) and FRUITFULL (FUL; Bowman et al., 1993; Ferrándiz et al., 2000). A more severe phenotype is also seen in lfy ap1 double mutants, in which all flowers are transformed into shoot-like structures, and in combinations of weak flo and squa mutations (Huala and Sussex, 1992; Weigel et al., 1992; Carpenter et al., 1995).
LFY/FLO and AP1/SQUA are strongly expressed in floral meristems, consistent with a direct role in promoting floral fate. This has been further demonstrated with transgenic plants in which either gene is overexpressed from the constitutive 35S viral promoter. These plants flower early, lateral inflorescence shoots are often transformed into solitary flowers, and the shoot apex, which in wild-type Arabidopsis remains indeterminate, terminates in a flower (Mandel and Yanofsky, 1995a; Weigel and Nilsson, 1995). The early-flowering phenotype indicates that both genes are targets of upstream floral inductive pathways, which has been confirmed in a variety of genetic and molecular studies (Simpson et al., 1999; Blázquez and Weigel., 2000). LFY and AP1 in turn bind directly to the promoters of downstream homeotic genes and activate their expression (Hill et al., 1998; Parcy et al., 1998; Tilly et al., 1998; Busch et al., 1999). There are no LFY-related genes in the Arabidopsis genome, and the DNA-binding protein encoded by LFY is unrelated to other classes of transcription factors (Weigel et al., 1992; Parcy et al., 1998). In contrast, AP1 belongs to the family of MADS box genes, many of which encode transcription factors regulating different aspects of flower development (Mandel et al., 1992). The majority of plant MADS domain proteins contain three conserved regions, the MADS, I, and K domains (together the M-I-K region); however, even between orthologs, the C-terminal domain can be highly divergent (Theissen et al., 1996).
Several monocot orthologs of dicot homeotic genes, which control organ identity within flowers, have recently been characterized (Mena et al., 1995; Ambrose et al., 2000). In contrast, less is known about monocot genes controlling floral meristem identity. Expression of RFL, a rice (Oryza sativa) homolog of FLO/LFY, has been reported to deviate from that of its dicot counterparts, and based on this finding a role in panicle branching has been proposed (Kyozuka, et al., 1998). An AP1-related gene from rice, OsMADS1, has been characterized by transgenic and mutant analysis (Jeon et al., 2000). However, OsMADS1 is more closely related to the SEPALLATA (SEP) genes of Arabidopsis, and its mutant phenotype, with defects in all floral organs except glumes, is similar to that of the SEP genes as well (Jeon et al., 2000; Pelaz et al., 2000).
Here, we report the isolation and functional characterization of LFY- and AP1-related genes from the long-day (LD) grass Lolium temulentum. Studies of the timing and localization of expression of LFY- and AP1-related genes at the shoot apex of L. temulentum has revealed both similarities and differences between these genes and their dicot homologs. Transgenic studies show that at least one of the AP1-related genes can partially substitute for its Arabidopsis homolog.
RESULTS
Characterization of Two AP1-Related Genes from L. temulentum
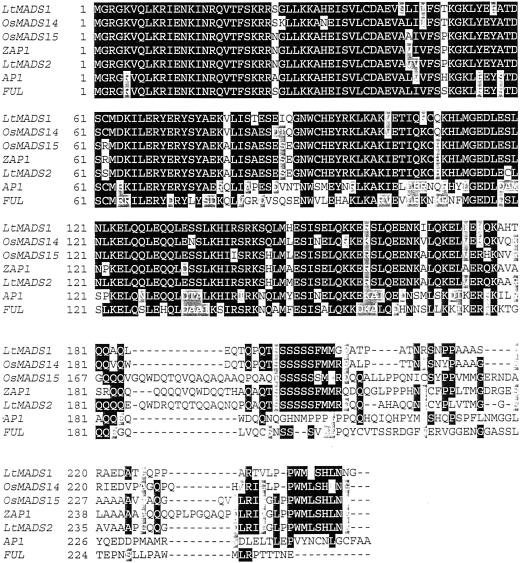
cDNAs of two genes termed L. temulentum MADS (LtMADS1 and LtMADS2; GenBank accession nos. AF035378 and AF035379) were isolated from a PCR-based cDNA library constructed from L. temulentum LD III shoot apex tissue. This library was screened using two 129-bp L. temulentum MADS probes that had been isolated from L. temulentum genomic DNA using primers described in “Materials and Methods.” At the RNA level LtMADS1 and LtMADS2 were 79% identical. The encoded proteins share 88% similar amino acids and 73% identical amino acids. An alignment of LtMADS1 and LtMADS2 with other MADS domain proteins from the AP1/SQUA clade is shown in Figure 1.
Figure 1.
Alignment of the predicted amino acid sequences for LtMADS1 and LtMADS2 with some other members of the AP1/SQUA branch: Arabidopsis, AP1 (accession no. Z16421) and FUL (accession no. U33473); Oryza sativa, OsMADS14 (accession no. AF058697) and OsMADS15 (accession no. AF058698); and Zea mays, ZAP1 (accession no. L46400). Dashes indicate gaps to maximize alignment. Amino acids that are identical in four or more of the proteins are blocked. The MADS box is amino acids 1 through 60, the I region is amino acids 61 through 93, and the K box is amino acids 94 through 170.
Fragments specific for the 3′ ends including the untranslated regions (UTR) of each of LtMADS1 and LtMADS2 hybridized to single bands in a genomic DNA gel blot, indicating that they are single-copy genes (data not shown).
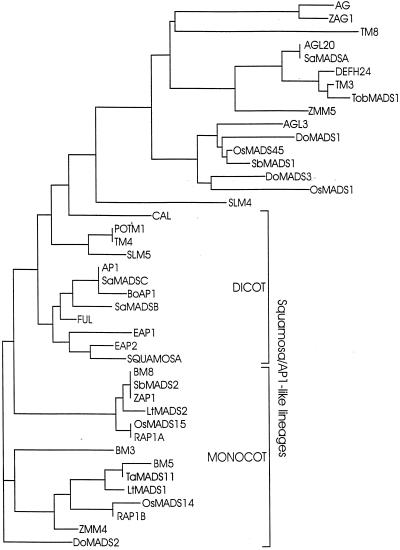
In comparisons involving a larger number of AP1-related genes, the phylogenetic trees were similar for alignments of amino acids over the four sequence regions detailed in “Materials and Methods.” The consensus of the most parsimonious tree for the MADS domain data set is presented in Figure 2. This tree matched results obtained by Theissen et al. (2000) but did not include sequences for several monocots. Up-to-date information on all sequences and their accession numbers are available on a Web site listed by Theissen et al. (2000).
Figure 2.
Phylogenetic tree based on complete MADS box domain sequences of selected members of the plant MADS box gene family. Apart from including more recent sequences the analysis and subfamily designations were based on Theissen et al. (1996). Sequences: Arabidopsis (AP1, CAL, FUL, AGL3, AG, and AGL20); snapdragon (SQUA and DEFH24); barley (Hordeum vulgare; BM3, BM5, and BM8); Brassica oleracea (BoAP1); Dendrobium globulus (DoMADS1, DoMADS2, and DoMADS3); Eucalyptus sp. (EAP1 and EAP2); L. temulentum (LtMADS1 and LtMADS2); maize (Zea mays; ZAG1, ZAP1, ZMM4, and ZMM5); potato (Solanum tuberosum; POTM1); rice (OsMADS1, OsMADS14 [also known as RAP1B], OsMADS15 [possibly the same as RAP1A], and OsMADS45); Silene latifolia (SLM4 and SLM5); Sinapis alba (SaMADSA, SaMADSB, and SaMADSC); sorghum (Sorghum bicolor; SbMADS1 and SbMADS2); tobacco (Nicotiana tabacum; TobMADS1), tomato (Lycopersicum esculentum; TM3, TM4, and TM8), and wheat (Triticum aestivum; TaMADS11; accession no. AB007504).
LtMADS1 and LtMADS2 are closely related members of the monocot AP1/SQUA branch of the plant MADS box gene family (Fig. 2), which includes BM3, BM5, and BM8 of barley; ZAP1 and ZMM4 of maize (for the latter, only partial sequence is available); and OsMADS14/RAP1B and OsMADS15/RAP1A from rice (Fischer et al., 1995; Mena et al., 1995; Theissen et al., 1996 and EST T12733; Moon et al., 1999; Kyozuka et al., 2000; Schmitz et al., 2000).
LtMADS2 is 88% identical to ZAP1 (Figs. 1 and 2) and has only six amino acid changes within the M-I-K region (the first 170 amino acid residues). In this same region, LtMADS1 has 16 differences compared with ZAP1. The high degree of sequence similarity between barley BM8, maize ZAP1, L. temulentum LtMADS2, and OsMADS15 of rice suggests that they are orthologs (RAP1A appears to be the same gene as OsMADS15). Likely LtMADS1 orthologs are BM5, OsMADS14/RAP1B, and ZMM4. Some of the AP1/SQUA-related proteins in dicots have a complete or partial prenylation motif at the carboxy terminus (Yalovsky et al., 2000), but this is absent from all known monocot AP1/SQUA-like proteins, including LtMADS1 and LtMADS2.
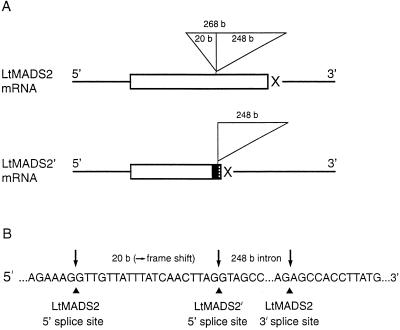
An alternatively spliced version of LtMADS2, designated LtMADS2′, was also isolated. This cDNA had a 20-bp insertion after the sequence coding for the K box as shown in Figure 3. Comparison with genomic DNA sequence in this region showed that the insertion resulted from differential processing of intron 4. This intron has two splice donor sites, separated by 20 bases, and a single acceptor site. The LtMADS2 transcript is produced when the entire intron of 268 bp is removed and contains an ORF with 261 codons. The LtMADS2′ transcript results when the downstream donor site is used, adding a 20-bp insertion, which changes the reading frame (Fig. 3).
Figure 3.
Scheme of alternative splicing events used to generate LtMADS2 and LtMADS2′ transcripts. A, The boxed sequence indicates the open reading frame (ORF) with stop codons indicated with an X and UTR as lines. Changes in the amino acid sequence of the putative translated protein from the LtMADS2′ transcript compared with the LtMADS2 transcript is indicated by the black (intron) and stippled boxes (frameshifted original ORF). Spliced introns are shown as triangles. B, Nucleotide sequence at the intron/exon 4 junction. Intron sequence was determined by genomic sequencing. Putative splice sites for LtMADS2 and LtMADS2′ transcripts are indicated with arrows. b, Bases.
These splice sites are conserved in AP1 and SQUA (Huijser et al., 1992; Mandel et al., 1992) and, although alternative splicing occurs in a Eucalyptus globulus AP1 homolog (EAP2), it occurs in intron 7 (Kyozuka et al., 1997).
L. temulentum LFY
LtLFY (GenBank accession no. AF321273) is almost identical at the nucleotide level to an LFY ortholog from Lolium perenne (C. Anderson, unpublished data). The encoded protein is highly similar to RFL from rice, with 84% amino acid identity (Kyozuka et al., 1998). Amino acid identity to Arabidopsis LFY is 54% (Weigel et al., 1992). As noticed before (Frohlich and Parker, 2000), the LFY homolog from common rush (Juncus effusus) is divergent from grass LFY homologs, it shares only 56% identical amino acid identity with LtLFY. As with other LFY orthologs, the carboxy-terminal region, which binds DNA (M.A. Busch and D.W., unpublished data) is more highly conserved than the amino terminal region. DNA gel-blot analysis showed LtLFY was present as a single-copy gene in L. temulentum.
Expression Patterns of LtMADS1, LtMADS2, and LtLFY
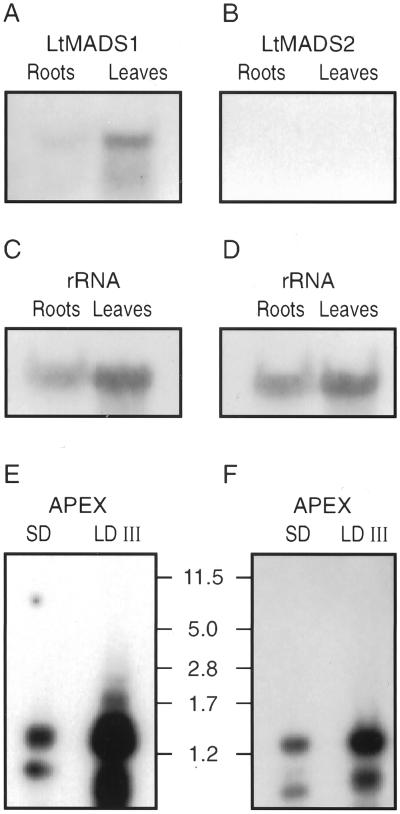
When blots of total RNA were probed with gene-specific probes, LtMADS1 but not LtMADS2 RNA was detected in leaves and roots (Fig. 4). At the shoot apex, detection of LtMADS1 and LtMADS2 transcripts was only possible after PCR amplification. Lower bands evident in these PCR products may represent alternative splice forms, but this has not yet been tested. Figure 4, E and F show that, compared with SD apices, expression of both genes increased within 30 h of the end of a single LD exposure. The expression of a control tubulin gene (see “Materials and Methods”) was not different between day length treatments.
Figure 4.
Gel-blot analysis of LtMADS1 and LtMADS2 expression in leaves, roots, and shoot apices of L. temulentum using the same gene specific probes for LtMADS1 (A) and LtMADS2 (B) as for in situ hybridization. Total RNA (20 μg) from leaves or roots was blotted and hybridized (A and B), then both blots were stripped and reprobed (C) and with a 9-kb wheat 26S rRNA clone, pTA71 (D; Gerlach and Bedbrook, 1979). For analysis of shoot apex mRNAs, PCR-based cDNA product (virtual northern) from mRNA of short day (SD; vegetative) or LD III (florally induced) apices was hybridized with the gene specific probe for LtMADS1 (E) or LtMADS2 (F). Sizes in kb are indicated between the blots.
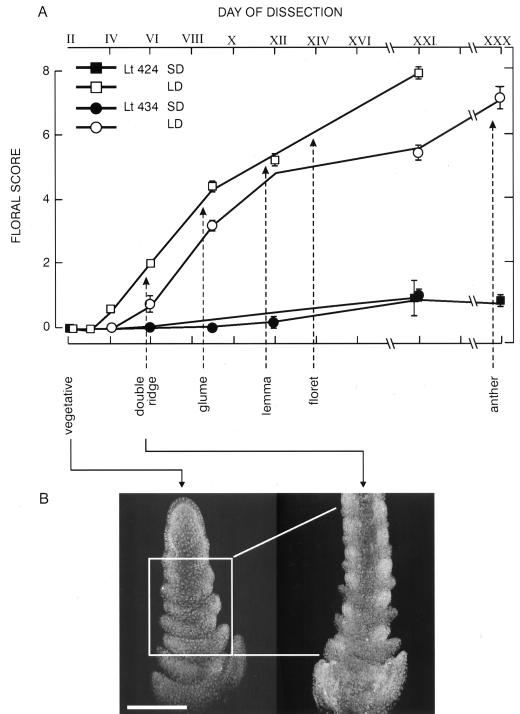
To provide a basis for relating mRNA expression and flowering, the timing of floral development was examined in two independent experiments. The SD apices (solid symbols) remained vegetative over the 30-d period of the experiment (Fig. 5A). The first morphologically definitive sign of flowering was the formation of double ridges, corresponding to developing spikelet sites overtopping regressing leaf primordia (stage 2), on Days V through VII. Thereafter, floral development progressed rapidly at least until floret formation at about Day XIV, timing comparable to our many earlier studies with L. temulentum (see McDaniel et al., 1991). In situ hybridization using longitudinal sections of apices showed that changes in LtMADS1 expression occurred by the time that flower formation was first visible on Day V (Fig. 5B). The three-dimensional characteristics of this early developmental change are illustrated here by a multidimensional reconstruction based on stacking/combining the digitized images of up to five sequential 10-μm-thick sections.
Figure 5.
A, Floral score, assessed by dissection, of L. temulentum shoot apices as a function of time after a single inductive LD (Day II). Values are means ± se, n = 10; some error bars are hidden by the symbols. Vertical arrows are positioned to indicate developmental stages. A three-dimensional pattern of expression of LtMADS1 mRNA (B) obtained by in situ hybridization was generated by apex reconstructions obtained by stacking/combining the images from four to five longitudinal sections up to the median one for a vegetative (SD) or LD IV early double-ridge stage apex (single median images shown in Fig. 6, B and C, respectively). Images were digitized and cropped using Adobe Photoshop and stacked using Confocal Assistant 4.02. The bar is 200 μm.
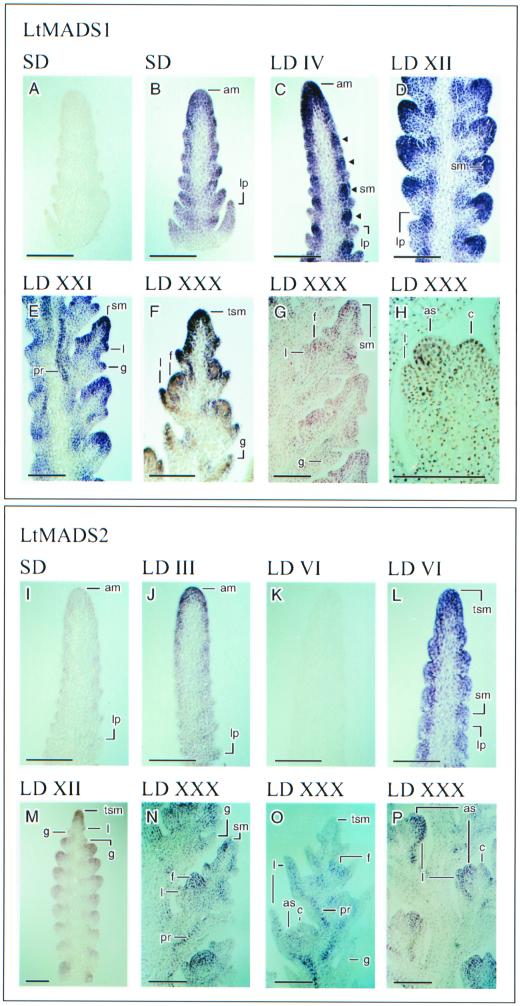
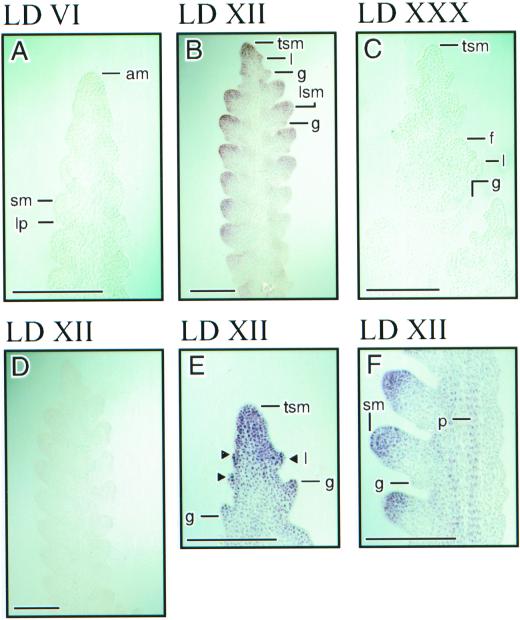
A detailed time course of LtMADS1, LtMADS2, and LtLFY expression in the single median longitudinal shoot apex sections is shown in Figures 6 and 7. Up to eight separate apices were sectioned and examined for each gene at each time, the results being reproducible between hybridizations. LtMADS1 and LtMADS2 were expressed in the shoot apical meristem prior to and at all stages following LD floral induction (Fig. 6; compare with Fig. 4). In contrast to the wide temporal range during which LtMADS1 and LtMADS2 were expressed, LtLFY was only expressed at the advanced double-ridge and glume stages of development (LD XII apex in Fig. 7). LtLFY was not detectable at earlier or later times and was not expressed in the vegetative apex (not shown). The expression of LtMADS1 was greater than that of LtMADS2 at all stages analyzed, the LtMADS2 probe requiring 2 d for color development, LtMADS1 requiring only 8 h at an equal probe concentration (among other things, equal sensitivity of the riboprobes is assumed). There was little or no hybridization with sense control probes for LtMADS1 (Fig. 6A, SD apex), LtMADS2 (Fig. 6K, LD VI apex), and LtLFY (Fig. 7D, LD XII apex).
Figure 6.
Expression of LtMADS1 and LtMADS2 RNA in L. temulentum shoot apices, assessed by in situ hybridization. All sections were hybridized with the antisense riboprobe except for A and K, which show representative sections hybridized with a sense control riboprobe. All sections were hybridized and processed together; therefore, the intensity of antisense probe staining reflects the expression of the genes. Apex stages: vegetative shoot apices (SD; A, B, I, and J); early double ridge (LD IV; C, K, and L); lateral spikelet sites on the flanks of the shoot apex at glume/lemma stage (LD XII; D and M) or the floret stage (LD XXI; E); terminal spikelet site subtended by two glume primordia with three floret meristems positioned alternately along the terminal spikelet axis subtended by lemma primordia (LD XXX; F and O); three lateral spikelet sites at floret stage (LD XXX; G and N); and a lemma, anterior stamen, and carpel primordium (left to right) on the flank of a floret site (LD XXX; H and P). The bars are 50 μm. am, Apical meristem; as, anterior stamen primordium; c, carpel; f, floret; g, glume; l, lemma; lp, leaf primordium; pr, provascular strand; sm, spikelet meristem; tsm, terminal spikelet meristem.
Figure 7.
Expression of LtLFY RNA in L. temulentum shoot apices assessed by in situ hybridization. Sections A through C, E, and F were hybridized with the antisense riboprobe. A representative section hybridized with the sense control riboprobe is shown in D. Magnified images of B are shown in E and F. The bars are 50 μm. Apex stages and other conditions as for Figure 6.
Increase in cell density potentially could give apparent increases in expression of these genes. However, this suggestion is clearly inappropriate for LtLFY given the limited time of its expression and the fact that cell density altered both prior to and after that time (Knox and Evans, 1966). A similar argument holds for expression patterns of the other two genes.
In the vegetative (SD) shoot apex, LtMADS1 was predominantly expressed in the epidermal and subepidermal layers (three or four cell layers deep) as well as in the tips of the developing leaf primordia (Fig. 6B), whereas LtMADS2 was expressed at a low level in the apical dome of the shoot apex (to a depth of three cell layers), but was largely excluded from the leaf primordia (Fig. 6I). Expression in leaf primordia of LtMADS1 but not of LtMADS2 is supported by the RNA gel-blot analysis (Fig. 4).
The first changes in expression pattern during inflorescence development were at LD IV for LtMADS1 and at LD III for LtMADS2. LtMADS1 was detected in spikelet sites forming in the axils of existing leaf primordia, whereas LtMADS2 expression extended into additional cell layers within the apical dome and into the epidermal and subepidermal layers along the length of the shoot apex. At the early double-ridge stage (Fig. 6C, LD IV), LtMADS1 expression remained concentrated around the edge of the shoot apical meristem in epidermal and subepidermal layers. However, different levels of LtMADS1 expression defined three distinct regions within the shoot apex. Highest levels were detected in the dome of the shoot apex as well as the developing spikelet primor dia (circular zones indicated with arrowheads), lower levels in the leaf primordia subtending the spikelet, and lowest levels in the center or core of the shoot apical meristem. At a somewhat later stage, LtMADS2 expression was only detected in the shoot apical meristem and within the spikelet primordia, with little expression in the regressing leaf primordia (Fig. 6L, LD VI apex).
At the advanced double-ridge stage, between LD IX (data not shown) and LD XII (Fig. 6M), each spikelet site (including the terminal dome) was elongated and more like the tip of the original SD apex. At this stage, LtMADS1 and LtMADS2 expression became localized to the tip (especially the abaxial side) of each spikelet (Fig. 6, C and M). Also, the expression of both genes was reduced in the region of the spikelet meristem nearest the rachis (adaxial side of the spikelet site; Fig. 6, D and M). From lemma stage until at least anther stage, LtMADS1 and LtMADS2 expression was localized to the epidermal and subepidermal layers (about three cell layers deep) of the apical dome of each spikelet (Fig. 6E, LD XXI apex; Fig. 6M, LD XXX apex). LtLFY expression was first detectable following the advanced double-ridge stage of development and it colocalized with that of LtMADS2 on the abaxial side of the spikelet site (Fig. 7B, LDXII apex).
In addition to being expressed in the apical dome of the shoot apex and spikelet sites, LtMADS1 (Fig. 6E) and LtMADS2 (Fig. 6M) were expressed at the tips of developing glume primordia, which are likely the counterparts of cauline leaf or bract primordia in dicots. LtLFY was also detected in the glume and lemma primordia (see Fig. 7, D and E). All three genes also showed provascular expression, with LtMADS1 and LtLFY being expressed, albeit transiently, in the core of the shoot apex at stages later than advanced double ridges (Figs. 6E and 7E). LtMADS1 and LtMADS2 were also detected in the provascular strand of the sterile glumes as they elongated (Fig. 6, F and N).
Whereas LtLFY expression was not detectable in either vegetative apices (not shown) or during and following floret meristem initiation (Fig. 7F), LtMADS1 and LtMADS2 were expressed throughout floret meristems, which form alternately along the rachilla axis of the spikelet (Fig. 6, F, G, and N, LD XXX apices). Both transcripts were present in the lemma, stamen, and carpel primordia. LtMADS1 expression persisted in the reproductive organ primordia, but at a lower level, until the latest stage examined (anther stage; see LD XXX apices in Fig. 6H), In contrast, LtMADS2 expression was first excluded from the carpel primordia (Fig. 6P) and later from the stamens, but persisted in the lemmas (Fig. 6O). It was difficult to adequately determine gene expression patterns in the palea and lodicule primordia because of their small size.
Activity of LtMADS1 and LtMADS2 in Arabidopsis
To determine whether the L. temulentum AP1-related genes have similar activities as Arabidopsis AP1 in vivo, we transformed into the Arabidopsis ap1-15 mutant LtMADS1, LtMADS2, and LtMADS2′ cDNAs under control of the 1.7-kb Arabidopsis AP1 promoter. The 1.7-kb promoter fragment has been previously shown to drive reporter gene expression that mimics activation of the endogenous gene upon floral induction (Hempel et al., 1997, 1998).
Wild-type flowers have four sepals in the first whorl, four petals in the second whorl, six stamens in the third whorl, and a gynoecium comprised of two fused carpels in the fourth whorl. ap1-15 is a likely null allele because it has a deletion within the MADS box that should truncate the protein (M. Yanofsky, personal communication). The ap1-15 mutant flowers typically have two or three leaf-like bracts in the first whorl. Second-whorl organs are either absent or petaloid bracts. The third whorl is largely normal, but there are often fewer than the canonical number of six stamens. The fourth whorl is the only whorl in which organ number is unaffected, and it consists as in wild type of two fused carpels. In addition, first-whorl bracts may produce secondary flowers, or occasionally secondary inflorescences, in the axils of first-whorl bracts (Table I; Bowman et al., 1993).
Table I.
Phenotype of ap1-15 mutants expressing LtMADS1, LtMADS2, or LtMADS2′ under the control of the AP1 promoter
| Transgene
|
||||
|---|---|---|---|---|
| None | LtMADS1 | LtMADS2 | LtMADS2′ | |
| Whorl 1 | ||||
| Total organs | 2.2 ± 0.3 | 2.5 ± 0.9 | 4.5 ± 0.3 | 2.8 ± 0.3 |
| Sepals | 0.0 | 0.2 ± 0.8 | 0.0 | 0.0 |
| Bracts | 2.0 ± 0.2 | 2.2 ± 1.0 | 4.3 ± 0.2 | 2.8 ± 0.3 |
| Filamentous organs | 0.1 ± 0.1 | 0.1 ± 0.3 | 0.2 ± 0.3 | 0.1 ± 0.1 |
| Whorl 2 | ||||
| Total organs | 1.0 ± 0.3 | 1.2 ± 1.1 | 2.6 ± 0.6 | 1.0 ± 0.2 |
| Petals | 0.0 | 0.0 | 0.0 | 0.0a |
| Petaloid bracts | 1.0 ± 0.3 | 1.2 ± 1.1 | 1.3 ± 0.6 | 1.0 ± 0.2 |
| Leaves | 0.0 | 0.0 | 1.3 ± 0.7 | 0.0 |
| Whorl 3 | ||||
| Total organs | 5.1 ± 0.3 | 5.2 ± 1.1 | 6.6 ± 0.5 | 5.7 ± 0.2 |
| Stamens | 4.1 ± 0.4 | 4.8 ± 1.1 | 5.7 ± 0.8 | 4.8 ± 0.3 |
| Mosaic stamens | 0.9 ± 0.4 | 0.3 ± 0.7 | 1.0 ± 0.7 | 0.9 ± 0.3 |
| Filamentous organs | 0.1 ± 0.1 | 0.2 ± 0.5 | 0.1 ± 0.2 | 0.1 ± 0.1 |
| Flower No. | 28 | 86 | 22 | 84 |
| 2° Flowers/inflorescences | 1.6 ± 0.3 | 1.6 ± 0.1 | 0.3 ± 0.3 | 1.8 ± 0.1 |
Averages with 2 × se of the mean are given. Increased organ no. in whorls 1 and 2 and reduced no. of secondary flowers/inflorescences in LtMADS2, when compared with the other three genotypes, are statistically significant (Student's t test, p < 0.001).
One petal in 84 flowers.
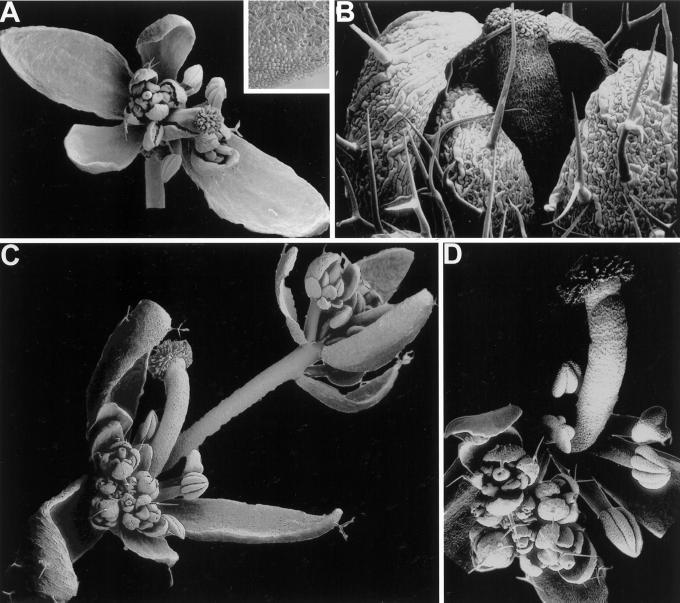
Only LtMADS2 had an obvious effect on the ap1-15 phenotype, and increased organ number in the first, second, and third whorls, whereas the number of axillary flowers and inflorescences was reduced (Table I). Most plants had four or five bracts or sepaloid bracts in the first whorl, and one to three bracts or petaloid bracts in their second whorl (Fig. 8B; Table I). It is interesting that these plants showed gain-of-function phenotypes similar to those of plants that overexpress AP1 under the control of the constitutive cauliflower mosaic virus 35S promoter (Mandel and Yanofsky, 1995a), suggesting that sequences in the LtMADS2 cDNA interacted with the AP1 promoter to cause ectopic expression. LtMADS2 plants flowered early, with 7.1 ± 1.0 total leaves compared with 11.9 ± 1.1 in untransformed ap1-15 plants (average ± 2 × se of the mean). In addition, side and main shoots formed terminal flowers, similar to 35S::AP1 plants (Mandel and Yanofsky, 1995a). The effect of LtMADS1 was more subtle, but about 10% of flowers had normal sepals, which were never observed in ap1-15 mutants (Fig. 8A; Table I).
Figure 8.
Scanning electron micrographs of flowers from plants expressing LtMADS1, LtMADS2, and LtMADS2′ under the control of a 1.7-kb fragment of the AP1 promoter. A, Flower from LtMADS1 transgenic plant with two petaloid bracts in the perianth (arrows). The inset in A shows petaloid cells on the margin of the lower bract-like petal at high magnification (size bar = 100 μm). B, Flower from LtMADS2 transgenic plant. A first-whorl bract has been removed to reveal a second-whorl bract (arrow). C, Inflorescence from LtMADS2′ transgenic plant. An axillary flower with an abnormally long pedicel is visible in the upper right hand corner. D, Enlargement of flower in lower left hand corner of C. Size bars are 1 mm, except 200 μm in C.
Floral organs of ap1-15 mutants carrying the LtMADS2′ transgene were similar to untransformed ap1-15 plants. One noteworthy feature was that eight out of 230 bracts were fused, which is normally not observed in ap1 mutants. The most obvious effect of LtMADS2′ was that almost one-third of structures in the axils of first-whorl organs developed into indeterminate inflorescences instead of secondary flowers. In addition, secondary flowers of LtMADS2′ plants had abnormally long pedicels as well as ectopic flowers which developed on their pedicels (Fig. 8C). Although a similar effect was not observed in LtMADS2 plants or the ap1-15 control population, it is unclear how significant this observation is because axillary inflorescences, albeit at a lower frequency, were also seen in LtMADS1 flowers.
DISCUSSION
The ABC model of floral organ development has been very successful in explaining floral development of higher dicots (Coen and Meyerowitz, 1991). Many of its tenets appear to apply to monocots as well (Ambrose et al., 2000), even though there may be some divergence in lower eudicots (Kramer and Irish, 1999). The function of the floral meristem identity gene FLO/LFY similarly is conserved in several dicots, as revealed by similar loss-of-function mutant phenotypes (Hofer et al., 1997; Souer et al., 1998; Molinero-Rosales et al., 1999) and by the ability of constitutively expressed LFY to promote the conversion of shoots into flowers (Weigel and Nilsson, 1995). Less is known, however, about the role of floral meristem identity genes in monocots. Although overexpression of Arabidopsis LFY reduces flowering time of rice, the effects are modest compared to those seen in dicots overexpressing LFY (Weigel and Nilsson, 1995; He et al., 2000). Constitutive overexpression of the LFY homolog RFL similarly has no dramatic effects on flowering of Arabidopsis (Kyozuka et al., 1998).
Here for the grass L. temulentum, we have taken advantage of extensive prior knowledge of the timing of its flowering in response to a single LD to examine expression of LFY- and AP1-related genes at different steps in floral evocation and development. Based on their expression early and in perianth-type organs, the L. temulentum AP1-like genes fit the concepts developed for dicots but there are difficulties when considering our findings for LtLFY, whose expression is not inextricably linked with the first events of floral evocation at the shoot apex.
LtMADS1 and LtMADS2 and Comparison with Other AP1/SQUA MADS Box Genes
The two L. temulentum MADS box genes, LtMADS1 and LtMADS2, share greatest similarity with the AP1/SQUA clade of the plant MADS box gene family and particularly with other monocot members of this branch (Figs. 1 and 2). The extensive similarity of LtMADS1 with LtMADS2 suggests that they have redundant functions, similar to AP1 and the two AP1-related genes in Arabidopsis, CAULIFLOWER (CAL) (Kempin et al., 1995) and FRUITFULL (FUL; Ferrándiz et al., 2000). There are also two AP1-like genes in rice (Moon et al., 1999; Kyozuka et al., 2000) and three in barley (Schmitz et al., 2000).
Whereas the M-I-K regions of members of the AP1/SQUA clade of plant MADS domain proteins share greater than 80% sequence identity, the carboxy terminus is less conserved. In addition to a Ser-rich motif in the monocot members of this clade, the sequence “LPPWMLSHL/IN” is conserved. A similar sequence “LPAWML” is found in FUL and SaMADSB from S. alba (Menzel et al., 1996). None of the monocot proteins contain the CaaX box that is present in several AP1-related proteins of dicots, and that allows prenylation of the protein by fanesyltransferases (Yalovsky et al., 2000).
Aside from similarity of the gene sequences, a function similar to SQUA and AP1 in dicots is supported by our studies of expression patterns of the two genes. In the shoot apex, LtMADS1 and LtMADS2 were detected in the apical dome of vegetative plants and, as for all AP1/SQUA-related genes, expression is strong in the floral meristem early in floral development. In common with maize (Mena et al., 1995) and rice (Kyozuka et al., 2000), later expression of LtMADS1 and LtMADS2 became restricted to perianth primordia, including the glumes, lemma, palea, and probably the lodicules. Expression in the epidermal and subepidermal layers and its exclusion from the central core of the spikelet sites indicates further similarity between LtMADS1, FUL, and SaMADSB, which are excluded from the pith of the inflorescence apex (Mandel and Yanofsky, 1995b; Menzel et al., 1996). Elsewhere in the plant the expression of LtMADS1 in leaves and roots is more similar to that of FUL and SaMADSB than to that of AP1, SQUA, or ZAP1. In contrast, there was no expression of LtMADS2 in leaf and root tissue that fits with RNA-blot data for ZAP1 (Mena et al., 1995), RAP1B in rice (Kyozuka et al., 2000), and BM8 in barley (Schmitz et al., 2000). The implication that LtMADS1 and LtMADS2 function in floral evocation and development is further supported by the ability of LtMADS2 to partially complement ap1-15 organ number defects as well as to transform the shoot into floral meristem (Fig. 8; Table I).
It is interesting that both LtMADS1 and LtMADS2 are expressed within the provascular strands in the central core of the meristem and in the glumes and lemmas. Provascular expression is also observed for SaMADSB (see Menzel et al., 1996). Any LtMADS2 expression in the carpel and stamen primordia was only transient early during their initiation. Overall, in L. temulentum, the intensive expression of LtMADS1 and LtMADS2 within the dome of the floral shoot apex, as well as in spikelet and floret meristems indicates that these genes may have a floral meristem identity function as well as a function in floral organ identity.
Comparison of LtLFY with Other LFY Homologs
Kyozuka and colleagues (1998) have previously described the expression pattern of RFL, the rice LFY homolog. Like LtLFY, RFL expresses in young panicles but in contrast, RFL is also expressed vegetatively. In this respect, RFL is similar to LFY and its dicot orthologs, with the exception of FLO from snapdragon (Coen et al., 1990; Bradley et al., 1996; Blázquez et al., 1997; Hofer et al., 1997; Pnueli et el., 1998; Souer et al., 1998). Vegetative expression has also been found for NEEDLY (NLY), a LFY homolog from gymnosperms. It is interesting that although NLY is more divergent from LFY than RFL both with respect to sequence similarity and origin, when ectopically expressed in Arabidopsis NLY is much more potent than RFL in transforming shoots into flowers (Kyozuka et al., 1998; Mouradov et al., 1998). Although we have not performed this experiment for LtLFY, its high sequence similarity with rice RFL suggests a similar outcome.
Timing of Expression of LtMADS1, LtMADS2, and LtLFY during Inflorescence Initiation
Characterization of the events of L. temulentum floral induction over the past 40 years has precisely defined the timing of biochemical changes at the shoot apex. These changes occur within hours to days of a single inductive LD and morphological change is first evident within several days (Fig. 5). Early expression of the L. temulentum AP1-like genes can now be added to this sequence of events then, much later, LtLFY expresses (compare Fig. 6 with Fig. 7). Up-regulated expression of LtMADS1 and LtMADS2 in lateral and terminal spikelet sites was evident by the afternoon of LD III (30 h after the end of the LD; Figs. 5 and 6) and this correlates with the time of commitment of the shoot apical meristem to flower as assessed by culturing induced apices (McDaniel et al., 1991). At the time of increased AP1 expression at the L. temulentum apex on LD III, there are also increases in cell division at the apical dome (Ormrod and Bernier, 1990) and RNA and DNA staining increases dramatically in the terminal and lateral spikelet sites (Knox and Evans, 1966) where LtMADS1 and LtMADS2 express (Figs. 5 and 6). In Arabidopsis AP1 is also expressed early during the floral transition (within 2 d) as it is in S. alba where, because the latter species flowers with exposure to a single LD, it is clear that, expression of the two S. alba AP1 genes correlates with commitment to flower (Menzel et al., 1996), as also in L. temulentum.
The much later expression of LtLFY compared with LtMADS1 and LtMADS2 contrasts with snapdragon and Arabidopsis, where increased FLO/LFY expression precedes AP1 activation (Coen et al., 1990; Huijser et al., 1992; Bradley et al., 1996; Blázquez et al., 1997; Hempel et al., 1997). Further, at least in early arising flowers, LFY is required for AP1 up-regulation (Liljegren et al., 1999) and LFY protein has been shown to be a direct regulator of AP1 (Parcy et al., 1998; Wagner et al., 1999). Our findings indicate, therefore, that this hierarchy of regulation involving floral meristem identity genes may not be conserved in monocots. As an alternative, however, when LtLFY does express it may act directly on the L. temulentum AP1-like genes but that, at earlier times during floral evocation, these same AP1-like genes may be acting independently of LtLFY.
MATERIALS AND METHODS
Plant Material
Lolium temulentum strain Ceres plants were grown as described elsewhere (Evans et al., 1990) in natural daylight of 8-h SD for 5 weeks. Tillers were removed and the plants transferred to SD in cabinets illuminated with fluorescent and incandescent lamps providing a total irradiance of 250 to 300 μmol m−2 s−1 photosynthetically active radiation. One week later the plants were exposed to a single inductive LD, designated as LD I, imposed as a 16-h extension of the 8-h SD photoperiod at a low irradiance (10–12 μmol m−2 s−1) from incandescent lamps. Two directly related measures of inflorescence development were employed: floral stage and shoot apex length (Evans et al., 1990). To allow comparison of the timing of floral development across experiments, floral stage was determined by daily dissections until at least Day XII.
For analysis of changes in mRNA, shoot apices (0.8 mm long on Day I) were harvested at various times following the single LD exposure (floral) or in parallel for the SD (vegetative) controls. The apices were immediately placed on dry ice and later stored at −70°C prior to RNA extraction.
Construction of PCR-Based cDNA Libraries
As described in Gocal (1997), PCR-based cDNA libraries were made from shoot apices from either SD vegetative or LD III plants (30 h after the end of the single LD). Primary libraries consisted of between 2 and 4 × 106 plaques with the portion of empty vector being less than 5%, and the average insert size being 1.2 kb. Although each library was constructed from a small amount of tissue (about 10 mg fresh weight), multiple PCR products from the same transcript were never isolated in our library screens. Of 27 full-length clones isolated to date from L. temulentum shoot apex PCR-based libraries, their poly-A tails ranged from four to 22 nucleotides and their 5′-UTRs range from 80 to 140 nucleotides.
Generation of MADS Box Fragments
A 129-bp fragment of the MADS box was PCR amplified from L. temulentum genomic DNA using a touchdown program with a degenerate oligonucleotide primer pair. MADS-1 [5′-d(CGGAATTCATGGGNMGNGGNAARRT)-3′] was identical to nucleotides 1 through 17 (underlined; restriction endonuclease sites indicated in bold), with MADS-43 [5′d(CGGGATCCIACYTGIGCRTCRCAIARIA-C)-3′] being complementary to nucleotides 111 through 129 (underlined; designed by Dr. Steve Strauss, Oregon State University, Corvallis). Single bands representing putative L. temulentum MADS box gene fragments were gel purified and cloned as EcoRI/BamHI fragments into pBluescript SK+ plasmid. The four inserts sequenced were from two genes.
Library Screening
Aliquots of the L. temulentum LD III apex-specific PCR-based cDNA library were screened with the two L. temulentum MADS fragments (see above) after 32P-dCTP labeling (Megaprime DNA labeling system, Amersham, Sydney). Hybridization was overnight in 5× sodium chloride/sodium dihydrogen phosphate/EDTA buffer (SSPE) and 0.5% (w/v) SDS, 5× Denhardt's solution, and 25 μg mL−1 sheared salmon sperm DNA at 65°C. Washes were for 30 min at 65°C in 2× SSPE and 0.1% (w/v) SDS (two washes) and a final high stringency wash in 0.1× SSPE and 0.2% (w/v) SDS.
Isolation of L. temulentum LEAFY (LtLFY)
Two approaches were used to isolate LtLFY. First, a 206-bp fragment of LFY was amplified from L. temulentum genomic DNA using the degenerate primers Glfy1 [5′-d(CGGAATTCATGCGVCACTACGTG/TCACTG)-3′] and Glfy2 [5′-d(CGGGATCCACATACCARATVGMGAGRCGVGG)-3′] from the conserved C terminus (amino acids 310–377 from exon 3) of LFY. A single band representing a putative L. temulentum LFY gene fragment was gel purified and cloned as an EcoRI/BamHI fragment into pBluescript SK+ plasmid. Second, a UniZAP Custom cDNA library (Stratagene, La Jolla, CA) was made from L. temulentum using mRNA isolated with Dynal beads. The source tissue included inflorescences, leaves, and stems. The library was screened at medium stringency (0.1× SSC, 0.1% [w/v] SDS; 2 × 30 min at 65°C) with the ORF from an LpLFY cDNA clone previously isolated from Lolium perenne (C. Andersen, unpublished data).
Sequencing and Sequence Analysis
Cloned DNA fragments were sequenced on both strands with either Dye Primer or DyeDeoxy Terminators using sequencers (PRISM 310 or 377, ABI, Foster City, CA). Sequence analysis was done using the University of Wisconsin GCG version 8 software package (Devereux et al., 1984), or Sequencer v4.0.2 software (Gene Codes Corporation, Ann Arbor, MI). PCR-based errors were detected by comparing the sequence for at least three clones over the entire ORF.
To determine the phylogenetic relationship of LtMADS1 and LtMADS2 within the plant MADS box gene family, protein sequences were first aligned using the PILEUP program of the GCG version 8 package. The various sequences examined were: (a) the MADS domain alone; (b) the MADS domain and I region; (c) the MADS domain, I region, and K domain; and (d) the entire protein sequence. Partial sequences were completed with ?s. Phylogenetic trees were constructed for each data set using the PROTPARS program of the phylogenetic inference package (PHYLIP) as described in Theissen et al. (1996). The input order of the sequences was randomized using the jumble option in the PROTPARS program and repeated ten times. Multiple most parsimonious trees were found in each instance and the consensus tree was determined by using the PHYLIP program CONSENSE. The consensus most parsimonious tree was drawn for each data set using the PHYLIP program DRAWTREE.
PCR-Based (Virtual) Northern-Blot Analysis
All PCR-based northern blots presented here were prepared from a single cDNA synthesis using 250 shoot apices from plants in SD or LD (Day III) Apices from this experiment (Lt424) reached the double-ridge stage on LD VI. The PCR-amplified cDNA was suspended in 65 μL of Tris/EDTA (pH 7.6) and 10 μL of each of the SD and LD III pools were electrophoresed in parallel on six 1.2% (w/v) agarose gels, blotted, hybridized, and washed as above. Virtual northern blots produced from different amplifications hybridized with the same probe always yielded similar results. To compare loading, each set of blots was also probed with a fragment of an α-tubulin gene from L. temulentum (not shown; G.F.W. Gocal, unpublished data).
RNA Gel-Blot Analysis
Leaf and root RNA were isolated by the method of Chandler et al. (1983). For RNA gel-blot analysis, 20 μg of total RNA was size fractionated by gel electrophoresis under denaturing conditions in a 1% (w/v) agarose gel with formaldehyde, transferred to a Hybond-N membrane (Amersham), and hybridized as above. To compare RNA loading across lanes, following autoradiography, RNA gel blots were stripped and reprobed with a 9-kb wheat rRNA clone, pTA71 (Gerlach and Bedbrook, 1979).
In Situ Hybridization
Shoot apices were processed according to Gocal (1997). To determine the temporal (vegetative through LD XXX) and spatial localization of the LtMADS1, LtMADS2, and LtLFY mRNAs within the apex following floral induction, 10-μm longitudinal sections were prepared. In vitro transcribed Digoxigenin-labeled riboprobes were synthesized for a 3′ gene-specific fragment of LtMADS1 (MADSc1/2; nucleotide 650–1062) or LtMADS2 (MADSr1/2; nucleotide 568–874) and for the conserved C-terminal region of LtLFY. Sense and antisense riboprobes for each gene were generated from the T7 promoter using PCR-generated 3′ gene-specific fragments, which for LtMADS1 and LtMADS2 were cloned as EcoRI/HindIII fragments and for LtLFY as an EcoRI/BamHI fragment in pBluescript SK+ or pBluescript KS+. Hybridizations were with an equivalent sense or antisense probe concentration and were performed simultaneously on each tissue set. Most of the in situ results presented were from material collected in one experiment (Lt434).
T-DNA Constructs
cDNAs of LtMADS1, LtMADS2, and its alternatively spliced version LtMADS2′, were cloned between a 1.7-kb fragment of the Arabidopsis APETALA1 promoter upstream of the ATG (HindIII/EcoICR I; Hempel et al., 1997) and the pea (Pisum sativum) RBCSE9 terminator (accession no. M21375) in a pBluescriptKS+ (Stratagene) derivative termed pGG62. Each pAP1:LtMADS:RBCSE9 construct was shuttled as a HindIII fragment into pCGN1547 (McBride and Summerfelt, 1990). These constructs were introduced via Agrobacterium tumefasciens-mediated vacuum infiltration into apetala1-15 mutant plants (Columbia ecotype; Bechtold et al., 1993). Transformants were selected on 0.5× Murashige and Skoog plates containing 50 μg mL−1 kanamycin. At least 20 T1 lines were obtained per construct and between four and six were chosen randomly for subsequent analysis. For LtMADS1 and LtMADS2′, floral organs were counted for the first four to eight flowers and for LtMADS2 all flowers were counted (up to four per plant). Inflorescences were fixed and examined using scanning electron microscopy as described (Weigel et al., 1992).
ACKNOWLEDGMENTS
The authors thank Drs. Liz Dennis, Lloyd Evans, Frank Gubler, Masumi Robertson (CSIRO, Plant Industry, Canberra) and Malcolm Whitecross (Australian National University, Canberra) for their support and helpful comments. Greg Gocal is also indebted to a number of scientists at the Australian National University, the Commonwealth Scientific and Industrial Research Organization, and the plant molecular biology community in San Diego.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–00–78277 to D.W.).
LITERATURE CITED
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. Molecular and genetic analysis of the Silky1 gene reveals conservation in floral organ specification between eudicots and monocots. Mol Cell. 2000;5:569–579. doi: 10.1016/s1097-2765(00)80450-5. [DOI] [PubMed] [Google Scholar]
- Bechtold J, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Weigel D. Integration of floral inductive signals in Arabidopsis. Nature. 2000;404:889–892. doi: 10.1038/35009125. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743. [Google Scholar]
- Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E. Control of inflorescence architecture in Antirrhinum. Nature. 1996;379:791–797. doi: 10.1038/379791a0. [DOI] [PubMed] [Google Scholar]
- Busch MA, Bomblies K, Weigel D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999;285:585–587. doi: 10.1126/science.285.5427.585. [DOI] [PubMed] [Google Scholar]
- Carpenter R, Coen ES. Floral homeotic mutations produced by transposon mutagenesis in Antirrhinum majus. Genes Dev. 1990;4:1483–1493. doi: 10.1101/gad.4.9.1483. [DOI] [PubMed] [Google Scholar]
- Carpenter R, Copsey L, Vincent C, Doyle S, Magrath R, Coen E. Control of flower development and phyllotaxy by meristem identity genes in Antirrhinum. Plant Cell. 1995;7:2001–2011. doi: 10.1105/tpc.7.12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Higgins TJV, Randall PJ, Spencer D. Regulation of legumin levels in developing pea seeds under conditions of sulfur deficiency. Plant Physiol. 1983;71:47–54. doi: 10.1104/pp.71.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliot R, Murphy G, Carpenter R. Floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LT, King RW, Chu A, Mander LN, Pharis RP. Gibberellin structure and florigenic activity in Lolium temulentum, a long-day plant. Planta. 1990;182:97–106. doi: 10.1007/BF00239990. [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–34. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Fischer A, Baum N, Saedler H, Theissen G. Chromosomal mapping of the MADS-box multigene family in Zea mays reveals dispersed distribution of allelic genes as well as transposed copies. Nucleic Acids Res. 1995;23:1901–1911. doi: 10.1093/nar/23.11.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich MW, Parker DS. The mostly male theory of flower evolution: from genes to fossils. Syst Bot. 2000;25:155–170. [Google Scholar]
- Gerlach WL, Bedbrook JR. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal GFW. Molecular biology of floral evocation in Lolium temulentum. PhD thesis. Canberra: Australian National University; 1997. [Google Scholar]
- He Z, Zhu Q, Dabi T, Li D, Weigel D, Lamb CJ (2000) Early heading of rice expressing the Arabidopsis floral regulator LEAFY. Transgenic Res (in press) [DOI] [PubMed]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF. Floral determination and expression of floral regulatory genes in Arabidopsis. Development. 1997;124:3845–3853. doi: 10.1242/dev.124.19.3845. [DOI] [PubMed] [Google Scholar]
- Hempel FD, Zambryski PC, Feldman LJ. Photoinduction of flower identity in vegetatively biased primordia. Plant Cell. 1998;10:1663–1675. doi: 10.1105/tpc.10.10.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF. Discrete temporal and spatial cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene apetala3. Development. 1998;125:1711–1721. doi: 10.1242/dev.125.9.1711. [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol. 1997;7:581–587. doi: 10.1016/s0960-9822(06)00257-0. [DOI] [PubMed] [Google Scholar]
- Huala E, Sussex IM. LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development. Plant Cell. 1992;4:901–913. doi: 10.1105/tpc.4.8.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Klein J, Lönnig W-E, Meijer H, Saedler H, Sommer H. Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 1992;11:1239–1249. doi: 10.1002/j.1460-2075.1992.tb05168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. Function of the apetala-1 gene during Arabidopsis flower development. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, Nam J, Kim C, Lee SH, Chung YY, Kim SR, Lee YH, Cho YG. leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell. 2000;12:871–84. doi: 10.1105/tpc.12.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF. Molecular basis of the cauliflower phenotype in Arabidopsis. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- Knox RB, Evans LT. Inflorescence initiation in Lolium temulentum L VIII Histochemical changes in the shoot apex at induction. Aust J Biol Sci. 1966;19:233–245. [Google Scholar]
- Kramer EM, Irish VF. Evolution of genetic mechanisms controlling petal development. Nature. 1999;399:144–148. doi: 10.1038/20172. [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Harcourt R, Peacock WJ, Dennis ES. Eucalyptus has functional equivalents of the Arabidopsis AP1 gene. Plant Mol Biol. 1997;35:573–584. doi: 10.1023/a:1005885808652. [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Kobayashi T, Morita M, Shimamoto K. Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B, and C genes. Plant Cell Physiol. 2000;41:710–718. doi: 10.1093/pcp/41.6.710. [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K. Down-regulation of RFL the FLO/LFY homolog of rice accompanied with panicle branch initiation. Proc Natl Acad Sci USA. 1998;95:1979–1982. doi: 10.1073/pnas.95.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF. Interactions among APETALA1, LEAGFY, and TERMINAL FLOWER specify meristem fate. Plant Cell. 1999;11:1007–1018. doi: 10.1105/tpc.11.6.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF. A gene triggering flower formation in Arabidopsis. Nature. 1995a;377:522–524. doi: 10.1038/377522a0. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF. The Arabidopsis AGL8 MADS-box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell. 1995b;7:1763–1771. doi: 10.1105/tpc.7.11.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KE, Summerfelt KR. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol Biol. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- McDaniel CN, King RW, Evans LT. Floral determination and in vitro floral differentiation in isolated shoot apices of Lolium temulentum L. Planta. 1991;185:9–16. doi: 10.1007/BF00194508. [DOI] [PubMed] [Google Scholar]
- Mena M, Mandel MA, Lerner DR, Yanofsky MF, Schmidt RJ. A characterization of the MADS-box gene family in maize. Plant J. 1995;8:845–854. doi: 10.1046/j.1365-313x.1995.8060845.x. [DOI] [PubMed] [Google Scholar]
- Menzel G, Apel K, Melzer S. Identification of two MADS box genes that are expressed in the apical meristem of the long-day plant Sinapis alba in transition to flowering. Plant J. 1996;9:399–408. doi: 10.1046/j.1365-313x.1996.09030399.x. [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N, Jamilena M, Zurita S, Gomez P, Capel J, Lozano R. The tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. 1999;20:685–693. doi: 10.1046/j.1365-313x.1999.00641.x. [DOI] [PubMed] [Google Scholar]
- Moon YH, Kang HG, Jung JY, Jeon JS, Sung SK, An G. Determination of the motif responsible for interaction between rice APETALA1/AGAMOUS-LIKE9 family proteins using a yeast two-hybrid system. Plant Physiol. 1999;120:1193–1204. doi: 10.1104/pp.120.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Glassick TV, Hamdorf BA, Murphy LC, Marla SS, Yang Y, Teasdale R. Family of MADS box genes expressed early in male and female reproductive structures of Monterey pine. Plant Physiol. 1998;117:55–62. doi: 10.1104/pp.117.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormrod JC, Bernier G. Cell cycle patterns in the shoot apex of Lolium temulentum L cv Ceres during the transition to flowering following a single long day. J Exp Bot. 1990;41:211–216. [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. A genetic framework for floral patterning. Nature. 1998;395:561–566. doi: 10.1038/26903. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Carmelgoren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E. The self-pruning gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125:1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Franzen R, Ngyuen TH, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W. Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol Biol. 2000;42:899–913. doi: 10.1023/a:1006425619953. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell. 1991;3:771–781. doi: 10.1105/tpc.3.8.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C. When to switch to flowering. Ann Rev Cell Develop Biol. 1999;15:519–550. doi: 10.1146/annurev.cellbio.15.1.519. [DOI] [PubMed] [Google Scholar]
- Souer E, van der Krol A, Kloos D, Spelt C, Bliek M, Mol J, Koes R. Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development. 1998;125:733–742. doi: 10.1242/dev.125.4.733. [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Munster T, Winter K-U, Saedler H. A short history of MADS-box genes in plants. Plant Mol Biol. 2000;42:115–149. [PubMed] [Google Scholar]
- Theissen G, Kim JT, Saedler H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol. 1996;43:484–516. doi: 10.1007/BF02337521. [DOI] [PubMed] [Google Scholar]
- Tilly J, Allen DW, Jack T. The CarG boxes in the promoter of the Arabidopsis floral organ identity gene apetala3 mediate diverse regulatory effects. Development. 1998;15:1647–1657. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- Wagner D, Sablowski RWM, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285:582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Rodriguez-Conception M, Bracha K, Toledo-Ortez G, Gruissem W. Prenylation of the floral traanscription factor APETALA1 modulates its function. Plant Cell. 2000;12:1257–1266. doi: 10.1105/tpc.12.8.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]