Abstract
Mitochondrial sequence variants affect phenotypic function, often through interaction with the nuclear genome. These “mitonuclear” interactions have been linked both to evolutionary processes and human health. The study of these interactions has focused on mechanisms regulating communication between mitochondrial and nuclear proteins; the role of mitochondrial (mt) RNAs has received little attention. Here, we show that small mt-RNAs bind to the nuclear protein Argonaute 2, and that nuclear miRNAs bind to mt-mRNAs. We identify one small mt-RNA that binds to Argonaute 2 in human tissues whose expression and sequence remain unchanged across vertebrates. Although analyses of CLEAR-CLIP sequencing data sets of human and mouse did not reveal consistent interactions between small mt-RNAs and nuclear mRNAs, we found that MT-ND4 and MT-ATP6 mRNAs are bound by different nuclear miRNAs in humans and mice. Our work homes in on previously unknown interactions between nuclear and small mt-RNAs, which may play key roles in intergenomic communication.
Keywords: mitonuclear communication, mtDNA, small RNAs, mitochondria, AGO2
Significance.
Communication between mitochondria and nucleus (mitonuclear communication) plays an important role in cell regulation and has been linked to evolutionary processes such as adaptation and speciation. Here, we study previously unknown mechanisms underpinning mitonuclear communication. We show that small RNAs encoded in the mitochondrial genome bind to one of the main proteins of RNA interference, suggesting mitochondrial involvement in the regulation of nuclear gene expression through RNAi.
Introduction
Interest in mitochondrial biology is increasing, with a growing number of studies highlighting the complex role of the mitochondria in cell regulation (Picard et al. 2016; Sloan et al. 2018; Sprenger and Langer 2019). Among these, numerous studies have found that sequence variation in the mitochondrial DNA (mtDNA) can affect the expression of a range of life-history and health-related traits, from fertility, to longevity and thermal tolerance (James and Ballard 2003; Rand et al. 2006; Song and Lewis 2008; Yee et al. 2013; Camus et al. 2017; Lajbner et al. 2018). Many of these mtDNA-mediated effects on phenotype appear to be moderated via interactions with the nuclear genetic background; as such mitonuclear interactions have been proposed to be key units shaping population evolutionary processes (Rand et al. 2004; Wolff et al. 2014). However, the mechanistic basis of the molecular interactions that underpin the interaction between mtDNA haplotypes and nuclear loci remain largely elusive.
One possible mechanism by which mitochondrial mutations could affect expression of physiological and life-history traits is through RNAs that are encoded within the mtDNA (mt-RNAs). The mt-RNAs represent plausible mediators of patterns of mitochondrial-mediated posttranscriptional regulation. Functional mt-RNAs are well-known in the form of the 22 tRNAs and two rRNAs encoded by the typical bilaterian mitochondrial genome. However, novel types of RNAs of mitochondrial origin have recently been identified. For example, Dhir et al. (2018) described a new class of double-stranded RNAs encoded in the mitochondria that are able to trigger antiviral signaling in humans (Dhir et al. 2018). Although to date these double-stranded RNAs have only been identified in humans, different types of novel small mitochondrial RNAs have been described in multiple species across two metazoan phyla, Chordata and Mollusca (Mercer et al. 2011; Ro et al. 2013; Bottje et al. 2017; Pozzi et al. 2017; Larriba et al. 2018; Riggs et al. 2018; Pozzi and Dowling 2019). Yet, despite increasing interest in the putative role these small mitochondrial RNAs may play in the regulation of cellular function, clear evidence of their functionality remains absent (Pozzi and Dowling 2021).
Given similarities in their length and sequence to microRNAs, Pozzi and Dowling (2021) recently hypothesized involvement of small mitochondrial RNAs in the regulation of mRNA translation through RNA interference (RNAi). RNAi is a process in which a microRNA (miRNA) leads a protein complex to block translation of a target mRNA (Ambros 2004; Ha and Kim 2014). miRNAs are partially complementary to a regulatory region of the mRNAs, and due to this close miRNA–mRNA affinity, the protein complex is able to precisely bind its target mRNA and hinder its binding to the ribosome (Ambros 2004; Cloonan 2015). Within this protein complex, the main protein binding the miRNAs is Argonaute 2 (AGO2), an endonuclease necessary for RNAi that is shared across multiple species (Ha and Kim 2014; Cloonan 2015). Intriguingly, AGO2 has previously been reported to colocalize with mitochondria (Bandiera et al. 2011; Zhang et al. 2014) and, moreover, to associate with mitochondrial tRNA (mt-tRNA Met) in the cytoplasm (Maniataki and Mourelatos 2005). Accordingly, AGO2 is an excellent candidate to further probe the hypothesis that the small mitochondrial RNAs may serve a similar role to nuclear-encoded miRNAs in regulating RNAi.
Here, we investigated whether a hitherto unrealized connection exists between mitochondria and RNAi, by screening for the presence of two main features of miRNAs in the mitochondrial small RNAs. Firstly, we sought to verify binding between the mitochondrial small RNAs and AGO2. To this end, we leveraged published data sets of RNA sequencing (RNA-seq) and RNA-binding-protein co-immunoprecipitation sequencing (RIP-seq, CLIP-seq, and CLEAR-CLIP-seq) (Townley-Tilson et al. 2006). These data sets come from published studies that reported novel mechanistic insights into RNAi. We repurposed these data sets to investigate the capacity for mtDNA-mediated involvement in RNAi. Secondly, given that miRNAs have been shown to be conserved across multiple clades, we explored levels of sequence conservation in the mitochondrial small RNAs. We compared the presence of mitochondrial small RNAs that exhibit features similar to nuclear miRNAs across multiple model organisms, using small RNA-seq data sets from several independent and taxonomically diverse studies. By combining multiple data sets, obtained using distinct approaches, our study thus has the capacity to provide new insights into whether a functional relationship exists between the mitochondrial small RNAs and RNAi; consistent with the hypothesis that these RNAs may function as mitochondrial-transcribed miRNAs.
Results
Small Mitochondrial RNAs Bind AGO2
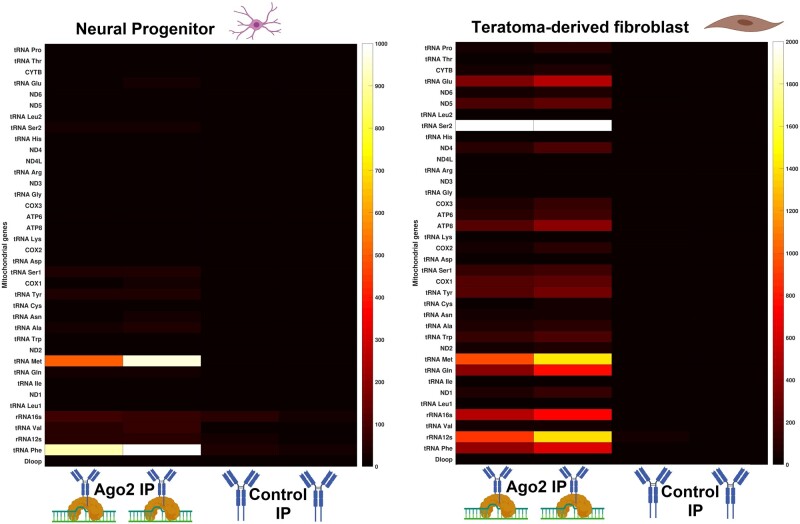
To investigate the ability of the small mitochondrial RNAs to bind AGO2, we analyzed an RNA immunoprecipitation (RIP-seq) data set of two different human cell lines, neural progenitor and teratoma-derived fibroblast (TDF) (GSE112006). The RIP-seq technique uses specific antibodies targeted to the protein of interest, to precipitate RNA–protein complexes. By using specific RNases to digest all unbound RNAs, the technique reveals the identity of RNAs that bind the target protein. This data set we used contained the small RNAs that bind to AGO2. However, our focus was only on the small mitochondrial RNAs, which have been previously ignored. We identified several small mitochondrial RNAs binding AGO2 in each of the two cell lines (fig. 1). Through this analysis, we found that these small mitochondrial RNAs are present almost exclusively in the AGO2 immunoprecipitations (IPs), mostly in the regions coding for mt-tRNAs; the control IP samples generally exhibited no binding of small mitochondrial RNAs across mt-genes (3 genes have very low expression and 35 have no expression). The expression of the small mitochondrial RNAs differs across the different cell line types, which are derived from different tissues and are thus suggestive of patterns of possible tissue-specific expression, consistent with previous studies demonstrating that mitochondrial transcription is usually tissue-specific (Scheibye-Alsing et al. 2007; Mercer et al. 2011; Pozzi and Dowling 2019). Aside from AGO2 RIP-seq, the authors of the original data set performed other treatments on their samples to verify genuine binding of their focal miRNAs with AGO2. One of these treatments is particularly significant for our study: RNase I treatment. RNase I is an endonuclease that digests only the RNAs that remain unbound to proteins, thus higher concentrations of RNAseq I can more efficiently eliminate RNAs not bound to AGO2 (contamination). We analyzed samples treated with different RNase I concentrations and found that higher concentrations of RNase I have no obvious effect on transcription levels of small mitochondrial RNAs, indicating true binding of these RNAs to AGO2 (see supplementary fig. S1, Supplementary Material online).
Fig. 1.
The small mitochondrial RNAs bind to AGO2. In two different cell lines, AGO2-IP samples are enriched in the expression of specific small mitochondrial RNAs when compared with a control IP, which contains only the IgG antibody. On the Y axis, all the canonical mitochondrial genes are listed. On the X axis, there are two biological replicates for each treatment. The two heatmaps are divided because they represent experiments from different cell lines. The sample type and treatments are illustrated through BioRender icons. As the small mitochondrial RNAs are an uncharacterized type of RNAs, we did not perform any normalization, thus the expression is measured in raw read counts. Gene-by-gene analyses of the transcriptional signatures are shown in supplementary figure S2, Supplementary Material online. See the Data Availability section to see from which studies the data come from.
Furthermore, we performed a gene-by-gene analysis on both AGO2-IP and control IP samples to verify whether the transcriptional signatures of the small mitochondrial RNAs are consistent with these representing functional RNAs or noise (Mercer et al. 2011; Pozzi et al. 2017). These analyses showed that the small mitochondrial RNAs with the highest levels of transcription are encoded within mt-tRNAs, confirming transcription patterns found by previous studies that observed the presence of small mitochondrial RNAs in other human cell lines (Mercer et al. 2011; Ro et al. 2013). Due to the large number of genes analyzed, the results of the gene-by-gene analysis are presented in the supplementary material (supplementary fig. S2, Supplementary Material online). This gene-by-gene analysis shows the transcriptional signature of each gene in each sample, showing which genes exhibit transcriptional signatures of small mitochondrial RNAs and which genes exhibit signals of noise only (i.e., in which sequences are spread through the gene). Our analysis of the AGO2-IP samples provides the first evidence that small mitochondrial RNAs consistently bind to AGO2, thus implicating their involvement in RNAi.
Small Mitochondrial RNAs Are Encoded in mt-tRNAs and Protein-Coding Genes
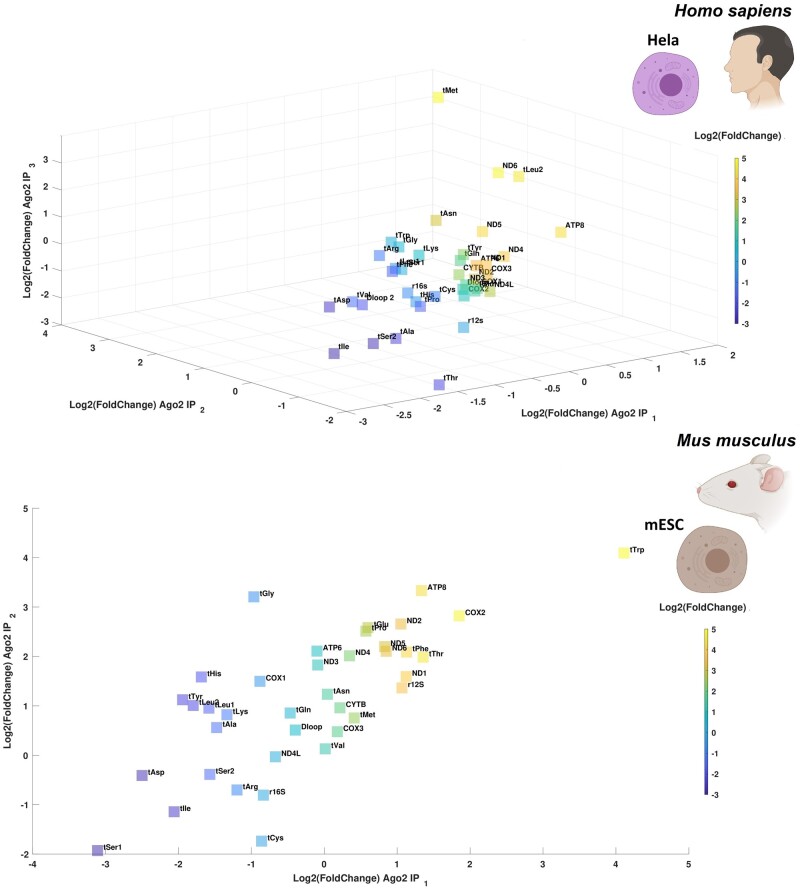
To further investigate the transcriptional signature of small mitochondrial RNAs across tissues and species, we expanded the number of data sets analyzed, by including more cell lines from humans and mice from other independent studies. These analyses further supported the presence of small mitochondrial RNAs binding to AGO2 in human (HeLa) and mouse embryonic cell lines (Scherer et al. 1953) (fig. 2). To verify the enrichment of small mitochondrial RNAs in AGO2-IP samples, we calculated the fold change between samples of the same cell line in which one sample had undergone the IP process, and the other had not (called input). Through this experiment, we identified enrichment in small mitochondrial RNAs across AGO2-IP samples in both species, however, we could test the significance of the enrichment only in the human samples (t-test paired test P = 0.0249) since we had only two mouse samples for each treatment. In HeLa cells, a mix of tRNAs and protein-coding genes are enriched for these small mitochondrial RNAs, providing the first evidence these small mitochondrial RNAs may be encoded within both mt-tRNAs and protein-coding genes. Nonetheless, the most enriched small mitochondrial RNA (5-fold higher expression) is encoded in the mt-tRNA Met. This finding supports a previous study which found that mt-tRNA Met binds to AGO2 outside the mitochondria (Maniataki and Mourelatos 2005). The high expression of small mitochondrial RNA Met in these samples (HeLa cells), and in our previous analysis (neural progenitor and TDF cell lines, fig. 1) suggests that although the small mitochondrial RNAs generally exhibit line-, and hence likely tissue-, specific expression, the expression of some small mitochondrial RNAs may well be stable across tissues. The analysis of the mouse samples provided similar results. Mouse embryonic stem cells are enriched for small mitochondrial RNAs encoded across multiple genes. Notably, the genes enriched for small mitochondrial RNAs in the mouse samples differed from those in the human HeLa cell lines, with MT-ATP8 the only small mitochondrial RNA exhibiting high expression in the AGO2-IP samples of both species.
Fig. 2.
The small mitochondrial RNAs are enriched in AGO2-IP samples. The plots show that AGO2-IP samples are enriched in expression for specific small mitochondrial RNAs when compared with the same sample without AGO2-IP treatment. The enrichment is calculated through log2-fold change, thus a fold change of two means that the gene has four times higher expression when compared with its counterpart without AGO2-IP treatment. As the small mitochondrial RNAs are an uncharacterized type of RNAs, we did not perform any normalization, thus the expression is measured in raw read counts. The species used in each plot is indicated through small BioRender icons. (A) The 3D-plot shows patterns of small mitochondrial RNA expression in AGO2-IP treated samples of HeLa cells. The three axes represent three different paired biological replicates for each of the two treatments (paired IP and non-IP). (B) The plot shows patterns of small mitochondrial RNA expression in the AGO2-IP treated samples derived from mouse embryonic stem cells. The two axes represent two different biological replicates for each of the two treatments (IP and non-IP). Gene-by-gene analyses of the transcriptional signatures are shown in supplementary figure S3, Supplementary Material online (human) and supplementary figure S4, Supplementary Material online (mouse). See the Data Availability section to see from which studies the data come from.
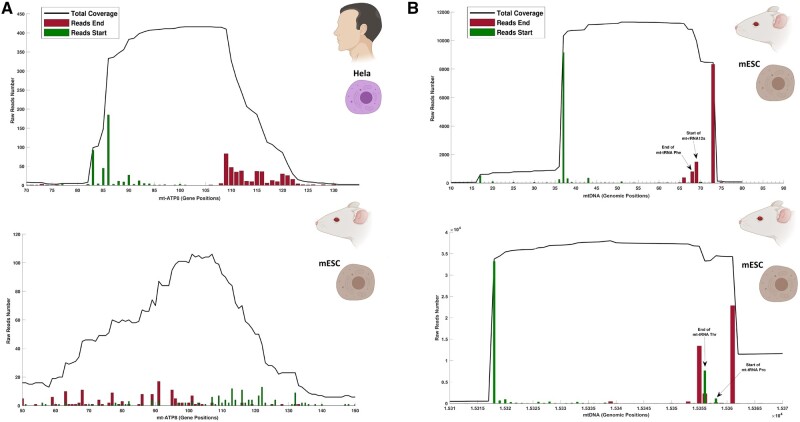
To better understand the differences and similarities in the results between human and mouse samples, we performed a gene-by-gene analysis of the transcriptional signatures of the small mitochondrial RNAs across the mouse embryonic stem cells and HeLa cell lines (see supplementary figs. S3 and S4, Supplementary Material online). In this gene-by-gene analysis, we performed a qualitative comparison of the transcriptional patterns of each gene, with the aim of distinguishing likely functional small mitochondrial RNAs from random alignments of small RNAs. For example, closer scrutiny of the putative small mitochondrial RNA at MT-ATP8 in both species showed that the expression of a conserved small RNA at this gene is only likely to exist in humans (fig. 3A). Although the human MT-ATP8 expresses a small RNA (32 nt long) with clear start and end, the same gene in mice does not, and is characterized by noise with no clear transcriptional signature. Moreover, other evidence suggests this putative small RNA found is perhaps most likely to be a target of AGO2 instead of acting as a small mitochondrial RNA. In particular, in humans the small RNA aligning to MT-ATP8, which was present in all AGO2-IP replicates, was absent from the RNA-seq libraries that had been through size selection for small RNAs (see supplementary fig. S3, Supplementary Material online), suggesting that the small RNA aligned to MT-ATP8 does not exist as a small RNA, but instead is part of the ATP8 mRNA and can be found only when we used the AGO2-IP protocol.
Fig. 3.
Small mitochondrial RNAs are encoded across genes. (A) shows the coverage of small RNAs within the gene mt-ATP8 in human HeLa cell lines (upper left-hand panel) and mice embryonic stem cells (lower left-hand panel), and highlights the difference between a transcriptional profile showing a genuine small mitochondrial RNA (human), and noise from the alignment of random small mitochondrial RNAs (mouse). The total coverage for each position in the gene is indicated with a black line, whereas the number of reads starting and ending at each position is indicated using green and red bars, respectively. This type of representation provides the resolution necessary to verify the presence or absence of a small mitochondrial RNA within a mitochondrial gene. As the small mitochondrial RNAs are an uncharacterized type of RNAs, we did not perform any normalization, thus the expression shown on the Y axis is measured in raw read counts. The X axis shows the gene positions of each read. (B) The panels represented on the right side of the figure highlight the transcriptional profile of two small mitochondrial RNAs encoded across genes in mice embryonic stem cells. The first small mitochondrial RNA (upper right-hand panel) is encoded mostly within the mt-tRNA Phenylalanine; however, a long isoform of this small mitochondrial RNA includes five nucleotides of the gene mt-rRNA12s. The second small mitochondrial RNA (lower right-hand panel) is encoded mostly within the mt-tRNA Threonine; however, a long isoform of this small mitochondrial RNA includes six nucleotides of the gene mt-tRNA Proline. Small black arrows indicate either the end or start of mt-genes where relevant. See the Data Availability section to see from which studies the data come from.
Furthermore, we identified unusual transcriptional signatures that might shed light on some aspects of the biogenesis of the small mitochondrial RNAs. Indeed, some small mitochondrial RNAs appear not to be fully encoded within a gene, but instead overlap across two different genes (fig. 3B). We identified this phenomenon only in the mouse samples, where both small mitochondrial RNA Phe and small mitochondrial RNA Thr partially overlap with the neighboring gene (an overlap of up to 6 bp). Moreover, both of these small mitochondrial RNAs feature two different isoforms of different lengths: the short isoform ends where the first gene ends, whereas the longer isoform overlaps with the second gene by several nucleotides. Interestingly, these isoforms are relatively long compared with other small RNAs such as miRNA (Ha and Kim 2014). In fact, the small mitochondrial RNA Phe isoforms are 32 and 37 nt long, respectively, whereas the small mitochondrial RNA Thr isoforms are 37 and 43 nt long. Due to the length of these small mitochondrial RNA isoforms, we suggest that the longer isoform may represent a transitional stage for the shorter mature form. Although the mechanism underpinning the generation of these small mitochondrial RNA isoforms remains unknown, our analysis provides the first report of small mitochondrial RNAs encoded across the boundaries of genes, and, in general, the presence of mitochondrial products encoded across genes.
Transcriptional Signature of Small Mitochondrial RNA Met Is Consistent across Chordata
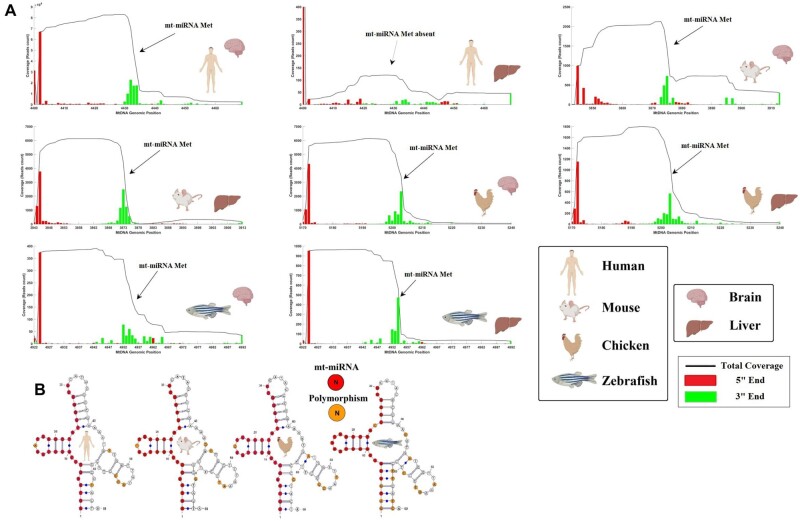
Nuclear miRNAs are usually conserved across species (Lee et al. 2007; Kenny et al. 2015), and we thus investigated whether small mitochondrial RNA Met, which is consistent across multiple tissues in humans, might be conserved in other model organisms. We analyzed the expression and primary sequence of small mitochondrial RNA Met across two tissues of four model organisms and verified its presence in Chordata, using RNA-seq data sets from multiple studies (fig. 4). The four model organisms (human, mouse, chicken, and zebrafish) show a very consistent transcriptional signature for small mitochondrial RNA Met across both brain and liver samples (fig. 4A). In fact, in all species, the small mitochondrial RNA Met 3′ end is at the beginning of the mt-tRNA Met gene, whereas the 5′ end follows approximately 32nt after. However, there was one exception exhibiting a deviation in this pattern. Samples from the human liver exhibit a noisy transcriptional signature (without clear start and end), suggesting that despite its broad presence across species, this small mitochondrial RNA might not be ubiquitously expressed across all tissues. Similar to the conserved transcriptional signature, we found that the primary sequence of small mitochondrial RNA Met is highly conserved (i.e., a very small proportion of polymorphic sites) across the four model organisms (fig. 4B). Indeed, this small RNA sequence is almost identical in all species, having only one polymorphic site shared by all Tetrapoda, and four additional mutated sites specific to zebrafish. When comparing the number of polymorphisms in small mitochondrial RNA Met to the polymorphisms in the rest of the mt-tRNA Met, we found that polymorphisms are underrepresented within the sequence particular to the small mitochondrial RNA Met. In fact, in human, mouse, and chicken there contain around four times more mutations in the rest of the mt-tRNA Met (5 of 39 bps) compared with within small mitochondrial RNA Met (1 of 30), whereas in zebrafish, there are around two times more mutations in the rest of the mt-tRNA Met (10 of 39) compared with within the small mitochondrial RNA Met (4 of 30). However, due to the small number of species under analysis, we could not statistically test if the region harboring the small mitochondrial RNAs has significantly less polymorphisms than the rest of the mt-tRNA. Nonetheless, this qualitative analysis suggests that the region harboring the small mitochondrial RNA Met might be under stronger purifying selection than its tRNA source, which we hypothesize may be due to the presence of overlapping selection due to the dual role of these regions in encoding both small mitochondrial RNA Met and mt-tRNA Met. Considering the level of conservation of this small mitochondrial RNA, and the consistency of expression and binding to AGO2, we believe it might act as a mitochondrial encoded miRNA (mt-miRNA), although evidence of its precise function is still lacking.
Fig. 4.
The small mitochondrial RNA encoded within the mt-tRNA Met (abbreviated to mt-miRNAMet) is conserved across species. In the figure panels, the expression and sequence of the mt-miRNAMet are represented for several major model organisms: human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), and zebrafish (Danio rerio). For each species, tissues from two different organs are considered; brain and liver. The species and tissue of each sample are indicated through BioRender icons. (A) The coverage (read count) in the mitochondrial genome, corresponding to the region of the mt-tRNA Met and mt-miRNAMet, is depicted. With exception of human liver tissue, the small mitochondrial RNA Met is present across all species and tissues. The Y axis represents the expression in raw reads count, whereas the X axis represents the genomic positions in the mitochondrial genome of reference for each species. (B) We represented the mt-tRNA Met of each species while highlighting in red the sequences corresponding to the mt-miRNAMet. We highlight in orange the presence of polymorphisms across the species, by comparing reference mitochondrial genomes of each species. See the Data Availability section to see a list of the studies that the data come from.
The Small Mitochondrial RNAs Do Not Bind mRNAs through AGO2
To further investigate whether the small mitochondrial RNAs can participate in RNAi through AGO2, we analyzed data from a CLEAR-CLIP sequencing experiment focused on AGO2. The CLEAR-CLIP is a modified version of CLIP-seq, which is itself a modified version of the RIP-seq. In CLIP-seq, prior to IP of the target protein, a UV treatment is used to cross-link proteins and RNAs, so that even RNAs with weak bonding to the proteins will be precipitated. As such, CLIP-seq is more sensitive than RIP-seq. Similarly, the CLEAR-CLIP has one main difference from the CLIP-seq, a ligase step. After IP, the CLEAR-CLIP method uses a ligase to connect both AGO2-bound miRNA and mRNA into a single chimeric RNA. Thus, the CLEAR-CLIP data set includes chimeric RNAs (2% of the library), partly miRNA and partly mRNA, and it is the most powerful tool for identification of miRNA targets currently available (Moore et al. 2015). Indeed, despite having only 2% chimeric RNAs, we can be confident that this 2% includes true miRNA–target interactions.
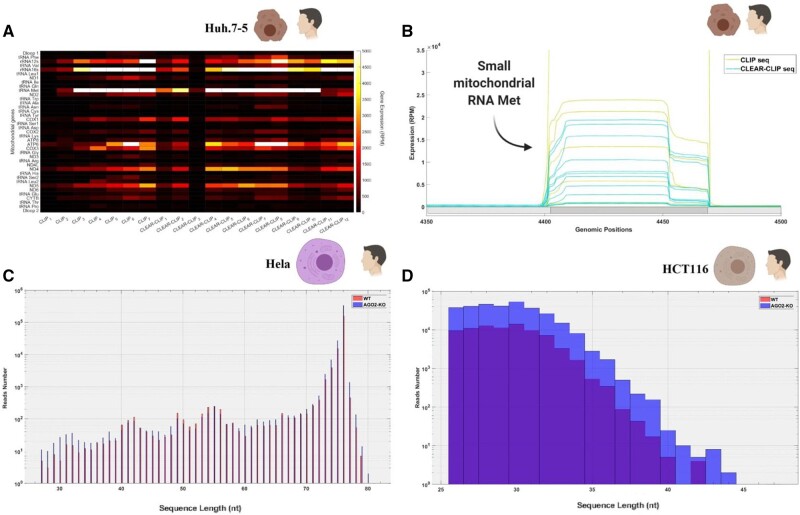
Prior to analyzing the chimeric RNAs within these samples, we aligned the sequences to the mtDNA, finding that a high percentage of sequences aligned to mt-genes (1–6% across libraries) and in particular to the gene for mt-tRNA Met (fig. 5A). As expected, the transcriptional signature of small mitochondrial RNA Met was conserved in this data set, despite the data set coming from another cell line (huh-7.5) than the analyses presented in the previous section. The transcriptional signature was almost identical across CLEAR-CLIP and CLIP samples (fig. 5B), albeit the CLIP samples tended to have higher expression levels of the small mitochondrial RNA Met (over 5,000 RPM). However, to our surprise, although the small mt-tRNA Met was abundant in our CLEAR-CLIP samples, we were not able to identify any chimeric RNA sequence involving this small RNA. Furthermore, the small mitochondrial RNA Met found in the data set were not small, being usually over 40 nt long, which suggests that AGO2 was bound to almost half of the mt-tRNA Met. This RNA is much longer than that which we observed in small RNA libraries and might indicate that AGO2 is not using this as a miRNA but rather that AGO2 might be involved in the processing of the mt-tRNA to small mitochondrial RNAs. To verify if the small mitochondrial RNA Met, or any other small mitochondrial RNA, was able to bind to mRNAs despite their length, we aligned the chimeric RNAs from the CLEAR-CLIP data sets to multiple sequence databases. First, we used a database including over 60,000 human transcripts, without being able to consistently identify chimeric RNAs (i.e., without finding the same binding pattern across three or more replicates) involving small mitochondrial RNAs and nuclear mRNAs. Likewise, we used databases for circular RNAs and long noncoding RNAs, without finding any consistent signatures of chimeric RNAs (i.e., signatures that were found across at least three independent replicates). To confirm that our inability to detect chimeric RNA involving small mt-RNA was not an artifact of the specific cell line used in our analysis, we repeated similar analyses using a separate CLEAR-CLIP data set from mouse brain samples. Again, we were unable to identify any consistent chimeric RNAs involving small mitochondrial RNAs and nuclear mRNAs across the 24 biological replicates in this data set. Thus, our results suggest that small mitochondrial RNAs do not bind AGO2 to participate in RNAi, and therefore alternative explanations need to be considered.
Fig. 5.
Expression of small mitochondrial RNAs expression in CLEAR-CLIP data sets and AGO2 knockouts. (A) The heatmap shows a comparison between CLIP and CLEAR-CLIP samples using Huh-7.5 cell cultures. The expression is normalized (reads per million), and we used a “hot” scale on Matlab to distinguish genes with high expression (white) from the ones with low expression (black). (B) Focuses on the area where the mt-tRNA Met is encoded (dark gray) to highlight that the expression of the small mitochondrial RNA encoded within this gene has the same transcriptional signature across all the samples. The light gray areas of different shades on the horizontal axis denote mitochondrial gene boundaries and are included only to highlight the boundaries of the mt-tRNA Met gene. The expression is normalized (reads per million). (C) The histogram compares the length of the reads in wild-type Hela cells (WT) and AGO2 knockout cells (AGO2-KO). The Y axis shows the raw number of reads in each treatment, whereas the X axis shows the range of lengths considered (25–80 nt). (D) The histogram compares the length of the reads in wild-type HCT116 cells (WT) and AGO2 knockout cells (AGO2-KO). The Y axis shows the raw number of reads in each treatment, whereas the X axis shows the range of lengths considered (25–45 nt).
AGO2 Knockout Does Not Affect the Transcription of Small Mitochondrial RNAs, but Affects the Transcription of Specific Mitochondrial Protein-Coding Genes
We explored an alternative hypothesis for why the small mitochondrial RNAs bind to AGO2, by further investigating the possibility that AGO2 may form part of the maturation process of the small mitochondrial RNAs. If AGO2 is required for the maturation of these small mitochondrial RNAs, then we would predict that knockdown of the AGO2 gene would lead to changes in the length or expression levels of these RNAs. To test this, we leveraged data from two independent RNA-seq data sets (Golden et al. 2017; Schuster et al. 2019). We used these data sets to perform a comparative analysis across wild-type and AGO2 knockout cells (Golden et al. 2017; Schuster et al. 2019), testing whether small mitochondrial RNAs have different expressions or lengths across the treatments. These two data sets were generated by independent research groups, utilizing two different cell cultures, HeLa and HCT116; where the first data set comprises total RNAs (non-poly A or size selection), and the second small RNAs (size selection). We found no differences in either the expression levels or length of small mitochondrial RNAs across the wild-type and knockout data sets (fig. 5C and D). In the small RNAs data set, the RNAs are between 21 and 45 nt long, and the size distribution of the small mitochondrial RNAs in the AGO2 knockout closely matches the distribution in the wild type of HeLa cells. Similarly, we observed no differences between wild type and knockout in the total RNAs data set, which includes mostly RNAs of approximately 70nt length, based on comparison of length distributions in histograms between samples with and without AGO2, suggesting that AGO2 is not involved in the processing of either small or long mitochondrial RNAs.
Nuclear miRNAs Bind to Mitochondrial mRNAs through Argonaute 2
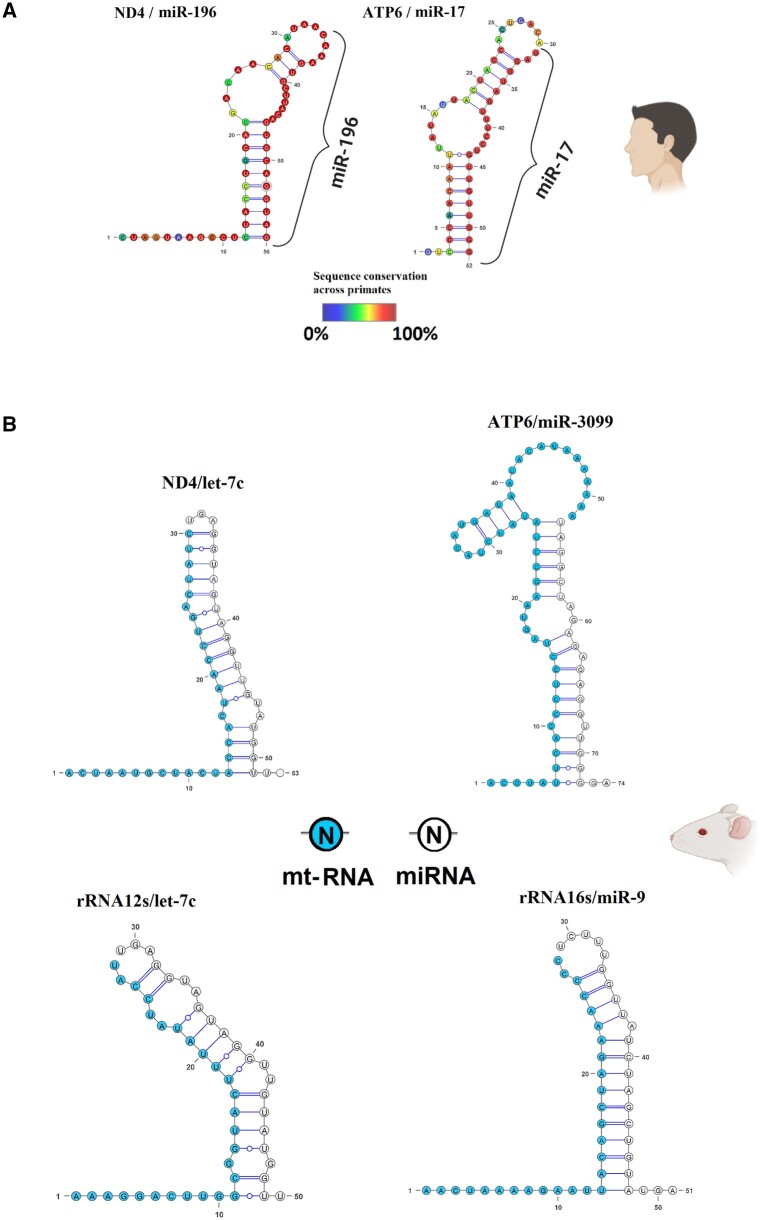
Notwithstanding the lack of interaction between small mitochondrial RNAs and nuclear mRNAs, we identified nuclear miRNAs that consistently bind to mitochondrial mRNAs through AGO2. Across the CLEAR-CLIP human data sets, we observed binding of hsa-miR-17-5p and hsa-miR-196a-5p to MT-ATP6 and MT-ND4, respectively (fig. 6A). The chimeric RNAs involving these two pairs of RNAs were found in multiple biological replicates, three replicates for the chimera ATP6/miR-17 and five replicates for ND4/miR196a. To further validate their genuine binding, we used bioinformatic prediction tools to estimate the RNA-binding energies, finding that the energy between the two RNAs in the chimera is predicted to be similar to other confirmed miRNA/target interactions (less than −20 kcal/mol). Although we found only hsa-miR-17-5p and no other isoforms can bind to MT-ATP6 mRNA, our analysis of MT-ND4 mRNA chimeras revealed both the hsa-miR-196a-5p and hsa-miR-196b-5p isoforms bind to MT-ND4 mRNA, although it should be noted that the -a and -b isoforms of miR-196 differ only by one nucleotide. We investigated if these interactions between nuclear miRNAs and mitochondrial mRNAs are conserved across primates, using the frequency of polymorphisms as a measure of conservation. Thus, we compared the binding regions of the human MT-ND4 and MT-ATP6 across all available primate mitochondrial genomes in the NCBI database (789 sequences). Through this comparative analysis, we found that, in primates, the binding regions of both mitochondrial genes are much less conserved than their binding miRNAs. Indeed, fewer than 50% of the nucleotides in the binding regions on the mitochondrial mRNAs are not polymorphic. For example, the MT-ND4 nucleotide in position 18 of the chimeric RNA is apparently important for the binding with the miRNA miR-196 but is mutated in approximately 70% of the primates considered. Similarly, positions such as 15 and 16 on the same mitochondrial mRNA describe important binding sites C = G, but are conserved only in about half of the species considered. On the contrary, the sequence of the two binding miRNAs (miR17 and miR196) is conserved across primates, with all species having the exact same sequence (no polymorphic sites). This result suggests that the binding between these specific nuclear miRNAs to the mitochondrial mRNAs is unlikely to be general beyond humans.
Fig. 6.
RNA chimeras found in the CLEAR-CLIP data set of human and mouse. Both figures have been made using VARNA, with the structure of the RNA dimer computed using RNAfold from the RNA Vienna 2.0 package. (A) Two RNA chimeras were found in human Huh-7.5 cell lines, involving MT-ND4 and MT-ATP6. The figure uses a color scale, spanning blue to red, which represents the conservation (percentage of how many species have a different nucleotide in that position) of each nucleotide across primates. The nuclear miRNAs are indicated using only their miRNA family, and the specific name of each miRNA is specified in the main text. (B) Four RNA chimeras were found in mouse brain samples, involving two mRNAs (MT-ND4, MT-ATP6) and the two rRNAs (rRNA12s and rRNA16s). The mitochondrial RNAs (mt-RNA) within the chimera are indicated in light blue, whereas the miRNAs are denoted in white. The nuclear miRNAs are indicated using only their miRNA family, and the specific name of each miRNA is specified in the main text.
We used the closest related species to humans with an available CLEAR-CLIP data set, Mus musculus, to verify if the binding we found in humans is present in other species (Moore et al. 2015). We analyzed a CLEAR-CLIP data set of 24 brain samples from mice, and confirmed nuclear miRNAs binding mitochondrial RNAs in this species as well (fig. 6B). We found that the miRNA mmu-miR-3099-3p can bind the mRNA of MT-ATP6, and that mmu-let-7c-5p can bind the mRNA of MT-ND4. Surprisingly, although the target mitochondrial mRNAs are the same as we observed in the human samples, the miRNAs interacting belong to different miRNA families. Moreover, the regions of interactions within the mt-mRNAs with the miRNAs are not the same across the two species. However, the nuclear miRNAs bind to more than mt-mRNAs. We discovered that mmu-let-7c-5p and mmu-miR-9-5p can interact with the mitochondrial rRNA 12S and rRNA 16S, respectively. Interactions between miRNA and rRNA have been observed before, although it has never been observed across RNAs originating from different genomes—nuclear and mitochondrial. The results of our analyses across humans and mice thus show that although the target mitochondrial genes regulated from the nucleus are the same, the miRNAs co-opted for this role change across species.
Discussion
This study sheds new light on previously unknown aspects of the small mitochondrial RNAs, on one hand highlighting shared features between these RNAs and nuclear miRNAs, such as the ability to bind AGO2, and on the other providing evidence that these small RNAs are unlikely participants in RNAi through AGO2. There are two main shared features between the small mitochondrial RNAs and the miRNAs. The first shared feature is the ability to bind to AGO2. We provided evidence that the small mitochondrial RNAs can bind AGO2, one of the key proteins in gene regulation through RNAi. Although we found some noise in the AGO2-IP samples, genes having clearly defined small mitochondrial RNAs were strongly upregulated in the comparison between AGO2-IP and control-IP treatments, thus supporting a genuine binding of these RNAs to AGO2. These patterns were verified across multiple independent studies, suggesting that technical differences in RNA preparation, or sequencing, did not tangibly affect the presence of the small mitochondrial RNAs in the samples. However, because the length of small mitochondrial RNAs is more variable than that of miRNAs, it is challenging to discern between genuine small mitochondrial RNAs and target RNAs using only AGO2-IP data sets. Thus, finding small RNAs in both AGO2 bound and unbound data sets, as we have shown for the RNA encoded in mt-tRNA Met, is key to identifying genuine small mitochondrial RNAs.
The second shared feature we observed pertains to the conservation of one particular small mitochondrial RNA, encoded within the mt-tRNA Met, across multiple species. Sequence conservation exists for many miRNAs (Ambros 2004; Lee et al. 2007), and has been used before for phylogenetic purposes (Sempere et al. 2006; Lee et al. 2007; Kenny et al. 2015). However, ours is the first study to show that a small mitochondrial RNA is conserved across multiple diverged species within Chordata. Although the conservation of miRNAs usually relies on verifying the presence of the miRNA sequence in the genome of the target species, this method was not possible for the small mitochondrial RNAs because their sequence lies cryptic within the DNA sequence of other host genes, such as the small mitochondrial RNA encoded within the first half of the mt-tRNA Met gene. Thus, the presence of this sequence across multiple species only proves that the host mt-tRNA Met gene is conserved. Nonetheless, by using small RNA expression data, we showed that this small mitochondrial RNA is expressed across multiple tissues and species with a conserved transcriptional signature, and moreover that the DNA sequence containing the small RNA is more conserved than the rest of the mt-tRNA Met gene sequence. Arguably, this level of evidence is more reliable than the benchmark generally used for the identification of miRNAs, because we not only demonstrated the presence of the sequences within the genome, but also showed a clear and conserved transcriptional signature. Therefore, we argue that these shared features suggest a conserved function for these RNAs, likely through AGO2.
We failed to find any evidence, however, for the hypothesis that the small mitochondrial RNAs participate in RNAi through AGO2. Despite evidence of binding between the small mitochondrial RNAs and AGO2, the CLEAR-CLIP data sets we analyzed clearly showed that the small mitochondrial RNAs do not bind to mRNAs in a consistent manner. We further investigated whether the small mitochondrial RNAs were binding to circular RNAs or long noncoding RNAs, but again found no such evidence. Moreover, we demonstrated that the transcriptional signatures of small mitochondrial RNAs are not changed in AGO2 knockouts relative to wild-type samples, suggesting that AGO2 is not involved in the maturation of these small RNAs, similarly to most nuclear miRNAs. We believe that there are two plausible explanations for our observations. First, it is possible that the CLEAR-CLIP sequencing is not able to find the consistently expressed small RNA–mRNA complexes due to their low abundances. Indeed, the publication (Moore et al. 2015) associated with the CLEAR-CLIP data we sourced and used in our analysis reported that only 2% of the reads in the data set are chimeric, thus likely to reveal only the most abundant small RNA–mRNA complexes in a cell at any specific moment. This suggests that the small mitochondrial RNAs binding nuclear mRNAs were simply not abundant enough to be detected by the CLEAR-CLIP method. The second possibility is that the small mitochondria RNAs do not use AGO2 to bind mRNAs. In fact, although AGO2 is mostly known for its ability to cleave mRNAs, this protein has other, less known, functions. For example, AGO2 has been observed to mediate transcriptional silencing, different from RNAi, where this protein uses a guide RNA to bind a DNA region and inhibit the transcription of a target gene (Janowski et al. 2006). Therefore, although the small mitochondrial RNAs are not binding AGO2 for posttranscriptional regulation, they might be binding to this protein for other purposes, such as pretranscriptional silencing (Nazer et al. 2018). These possibilities require future attention.
Finally, we draw attention to our findings of nuclear miRNAs binding to mitochondrial mRNAs. These cases represent the first such recorded cases of nuclear miRNAs involved in the regulation of mitochondrial genes, and we believe deserve further research attention because they suggest hitherto unknown molecular pathways by which the mitochondrion and nucleus can interact to shape organismal health and function. Taken together, our results highlight a previously neglected aspect of mitonuclear communication, demonstrating that the nucleus uses posttranscriptional regulation through miRNAs to affect the expression of mitochondrial genes, and providing new insights that provide some support for the prediction that the small mitochondrial RNAs may act as regulatory elements encoded within the mitochondrial genome. Considering the conservation of some of these RNAs, we suggest the small mitochondrial RNAs might be a previously overlooked site of selection within the mitochondrial genome, and play a key but as yet unknown, role in the regulation of cellular processes. Future studies should focus on resolving the function of these small RNAs, and understanding their evolutionary significance.
Materials and Methods
Cell Lines
According to Zhang et al. (2018), from which we sourced human cell line data for our analyses, HeLa cells were grown on 15 cm plates using MEDM plus 10% FBS. The neuronal progenitor (NP) and TDF cell culture were derived from human embryonic stem cells (hESC). The differentiation of hESC into NP was induced by replacing the original growing medium with DMEM/F12 supplemented with 2% B27, 100 ng/ml FGF, 100 ng/ml EGF, and 5 ng/ml heparin. Further details on the methods can be found at GSE115146 in the Gene Expression Omnibus (GEO) database and in the original article. The differentiation of hESC into TDF was induced by injection of resuspended cells into mice homozygous for severe combined immune deficiency spontaneous mutation (SCID). The tumors that grew were then removed after 6 weeks and cultured in a medium of 10% FBS, nonessential amino acids, 2 mM glutamine, 1% penicillin/streptomycin, and 0.55 μM β-mercaptoethanol. Further details on the methods can be found at GSE112006 in the Gene Expression Omnibus (GEO) database. According to Zamudio et al. (2014), mouse embryonic stem cell cultures (mESC) were grown on gelatinized tissue culture plates in Dulbecco’s Modified Essential Media supplemented with multiple other nutrients. The full list of nutrients and details about the growing method can be found at GSE50595 in the GEO database and in the original article.
Model Organisms
The experiments on mice sourced from the Jackson laboratory were carried out in compliance with their institutional protocols. According to Woo et al. (2017), the experiments on mice sourced by Stefan Somlo were carried out in compliance with the Animal Care and Use Committee (IACUC) rules at Sookmyung Women’s University. According to Vaz et al. (2015), zebrafish (Singapore strain) were maintained according to the Animal Care and Use Committee (IACUC) rules.
Human Samples
The BA9 samples were obtained by Hoss et al. (2015), and sample collection was exempt from ethics approval because the study involves only tissue collected postmortem, and consequently not classified as human subjects. This decision was taken by the Boston University School of Medicine Institutional Review Board (Protocol H-28974). The CHC samples were obtained by Butt et al. (2016), and they declared that written informed consent for the use of biological samples and clinical records was given by all the participants. Furthermore, they declared that their work was done in accordance with the ethical guidelines of the 1975 Declaration of Helsinki and the International Conference on Harmonization Guidelines for Good Clinical Practice.
AGO2-IP Sequencing
Although slightly different methods were used across studies, the studies all used CLIP protocols where the cross-linking between RNA and AGO2 was performed with UV-rays before cell lysis (Poria and Ray 2017). According to Zhang et al. (2018), before the AGO2 IP of HeLa cells, the cultures were UV irradiated at 400mj. The IP was performed using Anti-AGO2 (Abnova, H00027161-M01), and following the IP the RNA libraries were made and sequenced using Hi-Seq 2500. Further details can be found in the original article and in the GEO database (GSE115146). According to Kwon Y. (GEO database, GSE112006), before the cell lysis and AGO2-IP of the NP and TDF, the cultures were UV irradiated at 400mj. The IP was performed using Anti-AGO2 (ProteinTech) and following the IP the RNA libraries were made and sequenced using Hi-Seq 2000. Further details can be found in the GEO database (GSE112006). According to Zamudio et al. (2014), the mESC cells had been modified to leave only a modified AGO2 active in these cells. This modified AGO2 protein is known as FLAG-hemagglutinin (HA)-tagged hAGO2 (FHAGO2), which thus has a specific epitope that can be targeted for IP. Furthermore, contrary to the experiments in other cell lines, in this case the cell cultures were lysed before the UV cross-linking and IP. Further details can be found in the original article and in the GEO database (GSE50595).
CLEAR-CLIP Data Sets
We used two different CLEAR-CLIP data sets, both belonging to the same study where the CLEAR-CLIP method was initially presented (Moore et al. 2015). The first data set comprises Huh7.5 human hepatoma cells, whereas the second data set uses N2A mouse neuroblastoma. Although the full protocol used is described in the original article, here, we briefly describe the main difference between the previous IP experiments and this method. Contrary to the other CLIP methods, this protocol has a step where T4 Ligase I is used to bind together the cross-linked RNAs precipitated with AGO2, creating chimeric RNAs. These chimeric RNAs are approximately 70nt long RNAs where the first part of the sequence is the miRNA, and the second is the target of the miRNA present in the first section. Currently, this method is the gold standard to identify genuine miRNA targets.
AGO2 Knockout
We verified the effect of AGO2 knockout by analyzing the length distribution and abundance of small mitochondrial RNAs in two independent data sets. The first data set uses a total RNA-seq strategy, thus including mostly longer RNAs; whereas the second data set performed a size-selection step to sequence only smaller RNAs. The first data set includes wild-type and AGO2 knockout of human HCT116 cells (Golden et al. 2017). The protocol used to generate knockout cells relies on using lentiCRISPR v2, but full details are described in the original article. The second data set includes wild-type and AGO2 knockout of a human HeLa cells (Schuster et al. 2019). The protocol used to generate knockout cells relies on using pCRISPR-hCAS9-1xgRNA-Puro plasmid, but full details are described in the original article.
Small Mitochondrial RNAs Sequence Alignment
The alignment of the RNA-seq libraries, both with and without IP, was performed using BowTie2 (Langmead and Salzberg 2012). We did not remove the adapter for each library, and instead used the soft clipping provided by bowtie2 through the setting –local. Due to the possible differences between the reference genome used, and the sequence of the individual used for analysis, we used permissive parameters in the alignment. Thus, for the alignment, we set a seed of 20 nucleotides (-L 20) and we allowed up to one mismatch (-N 1) between the small RNA and the reference genome. To gauge the overall amount of small RNAs within one gene, we used samtools idxstats function (Li et al. 2009), which outputs the reads aligning to each feature (gene). Likewise, to obtain the coverage for each nucleotide, and thus identify the mitochondrial genes having the transcriptional signature of a small mitochondrial RNA, we used the function genomecov of bedtools (Quinlan and Hall 2010). To generate the total coverage for each gene, we used the setting genomecov -d–i, whereas for the 5′ and 3′ coverage we added the setting −5 and −3, respectively. To generate the correct format to plot the gene coverage in the R package circlize (Gu et al. 2014), we used the parameter -bg instead of -d on bedtools genomecov (Quinlan and Hall 2010). The alignment was done using the current reference genomes for Homo sapiens (NC_012920.1) and M. musculus (NC_005089.1).
Small Mitochondrial RNAs Met Polymorphism Identification
The identification of polymorphisms on the small mitochondrial RNA Met, and the mt-tRNA Met gene, across multiple species was performed by visual comparison of the sequences downloaded from the mt-tRNAs database mitotRNAdb (http://mttrna.bioinf.uni-leipzig.de/mtDataOutput/Welcome, last accessed February 15, 2022). We only used the reference sequences to establish the presence of polymorphisms, and we did not include any population data, because the quality of population data (i.e., the frequency of specific polymorphisms) is very different across the organisms considered. Therefore, we used the reference genome present in mitotRNAdb (Jühling et al. 2009) for Danio rerio (NC_002333.2), Gallus gallus (NC_001323.1), Homo sapiens (NC_012920.1), and M. musculus (NC_005089.1).
Analysis of RNA-Target Chimeras
The identification of the RNA chimeras present in the CLEAR-CLIP data set was performed using a custom pipeline. We created our own pipeline due to the lack of known methods suitable for analyzing mitochondrial transcripts and unconventional targets. A representation of the steps followed in the analysis can be found in supplementary figure S5, Supplementary Material online, however, we describe here the main steps of this pipeline. At first, we used Bowtie2 (Langmead and Salzberg 2012) with settings -local -N 1 -L 20 to identify all the small RNAs that align to the mitochondrial genome. Once we had identified all the mitochondrial sequences, we clustered the sequences using the function -cluster_fast from Vsearch (Rognes et al. 2016) with the following parameters: –id 0.9 –target_cov 0.9 -sizeout. This allowed us to group all the reads having mitochondrial sequences, bringing the several million reads to a mere few thousand. Next, by using BlastN, we tested if there were mtRNA–nuclear RNA chimeras among the sequences we selected. We investigated many possible types of nuclear RNAs that might be interacting with the small mitochondrial RNAs by making multiple databases to test with BlastN. We used BlastN locally with the following databases: long noncoding RNAs through Lncipedia (Volders et al. 2019); miRNAs through miRBase (Griffiths-Jones et al. 2006); human mRNAs through both the whole transcriptome and the selection of only 3′-UTRs from the ensembl Biomart (Hunt et al. 2018); circular RNAs through circbase (Rybak-Wolf et al. 2015; Maass et al. 2017); mitochondrial RNAs through the human mitochondrial reference genome (NC_012920.1). Using the tabular output of BlastN, we selected all the sequences that had a match on BlastN for more than 15 nt and less than 40 nt on both the mtDNA database and one of the others. We achieved this by using a combination of awk and comm functions in a Unix terminal, but it can be done in many other ways, including manually. In fact, we then visually verified the presence of genuine RNA chimeras, where a short RNA was bound to a longer RNA, usually with the shorter RNA being first. Lastly, we predicted the binding energy between the two RNAs in the chimera using RNAfold from the ViennaRNA2.0 package (Lorenz et al. 2011) and plotted the binding of the RNAs using VARNA. Links to the program used can be found on the supplementary table S1, Supplementary Material online.
In the following lines, we describe the sequences found in the chimeric RNAs described in figure 6. In the human samples, the mitochondrial sequences in the RNA chimeras are CTAGTAAGCCTCTACCTGCACGACAACACATAA (ATP6) and GTCCCAACAATTATATTACTACCACTGACA (ND4); the miRNA sequences are CAAAGUGCUUACAGUGCAGGUAG (miR-17) and UAGGUAGUUUCCUGUUGUUGGG (miR-196b). In the mouse samples, the mitochondrial sequences in the RNA chimeras are ACGTATTCACCCTCCTAGTAAGCCTATATCTACATGATAATACATAAAAAAA (ATP6) and ACTAATGCTACTACCACTAACCTGACTATC (ND4); the miRNA sequences are UGAGGUAGUAGGUUGUAUGGUU (Let-7) and UAGGCUAGAGAGAGGUUGGGGA (miR-3099).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We wish to acknowledge all researchers who made their data freely available, thus giving us the chance to perform this work. The study was supported through the Australian Research Council Discovery Program scheme to D.K.D. (Grant No. DP200100892) and an Australian Government Research Training Program (RTP) Scholarship to A.P.
Author Contributions
A.P. and D.K.D. conceived the study. A.P. performed the analyses. A.P. and D.K.D. discussed the results and wrote the manuscript.
Data Availability
This study is based on published data deposited in the Sequence Read Archive (SRA) in NCBI, thus the descriptions below are sourced from the original studies from which the data were obtained. All the accession IDs can be found on the supplementary table S1, within the Supplementary Material online.
Literature Cited
- Ambros V. 2004. The functions of animal microRNAs. Nature 431(7006):350–355. [DOI] [PubMed] [Google Scholar]
- Bandiera S, et al. 2011. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One 6(6):e20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottje WG, et al. 2017. Identification and differential abundance of mitochondrial genome encoding small RNAs (mitosRNA) in breast muscles of modern broilers and unselected chicken breed. Front Physiol. 8(October):816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AM, et al. 2016. Parallel expression profiling of hepatic and serum microRNA-122 associated with clinical features and treatment responses in chronic hepatitis C patients. Sci Rep. 6(February):21510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus MF, Wolff JN, Sgrò CM, Dowling DK.. 2017. Experimental support that natural selection has shaped the latitudinal distribution of mitochondrial haplotypes in Australian Drosophila melanogaster. Mol Biol Evol. 34(10):2600–2612. [DOI] [PubMed] [Google Scholar]
- Cloonan N. 2015. Re-thinking miRNA-mRNA interactions: intertwining issues confound target discovery. Bioessays 37(4):379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, et al. 2018. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560(7717):238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden RJ, et al. 2017. An argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542(7640):197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ.. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34(Database issue):D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B.. 2014. Circlize implements and enhances circular visualization in R. Bioinformatics 30(19):2811–2812. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN.. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 15(8):509–524. [DOI] [PubMed] [Google Scholar]
- Hoss AG, et al. 2015. miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med Genomics. 8(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ΘHunt SE, et al. 2018. Ensembl variation resources. Database 2018 (January). [Google Scholar]
- James AC, Ballard JWO.. 2003. Mitochondrial genotype affects fitness in Drosophila simulans. Genetics 164(1):187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, et al. 2006. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 13(9):787–792. [DOI] [PubMed] [Google Scholar]
- Jühling F, et al. 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37(Database issue):D159–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny NJ, et al. 2015. The phylogenetic utility and functional constraint of microRNA flanking sequences. Proc Biol Sci. 282(1803):20142983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajbner Z, Pnini R, Camus MF, Miller J, Dowling DK.. 2018. Experimental evidence that thermal selection shapes mitochondrial genome evolution. Sci Rep. 8(1):9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Langmead. 2013. Bowtie2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larriba E, Rial E, Del Mazo J.. 2018. The landscape of mitochondrial small non-coding RNAs in the PGCs of male mice, spermatogonia, gametes and in zygotes. BMC Genomics 19(1):634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-T, Risom T, Strauss WM.. 2007. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 26(4):209–218. [DOI] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, et al. 2011. ViennaRNA package 2.0. Algorithms Mol Biol. 6(November):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass PG, et al. 2017. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl). 95(11):1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z.. 2005. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the argonaute 2 protein. RNA 11(6):849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, et al. 2011. The human mitochondrial transcriptome. Cell 146(4):645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, et al. 2015. miRNA-target chimeras reveal miRNA 3’-end pairing as a major determinant of argonaute target specificity. Nat Commun. 6(November):8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazer E, Dale RK, Palmer C, Lei EP.. 2018. Argonaute2 attenuates active transcription by limiting RNA polymerase II elongation in Drosophila melanogaster. Sci Rep. 8(1):15685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, Wallace DC, Burelle Y.. 2016. The rise of mitochondria in medicine. Mitochondrion 30(September):105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poria DK, Ray PS.. 2017. RNA-protein UV-crosslinking assay. Bio Protocol. 7(6):e2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi A, Dowling DK.. 2019. The genomic origins of small mitochondrial RNAs: are they transcribed by the mitochondrial DNA or by mitochondrial pseudogenes within the nucleus (NUMTs)? Genome Biol Evol. 11(7):1883–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi A, Dowling DK.. 2021. Small mitochondrial RNAs as mediators of nuclear gene regulation, and potential implications for human health. BioEssays 43(6):e2000265. [DOI] [PubMed] [Google Scholar]
- Pozzi A, Plazzi F, Milani L, Ghiselli F, Passamonti M.. 2017. SmithRNAs: could mitochondria ‘bend’ nuclear regulation? Mol Biol Evol. 34(8):1960–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Fry A, Sheldahl L.. 2006. Nuclear–mitochondrial epistasis and Drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics 172(1):329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ.. 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 19(12):645–653. [DOI] [PubMed] [Google Scholar]
- Riggs CL, et al. 2018. Small non-coding RNA expression and vertebrate anoxia tolerance. Front Genet. 9(July):230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S, et al. 2013. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 23(6):759–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F.. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4(October):e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, et al. 2015. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 58(5):870–885. [DOI] [PubMed] [Google Scholar]
- Scheibye-Alsing K, Cirera S, Gilchrist MJ, Fredholm M, Gorodkin J.. 2007. EST analysis on pig mitochondria reveal novel expression differences between developmental and adult tissues. BMC Genomics 8(October):367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer WF, Syverton JT, Gey GO.. 1953. Studies on the propagation in vitro of poliomyelitis viruses: IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med. 97(5):695–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S, Overheul GJ, Bauer L, van Kuppeveld FJM, van Rij RP.. 2019. No evidence for viral small RNA production and antiviral function of argonaute 2 in human cells. Sci Rep. 9(1):13752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Cole CN, McPeek MA, Peterson KJ.. 2006. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zool B Mol Dev Evol. 306(6):575–588. [DOI] [PubMed] [Google Scholar]
- ΘSloan DB, et al. 2018. Cytonuclear integration and co-evolution. Nat Rev Genet. 19(10):635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song GJ, Lewis V.. 2008. Mitochondrial DNA integrity and copy number in sperm from infertile men. Fertil Steril. 90(6):2238–2244. [DOI] [PubMed] [Google Scholar]
- Sprenger H-G, Langer T.. 2019. The good and the bad of mitochondrial breakups. Trends Cell Biol. 29(11):888–900. [DOI] [PubMed] [Google Scholar]
- Townley-Tilson WHD, Pendergrass SA, Marzluff WF, Whitfield ML.. 2006. Genome-wide analysis of mRNAs bound to the histone stem–loop binding protein. RNA 12(10):1853–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz C, et al. 2015. Deep sequencing of small RNA facilitates tissue and sex associated microRNA discovery in zebrafish. BMC Genomics 16(November):950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders P-J, et al. 2019. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 47(D1):D135–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JN, Ladoukakis ED, Enríquez JA, Dowling DK.. 2014. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Philos Trans R Soc Lond B Biol Sci. 369(1646):20130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, et al. 2017. Profiling of miRNAs and target genes related to cystogenesis in ADPKD mouse models. Sci Rep. 7(1):14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee WKW, Sutton KL, Dowling DK.. 2013. In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Curr Biol. 23(2):R55–56. [DOI] [PubMed] [Google Scholar]
- Zamudio JR, Kelly TJ, Sharp PA.. 2014. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell 156(5):920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, et al. 2018. A novel class of microRNA-recognition elements that function only within open reading frames. Nat Struct Mol Biol. 25(11):1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. 2014. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158(3):607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study is based on published data deposited in the Sequence Read Archive (SRA) in NCBI, thus the descriptions below are sourced from the original studies from which the data were obtained. All the accession IDs can be found on the supplementary table S1, within the Supplementary Material online.