Abstract
Primary cutaneous CD8+ aggressive epidermotropic cytotoxic T-cell lymphoma (pcAECyTCL) is a rare variant of cutaneous T-cell lymphoma with an aggressive clinical course and a very poor prognosis. Until now, neither a systematic characterization of genetic alterations driving pcAECyTCL has been performed, nor effective therapeutic regimes for patients have been defined. Here, we present the first highresolution genetic characterization of pcAECyTCL by using wholegenome and RNA sequencing. Our study provides a comprehensive description of genetic alterations (i.e., genomic rearrangements, copy number alterations and small-scale mutations) with pathogenic relevance in this lymphoma, including events that recurrently impact genes with important roles in the cell cycle, chromatin regulation and the JAKSTAT pathway. In particular, we show that mutually exclusive structural alterations involving JAK2 and SH2B3 predominantly underlie pcAECyTCL. In line with the genomic data, transcriptome analysis uncovered upregulation of the cell cycle, JAK2 signaling, NF-κB signaling and a high inflammatory response in this cancer. Functional studies confirmed oncogenicity of JAK2 fusions identified in pcAECyTCL and their sensitivity to JAK inhibitor treatment. Our findings strongly suggest that overactive JAK2 signaling is a central driver of pcAECyTCL, and consequently, patients with this neoplasm would likely benefit from therapy with JAK2 inhibitors such as Food and Drug Adminstration-approved ruxolitinib.
Introduction
Primary cutaneous CD8+ aggressive epidermotropic cytotoxic T-cell lymphoma (pcAECyTCL) is a rare variant of cutaneous T-cell lymphoma (CTCL) still regarded as a provisional entity by the World Health Organization (WHO) and characterized by an abrupt onset and a highly aggressive clinical course.1,2 pcAECyTCL presents primarily in the skin with widespread plaques and tumors, often with hemorrhagic ulcerations and necrosis; however, dissemination to extracutaneous sites (especially the central nervous system, lungs, oral cavity and testes) is not uncommon.3,4 Malignant T cells causing pcAECyTCL typically express CD3, CD7, CD8, CD45RA, TCR-βF1, T-BET and one or more cytotoxic markers (e.g., granzyme B, perforin, TIA-1), which strongly suggests that neoplastic cells in this lymphoma derive from CD8+ T cells.2,3 Effective therapeutic regimes for pcAECyTCL are currently lacking, and consequently, patients have a poor prognosis with a median overall survival of 12 months.1
Thus far the study of the pathogenetic basis of this malignancy has been marginal due to its rarity. Recently, a study performed on tumors from 20 patients defined the copy number alteration (CNA) profile of pcAECyTCL by using array-based comparative genomic hybridization,5 and before this, two clinical case reports included the evaluation of CNA in single patients by using array-based methods as well.6,7 Recurrent CNA uncovered by these studies include losses within 1p, 9p, 13q and 16p as well as gains within 7q, 8q and 17q, with loss of the region containing CDKN2A/B being the most frequent CNA.5 However, aside from the aforementioned chromosomal imbalances, causative genetic changes in pcAECyTCL remain unknown.
Here, we present the first high-resolution genomic analysis of pcAECyTCL using whole-genome sequencing (WGS) and RNA sequencing (RNA-seq). We describe for the first time a number of genomic rearrangements, CNA and smallscale mutations with pathogenic relevance in this lymphoma. In particular, our results suggest that overactivation of JAK2 signaling due to oncogenic changes in JAK2 and SH2B3, two genes with key roles in this signaling pathway, underlie predominantly pcAECyTCL. These findings have important implications for patient standard of care.
Methods
Patient selection and sequencing
Frozen tumor biopsies (≥70% tumor cells) from 12 patients with pcAECyTCL (Online Supplementary Figure S1; Online Supplementary Table S1) were subjected to WGS. Six samples of this cohort (i.e., AEC2-4/6/8/12) were additionally subjected to RNA-seq. Sequencing, data processing and DNA/RNA analyses are described in the Online Supplementary Appendix (Online Supplementary Figures S2 and S3; Online Supplementary Tables S2 to S9). Diagnosis was performed by an expert panel of dermatologists/ pathologists in accordance with the WHO-EORTC classification for primary cutaneous lymphomas.1,2 Patient material was approved by the Institutional Review Boards of the Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico and Leiden University Medical Center. Informed consent was obtained from patients in accordance with the declaration of Helsinki.
Validation of structural genomic alterations and small-scale mutations
Select rearrangements, interstitial deletions and single nucleotide variants (SNV) were validated by Sanger sequencing, droplet digital polymerase chain reaction (ddPCR) and/or fluorescence in situ hybridization (FISH). Details of the validation experiments are included in the Online Supplementary Appendix (Online Supplementary Figures S4 to S9; Online Supplementary Table S10).
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) tissue sections were immunohistochemically stained with primary antibodies against phospho-STAT3 (Cell Signaling Technology, Cat.No. 9145) or phospho-STAT5 (Cell Signaling Technology, Cat.No. 9359) using Dako REAL detect system (Dako, Cat.No. K5005), counterstained in Mayer’s hematoxylin solution and coverslipped using Vectamount (Vector Laboratories, Cat.No. H5000).
Cell culture, fusion gene construction and viral transduction
Ba/F3 cells (DSMZ, Cat.No. ACC-300) were used for functional experiments. Parental Ba/F3 cells were cultured in RPMI-1640 (10% heat-inactivated fetal bovine serum, 10 ng/mL interleukin- 3 [IL3]) at 37° C with 5% CO2 in a humidified atmosphere. JAK2 fusions (i.e., TFG-JAK2, PCM1-JAK2, KHDRBS1-JAK2) and control genes (i.e., eGFP, TFG-MET) were constructed and inserted into a lentiviral vector using the method described by Lu et al.8 Primers, templates and vectors used for fusion gene construction are detailed in the Online Supplementary Appendix (Online Supplementary Tables S11 to S13). Lentiviral particles were produced in HEK-293T cells, quantified by p24 enzyme-linked immunosorbent assay (ELISA), and transduced into Ba/F3 cells at MOI-9 with lipofectamine. Successfully transduced Ba/F3 cells were selected with puromycin (2.5 mg/mL) for 3 days.
Ba/F3 cell viability and inhibitor assays
Cell viability of parental and transduced Ba/F3 cells was determined 7 days after IL3 withdrawal by MTT assay (Promega, Cat.No. G4000). Inhibitor assays were performed by treating IL3-independent Ba/F3 cells expressing fusion genes with ruxolitinib or AZD1480 at seven different concentrations for 72 hours and measuring cell viability by MTT assay.
Western blots
The effect of JAK1/2 inhibitors ruxolitinib and AZD1480 on JAK2 and STAT5 phosphorylation was evaluated by western blotting. Cells were washed to remove traces of serum and incubated with inhibitor for 90 minutes. Cells were lysed in SDS lysis buffer containing protease inhibitors and separated by SDSPAGE. Antibodies employed were anti-JAK2 (Abcam, Cat.No. ab108596), anti-phospho-JAK2 (Cell Signaling Technology, Cat.No. 3776), anti-STAT5 (Cell Signaling Technology, Cat.No. 94205), anti-phospho-STAT5 (Cell Signaling Technology, Cat.No. 9351) and anti-GAPDH (Cell Signaling Technology, Cat.No. 2118).
Results
JAK2 fusions are prominent in a complex landscape of rearrangements
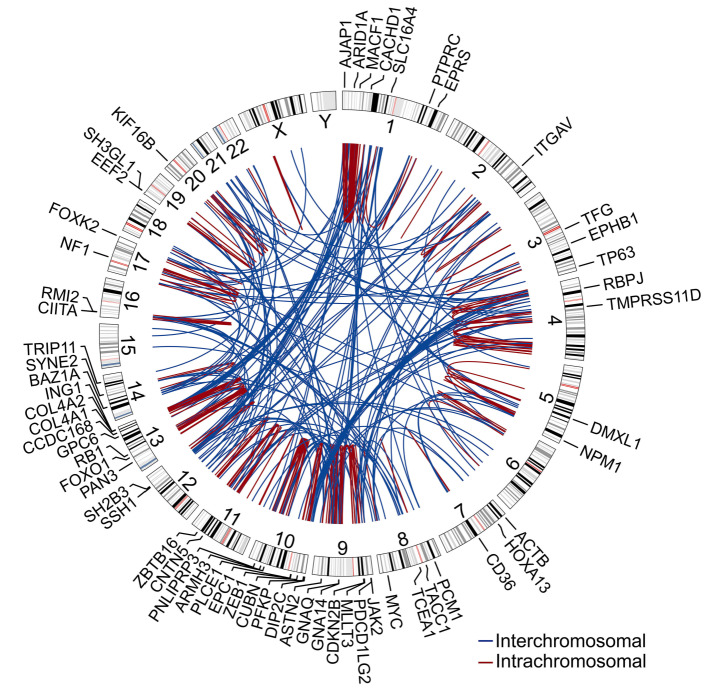
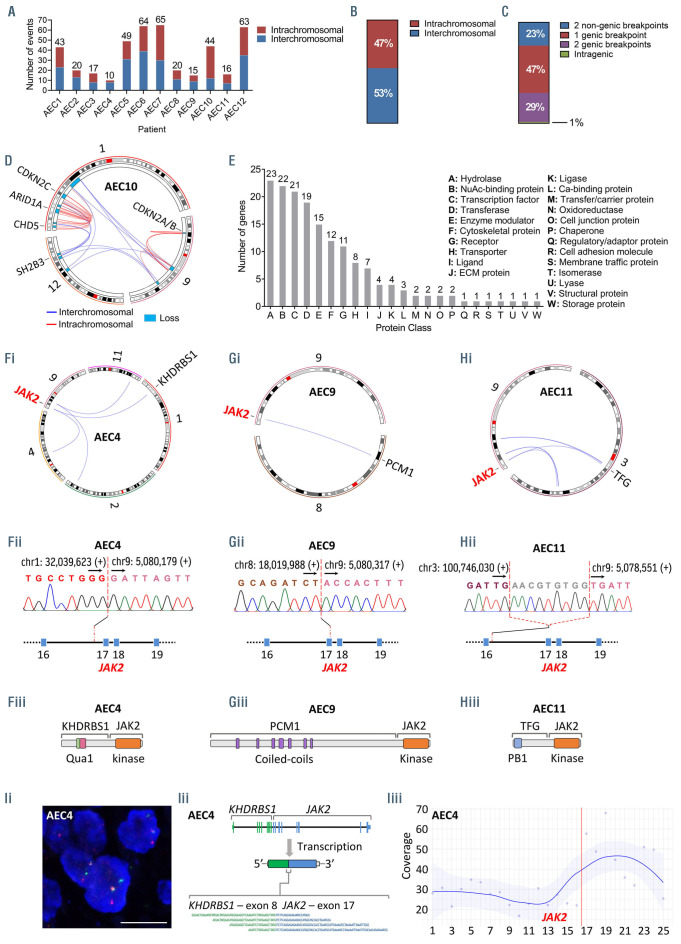
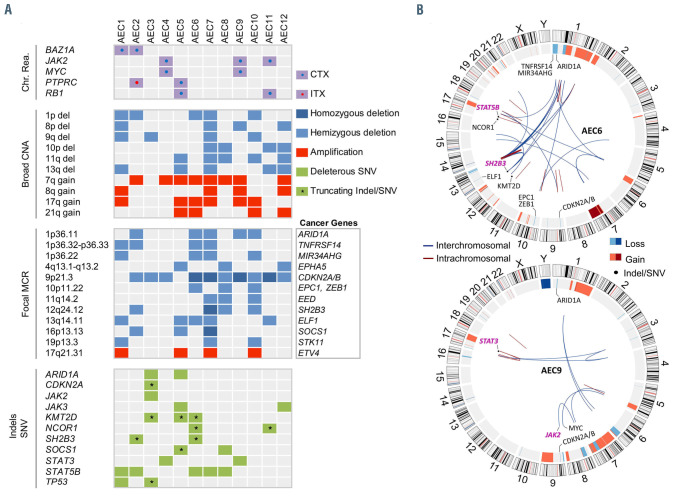
The analysis revealed a heterogeneous and complex landscape of genomic rearrangements (total events, 426; range, 10-65; mean/patient ± standard deviation [SD], 36±21) (Figure 1; Figure 2A; Online Supplementary Figure S3). Fifty-three percent of events were interchromosomal (range/patient, 27-80%) (Figure 2B). The majority of rearrangements (77%) disrupted either one or two annotated genes, while the rest (23%) disrupted nongenic regions (Figure 2C). Four patients, AEC6, AEC7, AEC10 and AEC12, displayed complex rearrangements (chromothripsis/ chromoplexy-like) affecting chromosomes 13, 10, 1/9/12 and 4, respectively (Figure 2D; Online Supplementary Figure S3 and S10). We observed a total of 305 rearranged genes, 59 of which are implicated in neoplasms at present (Online Supplementary Table S14). Gene ontology analysis revealed that rearranged genes encode principally (ngenes=91 of 305) proteins with roles in signal transduction (i.e., hydrolases, transferases, enzyme modulators, receptors) and transcriptional regulation (i.e., transcription factors, chromatin regulators) (Figure 2E; Online Supplementary Tables S15 and S16). Out of seventeen recurrently rearranged genes detected in our cohort (npatients =2 or 3) (Online Supplementary Table S17), six are established cancer genes with important functions in the regulation of the cell cycle (i.e., MYC, RB1), chromatin remodeling (i.e., BAZ1A) and the JAK-STAT pathway (i.e., JAK2, PTPRC, SH2B3) (Figure 1; Online Supplementary Figures S4 and S5).
The JAK-STAT pathway, a frequent driver of hematological neoplasms, was the only cytokine-elicited signal transduction pathway impacted by rearrangements in pcAECyTCL. Fusion genes involving JAK2 were detected in three of 12 patients (i.e., AEC4: KHDRBS1-JAK2; AEC9: PCM1-JAK2; AEC11: TFG-JAK2) (Figure 2F to H). These events fused the tyrosine kinase domain of JAK2 with one or more oligo/dimerization domains from the fusion partner (i.e., AEC4: Qua1 domain, AEC9: coiled-coil domains, AEC11: PB1 domain) (Figure 2F to H). The resulting chimeric proteins are predicted to self-oligo/dimerize and become activated without the need of cytokine-mediated receptor stimulation, ultimately overactivating JAK2 signaling. Of note, two of three patients carrying JAK2 fusions carried MYC fusions as well (i.e., AEC4: ACTBMYC, AEC9: NPM1-MYC) (Online Supplementary Figure S4). Interestingly, apart from acquiring the ability to selfactivate, JAK2 fusions under the transcriptional control of their partner’s promoter may also experience augmented expression in comparison to wild-type JAK2, as evidenced in patient AEC4 (Figure 2I). In contrast, rearrangements involving PTPRC and SH2B3, each observed in two of 12 patients, disrupted these two negative regulators of the JAK-STAT pathway.
Figure 1.
Landscape of genomic rearrangements in pcAECyTCL. Circos plot showing 426 genomic rearrangements detected in twelve pcAECyTCL genomes by wholegenome sequencing (WGS). The outer ring shows rearranged genes with established roles in cancer. The area at the center of the plot contains arcs representing interchromosomal (blue) and intrachromosomal (red) events. The ring between the gene labels and the arcs contains human chromosome ideograms arranged circularly end to end. pcAECyTCL: primary cutaneous CD8+ aggressive epidermotropic cytotoxic T-cell lymphoma.
JAK2 signaling inhibitor SH2B3 is focally deleted in pcAECyTCL
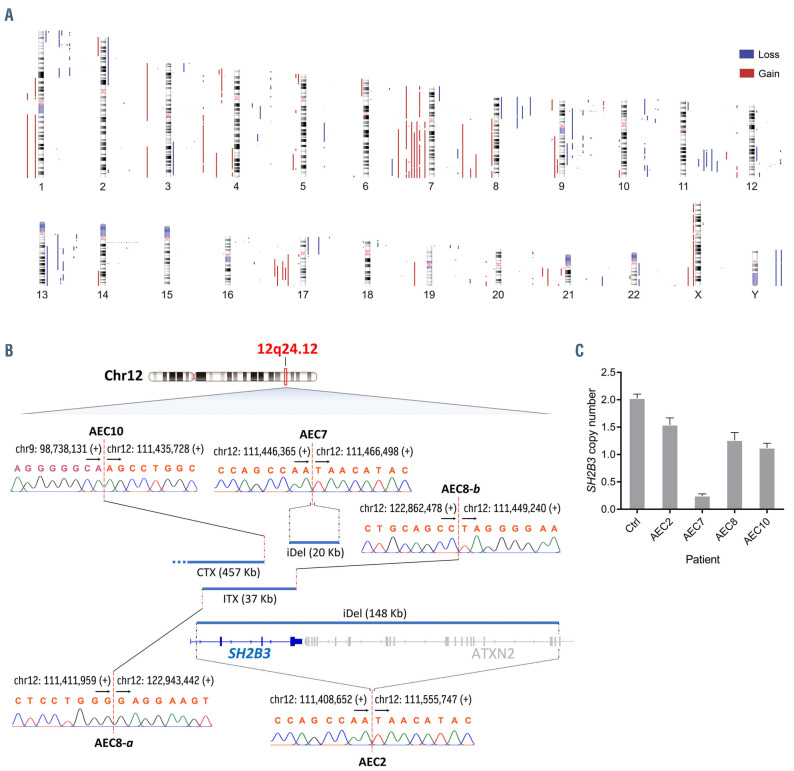
The most frequent broad chromosomal imbalances (npatients≥4; >3 Mb) were deletions within 1p, 8p, 9q, 10p, 11q and 13q and gains within 7q, 8q, 17q and 21q (Figure 3A; Figure 4A). We identified 24 recurrent focal (≤ 3 Mb) minimal common regions (MCR) shared by CNA between patients (npatients≥3; deletions: 19, gains: 5) (Online Supplementary Table S5), 12 of which contained cancer genes predominantly involved in the cell cycle, chromatin regulation and the JAK-STAT pathway (Figure 4A).
The most common focal MCR involving cancer genes was deletion at 9p21.3 (10 patients), which included cell cycle regulators CDKN2A/B (Online Supplementary Figure S7). Of note, CDKN2A/B were found to be inactivated by interstitial deletions, unbalanced rearrangements, SNV and presumably even the action of long non-coding RNA ANRIL (CDKN2B-AS1)9 (Figure 4A; Online Supplementary Figure S11). Five of 12 patients had deletions at 1p36.11 and 13q14.11, which contained chromatin remodeler ARID1A and candidate cancer gene ELF1,10 respectively. Deletions at 1p36.32-p36.33, 1p36.22 and 12q24.12, observed in four of 12 patients, involved tumor suppressors TNFRSF14, MIR34AHG and SH2B3, respectively. Finally, three of 12 patients had deletions at 4q13.1-q13.2, 10p11.22, 11q14.2, 16p13.13 and 19p13.3, which contained tumor suppressors EPHA5, EPC1 (alongside ZEB1), EED, SOCS1 and STK11, respectively. On the other hand, gain at 17q21.31 (four patients), which enclosed ETV4, was the only recurrent (npatients ≥3) focal gain containing a cancer gene.
Remarkably, deletions at 12q24.12 were strikingly focal in all affected patients (20 Kb – 457 Kb), leading to the loss of one or more functional domains of SH2B3 (i.e., DD, PH, SH2 domains) in these individuals (Figure 3B and C; Online Supplementary Figure S5). SH2B3 (LNK) encodes an adaptor protein that antagonizes JAK2 signaling as part of a negative feedback loop in various hematopoietic cell types (e.g., erythroid progenitors, hematopoietic stem cells, megakaryocytes, pre-B cells, etc.) by suppressing the kinase activity of JAK2 through its SH2 domain.11 Of note, structural alterations involving JAK2 and SH2B3 were mutually exclusive in our cohort, affecting altogether seven of 12 patients. In addition, we investigated the possibility of SH2B3 silencing by promoter hypermethylation in our patients using methylation-specific melting curve analysis (MS-MCA) and found no evidence of this inactivation mechanism (Online Supplementary Figure S9).
Figure 2.
JAK2 fusions are recurrent in a complex landscape of rearrangements. (A) Number of genomic rearrangements per patient. The distribution of inter- and intrachromosomal rearrangements per patient is shown too. (B) Distribution of inter- and intrachromosomal rearrangements (cohort). (C) Distribution of genomic rearrangements based on the type of DNA sequences (genic, nongenic) involved in the event (cohort). (D) Circos plot showing a chromoplexy-like event in patient AEC10 that mediated the loss of multiple genomic regions in chromosomes 1, 9 and 12, several of which enclosed established tumor suppressor genes. (E) Distribution of rearranged genes according to the protein class their encoded proteins belong to. (F, G and H) Genomic rearrangements generated self-activating JAK2 fusions in pcAECyTCL as evidenced in patients (F) AEC4, (G) AEC9 and (H) AEC11. (i) Circos plots showing interchromosomal rearrangements involving chromosome 9 in patients with pcAECyTCL. JAK2 rearrangements were the common denominator between chromosome 9 events observed in these individuals. (ii) Validation of translocation breakpoints at JAK2 by Sanger sequencing in pcAECyTCL patients. Breakpoints occurred between exon 16 and exon 17 in all cases. (iii) Rearrangements involving JAK2 led to the formation of fusion genes encoding the tyrosine kinase domain of JAK2 and the oligo/dimerization domains of the fusion partners (KHDRBS1: Qua1 domain, PCM1: coiled-coil domains, TFG: PB1 domain), conferring the resulting chimeric protein the ability to self-activate. (I) In addition to acquiring self-activation ability, JAK2 fusions can also experience increased expression in comparison to wild-type JAK2. (i) Image of break-apart fluorescence in situ hybridization (FISH) analysis showing a JAK2 rearrangement in patient AEC4. Scale bar, 10 mm. (ii) Active expression of fusion gene KHDRBS1-JAK2 in patient AEC4 was detected by RNA sequencing (chimeric reads shown in diagram). (iii) Plot showing mean read coverage across all exons of JAK2 in patient AEC4. RNA expression between exon 17 and exon 25, the part of JAK2 under the transcriptional control of KHDRBS1’s promoter and encoding its tyrosine kinase domain, is considerably higher compared to RNA expression between exon 1 and exon 16. The red line indicates the breakpoint position. pcAECyTCL: primary cutaneous CD8+ aggressive epidermotropic cytotoxic T-cell lymphoma.
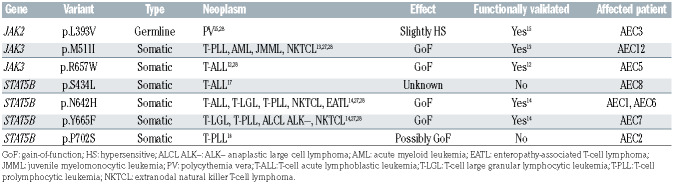
Table 1.
Identical variants in JAK and STAT proteins reported in other hematological malignancies.
Pathogenic small-scale mutations in JAK-STAT pathway genes predominate in pcAECyTCL
The discovery of recurrent structural alterations affecting principally genes involved in the cell cycle, chromatin regulation and the JAK-STAT pathway (via JAK2) prompted us to search for pathogenic indels and SNV in exonic sequences of genes with roles in the aforesaid cellular processes and additional signal transduction pathways (i.e., MAPK, NF-κB, PI-3-K/Akt and T-cell receptor [TCR] pathways) (Online Supplementary Table S7).
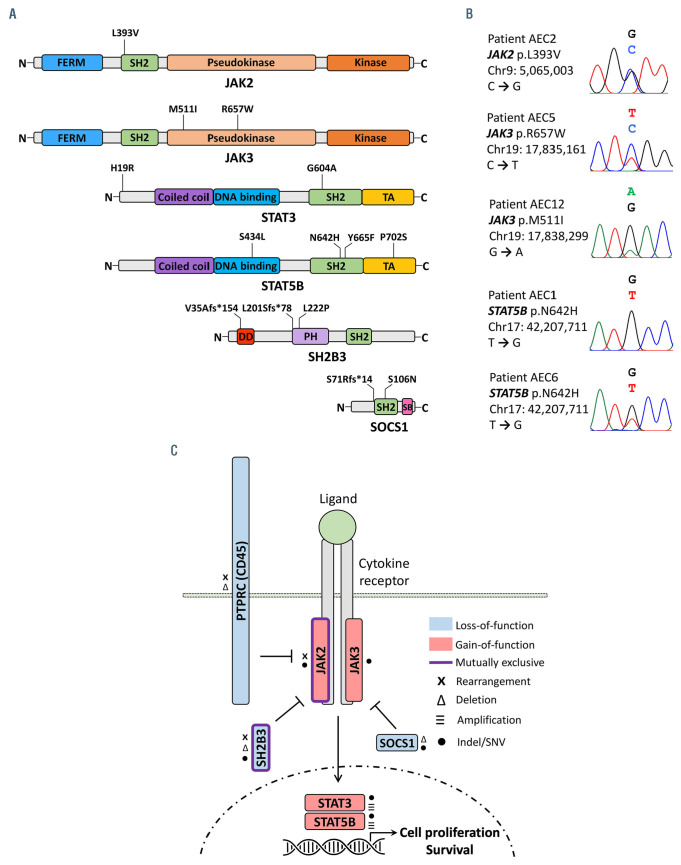
Besides the seven patients with structural alterations impacting the JAK2-SH2B3 signaling axis, four additional patients were found to carry bona fide gain-of-function SNV either in JAK3 (i.e., AEC5: p.R657W12; AEC12: p.M511I13) or STAT5B (i.e., AEC1 and AEC6: p.N642H14). Also, patient AEC3 bore a germline SNV in JAK2 (p.L393V15) which has been reported to render JAK2 slightly hypersensitive to cytokine stimulation (EPO ligand) (Figures 4A, 5A and B; Table 1). Moreover, two patients with JAK2 fusions and three patients with SH2B3 deletions also carried SNV affecting conserved residues in STAT3 (i.e., AEC4: p.H19R; AEC9: p.G604A) and STAT5B (i.e., AEC2: p.P702S16; AEC7: p.Y665F14; AEC8: p.S434L17), respectively (Online Supplementary Figure S6). Similarly, three patients carrying (putative) gain-of-function SNV in JAK or STAT genes also had indels leading to premature stop codons either in SH2B3 (i.e., AEC2: p.L201Sfs*78; AEC6: p.V35Afs*154) or SOCS1 (i.e., AEC5: p.S71Rfs*14) (Figures 4A and B, 5A).
Overall, nine of 12 patients had either structural or small-scale genetic alterations impacting the JAK2-SH2B3 signaling axis whereas the remaining three patients carried pathogenic indels/SNV in other JAK-STAT pathway genes (Figures 4A and B, 5C). In addition, cancer genes involved in the cell cycle (i.e., TP53) and chromatin regulation (i.e., ARID1A, KMT2D, NCOR1) were found to be recurrently impacted either by truncating mutations (i.e., nonsense, frameshift) or SNV predicted as deleterious (Figure 4A). We also observed 34 additional patient-specific small-scale mutations of unknown significance in reputable cancer genes (Online Supplementary Table S6).
Transcriptome analysis uncovers upregulation of JAK2 signaling in pcAECyTCL
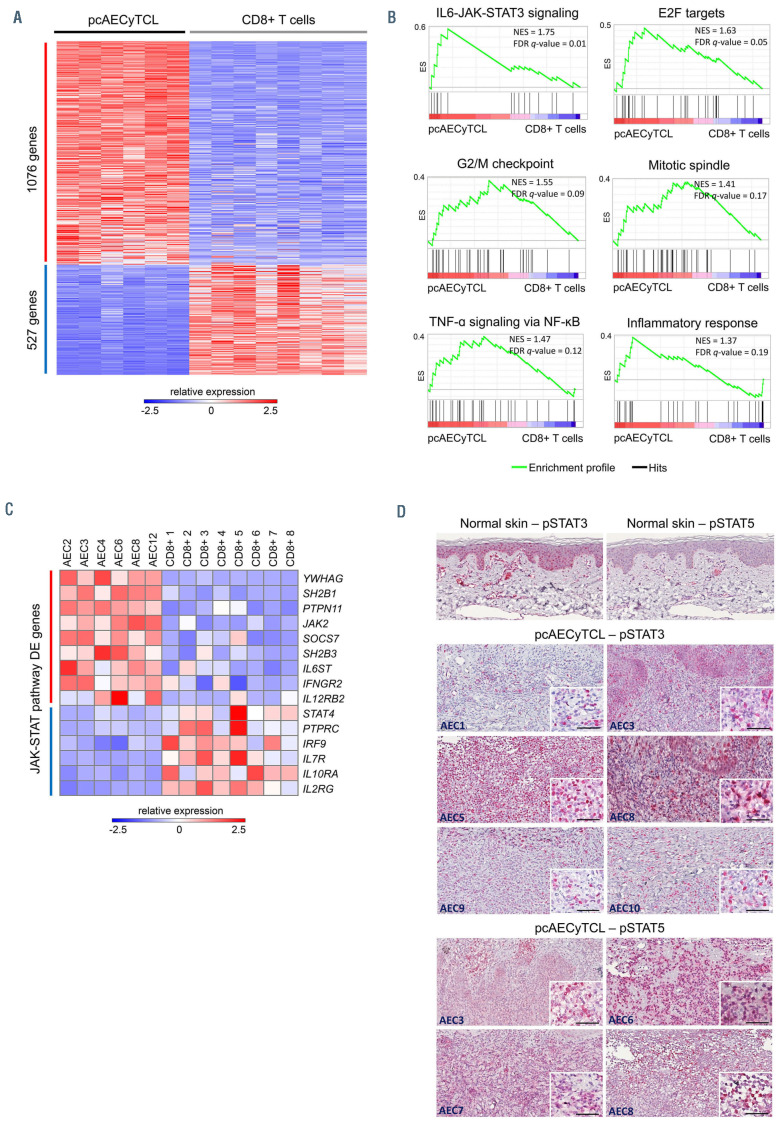
At present it is widely accepted that malignant T cells in pcAECyTCL derive from CD8+ T cells;1 however, to date no specific CD8+ T-cell subtype has been proposed as the cell of origin of this lymphoma. Since a hallmark of malignant T cells in pcAECyTCL is their distinctive epidermotropism, 3,4 we compared gene expression in pcAECyTCL with gene expression in normal skin-resident CD8+ T cells, which are characterized by a marked preferential tropism to the epidermal layer of the skin.18 This analysis identified 1,603 differentially expressed (DE) genes (1,076 upregulated, 527 downregulated, false discovery rate [FDR] <0.01) in the disease (Figure 6A; Online Supplementary Table S8). We next performed gene set enrichment analysis (GSEA) using annotated gene sets from MSigDB to search for deregulated pathways/ processes. Upregulated canonical signaling profiles included the JAK-STAT pathway (via STAT3, and to a lesser extent, via STAT5) and the TNF-α/NF-κB pathway. In addition, pcAECyTCL was characterized by the upregulation of the cell cycle (i.e., E2F targets, G2/M checkpoint, mitotic spindle) and high inflammatory response (Figure 6B; Online Supplementary Table S18).
Further examination of DE genes involved in the JAKSTAT pathway revealed that JAK2 signaling was specifically deregulated in pcAECyTCL. Upregulated genes included among others JAK2 itself, components of type I and II cytokine receptors that signal predominantly via JAK2 (i.e., IFNGR2, IL12RB2) and established enhancers of JAK2 signaling (i.e., PTPN11, SH2B1). In contrast, downregulated JAK-STAT pathway genes included PTPRC and genes encoding receptors exclusively associated with signal transduction via JAK1, JAK3 or TYK2 (Figure 6C).
In order to validate JAK-STAT pathway activation in pcAECyTCL, we investigated the presence of activated STAT proteins (pSTAT3 and pSTAT5) by immunohistochemistry (IHC) in eight sequenced patients with available tumor tissue. Robust activation of JAK-STAT signaling (via STAT3, STAT5 or both) was confirmed in all evaluated patients (Figure 6D).
JAK2 fusions identified in pcAECyTCL confer cytokineindependent survival ability to cells
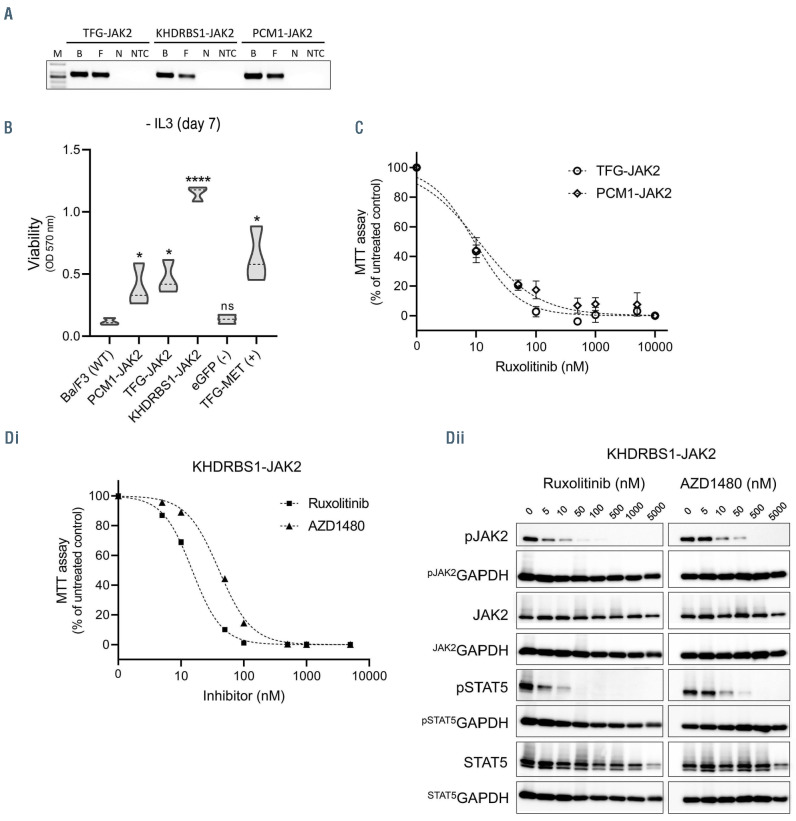
In order to validate the predicted effects of the JAK2 fusions found in pcAECyTCL (i.e., PCM1-JAK2, KHDRBS1-JAK2, TFG-JAK2) on cell survival, we engineered these fusion genes into murine pro-B Ba/F3 cells which die in the absence of exogenous IL3 (Figure 7A). Because self-oligo/dimerizing JAK2 fusions were predicted to activate downstream STAT proteins without the need of upstream cues elicited by cytokine stimulation, these chimeric proteins were expected to increase survival of Ba/F3 cells in the absence of IL3. Seven days after IL3 withdrawal, survival of Ba/F3 cells expressing each of the three engineered JAK2 fusions was noticeably higher (P<0.05, student’s t-test) than survival of the parental Ba/F3 cells (wild-type control) and Ba/F3 cells expressing eGFP (negative control) (Figure 7B).
Figure 3.
Landscape of copy number alterations reveals focal SH2B3 inactivation in pcAECyTCL. (A) Human chromosome ideograms showing regions of gain and loss detected through whole-genome sequencing (WGS) in twelve primary cutaneous CD8+ aggressive epidermotropic cytotoxic T-cell lymphoma (pcAECyTCL) genomes. Blue bars to the right of the chromosomes depict regions of loss whereas red bars to the left of the chromosomes depict regions of gain. (B) Deletions at 12q24.12 (blue bars), where SH2B3 resides, were the most focal (<500 Kb) copy number alteration (CNA) events in pcAECyTCL. Inactivation of SH2B3 was mediated by interstitial deletions and unbalanced rearrangements. Breakpoints of structural alterations at 12q24.12 in all affected patients were validated by Sanger sequencing. Genomic coordinates of breakpoints according to reference genome GRCh38. Arrows indicate the direction towards which genomic coordinate numbers increase. Plus (+) and minus (-) signs specify strand polarity. CTX: interchromosomal rearrangement; ITX: intrachromosomal rearrangement; iDel: interstitial deletion. (C) Copy number losses involving SH2B3 in patients with pcAECyTCL were validated by droplet digital polymerase chain reaction. Ctrl: control CD8+ T cells.
Figure 4.
Distribution of recurrent chromosomal rearrangements, copy number alterations and deleterious indels/single nucleotide variants in pcAECyTCL. (A) First panel: recurrent chromosomal rearrangements impacting cancer genes. Second panel: recurrent large-scale copy number alterations (CNA) (>3 Mb). Third panel: focal minimal common regions (MCR) (≤ 3 Mb) shared by CNA; bona fide cancer genes residing within focal MCR are specified. Fourth panel: Indels and single nucleotide variants (SNV) in cancer genes leading to protein truncations, reported as pathogenic in literature or predicted as disease-causing (SIFT and PolyPhen-2). Only genes altered in more than one patient are indicated. CTX: interchromosomal rearrangement; ITX: intrachromosomal rearrangement. (B) Circos plots showing genetic alterations in patients AEC6 and AEC9. Despite inter-patient heterogeneity, molecular abnormalities affecting genes with roles in the cell cycle, chromatin regulation and JAK2 signaling (genes in light purple) were recurrent in primary cutaneous CD8+ aggressive epidermotropic cytotoxic T-cell lymphoma (pcAECyTCL).
We next evaluated the effect of FDA-approved JAK1/2 inhibitor ruxolitinib on each of the three IL3-independent cell lines carrying JAK2 fusions. Ruxolitinib inhibited the growth of all cell lines in a dose-dependent manner (Figure 7C and D) with half maximal inhibitory concentration (IC50) values in the low nanomolar range (9–15 nM), in concordance with the reported inhibitory activity of this drug.19 Since fusion partners PCM1 and TFG have extensively been proven by others to confer chimeric kinases (including JAK2) the ability to trans-autophosphorylate via self-oligo/dimerization,20-25 we carried on further validation with JAK2 fusion containing novel kinase fusion partner KHDRBS1. For extra verification, we treated Ba/F3 cells expressing KHDRBS1-JAK2 with inhibitor AZD1480, which has higher specificity for JAK2 than ruxolitinib,26 and confirmed that cytokine-independent survival of these cells depends on JAK2 signaling (Figure 7D). Finally, we corroborated by western blotting that growth inhibition exerted by ruxolitinib and AZD1480 was accompanied by a dose-dependent inhibition of JAK2 and STAT5 phosphorylation in Ba/F3 cells driven by KHDRBS1-JAK2 (Figure 7D).
Discussion
This study describes the first high-resolution genetic profiling of pcAECyTCL using next-generation sequencing. The landscape of structural genomic alterations of pcAECyTCL was characterized by considerable genomic instability and inter-patient heterogeneity. Most rearrangements (328 of 426) identified in pcAECyTCL disrupted annotated genes, and approximately one-third of all rearranged genes (91 of 305) were found to play roles in signal transduction and transcriptional regulation. In addition, four of 12 patients experienced chromothripsis/chromoplexy- like events which mediated the deletion of relevant tumor suppressors (e.g., CDKN2C, CHD5, FAS, PTEN, etc.). In full agreement with previously published data,5 we found that gains within 7q and 17q as well as losses within 1p and 13q were the most common largescale chromosomal imbalances.
Figure 5.
Small-scale mutations in genes of the JAK-STAT pathway are predominant in pcAECyTCL. (A) Diagrams showing deleterious indels and single nucleotide variants (SNV) in JAK2, JAK3, STAT3, STAT5B, SH2B3 and SOCS1 detected in primary cutaneous CD8+ aggressive epidermotropic cytotoxic T-cell lymphoma (pcAECyTCL) by whole-genome sequencing (Table 1). (B) Sanger chromatograms confirming presence of bona fide pathogenic SNV in patients with pcAECyTCL. (C) Summary of genetic alterations affecting members of the JAK-STAT pathway in pcAECyTCL.
Figure 6.
RNA sequencing supports upregulation of JAK2 signaling in pcAECyTCL. (A) Heat map showing 1,603 differentially expressed genes (1,076 upregulated, 527 downregulated, false discovery rate [FDR] <0.01) in pcAECyTCL when compared to skin-resident CD8+ T cells. (B) Gene set enrichment analysis (GSEA) uncovered upregulation of the JAK-STAT pathway, the cell cycle (E2F targets, G2/M checkpoint, mitotic spindle), the NF-κB pathway and high inflammatory response in pcAECyTCL. NES: normalized enrichment score; FDR q-value: false discovery rate q-value. (See the Online Supplementary Table S18 for a complete list of GSEA signatures) (C) Examination of differentially expressed genes involved in the JAK-STAT pathway revealed that JAK2 itself, enhancers of JAK2 signaling and components of cytokine receptors that signal predominantly via JAK2 are upregulated in primary cutaneous CD8+ aggressive epidermotropic cytotoxic T-cell lymphoma (pcAECyTCL). (D) Activation of the JAK-STAT pathway (via STAT3 and/or STAT5) in pcAECyTCL was confirmed by immunohistochemistry (IHC) on tumor tissue from sequenced patients (i.e., AEC1/3/5-10). Neoplastic cells exhibited activated STAT3 and/or STAT5 in the nucleus. Normal skin (control) displayed STAT3 activation in keratinocytes and endothelial cells as well as STAT5 activation in melanocytes and endothelial cells. Scale bar, 50 mm.
Figure 7.
Oncogenicity validation of JAK2 fusions identified in pcAECyTCL. (A) Expression of JAK2 fusions in transduced Ba/F3 cells was verified by reverse transcriptase polymerase chain reaction. M: molecular-weight marker; B: Fusion DNA in backbone (positive control); F: cDNA from Ba/F3 cells transduced with JAK2 fusion gene; N: cDNA from Ba/F3 cells transduced with eGFP gene (negative control); NTC: non-template control (H2O). (B) Violin plots showing viability of Ba/F3 cells expressing fusion genes KHDRBS1-JAK2, PCM1-JAK2 or TFG-JAK2 seven days after interleukin-3 (IL3) withdrawal (mean OD, n=3). Viability of all cell lines expressing JAK2 fusions was noticeably higher (P<0.05, student’s t-test) compared to wild-type and negative control cells. Control samples: parental Ba/F3 cells (wild-type control), Ba/F3 cells expressing eGFP (negative control), Ba/F3 cells expressing fusion gene TFG-MET (positive control). *P<0.05; ****P<0.0001. (C) Dose-response curves of Ba/F3 cells expressing PCM1-JAK2 (half maximal inhibitory concentration [IC50]=11 nM) or TFG-JAK2 (IC50 =9 nM) when exposed to various concentrations of JAK1/2 inhibitor ruxolitinib (mean OD; error bars, standard deviation [SD], n=3). (D) Validation experiments with JAK2 fusion containing novel kinase fusion partner KHDRBS1. (i) Dose-response curves of Ba/F3 cells expressing KHDRBS1-JAK2 when exposed to various concentrations of JAK1/2 inhibitors ruxolitinib (IC50=15 nM) or AZD1480 (IC50=40 nM) (mean OD; error bars, SD, n=3). (ii) Western blot analysis of Ba/F3 cells expressing KHDRBS1-JAK2 showed a dose-response reduction in phosphorylation of JAK2 and STAT5 with increasing concentrations of ruxolitinib or AZD1480.
Our analysis identified a group of bona fide oncogenes and tumor suppressors with central roles in the cell cycle (i.e., CDKN2A/B, MIR34AHG, MYC, RB1, TP53), chromatin regulation (i.e., ARID1A, BAZ1A, EED, EPC1, KMT2D, NCOR1, ZEB1) and the JAK-STAT pathway (i.e., JAK2, JAK3, PTPRC, SH2B3, SOCS1, STAT3, STAT5B) whose copy number, sequence organization and/or nucleotide composition were found to be recurrently altered in our pcAECyTCL cohort. Genetic alterations involving JAK-STAT pathway genes were the most notable due to their predominance, likely proliferationpromoting effects and known causative roles in hematological cancers. A subset of SNV affecting JAK-STAT pathway genes in pcAECyTCL have confirmed oncogenic activity in other T-cell lymphomas (Table 1).27,28
JAK2 and SH2B3, which govern the activation and termination of JAK2 signaling in normal hematopoietic cells, respectively, underwent mutually exclusive alterations in nine of 12 patients from our cohort. Mutations in these two genes are associated with BCR-ABL1– myeloproliferative neoplasms (MPN), a group of myeloid malignancies driven by overactive JAK2 signaling.11,29 However, unlike BCR-ABL1– MPN where JAK2 and SH2B3 are mainly affected by pathogenic SNV and/or indels, these two genes experienced predominantly structural alterations in pcAECyTCL. On one hand, JAK2 formed fusion genes encoding self-activating chimeras. On the other hand, SH2B3 was inactivated by focal interstitial deletions and unbalanced rearrangements. The previous suggests that pcAECyTCL is mainly driven by aberrant JAK2 signaling resulting from oncogenic changes leading to JAK2 overactivation or SH2B3 deficiency. Moreover, we demonstrated that JAK2 fusions found in pcAECyTCL promote cytokine-independent cell survival and their oncogenic activity was shown to be successfully inhibited by ruxolitinib. Of note, JAK2 fusions functionally analogous to the ones identified in pcAECyTCL have been previously described and confirmed as oncogenic in other hematological malignancies (e.g., B- and T-cell acute leukemias, MPN).20 Also, recurrent deletion of SH2B3 has been reported in an aggressive subtype of B-cell precursor acute lymphoblastic leukemia.30
We found that genetic alterations involving JAK2 and SH2B3 co-existed with SNV predicted or confirmed as pathogenic in STAT3 or STAT5B in six of nine affected patients. Previous functional in vitro studies with cell lines have suggested that mutations in STAT proteins (especially dimerization-enhancing SNV) observed in T-cell lymphomas operate as aberrant amplifiers of upstream signals from cytokines, overactive receptors or deregulated JAK proteins, rather than as initiators of deregulated JAK-STAT signaling themselves.27 In this scenario, mutations in STAT3/5B would contribute to pcAECyTCL progression by making the pre-existing overactive JAK2 signaling more robust and severe. However, recent evidence derived from a murine model suggests that at least gainof- function mutation STAT5B (p.N642H), one of the most common pathogenic SNV in human T-cell lymphomas, 31 is sufficient by itself to promote the development of neoplasms primarily derived from mature CD8+ T cells.32 Remarkably, malignant CD8+ T cells in these animals showed preferential migration to the skin, lung and the central nervous system, all of which are commonly affected body sites in pcAECyTCL.32 Consistent with this evidence, patient AEC1, the only individual in our cohort who had a single JAK-STAT pathway gene mutated, carried the STAT5B (p.N642H) mutation biallelically.
Several pathogenetic features found in pcAECyTCL have also been reported in mycosis fungoides (MF) and/or Sézary syndrome (SS). Genetic alterations common to pcAECyTCL, MF and SS include recurrent inactivation of ARID1A, CDKN2A, CDKN2B, NCOR1, PTPRC, TP53 and ZEB1 as well as occasional activating mutations in JAK3, MYC and STAT3.33-39 Other genetic alterations observed in pcAECyTCL have been found before either in MF (e.g., SOCS1 and STK11 inactivation) or SS (e.g., RB1 inactivation, STAT5B mutations).33,34,39 By contrast, JAK2 fusions and SH2B3 inactivation have not been reported in other CTCL variants to the best of our knowledge and appear to be characteristic features of pcAECyTCL.
In agreement with the recurrent genetic alterations involving the JAK2-SH2B3 signaling axis observed in pcAECyTCL, transcriptome analysis revealed upregulation of JAK2 signaling. SH2B1 and PTPN11, which encode two proteins with the ability to enhance JAK2 signaling, 40,41 stood out among upregulated JAK-STAT pathway genes. Adaptor protein SH2B1 has been proven to bind to JAK2 and stimulate its kinase activity.42 Similarly, phosphatase PTPN11 (SHP-2) has been shown to positively regulate JAK2-mediated STAT5 phosphorylation.43 In contrast, phosphatase PTPRC (CD45), whose expression has been shown to attenuate JAK2 signaling in hematopoietic and lymphoma cells,44,45 was downregulated in pcAECyTCL. Yet, the exact molecular interactions underlying the action of these three regulators of JAK2 signaling remain to be fully elucidated.
Transcriptome analysis also revealed upregulation of the cell cycle, the TNF-α/NF-κB pathway and a high inflammatory response in pcAECyTCL. Notably, the co-activation (crosstalk) of JAK-STAT signaling (especially via STAT3) and NF-κB signaling is a well-documented phenomenon in cancer, and it has been shown to promote a pro-oncogenic inflammatory microenvironment in the tumor.46 For instance, aberrant JAK2 signaling (via STAT3) in MPN promotes chromatin changes that induce NF-κB signaling; and the resulting combined action of these two pathways, appear to drive the characteristic chronic inflammatory state observed in these neoplasms.47 Our data, in line with the previous, suggest that co-activation of JAK2 signaling and NF-κB signaling operates in pcAECyTCL as well, and their joint action might be responsible for the inflammatory state detected in pcAECyTCL tumors.
Taken together, our findings strongly suggest that overactivation of JAK2 signaling plays a pivotal role in the pathogenesis of pcAECyTCL. Therefore, patients with this lymphoma would likely benefit from treatment with JAK2 inhibitors (e.g., FDA-approved ruxolitinib). In addition, the potential combination of JAK2 inhibitors with NF-κB inhibitors (e.g. bortezomib,48 dimethyl fumarate49) represents an attractive possibility since targeting both pathways might have a synergistic effect and reduce the chance of resistance acquisition.
Supplementary Material
Acknowledgements
The authors thank Tim van Groningen and Yixin Luo for providing valuable technical support.
Funding Statement
Funding: This study was funded by the Dutch Cancer Society (KWF, grant UL2013-6104) and Associazione Amici di Sabrina Fadini Onlus (A.S.F.O., grant 0800000-PR-LUMC).
References
- 1.Berti E, Gaulard P, Willemze R, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues 4th ed. Lyon: IARC; 2017. [Google Scholar]
- 2.Willemze R, Jaffe ES, Burg G, et al. WHOEORTC classification for cutaneous lymphomas. Blood. 2005;105(10):3768-3785. [DOI] [PubMed] [Google Scholar]
- 3.Berti E, Tomasini D, Vermeer MH, Meijer CJLM, Alessi E, Willemze R. Primary cutaneous CD8-positive epidermotropic cytotoxic T cell lymphomas. Am J Pathol. 1999;155(2):483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guitart J, Martinez-Escala ME, Subtil A, et al. Primary cutaneous aggressive epidermotropic cytotoxic T-cell lymphomas: reappraisal of a provisional entity in the 2016 WHO classification of cutaneous lymphomas. Mod Pathol. 2017;30(5):761-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fanoni D, Corti L, Alberti-Violetti S, et al. Array-based CGH of primary cutaneous CD8+ aggressive EPIDERMO-tropic cytotoxic T-cell lymphoma. Genes Chromosomes Cancer. 2018;57(12):622-629. [DOI] [PubMed] [Google Scholar]
- 6.Kato K, Oh Y, Takita J, et al. Molecular genetic and cytogenetic analysis of a primary cutaneous CD8-positive aggressive epidermotropic cytotoxic T-cell lymphoma. Int J Hematol. 2016;103(2):196-201. [DOI] [PubMed] [Google Scholar]
- 7.Tomasini C, Novelli M, Fanoni D, Berti EF. Erythema multiforme-like lesions in primary cutaneous aggressive cytotoxic epidermotropic CD8+ T-cell lymphoma: A diagnostic and therapeutic challenge. J Cutan Pathol. 2017;44(10):867-873. [DOI] [PubMed] [Google Scholar]
- 8.Lu H, Villafane N, Dogruluk T, et al. Engineering and functional characterization of fusion genes identifies novel oncogenic drivers of cancer. Cancer Res. 2017;77(13):3502-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotake Y, Nakagawa T, Kitagawa K, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paczkowska J, Soloch N, Bodnar M, et al. Expression of ELF1, a lymphoid ETS domain-containing transcription factor, is recurrently lost in classical Hodgkin lymphoma. Br J Haematol. 2019;185(1):79-88. [DOI] [PubMed] [Google Scholar]
- 11.Maslah N, Cassinat B, Verger E, Kiladjian JJ, Velazquez L. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders. Leukemia. 2017;31(8):1661-1670. [DOI] [PubMed] [Google Scholar]
- 12.Degryse S, de Bock CE, Cox L, et al. JAK3 mutants transform hematopoietic cells through JAK1 activation, causing T-cell acute lymphoblastic leukemia in a mouse model. Blood. 2014;124(20):3092-3100. [DOI] [PubMed] [Google Scholar]
- 13.Yamashita Y, Yuan J, Suetake I, et al. Arraybased genomic resequencing of human leukemia. Oncogene. 2010;29(25):3723-3731. [DOI] [PubMed] [Google Scholar]
- 14.Rajala HL, Eldfors S, Kuusanmaki H, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121(22):4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanikova L, Babosova O, Swierczek S, et al. Coexistence of gain-of-function JAK2 germ line mutations with JAK2V617F in polycythemia vera. Blood. 2016;128(18):2266-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson EI, Putzer S, Yadav B, et al. Discovery of novel drug sensitivities in TPLL by high-throughput ex vivo drug testing and mutation profiling. Leukemia. 2018;32(3):774-787. [DOI] [PubMed] [Google Scholar]
- 17.Bandapalli OR, Schuessele S, Kunz JB, et al. The activating STAT5B N642H mutation is a common abnormality in pediatric T-cell acute lymphoblastic leukemia and confers a higher risk of relapse. Haematologica. 2014;99(10):e188-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takamura S. Niches for the Long-term maintenance of tissue-resident memory T cells. Front Immunol. 2018;9:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quintas-Cardama A, Vaddi K, Liu P, et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: therapeutic implications for the treatment of myeloproliferative neoplasms. Blood. 2010;115(15): 3109-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith CA, Fan G. The saga of JAK2 mutations and translocations in hematologic disorders: pathogenesis, diagnostic and therapeutic prospects, and revised World Health Organization diagnostic criteria for myeloproliferative neoplasms. Hum Pathol. 2008;39(6):795-810. [DOI] [PubMed] [Google Scholar]
- 21.Reiter A, Walz C, Watmore A, et al. The t(8;9)(p22;p24) is a recurrent abnormality in chronic and acute leukemia that fuses PCM1 to JAK2. Cancer Res. 2005;65(7): 2662-2667. [DOI] [PubMed] [Google Scholar]
- 22.Lierman E, Selleslag D, Smits S, Billiet J, Vandenberghe P. Ruxolitinib inhibits transforming JAK2 fusion proteins in vitro and induces complete cytogenetic remission in t(8;9)(p22;p24)/PCM1-JAK2-positive chronic eosinophilic leukemia. Blood. 2012;120(7):1529-1531. [DOI] [PubMed] [Google Scholar]
- 23.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greco A, Fusetti L, Miranda C, et al. Role of the TFG N-terminus and coiled-coil domain in the transforming activity of the thyroid TRK-T3 oncogene. Oncogene. 1998;16(6):809-816. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez L, Bea S, Bellosillo B, et al. Diversity of genomic breakpoints in TFGALK translocations in anaplastic large cell lymphomas: identification of a new TFGALK (XL) chimeric gene with transforming activity. Am J Pathol. 2002;160(4):1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedvat M, Huszar D, Herrmann A, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16(6):487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waldmann TA, Chen J. Disorders of the JAK/STAT pathway in T cell lymphoma pathogenesis: implications for immunotherapy. Annu Rev Immunol. 2017;35:533-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammarén HM, Virtanen AT, Raivola J, Silvennoinen O. The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine. 2019;118:48-63. [DOI] [PubMed] [Google Scholar]
- 29.Levine RL, Pardanani A, Tefferi A, Gilliland DG. Role of JAK2 in the pathogenesis and therapy of myeloproliferative disorders. Nat Rev Cancer. 2007;7(9):673-683. [DOI] [PubMed] [Google Scholar]
- 30.Baughn LB, Meredith MM, Oseth L, Smolarek TA, Hirsch B. SH2B3 aberrations enriched in iAMP21 B lymphoblastic leukemia. Cancer Genet. 2018;226-227:30-35. [DOI] [PubMed] [Google Scholar]
- 31.Pham HTT, Maurer B, Prchal-Murphy M, et al. STAT5BN642H is a driver mutation for T cell neoplasia. J Clin Invest. 2018;128(1):387-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Araujo ED, Erdogan F, Neubauer HA, et al. Structural and functional consequences of the STAT5B(N642H) driver mutation. Nat Commun. 2019;10(1):2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woollard WJ, Pullabhatla V, Lorenc A, et al. Candidate driver genes involved in genome maintenance and DNA repair in Sezary syndrome. Blood. 2016;127(26):3387-3397. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Ni X, Covington KR, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47(12):1426-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGirt LY, Jia P, Baerenwald DA, et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood. 2015;126(4):508-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiel MJ, Sahasrabuddhe AA, Rolland DC, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sezary syndrome. Nat Commun. 2015;6:8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.da Silva Almeida AC, Abate F, Khiabanian H, et al. The mutational landscape of cutaneous T cell lymphoma and Sezary syndrome. Nat Genet. 2015;47(12):1465-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi J, Goh G, Walradt T, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47(9):1011-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastidas Torres AN, Cats D, Mei H, et al. Genomic analysis reveals recurrent deletion of JAK-STAT signaling inhibitors HNRNPK and SOCS1 in mycosis fungoides. Genes Chromosomes Cancer. 2018;57(12):653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maures TJ, Kurzer JH, Carter-Su C. SH2B1 (SH2-B) and JAK2: a multifunctional adaptor protein and kinase made for each other. Trends Endocrinol Metab. 2007;18(1):38-45. [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Qu CK. Protein tyrosine phosphatase SHP-2 (PTPN11) in hematopoiesis and leukemogenesis. J Signal Transduct. 2011;2011:195239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rui L, Carter-Su C. Identification of SH2bbeta as a potent cytoplasmic activator of the tyrosine kinase Janus kinase 2. Proc Natl Acad Sci U S A. 1999;96(13):7172-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ali S, Nouhi Z, Chughtai N, Ali S. SHP-2 regulates SOCS-1-mediated Janus kinase-2 ubiquitination/degradation downstream of the prolactin receptor. J Biol Chem. 2003;278(52):52021-52031. [DOI] [PubMed] [Google Scholar]
- 44.Irie-Sasaki J, Sasaki T, Matsumoto W, et al. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature. 2001;409(6818):349-354. [DOI] [PubMed] [Google Scholar]
- 45.Wu L, Bijian K, Shen SH. CD45 recruits adapter protein DOK-1 and negatively regulates JAK-STAT signaling in hematopoietic cells. Mol Immunol. 2009;46(11-12):2167-2177. [DOI] [PubMed] [Google Scholar]
- 46.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11): 798-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleppe M, Koche R, Zou L, et al. Dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell. 2018;33(1):29-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinzani PL, Musuraca G, Tani M, et al. Phase II trial of proteasome inhibitor bortezomib in patients with relapsed or refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25(27):4293-4297. [DOI] [PubMed] [Google Scholar]
- 49.Nicolay JP, Muller-Decker K, Schroeder A, et al. Dimethyl fumarate restores apoptosis sensitivity and inhibits tumor growth and metastasis in CTCL by targeting NFkappaB. Blood. 2016;128(6):805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.